Abstract
Sphingolipid metabolism is known to play a role in cell death, survival, and therapy resistance in cancer. Sphingolipids, particularly dihydroceramide and ceramide, are associated with antiproliferative or cell death responses, respectively, and are central to effective cancer therapy. Within the last decade, strides have been made in elucidating many intricacies of sphingolipid metabolism. New information has emerged on the mechanisms by which sphingolipid metabolism is dysregulated during malignancy and how cancer cells survive and/or escape therapeutic interventions. This chapter focuses on three main themes: (1) sphingolipid enzymes that are dysregulated in cancer, particularly in prostate cancer; (2) inhibitors of sphingolipid metabolism that antagonize prosurvival responses; and (3) sphingolipid-driven escape mechanisms that allow cancer cells to evade therapies. We explore clinical and preclinical approaches to interdict sphingolipid metabolism and provide a rationale for combining strategies to drive the generation of antiproliferative ceramides with prevention of ceramide clearance.
1. INTRODUCTION
Prostate cancer is the most diagnosed cancer among American men and the second leading cause of cancer deaths in the United States. Over 160,000 new cases will be diagnosed in 2018 and nearly 30,000 deaths are anticipated (Siegel, Miller, & Jemal, 2018). Treatment for prostate cancer varies, but advanced disease typically involves surgical removal of the gland followed by androgen ablation therapy, radiation, or chemotherapy. Sphingolipid metabolism is known to play a role in cell death, survival, and therapy resistance in cancer, including prostate cancer (Beckham, Cheng, Marrison, Norris, & Liu, 2013; Ogretmen, 2018). Ceramide, a central molecule in sphingolipid metabolism, is generally associated with antiproliferative responses while sphingosine 1-phosphate (S1P) promotes survival, angiogenesis, inflammation, and resistance. Cancer therapy is therefore aimed at promoting ceramide generation and inhibiting S1P levels.
Within the last decade, strides have been made in elucidating the intricacies of sphingolipid metabolism, but even early studies identified correlations between therapy responses in prostate cancer and modulation of intracellular ceramides. For example, hormone withdrawal in androgensensitive LNCaP prostate cancer cells led to cell cycle arrest and apoptosis and was accompanied by an increase in C16-ceramide that could be blocked by inhibitors of the de novo pathway of sphingolipid metabolism (Eto et al., 2003). Nearly 20 years ago, researchers suggested that the relative levels of sphingolipid metabolites may also play a role in susceptibility to radiation therapy and that the enhancement of ceramide and sphingosine generation could be of therapeutic value (Nava et al., 2000). The role of sphingolipids in radiation therapy has been comprehensively reviewed (Hajj & Haimovitz-Friedman, 2013). Radiation resistance or treatment failure remains a major obstacle in advanced prostate cancer. While radiation exposure induces ceramide-mediated antitumor effects, ceramide itself induces a protective response in a subset of cells resulting not only in resistance to radiation therapy but also in cross-resistance to chemotherapy (Cheng et al., 2013; Mahdy et al., 2009). This chapter discusses sphingolipid enzymes, inhibitors, and mechanisms of therapy resistance with a focus on prostate cancer.
2. SPHINGOLIPID ENZYMES
2.1. Ceramide Synthases
Ceramide is a central molecule in sphingolipid metabolism and consists of a sphingoid base that, via an amide bond, is attached to an acyl chain. Differences in acyl chain length and saturation give rise to a variety of ceramide species that endow cells or tissues with a “ceramide profile.” The discovery of six different ceramide synthases (CerSs) with tissue-specific distribution and acyl chain preferences was key in beginning to understand the physiology and pathologies associated with alterations in ceramide profiles. Because several comprehensive reviews on CerSs have recently been published, we will only briefly introduce these enzymes (Mullen, Hannun, & Obeid, 2012; Park, Park, & Futerman, 2014; Wegner, Schiffmann, Parnham, Geisslinger, & Grosch, 2016).
CerSs are primarily localized to the endoplasmic reticulum, but some evidence suggests they can also be detected in mitochondria (and associated membranes) and the nucleus (and perinuclear structures). CerSs acetylate either dihydrosphingosine (dhSph) in the de novo pathway to generate dihydroceramide (dhCer) or sphingosine in the salvage pathway to generate ceramide (Fig. 1). Levy and Futerman (2010) analyzed mRNA distribution of CerSs across tissues. The mRNA for CerS1, which preferentially generates C18-ceramide, is strongly expressed in the brain and skeletal muscle, whereas mRNA for CerS3, which generates very-long-chain ceramides (C22eC26-ceramides), is primarily detected in the testis. The substrate preference of CerS2 is similar to CerS3, but in contrast to CerS3, CerS2 mRNA is widely expressed. CerS4 generates C18- and C20-ceramides and is strongly expressed in the skin, heart, and spleen. CerS5 and CerS6 both generate C16-ceramides, but their mRNAs differ in tissue distribution. Regulation of CerS activity is incompletely understood but complex. Members of the CerS family are regulated individually at the genetic and epigenetic level as well as through posttranslational mechanisms and stability. Activity can also be influenced through homodimer or heterodimer formation of family members (Laviad, Kelly, Merrill, & Futerman, 2012).
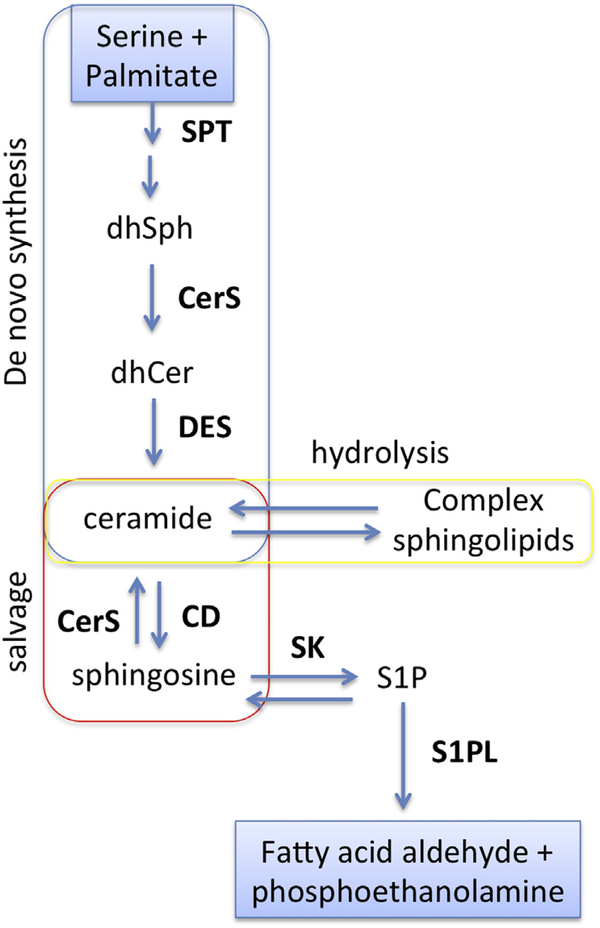
Figure 1. Sphingolipid metabolism and key enzymes.
Ceramide is central to sphingolipid metabolism and can be generated de novo (blue), through hydrolysis of complex sphingolipids (yellow) or via the salvage pathway (red). In the de novo pathway, serine palmitoyltransferase (SPT) catalyzes the first reaction resulting in 3-ketosphinganine, which is reduced to dihydrosphingosine (dhSph), further metabolized to dihydroceramide (dhCer) by ceramide synthases (CerSs), and desaturated by dihydroceramide desaturase (DES) to generate ceramide. Ceramide is used as a building block for complex sphingolipids such as sphingomyelin, glycosphingolipids, and gangliosides. These complex sphingolipids can be hydrolyzed to regenerate ceramide. Ceramide is hydrolyzed by ceramidases (CD) to generate sphingosine, which serves as substrates for CerSs or SK1 to regenerate ceramide via the salvage pathway or to generate sphingosine 1-phosphate (S1P). Further hydrolysis of S1P by S1P lyase represents the exit from sphingolipid metabolism. This schematic is focused on the enzymes discussed in the text and does not include all enzymes involved in sphingolipid metabolism.
2.1.1. Ceramide Synthase Expression in Prostate Cancer
Little is known about the role of CerSs in prostate cancer. However, some information on expression can be gleaned from the Protein Atlas (https://www.proteinatlas.org). Expression of CerS1, CerS3, and CerS6 was either not detected or not detected in the majority of prostate cancer samples examined. Although CerS6 immunohistochemistry yielded negative results, we have detected expression of this enzyme whole cell lysates of prostate cancer cells such as PPC1, 22Rv1, Du145, and LNCaP by Western blotting (Voelkel-Johnson, unpublished). However, in the prostate, mRNA for CerS5 is more prevalent than CerS6, suggesting a role for CerS5 rather than CerS6 in this organ (Levy & Futerman, 2010). Protein for CerS5 as well as CerS2 and CerS4 is detected at low to medium levels in all prostate cancers. Staining of these enzymes is primarily cytoplasmic/membranous. Nuclear staining was detected only for CerS5 in a fraction of samples (3/8). The relevance of CerS expression level and subcellular pattern remains to be determined.
2.2. Dihydroceramide Desaturase
Dihydroceramide (dhCer) generated by CerSs in the de novo pathway is further metabolized by dihydroceramide desaturase (DES), an enzyme that is responsible for inserting the 4,5-trans-double bond into the sphingolipid backbone of dihydroceramide to generate ceramide (Fig. 1). Two isoforms of DES have been identified. DES2 has dual functions as desaturase and hydroxylase and is preferentially expressed in the kidney, small intestine, and skin, where generation of phytoceramides is essential. In contrast, DES1 is ubiquitously expressed with high DES activity and very low hydroxylase activity (Siddique, Li, Chaurasia, Kaddai, & Summers, 2015). Expression of DES1 has been associated as an unfavorable marker in renal, liver, endometrial, and urothelial cancers (https://www.proteinatlas.org/ENSG00000143753-DEGS1/pathologys) and potentially plays an important role in prostate cancer.
2.2.1. Dihydroceramide Desaturase 1 in Prostate Cancer
DES1 has recently been identified as a target gene of the androgen receptor (McNair et al., 2017). Analysis by chromatin immunoprecipitation coupled to detection by quantitative real-time PCR confirmed that the androgen receptor binds to the DES1 regulatory region early in the G1 phase of the cell cycle and knockdown of DES1 significantly decreased migration of castration-resistant C4–2 prostate cancer cells. Cohort analysis of 780 patients with localized, high-risk prostate cancer revealed a correlation between high DES1 expression and poor prognosis (McNair et al., 2017). Although the recognition of DES1 as a potential therapeutic target in prostate cancer is novel, several compounds associated with antitumor activity in this disease, including fenretinide, celecoxib, resveratrol, and curcumin, were identified as inhibitors of DES1 (Siddique et al., 2015).
2.3. Ceramidases
Five ceramidases with different pH optimums hydrolyze ceramide to generate sphingosine (Fig. 1). There are three alkaline ceramidases (ACER1, ACER2, and ACER3), acid ceramidase (ASAH1), and neutral ceramidase (ASAH2) (Coant, Sakamoto, Mao, & Hannun, 2017). Among the ceramidases, ASAH1 and ASAH2 have been associated with cancer. ASAH2, which is localized to the plasma membrane and primarily expressed in the small intestine and colon, is involved in digestion and has been implicated in colon carcinogenesis (Camp, Patterson, Kester, & Voelkel-Johnson, 2017; Coant et al., 2017). ASAH1 is primarily found in the lysosomal compartment, and mutations in its gene result in rare genetic disorders and other diseases (Koch et al., 1996; Park & Schuchman, 2006). Knockout of ASAH1 leads to death during the two-cell stage of embryonic development, indicating that other ceramidases are unable to compensate for its enzymatic activity (Eliyahu, Park, Shtraizent, He, & Schuchman, 2007). The human ASAH1 gene is located in 8p22, a region that is frequently affected in cancer.
2.3.1. ASAH1 in Cancer
ASAH1 is dysregulated in many types of cancer with a majority of studies linking elevated ASAH1 expression to proliferation and cancer cell survival. Increased expression of ASAH1 in head and neck cancer reduces responsiveness to cisplatin (Mehta et al., 2000). Genetic or pharmacological inhibition of ASAH1 using shRNA or N-oleoyl-ethanolamine, respectively, increased susceptibility of head and neck cancer cells to cisplatin in vitro and in vivo (Roh, Park, Kim, & Jang, 2016). In the skin, melanocytes and melanoma cells express higher levels of ASAH1 than other cell types but with different subcellular localization between normal and malignant cells. In melanocytes, ASAH1 localizes to both the nucleus and the cytosol, whereas expression in melanoma cells was limited to the cytosol and associated with reduced ceramide levels (Realini et al., 2016). In MCF7 breast cancer cells, treatment with the ASAH1 inhibitor Ceranib-2 decreases viability in a dose- and time-dependent manner through modulation of the mitochondrial membrane potential (Vejselova, Kutlu, & Kus, 2016). Interestingly, in luminal subtypes of breast cancer and epithelial ovarian cancer, elevated expression of ASAH1 correlated with a better prognosis (Hanker et al., 2013; Sanger et al., 2014). The correlation between ASAH1 expression and prognosis may be context dependent and determined by the relative expression of CerSs and sphingosine kinases (SKs) that compete for ASAH1-generated sphingosine as a substrate (Camp et al., 2017).
2.3.2. ASAH1 in Prostate Cancer
A role for ASAH1 in prostate cancer was suggested as early as 2000 when Seelan et al. reported that this enzyme was frequently overexpressed but not mutated in prostate cancer. An increase in ASAH1 was found in almost half (15/36) of patient-matched prostate tumor tissues and was also detected in commonly used prostate cancer cell lines Du145, LNCaP, and PC3 (Seelan et al., 2000). Two other groups independently confirmed that ASAH1 expression is increased in prostate cancer (Camacho et al., 2013; Norris et al., 2006).
In prostate cancer ASAH1 can be induced transcriptionally or regulated at the posttranscriptional level by the androgen-stimulated deubiquitinase USP2, which stabilizes protein half-life (Cheng et al., 2013; Mizutani et al., 2015). Overexpression of ASAH1 in Du145 prostate cancer cells increases tumor burden in vivo and was associated with resistance to a wide range of chemotherapeutic agents (Saad et al., 2007). Turner et al. demonstrated that overexpression of ASAH1 increases lysosomal density and autophagy. The autophagy inhibitor 3-methyl-adenine restored susceptibility to exogenous ceramide, suggesting a connection between autophagy and resistance to ceramide (Turner et al., 2011). In human prostate tumors elevated expression of ASAH1 was associated with increased phosphorylation of the oncogenic kinase Akt. Mechanistic studies indicate that ASAH1 activates sphingosine kinase 1 (SK1) to generate S1P, which then via S1P receptor 2 stimulates PI3K (Beckham, Cheng, Lu, Shao, et al., 2013). In addition, S1P-mediated Akt signaling can promote nuclear export of the tumor suppressor PTEN (Beckham, Cheng, Lu, Marrison, et al., 2013). Increased expression of ASAH1 has also been shown to promote Ets1 nuclear expression and binding to the matrix-degrading protease cathepsin B. The increased abundance of cathepsin B in ASAH1 overexpressing prostate cancer cells enhanced their ability to invade through a collagen matrix, which was abrogated by cathepsin B inhibition (Beckham et al., 2012). The apparent link between ASAH1 and tumor aggression in prostate cancer prompted the pursuit of ASAH1 as a novel therapeutic target in this disease.
2.4. Sphingosine Kinases
As part of a superfamily of lipid signaling kinases, SKs phosphorylate sphingosine to generate the soluble lipid second messenger S1P (Wattenberg, Pitson, & Raben, 2006) (Fig. 1). Overexpression of SKs has been observed in the development and progression of many different cancer types (Haddadi, Lin, Simpson, Nassif, & McGowan, 2017). The two SK isoforms, SK1 and SK2, have multiple splice variants, although roles and functions of these variants are incompletely understood (Haddadi et al., 2017). SK1 and SK2 share high sequence similarity but have distinct subcellular distribution, regulation, and function. SK1 is a cytosolic enzyme that when activated translocates to the plasma membrane where its product S1P is released extracellularly and in an autocrine or a paracrine manner can bind to S1P receptors to promote tumorigenesis (Newton, Lima, Maceyka, & Spiegel, 2015). SK2 localizes to the nucleus and has been shown to bind and inhibit histone deacetylases (HDACs) (Fu et al., 2018; Neubauer & Pitson, 2013). The enzyme has also been detected in other subcellular compartments (Neubauer et al., 2016).
2.4.1. Sphingosine Kinase in Prostate Cancer
SK activity in prostate cancer is approximately twofold higher relative to patient-matched normal tissue and corresponded to higher levels of Prostate Specific Antigen. Analysis of SK1 in 30 patients revealed that elevated expression levels were associated with tumor volume, positive margins, and surgical failure (Malavaud et al., 2010). To gain additional insight into SK expression and its potential association with tumor aggressiveness, we analyzed a data set from Oncomine of a study that included samples from 59 patients with localized prostate cancer and 35 patients with metastatic castration-resistant disease (Grasso et al., 2012). Using the Manne—Whitney test we determined that mRNA for both SK1 and SK2 was significantly elevated in metastatic prostate cancer compared to localized disease (P = .0035 for SK1 and P = .00038 for SK2). Among the 35 patients with advanced disease 19 had an increase in both isoforms of SKs, 10 had an increase in either SK1 or SK2 but not both, and only 6 showed no increase relative to the median value of patients with localized disease. Although further studies at the protein level are needed, these data suggest that an increase in either one or both SK isoforms may contribute to tumor aggressiveness in prostate cancers.
The expression of SK1 in nonmalignant tissue has also been shown to play a role in cancer. In an elegant study Ponnusamy et al. demonstrated that systemic SK1-generated S1P is critical for communication with malignant cells. In the TRAMP model of prostate cancer, survival of mice was increased from 10 to 12.5 months in SK1-deficient hosts (Ponnusamy et al., 2012). Additional experiments in the MB49 bladder cancer model demonstrated that the growth and metastasis of SK1 + MB49 cells were significantly reduced in SK1-deficient hosts, suggesting an important role for nontumor-derived S1P in cancer promotion and dissemination (Ponnusamy et al., 2012). Thus although circulating S1P may not necessarily be tumor derived, targeting the generation of S1P remains a key strategy in cancer therapy.
3. SPHINGOLIPID ENZYMES AS TARGETS IN PROSTATE CANCER
3.1. Inhibition of Dihydroceramide Desaturase
Dihydroceramides (dhCers) do not have the biological effects of ceramide, which led to the dogma that they are inert sphingolipids. In 2006 a group led by Al Merrill developed novel technologies to measure sphingolipids and observed that dhCers rather than ceramides accumulated in Du145 prostate cancer cells treated with fenretinide (4-HPR) (Zheng et al., 2006). This seminal study was the first to associate dhCers with autophagy and led to increased interest in DES. Three comprehensive reviews on physiological and pathological functions of dhCers, DES biochemistry, and DES inhibitors have recently been published (Casasampere, Ordonez, Pou, & Casas, 2016; Rodriguez-Cuenca, Barbarroja, & Vidal-Puig, 2015; Siddique et al., 2015). The identification of DES1 as a target gene of the androgen receptor revealed a previously unknown link between these two proteins, and the observation that high levels of DES1 correspond to poor overall survival in prostate cancer suggests DES1 as a novel therapeutic target in this disease (McNair et al., 2017). Interestingly, compounds that inhibit DES activity exert antitumor effects in prostate cancer (Fig. 2).
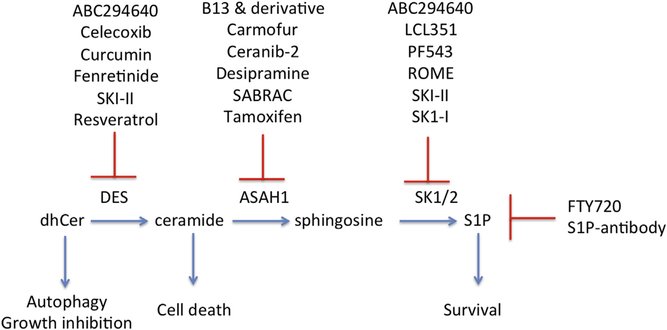
Figure 2. Inhibitors of sphingolipid metabolism.
Inhibitors of DES promote autophagy and growth inhibition. Drugs that target sphingolipid metabolism downstream of ceramide have the potential to result in accumulation of ceramide to promote cell death and/or to prevent sphingosine 1-phosphate (S1P)emediated survival signaling. Details of the inhibitors shown are discussed in the text. ROME, (R)-FTY720 methyl ether; SK1, sphingosine kinase 1.
3.1.1. Fenretinide
Fenretinide, a synthetic retinoid with chemopreventive effects on prostate cancer in rodent models, directly inhibits DES (Pienta, Nguyen, & Lehr, 1993; Pollard, Luckert, & Sporn, 1991; Rahmaniyan, Curley, Obeid, Hannun, & Kraveka, 2011; Slawin et al., 1993) Mechanistic studies in prostate cancer cell lines demonstrate that fenretinide interferes with FAK and Akt activation and promotes degradation of β-catenin (Benelli, Monteghirfo, Vene, Tosetti, & Ferrari, 2010). Fenretinide preferentially accumulates in fatty tissue and is effective against breast cancer (Formelli et al., 1993; Sabichi et al., 2003). However, accumulation of fenretinide in the human prostate was significantly lower than in mouse prostate or in human breast tissue, which may explain its lack of clinical success in prostate cancer patients (Cheung et al., 2009; Pienta, Esper, Zwas, Krzeminski, & Flaherty, 1997; Thaller et al., 2000). Subsequent efforts have focused on combining fenretinide with an inhibitor of acid ceramidase, which increased dhCer and reactive oxygen species resulting in cytotoxicity against PC3 and Du145 prostate cancer cells. However, the role of ceramide synthesis remains elusive because blocking de novo synthesis of ceramide did not block the cytotoxic response (Gouaze-Andersson et al., 2011).
3.1.2. Celecoxib
Celecoxib is a COX2 inhibitor that decreases the mean PSA velocity in prostate cancer patients (Pruthi et al., 2006; Smith et al., 2006). Analysis of gene expression in prostate cancer patients who had taken celecoxib for 4 weeks before prostatectomy revealed changes that were consistent with increased apoptosis and tumor suppressor function (Sooriakumaran, Macanas-Pirard, et al., 2009). A parallel study of tissue sections supported finding of the gene expression pattern and showed that celecoxib decreased tumor cell proliferation, microvessel density, and angiogenesis while enhancing apoptosis (Sooriakumaran, Coley, et al., 2009). In cell lines, celecoxib inhibits DES with a half maximal inhibitory concentration (IC50) of 78.9 ± 1.5 μM and significantly increases dhCers (C16, C24, C24:1) while decreasing C24- and C24:1-ceramides in a time- and dose-dependent manner (Schiffmann et al., 2009). Inhibitors of DES (8-CPPC) or CerS (FB1) prevented the increase in ceramides, and the serine palmitoyltransferase (SPT) inhibitor myriocin reduced the antiproliferative effect of celecoxib, suggesting that de novo synthesis of ceramide is required for the antiproliferative effect of celecoxib. Recently, Maeng et al. demonstrated that celecoxib induces SPT (mRNA, protein and activity) and CerS1/2/3/5/6 mRNA in HepG2 cells, whereas DES mRNA expression was unaltered. Changes in gene expression were accompanied by increased dhCers (C16, C24, C24:1), increases in long-chain ceramides (C14—C20), and a specific decrease in C16-sphingomyelin (SM) (Maeng et al., 2017). Celecoxib has also been shown to stimulate CerS activity nearly threefold and activates CerS6-mediated generation of C16-ceramide via the salvage pathway (Schiffmann et al., 2010). Taken together, these studies suggest a model in which celecoxib stimulates the expression and activity of SPT as well as CerSs to drive de novo synthesis of sphingolipids. However, without concomitant induction of DES mRNA potentially in combination with DES enzymatic inhibition, dhCers accumulate. Hydrolysis of complex sphingolipids such as SM or activation of the salvage pathway could be responsible for the increase in ceramide observed in response to celecoxib.
3.1.3. Natural Compounds
Analyses of dietary habits and prostate cancer have identified resveratrol and curcumin as natural compounds associated with reduced risk of prostate cancer (Bemis, Katz, & Buttyan, 2006). Resveratrol has numerous effects on cancer cells and signaling pathways, including sphingolipid enzymes (Casasampere et al., 2016). Inhibition of DES and induction of autophagy have been shown in a gastric cancer cells, although the mechanism may be mediated indirectly through modulation of redox status (Gagliostro et al., 2012; Shin et al., 2012; Signorelli et al., 2009). Resveratrol supplementation in animal models has shown variable results, and to date, there is no clinical evidence to make recommendations for cancer prevention or therapy (Carter, D’Orazio, & Pearson, 2014; Jasinski, Jasinska, & Ogrodowczyk, 2013).
Similar to COX2 inhibitors, curcumin can reduce PSA levels when used in conjunction with isoflavones (Ide et al., 2010). In PC3 prostate cancer cells, curcumin activated MAPK, JNK, and caspases, inducing the release of cytochrome c and AIF from mitochondria and resulted in accumulation of ceramides. Because neither kinase nor caspase inhibitors prevented curcumin-induced cytotoxicity, the authors speculated that the curcumin-induced increase in ceramide and caspase-independent cell death is responsible for the effects (Hilchie et al., 2010). Curcumin has been shown to induce rapid stimulation of ceramide synthesis through inducing dimerization of CerSs (Laviad et al., 2012). Curcumin can also reduce DES1 activity by 28% at 10 μM in a human gastric adenocarcinoma cell line (Fabrias et al., 2012). Because curcumin induces autophagy primarily in cancer models, it is possible that increased CerS activity through dimerization in combination with partial, indirect inhibition of DES leads to an accumulation of dhCer that promotes autophagy. The in vivo use of curcumin is hampered by poor bioavailability, although nanoparticle encapsulation was shown to enhance its therapeutic effect in prostate cancer (Gupta, Patchva, & Aggarwal, 2013; Thangavel, Yoshitomi, Sakharkar, & Nagasaki, 2015).
3.1.4. Sphingosine Kinase Inhibitors
Recently, two SK inhibitors, SKI-II and ABC294640, have been shown to potently inhibit DES (Cingolani et al., 2014; Venant et al., 2015). SKI-II was identified by the Smith laboratory in a high-throughput screen as a dual SK inhibitor of SK1 and SK2 (French et al., 2003, 2006). SKI-II inhibits the growth of PC3 prostate cancer cells, and analysis of structure-activity relationship suggests that DES rather than SK is responsible for this effect (Aurelio et al., 2016).
The Smith laboratory also identified the SK2-selective inhibitor ABC294640 and spearheaded its clinical translation (Britten et al., 2017; French et al., 2010). ABC294640 significantly inhibits DES activity in TRAMP prostate cancer cells and increases dhCer (Venant et al., 2015). The increase in dhCer was observed even in SK2-deficient murine embryonic fibroblasts (MEFs), suggesting the possibility that ABC294640 directly inhibits DES. Treatment of mice with 50 mg/kg ABC294640 also increased levels of dhCer in TRAMP subcutaneous tumors, and in the recent clinical trial, ABC294640 significantly elevated plasma dhC16-ceramide (the only dhCer measured) at 8 and 12 h (Britten et al., 2017; Venant et al., 2015). These data suggest that the drug may inhibit DES at clinically relevant concentrations.
3.2. Inhibitors of Acid Ceramidase
The observations that expression of ASAH1 is increased in several types of cancer and is associated with resistance have fueled an interest in targeting this enzyme therapeutically. The first-generation ceramidase inhibitors such as N-Oleoylethanolamide (NOE) were ceramide mimetics (Sugita, Willians, Dulaney, & Moser, 1975). However, low potency and poor selectivity precluded clinical development. The NOE analogue DM102 inhibits ASAH1 with an IC50 of ~15 μM in cells and can kill A549 lung cancer cells with a median lethal dose (LD50) of ~40 μM (Bedia et al., 2008). DM102 has been evaluated in combination with fenretinide in PC3 and Du145 prostate cancer cell lines, which synergistically decreased cell viability. Fenretinide, which inhibits DES, increased dhCer sixfold, and DM102 increased reactive oxygen species 30-fold compared to the control (Gouaze-Andersson et al., 2011). However, the mechanism of synergy remains elusive. Additional ASAH1 inhibitors have been identified in library screens or through structure-activity relationship analyses, some of which are further discussed below (Saied & Arenz, 2016).
3.2.1. SABRAC
PC3/Mc is a highly tumorigenic derivative of the PC3 prostate cancer cell line that has increased expression of ASAH1, metastatic potential, and chemoresistance. Genetic targeting of ASAH1 increased ceramide and decreased clonogenicity. SABRAC (N-[(2S,3R)-1,3-dihydroxyoctadecan-2-yl]2-bromoacetamide) is a novel ceramide analogue with an in vitro IC50 of 52 nM that irreversibly inhibits ASAH1. Treatment of PC3/Mc prostate cancer cells with SABRAC increased ceramide in a dose-dependent fashion and similar to genetic targeting of ASAH1 reduced growth and clonogenic potential (Camacho et al., 2013).
3.2.2. Ceranib-2
Ceranib-2 is a more potent analogue of Ceranib-1, a ceramidase inhibitor that was identified during a screen of a small-molecule library (Draper et al., 2011). This small-molecule inhibitor was identified as a quinolinone compound and has an IC50 of 28 μM. Ceranib-2 delayed tumor growth in a syngeneic model of breast cancer without apparent toxic side effects and decreased viability in LNCaP and Du145 prostate cancer cells in a time- and dose-dependent manner (Draper et al., 2011; Kus, Kabadere, Uyar, & Kutlu, 2015). Depending on the cell line studied, ceranib-2 induces apoptotic or nonapoptotic cell death, activates stress and mitogen-induced kinases JNK and p38 kinases, and inhibits the oncogenic kinase Akt (Kus et al., 2015; Vethakanraj et al., 2018).
3.2.3. B13 and Its Derivatives
B13 is a potent and selective ASAH1 inhibitor with an IC50 of about 10 μM and induces apoptosis in a variety of cancer cell lines (Bielawska, Linardic, & Hannun, 1992; Samsel et al., 2004). In PC3 prostate cancer xenografts, B13 also sensitized cells to radiation therapy (Samsel et al., 2004). However, as a neutral lipophilic molecule, B13 does not reach the acid lysosomal compartment where ASAH1 resides (Bai et al., 2014).
LCL204 is a lysosomotropic analogue of B13 and consistent with ASAH1 inhibition, treatment of Du145 prostate cancer cells selectively increased C14-, C16-, and C18-ceramides and decreased sphingosine. Exposure to LCL204 resulted in cathepsin-dependent degradation of ASAH1, rapid destabilization of lysosomes, and cytoplasmic release of lysosomal proteases, which led to depolarization of mitochondria, activation of caspases, and apoptotic cell death (Holman et al., 2008). LCL204 indirectly inhibited ASAH1 activity through lysosomal destabilization leading to proteolytic degradation of the enzyme, which prompted the development of a second series of lysosomotropic ω-N-aminoacyl analogues using the B13 scaffold (Bai et al., 2009). The ASAH1 inhibitor LCL385 sensitized PPC1 prostate cancer cells to radiation and significantly reduced xenograft growth in vivo (Mahdy et al., 2009).
A series of dimethylglycine (DMG)-B13 ester prodrugs (LCL521, LCL522, LCL596) have also been developed (Bai et al., 2014). The addition of the DMG ester significantly enhanced the cellular effectiveness of B13, and LCL521 was identified as the most promising ASAH1 inhibitor. Combination of radiation therapy with LCL521 completely inhibited radiation treatment failure in PPC1 prostate cancer xenografts (Cheng et al., 2013). Interestingly, the addition of LCL521 did not significantly improve the response to radiation treatment because tumors in all groups (radiation alone, radiation plus vehicle, or radiation plus LCL521) experienced a similar decrease in volume by day 40. However, mice treated with radiation or radiation plus vehicle relapsed with a survival rate of only 25% at week 35. In contrast, 100% of mice treated with radiation and LCL521 survived. LCL521 has also been evaluated in photodynamic therapy (PDT). Pretreatment with LCL521 greatly enhanced efficacy of PDT in vitro and an adjuvant in a vaccination approach. LCL521 was able to significantly reduce two populations of immune suppressor cells (T regulatory cells and myeloid-derived suppressor cells), which are known to interfere with the efficacy of PDT vaccines (Korbelik et al., 2016). In myeloid-derived suppressor cells, LCL521 increased autophagic vesicles and induced endoplasmic reticulum stress (Liu et al., 2016). Thus LCL521 may impact tumor cells directly or indirectly through modification of the tumor microenvironment (or both).
3.2.4. FDA-Approved Drugs With “Off-Target” ASAH1 Inhibitory Effects
Some drugs that were already used clinically were also identified to inhibit ASAH1. These include the tricyclic antidepressant desipramine, the prodrug carmofur, and the antiestrogen tamoxifen. Desipramine was shown to induce downregulation of ASAH1 in a time- and dose-dependent manner (Elojeimy et al., 2006). Carmofur is an orally administered prodrug that is taken up by the intestine to be intracellularly converted into 5-fluorouracil, which inhibits thymidylate synthase. This was believed to be the primary mechanism of action, and the use of carmofur as adjuvant chemotherapy for colorectal cancer patients in China, Japan, and Finland does extend survival (Sakamoto, Oba, Matsui, & Kobayashi, 2006). Interestingly, during screening of a commercial chemical library, carmofur was identified to inhibit recombinant rat ASAH1 with an IC50 of 29 nM (Realini et al., 2013). Carmofur increases intracellular levels of ceramide in cancer cells, including in LNCaP prostate cancer cells, and the uracil carboxamide scaffold was subsequently used as a platform for the development of novel ASAH1 inhibitors (Realini et al., 2013). A group led by Myles Cabot demonstrated that a low micromolar dose of tamoxifen inhibited ASAH1 in a spectrum of cancer cells, including prostate cancer. Further analysis of the mechanism by which tamoxifen inhibits ASAH1 pointed to an indirect effect because enzyme activity was not affected in cell-free assays. In intact cells, tamoxifen increased lysosomal permeability, which in a cathepsin B—dependent manner resulted in downregulation of ASAH1 protein levels. The structurally similar antiestrogen toremifene similarly inhibited ASAH1 (Morad et al., 2013).
3.3. Inhibition of Sphingosine Kinases
SKs generate the soluble, bioactive S1P that plays an important role in angiogenesis, tumor cell survival, metastasis, and therapy resistance. Thus targeting SK activity has been a major therapeutic effort. However, interpreting the effects of SK inhibitors in cells or in preclinical models is complicated by lack of specificity for SK isoforms and off-target effects. SKI-II was identified as a dual SK inhibitor (Gao, Peterson, Smith, & Smith, 2012). Recently, this inhibitor was shown to also target DES1, and structure-activity relationship analysis of SKI-II targets suggests that DES1 rather than SK inhibition is primarily responsible for the antiproliferative effects of SKI-II and its analogues (Aurelio et al., 2016; Cingolani et al., 2014). Additional caution is warranted because SKI-II is sometimes referred to as an SK1 inhibitor in the literature. Because SK1 and SK2 exhibit structural differences in their binding pocket, attempts have been made to generate isoform-specific inhibitors (Gao et al., 2012).
3.3.1. Sphingosine Kinase 1 Inhibitors
SK1-I is a sphingosine analogue and a competitive, specific inhibitor of SK1 that is not to be confused with the dual SK inhibitor SKI-II (Paugh et al., 2008). SK1-I decreases S1P, increases ceramide, and inhibits tumor cell growth in vitro and in vivo (Kapitonov et al., 2009; Nagahashi et al., 2012; Paugh et al., 2008; Song et al., 2011). SK1-I reduces phosphorylation of Akt in glioblastoma, sensitizes lungs cancer cells to chemotherapy, and decreases lymph node and lung metastasis in a breast cancer model through inhibition of hemangiogenesis and lymphangiogenesis.
PF543 is a potent inhibitor of SK1 with an IC50 of 2—3.6 nM (vs. 356 nM for SK2) and prevents the phosphorylation of sphingosine in cancer cells. At 10 μM, PF543 did not significantly inhibit the activity of other protein and lipid kinases or bind to S1P receptors (Schnute et al., 2012). Cingolani et al. compared the effects observed with SKI-II and PF543. Although both SKI-II and PF543 decreased S1P to nearly undetectable levels, only SKI-II but not PF543 reduced proliferation, G0/G1 inhibition, and autophagy (Cingolani et al., 2014). Rex et al. (2013) also failed to detect a cytotoxic effect of SK inhibitors on cancer cells. The lack of anticancer efficacy by specific SK inhibitors may have been an underlying factor for abandoning the pursuit of patents (Lynch, Thorpe, & Santos, 2016).
Nevertheless, as a soluble mediator that can be produced by noncancerous cells to influence malignant cell behavior, extracellular S1P generated by SK1 remains an important target. For example, a recent study using cell-specific SK1 knockout mice demonstrated that macrophage-derived S1P plays an important role in colon carcinogenesis (Furuya et al., 2017). In prostate and bladder cancer, nontumor-derived S1P plays an important role in metastasis (Ponnusamy et al., 2012). The novel SK1-specific inhibitor LCL351 decreases the migration rate of human Du145 prostate cells (Sharma, 2011). LCL351 also decreases the inflammatory response and reduces colitis-induced development of colorectal cancer (Furuya et al., 2017).
3.3.2. SK2 Inhibitors
The SK2-selective inhibitor, (R)-FTY720 methyl ether (ROME), has been evaluated in LNCaP prostate cancer cells. Treatment with ROME increases sphingosine and decreases S1P without affecting ceramide levels or inducing apoptosis (Watson et al., 2013). ROME was also compared with SKI-II for its ability to reduce expression of the androgen receptor but no SK2-specific effect was found (Tonelli et al., 2013). The effect of SKI-II on the androgen receptor was independent of inhibitor-induced proteasomal degradation of SK1 and could not be reversed with the pan-CerS inhibitor Fumonisin B1.
Another SK2-selective inhibitor is ABC294640, which, similar to SKI-II, inhibits the activity of DES but not SK1 or other kinases (Gao et al., 2012; Venant et al., 2015). Although ABC294640 has been reported to induce apoptosis in some cell models, in prostate cancer, it promotes lysosomal acidification, autophagy and growth inhibition (Beljanski, Knaak, & Smith, 2010; Qin et al., 2014; Venant et al., 2015). It also downregulates signaling pathways including STAT3, Akt, ERK, p21, p53, and FAK (Gao et al., 2012). Two studies have shown that ABC294640 decreases c-Myc and androgen receptor expression in prostate cancer (Schrecengost, Keller, Schiewer, Knudsen, & Smith, 2015; Venant et al., 2015). Genetic targeting of SK2 confirmed that the effect of ABC294640 on c-Myc requires SK2 expression. However, the effect of ABC294640 on the androgen receptor remains to be elucidated and could be mediated through inhibition of DES or indirectly via c-Myc (Schrecengost et al., 2015; Venant et al., 2015). ABC294640 exerts antitumor efficacy in numerous preclinical models (Antoon et al., 2010; Chumanevich et al., 2010; Dai, Smith, Foroozesh, Miele, & Qin, 2017; Lewis, Voelkel-Johnson, & Smith, 2016; Qin et al., 2014; Schrecengost et al., 2015; Venkata et al., 2014) and has recently been evaluated in a Phase I clinical trial. The study, which involved 21 patients with advanced solid tumors, demonstrated that a dose of 500 mg ABC294640 was well tolerated and resulted in plasma sphingolipid changes that may have utility as pharmacodynamics markers of drug activity (Britten et al., 2017). The Phase I trial did not include any prostate cancer patients. However, ABC294640 may be particularly relevant for the treatment of prostate cancer because both c-Myc and the androgen receptor are frequently dysregulated in this disease.
3.3.3. Targeting the Downstream Effects of Sphingosine Kinase
The primary goal of targeting SK is to prevent the protumor function of its product S1P. Two strategies that interfere with S1P and S1P signaling are S1P antibodies and SK substrate analogues. An antibody against S1P known as Sphingomab has been evaluated in prostate cancer models. Prostate cancer preferentially metastasizes to the bone, and S1P generated by osteoblasts has been hypothesized to influence prostate cancer cell behavior. Brizuela et al. (2014) investigated whether paracrine S1P signaling from osteoblastic MC3T3 cells played a role in the proliferation of PC3 and C4–2 prostate cancer cells and demonstrated that genetic knockdown of SK1 or neutralization of S1P using Sphingomab significantly inhibited cancer cell proliferation. A subsequent study revealed that Sphingomab was able to transiently normalize tumor vasculature, reducing tumor hypoxia, and increasing efficacy of chemotherapy as evidenced by inhibiting tumor growth and metastasis (Ader et al., 2015). However, despite proof of concept, promising results in preclinical models, and safety of a humanized version of the antibody, therapeutic efficacy of S1P antibodies fell short of expectations in clinical trials for the treatment of macular degeneration (NCT01334255) and advanced renal cell carcinoma (NCT01762033).
FTY720 (Fingolimod, Gilenya) is a sphingosine analogue that is phosphorylated by SK to FTY720-P, which unlike S1P cannot signal S1P receptors. As a S1P receptor modulator, FTY720 is currently approved as an immune-modulating drug for the treatment of multiple sclerosis. Depending on culture conditions, FTY720 can also modulate ceramide synthesis in cells. When dhSph concentrations are high (500 nM—5 μM), FTY720 can significantly inhibit ceramide synthesis, but this effect was not observed when dhSph was <200 nM (Lahiri et al., 2009). In contrast, treatment of cells with 25 μM FTY720 increased C18—C22-ceramides within 90 min, which points to a potentially complex interaction between FTY720 and CerSs (Lahiri et al., 2009). FTY720 may also exert antitumor effects in a S1P receptor—independent manner because silencing of S1P receptors had no effect on cell death in PC3 cells (Pchejetski et al., 2010).
Although the exact mode of action for FTY720 remains to be further elucidated, several studies point to an anticancer effect in models of prostate cancer. High doses of FTY720 induce apoptosis (40 μM) or G1-cell cycle arrest (20 μM) in Du145 prostate cancer cells (Permpongkosol et al., 2002; Wang et al., 1999). In CWR22R prostate tumors, administration of 10 mg/kg FTY720 suppressed tumor growth in vivo and corresponded to a reduction in serum PSA levels, suppression of angiogenesis, reversal of epithelial mesenchymal transition through restoration of E-cadherin expression, and an increase in apoptotic cells (Chua et al., 2005). A dose of 1.5 μM FTY720 was sufficient to decrease prostate cancer cell migration and invasion in vitro and caused a significant reduction in actin stress fibers and filopodia through inhibition of RhoA-GTPase activity (Zhou et al., 2006). A low dose of FTY720 also decreased phosphorylation of Akt, reduced Runx2 and N-cadherin, which plays an important role in prostate cancer metastasis (Chua et al., 2009). Similar to other modulators of sphingolipid metabolism, FTY720 can sensitize tumor cells to radiation therapy in vitro and in vivo (Pchejetski et al., 2010).
4. ESCAPE FROM THERAPEUTIC TARGETING
It is known that the malignant cell population is heterogeneous and genomically unstable. Transformed cells acquire the ability to evade immune surveillance and survive in the hostile tumor microenvironment eventually developing into detectable malignant lesions. Cellular stress induced by cancer therapy, such as radiation or chemotherapy, increases intracellular levels of ceramide to induce cell death. However, invariably, a small, resistant subset of malignant cells cannot be eradicated and the cancer stem cell (CSC) hypothesis posits that this small cell population can regenerate tumors with even more heterogeneity and resistance, which makes recurrent cancer extremely difficult to treat.
Therapy may select for a preexisting resistant subset of cells, but evidence also points to therapy-induced resistance as a consequence of ceramide stress. Chronic exposure to ceramide or glucosylceramide increases the expression of the multidrug resistance gene that codes for P-glycoprotein, which enhances resistance to doxorubicin and paclitaxel (Gouaze-Andersson et al., 2007). Daunorubicin has been shown to posttranscriptionally activate ASAH1 in liver cancer cells (Morales et al., 2007). In colon cancer cells overexpression of CerS6 increases intracellular C16-ceramide and transcriptionally induces ASAH1 through a c-Jun N-terminal kinase—mediated mechanism (Tirodkar et al., 2015). Cheng et al. showed that radiation-induced ceramide stress also transcriptionally activates the expression of ASAH1 in a wide variety of other cancer cell types. Clinical data from prostate cancer patients for whom a preradiation and postradiation biopsy was available support the hypothesis that ASAH1 expression becomes elevated following treatment (Cheng et al., 2013).
4.1. Sphingolipids in Epithelial Mesenchymal Transition
A key to cancer progression in solid tumors is epithelial mesenchymal transition (EMT), a process during which epithelial cells lose polarization and adhesion, gain the ability to migrate and invade, and acquire pluripotency (Nieto, Huang, Jackson, & Thiery, 2016). The EMT program is driven by several families of transcription factors that directly or indirectly suppress the expression of E-cadherin resulting in a so-called “cadherin switch” during which E-cadherin is lost and expression of the mesenchymal filament protein vimentin is gained. Treatment of prostate cancer with androgen deprivation therapy on the one hand promotes accumulation of C16-ceramide and apoptosis but on the other hand induces EMT through loss of androgen receptor—mediated suppression of the chemokine CCL2 (Eto et al., 2003; Tsai et al., 2018). Whether a direct cause and effect exists between androgen withdrawal—induced ceramide and EMT is not yet known, but a role of sphingolipids in EMT has recently been explored. A study using a 35-gene sphingolipid-EMT signature linked EMT to a significant reduction in ceramide and an increase in S1P (Meshcheryakova et al., 2016).
4.1.1. Reduced Ceramide Generation
CerS4 and CerS6 mRNA was found to be decreased with induction of EMT (Meshch-eryakova et al., 2016). CerS6 preferentially generates C16-ceramide, a ceramide species that has shown to be preferentially increased during apoptosis (Tirodkar & Voelkel-Johnson, 2012). Reduced expression of CerS6 enhances therapy resistance in colon cancer cells and increases invasive potential in melanoma cells (Tang et al., 2016; White-Gilbertson et al., 2009). Both cell death resistance and elevated metastatic potential are characteristics of EMT. Edmond et al. investigated CerS6 expression in the context of EMT in the NCI-60 panel of cancer cells. Grouping cells into “epithelial” or “mesenchymal” phenotypes based on E-cadherin/vimentin ratios, they showed a significant decrease in CerS6 mRNA in the mesenchymal phenotype (Edmond et al., 2015). Analysis of CerS6 expression at the protein level in a molecularly defined model of EMT confirmed that reduced CerS6 expression coincides with the “cadherin switch” (Edmond et al., 2015). Thus a reduction in ceramide, perhaps specifically C16-ceramide, may be associated with EMT.
4.1.2. Mechanisms of Ceramide Clearance
Ceramide levels can also be decreased through enhanced clearance. Enzymes that clear ceramide and have been associated with therapy resistance include glucosylceramide synthase (GCS) in breast cancer, ASAH1 in prostate cancer, and ASAH2 in colorectal cancer. GCS clears ceramide through adding glucose, which is the first step in the synthesis of glycosphingolipids. Glucosylceramide forms the backbone of hundreds of structurally different glycosphingolipids including gangliosides that are found on the outer leaflet of the plasma membrane where they interact with various other molecules and play a role in adhesion, proliferation, and cell recognition. Expression of GCS is elevated in metastatic breast cancer, and tamoxifen, which inhibits ceramide glycosylation, plays an important role as part of breast cancer therapy (Liu et al., 2011; Morad & Cabot, 2015).
The ceramidase product sphingosine can either be recycled to ceramide by CerS in the salvage pathway or serve as substrate for SKs. A shift in the EMT sphingolipid signature, including a strong increase in SK1, suggests that sphingosine would be preferentially metabolized to S1P (Meshcheryakova et al., 2016). Indeed cells with a mesenchymal phenotype have a twofold increase in S1P, and analysis of the sphingolipid-EMT signature could be used to differentiate the epithelial cancer stage from cells that had undergone EMT. Recent studies suggest a key role for S1P in the maintenance of CSCs (Lewis, Powell, & Pitson, 2017).
4.2. Sphingolipids and Stemness
The concept of CSCs is still debated within the scientific community, but it is generally accepted that a small population of malignant cells can lie dormant, even for many years. Unless this population is eradicated, the cancer has the potential to recur. There is now ample evidence that sphingolipid metabolism has multiple intersections with stem cell signaling and function in cancer (Lewis et al., 2017). The literature supports a model in which cancer therapy induces the generation of ceramide and cell death in a majority of cells but also induces a shift toward ceramide clearance and enhanced S1P signaling in a fraction of cells. S1P plays an important role in the tumor microenvironment that acts as a chemotactic factor and promoter of metastasis.
The rational combination of agents that interfere with ceramide clearance should in theory be able to improve therapy outcomes. Proof of principal for this hypothesis comes from combining tamoxifen, which has an off-target effect on GCS, with chemotherapy in breast cancer. In mice, targeting GCS can prevent the accumulation of breast CSCs during doxorubicin chemotherapy (Morad & Cabot, 2015). A similar approach may be of value for the treatment of prostate cancer in which ASAH1 is strongly upregulated. As discussed previously, genetic targeting or pharmacological inhibition of ASAH1 completely abrogated radiation treatment failure in a preclinical model of prostate cancer (Cheng et al., 2013). Prostate cancer is among the malignancies described to harbor CSCs, and the presence of CSCs in the PPC1 model used by Cheng et al. has been established (Liu et al., 2015). Although the effect on PPC1 CSCs was not directly tested, the lack of relapse suggests that all malignant cells were eradicated. To date, only two studies specifically link ASAH1 and CSCs. Lai et al. (2017) targeted ASAH1 using CRISPR-Cas9-mediated approach and demonstrated that the formation of cancer-initiating cells in melanoma cells could be prevented. In glioblastoma, S1P in the tumor microenvironment has been shown to fuel the survival of tumor stem cells (Marfia et al., 2014). Three different inhibitors of ASAH1 (carmofur, ARN14988, and NOE) were able to kill the tumor stem cell population in four different glioblastoma cell lines (Doan et al., 2017).
5. CONCLUSIONS
In prostate cancer, all standard therapeutic modalities such as androgen ablation therapy, radiation, and chemotherapy increase ceramide. ASAH1 is frequently upregulated in prostate cancer, and treatment-induced generation of ceramide has the potential to further increase ASAH1 expression, which could lead to cross-resistance. Effective therapy for advanced castration-resistant prostate cancer remains a major clinical challenge, and combination with agents that interfere with ASAH1 should be considered. Future therapies for cancer should consider not only promoting the elevation of intracellular ceramide to drive cell death but also combination with strategies that prevent ceramide clearance.
ACKNOWLEDGMENTS
Financial support for the authors was provided by grant P01 CA203628. James S. Norris has a financial interest in Sphingogene.
REFERENCES
- Ader I, Gstalder C, Bouquerel P, Golzio M, Andrieu G, Zalvidea S, et al. (2015). Oncotarget, 6, 13803–13821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoon JW, White MD, Meacham WD, Slaughter EM, Muir SE, Elliott S, et al. (2010). Endocrinology, 151, 5124–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aurelio L, Scullino CV, Pitman MR, Sexton A, Oliver V, Davies L, et al. (2016). Journal of Medicinal Chemistry, 59, 965–984. [DOI] [PubMed] [Google Scholar]
- Bai A, Szulc ZM, Bielawski J, Mayroo N, Liu X, Norris J, et al. (2009). Bioorganic & Medicinal Chemistry, 17, 1840–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai A, Szulc ZM, Bielawski J, Pierce JS, Rembiesa B, Terzieva S, et al. (2014). Bioorganic & Medicinal Chemistry, 22, 6933–6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham TH, Cheng JC, Lu P, Marrison ST, Norris JS, & Liu X (2013). PLoS One, 8, e76593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham TH, Cheng JC, Lu P, Shao Y, Troyer D, Lance R, et al. (2013). Oncogenesis, 2, e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham TH, Cheng JC, Marrison ST, Norris JS, & Liu X (2013). Advances in Cancer Research, 117, 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham TH, Lu P, Cheng JC, Zhao D, Turner LS, Zhang X, et al. (2012). International Journal of Cancer, 131, 2034–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedia C, Canals D, Matabosch X, Harrak Y, Casas J, Llebaria A, et al. (2008). Chemistry and Physics of Lipids, 156, 33–40. [DOI] [PubMed] [Google Scholar]
- Beljanski V, Knaak C, & Smith CD (2010). The Journal of Pharmacology and Experimental Therapeutics, 333, 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis DL, Katz AE, & Buttyan R (2006). Expert Opinion on Investigational Drugs, 15, 1191–1200. [DOI] [PubMed] [Google Scholar]
- Benelli R, Monteghirfo S, Vene R, Tosetti F, & Ferrari N (2010). Molecular Cancer, 9, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielawska A, Linardic CM, & Hannun YA (1992). Journal of Biological Chemistry, 267, 18493–18497. [PubMed] [Google Scholar]
- Britten CD, Garrett-Mayer E, Chin SH, Shirai K, Ogretmen B, Bentz TA, et al. (2017). Clinical Cancer Research, 23, 4642–4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizuela L, Martin C, Jeannot P, Ader I, Gstalder C, Andrieu G, et al. (2014). Molecular Oncology, 8, 1181–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho L, Meca-Cortes O, Abad JL, Garcia S, Rubio N, Diaz A, et al. (2013). The Journal of Lipid Research, 54, 1207–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp ER, Patterson LD, Kester M, & Voelkel-Johnson C (2017). Cancer Biology & Therapy, 18, 640–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter LG, D’Orazio JA, & Pearson KJ (2014). Endocrine-Related Cancer, 21, R209–R225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casasampere M, Ordonez YF, Pou A, & Casas J (2016). Chemistry and Physics of Lipids, 197, 33–44. [DOI] [PubMed] [Google Scholar]
- Cheng JC, Bai A, Beckham TH, Marrison ST, Yount CL, Young K, et al. (2013). Journal of Clinical Investigation, 123, 4344–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung E, Pinski J, Dorff T, Groshen S, Quinn DI, Reynolds CP, et al. (2009). Clinical Genitourinary Cancer, 7, 43–50. [DOI] [PubMed] [Google Scholar]
- Chua CW, Chiu YT, Yuen HF, Chan KW, Man K, Wang X, et al. (2009). Clinical Cancer Research, 15, 4322–4335. [DOI] [PubMed] [Google Scholar]
- Chua CW, Lee DT, Ling MT, Zhou C, Man K, Ho J, et al. (2005). International Journal of Cancer, 117, 1039–1048. [DOI] [PubMed] [Google Scholar]
- Chumanevich AA, Poudyal D, Cui X, Davis T, Wood PA, Smith CD, et al. (2010). Carcinogenesis, 31, 1787–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani F, Casasampere M, Sanllehi P, Casas J, Bujons J, & Fabrias G (2014). The Journal of Lipid Research, 55, 1711–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coant N, Sakamoto W, Mao C, & Hannun YA (2017). Advances in Biological Regulation, 63, 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Smith CD, Foroozesh M, Miele L, & Qin Z (2017). International Journal of Cancer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan NB, Alhajala H, Al-Gizawiy MM, Mueller WM, Rand SD, Connelly JM, et al. (2017). Oncotarget, 8, 112662–112674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper JM, Xia Z, Smith RA, Zhuang Y, Wang W, & Smith CD (2011). Molecular Cancer Therapeutics, 10, 2052–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmond V, Dufour F, Poiroux G, Shoji K, Malleter M, Fouque A, et al. (2015). Oncogene, 34, 996–1005. [DOI] [PubMed] [Google Scholar]
- Eliyahu E, Park JH, Shtraizent N, He X, & Schuchman EH (2007). The FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology, 21, 1403–1409. [DOI] [PubMed] [Google Scholar]
- Elojeimy S, Holman DH, Liu X, El-Zawahry A, Villani M, Cheng JC, et al. (2006). FEBS Letters, 580, 4751–4756. [DOI] [PubMed] [Google Scholar]
- Eto M, Bennouna J, Hunter OC, Hershberger PA, Kanto T, Johnson CS, et al. (2003). The Prostate, 57, 66–79. [DOI] [PubMed] [Google Scholar]
- Fabrias G, Munoz-Olaya J, Cingolani F, Signorelli P, Casas J, Gagliostro V, et al. (2012). Progress in Lipid Research, 51, 82–94. [DOI] [PubMed] [Google Scholar]
- Formelli F, Clerici M, Campa T, Di Mauro MG, Magni A, Mascotti G, et al. (1993). Journal of Clinical Oncology, 11, 2036–2042. [DOI] [PubMed] [Google Scholar]
- French KJ, Schrecengost RS, Lee BD, Zhuang Y, Smith SN, Eberly JL, et al. (2003). Cancer Research, 63, 5962–5969. [PubMed] [Google Scholar]
- French KJ, Upson JJ, Keller SN, Zhuang Y, Yun JK, & Smith CD (2006). The Journal of Pharmacology and Experimental Therapeutics, 318, 596–603. [DOI] [PubMed] [Google Scholar]
- French KJ, Zhuang Y, Maines LW, Gao P, Wang W, Beljanski V, et al. (2010). The Journal of Pharmacology and Experimental Therapeutics, 333, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu P, Ebenezer DL, Ha AW, Suryadevara V, Harijith A, & Natarajan V (2018). Journal of Cellular Biochemistry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya H, Tamashiro PM, Shimizu Y, Iino K, Peres R, Chen R, et al. (2017). Carcinogenesis, 38, 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliostro V, Casas J, Caretti A, Abad JL, Tagliavacca L, Ghidoni R, et al. (2012). International Journal of Biochemistry & Cell Biology, 44, 2135–2143. [DOI] [PubMed] [Google Scholar]
- Gao P, Peterson YK, Smith RA, & Smith CD (2012). PLoS One, 7, e44543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouaze-Andersson V, Flowers M, Karimi R, Fabrias G, Delgado A, Casas J, et al. (2011). The Prostate, 71, 1064–1073. [DOI] [PubMed] [Google Scholar]
- Gouaze-Andersson V, Yu JY, Kreitenberg AJ, Bielawska A, Giuliano AE, & Cabot MC (2007). Biochimica et Biophysica Acta, 1771, 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. (2012). Nature, 487, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SC, Patchva S, & Aggarwal BB (2013). The AAPS Journal, 15, 195–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddadi N, Lin Y, Simpson AM, Nassif NT, & McGowan EM (2017). International Journal of Molecular Sciences, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajj C, & Haimovitz-Friedman A (2013). Handbook of Experimental Pharmacology, 115–130. [DOI] [PubMed] [Google Scholar]
- Hanker LC, Karn T, Holtrich U, Gatje R, Rody A, Heinrich T, et al. (2013). International Journal of Gynecological Pathology, 32, 249–257. [DOI] [PubMed] [Google Scholar]
- Hilchie AL, Furlong SJ, Sutton K, Richardson A, Robichaud MR, Giacomantonio CA, et al. (2010). Nutrition and Cancer, 62, 379–389. [DOI] [PubMed] [Google Scholar]
- Holman DH, Turner LS, El-Zawahry A, Elojeimy S, Liu X, Bielawski J, et al. (2008). Cancer Chemotherapy and Pharmacology, 61, 231–242. [DOI] [PubMed] [Google Scholar]
- Ide H, Tokiwa S, Sakamaki K, Nishio K, Isotani S, Muto S, et al. (2010). The Prostate, 70, 1127–1133. [DOI] [PubMed] [Google Scholar]
- Jasinski M, Jasinska L, & Ogrodowczyk M (2013). Central European Journal of Urology, 66, 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov D, Allegood JC, Mitchell C, Hait NC, Almenara JA, Adams JK, et al. (2009). Cancer Research, 69, 6915–6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch J, Gartner S, Li CM, Quintern LE, Bernardo K, Levran O, et al. (1996). Journal of Biological Chemistry, 271, 33110–33115. [DOI] [PubMed] [Google Scholar]
- Korbelik M, Banath J, Zhang W, Saw KM, Szulc ZM, Bielawska A, et al. (2016). International Journal of Cancer, 139, 1372–1378. [DOI] [PubMed] [Google Scholar]
- Kus G, Kabadere S, Uyar R, & Kutlu HM (2015). In Vitro Cellular & Developmental Biology Animal, 51, 1056–1063. [DOI] [PubMed] [Google Scholar]
- Lahiri S, Park H, Laviad EL, Lu X, Bittman R, & Futerman AH (2009). Journal of Biological Chemistry, 284, 16090–16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M, Realini N, La Ferla M, Passalacqua I, Matteoli G, Ganesan A, et al. (2017). Scientific Reports, 7, 7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laviad EL, Kelly S, Merrill AH Jr., & Futerman AH (2012). Journal of Biological Chemistry, 287, 21025–21033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, & Futerman AH (2010). IUBMB Life, 62, 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CL, Powell JA, & Pitson SM (2017). In Pebay A, & Wong RCB (Eds.), Lipidomics of stem cells. [Google Scholar]
- Lewis CS, Voelkel-Johnson C, & Smith CD (2016). Oncotarget, 7, 60181–60192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Li X, Lu C, Bai A, Bielawski J, Bielawska A, et al. (2016). Oncotarget, 7, 83907–83925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chen X, Rycaj K, Chao HP, Deng Q, Jeter C, et al. (2015). Oncotarget, 6, 23959–23986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YY, Patwardhan GA, Xie P, Gu X, Giuliano AE, & Cabot MC (2011). International Journal of Oncology, 39, 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KR, Thorpe SB, & Santos WL (2016). Expert Opinion on Therapeutic Patents, 26, 1409–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeng HJ, Song JH, Kim GT, Song YJ, Lee K, Kim JY, et al. (2017). BMB Reports, 50, 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdy AE, Cheng JC, Li J, Elojeimy S, Meacham WD, Turner LS, et al. (2009). Molecular Therapy: The Journal of the American Society of Gene Therapy, 17, 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malavaud B, Pchejetski D, Mazerolles C, de Paiva GR, Calvet C, Doumerc N, et al. (2010). European Journal of Cancer, 46, 3417–3424. [DOI] [PubMed] [Google Scholar]
- Marfia G, Campanella R, Navone SE, Di Vito C, Riccitelli E, Hadi LA, et al. (2014). Glia, 62, 1968–1981. [DOI] [PubMed] [Google Scholar]
- McNair C, Urbanucci A, Comstock CE, Augello MA, Goodwin JF, Launchbury R, et al. (2017). Oncogene, 36, 1655–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S, Blackinton D, Omar I, Kouttab N, Myrick D, Klostergaard J, et al. (2000). Cancer Chemotherapy and Pharmacology, 46, 85–92. [DOI] [PubMed] [Google Scholar]
- Meshcheryakova A, Svoboda M, Tahir A, Kofeler HC, Triebl A, Mungenast F, Heinze G, Gerner C, Zimmermann P, Jaritz M, & Mechtcheriakova D (2016). Oncotarget, 7, 22295–22323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani N, Inoue M, Omori Y, Ito H, Tamiya-Koizumi K, Takagi A, et al. (2015). Journal of Biochemistry, 158, 309–319. [DOI] [PubMed] [Google Scholar]
- Morad SA, & Cabot MC (2015). Biochimica et Biophysica Acta, 1851, 1134–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morad SA, Levin JC, Tan SF, Fox TE, Feith DJ, & Cabot MC (2013). Biochimica et Biophysica Acta, 1831, 1657–1664. [DOI] [PubMed] [Google Scholar]
- Morales A, Paris R, Villanueva A, Llacuna L, Garcia-Ruiz C, & Fernandez-Checa JC (2007). Oncogene, 26, 905–916. [DOI] [PubMed] [Google Scholar]
- Mullen TD, Hannun YA, & Obeid LM (2012). The Biochemical Journal, 441, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahashi M, Ramachandran S, Kim EY, Allegood JC, Rashid OM, Yamada A, et al. (2012). Cancer Research, 72, 726–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava VE, Cuvillier O, Edsall LC, Kimura K, Milstien S, Gelmann EP, et al. (2000). Cancer Research, 60, 4468–4474. [PubMed] [Google Scholar]
- Neubauer HA, Pham DH, Zebol JR, Moretti PA, Peterson AL, Leclercq TM, et al. (2016). Oncotarget, 7, 64886–64899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer HA, & Pitson SM (2013). The FEBS Journal, 280, 5317–5336. [DOI] [PubMed] [Google Scholar]
- Newton J, Lima S, Maceyka M, & Spiegel S (2015). Experimental Cell Research, 333, 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA, Huang RY, Jackson RA, & Thiery JP (2016). Cell, 166, 21–45. [DOI] [PubMed] [Google Scholar]
- Norris JS, Bielawska A, Day T, El-Zawahri A, Elojeimy S, Hannun Y, et al. (2006). Cancer Gene Therapy, 13, 1045–1051. [DOI] [PubMed] [Google Scholar]
- Ogretmen B (2018). Nature Reviews Cancer, 18, 33–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, & Schuchman EH (2006). Biochimica et Biophysica Acta, 1758, 2133–2138. [DOI] [PubMed] [Google Scholar]
- Park JW, Park WJ, & Futerman AH (2014). Biochimica et Biophysica Acta, 1841, 671–681. [DOI] [PubMed] [Google Scholar]
- Paugh SW, Paugh BS, Rahmani M, Kapitonov D, Almenara JA, Kordula T, et al. (2008). Blood, 112, 1382–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pchejetski D, Bohler T, Brizuela L, Sauer L, Doumerc N, Golzio M, et al. (2010). Cancer Research, 70, 8651–8661. [DOI] [PubMed] [Google Scholar]
- Permpongkosol S,Wang JD,Takahara S,Matsumiya K,Nonomura N, Nishimura K, et al. (2002). International Journal of Cancer, 98, 167–172. [DOI] [PubMed] [Google Scholar]
- Pienta KJ, Esper PS, Zwas F, Krzeminski R, & Flaherty LE (1997). American Journal of Clinical Oncology, 20, 36–39. [DOI] [PubMed] [Google Scholar]
- Pienta KJ, Nguyen NM, & Lehr JE (1993). Cancer Research, 53, 224–226. [PubMed] [Google Scholar]
- Pollard M, Luckert PH, & Sporn MB (1991). Cancer Research, 51, 3610–3611. [PubMed] [Google Scholar]
- Ponnusamy S, Selvam SP, Mehrotra S, Kawamori T, Snider AJ, Obeid LM, et al. (2012). EMBO Molecular Medicine, 4, 761–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruthi RS, Derksen JE, Moore D, Carson CC, Grigson G, Watkins C, et al. (2006). Clinical Cancer Research, 12, 2172–2177. [DOI] [PubMed] [Google Scholar]
- Qin Z, Dai L, Trillo-Tinoco J, Senkal C, Wang W, Reske T, et al. (2014). Molecular Cancer Therapeutics, 13, 154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmaniyan M, Curley RW Jr., Obeid LM, Hannun YA, & Kraveka JM (2011). Journal of Biological Chemistry, 286, 24754–24764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Realini N, Palese F, Pizzirani D, Pontis S, Basit A, Bach A, et al. (2016). Journal of Biological Chemistry, 291, 2422–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Realini N, Solorzano C, Pagliuca C, Pizzirani D, Armirotti A, Luciani R, et al. (2013). Scientific Reports, 3, 1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex K, Jeffries S, Brown ML, Carlson T, Coxon A, Fajardo F, et al. (2013). PLoS One, 8, e68328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Cuenca S, Barbarroja N, & Vidal-Puig A (2015). Biochimica et Biophysica Acta, 1851, 40–50. [DOI] [PubMed] [Google Scholar]
- Roh JL, Park JY, Kim EH, & Jang HJ (2016). European Journal of Cancer, 52, 163–172. [DOI] [PubMed] [Google Scholar]
- Saad AF, Meacham WD, Bai A, Anelli V, Elojeimy S, Mahdy AE, et al. (2007). Cancer Biology & Therapy, 6, 1455–1460. [DOI] [PubMed] [Google Scholar]
- Sabichi AL, Modiano MR, Lee JJ, Peng YM, Xu MJ, Villar H, et al. (2003). Clinical Cancer Research, 9, 2400–2405. [PubMed] [Google Scholar]
- Saied EM, & Arenz C (2016). Chemistry and Physics of Lipids, 197, 60–68. [DOI] [PubMed] [Google Scholar]
- Sakamoto J, Oba K, Matsui T, & Kobayashi M (2006). Diseases of the Colon and Rectum, 49, S82–S91. [DOI] [PubMed] [Google Scholar]
- Samsel L, Zaidel G, Drumgoole HM, Jelovac D, Drachenberg C, Rhee JG, et al. (2004). The Prostate, 58, 382–393. [DOI] [PubMed] [Google Scholar]
- Sanger N, Ruckhaberle E, Gyorffy B, Engels K, Heinrich T, Fehm T, et al. (2014). Molecular Oncology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffmann S, Sandner J, Schmidt R, Birod K, Wobst I, Schmidt H, et al. (2009). The Journal of Lipid Research, 50, 32–40. [DOI] [PubMed] [Google Scholar]
- Schiffmann S, Ziebell S, Sandner J, Birod K, Deckmann K, Hartmann D, et al. (2010). Biochemical Pharmacology, 80, 1632–1640. [DOI] [PubMed] [Google Scholar]
- Schnute ME, McReynolds MD, Kasten T, Yates M, Jerome G, Rains JW, et al. (2012). The Biochemical Journal, 444, 79–88. [DOI] [PubMed] [Google Scholar]
- Schrecengost RS, Keller SN, Schiewer MJ, Knudsen KE, & Smith CD (2015). Molecular Cancer Research, 13, 1591–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelan RS, Qian C, Yokomizo A, Bostwick DG, Smith DI, & Liu W (2000). Genes Chromosomes & Cancer, 29, 137–146. [DOI] [PubMed] [Google Scholar]
- Sharma AK (2011). Expert Opinion on Therapeutic Patents, 21, 807–812. [DOI] [PubMed] [Google Scholar]
- Shin KO, Park NY, Seo CH, Hong SP, Oh KW, Hong JT, et al. (2012). Biomolecules & Therapeutics (Seoul), 20, 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique MM, Li Y, Chaurasia B, Kaddai VA, & Summers SA (2015). Journal of Biological Chemistry, 290, 15371–15379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, & Jemal A (2018). CA: A Cancer Journal for Clinicians, 68, 7–30. [DOI] [PubMed] [Google Scholar]
- Signorelli P, Munoz-Olaya JM, Gagliostro V, Casas J, Ghidoni R, & Fabrias G (2009). Cancer Letters, 282, 238–243. [DOI] [PubMed] [Google Scholar]
- Slawin K, Kadmon D, Park SH, Scardino PT, Anzano M, Sporn MB, et al. (1993). Cancer Research, 53, 4461–4465. [PubMed] [Google Scholar]
- Smith MR, Manola J, Kaufman DS, Oh WK, Bubley GJ, & Kantoff PW (2006). Journal of Clinical Oncology, 24, 2723–2728. [DOI] [PubMed] [Google Scholar]
- Song L, Xiong H, Li J, Liao W, Wang L, Wu J, et al. (2011). Clinical Cancer Research, 17, 1839–1849. [DOI] [PubMed] [Google Scholar]
- Sooriakumaran P, Coley HM, Fox SB, Macanas-Pirard P, Lovell DP, Henderson A, et al. (2009). AntiCancer Research, 29, 1483–1488. [PubMed] [Google Scholar]
- Sooriakumaran P, Macanas-Pirard P, Bucca G, Henderson A, Langley SE, Laing RW, et al. (2009). Cancer Genomics Proteomics, 6, 93–99. [PubMed] [Google Scholar]
- Sugita M, Willians M, Dulaney JT, & Moser HW (1975). Biochimica et Biophysica Acta, 398, 125–131. [DOI] [PubMed] [Google Scholar]
- Tang Y, Cao K, Wang Q, Chen J, Liu R, Wang S, et al. (2016). Oncology Reports, 35, 2907–2915. [DOI] [PubMed] [Google Scholar]
- Thaller C, Shalev M, Frolov A, Eichele G, Thompson TC, Williams RH, et al. (2000). Journal of Clinical Oncology, 18, 3804–3808. [DOI] [PubMed] [Google Scholar]
- Thangavel S, Yoshitomi T, Sakharkar MK, & Nagasaki Y (2015). Journal of Controlled Release, 209, 110–119. [DOI] [PubMed] [Google Scholar]
- Tirodkar TS, Lu P, Bai A, Scheffel MJ, Gencer S, Garrett-Mayer E, et al. (2015). Journal of Biological Chemistry, 290, 13157–13167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirodkar TS, & Voelkel-Johnson C (2012). Experimental Oncology, 34, 231–242. [PubMed] [Google Scholar]
- Tonelli F, Alossaimi M, Williamson L, Tate RJ, Watson DG, Chan E, et al. (2013). British Journal of Pharmacology, 168, 1497–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YC, Chen WY, Abou-Kheir W, Zeng T, Yin JJ, Bahmad H, et al. (2018). Biochimica et Biophysica Acta. [DOI] [PubMed] [Google Scholar]
- Turner LS, Cheng JC, Beckham TH, Keane TE, Norris JS, & Liu X (2011). Prostate Cancer and Prostatic Diseases, 14, 30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vejselova D, Kutlu HM, & Kus G (2016). Cytotechnology, 68, 2721–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venant H, Rahmaniyan M, Jones EE, Lu P, Lilly MB, Garrett-Mayer E, et al. (2015). Molecular Cancer Therapeutics, 14, 2744–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkata JK, An N, Stuart R, Costa LJ, Cai H, Coker W, et al. (2014). Blood, 124, 1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vethakanraj HS, Sesurajan BP, Padmanaban VP, Jayaprakasam M, Murali S, & Sekar AK (2018). Anti-cancer Drugs, 29, 50–60. [DOI] [PubMed] [Google Scholar]
- Wang JD, Takahara S, Nonomura N, Ichimaru N, Toki K, Azuma H, et al. (1999). The Prostate, 40, 50–55. [DOI] [PubMed] [Google Scholar]
- Watson DG, Tonelli F, Alossaimi M, Williamson L, Chan E, Gorshkova I, et al. (2013). Cellular Signalling, 25, 1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattenberg BW, Pitson SM, & Raben DM (2006). The Journal of Lipid Research, 47, 1128–1139. [DOI] [PubMed] [Google Scholar]
- Wegner MS, Schiffmann S, Parnham MJ, Geisslinger G, & Grosch S (2016). Progress in Lipid Research, 63, 93–119. [DOI] [PubMed] [Google Scholar]
- White-Gilbertson S, Mullen T, Senkal C, Lu P, Ogretmen B, Obeid L, et al. (2009). Oncogene, 28, 1132–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Kollmeyer J, Symolon H, Momin A, Munter E, Wang E, et al. (2006). Biochimica et Biophysica Acta, 1758, 1864–1884. [DOI] [PubMed] [Google Scholar]
- Zhou C, Ling MT, Kin-Wah Lee T, Man K, Wang X, & Wong YC (2006). Cancer Letters, 233, 36–47. [DOI] [PubMed] [Google Scholar]