Abstract
Electron cryo-microscopy, or simply cryo-EM, refers mainly to three very different yet closely related techniques: electron crystallography, single particle cryo-EM and electron cryo-tomography. In the past few years, single particle cryo-EM in particular has triggered a revolution in structural biology and has become a newly dominant discipline. This review examines the fascinating story of its start and evolution over the past forty plus years, delves into how and why the recent technological advances have been so ground-breaking, and briefly considers where the technique may be headed in the future.
Introduction
Physicist and Nobel Laureate Richard Feynman once famously stated, “It is very easy to answer many fundamental biological questions; you just look at the thing!” (1). Indeed, the central idea behind structural biology is that once we are able to “look” at “things” in great enough detail to discern their atomic structures, we will naturally be able to answer how and why the components and players of complex biological processes work the way they do. True to this aim, structural biology has contributed significantly to major biological discoveries throughout history (2). It has also and will continue to facilitate developments of therapeutic agents to cure diseases or ameliorate pathological symptoms.
The major techniques available to structural biologists are X-ray crystallography, nuclear magnetic resonance (NMR) spectroscopy and electron microscopy (EM). Among them, X-ray crystallography contributes most of the atomic coordinates of biological macromolecules deposited in the Protein DataBank (PDB) (3). In this method, structures are determined from diffraction patterns generated from well-ordered three-dimensional (3D) crystals of biological macromolecules. The resolutions of structures determined depend largely upon the quality of the crystals; in short, obtaining well-ordered 3D crystals of sufficient size is usually a prerequisite for atomic structure determination of any biological macromolecules using this technique. It works well for many proteins or stable complexes; however, for certain categories of biological macromolecules, growing large and well-ordered 3D crystals is a very difficult or impossible task. For examples, crystallizing integral membrane proteins or large and dynamic complexes and machineries can be challenging. An extension of the X-ray crystallography is the X-ray Free Electron Laser (XFEL), whose ultimate goal is to determine atomic structures without crystals, but currently still requires a large amount of small crystals (4).
Can atomic structures of biological macromolecules be determined without crystallization? Or is it possible, as Feynman once suggested, to determine their structures by “looking” at them using a powerful electron microscope? Early pioneers pursued this question in the 1970s and developed a new EM-based method known today as single particle cryo-EM (5, 6). At the beginning, the method yielded rather low-resolution results. Drawn to the promise of being able to study biological macromolecules without crystallizing them, however, the cryo-EM community dedicated themselves to perfecting the technique over more than four decades, yielding steady improvements in both the technique’s applicability and the resolution of its results. Gradually, it has become a major tool in structural biology, complementary to X-ray crystallography and widely used to study large macromolecular complexes that are difficult to be crystallized. A few years ago, some amazing technological breakthroughs further enabled routine atomic resolution structure determinations by this method. Today, single particle cryo-EM is no longer a complementary technique but a dominant one, changing the field of structural biology in a profound and unprecedented way and facilitating significant new discoveries.
A brief history of single particle cryo-EM
It is actually quite difficult to “look” at biological macromolecules in an electron microscope. Determining their atomic structures from electron micrographs is even more complicated. Firstly, EM images are two-dimensional (2D) projections of biological macromolecules, but not their 3D structures. This was resolved by DeRosier and Klug, who demonstrated that a 3D structure can be reconstructed by combining 2D projection images of the same object along different directions (7).
Secondly, because of strong scattering, the electron beam has to be confined in a high-vacuum and all EM samples need to be placed inside said vacuum. This is not a problem for inorganic materials. But if one simply places a biological sample inside an electron microscope, vacuum caused dehydration would destroy sample’s structural integrity. The seemingly impossible task of keeping protein samples hydrated in high vacuum was accomplished by Taylor and Glaeser. They recorded better than 3 Å resolution electron diffraction patterns from frozen hydrated catalase crystals, demonstrating that the structural integrity of biological macromolecules in high vacuum can be maintained by frozen hydration (Figure 1A) (8, 9). The practical implication of this approach, however, was not easy until a plunge freezing technique was developed by Dubochet and colleagues in the 1980s (10, 11). They applied purified protein samples in solution to an EM grid covered with a thin layer of carbon holey film, blotted the grid with a filter paper removed most of the solution, surface tension drove the remaining solution into a thin liquid film across holes in the carbon film. Plunging the grid rapidly into liquid ethane cooled by liquid nitrogen froze it into a thin layer of amorphous ice with protein sample embedded within it in random orientations. After that, the frozen grid was transferred into an electron microscope and kept at near liquid nitrogen temperature for imaging (Figure 1B). This method is still used routinely without major changes, except we now use a machine to blot and plunge grids with tunable parameters.
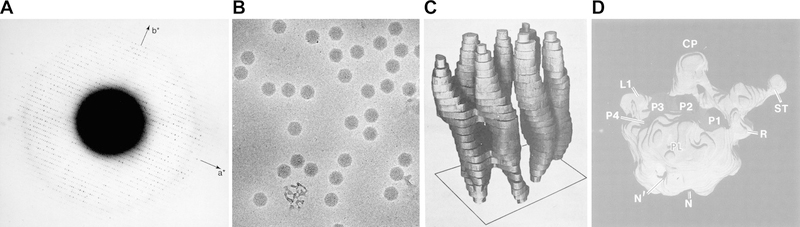
Figure 1. Establishing of single particle cryo-EM.
A. An electron diffraction pattern of frozen hydrated catalase crystal (9). The diffraction spots are visible at beyond 3 Å resolution. This experiment established the concept of cryo-EM. B. An electron micrograph of frozen hydrated adenovirus particle recorded from a frozen hydrated grid prepared by plunge freezing. The micrograph is from the same dataset described in (11) and reproduced from (9). C. The first 3D reconstruction of bacteriorhodopsin determined by electron crystallography (15). D. A 3D model of the 50S ribosome subunit determined by single particle reconstruction of negatively stained large ribosomal subunit from Escherichia coli (22).
Thirdly, radiation damage by the high-energy electron beam limits the total electron dose that can be used to image biological samples. The consequence is that images recorded with such a low electron dose have very poor signal-to-noise ratios (SNR). Cooling the sample to liquid nitrogen or even liquid helium temperature can reduce the radiation damage and allow the sample to tolerate a higher electron dose (12, 13), but still far from enough to be able to directly visualize high-resolution details from raw micrographs. Henderson and Unwin overcame this problem by employing a crystallographic approach, averaging images of many identical proteins packed as 2D crystals (14). Images of glucose embedded 2D crystals recorded with very low electron doses show no visible features but their Fourier transforms show clear reflections. Similarly, electron diffraction from such 2D crystals at very low electron doses produced good quality diffraction patterns. Combining the phases calculated from the Fourier transformations of images and the amplitudes obtained from diffractions produced a high-resolution projection map of the specimen (14). Combining data collected from specimens tilted at different angles produced a 3D reconstruction similar to the density map of an X-ray crystal structure. This approach produced the first structure of an integral membrane protein, initially at ~7 Å resolution (Figure 1C) (15) and finally at atomic resolution (16). The method became known as electron crystallography, which relies upon well-ordered 2D crystals. This method has produced atomic structures of several integral membrane proteins (17, 18) and one soluble protein (19). The highest resolution achieved was 1.9 Å, resolving a lipid bilayer surrounding AQP0 water channel (20). But the difficulty of growing well-ordered 2D crystals hinders the broad application of the method.
In parallel, Frank proposed an idea to determine protein structures without crystallization: computationally combining images of many individual protein particles of the same type (5). This conceptually novel idea was first tested using protein samples that were negatively stained for EM observation (21, 22). The later combination of this approach with the plunge freezing sample preparation became what we now call “single particle cryo-EM”. It does not require growing proteins into crystals of any form. Instead, it determines structure by computationally aligning and combining cryo-EM images of many biological molecules randomly oriented within a thin layer of vitreous ice. A large number of images is needed to both enhance SNR and to provide all different views needed for 3D reconstruction (Figure 1D). More detailed technical descriptions of this method can be found in many recent reviews, for example (23, 24).
Furthermore, what electron micrographs record are projections of the specimen convoluted by a contrast transfer function (CTF). CTF is a sine function oscillating in a frequency dependent manner that modulates both phase and amplitude of an image in frequency space (25). In addition to a number of microscope dependent parameters, CTF is determined by how much off the focus an image is recorded, the so-called defocus. To retain the highest resolution, images must be recorded very close to focus. Such images, however, have very limited contrast. This is not a problem for radiation insensitive inorganic materials, which can be imaged with very high electron doses to generate sufficient contrast while retaining a high-resolution signal. However, images of radiation sensitive frozen hydrated biological samples have to be recorded with a large defocus to generate sufficient contrast, which in turn dampens the high-resolution signal.
The resolution of a single particle cryo-EM structure depends on many factors, including the resolution and contrast of individual particle images, accuracy of aligning these images with each other, obtaining a sufficient number of images from all necessary views of the macromolecule within a reasonable timeframe, the conformational and compositional homogeneity of these particles, and access to powerful enough computers to process images efficiently. Less than optimal conditions for any of these factors, for example, instability of the electron microscope, relatively poor performance of the image recording device and beam-induced sample motions, accuracy of classifying and aligning particle images limited by image quality and computational algorithms, and limited computer power to process very large number of particle images, etc., can limit the resolution. Because of these many technical challenges, the resolution achieved was, for a long time, limited to levels far from sufficient for deriving de novo atomic models.
At a time when the resolution of some best reconstructions were at the 30 ~ 50Å range (for example, the mammalian 40S ribosome (26) and ryanodine receptor (27)) microscopists were nevertheless encouraged by the prediction that single particle cryo-EM could, theoretically, achieve atomic resolution (28). The prediction was made by considering how much electron scattering a biological sample could tolerate, how much structural information or SNR such scattering can produce, and—in a perfect situation—how many images would be needed to produce a reconstruction at a given resolution. Although the claim was bold, the theories behind are solid and this motivated the cryo-EM community to push the methodology towards its perfection. Finally, twenty years after Henderson made the prediction, the necessary breakthrough came when direct electron detection cameras were developed and became commercially available (29). The expanded capabilities of the new cameras coupled with unprecedented and ever-increasing computational power fueled the development of new computational algorithms so that cryo-EM images could be reliably produced with sufficient quality for atomic structure determination (30, 31). The resolution potential of single particle cryo-EM at long last became a reality (32).
Transformative technological breakthroughs
The direct electron detection camera:
Since the beginning of cryo-EM, electron micrographs were recorded on photographic films, which were subsequently processed in dark rooms and digitized for computational processing. For single particle cryo-EM, the performance of film was sufficient to produce 3D reconstructions at sub-nanometer resolutions (33), at which α-helices are resolved, but atomic models cannot be built. It was difficult to push the resolution further because photographic film is particularly poor at retaining low frequency signals. Images were therefore recorded with high defocuses in order to generate sufficient contrast for particle picking or alignment, at the price of losing high-resolution signal. For large icosahedral viruses, it was possible to record two images of the same specimen area, the first with a low defocus to retain the high-resolution signal, and the second with a high defocus to generate sufficient contrast (34). With substantial effort, the resolution of some virus particles reached near atomic level (35), but the whole process is slow and tedious, limiting the throughput of structure determination.
In the late 1990s, charge coupled device (CCD) cameras were introduced to record EM micrographs digitally. CCD cameras cannot detect electrons directly and require a phosphor scintillator to convert electrons into photon signals. Such conversion blurs a point event of a single electron striking the sensor into a blob of photons of much larger size and reduces high-resolution signals (36). Characterized by detective quantum efficiency (DQE), which measures the level of signals retained by a camera in spatial frequency (36, 37), CCD cameras are not suitable for routine high-resolution structures determinations (Figure 2A). The impact of introducing CCD cameras into cryo-EM was nonetheless significant because it facilitated automated image acquisition (38).
Figure 2. Direct electron detection camera enabled atomic structure determination.
A. Comparison of DQE curves of a scintillator based CCD camera (black), and direct electron detection camera K2 operating in base mode (blue) and super-resolution counting mode (red) (31). B. A typical electron micrograph of archaeal 20S proteasome (~700kDa in molecular weight) recorded using direct electron detection camera (60). C. Fourier power spectrum calculated from the image (B) (60). High-resolution signal at ~3Å resolution is clearly visible (60). D. A portion of density map from the 3D reconstruction of archaeel 20S proteasome (31).
A major breakthrough that elevated single particle cryo-EM from “blobology” to a practical technique for atomic structure determination was the introduction of direct electron detection cameras (29). Such cameras detect charges generated directly from electrons striking the camera sensor, thus localizing the electron with much greater precision and resulting in significantly higher DQE than scintillator-based cameras. These sensors also run at high frame rates, enabling cryo-EM images to be recorded as a stack of movie frames that each is recorded in a short period of time (39). Certain cameras can even count individual electron events on every single frame. The DQE of such single electron counting cameras is even higher (31) (Figure 2A). Images recorded with direct electron detection cameras retain signal both at high frequency, for high-resolution structure determination, and at low frequency, for contrast required for image alignment (Figure 2B and C).
Being able to record images as movie stacks facilitated many new imaging approaches that are critical for maximizing the achievable resolution. Most importantly, it allows the correction of beam induced image motion (30, 31, 39) and partially mitigates radiation damage (31, 40, 41), solving the two most difficult problems in cryo-EM. The use of the direct detection camera, improved motion correction, and the ability to record images at a high electron dose make the reconstruction of 3D density maps at atomic resolution possible for many proteins.
New image processing algorithms:
The resolution of a reconstruction also depends on the conformational homogeneity of the sample and accuracy of image alignment of all particles used to reconstruct the density map. Image classification and alignment are thus the two most critical steps in the computational image processing. Because each particle image has a poor SNR, individual particles cannot be classified and assigned to a specific class and orientation with certainty, making a probabilistic approach better suited for image classification and alignment. Early attempts to use a maximum likelihood-based probabilistic approach in cryo-EM image processing were made in the late 1990s (42). Later, a Bayesian approach to cryo-EM reconstruction was described (43), and implemented in RELION (44), a user-friendly program that soon became very popular in single particle cryo-EM image processing. This approach is more powerful and robust than traditional deterministic approaches, particularly in classifying a subset of particle images with homogeneous conformations out of a larger and more heterogeneous dataset. Coincidentally, this development happened at around the same time that direct electron detection cameras were becoming widely used in cryo-EM. Together, they allowed the full potential of single particle cryo-EM, as theoretically predicted more than 20 years earlier, to be realized!
Automation in electron microscopy:
Technically, single particle cryo-EM had been a rather complicated and tedious technique. A very large number of high-quality cryo-EM images are required for each reconstruction. Such images were mainly collected by individuals who had many years of training and experience in operating complicated electron microscopes. The technical requirement on users was high because they needed a good understanding of not only the electron optic system, but also many fundamental technical issues related to cryo-EM data acquisition. They also need to be very patient, sitting in front of a microscope for many hours repeating the same procedure to collect large numbers of micrographs. The level of expertise required made single particle cryo-EM inaccessible to the wider structural biology community. Fortunately, pioneered by Carragher and Potter, who recognized this problem early on, automation of high-quality data acquisition was developed (38). The electron microscope itself also evolved to better suited for automated data acquisition. Nowadays, many cryo-EM facilities are operated in a way similar to X-ray synchrotron beam-lines—supported by a few highly trained staff scientists. In such facilities, regular users with minimal training can also acquire high-quality cryo-EM data, even remotely, using automated procedures.
A new era of structural biology
Thanks to these technological breakthroughs in single particle cryo-EM, structural biology has entered a new era. Structures of many difficult crystallization targets are now within the reach. One such area is integral membrane proteins. A specific example is the transient receptor potential (TRP) ion channel. The TRP channel superfamily contains seven subfamilies with a total of 27 members in humans (45, 46). Each of these channels play different physiological roles; some of them are drug targets for treatment of various human diseases (47). With the exception of some small domains, attempts to crystallize any member of the TRP channel superfamily had failed (48). This lack of structural information ended when atomic structures of the TPRV1 ion channel, a capsaicin receptor that plays a physiological role in sensing heat and activating pain pathways, were determined by single particle cryo-EM in three different functional states (49, 50). This discovery demonstrated the power of single particle cryo-EM and showed that it rivaled X-ray crystallography in determining atomic structures of challenging protein complexes that resist crystallization. The TRPV1 structures prompted many crystallographers to think seriously about cryo-EM, and many quickly seized the opportunity to apply it to their favorite difficult targets. With the roadblock of crystallization removed, atomic structures of integral membrane proteins are now being determined at a rapid pace. In less than five years, atomic structures of at least one member of each of the seven TRP channel subfamilies have now been determined (Figure 3).
Figure 3. Single particle cryo-EM enables atomic structure determination of TRP channels.
Ribbon diagrams of atomic structures from each subfamily of TRP channel superfamily. They are TRPV1 (49), TRPA1 (61), TRPM4 (62), TRPC4 (63), NOMOPC (also named TRPN1) (64), PKD2 (or TRPP2) (65) and TRPML (66). The rapid pace of integral membrane protein structure determination is enabled by single particle cryo-EM and is unprecedented.
Single particle cryo-EM is also game-changing for structural studies of many large and dynamic complexes and machineries that were impossible for crystallization. A traditional approach was to fit crystal structures of individual components or domains into lower resolution cryo-EM density maps of the whole complex (51). Now, atomic structures of many large complexes are determined directly using single particle cryo-EM. An excellent example is the spliceosome complex. Past efforts produced atomic structures of some small components (52), as well as low-resolution cryo-EM structures of the whole complex (53). Now, in only a few short years, atomic structures of the spliceosome in different functional states have been determined (54, 55).
One can easily call forth many more examples to illustrate how the recent technological breakthroughs in single particle cryo-EM have changed how we tackle complex biological problems. The rapid pace of advancement in structural biology is unprecedented. It has also attracted major pharmaceutical companies, with many hoping to implement cryo-EM into structure-based drug discovery and optimization.
The future
Single particle cryo-EM continues to move forward fast, with many new technologies being developed. One such example is the Volta phase plate, which is a thin carbon film placed in the back focal plane of the microscope’s objective lens. It adds a phase shift to the CTF so that image recorded at near focus has good contrast (56), facilitating studying of very small proteins (57). Another example is the development of a new sample preparation technology that is fundamentally different than the current blotting method (58). The new method promises many benefits including reducing the total amount of sample needed from microliters to nanoliters.
As impressive as these innovations have been, there are still many technical details that can be improved. Regularly achieving resolutions close to 2 Å and beyond is one goal; improving robustness and throughput is another. Once very high resolutions can be achieved reliably, pharmaceutical companies will be able to routinely use EM to speed up structure-based drug discovery. The range of what can be studied by single particle cryo-EM can also be expanded to include smaller or bigger targets with higher resolution, as well as more dynamic complexes or assemblies with irregular shapes.
There is still more that single particle cryo-EM can offer to further biological discoveries beyond structure determination if we continue to push the boundary of this technology. For example, in theory, image classification can sort a cryo-EM dataset that contains particles of heterogeneous conformation or composition into multiple classes, each corresponding to a different functional state. By freezing cryo-EM grids at specific time points, it could be possible to derive structural information in a time dependent manner (59). It may therefore be possible to understand and break down the complex cycles, movements, and processes of biological macromolecular complexes and machineries step by step, in complete atomic detail. In the future, it may also be possible to study both physiological and pathological protein states by affinity purifying specific proteins directly from cells onto EM grids for single particle cryo-EM studies.
The past forty years have seen the evolution of single particle cryo-EM from blobology to a routine and powerful method for atomic structure determination. The technique’s new popularity is attracting not only more users, but also talents from different fields ranging from physics, material science, mathematics, and machine learning. This infusion of new ideas and a larger community heralds an even brighter future for single particle cryo-EM full of method development, collaboration, and most importantly, wonderful new biological discoveries.
Acknowledgement
This is a brief story of single particle cryo-EM but not a comprehensive historical review. I apologize to many colleagues whose important works are not cited here. I thank many of my colleagues, all past and present members of my laboratory for sharing their thoughts about single particle cryo-EM with me, and Linda Wang for editing. The cryo-EM work in my laboratory is funded by various NIH grants: R01GM082893, R01GM098672, R01HL134183, P50GM082250 (Nevan Krogan), P01GM111126 (Robert Stroud), and S10OD020054 and S10OD021741. I am a Howard Hughes Medical Institute Investigator.
Reference
- 1.Feynman RP, in Feynman and computation, Hey AJG, Ed. (Perseus Books Cambridge, MA, USA, 1999), pp. 63–76. [Google Scholar]
- 2.Shi Y, A glimpse of structural biology through X-ray crystallography. Cell 159, 995–1014 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Jones N, Crystallography: Atomic secrets. Nature 505, 602–603 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Kang Y et al. , Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature 523, 561–567 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frank J, Averaging of low exposure electron micrographs of non-periodic objects. Ultramicroscopy 1, 159–162 (1975). [DOI] [PubMed] [Google Scholar]
- 6.Dubochet J et al. , Cryo-electron microscopy of vitrified specimens. Q Rev Biophys 21, 129–228 (1988). [DOI] [PubMed] [Google Scholar]
- 7.De Rosier DJ, Klug A, Reconstruction of three dimensional structures from electron micrographs. Nature 217, 130–134 (1968). [DOI] [PubMed] [Google Scholar]
- 8.Taylor KA, Glaeser RM, Electron diffraction of frozen, hydrated protein crystals. Science 186, 1036–1037 (1974). [DOI] [PubMed] [Google Scholar]
- 9.Taylor KA, Glaeser RM, Retrospective on the early development of cryoelectron microscopy of macromolecules and a prospective on opportunities for the future. J Struct Biol 163, 214–223 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubochet J, Chang JJ, Freeman R, Lepault J, McDowall AW, Frozen aqueous suspensions. Ultramicroscopy 10, 55–61 (1982). [Google Scholar]
- 11.Adrian M, Dubochet J, Lepault J, McDowall AW, Cryo-electron microscopy of viruses. Nature 308, 32–36 (1984). [DOI] [PubMed] [Google Scholar]
- 12.Stark H, Zemlin F, Boettcher C, Electron radiation damage to protein crystals of bacteriorhodopsin at different temperature. Ultramicroscopy 63, 75–79 (1996). [Google Scholar]
- 13.Fujiyoshi Y, The structural study of membrane proteins by electron crystallography. Adv Biophys 35, 25–80 (1998). [DOI] [PubMed] [Google Scholar]
- 14.Unwin PN, Henderson R, Molecular structure determination by electron microscopy of unstained crystalline specimens. J Mol Biol 94, 425–440 (1975). [DOI] [PubMed] [Google Scholar]
- 15.Henderson R, Unwin PN, Three-dimensional model of purple membrane obtained by electron microscopy. Nature 257, 28–32 (1975). [DOI] [PubMed] [Google Scholar]
- 16.Henderson R et al. , Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol 213, 899–929 (1990). [DOI] [PubMed] [Google Scholar]
- 17.Kuhlbrandt W, Wang DN, Fujiyoshi Y, Atomic model of plant light-harvesting complex by electron crystallography. Nature 367, 614–621 (1994). [DOI] [PubMed] [Google Scholar]
- 18.Murata K et al. , Structural determinants of water permeation through aquaporin-1. Nature 407, 599–605 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Nogales E, Wolf SG, Downing KH, Structure of the alpha beta tubulin dimer by electron crystallography. Nature 391, 199–203 (1998). [DOI] [PubMed] [Google Scholar]
- 20.Gonen T et al. , Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature 438, 633–638 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frank J, Goldfarb W, Eisenberg D, Baker TS, Reconstruction of glutamine synthetase using computer averaging. Ultramicroscopy 3, 283–290 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radermacher M, Wagenknecht T, Verschoor A, Frank J, Three-dimensional structure of the large ribosomal subunit from Escherichia coli. EMBO J 6, 1107–1114 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng Y, Grigorieff N, Penczek PA, Walz T, A primer to single-particle cryo-electron microscopy. Cell 161, 438–449 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fernandez-Leiro R, Scheres SH, Unravelling biological macromolecules with cryo-electron microscopy. Nature 537, 339–346 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wade RH, A brief look at imaging and contrast transfer. Ultramicroscopy 46, 145–156 (1992). [Google Scholar]
- 26.Srivastava S, Verschoor A, Radermacher M, Grassucci R, Frank J, Three-dimensional reconstruction of mammalian 40 S ribosomal subunit embedded in ice. J Mol Biol 245, 461–466 (1995). [DOI] [PubMed] [Google Scholar]
- 27.Radermacher M et al. , Cryo-electron microscopy and three-dimensional reconstruction of the calcium release channel/ryanodine receptor from skeletal muscle. J Cell Biol 127, 411–423 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henderson R, The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q Rev Biophys 28, 171–193 (1995). [DOI] [PubMed] [Google Scholar]
- 29.McMullan G, Faruqi AR, Henderson R, Direct Electron Detectors. Methods Enzymol 579, 1–17 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Bai XC, Fernandez IS, McMullan G, Scheres SH, Ribosome structures to near-atomic resolution from thirty thousand cryo-EM particles. Elife 2, e00461 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li X et al. , Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods 10, 584–590 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng Y, Single-Particle Cryo-EM at Crystallographic Resolution. Cell 161, 450–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bottcher B, Wynne SA, Crowther RA, Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature 386, 88–91 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Ludtke SJ, Chiu W, Focal pair merging for contrast enhancement of single particles. J Struct Biol 144, 73–78 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Zhang X, Jin L, Fang Q, Hui WH, Zhou ZH, 3.3 A cryo-EM structure of a nonenveloped virus reveals a priming mechanism for cell entry. Cell 141, 472–482 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mooney P, Optimization of image collection for cellular electron microscopy. Methods Cell Biol 79, 661–719 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Samei E, Flynn MJ, Reimann DA, A method for measuring the presampled MTF of digital radiographic systems using an edge test device. Med Phys 25, 102–113 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Suloway C et al. , Automated molecular microscopy: the new Leginon system. J Struct Biol 151, 41–60 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Brilot AF et al. , Beam-induced motion of vitrified specimen on holey carbon film. J Struct Biol 177, 630–637 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheres SH, Beam-induced motion correction for sub-megadalton cryo-EM particles. Elife 3, e03665 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grant T, Grigorieff N, Measuring the optimal exposure for single particle cryo-EM using a 2.6 A reconstruction of rotavirus VP6. Elife 4, e06980 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sigworth FJ, A maximum-likelihood approach to single-particle image refinement. J Struct Biol 122, 328–339 (1998). [DOI] [PubMed] [Google Scholar]
- 43.Scheres SH, A Bayesian view on cryo-EM structure determination. J Mol Biol 415, 406–418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheres SH, RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clapham DE, TRP channels as cellular sensors. Nature 426, 517–524 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Julius D, TRP channels and pain. Annu Rev Cell Dev Biol 29, 355–384 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Moran MM, TRP Channels as Potential Drug Targets. Annu Rev Pharmacol Toxicol 58, 309–330 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Li M, Yu Y, Yang J, Structural biology of TRP channels. Adv Exp Med Biol 704, 1–23 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao M, Cao E, Julius D, Cheng Y, Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao E, Liao M, Cheng Y, Julius D, TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lander GC et al. , Complete subunit architecture of the proteasome regulatory particle. Nature 482, 186–191 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galej WP, Nguyen TH, Newman AJ, Nagai K, Structural studies of the spliceosome: zooming into the heart of the machine. Curr Opin Struct Biol 25, 57–66 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohi MD, Ren L, Wall JS, Gould KL, Walz T, Structural characterization of the fission yeast U5.U2/U6 spliceosome complex. Proc Natl Acad Sci U S A 104, 3195–3200 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fica SM, Nagai K, Cryo-electron microscopy snapshots of the spliceosome: structural insights into a dynamic ribonucleoprotein machine. Nat Struct Mol Biol 24, 791–799 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi Y, Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat Rev Mol Cell Biol 18, 655–670 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Danev R, Buijsse B, Khoshouei M, Plitzko JM, Baumeister W, Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc Natl Acad Sci U S A 111, 15635–15640 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khoshouei M, Danev R, Plitzko JM, Baumeister W, Revisiting the Structure of Hemoglobin and Myoglobin with Cryo-Electron Microscopy. J Mol Biol 429, 2611–2618 (2017). [DOI] [PubMed] [Google Scholar]
- 58.Dandey VP et al. , Spotiton: New features and applications. J Struct Biol 202, 161–169 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaledhonkar S, Fu Z, White H, Frank J, Time-Resolved Cryo-electron Microscopy Using a Microfluidic Chip. Methods Mol Biol 1764, 59–71 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Li X, Zheng SQ, Egami K, Agard DA, Cheng Y, Influence of electron dose rate on electron counting images recorded with the K2 camera. J Struct Biol 184, 251–260 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Paulsen CE, Armache JP, Gao Y, Cheng Y, Julius D, Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature 520, 511–517 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Autzen HE et al. , Structure of the human TRPM4 ion channel in a lipid nanodisc. Science 359, 228–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vinayagam D et al. , Electron cryo-microscopy structure of the canonical TRPC4 ion channel. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin P et al. , Electron cryo-microscopy structure of the mechanotransduction channel NOMPC. Nature 547, 118–122 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen PS et al. , The Structure of the Polycystic Kidney Disease Channel PKD2 in Lipid Nanodiscs. Cell 167, 763–773 e711 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Q et al. , Structure of mammalian endolysosomal TRPML1 channel in nanodiscs. Nature 550, 415–418 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]