Abstract
Background
Reverse transcription quantitative PCR (RT-qPCR) is widely used for gene expression analysis in various organisms. Its accuracy largely relies on the stability of reference genes, making reference gene selection a vital step in RT-qPCR experiments. However, previous studies in mollusks only focused on the reference genes widely used in vertebrates.
Results
In this study, we conducted the transcriptome-wide identification of reference genes in the bivalve mollusk Mizuhopecten yessoensis based on 60 transcriptomes covering early development, adult tissues and gonadal development. A total of 964, 1210 and 2097 candidate reference genes were identified, respectively, resulting in a core set of 568 genes. Functional enrichment analysis showed that these genes are significantly overrepresented in Gene Ontology (GO) terms or Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways related to ribosomes, energy production, etc. Six genes (RS23, EF1A, NDUS4, SELR1, EIF3F, and OLA1) were selected from the candidate genes for RT-qPCR validation, together with 6 commonly used reference genes (ACT, CYTC, HEL, EF1B, GAPDH and RPL16). Stability analyses using geNorm, NormFinder and the comparative delta-Ct method revealed that the new candidate reference genes are more stable than the traditionally used genes, and ACT and CYTC are not recommended under either of the three circumstances. There was a significant correlation between the Ct of RT-qPCR and the log2(TPM) of RNA-Seq data (Ct = − 0.94 log2(TPM) + 29.67, R2 = 0.73), making it easy to estimate the Ct values from transcriptome data prior to RT-qPCR experiments.
Conclusion
Our study represents the first transcriptome-wide identification of reference genes for early development, adult tissues, and gonadal development in the Yesso scallop and will benefit gene expression studies in other bivalve mollusks.
Electronic supplementary material
The online version of this article (10.1186/s12864-019-5661-x) contains supplementary material, which is available to authorized users.
Keywords: Reference genes, Transcriptome-wide, Scallop, Early development, Adult tissues, Gonadal development
Background
Reverse transcription quantitative PCR (RT-qPCR) is the most frequently used method in gene expression analysis due to its high sensitivity, specificity, reproducibility and broad dynamic range [1]. To obtain reliable RT-qPCR results, many considerations need to be taken [2]. One essential component of a reliable RT-qPCR assay is normalization, which controls for variations in the amount and quality of RNA between different samples, thus enabling comparisons of the expression of a gene of interest among different samples. Reference genes are most commonly used for normalization. Choosing a reference gene that shows stable expression across all samples is critical for relative quantification in RT-qPCR assays.
Despite the awareness of the importance of reference genes, most previous studies directly applied commonly used reference genes for gene expression quantification without validation or conducted reference gene selection from a few traditionally used reference genes [3–6]. However, an increasing number of recent studies have shown that the frequently used reference genes are not always stable under different conditions [7, 8]. Housekeeping genes, which maintain constant expression levels in all cells and under all conditions, are an ideal source for reference gene selection. In mammals, housekeeping genes have been detected using various large-scale gene expression profiling methods, such as expressed sequence tags (ESTs), serial analysis of gene expression (SAGE) and microarray analysis [9–11]. The advent of RNA-Seq enables the identification of housekeeping genes in well-studied model organisms and less-studied nonmodel organisms. To date, housekeeping or reference genes have been successfully identified from transcriptome data in various organisms, such as humans [12], mice [13], zebrafish [14], tomato [15], seaweed [16] and kiwi [17].
Bivalve mollusks comprise approximately 14,000 existing species that are widely distributed worldwide. Many of these species are economically important aquaculture shellfish; therefore, extensive studies on the genes related to immunity [18–20], growth [21–23] and reproduction [24, 25] have been conducted in this group of animals. Gene expression quantification via RT-qPCR has been extensively conducted, but mostly using a single reference gene, such as ACT and CYTC [26–28]. To eliminate misleading effects due to the use of inappropriate reference genes, reference genes have been selected but generally focused on some traditionally used genes [29–31]. To our knowledge, the transcriptome-wide identification of reference genes has only been conducted in the Pacific oyster [32, 33], despite the availability of extensive transcriptome data in various organisms [34–38].
In this study, we carried out systematic analyses of the 60 transcriptomes covering early development, adult tissues and gonadal development in the Yesso scallop M. yessoensis. The candidate reference genes were identified for each dataset, and a core set was generated. Six selected candidate reference genes were compared with 6 commonly used reference genes for the RT-qPCR validation of stability, and the relationship between the Ct of the RT-qPCR and transcripts per million (TPM) of the RNA-Seq was examined. Our study provides a valuable resource of the reference genes for the scallop. This information will contribute to the accurate quantification of gene expression in future studies.
Methods
Sample collection
The Yesso scallops used for RT-qPCR were sampled from the hatchery of the Zhangzidao Group Co., Ltd. (Dalian, China).
Embryos (two to eight cells, blastulae, and gastrulae) and larvae (trochophore larvae, D-stage larvae, pediveliger larvae, and juvenile) were obtained as described in Sun et al. [39] and Wang et al. [34]. Then, the samples were preserved in RNAlater and stored at − 80 °C until further use.
Two-year-old adult scallops were maintained in aerated seawater for a week. After acclimation, the following tissues were dissected: digestive gland, eyes, foot, gill, gonad, hemocytes, mantle, striated muscle, smooth muscle and nerve ganglia. The ovaries and testes of mollusks at four developmental stages (resting stage, proliferative stage, growing stage and maturation stage) were collected as described in Li et al. [40]. Three biological replicates were collected for each tissue, frozen in liquid nitrogen, and then stored at − 80 °C prior to RNA isolation.
RNA-Seq datasets
To identify reference genes in early development, the RNA-Seq data from 7 embryo/larva stages (two to eight cells, blastulae, gastrulae, trochophore larvae, D-stage larvae, pediveliger larvae and juvenile) were obtained from Wang et al. (NCBI Bioproject ID: PRJNA259405) [34]. Twenty-nine RNA-Seq datasets for the eyes, mantle, gill, hemocytes, digestive gland, striated muscle, smooth muscle, foot, ganglia, ovaries and testes (NCBI Bioproject ID: PRJNA259405 and PRJNA423107) [34, 36, 41] were used to identify the reference genes across tissues. Reference genes during gonadal development were evaluated using 24 RNA-Seq datasets from four developmental stages of ovaries and testes (NCBI Bioproject ID: PRJNA516336) [42]. Detailed information on all RNA-Seq data is available in Additional file 1: Table S1.
Data quantification and analysis
All raw reads downloaded from NCBI (https://www.ncbi.nlm.nih.gov) were first filtered using a homemade Perl script to obtain high-quality reads by removing reads containing undetermined bases (‘N’) or excessive numbers of low-quality positions (> 10 positions with quality scores < 20). Then the high-quality reads were mapped to the M. yessoensis genome [34] using STAR [43] with the parameters of ‘STAR --runThreadN 8 --genomeDir --readFilesIn R1.fq R2.fq --sjdbGTFtagExonParentGene --outFilterMultimapNmax 100000 --outFileNamePrefix --outReadsUnmapped Fasta >STAR_sample.log’. The raw counts for each gene were converted into TPM using the RNA-Seq by Expectation Maximization (RSEM) software package.
Identification of reference genes from transcriptomes
The reference genes were detected using the method provided by Eisenberg and Levanon [12], with minor modifications. Briefly, considering that RPKM is not appropriate for comparisons between samples [44–46], we used TPM values to measure gene expression levels. After calculating the TPM values for each gene in all samples, TPMs from biological replicates were averaged for subsequent analysis. Finally, four criteria were adopted for the detection of reference genes: (I) expression observed in all tissue types or developmental stages; (II) low variance over tissues or developmental stages by requiring standard-deviation [log2(TPM)] < 1; (III) no exceptional expression in any single tissue or developmental stage by requiring no log2(TPM) differed from the mean log2(TPM) by two or more; and (IV) medium to high expression level by requiring mean [log2(TPM)] > 5. The stability of the candidate reference genes was further evaluated according to the coefficient of variation (CV = stdev/mean). Here, all candidate reference genes have a mean [log2(TPM)] > 5 and a standard-deviation [log2(TPM)] < 1, so their CV values are always less than 0.2.
Functional enrichment analysis
To better understand the function of the reference genes, GO and KEGG enrichment analyses were performed. The Swiss-Prot Blast results for all genes were imported into Blast2GO, and GO terms at level 3 were assigned to produce a broad overview of the groups of genes for biological processes, cellular component, and molecular function categories. The KEGG annotation was conducted by the KEGG automatic annotation server (KAAS). The significance of GO and KEGG enrichment was determined by using the algorithm implemented in GOstats with an FDR cutoff of 0.05 [47].
RNA extraction and cDNA synthesis
For all samples, total RNA was extracted using the conventional guanidinium isothiocyanate method, and potential DNA contamination was eliminated by DNase I (Takara Bio, Shiga, Japan) digestion. The obtained RNA samples were then assessed on 1.5% agarose gel electrophoresis and determined by using Nanovue Plus (GE Healthcare, Piscataway, United States) to detect the RNA concentration and purity. cDNA synthesis was carried out using MMLV Reverse Transcriptase (TaKaRa Bio, Shiga, Japan) and oligo(dT)18. Finally, the cDNA products were diluted 20-fold with nuclease-free water for RT-qPCR and kept at − 20 °C until further use.
RT-qPCR validation
Twelve genes, including 6 new candidate and 6 commonly used reference genes, were selected for RT-qPCR validation. Their primers were designed using Primer Premier 5.0 software. Two additional criteria need to be met, including PCR products between 80 and 120 bp in size and an annealing temperature of approximately 63 °C. The primer sequences are listed in Table 1. The amplification efficiency of each primer pair was calculated based on the standard curve generated from a two-fold dilution series of cDNA spanning eight orders of magnitude (0.78 ng ~ 100 ng).
Table 1.
List of primers used for RT-qPCR analysis
| Gene ID | Accession No. | Gene Name | Primer Sequence (5′-3′) | Amplicon Length (bp) | Amplification efficiency |
|---|---|---|---|---|---|
| PYT16215 | XM_021518643.1 | RS23 | F:TTACACGAATATCCGCCATCA | 100 | 1.05 |
| R:GTAATCGTTATCGTGCCACTT | |||||
| PYT23375 | XM_021500266.1 | EF1A | F:GCGGTGGTATTGACAAGAGA | 113 | 1.04 |
| R:GTTCACGTTCAGCCTTCAGT | |||||
| PYT15332 | XM_021490269.1 | NDUS4 | F:TGTGAGAAGCTGGTGTGCTA | 108 | 1.00 |
| R:TGTGTCTTCCGTCCATTCTAT | |||||
| PYT08134 | XM_021506528.1 | SELR1 | F:AAGGCTGGATCAGCGTACTT | 97 | 1.04 |
| R:CCGTTCTCACACGCCTTAAT | |||||
| PYT12324 | XM_021496512.1 | EIF3F | F:TGCTTCGCTGTGCCTCATAA | 105 | 1.04 |
| R:TGACTTCAGCTGCATTGACTT | |||||
| PYT24193 | XM_021485429.1 | OLA1 | F:AAGGTCAAGGTCTCGGCAAT | 100 | 1.02 |
| R:CACGTGGACAATCTCTTCGT | |||||
| PYT04221 | XM_021511578.1 | ACT | F:GACAGCTACGTAGGAGATGA | 112 | 1.02 |
| R:TGATGCCAGATCTTCTCCATA | |||||
| PYT19618 | XM_021517608.1 | CYTC | F:TGTATGTGACAGTCCTTGGTT | 126 | 1.01 |
| R:GTGTGGCATTGAGCACACTT | |||||
| PYT19918 | XM_021485125.1 | HEL | F:CAGGAAGCAGTGGACTTACA | 100 | 1.02 |
| R:TCATAACGGTCACCGTAAGAA | |||||
| PYT19808 | XM_021522100.1 | EF1B | F:TAGGCCAGTATGGACCTTCA | 105 | 1.05 |
| R:TCTTCTTCCTCTTCAGAGTCT | |||||
| PYT21544 | XM_021497981.1 | GAPDH | F:GTGTACATGCTGAAGTACGAT | 115 | 1.04 |
| R:CGCTCCATGAAGACAGAGAT | |||||
| PYT03203 | XM_021484929.1 | RPL16 | F:GTTATCTTCAGTACGCTCACA | 103 | 1.00 |
| R:ACGCCATGGATATTCTATCCT |
RT-qPCR reactions were conducted in a 384-well plate using a Light Cycler 480 Real-time PCR System (Roche Diagnostics, Mannheim, Germany). Each reaction contained 2 μl 20-fold diluted cDNA, 10 μl Light Cycler 480 SYBR Green I Master and 4 μl 2 μM primers. The cycling conditions were 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 63 °C for 1 min. Each RT-qPCR analysis was performed in triplicate. For each gene, the melting curve and gel picture were analyzed to confirm that a single PCR product had been amplified.
The expression stabilities of the twelve reference genes were assessed using three statistical approaches, geNorm [48], NormFinder [49], and comparative delta-Ct method [50]. The geometric mean was calculated for the ranking values in the three methods to guarantee a comprehensive analysis.
Results
Identification of the candidate reference genes from RNA-Seq data
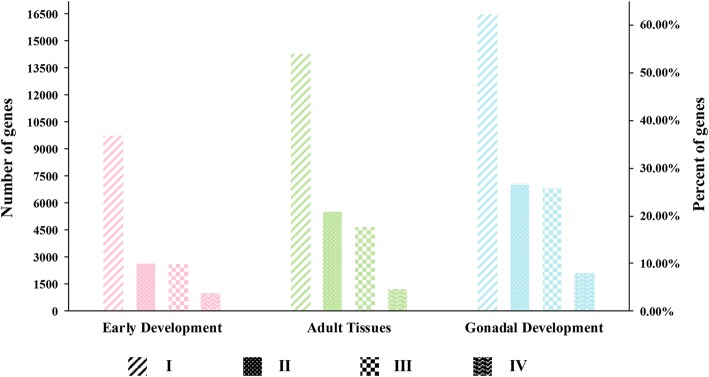
A total of 60 RNA-Seq datasets from scallop embryos/larvae and adult tissues were used to select candidate reference genes for early development, adult tissues and gonadal development. As shown in Fig. 1, among the 26415 genes in the scallop genome, 2560 (9.69%), 4638 (17.56%) and 6780 (25.67%) genes met the first three filter criteria. To obtain the medium expression level for the fourth criterion, we investigated the expression levels of the genes screened by the first three steps and found that the median log2(TPM) values for these genes are approximately 5 for all three datasets (Fig. 2a). Thus, a final criterion was applied to obtain reference genes with an average log2(TPM) value higher than 5, resulting in 964 (3.65%), 1210 (4.58%) and 2097 (7.94%) candidate reference genes for early development, adult tissues, and gonadal development, respectively (Additional file 2: Table S2). The 10 most stable genes with the lowest CV values in early development, adult tissues, and gonadal development are listed in Table 2. According to the results, the ten most stable genes in early development have log2(TPM) values ranging from 5.48 to 10.72 and CV values between 0.016 and 0.027. Six genes were annotated, of which four (ATPK, ATP5J, ATP5L and NDUAB) encode proteins related to energy production. In contrast, the 10 most stable genes in adult tissues possess higher expression levels, with log2(TPM) values ranging from 11.15 to 12.08. All of these genes were annotated as ribosomal proteins. The ten most stable genes in gonadal development showed a wide range of expression, with log2(TPM) values ranging from 5.76 to 13.74. All of these genes were annotated, of which 5 (RS23, EF1A, RLA1, IF4A1 and RM54) function in protein synthesis.
Fig. 1.
Results of each screening procedure for all three datasets. Both the number and percentage of genes that met the four criteria are shown. (I) TPM > 0; (II) standard-deviation [log2(TPM)] < 1; (III) no log2(TPM) differed from the mean log2(TPM) by two or more; (IV) mean [log2(TPM)] > 5. Different colors represent different datasets, pink represents early development, green represents adult tissues and blue represents gonadal development.
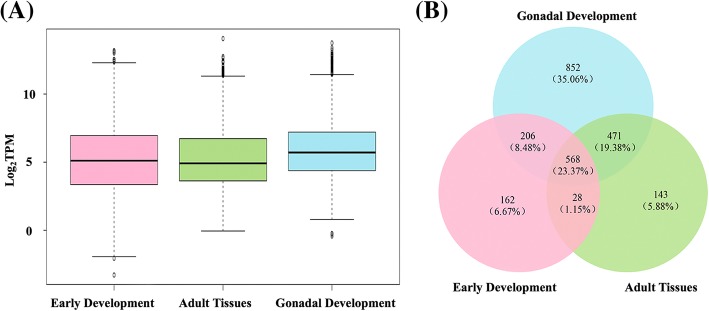
Fig. 2.
Information on the stable genes in early development, adult tissues and gonadal development. a Boxplot showing the log2(TPM) for genes that passed criteria I to III in the three datasets. b A Venn diagram showing the relationships of candidate reference genes that passed criteria I to IV in the three datasets. Pink: early development; Green: adult tissues; Blue: gonadal development
Table 2.
The top 10 candidate reference genes in the early development, adult tissues and gonadal development of the scallop M. yessoensis
| Gene ID | Gene Symbol | Gene Name | CV | Mean | |
|---|---|---|---|---|---|
| Early Development | PYT09804 | – | – | 0.016 | 9.68 |
| PYT05988 | ATPK | Putative ATP synthase subunit f | 0.024 | 10.65 | |
| PYT22252 | ATP5J | ATP synthase-coupling factor 6 | 0.025 | 10.58 | |
| PYT08134 | SELR1 | Sel1 repeat-containing protein 1 | 0.025 | 5.48 | |
| PYT07181 | – | – | 0.025 | 8.22 | |
| PYT20584 | TBB4B | Tubulin beta-4B | 0.026 | 7.88 | |
| PYT12567 | – | – | 0.026 | 10.23 | |
| PYT01876 | ATP5L | ATP synthase subunit g | 0.026 | 10.72 | |
| PYT08398 | NDUAB | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex | 0.027 | 8.83 | |
| PYT17204 | – | – | 0.027 | 10.60 | |
| Adult Tissues | PYT25527 | RL21 | 60S ribosomal protein L21 | 0.030 | 12.08 |
| PYT22182 | RL40 | Ubiquitin-60S ribosomal protein L40 | 0.031 | 11.85 | |
| PYT16215 | RS23 | 40S ribosomal protein S23 | 0.032 | 11.15 | |
| PYT05179 | RS2 | 40S ribosomal protein S2 | 0.032 | 11.38 | |
| PYT03250 | RL8 | 60S ribosomal protein L8 | 0.032 | 11.60 | |
| PYT05293 | RS24 | 40S ribosomal protein S24 | 0.032 | 11.56 | |
| PYT21874 | RS20 | 40S ribosomal protein S20 | 0.032 | 11.81 | |
| PYT12227 | RS3 | 40S ribosomal protein S3 | 0.032 | 11.54 | |
| PYT19800 | RL27 | 60S ribosomal protein L27 | 0.033 | 11.73 | |
| PYT23759 | RL19 | 60S ribosomal protein L19 | 0.033 | 11.89 | |
| Gonadal Development | PYT16215 | RS23 | 40S ribosomal protein S23 | 0.012 | 11.30 |
| PYT23375 | EF1A | Elongation factor 1-alpha | 0.013 | 13.74 | |
| PYT15332 | NDUS4 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 4 | 0.014 | 8.51 | |
| PYT24062 | RLA1 | 60S acidic ribosomal protein P1 | 0.017 | 13.36 | |
| PYT20779 | IF4A1 | Eukaryotic initiation factor 4A-I | 0.018 | 10.28 | |
| PYT10011 | LARK | RNA-binding protein lark | 0.018 | 9.31 | |
| PYT14375 | SMD1 | Small nuclear ribonucleoprotein Sm D1 | 0.019 | 10.89 | |
| PYT01777 | RM54 | 39S ribosomal protein L54 | 0.019 | 5.76 | |
| PYT24327 | CSN8 | COP9 signalosome complex subunit 8 | 0.019 | 7.23 | |
| PYT10755 | CYP1 | Peptidyl-prolyl cis-trans isomerase 1 | 0.020 | 9.86 |
Generation and functional enrichment analysis of the core set of reference genes
To obtain a core set of reference genes, we compared the three candidate reference gene sets. As shown in Fig. 2b, the candidate reference genes identified from different datasets resulted in a total of 2430 genes, of which 568 (23.37%) were shared. GO enrichment analysis (Table 3) revealed that the 568 genes were highly overrepresented in biological process (BP) terms associated with biosynthetic process (FDR = 1.07E-34) and cellular metabolic process (FDR = 4.43E-32) and in molecular functions (MF) terms related to structural constituent of ribosome (FDR = 4.73E-252). These genes were also enriched in cellular components (CC) terms associated with various protein complexes and organelles. KEGG pathway enrichment analysis (Table 3) revealed that the core set reference genes was significantly enriched in 6 pathways, with ribosome (FDR = 5.41E-197) being the most significant, followed by oxidative phosphorylation (FDR = 1.20E-54).
Table 3.
GO terms and KEGG pathways enriched in the core reference genes of the scallop M. yessoensis
| ID code | Term/Pathway | Gene Number | P value | FDR |
|---|---|---|---|---|
| GO | ||||
| GO:0009058 | biosynthetic process | 119 | 2.64E-36 | 1.07E-34 |
| GO:0044237 | cellular metabolic process | 184 | 1.18E-33 | 4.43E-32 |
| GO:0009056 | catabolic process | 24 | 1.48E-03 | 6.50E-03 |
| GO:0045184 | establishment of protein localization | 17 | 3.45E-03 | 1.38E-02 |
| GO:0030529 | ribonucleoprotein complex | 83 | 1.77E-218 | 4.30E-216 |
| GO:0043228 | nonmembrane-bounded organelle | 89 | 5.81E-95 | 5.14E-93 |
| GO:0043234 | protein complex | 67 | 8.58E-34 | 3.34E-32 |
| GO:0016469 | proton-transporting two-sector ATPase complex | 13 | 1.12E-13 | 1.95E-12 |
| GO:0043227 | membrane-bounded organelle | 56 | 2.46E-12 | 3.86E-11 |
| GO:0044455 | mitochondrial membrane part | 9 | 8.59E-11 | 1.06E-09 |
| GO:0003735 | structural constituent of ribosome | 81 | 4.85E-255 | 4.73E-252 |
| GO:0004129 | cytochrome-c oxidase activity | 5 | 1.48E-07 | 1.33E-06 |
| GO:0051540 | metal cluster binding | 10 | 1.73E-07 | 1.52E-06 |
| GO:1901363 | heterocyclic compound binding | 106 | 2.72E-04 | 1.43E-03 |
| KEGG | ||||
| map03010 | Ribosome | 87 | 2.89E-199 | 5.41E-197 |
| map00190 | Oxidative phosphorylation | 58 | 3.20E-56 | 1.20E-54 |
| map03050 | Proteasome | 25 | 2.22E-24 | 5.94E-23 |
| map03013 | RNA transport | 25 | 3.32E-06 | 6.90E-05 |
| map04141 | Protein processing in endoplasmic reticulum | 21 | 1.15E-05 | 2.14E-04 |
| map00020 | Citrate cycle (TCA cycle) | 7 | 3.26E-03 | 4.06E-02 |
Comparisons between candidate and commonly used reference genes by RNA-Seq analysis and RT-qPCR validation
To compare the stability between the candidate and commonly used reference genes, 6 candidate and 6 commonly used reference genes were selected for further analysis. All six candidate reference genes were selected from the core set of reference genes. Four of these genes were chosen from the top 10 candidates shown in Table 2, including RS23, EF1A, NDUS4, and SELR1. The other two genes, EIF3F, and OLA1, were selected from the less stable candidates. The 6 commonly used reference genes include ACT, CYTC, HEL, EF1B, GAPDH and RPL16 (Table 4).
Table 4.
Detailed information on the six selected candidate reference genes and the six commonly used reference genes
| Gene ID | Gene Symbol | Gene Name | Embryos/Larvae Development | Adult Tissues | Gonadal Development | |||
|---|---|---|---|---|---|---|---|---|
| CV | Mean | CV | Mean | CV | Mean | |||
| PYT16215 | RS23 | 40S ribosomal protein S23 | 0.037 | 11.66 | 0.032 | 11.15 | 0.012 | 11.30 |
| PYT23375 | EF1A | Elongation factor 1-alpha | 0.028 | 12.43 | 0.033 | 12.75 | 0.013 | 13.74 |
| PYT15332 | NDUS4 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 4 | 0.052 | 8.24 | 0.069 | 7.79 | 0.014 | 8.51 |
| PYT08134 | SELR1 | Sel1 repeat-containing protein 1 | 0.025 | 5.48 | 0.098 | 5.04 | 0.032 | 5.74 |
| PYT12324 | EIF3F | Eukaryotic translation initiation factor 3 subunit F | 0.035 | 8.13 | 0.050 | 7.80 | 0.027 | 8.75 |
| PYT24193 | OLA1 | Obg-like ATPase 1 | 0.051 | 6.87 | 0.042 | 6.96 | 0.057 | 7.07 |
| PYT04221 | ACT | Beta-actin | 0.523 | 8.29 | 0.223 | 12.45 | 0.065 | 10.08 |
| PYT19618 | CYTC | Cytochrome c | 0.209 | 9.33 | 0.106 | 8.33 | 0.181 | 6.96 |
| PYT19918 | HEL | DEAD-box RNA helicase | 0.277 | 3.98 | 0.163 | 4.79 | 0.058 | 6.37 |
| PYT19808 | EF1B | Elongation factor 1-beta | 0.074 | 10.02 | 0.045 | 9.86 | 0.029 | 10.25 |
| PYT21544 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 0.230 | 7.66 | 0.083 | 9.92 | 0.026 | 10.26 |
| PYT03203 | RPL16 | 39S ribosomal protein L16 | 0.096 | 6.32 | 0.155 | 4.86 | 0.061 | 6.61 |
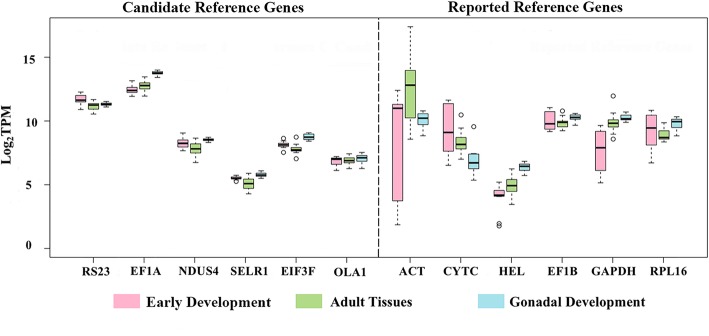
The log2(TPM) values of the twelve genes in the three RNA-Seq datasets are shown in Fig. 3. The six candidate reference genes have smaller variances in contrast to the commonly used reference genes in all three datasets, suggesting that these genes are more stably expressed, irrespective of developmental stages or tissue types. The variances differ among datasets for the commonly used reference genes. Most of these genes, including ACT, CYTC, GAPDH and RPL16, are highly unstable during early development. In adult tissues, the expression of ACT is most variable, and during gonadal development, CYTC is most unstable. EF1B is relatively stable compared to the other commonly used reference genes in all three datasets, and the only gene that falls within the core set candidate reference gene list.
Fig. 3.
Evaluation of the reference gene candidates and reported reference genes based on RNA-seq analysis. A boxplot showing the log2(TPM) values of the 6 candidate reference genes and 6 reported reference genes in early development (pink), adult tissues (green) and gonadal development (blue)
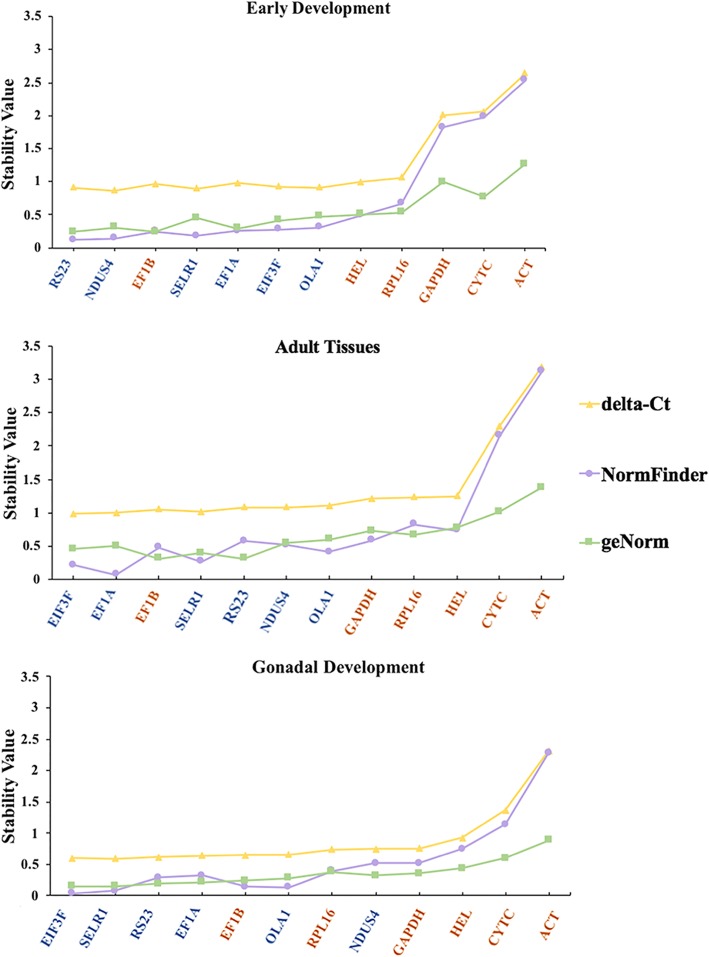
To validate the results, RT-qPCR was conducted, and the expression stability of the twelve genes was evaluated using three methods (geNorm, NormFinder, and comparative delta-Ct). As shown in Fig. 4, the three methods gave very similar results for the unstable genes. Specifically, three genes, including ACT, CYTC and GAPDH, are relatively unstable during early development. Two genes, ACT and CYTC, are unstable in adult tissues. These two genes are also unstable during gonadal development, with ACT exhibiting higher expression variation than CYTC. Although stability rankings are different among methods for the highly and medium stable genes, the candidate reference genes tend to have higher comprehensive ranking values than those of the commonly used reference genes. Among the twelve genes, the most stable gene for early development is RS23, and EIF3F is the most stable in adult tissues and during gonadal development.
Fig. 4.
Expression stability of the twelve genes in early development (a), adult tissues (b) and gonadal development (c) based on RT-qPCR experiments. The stability was evaluated based on geNorm, NormFinder and comparative delta-Ct analyses of the RT-qPCR data. The genes are arranged in descending order of comprehensive stability from left to right
Relationship between the Ct of RT-qPCR and the TPM of RNA-Seq
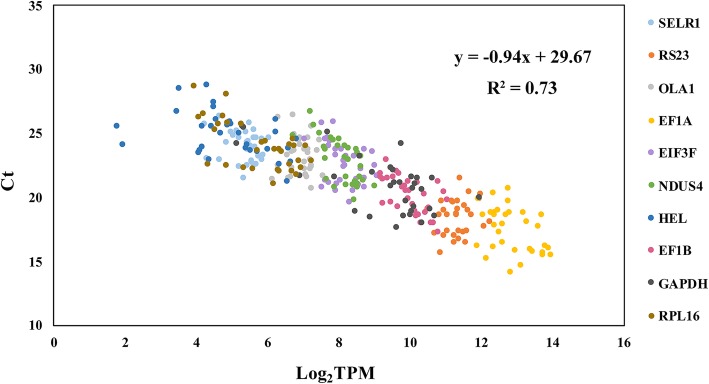
To facilitate an estimation of the Ct of RT-qPCR based on RNA-Seq data, we further evaluated the relationship of the gene expression levels between RNA-Seq and RT-qPCR data using the 10 reference genes, excluding ACT and CYTC. As shown in Fig. 5, the log2(TPM) values mainly range from 4 to 14, and the Ct values of RT-qPCR fall between 14 and 29. There is a significantly negative correlation between the two values (r = − 0.85, P < 0.01), with the formula Ct = − 0.94 log2(TPM) + 29.67.
Fig. 5.
Gene expression correlation between RT-qPCR and RNA-Seq data
Discussion
In this study, an improved method for the systematic identification of reference genes is proposed. Compared with the study by Eisenberg and Levanon [12], there are two modifications in the reference gene screening procedures. First, we used TPM instead of RPKM for gene expression measurement because the widely used RPKM measures can differ substantially between samples and thus potentially cause inflated statistical significance values. In contrast, TPM, as a slight modification of RPKM, eliminates this inconsistency and is suitable for the comparison of RNA abundance among samples [45]. Second, the method provided by Eisenberg and Levanon [12] is developed for the identification of housekeeping genes, but here we aimed to identify reference genes that can be easily detected in RT-qPCR assays. Therefore, the medium to high expression level is used as the fourth criterion. Notably, together with the requirement of low variance among samples (criterion II), the candidate reference genes will have a CV values within 0.2 in our case, which agrees with the standard for reference gene selection in other studies [14, 15, 51]. After applying the four criteria, we obtained candidate reference genes from three separate RNA-Seq datasets and found that the number of candidate reference genes differs among datasets (gonadal development > adult tissues > early development). Further analysis reveals that steps 2 and 4 are the most stringent, which filter out approximately 60~75% of the genes from the previous step. Step 3 is relatively loose and retains 84~97% of the genes. The stringency of step 1 differs among datasets, with a screening rate of 37% for early development, 54% for adult tissues and 62% for gonadal development, indicating that step 1 is the main reason for the difference in the reference gene number among datasets. As criterion 1 requires ubiquitous expression across all samples, the difference in reference gene numbers among datasets can be explained by the variation of sample heterogeneity, i.e., samples of early development are more distinct than those from different tissues or gonadal development. This result is expected because during early development, dramatic changes in gene expression can occur in a short span of time, which has been well reported in other organisms [52, 53]. The four criteria reported in our study facilitate the efficient detection of reference genes from RNA-Seq data.
Functional analysis of the candidate reference genes gives some interesting results. The top 10 most stable genes for adult tissues and gonadal development are all annotated proteins. Although the two gene lists only share a single gene (RS23), most of these genes are involved in protein synthesis. In contrast, 4 of the top 10 most stable genes for early development are unannotated. This result is not expected because housekeeping genes are supposed to play important roles in the maintenance of basal cellular functions, and most of these genes are well studied. We assume these unannotated genes in the top stable gene list could be novel genes that are critically important for the early development of scallops, bivalves or mollusks but not for other animals. Among the six annotated genes, four are related to energy production. This result is different from the function of the top reference genes for adult tissues and gonadal development, suggesting that embryos/larvae and adults may emphasize different biological processes or pathways. Consistent with the function of the top stable genes, the 568 core set reference genes are enriched in GO terms and KEGG pathways related to ribosomes, energy production, etc. Similar results are also reported for housekeeping or reference genes identified by genome-wide analysis in other organisms, such as humans [54], mice [13], and maize [55]. These processes should be critically important for life maintenance in various organisms.
The results of RNA-Seq were validated by RT-qPCR for 6 candidate and 6 commonly used reference genes. Although the samples used in RT-qPCR differ from those used in RNA-Seq, there seems to be high consistency between the two results, with candidate reference genes being more stable than most of the traditionally used reference genes. EF1B is more stable than the other five commonly used reference genes in early development, adult tissues and gonadal development, which is expected because EF1B is in our candidate reference gene list. ACT is the least stable gene in early development and adult tissues, which is consistent with the results from a previous report in the same species [29]. However, there is also some inconsistency between our study and the previous study. For example, CYTC, which showed medium stable expression in adult tissues in the previous study, is not recommended based on our analysis. This result could be due to a difference in the number of tissues investigated, 6 in the previous study and 11 in our study. The stable expression of CYTC and GAPDH during early development reported by Feng et al. also deviates from our result, primarily because Feng et al. only evaluated the expression stability until D-stage larvae, but there is obvious CYTC downregulation and GAPDH upregulation thereafter (Additional file 3: Figure S1). Similarly, the three traditionally used reference genes (ACT, CYTC and GAPDH) have been widely reported to be unsuitable as internal controls for RT-qPCR in various organisms, such as oysters, salmon, mice, and humans [32, 56, 57].
For bivalves of economic importance, some research has been conducted on genes related to reproduction [58–60]. Examining their expression dynamics during gonadal development is an important task in these studies. However, several traditionally used reference genes, such as EF1A [36, 40, 61], ACT [62] and GAPDH [63] have been applied, and until now, no study on the transcriptome-wide identification of reference genes in bivalves has been reported. According to our results from RNA-Seq analysis and RT-qPCR validation, ACT is not an appropriate reference gene and may result in misleading conclusions. Based on RT-qPCR analysis, EF1A ranks fourth after EIF3F, SELR1 and RS23, indicating that these genes are all suitable reference genes. The four genes exhibit different expression levels, with EF1A being most abundant, followed by RS23, EIF3F and SELR1; thus, researchers can select a reference gene according to the expression level of the genes of interest.
Although a high correlation between RNA-Seq data and RT-qPCR results has been reported in previous studies [36] [64], there is no formula for the relationship between the Ct of RT-qPCR and the TPM of RNA-Seq. The formula presented in our study will facilitate an estimation of the Ct prior to RT-qPCR experiments based on RNA-Seq data in Yesso scallop and might be applied to other organisms.
Conclusions
In this study, by taking advantage of 60 transcriptomes of the Yesso scallop, we identified candidate reference genes for early development, adult tissues, and gonadal development and found a core set of 568 reference genes. We also conducted stability comparisons of 6 candidate and 6 commonly used reference genes based on RNA-Seq data and RT-qPCR validation and found several traditionally used reference genes that were inappropriate as internal controls. Our study will benefit gene expression studies in bivalve mollusks.
Additional files
Table S1. Detailed information on the RNA-Seq datasets (XLSM 9 kb)
Table S2. Detailed information on the expression variability and annotation of the candidate reference genes in early development, adult tissues and gonadal development of the Yesso scallop (XLSM 330 kb)
Figure S1. The expression levels of CYTC and GAPDH during the early development of the Yesso scallop (PDF 33 kb)
Acknowledgements
We would like to thank all members of the laboratory for valuable discussions. We also would like to thank Dalian Zhangzidao Group for providing scallop materials and facilities.
Funding
This work was supported by the National Key Research and Development Project (2018YFD0900206), the National Natural Science Foundation of China (U1706203), the key Research and Development Program of Shandong Province (2016ZDJQ0208), and Fundamental Research Funds for the Central Universities (201762001 and 201841001). The funding bodies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Abbreviations
- ACT
Beta-actin
- ATP5J
ATP synthase-coupling factor 6
- ATP5L
ATP synthase subunit g
- ATPK
Putative ATP synthase subunit f
- CV
Coefficient of variation
- CYTC
Cytochrome c
- EF1A
Elongation factor 1-alpha
- EF1B
Elongation factor 1-beta
- EIF3F
Eukaryotic translation initiation factor 3 subunit F
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- GO
Gene Ontology
- HEL
DEAD-box RNA helicase
- IF4A1
Eukaryotic initiation factor 4A-I
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- NDUAB
NADH dehydrogenase [ubiquinone] 1 alpha subcomplex
- NDUS4
NADH dehydrogenase [ubiquinone] iron-sulfur protein 4
- OLA1
Obg-like ATPase 1
- RLA1
60S acidic ribosomal protein P1
- RM54
39S ribosomal protein L54
- RPKM
Reads Per Kilobase Million
- RPL16
39S ribosomal protein L16
- RS23
40S ribosomal protein S23
- RT-qPCR
Reverse transcription quantitative PCR
- SELR1
Sel1 repeat-containing protein 1
- TPM
Transcripts per million
Authors’ contributions
LZ and ZB conceived and designed the project. RL, MZ, and YPL collected the samples. YL performed the experiments. YL, LZ, and HW analyzed and interpreted the data. LZ, YL, SW drafted and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Animal materials of M. yessoensis used in this study were obtained from Zhangzidao Group Co., Ltd., Dalian Province, China. No field permissions were necessary to collect the animal samples for this study. The authors declared that the experimental research on the animals described in this paper was in compliance with institutional, national and international guidelines.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yajuan Li, Email: lyj930722@126.com.
Lingling Zhang, Phone: +86 532 82031969, Email: lingling80@ouc.edu.cn.
Ruojiao Li, Email: lrj2057@163.com.
Meiwei Zhang, Email: meiwzh@163.com.
Yangping Li, Email: liyangping2009@163.com.
Hao Wang, Email: wanghao5533@126.com.
Shi Wang, Email: swang@ouc.edu.cn.
Zhenmin Bao, Email: zmbao@ouc.edu.cn.
References
- 1.Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K. The real-time polymerase chain reaction. Mol Asp Med. 2006;27(2–3):95–125. doi: 10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Johnson G, Nour AA, Nolan T, Huggett J, Bustin S. Minimum information necessary for quantitative real-time PCR experiments. Methods Mol Biol. 2014;1160(1160):5. doi: 10.1007/978-1-4939-0733-5_2. [DOI] [PubMed] [Google Scholar]
- 3.Jarošová J, Kundu JK. Validation of reference genes as internal control for studying viral infections in cereals by quantitative real-time RT-PCR. BMC Plant Biol. 2010;10(1):146. doi: 10.1186/1471-2229-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang R, Dodd A, Lai D, McNabb WC, Love DR. Validation of zebrafish (Danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Biochim Biophys Sin. 2007;39(5):384–390. doi: 10.1111/j.1745-7270.2007.00283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mamo S, Gal AB, Bodo S, Dinnyes A. Quantitative evaluation and selection of reference genes in mouse oocytes and embryos cultured in vivo and in vitro. BMC Dev Biol. 2007;7(1):14. doi: 10.1186/1471-213X-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willems E, Mateizel I, Kemp C, Cauffman G, Sermon K, Leyns L. Selection of reference genes in mouse embryos and in differentiating human and mouse ES cells. Int J Dev Biol. 2004;50(7):627–635. doi: 10.1387/ijdb.052130ew. [DOI] [PubMed] [Google Scholar]
- 7.Ghani M, Sato C, Rogaeva E. Segmental duplications in genome-wide significant loci and housekeeping genes; warning for GAPDH and ACTB. Neurobiol Aging. 2013;34(6):1710–17e1. doi: 10.1016/j.neurobiolaging.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Olsvik PA, Søfteland L, Lie KK. Selection of reference genes for qRT-PCR examination of wild populations of Atlantic cod Gadus morhua. BMC Res Notes. 2008;1(1):47. doi: 10.1186/1756-0500-1-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu J, He F, Song S, Wang J, Yu J. How many human genes can be defined as housekeeping with current expression data? BMC Genomics. 2008;9(1):172. doi: 10.1186/1471-2164-9-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Velculescu VE, Madden SL, Zhang L, Lash AE, Yu J, Rago C, et al. Analysis of human transcriptomes. Nature Genet. 1999;23(4):387. doi: 10.1038/70487. [DOI] [PubMed] [Google Scholar]
- 11.Eisenberg E, Levanon EY. Human housekeeping genes are compact. Trends Genet. 2003;19(7):362–365. doi: 10.1016/S0168-9525(03)00140-9. [DOI] [PubMed] [Google Scholar]
- 12.Eisenberg E, Levanon EY. Human housekeeping genes, revisited. Trends Genet. 2013;29(10):569–574. doi: 10.1016/j.tig.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 13.Zeng J, Liu S, Zhao Y, Tan X, Aljohi HA, Liu W, Hu S. Identification and analysis of house-keeping and tissue-specific genes based on RNA-seq data sets across 15 mouse tissues. Gene. 2016;576(1):560–570. doi: 10.1016/j.gene.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 14.Xu H, Li C, Zeng Q, Agrawal I, Zhu X, Gong Z. Genome-wide identification of suitable zebrafish Danio rerio reference genes for normalization of gene expression data by RT-qPCR. J Fish Biol. 2016;88(6):2095–2110. doi: 10.1111/jfb.12915. [DOI] [PubMed] [Google Scholar]
- 15.Pombo MA, Zheng Y, Fei Z, Martin GB, Rosli HG. Use of RNA-seq data to identify and validate RT-qPCR reference genes for studying the tomato-Pseudomonas pathosystem. Sci Rep. 2017;7:44905. doi: 10.1038/srep44905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao D, Kong F, Sun P, Bi G, Mao Y. Transcriptome-wide identification of optimal reference genes for expression analysis of Pyropia yezoensis responses to abiotic stress. BMC Genomics. 2018;19(1):251. doi: 10.1186/s12864-018-4643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Huang S, Niu X, Chen D, Chen Q, Tian L, et al. Genome-wide identification and validation of new reference genes for transcript normalization in developmental and post-harvested fruits of Actinidia chinensis. Gene. 2018;645:1–6. doi: 10.1016/j.gene.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, He X, Yu F, Xiang Z, Li J, Thorpe KL, Yu Z. Characteristic and functional analysis of toll-like receptors (TLRs) in the lophotrocozoan, Crassostrea gigas, reveals ancient origin of TLR-mediated innate immunity. PLoS One. 2013;8(10):e76464. doi: 10.1371/journal.pone.0076464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chi C, Giri SS, Jun JW, Kim HJ, Kim SW, Yun S, Park SC. Effects of algal toxin okadaic acid on the non-specific immune and antioxidant response of bay scallop (Argopecten irradians) Fish Shellfish Immunol. 2017;65:111–117. doi: 10.1016/j.fsi.2017.03.031. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Wang R, Wang S, Zhang M, Ma X, Liu P, et al. Genome-wide identification and characterization of TRAF genes in the yesso scallop (Patinopecten yessoensis) and their distinct expression patterns in response to bacterial challenge. Fish Shellfish Immunol. 2015;47(1):545–555. doi: 10.1016/j.fsi.2015.09.050. [DOI] [PubMed] [Google Scholar]
- 21.Rico-Villa B, Pouvreau S, Robert R. Influence of food density and temperature on ingestion, growth and settlement of Pacific oyster larvae, Crassostrea gigas. Aquaculture. 2009;287(3–4):395–401. [Google Scholar]
- 22.Feng L, Li X, Yu Q, Ning X, Dou J, Zou J, et al. A scallop IGF binding protein gene: molecular characterization and association of variants with growth traits. PLoS One. 2014;9(2):e89039. doi: 10.1371/journal.pone.0089039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Sun G, Zeng Q, Chen Z, Hu X, Li H, et al. Predicting growth traits with genomic selection methods in Zhikong scallop (Chlamys farreri) Mar Biotechnol. 2018;1:1–11. doi: 10.1007/s10126-018-9847-z. [DOI] [PubMed] [Google Scholar]
- 24.Nagasawa K, Oouchi H, Itoh N, Takahashi KG, Osada M. In vivo administration of scallop GnRH-like peptide influences on gonad development in the yesso scallop, Patinopecten yessoensis. PloS One. 2015;10(6):e0129571. doi: 10.1371/journal.pone.0129571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.PAULET YM, BOUCHER J. Is reproduction mainly regulated by temperature or photoperiod in Pecten maximus? Invertebr Reprod Dev. 1991;19(1):61–70. [Google Scholar]
- 26.Qiu L, Song L, Xu W, Ni D, Yu Y. Molecular cloning and expression of a toll receptor gene homologue from Zhikong scallop, Chlamys farreri. Fish Shellfish Immunol. 2007;22(5):451–466. doi: 10.1016/j.fsi.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 27.Bao Y, Li L, Wu Q, Zhang G. Cloning, characterization, and expression analysis of extracellular copper/zinc superoxide dismutase gene from bay scallop Argopecten irradians. Fish Shellfish Immunol. 2009;27(1):17–25. doi: 10.1016/j.fsi.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 28.Guo H, Li Y, Zhang M, Li R, Li W, Lou J, et al. Expression of Cathepsin F in response to bacterial challenges in yesso scallop Patinopecten yessoensis. Fish Shellfish Immunol. 2018;80:141–147. doi: 10.1016/j.fsi.2018.06.005. [DOI] [PubMed] [Google Scholar]
- 29.Feng L, Yu Q, Li X, Ning X, Wang J, Zou J, et al. Identification of reference genes for qRT-PCR analysis in yesso scallop Patinopecten yessoensis. PLoS One. 2013;8(9):e75609. doi: 10.1371/journal.pone.0075609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siah A, Dohoo C, Mckenna P, Delaporte M, Berthe FCJ. Selecting a set of housekeeping genes for quantitative real-time PCR in normal and tetraploid haemocytes of soft-shell clams, Mya arenaria. Fish Shellfish Immunol. 2008;25(3):202–207. doi: 10.1016/j.fsi.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 31.Morga B, Arzul I, Faury N, Renault T. Identification of genes from flat oyster Ostrea edulis as suitable housekeeping genes for quantitative real time PCR. Fish Shellfish Immunol. 2010;29(6):937–945. doi: 10.1016/j.fsi.2010.07.028. [DOI] [PubMed] [Google Scholar]
- 32.Du Y, Zhang L, Xu F, Huang B, Zhang G, Li L. Validation of housekeeping genes as internal controls for studying gene expression during Pacific oyster (Crassostrea gigas) development by quantitative real-time PCR. Fish Shellfish Immunol. 2013;34(3):939–945. doi: 10.1016/j.fsi.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 33.Dheilly NM, Lelong C, Huvet A, Favrel P. Development of a Pacific oyster (Crassostrea gigas) 31,918-feature microarray: identification of reference genes and tissue-enriched expression patterns. BMC Genomics. 2011;12(1):468. doi: 10.1186/1471-2164-12-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang S, Zhang J, Jiao W, Ji L, Xun X, Sun Y, et al. Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat Ecol Evol. 2017;1(5):0120. doi: 10.1038/s41559-017-0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Sun X, Hu X, Xun X, Zhang J, Guo X, et al. Scallop genome reveals molecular adaptations to semi-sessile life and neurotoxins. Nat Commun. 2017;8(1):1721. doi: 10.1038/s41467-017-01927-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Zhang L, Sun Y, Ma X, Wang J, Li R, et al. Transcriptome sequencing and comparative analysis of ovary and testis identifies potential key sex-related genes and pathways in scallop Patinopecten yessoensis. Mar Biotechnol. 2016;18(4):453–465. doi: 10.1007/s10126-016-9706-8. [DOI] [PubMed] [Google Scholar]
- 37.Leung PT, Ip JC, Mak SS, Qiu JW, Lam PK, Wong CK, et al. De novo transcriptome analysis of Perna viridis highlights tissue-specific patterns for environmental studies. BMC Genomics. 2014;15(1):804. doi: 10.1186/1471-2164-15-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clark MS, Thorne MA, Vieira FA, Cardoso JC, Power DM, Peck LS. Insights into shell deposition in the Antarctic bivalve Laternula elliptica: gene discovery in the mantle transcriptome using 454 pyrosequencing. BMC Genomics. 2010;11(1):362. doi: 10.1186/1471-2164-11-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Y, Zhang Y, Fu X, Zhang R, Zou J, Wang S, et al. Identification of two secreted ferritin subunits involved in immune defense of yesso scallop Patinopecten yessoensis. Fish Shellfish Immunol. 2014;37(1):53–59. doi: 10.1016/j.fsi.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 40.Li R, Zhang L, Li W, Zhang Y, Li Y, Zhang M, et al. FOXL2 and DMRT1L are yin and Yang genes for determining timing of sex differentiation in the bivalve mollusk Patinopecten yessoensis. Front Physiol. 2018;9:1166. doi: 10.3389/fphys.2018.01166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang M, Wang Y, Li Y, Li W, Li R, Xie X, et al. Identification and characterization of neuropeptides by transcriptome and proteome analyses in a bivalve mollusc Patinopecten yessoensis. Front Genet. 2018;9:197. doi: 10.3389/fgene.2018.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li R. Molecular basis of sex differentiation in Patinopecten yessoensis. Qingdao: Doctoral dissertation, Ocean University of China; 2018.
- 43.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin H, Wan YW, Liu Z. Comprehensive evaluation of RNA-seq quantification methods for linearity. BMC Bioinf. 2017;18(4):117. doi: 10.1186/s12859-017-1526-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 2012;131(4):281–285. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- 46.Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17(1):13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beißbarth T, Speed TP. GOstat: find statistically overrepresented gene ontologies within a group of genes. Bioinformatics. 2004;20(9):1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 48.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:1–11. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 50.Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006;7(1):33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Narsai R, Ivanova A, Ng S, Whelan J. Defining reference genes in Oryza sativa using organ, development, biotic and abiotic transcriptome datasets. BMC Plant Biol. 2010;10(1):56. doi: 10.1186/1471-2229-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang Z, Sun J, Dong H, Luo O, Zheng X, Obergfell C, et al. Transcriptional profiles of bovine in vivo pre-implantation development. BMC Genomics. 2014;15(1):756. doi: 10.1186/1471-2164-15-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hofmann Falko, Schon Michael A., Nodine Michael D. The embryonic transcriptome of Arabidopsis thaliana. Plant Reproduction. 2019;32(1):77–91. doi: 10.1007/s00497-018-00357-2. [DOI] [PubMed] [Google Scholar]
- 54.She X, Rohl CA, Castle JC, Kulkarni AV, Johnson JM, Chen R. Definition, conservation and epigenetics of housekeeping and tissue-enriched genes. BMC Genomics. 2009;10(1):269. doi: 10.1186/1471-2164-10-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin F, Jiang L, Liu Y, Lv Y, Dai H, Zhao H. Genome-wide identification of housekeeping genes in maize. Plant Mol Biol. 2014;86(4–5):543–554. doi: 10.1007/s11103-014-0246-1. [DOI] [PubMed] [Google Scholar]
- 56.Jorgensen SM, Kleveland EJ, Grimholt U, Gjoen T. Validation of reference genes for real-time polymerase chain reaction studies in Atlantic salmon. Mar Biotechnol. 2006;8(4):398–408. doi: 10.1007/s10126-005-5164-4. [DOI] [PubMed] [Google Scholar]
- 57.De Jonge HJ, Fehrmann RS, de Bont ES, Hofstra RM, Gerbens F, Kamps WA, et al. Evidence based selection of housekeeping genes. PLoS One. 2007;2(9):e898. doi: 10.1371/journal.pone.0000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu J, Zhang L, Li Y, Li R, Zhang M, Li W, et al. Genome-wide identification and expression profiling of the SOX gene family in a bivalve mollusc Patinopecten yessoensis. Gene. 2017;627:530–537. doi: 10.1016/j.gene.2017.07.013. [DOI] [PubMed] [Google Scholar]
- 59.Liu XL, Zhang ZF, Shao MY, Liu JG, Muhammad F. Sexually dimorphic expression of foxl2 during gametogenesis in scallop Chlamys farreri, conserved with vertebrates. Dev Genes Evol. 2012;222(5):279–286. doi: 10.1007/s00427-012-0410-z. [DOI] [PubMed] [Google Scholar]
- 60.Guévélou E, Huvet A, Galindo-Sánchez CE, Milan M, Quillien V, Daniel JY, et al. Sex-specific regulation of AMP-activated protein kinase (AMPK) in the Pacific oyster Crassostrea gigas. Biol Reprod. 2013;89(4):100–101. doi: 10.1095/biolreprod.113.109728. [DOI] [PubMed] [Google Scholar]
- 61.Yue C, Li Q, Yu H. Gonad transcriptome analysis of the Pacific oyster Crassostrea gigas identifies potential genes regulating the sex determination and differentiation process. Mar Biotechnol. 2018;20(2):206–219. doi: 10.1007/s10126-018-9798-4. [DOI] [PubMed] [Google Scholar]
- 62.Liang S, Zhang Z, Yang D, Chen Y, Qin Z. Different expression of sox17 gene during gametogenesis between scallop Chlamys farreri and vertebrates. Gene Expr Patterns. 2017;25:102–108. doi: 10.1016/j.gep.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 63.Teaniniuraitemoana V, Huvet A, Levy P, Klopp C, Lhuillier E, Gaertner-Mazouni N, et al. Gonad transcriptome analysis of pearl oyster Pinctada margaritifera: identification of potential sex differentiation and sex determining genes. BMC Genomics. 2014;15(1):491. doi: 10.1186/1471-2164-15-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Everaert C, Luypaert M, Maag JL, Cheng QX, Dinger ME, Hellemans J, Mestdagh P. Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci Rep. 2017;7(1):1559. doi: 10.1038/s41598-017-01617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Detailed information on the RNA-Seq datasets (XLSM 9 kb)
Table S2. Detailed information on the expression variability and annotation of the candidate reference genes in early development, adult tissues and gonadal development of the Yesso scallop (XLSM 330 kb)
Figure S1. The expression levels of CYTC and GAPDH during the early development of the Yesso scallop (PDF 33 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.