Despite the prokaryotic origin of ATP-dependent proteases, Lon in plant organelles acquired distinct features in terms of gene expression, protein targeting, and functionalization through independent gene duplication and evolution events.
Keywords: Expression divergence, gene duplication, gene evolution, gene paralogs, Lon protease, protein dual-targeting, protein functionalization, protein quality control, proteostasis
Abstract
The degradation of damaged proteins is essential for cell viability. Lon is a highly conserved ATP-dependent serine-lysine protease that maintains proteostasis. We performed a comparative genome-wide analysis to determine the evolutionary history of Lon proteases. Prokaryotes and unicellular eukaryotes retained a single Lon copy, whereas multicellular eukaryotes acquired a peroxisomal copy, in addition to the mitochondrial gene, to sustain the evolution of higher order organ structures. Land plants developed small Lon gene families. Despite the Lon2 peroxisomal paralog, Lon genes triplicated in the Arabidopsis lineage through sequential evolutionary events including whole-genome and tandem duplications. The retention of Lon1, Lon4, and Lon3 triplicates relied on their differential and even contrasting expression patterns, distinct subcellular targeting mechanisms, and functional divergence. Lon1 seems similar to the pre-duplication ancestral gene unit, whereas the duplication of Lon3 and Lon4 is evolutionarily recent. In the wider context of plant evolution, papaya is the only genome with a single ancestral Lon1-type gene. The evolutionary trend among plants is to acquire Lon copies with ambiguous pre-sequences for dual-targeting to mitochondria and chloroplasts, and a substrate recognition domain that deviates from the ancestral Lon1 type. Lon genes constitute a paradigm of dynamic evolution contributing to understanding the functional fate of gene duplicates.
Introduction
Energy metabolism relies on the decomposition of polyunsaturated fatty acids and carbohydrates during storage reserve mobilization. This process is performed by intricately interconnected biochemical pathways, functional in more than one plant organelle including mitochondria, chloroplasts, and peroxisomes. While energy production is efficient, it often results in harmful by-products. In these eukaryotic organelles, the accumulation of oxidants creates an oxidative environment and causes irreversible modification of proteins (Møller et al., 2007, 2011). Proteome homeostasis in plant organelles is preserved by energy-driven processive proteases that maintain protein quality control by removing defective or damaged proteins, preventing the formation of insoluble and deleterious protein aggregates.
The Lon (named from the long filament phenotype of bacterial mutant cells), Clp (caseinolytic protease), and FtsH (filament-forming temperature-sensitive) members of the ATP-dependent proteases of the AAA+ (ATPases associated with a variety of cellular activities) superfamily, evolved to degrade non-functional or short-lived regulatory proteins in plant organelles (Sakamoto, 2006; Rigas et al., 2012; Janska et al., 2013; Smakowska et al., 2014; van Wijk, 2015). While FtsH proteases, also known as AAA proteases, are membrane integrated, Lon and Clp are soluble. Lon consists of a long N-terminal domain that possibly together with the central AAA+ module selectively interacts with target proteins and the C-terminal proteolytic domain with a serine-lysine catalytic dyad (Botos et al., 2004; Rigas et al., 2012). The AAA+ module regulates ATP hydrolysis and protein substrate remodeling, and is adjacent to the sensor and substrate discrimination (SSD) domain (Neuwald et al., 1999; Iyer et al., 2004). In both FtsH and Lon, a single polypeptide contains the ATPase and proteolytic domains, whereas in Clp these domains are kept in distinct subunits that assemble to form the proteolytic holoenzyme complex. Lon subunits of Escherichia coli assemble forming a ring-shaped homohexamer enclosing an internal degradation chamber accessible via an axial pore in the AAA+ ring, which can also interact to form a dodecamer complex (Park et al., 2006; Vieux et al., 2013). In contrast, the mitochondrial Lon of yeast forms a seven-membered, ring-shaped structure (Stahlberg et al., 1999).
Molecular genetics in the model Arabidopsis thaliana has contributed to unraveling the role of Lon1 and Lon2 proteases in maintenance of organelle biogenesis and function. The lon1 mutants exhibit growth retardation, delayed seedling establishment, aberrant mitochondrial morphology, and aberrant function of the tricarboxylic acid cycle and respiration enzymes (Rigas et al., 2009a; Solheim et al., 2012). Recently, the multiple roles of Lon1 in mitochondrial protein homeostasis, as a chaperone aiding the proper folding and stabilization of newly synthesized/imported proteins and as a protease to degrade protein aggregates, have been revealed (Li et al., 2017). The lon2 mutants show growth retardation and peroxisome-deficient phenotypes including enlargement of peroxisomes upon transition from germinating seedlings to mature plants (Lingard and Bartel, 2009). Lon2 is targeted to peroxisomes, facilitates protein import into the matrix, and controls pexophagy that is enhanced upon Lon2 deficiency (Farmer et al., 2013; Bartel et al., 2014; Young and Bartel, 2016). While Lon3 targeting remains unknown, it is probably expressed in sperm cells (Borges et al., 2008; Rigas et al. 2014).
On the basis of transcriptional and translational regulation, Lon1 is dual-targeted to both mitochondria and chloroplasts by encoding two distinct protein isoforms due to twin N-terminal pre-sequences (Daras et al., 2014). In contrast, Lon4 bears an ambiguous pre-sequence generating a single protein isoform with a targeting peptide recognized by the import apparatus of mitochondria and chloroplasts (Sakamoto, 2006; Ostersetzer et al., 2007; Rigas et al., 2014). Moreover, as predicted by molecular modeling, the architecture of the Lon1 protease core structure shows different internal geometry compared with Lon4 (Rigas et al., 2014). The internal loop of Lon4 SSD shows a right-handed extension, whereas in Lon1 this domain is left-handed.
Given the progress made on the functional characterization of Lon genes, Arabidopsis represents a promising model system to determine gene duplication and functional divergence within the Lon proteases in plants (Koch, 2019). Our previous reports show that Lon1 and Lon4 discriminate in terms of subcellular dual-targeting mechanisms and functionalization domains (Rigas et al., 2009b; Daras et al., 2014; Rigas et al., 2014). This work shows that Arabidopsis Lon (AtLon) genes present differential spatiotemporal gene expression profiles. In contrast to AtLon1 that is widely expressed, AtLon3 and AtLon4 expression is exclusively restricted to male and female reproductive tissues, respectively. This mode of Lon paralogous gene evolution is evident only in the Arabidopsis genus emanating from the most recent whole-genome duplication (WGD) that occurred close to the base of the Brassicaceae family, also known as the α-WGD (Blanc et al., 2003; Barker et al., 2009), and a subsequent tandem duplication also specific to the Brassicaceae family (Haberer et al., 2004; Audemard et al., 2012). In the wide context of land plants, comparative analysis of Lon phylogeny and protein structural properties provides evidence that distinct gene duplication events were the primary evolutionary force pinpointing the gradual co-evolution of alternative Lon members in embryophytes.
Materials and methods
Identification of Lon protease gene family members
A workflow of the genome-wide analysis to determine the evolutionary history of Lon proteases is presented in Supplementary Fig. S1 at JXB online. Lon protease gene information from the model plant A. thaliana was downloaded from The Arabidopsis Information Resources database (TAIR: http://www.arabidopsis.org/;Lamesch et al., 2012). To identify Lon genes of unknown species, BLASTP searches were conducted using orthologous A. thaliana protein sequences as the query search in the publicly available phytozome database (http://www.phytozome.net/;Goodstein et al., 2012). Homologs from gymnosperm species were obtained from the 1000 Plants (1KP) project (www.onekp.com;Matasci et al., 2014). Homologs of animal and fungal species were obtained from the NCBI. All the data obtained are listed in Supplementary Table S1. The phylogenetic relationship of the 99 selected species was determined by the common tree taxonomy tool at NCBI (http://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi).
Sequence alignment, structural modeling, and subcellular targeting prediction
Multiple sequence alignment of the identified amino acid sequences of Lon protease genes were performed by the ClustalX 1.83 software. The results were exported as GCG/MSF format and analyzed using the GeneDoc MFC Application version 2.6.0.2. The construction of all the phylogenetic trees was made by the Interactive Tree of Life (http://itol.embl.de) online platform. The alignment logos of the protein conserved domains or nucleotides were generated with WebLogo (http://weblogo.berkeley.edu/). Subcellular localization predictions of land plant Lon proteases were carried out using the TargetP 1.1 (http://www.cbs.dtu.dk/services/TargetP/), WolfPSORT (https://wolfpsort.hgc.jp/), ChloroP 1.1 (http://www.cbs.dtu.dk/services/ChloroP/), MITOPROT (https://ihg.gsf.de/ihg/mitoprot.html), and Predotar v1.04 (https://urgi.versailles.inra.fr/predotar/) servers. Structural modeling of the SSD domain of mitochondrial Lon homologs was carried out with Phyre2 using the intensive mode. Modeling was performed on the basis of known crystallographic data mainly available from AAA+ proteins and bacterial Lon proteases. The ribbon model was generated in PyMol (www.pymol.org). All SSDs were aligned to EcLon and then were classified according to their structure into three groups. The length and the sequences of the SSD domain of each homolog are listed in Supplementary Table S2.
Phylogeny reconstruction
The evolutionary history was inferred by using the maximum likelihood method based on the JTT matrix-based model (Jones et al., 1992). The initial tree for the heuristic search was obtained automatically by applying Neighbor–Joining and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with the superior log likelihood value. The analysis was performed using the MEGA 7.0 software (Kumar et al., 2016).
Synteny analysis
Synteny analyses of duplicate gene pairs and the WGD data for A. thaliana were achieved using the Plant Genome Duplication Database (PGDD; http://chibba.agtec.uga.edu/duplication/index/locus;Lee et al., 2017).
In silico expression profiles
For the expression profile analysis of Lon genes in Arabidopsis, ATH1 22k microarray data in the Genevestigator V3 database (Zimmermann et al., 2004) and RNA sequencing (RNA-seq) data from the TravaDB database (Klepikova et al., 2016) were used, and then the heat maps were constructed using the obtained gene expression data sets. Heat maps were generated using the Perseus software (version 1.5.3.2).
cis-element identification and co-expression network construction
The identification of cis-regulatory elements was performed using the web-based analysis tool AthaMap (www.athamap.de) analyzing the promoter region extending 2000 bp upstream of the translation initiation site. The co-expression network of Lon4 was generated by the ATTED II application (http://atted.jp/). The gene expression map was drawn by the Arabidopsis eFP browser (Winter et al., 2007).
Results
Members of the Arabidopsis Lon family show gene expression divergence
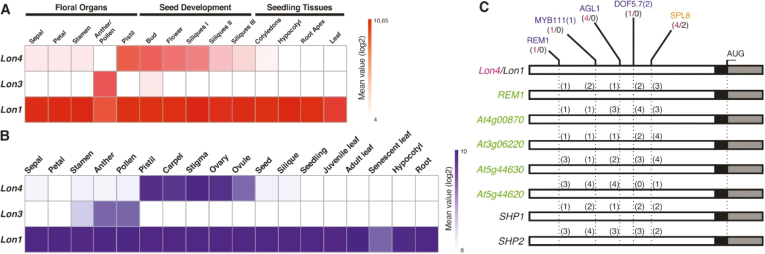
Eukaryotic genomes have large numbers of duplicated genes that can evolve expression divergence, which might represent an important evolutionary mechanism for retention of duplicated genes (Haberer et al., 2004; Liu et al., 2011). To test whether Lon genes differentiate in terms of the expression profile, we analyzed data obtained by independent high-throughput approaches of the Arabidopsis transcriptome. The Lon genes showed reciprocal expression patterns. Lon1 was broadly expressed across multiple organ types, whereas Lon4 and Lon3 appeared to have acquired a very restricted expression pattern after duplication from the common ancestor. While Lon4 was exclusively expressed in female reproductive organs including the stigma, pistil, ovary, and ovules, Lon3 expression was restricted to the male floral organs such as the stamen, anther, and pollen (Fig. 1A). The differential expression pattern between the Lon genes was also confirmed by meta-analysis of microarray data for Arabidopsis (Fig. 1B; Zimmermann et al., 2004). The organ-specific expression pattern between Lon4 and Lon3 could only result from regulatory neofunctionalization and both genes probably represent rapidly evolving copies (Liu et al., 2011). In contrast to Lon4 and Lon3, Lon1 showed a wide expression pattern, suggesting that Lon1 retained the expression profile of the ancestral gene. Regulatory subfunctionalization is most likely to be the evolutionary fate that discriminates Lon1 expression from that of Lon3 and Lon4 (Lynch and Force, 2000).
Fig. 1.
Lon genes in Arabidopsis show distinct expression profiles derived after gene duplication. (A and B) Heatmap plot of Lon gene expression obtained by (A) RNA-seq and (B) microarray data analysis. (C) The evolutionary context of CREs in Lon4 and Lon1 promoters contributes to their differential expression pattern in floral organs. Lon4 contains specific CREs, depicted in blue, associated with expression restricted to pistils, which are absent from Lon1. Numbers in red and black depict the plethora of CREs in Lon4 and Lon1 promoters, respectively. Genes in green were identified by Lon4 gene co-expression analysis (Supplementary Fig. S2A). Graphical representations of CREs do not correlate with their exact position.
Α comparison of the genomic regulatory regions, which cover 2000 bp upstream of the translation start site, was performed to understand the molecular basis of gene expression divergence between Lon1 and Lon4. The analysis led to the identification of four cis-regulatory elements (CREs), namely REM1, MYB111(1), AGL1, and DOF5.7(2), that exist only in the Lon4 promoter but not in Lon1 (Fig. 1C). However, both Lon1 and Lon4 share SPL8 as a common CRE, albeit SPL8 exists four times in the Lon4 promoter and just twice in Lon1. The over-representation of SPL8 in the Lon4 promoter is most probably associated with enhancement of its expression in pistils, since SPL8 has been reported to be crucial for gynoecium development and maintenance of female fertility (Xing et al., 2013). This divergence in CRE constitution further supports the differential expression pattern of the two genes.
The tissue specificity of Lon4 expression was an impetus to constitute the gene co-expression network using available public resources. The results showed a significant co-expression relationship between Lon4 and REPRODUCTIVE MERISTEM1 (REM1), At4g00870, At5g44630, At3g06220, and At5g44620 sharing common CREs (Fig. 1C; Supplementary Fig. S2A). Despite the fact that At5g44620 is missing the DOF5.7(2) element, the other four genes with a tight expression profile carried all four Lon4-specific elements. Remarkably, SHP1 and SHP2 responsible for carpel specification (Ó’Maoiléidigh et al., 2014), which were recently experimentally characterized as being solely expressed in pistils (Villarino et al., 2016), also contained the four Lon4-specific CREs in their promoter regions (Fig. 1C). On the basis of their electronic fluorescent pictograph (Winter et al., 2007), these genes have a similar spatiotemporal expression pattern to Lon4, which is exclusive in pistils (Supplementary Fig. S2B). Since certain gains and losses of CREs occur in both promoters, after gene duplication, Lon1 and Lon4 in addition to the dual-targeting mechanism and SSD divergence, experienced different modes of evolution of their regulatory regions that explain the differences in the expression pattern.
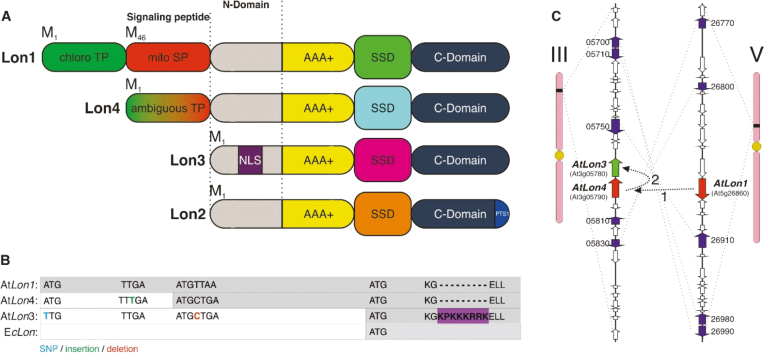
Amorphic mutations caused Lon4 and Lon3 differentiation of targeting domains from Lon1
In addition to the differential expression pattern, Lon genes in Arabidopsis evolved distinct mechanisms for subcellular targeting and substrate recognition (Fig. 2A). Apart from Lon2 that carries a PTS1 motif at the C-terminus for peroxisomal localization, it seems interesting to understand whether there is a sequence of mutation events contributing to the distinct targeting properties of Lon1, Lon3, and Lon4. Strikingly, Lon3 and Lon4 genes showed accelerated sequence evolution compared with their Lon1 paralog. While Lon4 has both ATG contexts similar to Lon1 twin pre-sequences, there is a thymine (T) insertion downstream of the vicinity of the first AUG context that disrupts the ORF permitting the translation to initiate only from the second ATG (Fig. 2B; Supplementary Fig. S3A). In Lon3, the first ATG is mutated due to a nucleotide transition of adenine (A) to T, which restricts the initiation of translation, whereas the first nucleotide after the second ATG, that is most probably a cytosine (C), is deleted, resulting in initiation of translation from the downstream start codon (Fig. 2B; Supplementary Fig. S3A). These are all amorphic mutations, which either prevent the initiation of translation due to mutations on the initiation codon or disrupt the translation process at the region encoding the N-terminal pre-sequence permitting the translation to initiate from downstream ATG codons.
Fig. 2.
Accelerated sequence evolution after gene duplication contributed to the distinct targeting properties of Arabidopsis Lon genes. (A) The proteases encoded by Arabidopsis Lon genes acquired distinct structural domains for subcellular targeting and substrate recognition. Lon1 is dual-targeted to mitochondria and chloroplasts by twin pre-sequences, whereas Lon4 evolved an ambiguous pre-sequence at the N-terminus. Lon2 targets to peroxisomes due to a PTS1 motif at the C-terminus. The N-terminal domain of Lon3 for organellar targeting is apparently depleted and carries an NLS motif potentially for nuclear localization highlighted in purple. (B) A series of nucleotide mutations prevent the initiation of translation or abort protein synthesis upstream of the annotated ATG of Lon3 and Lon4. The ORF is highlighted in gray. (C) Sequential gene duplication events and rapid evolution led to preservation of Lon3 and Lon4 duplicates from their common ancestor.
Intriguingly, the reconstitution of the mutated nucleotides reversed evolution and led to ORFs for both Lon4 and Lon3. To be precise, the removal of T in Lon4 opened the frame downstream of the first ATG and resulted in a chloroplast transit peptide (Supplementary Fig. S3B, C). In Lon3, the reconstitution of the first ATG and insertion of C immediately downstream of the second ATG opened a reading frame with two ATG initiation codons in-frame (Supplementary Fig. S3B). By reversion of nucleotide sequence evolution, Lon4 and Lon3 acquired Lon1-like twin pre-sequences encoding two distinct subcellular-targeted isoforms (Supplementary Fig. S3C). These—hidden by evolution—ancestral subcellular targeting properties of Lon4 and Lon3 revealed their common origin, suggesting that both are duplicates of Lon1. To support this notion further, we used the Plant Genome Duplication Database (PGDD) that provides intraplant genome alignment information to investigate evolutionary consequences of genome duplication (Lee et al., 2017). The analysis revealed that Lon4 originated from a WGD event that occurred in the herbaceous annual genus Arabidopsis and that Lon3 is a tandem duplicate of Lon4 as both are repeated genes located in close physical proximity to each other (Fig. 2C). Overall, the two gene copies, Lon3 and Lon4, have experienced an accelerated sequence evolution in the proximity of the protein targeting region and deviated extensively from their common ancestral gene.
Phylogenetic classification and diversification of Lon homologs
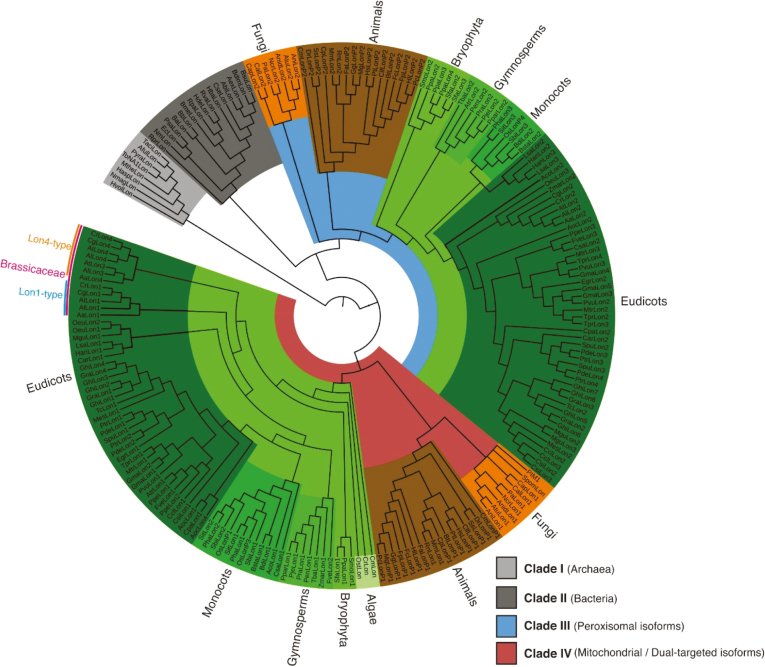
Unlike other eukaryotic genomes, plant genomes tend to evolve at higher rates, presenting higher genome diversity and organizing genes in multicopy families (Panchy et al., 2016). To explore and understand the evolutionary history of Lon proteases, we examined their phylogenetic classification in several taxa (kingdoms) including archaea, bacteria, fungi, animals, algae, and land plants. All gene entries were obtained from fully sequenced and annotated genomes (Supplementary Table S1). Maximum likelihood phylogeny reconstruction categorized the Lon homologs into four robust phylogenetic clades (Fig. 3). The Lon homologs of Archaea (Clade I) and Bacteria (Clade II) grouped separately from eukaryotes. Homologs of Archaea formed a monophyletic clade at the base of the phylogenetic tree due to the membrane-anchoring region emerging from the ATPase domain that discriminates the Archaea-specific LonB subfamily from the typical LonA structure of Lon homologs (Rigas et al., 2012). Nonetheless, the two clades of prokaryotes were comprised of Lon homologs lacking an N-terminal targeting domain.
Fig. 3.
The maximum likelihood phylogeny categorized Lon homologs in four major clades. Clades I and II contain Lon of prokaryotic origin diverged in Archaea and bacteria, respectively. Clade III includes the peroxisomal Lon homologs, whereas Clade IV contains the bona fide mitochondrial Lon homologs of fungi, animals, and algae, and Lon homologs from land plants dual-targeted to mitochondria and chloroplasts.
Based on the analysis, Lon homologs of eukaryotes with peroxisome-targeting properties comprised the third major clade (Clade III; Fig. 3). This clade was further diverged, resulting in three separate subclades of fungi, animals, and plants. This classification of Lon between the three kingdoms is indicative of the differences in the peroxisomal biochemical pathways between fungi, animals, and the green lineage. The nodes of Lon peroxisomal divergence within plants were particularly apparent with respect to their taxonomic events. Homologs from bryophyta, gymnosperms, monocots, and eudicots including Brassicaceae formed distinct subgroups. This classification of Lon peroxisomal proteases was evident by analyzing the conservation of the PTS1 motifs (Supplementary Fig. S4). The typical tripeptide serine–lysine–leucine (SKL motif) was conserved in peroxisomal Lon homologs of animal origin and mainly in higher plants including the eudicots, monocots, and conifers. The motif of bryophytic Lon homologs of lower land plants together with those of fungi deviated extensively from the typical PTS1 motif consensus. In the green lineage, there is great variability concerning the four amino acid residues upstream of the SKL motif and before the highly conserved proline (P) with respect to their taxonomic rank. From this analysis, we can suggest that plant speciation had a great impact on the diversification of the peroxisomal Lon proteases.
The clade including the non-peroxisomal-targeted Lon proteases of eukaryotes retained the largest number of homologs (Fig. 3). The phylogenetic clustering pattern divided these Lon homologs into two subclades. The first subclade included the bona fide mitochondrial-targeted homologs from algae, fungi, and animals. Concerning the biological significance of this finding, it is worth noting that algae lack a peroxisomal homolog, which is also missing from fission yeast and budding yeast (Supplementary Table S1). In contrast to hyphae-forming multicellular fungi that retained both a mitochondrial and a peroxisomal Lon isoform, yeasts, which are unicellular organisms, preserved a single mitochondrial homolog. In striking contrast, animals that like algae and fungi possess a bona fide mitochondrial Lon homolog also retained a peroxisomal copy. A possible explanation could be that animals and hyphae-forming fungi develop complex multicellular structures that require adequate energy support in contrast to unicellular organisms such as yeast or algae. Peroxisomes have an important role in lipid metabolism (Wanders et al., 2010) and are implicated in ubiquitous energy metabolic pathways throughout eukaryotes, such as the β-oxidation of fatty acids (Poirier et al., 2006). In filamentous fungi characterized by a multicellular architecture, the peroxisomal Lon displays a dual function as an ATP-stimulated protease and molecular chaperone to maintain proteostasis by protein quality control in the matrix (Bartoszewska et al., 2012).
The phylogeny of Lons in the green lineage is reflected by their taxonomy (Fig. 3). Strikingly, the plant homologs acquired N-terminal domains for dual-targeting to mitochondria and chloroplasts (Supplementary Table S2). Like animal species, land plants evolved multicellular organs and thus they all contained peroxisomal Lon homologs (Supplementary Table S1). Taken together, we may infer that Lon first appeared as a single gene in prokaryotes. Then, in lower eukaryotes, Lon evolved to a single mitochondrial isoform and subsequently underwent duplication to generate an additional peroxisomal-targeted homolog in multicellular organisms. This event led to the emergence of Lon paralogs dual-targeted to mitochondria and chloroplasts in land plants.
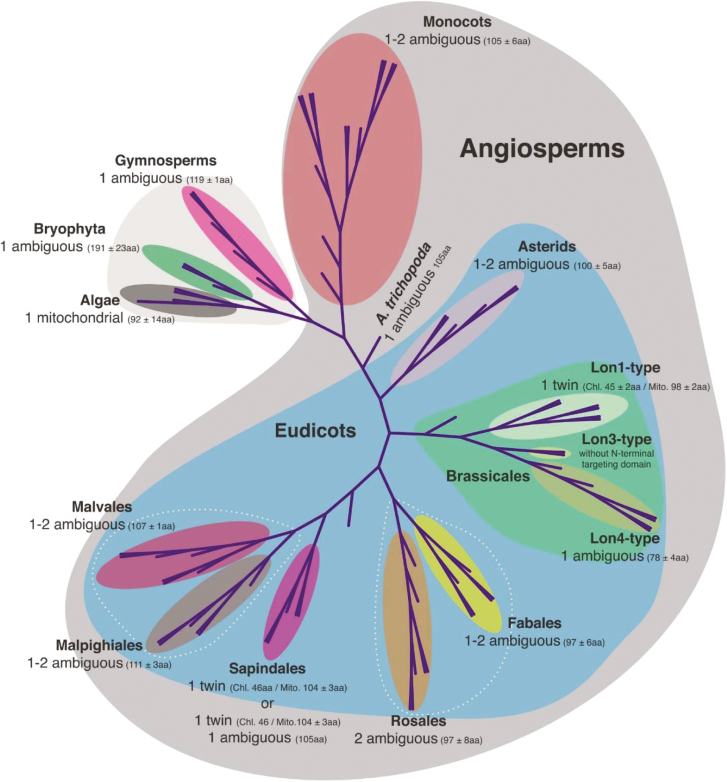
The majority of Lon proteases in the land plant lineage acquired an ambiguous pre-sequence
To gain further insight into the evolution of the subcellular targeting properties of Lon homologs from land plants (Clade IV; Fig. 3), we focused on the analysis of the N-terminal targeting domains. The analysis divided algae, bryophytes, and gymnosperms from the rest of the higher plants due to the presence of only one Lon gene copy (Fig. 4). Nevertheless, the Lon isoform of algae was mitochondrial with a short pre-sequence, with an average polypeptide length of 92 amino acids, whereas both bryophytes and gymnosperms evolved a dual-targeted Lon homolog with an ambiguous pre-sequence with a longer polypeptide length. In the angiosperms, monocots retained up to two copies both with an ambiguous pre-sequence, which is relatively shorter than the single homolog of bryophytes and gymnosperms. While Amborella trichopoda is a lower angiosperm that emerged separately from the other species of monocots and eudicots, it has evolved a single Lon homolog with an ambiguous pre-sequence.
Fig. 4.
An evolutionary classification of Lon N-terminal targeting domains in plants.
In eudicots, the evolution of the N-targeting domains categorized the Lon homologs into four groups (Fig. 4). The first group consists of four orders, namely Asterids, Fabales, Malvales, and Malpighiales, that acquired up to two Lon copies with an ambiguous pre-sequence. Rosales is a unique order which always has two Lon homologs dual-targeted to mitochondria and chloroplasts with an ambiguous pre-sequence. The orders Sapindales and Brassicales differentiated from the others due to the presence of at least one Lon homolog with twin pre-sequences. As a third group, the Sapindales lineage contained either a single Lon gene with twin pre-sequences or an additional gene copy with an ambiguous pre-sequence. The last group contains the Lon gene families of Brassicales with three members and distinct targeting mechanisms. While the Lon1-like and Lon4-like homologs have dual-targeting properties, they acquired different mechanisms for protein subcellular localization. The Lon1-like copy evolved twin pre-sequences, whereas Lon4-like has an ambiguous pre-sequence. In contrast, the N-terminal targeting domain was missing from the Lon3-like homologs. Overall, the analysis supports the notion that Lon homologs in land plants were formed by duplications. This is particularly evident in the Arabidopsis lineage that was derived from the Brassicaceae-specific α-WGD event leading to the formation of multigene families (Barker et al., 2009).
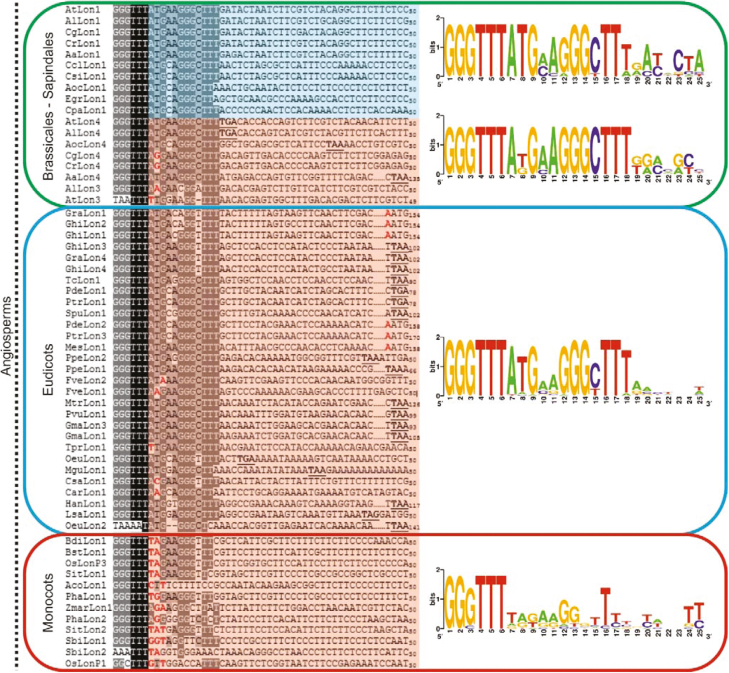
The evolutionary history of Lon homologs in terms of their targeting properties was further supported by analyzing the conservation of the first ATG context in angiosperms. To identify the Lon homologs with twin pre-sequences, the nucleotide sequence upstream of the annotated ATG, which hereafter is called the second initiation codon, was analyzed to confirm the ORF upstream of the first ATG. This approach led to the identification of 10 out of 59 Lon homologs with twin pre-sequences (Supplementary Fig. S5), whereas the majority of the other 49 genes evolved an ambiguous pre-sequence. Multiple sequence alignment analysis revealed that in the angiosperms the first ATG was present but it was gradually deteriorated in Lon homologs that lost their chloroplast transit peptide upon evolution of the ambiguous pre-sequence (Fig. 5).
Fig. 5.
The first ATG context is highly conserved in angiosperms. In Lon1-like genes of Sapindales and Brassicales, the chloroplast transit peptide was retained for dual-targeting with twin pre-sequences through ORF preservation (highlighted in blue). In the Lon genes with an ambiguous pre-sequence for dual-targeting, the accumulation of mutational modifications led to deterioration of the chloroplast transit peptide (highlighted in red). The mutations that result in a stop codon are displayed in black underlined bold triplets, whereas the nucleotides that modify the translation initiation codon or shift the reading frame are marked in bold red.
The highest level of conservation was evident in the orders Brassicales and Sapindales, that contain the Lon genes with functional twin pre-sequences due to the N-terminal extension for chloroplast targeting (Fig. 5; Supplementary Fig. S5). While the first ATG context remained highly conserved in Lon homologs from eudicots with an ambiguous pre-sequence, the mutational changes either modified the initiation codon or closed the reading frame only at the region encoding the N-terminal pre-sequence, resulting in initiation of translation from the downstream start codon (Fig. 5). These amorphic mutations were highly frequent in monocots.
Lon homologs of land plants show a strong trend to acquire a Lon4-like SSD domain
The SSD domain is possibly involved in modulating selective substrate recognition by Lon proteases and thereby exhibits substantial diversity within the Lon homologs (Rigas et al., 2009b). To gain a deeper insight into the evolution and functional diversification of Lon proteases, we analyzed the structure of the SSD domain (Fig. 6; Supplementary Table S3; Supplementary Fig. S6). The genome-wide study confirmed that the SSD domains of bacteria and animals are highly similar, preserving the minimal structure of E. coli Lon. In the green lineage, three categorization domains were formed. The first included only two species, Helianthus annuus and Carica papaya, both with a single Lon gene bearing an SSD domain structurally similar to AtLon1. Apart from these plants, this domain also included Lon homologs from fungi and most homologs from algae. The second domain included Lon homologs with more than one gene, preserving the SSD domain structure of both AtLon1 and AtLon4 from Rosids including the Brassicales and one monocot, Sorghum bicolor. Most land plants, namely bryophytes, gymnosperms, and angiosperms, including monocots and eudicots, were included in the third domain, which contained the Lon homologs with the SSD domain structure of only AtLon4. The results show a dynamic evolutionary process of Lon proteases in plants to acquire and preserve the SSD domain structure of AtLon4. The only exceptions to this rule are helianthus and papaya that preserved the SSD domain of AtLon1 and retained a single gene copy with dual-targeting properties to mitochondria and chloroplasts (Supplementary Table S2).
Fig. 6.
Classification of Lon homologs based on the structure of the SSD domain. Bacteria and animal SSD domains out-group from plants and fungi preserving a minimal structure. In the plant lineage, only helianthus and papaya that grouped together with most algae and fungi preserved a single gene with a Lon1-type SSD domain. Among Rosids, Brassicales in particular retained one gene copy with a Lon1-type structure and evolved copies with the Lon4-type SSD domain. The majority of land plants acquired the Lon4-type SSD domain.
Given that evolution had a great impact on the structural properties of Lon proteases in plants, we constructed the phylogeny of Lons in accordance with both domains that modulate subcellular targeting and substrate recognition (Fig. 7A). The base began with Lon homologs from algae having a mitochondrial pre-sequence and mostly preserving a Lon1-type SSD domain. The homologs from bryophytes, gymnosperms, monocots, and eudicots, mainly asterids, rosales, and fabales, almost exclusively had a typical Lon4 structure combining an ambiguous pre-sequence with a Lon4-type SSD domain.
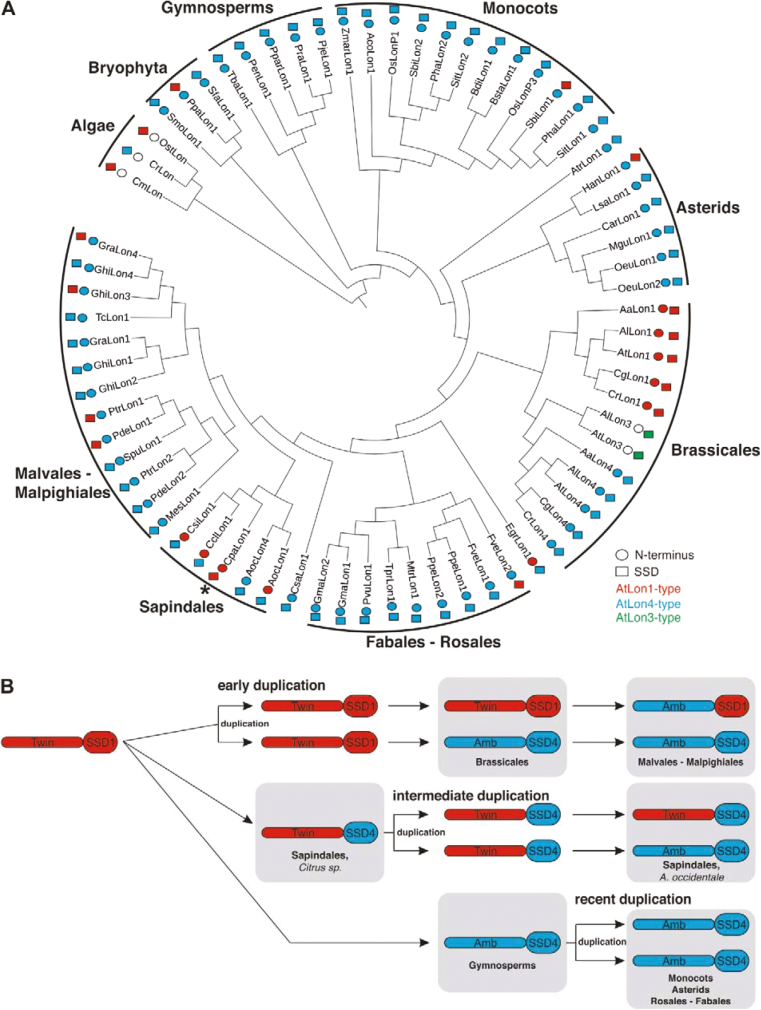
Fig. 7.
Evolutionary analysis of the structural properties and model of evolution of Lon paralogs in the plant kingdom. (A) Phylogenetic classification of Lon paralogs in the plant lineage showing the evolution of the N-terminal dual-targeting domains (squares) and the SSD domains (circles). The asterisk indicates papaya Lon that retained a single Lon1-type gene copy. (B) Model of gene duplication events driving the molecular evolution of Lon genes in land plants. Lon genes might have evolved from an ancestral gene with twin pre-sequences for dual-targeting and a Lon1-type SSD domain. Independent events of gene duplications, classified as early, intermediate, and recent, occurred together with mutational changes evolving the modern Lon genes in embryophytes.
Βrassicales, especially the Arabidopsis lineage, have evolved three genes with different subcellular targeting mechanisms and distinct SSD domain structures. Chimeric Lon homologs were apparent in Malpighiales, Malvales, and Sapindales, highlighting the alternation of the Lon1-type and Lon4-type structural features. This analysis yielded four major findings. First, the Arabidopsis lineage—probably due to a specific sequence of genome duplication events—evolved three Lon paralogs. Secondly, papaya, though it belongs in Brassicales, remained the only species with a single Lon1-type gene, providing evidence that unlike Arabidopsis taxa, papaya missed critical duplication events. Thirdly, in most land plants excluding algae, the Lon4 type is dominant. Finally, the chimeric Lon genes demonstrated rapid evolution to combine both Lon1-type and Lon4-type features.
Discussion
Arabidopsis Lon duplicates acquired diverged targeting mechanisms, asymmetric sequence evolution, and gene expression divergence
Gene duplication has contributed to the evolution of novel structures or agronomic traits, which are thought to be important in plant adaptation to variable abiotic and biotic environments, in speciation and diversification (Soltis et al., 2009; Panchy et al., 2016). Polyploidy, or WGD, is the most extreme gene duplication mechanism that results in a vast increase in genome size and gene sets. Apart from WGD, subgenomic mechanisms also contribute to gene duplications (Zhang, 2003). Tandem (or local) duplication is attributed to unequal crossing-over events and leads to a cluster of at least two paralogous sequences with no or a few intervening gene sequences. While WGD leads to a dramatic increase of the gene content, it is a rare phenomenon. In contrast, tandem duplication, although affecting a limited number of genes, can modify the gene number upon every meiotic division. Both WGDs and tandem duplications account for the majority of gene duplicates in plants. However, the predominant fate of most duplicates is either to become a non-functional pseudogene or gene loss through the diploidization process that ultimately leads to the return of many genes to single copy (gene fractionation). Genome-wide studies have revealed that the preservation of duplicated genes depends on nucleotide sequence modifications resulting in a differential expression pattern, distinct subcellular localization properties, and functional divergence (Ganko et al., 2007; Carretero-Paulet and Fares, 2012; Byun and Singh, 2013; Jiang et al., 2013).
Interestingly, Lon genes of Arabidopsis were preserved as paralogs after duplication because they fulfill these criteria. In terms of expression pattern, Lon1 is expressed in all organs, whereas Lon4 expression is mainly detected in pistils, buds, and at the first stages of silique development, suggestive of a highly specific function. The differences in expression pattern can be explained by the presence or absence of certain portions of the regulatory regions. After duplication, Lon4 experienced evolutionary changes in the regulatory region, gaining specific CREs associated with gene expression specificity in pistils. In particular, CREs such as REM1, MYB111, AGL1, and DOF5.7 in the promoter region of Lon4 seem to be the key to understanding the mechanisms that underlie the expression differences between Lon1 and Lon4. These elements are Lon4 specific, as they do not exist in the Lon1 promoter, and presumably play an important role in shaping the expression pattern of Lon4. Furthermore, these regulatory sites are shared by the promoters of three genes including REM1 that are co-expressed with Lon4. They are also present in the promoters of SHATTERPROOF1 (SHP1) and SHATTERPROOF2 (SHP2) identified by a genome-wide transcriptional analysis of genes involved in the development of pistils (Villarino et al., 2016). The fact that the four CREs are highly conserved among these genes that show specific expression in pistils further implies a minimal context of CREs necessary to drive gene expression specificity. In contrast to Lon4, the expression of Lon3 was restricted to the male reproductive parts of the flower. On the basis of these findings, the pre-duplication expression pattern is likely to be a broad expression pattern similar to that of Lon1. Furthermore, it is widely appreciated that as younger duplicates, Lon4 and Lon3 evolved rapidly, acquiring a more divergent and restricted expression pattern compared with Lon1 (Casneuf et al., 2006; Ganko et al., 2007; Liu et al., 2011). Hence, Lon3 and Lon4 gene duplicates were preserved by dividing the ancestral expression pattern of Lon1 between the duplicates, whereas Lon3 and Lon4 differentiated by each gaining a discrete expression pattern.
The distinct subcellular targeting properties of Arabidopsis Lon genes increased the retention of duplicates. Both Lon4 and Lon3 gene copies showed accelerated sequence evolution compared with their Lon1 paralog. A series of amorphic mutations gradually modified the first ATG context, inhibiting the initiation of translation or disrupting the reading frame. Consequently, Lon4 acquired an ambiguous pre-sequence for dual-targeting to chloroplasts and mitochondria, whereas Lon1 preserved both ATGs in-frame. The mutational changes of Lon3 were more severe in the vicinity of both ATGs, probably resulting in the loss of a functional N-terminal targeting domain.
Homology modeling revealed that structurally the Arabidopsis Lon proteases deviate from the hexameric bacterial and human Lon proteases, fitting best with the heptameric yeast structure (Rigas et al., 2014). This is most probably attributed to the primary amino acid sequences of Arabidopsis Lon sequences, which are significantly longer than the bacterial and human Lon proteases (Rigas et al., 2009b; Venkatesh et al., 2012). Moreover, the internal geometry is different between AtLon1 and AtLon4. The internal loop domain of AtLon4 shows a right-handed extension, whereas in AtLon1 it is left-handed. The Arabidopsis Lon genes showed an asymmetric rate of amino acid sequence evolution at the highly variable SSD domain (Rigas et al. 2009b; Daras et al., 2014). The accumulation of neomorphic mutations led to non-synonymous amino acid substitutions contributing to the modification of the core domain likely to be involved in substrate recognition. Taking these points into consideration, it appears reasonable to suggest that evolution of Lon genes in Arabidopsis is an ongoing process contributing to better fitness, selection, and adaptation of gene duplicates. Lon1 has probably retained the ancestral form, whereas Lon3 and Lon4 have evolved independently.
Lon genes were formed by duplication events during the evolution of land plants
The phylogenetic analysis of Lon genes provided evidence of evolutionary divergence among species. The prokaryotes together with unicellular eukaryotic species, namely yeasts and algae, retained a single gene copy, despite the divergence of subcellular targeting and substrate recognition domains. Unlike these eukaryotic organisms, hyphae-forming fungi, animal species, and land plants acquired an additional peroxisomal copy. While animals strictly preserved one mitochondrial and one peroxisomal gene copy, the land plants evolved small Lon gene families with members acquiring dual-targeting properties to chloroplasts and mitochondria and differentiated functional domains. It is worth mentioning that bacteria and animals preserved a minimal structure of the SSD domain, whereas plants and fungi evolved an extended domain. These results coincide with the concept that plants, as sessile organisms, are constantly exposed to acute environmental conditions and thus, through adaptive evolution, preserved the duplicated Lon genes to maintain proteostasis in organelles. Furthermore, gene expression of Arabidopsis Lon duplicates specifically in floral organs supports the view of Lon proteases having evolved to satisfy the growing demands for energy and carbon during sexual reproduction.
Our analysis also revealed that the driving force towards the evolution of Lon proteases in land plants was a series of independent gene duplication events together with the accumulation of asymmetric mutations in the duplicates. Even though the exact phylogenetic placement of these events is an issue that cannot be easily resolved, we propose a model that discriminates Lon evolutionary fate in three phases, hereafter called early, intermediate, and recent gene duplication events (Fig. 7B). Early duplication in Arabidopsis led to the differentiation of gene duplicates. Consistent with previous reports (Liu et al., 2014), we provide evidence that Lon4 was generated by a WGD event and Lon3 by a tandem duplication that placed it next to Lon4. The broad expression pattern, the subcellular targeting mechanism by twin pre-sequences, and the divergence within functional domains, reveal that the pre-duplication ancestral gene unit is likely to be similar to Lon1.
These observations agree with the widely appreciated concept that the Arabidopsis lineage has experienced three WGD events (Blanc et al., 2003; Bowers et al., 2003; Barker et al., 2009). The most recent event is called the At-α (alpha), the intermediate event is referred to as the At-β (beta), and the oldest is the At-γ (gamma). While At-γ is shared with other Rosids, including papaya (Carica), poplar (Populus), and grape (Vitis), the most recent WGD, At-α, is specific to the Brassicaceae family (Schranz and Mitchell-Olds, 2006) and accounts for ~2500 pairs of duplicated genes in the Arabidopsis genome (Blanc et al., 2003; Bowers et al., 2003). Even though Carica papaya belongs together with Arabidopsis in the same order, Brassicales, its genome did not undergo the two most recent WGD events, At-α and At-β (Barker et al., 2009). Surprisingly, the ancestral genome of papaya is the only one among the land plants containing a single-copy Lon gene that shares outstanding similarities with Arabidopsis Lon1 with respect to the twin pre-sequences and the structure of the SSD domain. These observations suggest that among the order Brassicales, papaya carries the pre-duplication ancestral Lon gene, which is placed at the beginning of the model for Lon evolution in plants (Fig. 7B). Moreover, the duplication of Lon4 is evolutionarily recent because it was derived from the Arabidopsis-specific At-α WGD after divergence from the Brasssica lineage.
Nevertheless, papaya seems to be the sole exception to the rule of Lon evolutionary history. The analysis revealed that the majority of plant species evolved genes that are similar to the Lon4 type (Fig. 7B). These duplicates acquired an ambiguous pre-sequence to drive the protein isoform to both chloroplasts and mitochondria and an SSD domain that deviates from the Lon1 structure. Although this gene form dominates, Citrus sp. retained an intermediate form, which preserved the twin pre-sequences but lost the Lon1-type structure of the SSD domain.
In conclusion, our results provide evidence of the complex dynamics underlying the evolution of Lon proteases by gene duplication and mutational events. Despite the complexity, this genome-wide study sheds light on the ancestral Lon gene form in plants and pinpoints the trend of evolutionary history to enhance plant adaptability to harsh environmental conditions, counteracting the organellar oxidative environment and irreversible modification of proteins. Overall, the results provide valuable information for further studies on the roles of Lon proteases in the protein quality control of plant organelles.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Schematic presentation of the workflow applied to determine the evolutionary history of Lon proteases.
Fig. S2. Lon4 is co-expressed with pistil-specific genes.
Fig. S3. Correction of Lon3 and Lon4 mutations reconstitutes the N-terminal targeting domain to Lon1 twin pre-sequences.
Fig. S4. Conservation and dissimilarities of the PTS1 motif in Lon peroxisomal homologs.
Fig. S5. The N-terminal extension of Lon1-like homologs from Sapindales and Brassicales encoding a chloroplast transit peptide for dual-targeting with twin pre-sequences.
Fig. S6. Molecular modeling of the sensor and substrate discrimination (SSD) domain reveals the functional features of Lon proteases.
Table S1. List of Lon genes used in the study and their accessions.
Table S2. Dual-targeting properties and sensor and substrate discrimination (SSD) domain structure of Lon homologs from land plants.
Table S3. Protein length and the co-ordinates of the SSD domain of the bacterial and eukaryotic Lon homologs targeted to mitochondria (fungi and animals) or dual-targeted to chloroplasts and mitochondria (plants).
Acknowledgements
This work was supported by COST Action PROTEOSTASIS BM1307. D. Tsitsekian, GD, and AA are indebted for funding to IKY fellowships of excellence for postgraduate studies in Greece-Siemens program. D. Templalexis is grateful to Bodossaki Foundation for support.
References
- Audemard E, Schiex T, Faraut T. 2012. Detecting long tandem duplications in genomic sequences. BMC Bioinformatics 13, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B, Farmer LM, Rinaldi MA, Young PG, Danan CH, Burkhart SE. 2014. Mutation of the Arabidopsis LON2 peroxisomal protease enhances pexophagy. Autophagy 10, 518–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoszewska M, Williams C, Kikhney A, Opaliński Ł, van Roermund CW, de Boer R, Veenhuis M, van der Klei IJ. 2012. Peroxisomal proteostasis involves a Lon family protein that functions as protease and chaperone. Journal of Biological Chemistry 287, 27380–27395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, Vogel H, Schranz ME. 2009. Paleopolyploidy in the Brassicales: analyses of the Cleome transcriptome elucidate the history of genome duplications in Arabidopsis and other Brassicales. Genome Biology and Evolution 1, 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH. 2003. A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Research 13, 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijó JA, Becker JD. 2008. Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiology 148, 1168–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botos I, Melnikov EE, Cherry S, et al. 2004. The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site. Journal of Biological Chemistry 279, 8140–8148. [DOI] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. 2003. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 433–438. [DOI] [PubMed] [Google Scholar]
- Byun SA, Singh S. 2013. Protein subcellular relocalization increases the retention of eukaryotic duplicate genes. Genome Biology and Evolution 5, 2402–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carretero-Paulet L, Fares MA. 2012. Evolutionary dynamics and functional specialization of plant paralogs formed by whole and small-scale genome duplications. Molecular Biology and Evolution 29, 3541–3551. [DOI] [PubMed] [Google Scholar]
- Casneuf T, De Bodt S, Raes J, Maere S, Van de Peer Y. 2006. Nonrandom divergence of gene expression following gene and genome duplications in the flowering plant Arabidopsis thaliana. Genome Biology 7, R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daras G, Rigas S, Tsitsekian D, Zur H, Tuller T, Hatzopoulos P. 2014. Alternative transcription initiation and the AUG context configuration control dual-organellar targeting and functional competence of Arabidopsis Lon1 protease. Molecular Plant 7, 989–1005. [DOI] [PubMed] [Google Scholar]
- Farmer LM, Rinaldi MA, Young PG, Danan CH, Burkhart SE, Bartel B. 2013. Disrupting autophagy restores peroxisome function to an Arabidopsis lon2 mutant and reveals a role for the LON2 protease in peroxisomal matrix protein degradation. The Plant Cell 25, 4085–4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganko EW, Meyers BC, Vision TJ. 2007. Divergence in expression between duplicated genes in Arabidopsis. Molecular Biology and Evolution 24, 2298–2309. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Research 40, D1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberer G, Hindemitt T, Meyers BC, Mayer KFX. 2004. Transcriptional similarities, dissimilarities, and conservation of cis-elements in duplicated genes of Arabidopsis. Plant Physiology 136, 3009–3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L. 2004. Evolutionary history and higher order classification of AAA+ ATPases. Journal of Structural Biology 146, 11–31. [DOI] [PubMed] [Google Scholar]
- Janska H, Kwasniak M, Szczepanowska J. 2013. Protein quality control in organelles—AAA/FtsH story. Biochimica et Biophysica Acta 1833, 381–387. [DOI] [PubMed] [Google Scholar]
- Jiang WK, Liu YL, Xia EH, Gao LZ. 2013. Prevalent role of gene features in determining evolutionary fates of whole-genome duplication duplicated genes in flowering plants. Plant Physiology 161, 1844–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Computer Applications in the Biosciences 8, 275–282. [DOI] [PubMed] [Google Scholar]
- Klepikova AV, Kasianov AS, Gerasimov ES, Logacheva MD, Penin AA. 2016. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. The Plant Journal 88, 1058–1070. [DOI] [PubMed] [Google Scholar]
- Koch MA. 2019. The plant model system Arabidopsis set into an evolutionary, systematic and spatio-temporal context. Journal of Experimental Botany 70 (in press). [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, et al. 2012. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Research 40, D1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Kim J, Robertson JS, Paterson AH. 2017. Plant Genome Duplication Database. Methods in Molecular Biology 1533, 267–277. [DOI] [PubMed] [Google Scholar]
- Li L, Nelson C, Fenske R, Trösch J, Pružinská A, Millar AH, Huang S. 2017. Changes in specific protein degradation rates in Arabidopsis thaliana reveal multiple roles of Lon1 in mitochondrial protein homeostasis. The Plant Journal 89, 458–471. [DOI] [PubMed] [Google Scholar]
- Lingard MJ, Bartel B. 2009. Arabidopsis LON2 is necessary for peroxisomal function and sustained matrix protein import. Plant Physiology 151, 1354–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Baute GJ, Adams KL. 2011. Organ and cell type-specific complementary expression patterns and regulatory neofunctionalization between duplicated genes in Arabidopsis thaliana. Genome Biology and Evolution 3, 1419–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Pan AQ, Adams KL. 2014. Protein subcellular relocalization of duplicated genes in Arabidopsis. Genome Biology and Evolution 6, 2501–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Force A. 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154, 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matasci N, Hung LH, Yan Z, et al. 2014. Data access for the 1,000 Plants (1KP) project. Gigascience 3, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller IM, Jensen PE, Hansson A. 2007. Oxidative modifications to cellular components in plants. Annual Review of Plant Biology 58, 459–481. [DOI] [PubMed] [Google Scholar]
- Møller IM, Rogowska-Wrzesinska A, Rao RS. 2011. Protein carbonylation and metal-catalyzed protein oxidation in a cellular perspective. Journal of Proteomics 74, 2228–2242. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. 1999. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Research 9, 27–43. [PubMed] [Google Scholar]
- Ó’Maoiléidigh DS, Graciet E, Wellmer F. 2014. Gene networks controlling Arabidopsis thaliana flower development. New Phytologist 201, 16–30. [DOI] [PubMed] [Google Scholar]
- Ostersetzer O, Kato Y, Adam Z, Sakamoto W. 2007. Multiple intracellular locations of Lon protease in Arabidopsis: evidence for the localization of AtLon4 to chloroplasts. Plant & Cell Physiology 48, 881–885. [DOI] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu SH. 2016. Evolution of gene duplication in plants. Plant Physiology 171, 2294–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SC, Jia B, Yang JK, Van DL, Shao YG, Han SW, Jeon YJ, Chung CH, Cheong GW. 2006. Oligomeric structure of the ATP-dependent protease La (Lon) of Escherichia coli. Molecules and Cells 21, 129–134. [PubMed] [Google Scholar]
- Poirier Y, Antonenkov VD, Glumoff T, Hiltunen JK. 2006. Peroxisomal beta-oxidation—a metabolic pathway with multiple functions. Biochimica et Biophysica Acta 1763, 1413–1426. [DOI] [PubMed] [Google Scholar]
- Rigas S, Daras G, Laxa M, Marathias N, Fasseas C, Sweetlove LJ, Hatzopoulos P. 2009a Role of Lon1 protease in post-germinative growth and maintenance of mitochondrial function in Arabidopsis thaliana. New Phytologist 181, 588–600. [DOI] [PubMed] [Google Scholar]
- Rigas S, Daras G, Sweetlove LJ, Hatzopoulos P. 2009b Mitochondria biogenesis via Lon1 selective proteolysis: who dares to live for ever? Plant Signaling and Behavior 4, 221–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigas S, Daras G, Tsitsekian D, Hatzopoulos P. 2012. The multifaceted role of Lon proteolysis in seedling establishment and maintenance of plant organelle function: living from protein destruction. Physiologia Plantarum 145, 215–223. [DOI] [PubMed] [Google Scholar]
- Rigas S, Daras G, Tsitsekian D, Alatzas A, Hatzopoulos P. 2014. Evolution and significance of the Lon gene family in Arabidopsis organelle biogenesis and energy metabolism. Frontiers in Plant Science 5, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W. 2006. Protein degradation machineries in plastids. Annual Review of Plant Biology 57, 599–621. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Mitchell-Olds T. 2006. Independent ancient polyploidy events in the sister families Brassicaceae and Cleomaceae. The Plant Cell 18, 1152–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smakowska E, Czarna M, Janska H. 2014. Mitochondrial ATP-dependent proteases in protection against accumulation of carbonylated proteins. Mitochondrion 19, 245–251. [DOI] [PubMed] [Google Scholar]
- Solheim C, Li L, Hatzopoulos P, Millar AH. 2012. Loss of Lon1 in Arabidopsis changes the mitochondrial proteome leading to altered metabolite profiles and growth retardation without an accumulation of oxidative damage. Plant Physiology 160, 1187–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, Zheng C, Sankoff D, Depamphilis CW, Wall PK, Soltis PS. 2009. Polyploidy and angiosperm diversification. American Journal of Botany 96, 336–348. [DOI] [PubMed] [Google Scholar]
- Stahlberg H, Kutejová E, Suda K, Wolpensinger B, Lustig A, Schatz G, Engel A, Suzuki CK. 1999. Mitochondrial Lon of Saccharomyces cerevisiae is a ring-shaped protease with seven flexible subunits. Proceedings of the National Academy of Sciences, USA 96, 6787–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk KJ. 2015. Protein maturation and proteolysis in plant plastids, mitochondria, and peroxisomes. Annual Review of Plant Biology 66, 75–111. [DOI] [PubMed] [Google Scholar]
- Venkatesh S, Lee J, Singh K, Lee I, Suzuki CK. 2012. Multitasking in the mitochondrion by the ATP-dependent Lon protease. Biochimica et Biophysica Acta 1823, 56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieux EF, Wohlever ML, Chen JZ, Sauer RT, Baker TA. 2013. Distinct quaternary structures of the AAA+ Lon protease control substrate degradation. Proceedings of the National Academy of Sciences, USA 110, E2002–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarino GH, Hu Q, Manrique S, et al. 2016. Transcriptomic signature of the SHATTERPROOF2 expression domain reveals the meristematic nature of Arabidopsis gynoecial medial domain. Plant Physiology 171, 42–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanders RJ, Ferdinandusse S, Brites P, Kemp S. 2010. Peroxisomes, lipid metabolism and lipotoxicity. Biochimica et Biophysica Acta 1801, 272–280. [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An ‘Electronic Fluorescent Pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Salinas M, Garcia-Molina A, Höhmann S, Berndtgen R, Huijser P. 2013. SPL8 and miR156-targeted SPL genes redundantly regulate Arabidopsis gynoecium differential patterning. The Plant Journal 75, 566–577. [DOI] [PubMed] [Google Scholar]
- Young PG, Bartel B. 2016. Pexophagy and peroxisomal protein turnover in plants. Biochimica et Biophysica Acta 1863, 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. 2003. Evolution by gene duplication: an update. Trends in Ecology and Evolution 18, 292–298. [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. 2004. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology 136, 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.