Abstract
Aim:
The purpose of this study was to isolate and characterize the Mannheimia haemolytica and Pasteurella multocida from blood, nasal discharge, and lung tissue of pneumonic goats.
Materials and Methods:
A total of 14 goats were investigated for pneumonic pasteurellosis. Of 14 goats, nasal swabs and blood samples were collected from 10 clinically diseased animals. Moreover, lung tissue and heart blood samples were collected during necropsy of four goats died with pneumonia. All the samples were processed for the isolation of M. haemolytica and P. multocida in the laboratory. Bacterial isolates were identified by cultural and biochemical characters and 16S rRNA sequence analysis. All the isolates were subjected to susceptibility testing using commonly used antimicrobials. M. haemolytica isolates were characterized by PHSSA gene detection. P. multocida isolates were characterized by KMT1 gene detection and capsule typing.
Results:
On necropsy of dead goats, the pneumonia was characterized as acute fibrinous bronchopneumonia. Bacterial culture revealed the isolation of M. haemolytica (7) and P. multocida (5) of 10 clinical cases. Moreover, M. haemolytica and P. multocida were coisolated from two of the lung tissues. Furthermore, one of the other two lung tissues showed the isolation of M. haemolytica while the other showed recovery of P. multocida. Bacterial isolates were specifically identified by the 16S rRNA sequence analysis. The isolates showed reduced susceptibility to β-lactams, aminoglycosides, and fluoroquinolones. Moreover, the PHSSA and KMT1 genes were specifically detected among M. haemolytica, and P. multocida isolates, respectively. All P. multocida isolates belonged to serogroup A.
Conclusion:
The present study reported an occurrence of pneumonic pasteurellosis caused by M. haemolytica and P. multocida in a goat flock.
Keywords: bronchopneumonia, goat, Mannheimia haemolytica, Pasteurella multocida, pneumonic pasteurellosis
Introduction
Pneumonic pasteurellosis, also known as respiratory mannheimiosis, is most common among the respiratory tract infections with a wide prevalence in ruminant animals. Small ruminants such as goats are fairly susceptible and contract the disease due to exposure to physical stress or unfavorable environmental conditions [1]. It is one of the most common respiratory illnesses in goats throughout the world [2]. Pneumonia in goat is clinically characterized by anorexia, fever (40-41°C), painful coughing, dyspnea, mucopurulent nasal discharge, and depression.
Bacterial pneumonia is most often caused by Mannheimia haemolytica and Pasteurella multocida which are more frequently associated with the outbreak of acute pneumonia and death of goats in all age groups [3]. P. multocida, an opportunistic pathogen, is analogous to M. haemolytica [4] and is one of the most important respiratory pathogens of domestic ruminants in which it causes serious outbreaks of acute pneumonia [2,5]. This syndrome is caused by a complex interaction between the environmental stress, microorganisms, and immunity of the host. Although pneumonia in small ruminants is primarily caused by certain viral agents and is predisposed by the extremes of environmental insults, M. haemolytica is the most frequently detected bacterial pathogen [6]. Furthermore, M. haemolytica promotes P. multocida colonization in lung tissue, leading to more severe disease [7].
Presumptive bacterial identification may be made on the basis of cultural and biochemical features. However, molecular methods are required for confirmatory bacterial identification [8]. The goat is an important food animal for sustaining the livelihood of various rural families in Chhattisgarh, India. Therefore, it was planned to isolate and characterize M. haemolytica and P. multocida associated with clinical pneumonia in goats.
Materials and Methods
Ethical approval
The approval from the Institutional Animal Ethics Committee was not required for the present study since the samples were collected from the animals without animal experimentation and the dead animals during the necropsy.
Collection and processing of samples
A total of 14 goats (six adults, two hogget, and six suckling kids) in a backyard flock of 25 goats, maintained in one of the villages of Dongargaon block of district Rajnandgaon in the state of Chhattisgarh, were clinically ill and showing anorexia, fever, mucoid nasal discharge, and coughing. Moreover, four goats including one adult and three suckling kids suddenly died after an episode of fever and respiratory signs. All the animals were investigated for the occurrence of pneumonic pasteurellosis. The nasal swabs were collected from all the diseased animals (n=10) showing respiratory symptoms. Lung tissue samples were collected from dead animals (n=04) at the time of necropsy. For sampling during necropsy of the dead animals, the outer surface of the pneumonic lungs was first cleaned by touching the lung surface with a heated spatula before cutting their inner surface. The inner part of lung tissue from each animal was collected with aseptic measures for the isolation of M. haemolytica and P. multocida. Simultaneously, the heart blood samples were collected directly in the sterile syringes. The lung tissue samples were aseptically processed by making 20% w/v tissue homogenate using sterilized phosphate buffer saline (PBS) in laminar air flow cabinet near flame. The nasal swabs were suspended in 0.5 ml of the sterilized PBS.
Bacterial isolation and identification
All the samples were directly streaked on blood agar base (HiMedia, Mumbai) supplemented with 7% defibrinated goat blood and incubated aerobically for 24-48 h at 37°C. The plates were then examined for bacterial growth and the colonies examined for colony morphology, color, and odor. The suspected colonies were stained by Gram staining, observed, and tested for the following characteristics: presence or absence of hemolysis on blood agar, motility, indole and oxidase production, and growth on MacConkey agar (HiMedia, Mumbai). Gram-negative coccobacilli identified by the standard cultural and biochemical tests above were selected for further identification of P. multocida or M. haemolytica [9]. The pure bacterial colonies were maintained in brain heart infusion (BHI) agar (HiMedia, Mumbai) slants for further use.
Antimicrobial susceptibility testing
All of P. multocida and M. haemolytica isolates were tested for their susceptibility to amoxicillin (30 µg), cefotaxime (30 µg), amikacin (30 µg), gentamicin (10 µg), ciprofloxacin (5 µg), enrofloxacin (5 µg), tetracycline (30 µg), and chlortetracycline (30 µg) disks (HiMedia, Mumbai) using disk diffusion method [10]. Escherichia coli ATCC 25922 was used as a quality control strain. The interpretation of the results was based on the Clinical and Laboratory Standards Institute breakpoints [11].
Isolation of DNA from bacterial colonies
The isolates presumptively identified as M. haemolytica and P. multocida were streaked on BHI agar (HiMedia, Mumbai) and incubated aerobically at 37°C for 48 h to isolate the genomic DNA. About 5-10 colonies of each of the pure isolates of M. haemolytica and P. multocida were transferred into 1.5 ml Eppendorf tubes. The bacterial colonies were washed twice in nuclease-free water by centrifugation at 10,000 rpm for 3 min in a microcentrifuge. The genomic DNA was isolated using DNeasy Blood and Tissue Kit (Qiagen, USA) as per the manufacturer’s instructions.
Polymerase chain reaction (PCR) amplification
PCR amplification was carried out using the specific primers (Sigma-Aldrich, USA) to identify the bacterial isolates (Table-1) [12-15]. The reaction mixtures and amplification conditions were optimized for all the genes. M. haemolytica and P. multocida isolates were specifically identified based on the amplification of 16S rRNA using universal primers. Further, M. haemolytica and P. multocida were identified by species-specific amplification of the PHSSA and KMT1 gene, respectively. P. multocida isolates were subjected to capsule typing. For PCR amplification, 50 ng of DNA was added to 25 µl reaction mixture containing 200 µM of dNTPs, 0.2 µM of each primer, 1.875 mM of MgCl2, and 1.25 U of Taq DNA polymerase (Sigma-Aldrich, USA) in 1×PCR buffer. PCR conditions for PHSSA included the initial denaturation at 95°C for 3 min, followed by 35 cycles consisted of denaturation at 95°C for 1 min, annealing at 48°C for 1 min, extension at 72°C for 30 s, and the final extension at 72°C for 5 min. PCR conditions for the KMT1 gene comprised initial denaturation at 95°C for 3 min, followed by 35 cycles consisted of denaturation at 95°C for 45 s, annealing at 56°C for 45 s, extension at 72°C for 1 min, and the final extension at 72°C for 5 min. The amplified PCR products (5 µl) were separated in agarose gel (1.5% w/v) stained with ethidium bromide (0.5 µg/ml) by running in horizontal submarine electrophoresis unit using 1×TAE as running buffer and examined under the gel documentation system (UVP, USA).
Table-1.
Primer sequence of different genes.
| Gene | Primer sequence | Amplicon size (bp) | Annealing temp (°C) | References |
|---|---|---|---|---|
| 16S rRNA | 27F 5`AGAGTTTGATCMTGGCTCAG3` | ~1466 | 52 | [12] |
| 1492R 5`CGGTTACCTTGTTACGACTT3` | ||||
| PHSSA | F 5` TTCACATCTTCATCCTC3` | 327 | 48 | [13] |
| R 5` TTTTCATCCTCTTCGTC3 | ||||
| KMT1 | F 5`ATCCGCTATTTACCCAGTGG3` | 457 | 56 | [14] |
| R 5`GCTGTAAACGAACTCGCCAC3` | ||||
| hyaD-hyaC | F 5`TGCCAAAATCGCAGTCAG3` | 1046 | 55 | [15] |
| R 5`TTGCCATCATTGTCAGTG3 |
Nucleotide sequencing
The amplified PCR products (50 µl) were purified using gel extraction kit (Qiagen, USA) as per the manufacturer’s instructions. The purified PCR products were quantified (~100 ng/µl concentration) and used for sequencing by bidirectional Sanger`s sequencing method.
Results
The clinically ill animals showed anorexia, fever (104-105°F), mucoid nasal discharge, and coughing for up to two days. A few animals showed mild diarrhea with semi-solid fecal consistency. Of the 14 sick goats, four goats died after an episode of fever and respiratory signs. On necropsy of the dead goats, pneumonia was diagnosed among all the cases and classified as acute fibrinous bronchopneumonia characterized by serofibrinous exudation in the bronchiolar lumen. The pneumonic lung tissues showed congestion and petechial and ecchymotic type of hemorrhages (Figure-1). Consolidation was recorded in whole of the apical and parts of the diaphragmatic lobes. The tracheal rings were hemorrhagic and the tracheal lumen was filled with the mucus and froth. Bacterial culture revealed isolation of Gram-negative coccobacilli and small bacilli suspected to P. multocida and/or M. haemolytica. The isolates were presumptively identified by cultural and biochemical characters. The small, smooth, white-creamy, mucoid, and hemolytic, as well as non-hemolytic colonies, were developed on the blood agar. The hemolytic colonies were further streaked on the MacConkey agar which showed a very mild growth after 48 h. The non-hemolytic colonies failed to grow on the MacConkey agar. All the isolates did not show any growth on the Deoxycholate citrate agar (HiMedia, Mumbai). P. multocida suspected isolates revealed Gram-negative small coccobacilli by Gram’s staining. However, the colonies suspected to M. haemolytica showed Gram-negative, pleomorphic, coccobacillary-to-small bacillary morphology. All the isolates were positive for the production of catalase and oxidase. The indole was produced by P. multocida isolates. Bacterial isolation showed the recovery of M. haemolytica (5) and P. multocida (3) of eight clinical cases. Two clinical cases were found to be complicated by both M. haemolytica and P. multocida. Moreover, M. haemolytica and P. multocida were coisolated from two of the lung tissues. Furthermore, one of the other two lung tissues showed the isolation of M. haemolytica while the other showed recovery of P. multocida. The heart blood of the dead animals showed bacterial isolation similar to the respective lung tissues. Thus, a total of 18 distinct isolates including M. haemolytica (n=10) and P. multocida (n=08) were recovered from all 14 cases.
Figure-1.
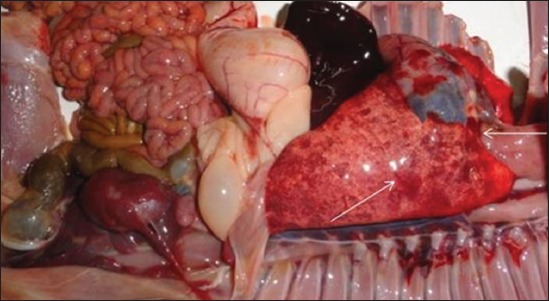
Characteristic hemorrhagic lesions on the diaphragmatic lobe of the pneumonic lung.
The isolates showed susceptibility to tetracycline and chlortetracycline. However, an intermediate sensitivity was recorded against amikacin (33.3%), gentamicin (22.2%), amoxicillin (16.6%), cefotaxime (16.6%), ciprofloxacin (16.6%), and enrofloxacin (16.6%).
PCR amplification of 16S rRNA using universal primers followed by sequencing confirmed M. haemolytica and P. multocida bacterial isolates. M. haemolytica and P. multocida isolates were further identified by specific amplification of PHSSA (327 bp) (Figure-2) and KMT1 (457 bp) (Figure-3) gene, respectively. Capsule typing of P. multocida isolates showed the presence of hyaD-hyaC gene specific to serogroup A (1046 bp) (Figure-4). Nucleotide sequences have been submitted to NCBI GenBank database with their respective accession numbers (MH068780.1, MH068781.1, MH068782.1, MH068783.1, and MK295774.1).
Figure-2.
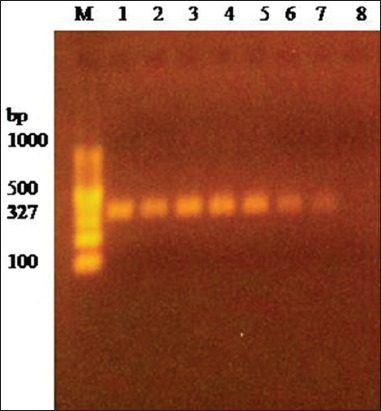
Amplification of PHSSA gene (327 bp) of Mannheimia haemolytica. Lane 1-7: Test positive. Lane 8: Test negative. Lane M: 100 bp DNA Ladder.
Figure-3.
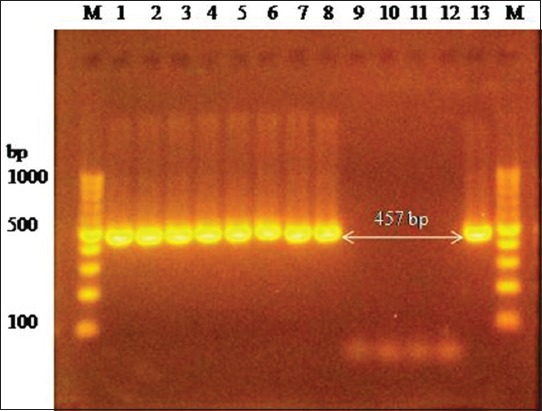
Amplification of KMT1 gene (457 bp) specific to Pasteurella multocida. Lane 1-8: Test positive. Lane 9-11: Test negative. Lane 12: Negative control. Lane 13: Positive control. Lane M: 100 bp DNA Ladder.
Figure-4.
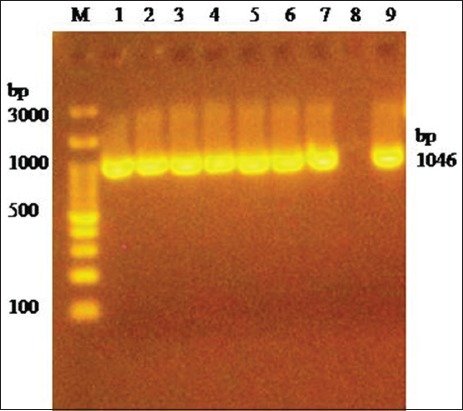
Amplification of hyaD-hyaC gene (1046 bp) specific to Pasteurella multocida serogroup A capsule. Lane 1-7: Test positive. Lane 8: Negative control. Lane 9: Positive control. Lane M: 100 bp DNA Ladder.
Discussion
Globally, the goat is reared as a food animal which provides meat, leather, milk, and manure and inhabited mainly to semi-arid and arid climate. Goat farming plays an important role for sustaining the livelihood of various rural families in Chhattisgarh, India. However, variable environmental factors predispose the goats to the development of respiratory diseases including pneumonia. Pneumonic pasteurellosis is important to sheep and goats throughout the world and can be acute with fever, listlessness, dyspnea, poor appetite, and sudden death [16]. Physical environmental stresses predispose the goats to respiratory infections by means of lowering their innate resistance to P. haemolytica infection [16]. There is a direct correlation of sudden environmental changes in terms of rainfall, wind, temperature etc. to the occurrence, and development of pneumonia in small ruminants [17]. A confluence of important factors such as production stress in prevailing environmental conditions in a particular geographical area and changing weather patterns predispose these animals to pasteurellosis (mannheimiosis) [18]. A survey by 126 member countries of the World Organization for Animal Health (OIE) remarked that pasteurellosis has emerged as one of the major animal diseases affected by climate change [19].
The necropsy-based gross pathological observations are important to diagnose the diseases like pneumonia among animals in rural areas. Such a diagnosis of infectious diseases is very important for record keeping and National Disease Reporting System. Necropsy-based observation of fibrinous bronchopneumonia with congestion and hemorrhages of the lungs and trachea were similar to the earlier studies [20]. Among pneumonic lungs, the consolidation was recorded in whole of the apical lobe [21]. The presence of hemorrhagic lesions on the different organs, namely heart epicardium, spleen, and kidneys indicated septicemia due to these bacteria.
Pneumonia in small ruminants is primarily caused by parainfluenza-3 virus and respiratory syncytial virus and mycoplasma infection [22] and is predisposed by variable weather conditions. Respiratory viral infections affect mucociliary clearance mechanisms in lungs for removing the pathogens that reach the lower respiratory tract and thus increase the susceptibility of sheep and goats to secondary bacterial infections [16]. The respiratory viral agents create a favorable environment in the lungs supporting the bacterial growth by interfering with the mucociliary clearance mechanism of the respiratory tract and by downregulating the phagocytosis by the pulmonary macrophages [23]. However, M. haemolytica, the most frequently isolated bacterial pathogen from the lungs, is considered as the main cause of the pneumonic pasteurellosis/respiratory mannheimiosis [6]. Further, the respiratory syncytial virus and parainfluenza-3 virus can cause non-fatal pneumonia but are not necessary predisposing agents for M. haemolytica-caused pneumonia of bighorn sheep [24]. However, M. ovipneumoniae, a cause of non-fatal pneumonia in bighorn sheep, can predispose them to fatal pneumonia due to M. haemolytica infection [22]. The viral and mycoplasma investigation was not done in the present study. M. haemolytica significantly enhances the colonization of P. multocida, leading to more severe disease [7].
The isolates were susceptible to tetracycline and chlortetracycline, which might probably be due to the controlled use of these antimicrobials in the flock. Susceptibility results in the present study were more or less similar to the previous studies [25,26]. Reduced susceptibility to β-lactams (amoxicillin and cefotaxime), aminoglycosides (amikacin and gentamicin), and fluoroquinolones (ciprofloxacin and enrofloxacin) might be due to their selective use in the flock.
The presence of the virulent strains of M. haemolytica and/or P. multocida indicated their role for the pathogenesis and development of pneumonia in goats. The PHSSA is homologous to virulence-associated genomic fragments of M. haemolytica, and it could have significant pathobiological effect in the progression of pneumonic pasteurellosis [27]. P. multocida Type A involvement to pneumonic sheep has been reported from Tamil Nadu, India [28], and thus supported the findings of the present study. P. multocida lipopolysaccharide capsule plays an important role in the pathogenesis of the disease. It stimulates humoral immunity and is considered as a protective antigen [29]. It is observed that the capsule plays a significant role in the resistance to phagocytosis [30] and P. multocida requires a complete lipopolysaccharide structure to replicate in vivo and to establish the disease.
Isolation and identification of M. haemolytica and P. multocida based on the cultural and biochemical characters and by the PCR assays targeting the specific genes have been reported earlier similar to the present study [13,31]. Moreover, the specific identification of P. multocida by 16S rRNA and KMT1 gene and the characterization by capsular typing has a significant impact on the epidemiology and control of pasteurellosis in domestic animals [8,32,33]. It is also proposed that the molecular typing methods provide rapid bacterial identification and have proved to be more specific over the cultural and biochemical tests [13,34].
Conclusion
High prevalence of pneumonic pasteurellosis that leads to high goat mortality was recorded in the backyard flock of a village in Chhattisgarh. The occurrence of goat pneumonia might have been predisposed due to environmental and nutritional stress. Acute fibrinous bronchopneumonia was predominantly recorded among the dead goats. Isolation and molecular characterization of M. haemolytica and P. multocida confirmed the occurrence of pneumonic pasteurellosis or respiratory mannheimiosis in the affected goats.
Authors’ Contributions
NR was involved in the disease investigation in the field, planning, sampling, and bacterial isolation; VRG, FFK, KKK, and RKS performed sample processing and microbiological analysis; DDH and DKJ performed molecular analysis and manuscript writing. All authors read and approved the final manuscript.
Acknowledgments
The authors are thankful to the Dean of the College of Veterinary Science and Animal Husbandry, Anjora, Durg, Chhattisgarh, for providing the necessary facilities to conduct the present research work. The authors also thank Dr. Fateh Singh, Scientist, ICAR-Central Sheep and Wool Research Institute, Avikanagar, Tonk, Rajasthan, for his help in the molecular analysis of the bacterial isolates. The authors did not receive any fund for this study.
Competing Interests
The authors declare that they have no competing interests.
Publisher’s Note
Veterinary World remains neutral with regard to jurisdictional claims in published institutional affiliation.
References
- 1.Mohamed R.A, Abdelsalam E.B. A review on pneumonic pasteurellosis (respiratory mannheimiosis) with emphasis on pathogenesis, virulence mechanism and predisposing factors. Bulg. J. Vet. Med. 2008;11(3):139–160. [Google Scholar]
- 2.Marru H.D, Anijajo T.T, Hassen A.A. A study on ovine pneumonic pasteurellosis:Isolation and identification of pasteurellae and their antibiogram susceptibility pattern in Haramaya district, Eastern Hararghe, Ethiopia. BMC Vet. Res. 2013;9(1):239. doi: 10.1186/1746-6148-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falade S. Further Pasteurella isolates from the Republic of Zambia. Trop. Vet. 2002;20(3):130–131. [Google Scholar]
- 4.Dowling A, Hodgson J.C, Schock A, Donachie W, Eckersall P.D, Mckendrick I.J. Experimental induction of pneumonic pasteurellosis in calves by intratracheal infection with Pasteurella multocida biotype A:3. Res. Vet. Sci. 2002;73(1):37–44. doi: 10.1016/s0034-5288(02)00037-1. [DOI] [PubMed] [Google Scholar]
- 5.Abdullah F.F.J, Tijjani A, Adamu L, Chung E.L.T, Abba Y, Mohammed K, Saharee A.A, Haron A.W, Sadiq M.A, Mohd A.M.L. Pneumonic pasteurellosis in a goat. Iran. J. Vet. Med. 2014;8(4):293–296. [Google Scholar]
- 6.Shanthalingam S, Goldy A, Bavananthasivam J, Subramaniam R, Batra S.A, Kugadas A, Raghavan B, Dassanayake R.P, Jennings-Gaines J.E, Killion H.J, Edwards W.H, Ramsey J.M, Anderson N.J, Wolff P.L, Mansfield K, Bruning D, Srikumaran S. PCR assay detects Mannheimia haemolytica in culture-negative pneumonic lung tissues of bighorn sheep (Ovis canadensis) from outbreaks in the Western USA, 2009-2010. J. Wildl. Dis. 2014;50(1):1–10. doi: 10.7589/2012-09-225. [DOI] [PubMed] [Google Scholar]
- 7.Dabo S.M, Taylor J.D, Confer A.W. Pasteurella multocida and bovine respiratory disease. Anim. Health Res. Rev. 2007;8(2):129–150. doi: 10.1017/S1466252307001399. [DOI] [PubMed] [Google Scholar]
- 8.Hassan G.M, El-Feky Z.A, Eissa E.A, Teleb A.A. Rapid diagnosis of virulent Pasteurella multocida isolated from farm animals with clinical manifestation of pneumonia respiratory infection using 16S rDNA and KMT1gene. Asian Pac. J. Trop. Dis. 2016;6(1):21–26. [Google Scholar]
- 9.Quinn P.J, Markey B.K, Carter M.E, Donnelly W.J, Leonard F.C. Veterinary Microbiology and Microbial Disease. 1st ed. USA: Wiley-Blackwell; 2001. [Google Scholar]
- 10.CLSI (Clinical and Laboratory Standards Institute) Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals-Document M31-A3. Wayne, PA, USA. 2008 [Google Scholar]
- 11.CLSI (Clinical and Laboratory Standards Institute) Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals;Second Informational Supplement-Document VET01-S2. Wayne, PA, USA. 2013 [Google Scholar]
- 12.Lane D.J. 16S/23S rRNA sequencing. In: Stackenbrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. Chichester, United Kingdom: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- 13.Hawari A.D, Hassawi D.S, Sweiss M. Isolation and identification of Mannheimia haemolytica and Pasteurella multocida in sheep and goats using biochemical tests and random amplified polymorphic DNA (RAPD) analysis. J. Biol. Sci. 2008;8(7):1251–1254. [Google Scholar]
- 14.Townsend K.M, Frost A.J, Lee C.W, Papadimitriou J.M, Dawkins H.J.S. Development of PCR assays for species and type specific identification of Pasteurella multocida isolates. J. Clin. Microbiol. 1998;36(4):1096–1100. doi: 10.1128/jcm.36.4.1096-1100.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Townsend K.M, Boyce J.D, Chung J.Y, Frost A.J, Adler B. Genetic organization of Pasteurella multocida cap loci and development of a multiplex capsular PCR typing system. J. Clin. Microbiol. 2001;39(3):924–929. doi: 10.1128/JCM.39.3.924-929.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brogden K.A, Lehmkuhl H.D, Cutlip R.C. Pasteurella haemolytica complicated respiratory infections in sheep and goats. Vet. Res. 1998;29:3–4. 233–254. [PubMed] [Google Scholar]
- 17.Galapero J, Fernández S, Pérez C.J, Calle-Alonso F, Rey J, Gomez L. Identifying risk factors for ovine respiratory processes by using Bayesian networks. Small Rumin. Res. 2016;136(3):113–120. [Google Scholar]
- 18.Kumar J, Dixit S.K, Kumar R. Rapid detection of Mannheimia haemolytica in lung tissues of sheep and from bacterial culture. Vet. World. 2015;8(9):1073–1077. doi: 10.14202/vetworld.2015.1073-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Black P, Nunn M. Impact of Climate Change and Environmental Change on Emerging and Re-Emerging Animal Diseases and Animal Production. In:Compendium of Technical Items Presented in 77thGeneral Session of the OIE World Assembly of Delegates, Paris, France, 24th-29thMay. 2009:15. [Google Scholar]
- 20.Sadeghian S, Dezfouli M.R.M, Kojouri G.A, Bazargani T.T, Tavasoli A. Pasteurella multocida pneumonic infection in goat:Hematological, biochemical, clinical and pathological studies. Small Rumin. Res. 2011;100:2–3. 189–194. [Google Scholar]
- 21.Akloul K, Menoueri M.N. Pneumonia in Algerian Ouled Djellal sheep:Bacteriological study and macroscopic aspect of lung lesions. Afr. J. Microbiol. Res. 2016;10(40):1685–1693. [Google Scholar]
- 22.Dassanayake R.P, Shanthalingam S, Herndon C.N, Subramaniam R, Lawrence P.K, Bavananthasivam J, Cassirer E.F, Haldorson G.J, Foreyt W.J, Rurangirwa F.R, Knowles D.P, Besser T.E, Srikumaran S. Mycoplasma ovipneumoniae can predispose bighorn sheep to fatal Mannheimia haemolytica pneumonia. Vet. Microbiol. 2010;145:3–4. 354–359. doi: 10.1016/j.vetmic.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 23.Jakab G.J. Viral-bacterial interactions in pulmonary infection. Adv. Vet. Sci. Comp. Med. 1982;26:155–171. [PubMed] [Google Scholar]
- 24.Dassanayake R.P, Shanthalingam S, Subramaniam R, Herndon C.N, Bavananthasivam J, Haldorson G.J, Foreyt W.J, Evermann J.F, Herrmann-Hoesing L.M, Knowles D.P, Srikumaran S. Role of Bibersteinia trehalosi respiratory syncytial virus, and parainfluenza-3 virus in bighorn sheep pneumonia. Vet. Microbiol. 2013;162(1):166–172. doi: 10.1016/j.vetmic.2012.08.029. [DOI] [PubMed] [Google Scholar]
- 25.Kumar P, Singh V.P, Agrawal R.K, Singh S. Identification of Pasteurella multocida isolates of ruminant origin using polymerase chain reaction and their antibiogram study. Trop. Anim. Health Prod. 2009;41(4):573–578. doi: 10.1007/s11250-008-9226-2. [DOI] [PubMed] [Google Scholar]
- 26.Sarangi L.N, Thomas P, Gupta S.K, Priyadarshini A, Kumar S, Nagaleekar V.K, Kumar A, Singh V.P. Virulence gene profiling and antibiotic resistance pattern of Indian isolates of Pasteurella multocida of small ruminant origin. Comp. Immunol. Microbiol. Infect. Dis. 2015;38(1):33–39. doi: 10.1016/j.cimid.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez C, Murtaugh M.P, Maheswaran S.K. Genomic distribution of a serotype 1-specific antigen coding DNA fragment of Pasteurella haemolytica. J. Vet. Med. B Infect. Dis. Vet. Public Health. 1991;38(8):599–609. doi: 10.1111/j.1439-0450.1991.tb00917.x. [DOI] [PubMed] [Google Scholar]
- 28.Prabhakar T.G, Prabhakar P, Venkadabadi Molecular characterization of Pasteurella multocida isolated from an incidence of sheep pasteurellosis in Karamadai hill tract of Tamil Nadu. Tamilnadu J. Vet. Anim. Sci. 2010;6(2):81–87. [Google Scholar]
- 29.Harper M, Boyce J.D, Adler B. Pasteurella multocida pathogenesis:125 years after Pasteur. FEMS Microbiol. Lett. 2006;265(1):1–10. doi: 10.1111/j.1574-6968.2006.00442.x. [DOI] [PubMed] [Google Scholar]
- 30.Harmon B.G, Glisson J.R, Latimer K.S, Steffens W.L, Nunnally J.C. Resistance of Pasteurella multocida A:3, 4 to phagocytosis by turkey macrophages and heterophils. Am. J. Vet. Res. 1991;52(9):1507–1511. [PubMed] [Google Scholar]
- 31.Aski H.S, Tabatabaei M. Occurrence of virulence-associated genes in Pasteurella multocida isolates obtained from different hosts. Microb. Pathog. 2016;96(7):52–57. doi: 10.1016/j.micpath.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 32.Singh F, Sonawane G.G, Meena R.K. Molecular detection of virulent Mannheimia haemolytica and Pasteurella multocida in lung tissues of pneumonic sheep from semiarid tropics, Rajasthan, India. Turk. J. Vet. Anim. Sci. 2018;42(6):556–561. [Google Scholar]
- 33.Singh F, Sonawane G.G, Meena R.K. Pathology, isolation and characterisation of virulent and diverse Mannheimia haemolytica and Pasteurella multocida associated with fatal pneumonia in sheep, Rajasthan, India. Comp. Clin. Pathol. 2019 Doi:org/10.1007/s00580-019-02916-z. [Google Scholar]
- 34.Tabatabaei M, Abdollahi F. Isolation and identification of Mannheimia haemolytica by culture and polymerase chain reaction from sheep's pulmonary samples in Shiraz, Iran. Vet. World. 2018;11(5):636–641. doi: 10.14202/vetworld.2018.636-641. [DOI] [PMC free article] [PubMed] [Google Scholar]


