SUMMARY
Natural selection is generally expected to favor one form of a given trait within a population. The presence of multiple functional variants of traits involved in activities such as feeding, reproduction, or the defense against predators is relatively uncommon within animal species. The genetic architecture and evolutionary mechanisms underlying the origin and maintenance of such polymorphisms are of special interest. Among rattlesnakes, several instances of the production of biochemically distinct neurotoxic or hemorrhagic venom types within the same species are known. Here, we investigated the genetic basis of this phenomenon in three species and found that neurotoxic and hemorrhagic individuals of the same species possess markedly different haplotypes at two toxin gene complexes. For example, neurotoxic and hemorrhagic Crotalus scutulatus individuals differ by 5 genes at the phospholipase A2 (PLA2) toxin gene complex and by 11 genes at the metalloproteinase (MP) gene complex. A similar set of extremely divergent haplotypes also underlies alternate venom types within C. helleri and C. horridus. We further show that the MP and PLA2 haplotypes of neurotoxic C. helleri appear to have been acquired through hybridization with C. scutulatus—a rare example of the horizontal transfer of a potentially highly adaptive suite of genes. These large structural variants appear analogous to immunity gene complexes in host-pathogen arms races and may reflect the impact of balancing selection at the PLA2 and MP complexes for predation on different prey.
In Brief
Individuals within several rattlesnake species produce alternative types of venoms composed of distinct neurotoxins or hemorrhagic metalloproteinases. Dowell et al. show that haplotypes differing by 5–11 genes at two gene complexes encode these alternative venom types, which may be maintained by balancing selection for predation on different prey.
INTRODUCTION
A central quest in biology is to understand the genetic mechanisms and ecological factors that generate species diversity. The adaptation of species most often entails the specialization of traits—such that a single functional form occurs within a given species. Even very closely related species—such as the Galapagos finches, East African cichlid fish, or Caribbean Anolis lizards—have evolved distinct species-specific characters for acquiring food or mates. However, in some cases, two or more functionally differentiated variants of particular traits may co-occur at significant frequencies within species [1]. Well-studied examples of such polymorphisms include wing coloration in Heliconius butterflies [2, 3], body coloration in side-blotched lizards [4], and horn size in Soay sheep [5]. The co-occurrence of multiple forms of traits raises numerous questions, including how such traits arise, the genetic architecture underlying trait divergence, and the ecological mechanisms that favor their persistence.
Pit vipers (Crotalinae) underwent an expansive radiation into numerous genera after arriving in the Americas from Asia ~22 mya [6]. Viperid venoms are composed of proteins from a limited set of gene families but are distinguished by which family members are expressed and their levels of gene expression. One group, the rattlesnakes (Crotalus and Sistrurus), consists of 36 extant species that radiated over the last 12–14 million years [6, 7]. In general, each rattlesnake species’ venom is one of two main types with distinct protein toxins and modes of subduing prey: a hemorrhagic type as in the North American eastern and western diamondback (C. atrox and C. adamanteus) rattlesnakes or a neurotoxic type as in the South American rattlesnake (C. durissus). Curiously, however, these venom types are not always mutually exclusive, as certain species have been shown to include both neurotoxic and hemorrhagic individuals.
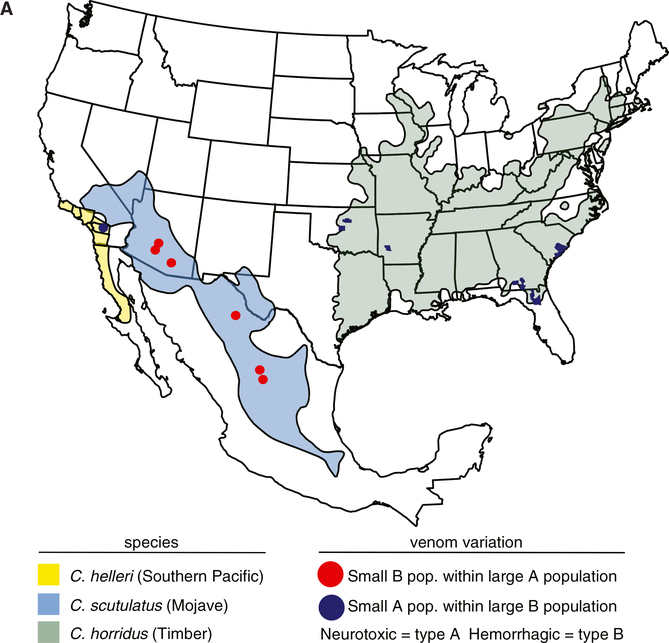
For example, the widely dispersed, predominantly neurotoxic Mojave rattlesnake (C. scutulatus) includes geographically restricted, non-neurotoxic, hemorrhagic populations (Figure 1A [red dots on light blue]) [10–12], whereas the Southern Pacific rattlesnake (C. helleri [14]) is predominantly hemorrhagic but includes a geographically restricted neurotoxic population (Figure 1A [blue dot on yellow]) [8, 9] and the timber rattlesnake (C. horridus) includes widely dispersed individuals of both types (Figure 1A [blue dots on green]) [13]. In general, neurotoxic individuals express high levels of a heterodimeric neurotoxin (composed of acidic and basic phospholipase A2 [PLA2] polypeptides) and have been designated type A venoms [11], while hemorrhagic individuals produce undetectable levels of neurotoxin [11, 15] and express high levels of venom metalloproteinases (MPs) [15, 16], an abundant component of hemorrhagic venoms [17, 18]. The latter has been designated type B venom. Several other groups, including C. viridis [19], C. durissus [20], and S. catenatus [21, 22], exhibit similar polymorphisms in which neurotoxicity varies among individuals or among subspecies or during development [23].
Figure 1. Geographic Distribution of Three Rattlesnake Species with Polymorphic Venom.
The modern North American ranges of C. helleri (yellow), C. scutulatus (light blue), and C. horridus (green) are shown as polygons. C. helleri individuals expressing neurotoxic venom have been reported in California near the San Jacinto mountains (navy blue circle) [8, 9]. C. scutulatus individuals expressing hemorrhagic venom have been identified in three Arizona locales and in Mexico (red circles) [10–12]. C. horridus individuals expressing neurotoxic venom have been identified in Oklahoma, Arkansas, Georgia, Florida, and South Carolina counties (navy blue polygons) [13]. See also Table S1.
The genetic, evolutionary, and ecological mechanisms underlying the alternative venom types within species are not understood. One key issue is the genetic basis of the A/B polymorphisms and whether differences in venom type are due to differences in toxin gene structure, number, or regulation. The failure to amplify one or more neurotoxin gene subunits in type B C. scutulatus [24, 25]and type B C. horridus [26] have raised the possibility that gene content varies between venom types. On the other hand, structural differences in MP genes have also been proposed to underlie alternative venom types [27]. Resolving these possibilities requires a genomic picture of toxin gene family content and arrangement. However, the complete genomic arrangement of toxin genes is only known for type A C. scutulatus PLA2 genes [28], and nothing is known about the genomic organization of rattlesnake MP genes. Understanding the genetic basis of the polymorphisms is also key to unraveling their evolutionary origin and whether and how they have evolved independently in different species or could have a common origin.
Here, we explored the genetic and evolutionary bases of this phenomenon by isolating and examining the PLA2 and MP toxin gene complexes of type A and B C. scutulatus, C. helleri, and C. horridus animals. We discovered that neurotoxic and hemorrhagic individuals of each species possess markedly different haplotypes at each of two gene complexes that encode the major toxins for each venom type. This divergence is not due to the typical alternative alleles at shared loci that underlie most genetic variation in animals. Rather, it entails the presence/absence of up to as many as 16 genes among the 2 complexes. We suggest that these unusual polymorphisms may be maintained by selection for different available prey across the species’ ranges.
RESULTS
Structurally Distinct PLA2 Gene Haplotypes Underlie Variation in Neurotoxin Expression in C. scutulatus
The majority of C. scutulatus individuals express high levels of the heterodimeric Mojave neurotoxin [11], while some individuals produce undetectable levels of neurotoxin [11, 12, 15] and express high levels of venom MPs [12, 15, 16] Some individuals also express both types of toxins [12, 15].
In order to explore the genetic basis of the different venom types, we compared the PLA2 gene complexes from C. scutulatus animals expressing either type A (neurotoxic) or type B (non-neurotoxic/hemorrhagic) venom. In previous work, we established that the most recent common ancestor of rattlesnakes possessed at least seven distinct group G PLA2 genes (a tandem array of duplicated genes within a complex of other type II Pla2 genes that has specifically expanded in Crotalids). This ancestral set included the Pla2-gA2 and Pla2-gB2 genes, homologs of which encode the protein subunits of the Mojave toxin (Mtx), MtxA and MtxB, respectively (Figures 2A and S1) [28]. These two genes constitute the majority of venom gland Pla2 mRNA expression originating from the PLA2 complex in type A (neurotoxic) individuals (Figures S2A-S2C). Thus, one simple model to explain alternative venom phenotypes within this species could be the selective expression of distinct orthologs from a large PLA2 complex.
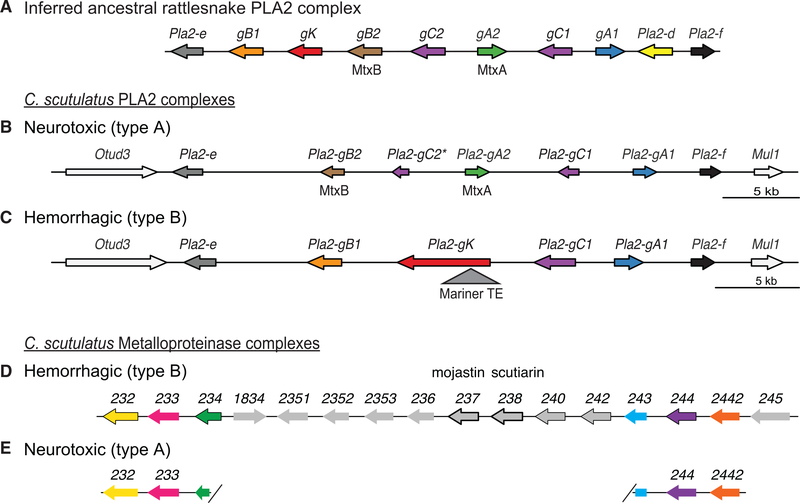
Figure 2. Extremely Divergent PLA2 and MP Gene Haplotypes Encode Alternative Venom Types in C. scutulatus.
(A) The last common ancestor of rattlesnakes Is Inferred to have possessed at least seven distinct group G PLA2 genes, Including Pla2-gB2 (brown arrow) and Pla2-gA2 (green arrow), which encode the subunits of the heterodimer neurotoxin found in several species.
(B and C) Type A and type B C. scutulatus differ by five genes at their PLA2 gene complexes. (B) The type A (neurotoxic) PLA2 complex contains the following distinct genes: Pla2-gB2 (brown arrow) and Pla2-gA2 (green arrow), which encode the subunits of the Mojave toxin, MtxB and MtxA, respectively, and Pla2-gC2 (purple arrow), which contains a coding substitution that likely renders this gene non-functional (asterisk). (C) The type B (non-neurotoxic) PLA2 complex contains the distinct Pla2-gB1 (orange arrow) and Pla2-gK (red arrow) genes. There is a TE from the mariner family inserted into the first intron of Pla2-gK (gray triangle). Two genes, Pla2-gC1 (purple arrow) and Pla2-gA1 (blue arrow), are present in complexes from both type A and type B individuals. Pla2-e and Pla2-f are Pla2 group II genes that are not expressed in venom.
(D and E) Type A and type B C. scutulatus differ by 11 genes at their MP gene complexes. (D) In the type B (hemorrhagic) C. scutulatus genome, 16 MP genes, depicted schematically as arrows oriented in the coding direction, are present in a tandem array. MP genes with high expression in venom glands (>10000 transcripts per million [TPM]) are outlined in black. The computationally generated numeric gene identifiers above each gene arrow are used to distinguish genes within the cluster; previously established names for two individual venom proteins are listed above their numerical identifiers. (E) The type A (neurotoxic) C. scutulatus genome has a large deletion at the MP complex. The only MP orthologs present are genes 232, 233, 244, and 2442, and the C-terminal exons of gene 234 (pink half arrow) and the N-terminal exons three, four, and five of gene 243 (gray rectangle) are fused (exons one and two are absent). The black forward slashes depict the borders of the large genomic region spanning the nine full-length genes that is deleted.
See also Figures S1, S2, S3, S4, S5, and S6 and Tables S2 and S3.
However, we found that the PLA2 complex from a type B specimen contains no trace of the Pla2-gA2, Pla2-gB2, or Pla2-gC2 genes (which is consistent with and explains prior studies that were unable to amplify Pla2-gA2 and Pla2-gB2 in type B animals [18, 19]) (Figures 2B and S1). The type B complex does contain and express Pla2-gB1 and Pla2-gK homologs, which are absent from the type A PLA2 complex, as well as Pla2-gC1, which is also found in type A animals but is expressed only at low background levels (Figures 2B and S1). Thus, the two venom phenotypes are encoded by haplotypes that differ by the presence/absence of a total of five genes. In fact, the arrangement and composition of the PLA2 complex in the type B C. scutulatus is more similar to that from type B hemorrhagic venoms of other species, such as C. atrox and C. adamanteus, than it is to the type A complex within C. scutulatus ([28]). We note that because each haplotype contains members of distinct ortholog groups that are not present in the other and that predate the origin of the species [28], neither haplotype could be simply derived directly from the other by gene duplication or deletion.
The discovery of a Pla2-gK gene in type B C. scutulatus (confirmed in three of three specimens; Figure S5G) was surprising because the gene, while present in certain Asian crotalids [29], has previously been reported in only two species of rattlesnakes: C. atrox [28] and C. molossus [30], which are not the closest relatives of C. scutulatus [31]. The Pla2-gK locus encodes a protein with myo- and hemotoxicity [32] that is abundant in C. atrox venom, but no PLA2gK polypeptides have been detected in mass spectrometry of C. scutulatus venom proteomes [15]. The C. scutulatus Pla2-gK gene differs from the C. atrox ortholog by the insertion of a mariner-class transposable element (TE) into the first intron (Figure 2B). This insertion could account for the relatively low expression of C. scutulatus Pla2-gK transcript as compared to Pla2-gB1 expression (Figures S2A-S2C). Why a gene that does not contribute to venom would be present and retained in type B C. scutulatus is not clear but, as we discuss later, may be informative as to the evolutionary origin of the type B PLA2 haplotypes. The absence of the neurotoxin genes Pla2-gB2 and Pla2-gA2 and lack of expression of PLA2gK raised the question: what toxin genes are present and being expressed in type B animals?
The C. scutulatus MP Gene Complex Also Varies in Gene Content
MP are abundant components of hemorrhagic venoms such as those of C. atrox. While metalloproteinase peptides are typically absent from type A C. scutulatus venom, they are abundant in type B C. scutulatus venom [15, 16]. The number and genomic organization of Crotalid MP genes has not been elucidated previously [27]. To investigate the potential genetic basis for MP diversity, we annotated the MP complex of the same type B C. scutulatus individual described above.
We identified a tandem array of 16 MP genes located between 2 flanking non-MP genes, the NEF-M and STC1 loci that are conserved among vertebrates (Figure 2C). Within this large array of highly similar MP genes, the majority of venom gland MP mRNA expression is the product of only two genes (designated 237 and 238 in Figures 2C and S2E-S2F) that encode the previously identified mojastin (237) and scutiarin (238) proteins [16]. Phylogenetic analysis indicates that these proteins are orthologs of the C. atrox MPs crotatroxin [33] and VAP2B [34], respectively (Figure S3). Functional studies of the C. atrox proteins have revealed that they disrupt hemostasis and inhibit platelet aggregation [35, 36], which would indicate that the type B C. scutulatus venom exerts similar hemotoxic effects on prey.
By contrast, examination of the MP complex in a type A C. scutulatus genome revealed only four intact MP genes: 232, 233, 244, and 2442 (Figure 2D), with a fifth apparent fusion gene composed of nine exons from MP 234 (Figure 2D [exons 7–17 shown as partial green arrows) and four exons from MP 243 (Figure 2D [exons 3–6 shown as blue rectangles and exons 1 and 2 are deleted). Thus, there appeared to be an 11-gene deletion within the MP complex of this individual that removes the predominant MP genes expressed in type B animals (Figure 2D [forward slashes]).
To confirm that 11 genes were completely or partially deleted from the type A genome, and to exclude the possibility of large-scale genome rearrangements shuffling the MP complex to a different genomic region, we screened a type A C. scutulatus whole-genome bacterial artificial chromosome (BAC) library for additional clones containing MP gene sequences. We did not detect any of the intervening genes, but we did isolate an additional nine distinct clones that also spanned the ~330-kb genomic interval from MP genes 234–243 (Figure S4). We further confirmed the presence of the large deletion in two additional type A specimens by PCR analysis using primers for sequences flanking or located within the deleted segment (Figure S5C). These data reveal that, just as within the PLA2 complex, markedly different haplotypes exist at the MP complex, and these alternative haplotypes account for the alternative venom types in C. scutulatus.
The existence of haplotypes differing in the presence/absence of numerous genes within C. scutulatus raised the question of whether differences in gene content also underlie alternative venom types in other rattlesnakes.
Structural Variation in PLA2 Gene Complex Haplotypes Underlies Variation in Neurotoxicity in C. horridus and C. helleri
Most C. horridus animals produce a hemorrhagic type B venom, but neurotoxic (type A) individuals have also been reported in many locales within their broad range (Figure 1 [counties colored dark navy blue]) [13]. Examination of the PLA2 complexes of alternative types of timber rattlesnakes also revealed haplotypes that differed greatly in gene content and that were distinct from those found in C. scutulatus. The type A C. horridus PLA2 complex contained six snake-specific PLA2 genes, including the genes encoding the neurotoxin subunits (Pla2-gA2 and Pla2-gB2), as well as an additional Pla2-gB2 gene (Figure 3A). We also identified an atypical gene (Pla2-gA3) that encodes a putative acidic Pla2 protein that is distinct from the Pla2-gA1 gene found in the same relative position in other rattlesnake PLA2 complexes (Figures 3 and S1) and is expressed in venom [26, 37]. We note that the C. horridus type A complex also contained the Pla2-d locus that is conserved among tetrapods but deleted from several other rattlesnake species (Figure 3A).
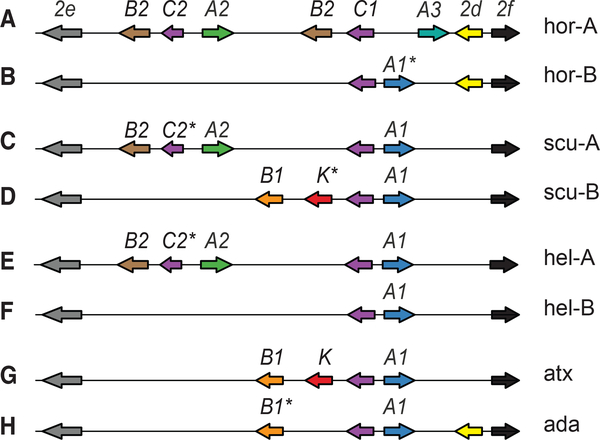
Figure 3. Divergent PLA2 Gene Complex Haplotypes Underlie Variation in C. horridus and C. heilen Venom.
(A–F) Type A and type B individuals of each of the three species examined have divergent haplotypes at the PLA2 complex (compare A to B, C to D, and E to F).
(A) The type A C. horridus type-A complex contains the neurotoxin-encoding genes (Pla2-gB2 and Pla2-gA2) but also has an additional Pla2-gB2 and an atypical Pla2-gA3 gene, both of which are absent in the other species type A complexes (compare A, C, and E).In contrast, type A C. scutulatus and type A C. helleri complexes have the identical set of genes and share a putative inactivating mutation in Pla2-gC2 (C and E; asterisk), while the type B C. scutulatus and type B C. helleri haplotypes differ by two genes (Pla2-gB1 [orange arrow] and Pla2-gK [red arrow]; compare D and F). Pla2-e, Pla2-f, and Pla2-d are Pla2 group II genes that are not expressed in venom. Species names are abbreviated in the following manner: C. horridus, hor; C. scutulatus, scu; C. helleri, hel; C. atrox, atx; C. adamanteus, ada.
The C. horridus type B PLA2 complex lacks five genes present in the type A complex (Pla2-gA2, Pla2-gC2, both Pla2-gB2 copies, and Pla2-gA3), which is consistent with a previous study that was unable to amplify Pla2-gA2 or Pla2-gB2 in type B animals [26], and shares only the Pla2-gC1 locus with the type A complex (Figure 3B). Like most other complexes we have analyzed (except for the C. horridus type A complex), the type B complex contains a Pla2-gA1 gene; however, this locus is a likely pseudogene due to a stop codon substitution in the coding sequence, which has been described previously [37]. Altogether, the C. horridus and C. scutulatus A haplotypes differ from one another by the presence/absence of four genes (Figures 3A and 3C), and the B haplotypes differ by the presence/absence of three genes (Figures 3B and 3D).
In contrast to the distinct neurotoxic PLA2 complex haplotypes of the geographically distant C. horridus and C. scutulatus species, we found that the C. helleri type A (neurotoxic) PLA2 complex was strikingly similar to the type A (neurotoxic) C. scutulatus PLA2 complex in that it contained the same five snake-specific PLA2 genes in a syntenic arrangement (Figures 3C, 3E, and S1). Furthermore, the Pla2-gC2 gene of both PLA2 complexes contained an identical substitution that introduced a stop codon. The C. horridus Pla2-gC2 gene does not contain this mutation.
However, the type B C. helleri PLA2 complex (Figure 3F) differs from both the type A C. helleri complex (Figure 3E) and the type B C. scutulatus complex (Figure 3D). The C. helleri type B complex lacks three genes present in the type A C. helleri complex (the neurotoxin subunit genes Pla2-gA2 and Pla2-gB2, as well as Pla2-gC2; Figures 3F and 3E) and also lacks the Pla2-gB1 and Pla2-gK genes present in type B C. scutulatus animals (Figure 3F and 3D). Indeed, it contains only two snake-specific PLA2 genes (Pla2-gC1 and Pla2-gA1) and is the most reduced rattlesnake PLA2 complex that we have discovered. (Figures 3A-3H; [28])
Thus, the presence or absence of neurotoxicity within each species is due to extremely divergent A and B PLA2 complex haplotypes that are largely distinct in each species—with the exception of the very similar A haplotypes of C. scutulatus and C. helleri. We next examined whether divergent and distinct MP haplotypes also exist in these species.
Both Shared and Distinct Haplotypes Occur at the MP Complex
We found large MP complex haplotypes in type B C. horridus (13 genes; Figure 4A) and type B C. helleri (21 genes; Figure 4B) that had similarities to, but were distinct from, the type B haplotype in C. scutulatus (16 genes; Figure 2C). The differences in gene number between the species’ type B complexes appear to be the result of lineage-specific gains or losses of individual MP genes.
Figure 4. Both Shared and Distinct Haplotypes Occur at the MP Complex.
(A–D) The arrangement of the MP genes (arrows) in four genomes. Solid lines indicate a gap-free nucleotide sequence. Dotted lines indicate the gaps between the non-overlapping contigs. A forward slash shows where contigs overlap. Solid boxes denote genomic regions absent from particular complexes relative to the C. scutulatus type BMP complex. Candidate duplicated genes (arrows) are shown below the putative ortholog. The brackets indicate the sequence region that is not present in the C. helleri type A clone.
(A) The C. horridus hemorrhagic (type B)MP complex is reduced relative to the 16-MP gene C. scutulatus type B complex. The approximate overlapping ranges for the annotated BAC clones are shown below the gene complex schematic. Note that the regions (solid boxes) with absent genes (1834, 242, and 243) are spanned completely by single clones. The duplicated 232 is on a clone (31E6) that does not overlap with clone 27I3. We identified MP 7* in the C. horridus type B complex, but it contains a small deletion in exon 10, resulting in an early stop codon (*) and represents a likely pseudogene.
(B) The C. helleri type BMP complex is expanded compared to the C. scutulatus type BMP complex. While some genes are absent in C. helleri type B (see also Table S4), at least three C. helleri genes (V, 80, and 13) lack C. scutulatus type B homologs.
(C) The C. horridus type A MP complex contains a similar set of genes identified in the C. horridus type B complex but lacks the duplicated 232 (compare A with C). The complex is reduced relative to the C. scutulatus type B complex (solid boxes highlight the absence of 1834, 242, and 243) but is expanded relative to the C. scutulatus type A complex (it contains a complete 234 gene and MP genes 2351 to 240). Three gap-free contigs (identifiers and lengths in (kb) from a draft genome assembly of C. horridus were used to identify the MP genes shown here.
(D) The C. helleri type A MP complex bears the same deletion as the C. scutulatus type A complex. The 3′ exons of 234 (green arrow) are adjacent to the 5′ exons of 243 (blue rectangle) in the genomic sequence. The brackets and intervening white space highlight the deleted genomic segment.
(E) The C. helleri and C. scutulatus type A complexes share an MP haplotype as indicated by the identical nucleotide sequences flanking their respective ~330-kb deletions. Assembled BAC clones from C. helleri type A (46A6) and C. scutulatus type A (133D14) individuals were aligned to the C. scutulatus type B MP complex. The top row is the scale bar for the C. scutulatus type BMP sequence. The second row (scu-B genes) shows the annotated genes in the C. scutulatus type BMP complex. The highly expressed venom gland MP genes are outlined in black. Gene identifiers are shown (232,233,234,243,244,2442) for genes (colored arrows) completely or partially present in both type Band type A individuals (scu-Aand hel-Agenes). Gray rectangles represent aligned nucleotides with gaps depicted as thin connecting lines. Azoomed view of the exons (MP exons row) for the genes(234 and 243) that flank the deletion break point is called out with slanted dashed lines. An additional zoomed view (slanted dashed lines) shows the nucleotide sequences in C. helleri (hel-A) and C. scutulatus (scu-A) that flank the deletion boundaries (underlined). The C. scutulatus (scu-B) sequence extends across the gap (ellipses). The filled black circles show the SNPs that are shared by C. helleri and C. scutulatus type A individuals.
Among type A animals, however, we found instances of MP haplotypes that were shared within and between species. In the case of C. horridus, the type A MP complex contained the same genes as the 13-gene type B complex with the exception of the duplicated 232 and 2351 and the deleted 243 genes (compare Figures 4A and 4C). But this complex is distinct from the C. scutulatus type A MP complex in that it contains many genes that are absent from C. scutulatus type A individuals (Figure 2D). Thus, the C. horridus MP complex haplotype is similar within but not between species.
However, in the C. helleri type A MP complex, we discovered the exact same five genes found in the C. scutulatus type A MP complex: 232,233,244,2442 and the fusion of 234 and 243 (Figure 4D [green arrow and blue rectangle flanked by square brackets]). This arrangement indicated that the type A C. helleri animal carried the same deletion/gene fusion of the MP complex as type A C. scutulatus individuals. Indeed, we found the identical genome sequences at the boundaries of the deletions in the neurotoxic C. helleri and C. scutulatus animals (Figure 4E [underlined sequence]). In addition, we noted single-nucleotide polymorphisms (SNPs) in the sequence flanking the deletions that are shared by type A C. helleri and C. scutulatus individuals, but not with the type B C. scutulatus haplotype (Figure 4E [closed black circles below sequence]). These data reveal that the same MP haplotype is shared across species by type A individuals, but not type B C. scutulatus and C. helleri individuals.
C. helleri Acquired Some Haplotypes via Hybridization with C. scutulatus
The nearly identical C. helleri MP haplotype and highly similar PLA2 haplotype (including the identical nonsense mutation in Pla2-gC2) shared with type A C. scutulatus are extremely unlikely to have arisen independently within each species. Rather, they are consistent with either an origin via hybridization of C. helleri with C. scutulatus or with each being ancestral polymorphisms that predate the divergence of C. helleri and C. scutulatus.
In order to try to distinguish between these two possibilities, we utilized the “ABBA-BABA” test [38, 39] to examine the distribution of shared derived sites among C. helleri and C. scutulatus haplotypes, using C. horridus as an outgroup. In this test, where a derived site (B) has evolved via mutation from an ancestral site (A), it is expected that lineage sorting will produce an equal distribution of ABBA and BABA sites among descendant lineages (Figure 5A). An unequal distribution of sites is observed when there has been gene flow among two ingroup lineages.
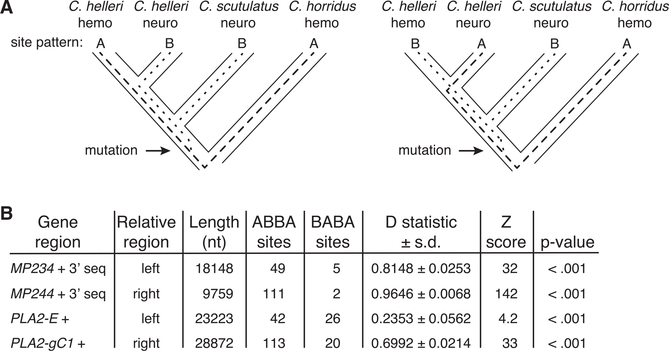
Figure 5. The Evolutionary Origin of the C. helleri Neurotoxic Venom Haplotypes.
(A) The four-taxon ABBA/BABA test of introgression was employed to test for introgression of the C. scutulatus—neurotoxic haplotypes into C. helleri with C. horridus as the outgroup. Under the null hypothesis of no introgression (vertical inheritance of an ancestral polymorphism), the expectation is an equal distribution ABBA and BABA sites, whereas a skew toward one site pattern is support for an introgression event. Gene genealogies are shown as dashed and dotted lines within the species tree.
(B) ABBA/BABA analysis using the sequences flanking the PLA2 and MP gene complexes. At all four regions, we observe a skew of ABBA sites compared to BABA sites and the D statistic differs significantly (p < 0.001) from zero, indicating introgression of both the PLA2 and MP neurotoxic haplotypes from C. scutulatus into C. helleri.
Here, we configured the analysis to test the hypothesis of gene flow between neurotoxic C. scutulatus and C. helleri (we use the designations neurotoxic and hemorrhagic here to avoid confusing venom type with the ABBA-BABA nomenclature). Because haplotypes differ in the presence/absence of toxin genes, we examined the distribution of ABBA and BABA sites in flanking sequences of the PLA2 and MP complex that are shared among all genotypes. We found a significant excess of ABBA sites in both flanking regions of the MP complex (Figure 5B [left: Dhelleri hemo, helleri neuro, scutulatus neuro, horridus 0.8148 ± 0.0253; p < < 0.001; right Dhelleri hemo, helleri neuro, scutulatus neuro, horridus 0.9646 ± 0.0068, p < < 0.001) and the PLA2 complex (Figure 5B [left: Dhelleri hemo, helleri neuro, scutulatus neuro, horridus 0.2353 ± 0.0562; p < < 0.001; right Dhelleri hemo, helleri neuro, scutulatus neuro, horridus 0.6992 ± 0.0214, p < < 0.001). These results are evidence for the origin of neurotoxic type A C. helleri MP and PLA2 haplotypes occurring via hybridization with C. scutulatus.
DISCUSSION
We have shown that in all three species examined, type A (neurotoxic) and type B (hemorrhagic) individuals possess markedly different haplotypes at the PLA2 complex, and two species also exhibit different haplotypes at their MP gene complexes. The nature and extent of genetic divergence observed within species is very unusual for vertebrates in that variation is not due to alternative alleles at shared loci but entails the presence/absence of many entire genes. It is of particular interest, then, to understand the evolutionary origin of such distinct haplotypes and why they persist in nature.
The Evolutionary Origin and Maintenance of Divergent Toxin Gene Complex Haplotypes
In principle, we envision three possible mechanisms that could account for the origin of structurally distinct haplotypes within each species: (1) the haplotypes arose independently within each lineage, (2) alternate haplotypes predate the divergence of certain species and are ancestral polymorphisms, or (3) one or more haplotypes were acquired by a hybridization event, perhaps between neurotoxic and hemorrhagic members of two different species (Figure 6).
Figure 6. Models for the Evolutionary Origins of Alternative Venom Haplotypes within Different Species.
(A–C) A schematic species tree is shown for four hypothetical species (tip labels: 1–4) in which species 1 and 4 are dimorphic for venom type and share identical venom haplotypes (black and white squares) and species 2 and 3 are monomorphic for venom type and associated haplotypes. The ancestral haplotype (gray rectangle) on the branch leading to the most recent common ancestor for these four species is larger compared to extant haplotypes (white and black squares). Three possible models for the origin of dimorphic species are shown.
(A) The alternative haplotypes could have originated independently within each lineage (forward or reverse slashes), and the expression of dimorphic venom in species 1 and 4 represents a case of convergent evolution.
(B) The alternative haplotypes could have arisen as a polymorphism in a common ancestor (reverse slashes), and the ancestral variation was maintained through vertical inheritance during some speciation events (e.g., leading to species 1 and 4) but was lost on the lineages yielding species 2 and 3.
(C) Shared haplotypes could also have been acquired through a hybridization event (dashed arrow and white box) followed by selection to retain the newly acquired haplotype. In this schematic, one haplotype (black box) was vertically inherited from a common ancestor, whereas the second haplotype (white box) was horizontally transmitted via a hybridization event.
We have found evidence that the origin of neurotoxic type A C. helleri individuals in the otherwise largely hemorrhagic type B species occurred via hybridization with C. scutulatus. This appears to be a very plausible explanation in light of the present range of neurotoxic C. helleri being very close to the present extent of the range of C. scutulatus [8, 14], and viable hybrids having been obtained between C. scutulatus and C. helleri in captivity [40]. We note that these two species are estimated to have shared a common ancestor 2–4 mya [31]. Given similar, relatively recent divergence times among many rattlesnake species, our finding highlights the potential for the exchange of toxin genes and other traits among this group by hybridization.
However, we have no reason to suspect that the explanation for polymorphism in all species will be the same. Rather, each polymorphic species, and even perhaps each gene complex, will have to be sorted out case by case, and the confirmation of certain mechanisms (e.g., ancestral polymorphism, hybridization) will require population genomic data from many more specimens than are presently available. Nonetheless, with current data, we can point to the most likely alternatives.
The origin of the hemorrhagic type B C. scutulatus individuals in an otherwise largely neurotoxic type A species requires consideration of the evolutionary history of neurotoxins in rattlesnakes. We have previously inferred that the neurotoxin subunit genes were present in the most recent common ancestor of the clade containing the species examined here—indeed, of all rattlesnakes [28]. The absence of neurotoxins in type B animals could therefore arise in principle by the deletion of neurotoxin subunit genes from a type A haplotype. However, the C. scutulatus type B PLA2 haplotype cannot be derived directly from the type A PLA2 haplotype because it contains two genes (Pla2-gB1 and Pla2-gK) that are not present in the A haplotype. Indeed, we are struck by the specific discovery of the Pla2-gK gene because the gene has been found in only two other rattlesnake species (C. atrox and C. molossus) and is absent from several close relatives [28]. We also note the uniquely shared syntenic arrangement of PLA2 genes in type B C. scutulatus and C. atrox (Figure 3). Two plausible explanations that could account for these observations are (1) the Band APLA2 haplotypes were both present in a polymorphic common ancestor of C. atrox and C. scutulatus and sorted differently between the lineages, or (2) the type B haplotype may have been introduced into the C. scutulatus lineage from C. atrox or some other hemorrhagic species at some time in the past. We note that viable hybrids between C. scutulatus with C. atrox have been obtained in captivity [41] and that hybridization between C. scutulatus and C. viridis in the wild has been well documented [25, 42]. However, incomplete lineage sorting and past hybridization events are notoriously difficult to disentangle [43–46] and will require more extensive sampling of both species to assess.
The third instance of alternate venom types that we examined in C. horridus revealed type A and B haplotypes that appear genetically distinct from those of C. scutulatus or C. helleri and also do not bear any striking resemblances to haplotypes in other species. For example, the C. horridus type A PLA2 haplotype contains three genes not found in any other species A haplotypes, and the type B PLA2 haplotype differs from the A haplotype by the absence of five genes and the presence of one gene. In addition, the type A C. horridus Pla2-gC2 gene lacks the inactivating mutation shared by the C. scutulatus and C. helleri.
As for the specific origin of the type A/B polymorphism in C. horridus, the present data do not allow us to ascertain whether it might be due to an ancestral polymorphism or the result of interspecific hybridization. It is important to note that, unlike the restricted geographic distributions of type A C. helleri and type B C. scutulatus, or the narrow hybrid zone between C. scutulatus and C. viridis [25], type A C. horridus individuals occur in numerous, widely dispersed locations across much of the species’ large range. Rokyta et al. (2015) have proposed an intergeneric hybridization with neurotoxic Sistrurus for the origin of type A C. horridus; however, these authors did not consider the possibility of an ancestral polymorphism [26]. Whatever the origin of the A/B polymorphism in C. horridus, such a widely dispersed, polymorphic species as C. horridus highlights the potential for speciation and incomplete lineage sorting of toxin gene complex haplotypes to generate descendant lineages that may be type A, type B, or a mixture of haplotypes.
Arms Races and the Maintenance of Genetic Diversity
Apart from their evolutionary origins, the presence of such divergent haplotypes at two gene complexes in each species also raises questions about how and why they persist. At the genetic level, one issue is whether there are any mechanisms that ensure that an animal is either type A or type B. For example, the MP and PLA2 complexes could be physically linked such that A or B haplotypes co-segregate and offspring inherit either neurotoxin genes or hemorrhagic toxin genes. All C. scutulatus individuals examined in our study were homozygous type A or type B individuals (Figures S5 and S6), but phenotypic studies of C. scutulatus have revealed individuals that contain both kinds of toxins and in different relative proportions (high neurotoxin/low MP; low neurotoxin/high MP; [15]) which indicates that the PLA2 and MP haplotypes are unlinked and assort independently and that heterozygotes are viable. Type A/B individuals have also been reported in C. horridus [13].
One precedent for such extremely divergent gene complex haplotypes in vertebrates involves hominid natural killer cell immunoglobulin-like receptors (KIRs) and their roles in immunity and reproduction. Numerous KIR haplotypes have evolved in apes and humans that contain different sets of KIR genes with distinct and complementary functions in immune defense and pregnancy [47]. The divergent and polymorphic KIR haplotypes appear to be maintained by balancing selection for immunity to the varying and evolving pathogens across human populations [48].
The host-pathogen arms race that has driven and maintained KIR haplotype diversity may be analogous to the arms race that exists between rattlesnake predators and their prey. Rattlesnakes eat a variety of evolutionarily disparate prey (e.g., birds, mammals, reptiles) that vary in their susceptibility to venom [49]. For example, among four tax a of Sistrurus rattlesnakes, a correlation was found between variation in venom composition and the incidence of mammals in the snakes’ diet [49]. Furthermore, several instances are known of prey evolving resistance to rattlesnake [50, 51] or other snake venoms [52]. The simplest rationale for the persistence of alternative venom types and PLA2 and MP haplotypes within species would appear to be selection for the ability to subdue alternate prey. If so, this would also constitute a mode of balancing selection to maintain venom polymorphism and a very rare instance of balancing selection acting on multiple loci at each of two unlinked gene complexes [1, 53].
STAR⋆METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sean B. Carroll (sbcarrol@wisc.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
For this study we aimed to genetically compare individuals within species that produce different venom types. This required expert field collection of wild animals to identify and capture the relevant species and transport of the specimens to the Natural Toxin Research Center. Venom was extracted from adult Crotalus scutulatus and Crotalus oreganus helleri animals housed in the serpentarium at the Texas A&M – Kingsville National Natural Toxins Research Center (NNTRC). The collection efforts often yielded animals expressing the standard venom type (C. scutulatus – type A and C. helleri – type B) but we did identify specimens expressing the atypical venom (C. scutulatus – type B; 3 animals and C. helleri – type A; 1 animal). The locations for each specimen are in Table S1.
The snakes were euthanized (IACUC approval # 2010–09-01A) and venom glands, blood, liver and kidney tissue samples were dissected and snap frozen for preservation until nucleic acid extractions. The Kentucky Reptile zoo provided blood samples from adult Crotalus horridus animals that express non-neurotoxic (female specimen) or neurotoxic (male specimen) venom. The venom classification of the C. horridus neurotoxic specimen was made on the basis of the individual being a descendent of a neurotoxin-producing parent.
The distribution data for C. scutulatus, C. oreganus, and C. horridus geographic ranges was downloaded from the Internaltional Union for Conservation of Nature (iucnredlist.org). The C. oreganus dataset contained multiple species, including C. helleri, and was processed to show the known C. helleri range that has been described previously [14].
METHOD DETAILS
Draft de novo genome assembly using long reads
High quality genomic DNA from C. scutulatus (type B), C. helleri (type B) and C. horridus (type A) was extracted from blood and sent to Great Lakes Sequencing Center (Milwaukee, WI), EA Genomics (Durham, North Carolina) and University of Washington Sequencing Core (Seattle, WA), respectively. Multiple long insert sequencing libraries were made using a BluePippin cutoff of 7 – 40 kbp. These libraries were sequenced in several SMRTcells to determine if the insert lengths and raw read yields were satisfactory. Libraries of sufficient quality (length and yield) were then sequenced in 40–60 SMRTcells to obtain 35 – 45X genome coverage with raw sequence reads.
The raw reads were assembled using versions of the Celera assembler [54] (Whole Genome Shotgun Assembler (WGS) version 8.3rc1 and CANU version 1.0) that have been adapted to assembly of single molecule real – time (SMRT) sequencing reads with low (85 – 88%) single nucleotide accuracy. C. helleri raw reads were assembled using the WGS (version 8.3rc1) [55] and C. scutulatus and C.horridus reads were assembled using CANU (version 1.0) [56]. Quiver was used to re-align raw reads to the assembly and correct potentially misassembled regions. See Table S3 and Figure S6 for sequencing and assembly statistics.
To identify putative PLA2 and MP contigs venom gland PLA2 or MP transcripts were aligned using LAST [60] to the genome assemblies. The C. scutulatus type B MP and PLA2 complexes were on single contigs. The C. horridus type A PLA2 complex was split across two contigs and the MP complex was split across three contigs. The C. helleri type B PLA2 complex was on single contig but the MP genes were split across twenty-two non-overlapping contigs making gene annotation a challenge. For this study, our annotation and phylogenetic analysis efforts focused on the conserved metalloproteinase domain [69] (exons 7 –12) thus excluding some partial MP genes found across the twenty-two contigs. C. helleri duplicated MP genes with annotatable metalloproteinase domains are shown below the main complex (Figure 4B). See Table S4 for the MP genes identified on the C. helleri contigs, including the annotated genes containing a metalloproteinase domain. The available database MP proteins for C. helleri are SVMP-CohPH-3 (238), SVMP-ChoPH-2 (240), SVMP-CoPH-1 (232) [9].
BAC library construction and screening
Whole blood (C. scutulatus and C. helleri) was sent to Amplicon Express (Pullman, WA) for high molecular weight genomic DNA extraction and library construction. The resulting genomic libraries consisted of ~73,000 arrayed clones (5–7X genome coverage) with each clone containing an insert length of 80–150 kb. The complete library was pooled in a combinatorial manner that facilitated PCR-based screening [70]. A list of PCR primer sequences used for screening are at the following link: https://figshare.com/s/d00e4d7fb95085d945b2.
PCR-positive clones were picked from the library, streaked on plates and single colonies grown overnight at 3700B0030C in 500 mL LB containing chloramphenicol and processed using the standard QIAGEN midi-prep protocol.
BAC clone library sequencing and assembly
The University of Michigan DNA sequencing core prepared the Pacific Biosciences sequencing libraries using ten micrograms (mg) of BAC DNA according to the standard protocol with a size selection of large (> 10000 bp) DNA fragments. The library for single BAC clones was sequenced in a single SMRT cell. The raw reads were assembled using the accuracy optimized HGAP2 (Hierarchical Genome Assembly Protocol) algorithm [57]. See Table S2 for summary of the BAC clones presented in this study.
Assembly evaluation
First, potential contaminating bacterial contigs were identified using BLAST [58] and the NCBI bacterial genome database and removed from further analysis. Next, the vector sequence was removed from the contig and the appropriate ends (non-overlapping) were stitched together to yield a single contig representing the BAC clone insert. The corrected reads were aligned to the assembled genomic sequence (BAC insert) with LAST [60] and the read alignments and coverage were inspected for approximate evenness across the contig.
Annotation of venom loci
The MAKER annotation software [61] was used for ab initio gene prediction on the raw sequence. The MAKER output protein sequences were BLASTed against the NCBI database of human and rattlesnake proteins to identify the candidate genes. This approach accurately identified exon coordinates and full-length proteins for most genes. Genes were identified as venom genes if the BLAST hit was to a known venom sequence and if phylogenetic analysis (see below) confirmed the BLAST result.
However, computational annotation of venom genes is often insufficient because of the high sequence identity among family members and the presence of putative pseudo-genes. Therefore, the predicted gene models were inspected for missing or frame-shifted exons. For these challenging gene models accurate coordinate determination relied upon manual annotation with the following pieces of evidence: i) BLAST hit coordinates using known venom proteins and exons as query sequences, ii) venom gland transcript alignments and iii) HISAT2 exon junction predictions using splice sites that are based on venom gland read alignments [63].
RNA isolation, sequencing and expression
Venom gland RNA was isolated using the standard Trizol method. One microgram of total RNA was provided to the University of Wisconsin - Madison DNA sequencing core for strand-aware Illumina TruSeq RNA-sequencing library preparation. The sequencing libraries were created using size selected RNA (300–800 bp). The venom gland libraries were sequenced in a single HiSeq2500 lane for 2 × 150 cycles producing paired reads 150 nucleotides in length.
The RNA-seq reads were pre-processed by trimming both at the 5′ end (7 nt) and any Illumina adaptor sequence using Trimmomatic [62] [command options: ILLUMINACLIP:/path/to/adapterSeqs/TruSeq3-PE-2.fa:2:30:12 HEADCROP:7 MINLEN:50]. After alignment of venom gland reads to the annotated genomic sequences using STAR [64], gene-level abundance measurements were obtained with RSEM [65]. To do this, the genome was indexed using the ‘rsem-prepare-reference’ command with the ‘star’ option set, while passing in the gene annotations as a GFF3 file. Next, the trimmed reads were aligned and gene expression calculated using the ‘rsem-calculate-expression’ command.
Biological replication of our venom gland gene expression was obtained by dissecting C. scutulatus venom glands from two type A (specimens 993 and 927) and two type B (specimens 983 and 928) animals. These four samples were processed in parallel for total RNA extraction, sequencing and data analysis (Figure S2).
Phylogenetic analysis
Sequences used in our protein phylogenies were deduced from hypothetical translation of genomic regions or from the UniProt and NCBI databases. To construct protein phylogenies we first performed multiple protein sequence alignment with the program MUSCLE [66]. Phylogenetic trees were calculated using maximum likelihood (phyML) [67] in the program SeaView [68] (v4.4.2). Bootstrap analysis to assess node support was based on 1000 replicate trees. Trees were then formatted with FigTree (v1.4.0; http://tree.bio.ed.ac.uk/software/Figuretree/).
ABBA-BABA analysis
Genomic sequence flanking the SVMP or PLA2 venom gene complexes from C. helleri – hemo, C. helleri – neuro, C. scutulatus – neuro, C. horridus – hemo was aligned using CLUSTAL W [59] and Patterson’s D statistic [38, 39] was calculated using the evobiR package (http://coleoguy.github.io/).
QUANTIFICATION AND STATISTICAL ANALYSIS
Venom toxicity was measured using median lethal dose (LD50) of BALB/C mice (animal protocol # 2015–12-09-A6). LD50 values were used to classify individuals as neurotoxic (type A) if the LD50 was between 0.3 – 0.9 mg/kg or as hemorrhagic (type B) if the LD50 was > 3.0 mg/kg [15].
The test for significance in the skew of the ABBA/BABA site observations was assessed using a standard block jackknife procedure (1000 jackknifes with a block size of 1000).
DATA AND SOFTWARE AVAILABILITY
The accession number for clone 125O12 reads reported in this paper is SRA: SRR5858077. The accession number for clone 133D14 reads reported in this paper is SRA: SRR5858076. The accession number for clone 85A17 reads reported in this paper is SRA: SRR5858079. The accession number for clone 46A6 reads reported in this paper is SRA: SRR5858078. The accession number for clone 136F17 reads reported in this paper is SRA: SRR5858071. The accession number for clone 190H17 reads reported in this paper is SRA: SRR5858070. The accession number for clone 27I3 reads reported in this paper is SRA: SRR5858069. The accession number for clone 145P9 reads reported in this paper is SRA: SRR5858068. The accession number for clone 132M5 reads reported in this paper is SRA: SRR5858067. The accession number for C. scutulatus 927 type A RNA-seq reads reported in this paper is SRA: SRR5858073; The accession number for C. scutulatus 993 type A RNA-seq reads reported in this paper is SRA: SRR5858072. The accession number for C. scutulatus 928 type B RNA-seq reads reported in this paper is SRA: SRR5858075. The accession number for C. scutulatus 983 type B RNA-seq reads reported in this paper is SRA: SRR5858074. See Table S2. See Table S2. The assembled BAC clones and genomic sequences are available on FigShare:
SVMP genomic sequence files: https://figshare.com/articles/SVMP_genomic_sequences/
PLA2 genomic sequence files: https://figshare.com/articles/SVMP_genomic_sequences/
SVMP BAC clone sequence files: https://figshare.com/articles/SVMP_genomic_sequences/
PLA2 BAC clone sequence files: https://figshare.com/articles/SVMP_genomic_sequences/
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| E. coli DH10b | ThermoFisher | Cat. # 18297–010 |
| Biological Samples | ||
| C. scutulatus neurotoxic (type A) venom glands (see Table S1) | Natural Toxins Research Center (NTRC) | 927, 993 |
| C. scutulatus neurotoxic (type A) blood | NTRC | NA |
| C. scutulatus hemorrhagic (type B) venom glands | NTRC | 928, 983 |
| C. scutulatus hemorrhagic (type B) blood | NTRC | 928 |
| C. helleri neurotoxic (type A) blood | NTRC | 677 |
| C. helleri hemorrhagic (type B) blood | NTRC | 789 |
| C. horridus neurotoxic (type A) blood | Kentucky Reptile Zoo | CH-A090108–06 |
| C. horridus hemorrhagic (type B) blood | Kentucky Reptile Zoo | CHO114KY |
| Critical Commercial Assays | ||
| TruSeq stranded mRNA kit | Illumina | Cat. # 20020594 |
| QIAGEN Genome-tip 100/G DNA purification kit | QIAGEN | Cat. # 10243 |
| Deposited Data | ||
| Raw reads for 10 BAC clones. See Table S2 for details. | This study | SRR5858067 −77 |
| Raw reads for C. scutulatus (type B) genome sequencing. | This study | SRR6410430 |
| Raw reads for C. helleri (type B) genome sequencing. | This study | SRR6410429 |
| Raw reads for C. horridus (type A) genome sequencing. | This study | SRR6410431 |
| Assembled genomic sequence for SVMP regions. | This study | https://figshare.com/articles/SVMP_genomic_sequences/5450095 |
| Assembled genomic sequence for PLA2 regions. | This study | https://figshare.com/articles/SVMP_genomic_sequences/5450095 |
| Assembled BAC clone sequences for SVMP regions. | This study | https://figshare.com/articles/SVMP_genomic_sequences/5450095 |
| Assembled BAC clone sequences for PLA2 regions. | This study | https://figshare.com/articles/SVMP_genomic_sequences/5450095 |
| Experimental Models: Organisms/Strains | ||
| C. scutulatus neurotoxic (type A) | NTRC | 927, 993 |
| C. scutulatus hemorrhagic (type B) | NTRC | 928, 931, 983 |
| C. helleri neurotoxic (type A) | NTRC | 677 |
| C. helleri hemorrhagic (type B) | NTRC | 789 |
| C. horridus neurotoxic (type A) | Kentucky Reptile Zoo | CH-A090108–06 |
| C. horridus hemorrhagic (type B) | Kentucky Reptile Zoo | CHO114KY |
| Oligonucleotides | ||
| BAC library screening primers | This study | https://figshare.com/s/d00e4d7fb95085d945b2 |
| Recombinant DNA | ||
| CopyControl pCCC1BAC vector | Epicenter | Cat. # CBAC311B |
| Software and Algorithms | ||
| Whole Genome Shotgun Assembler (WGS) version 8.3rc1 | [54, 55] | RRID: SCR_010750 |
| CANU version 1.0 | [56] | RRID: SCR_015880 |
| Hierarchical Genome Assembly Protocol (HGAP2) | [57] | https://github.com/PacificBiosciences/Bioinformatics-Training/wiki |
| BLAST | [58] | RRID: SCR_001598 |
| CLUSTAL W | [59] | RRID: SCR_002909 |
| LAST | [60] | RRID: SCR_006119 |
| MAKER | [61] | RRID: SCR_005309 |
| Trimmomatic | [62] | RRID: SCR_011848 |
| HISAT2 | [63] | RRID: SCR_015530 |
| STAR | [64] | RRID: SCR015899 |
| RSEM | [65] | RRID: SCR_013027 |
| MUSCLE | [66] | RRID: SCR_011812 |
| phyML | [67] | RRID: SCR_014629 |
| FigTree v1.4.0 | http://tree.bio.ed.ac.uk/software/figtree | RRID: SCR_008515 |
| SeaView | [68] | RRID: SCR_015059 |
| ABBA/BABA D statistic | [38, 39] | |
| evobiR | http://coleoguy.github.io/#resources | http://coleoguy.github.io/ |
| R | http://www.r-project.org/ | RRID:SCR_001905 |
Highlights.
Members of some rattlesnake species produce either hemorrhagic or neurotoxic venoms
This polymorphism is due to variation in toxin gene number at two gene complexes
Neurotoxin genes were transferred into one rattlesnake species by hybridization
ACKNOWLEDGMENTS
We thank Frances Brodsky for pointing out the similarities between venom and KIR gene complexes, Mark Hockmuller for expert care and handling of the venomous snakes, Adam Phillippy and Michael Schatz for genome assembly advice, and past and present Carroll lab members for stimulating discussions. This work was supported by the Howard Hughes Medical Institute(S.B.C.) and by the Office of the Director, National Institutes of Health award number P40OD010960 (E.E.S.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and four tables and can be found with this article online at https://doi.org/10.1016/j.cub.2018.02.031.
REFERENCES
- 1.Llaurens V, Whibley A, and Joron M (2017). Genetic architecture and balancing selection: the life and death of differentiated variants. Mol. Ecol 26, 2430–2448. [DOI] [PubMed] [Google Scholar]
- 2.Reed RD, Papa R, Martin A, Hines HM, Counterman BA, Pardo-Diaz C, Jiggins CD, Chamberlain NL, Kronforst MR, Chen R, et al. (2011). Optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science 333, 1137–1141. [DOI] [PubMed] [Google Scholar]
- 3.Martin A, Papa R, Nadeau NJ, Hill RI, Counterman BA, Halder G, Jiggins CD, Kronforst MR, Long AD, McMillan WO, and Reed RD (2012). Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proc. Natl. Acad. Sci. USA 109, 12632–12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinervo B, and Lively CM (1996). The rock-paper-scissors game and the evolution of alternative male strategies. Nature 380, 240–243. [Google Scholar]
- 5.Johnston SE, Gratten J, Berenos C, Pilkington JG, Clutton-Brock TH, Pemberton JM, and Slate J (2013). Life history trade-offs at a single locus maintain sexually selected genetic variation. Nature 502, 93–95. [DOI] [PubMed] [Google Scholar]
- 6.Wuster W, Peppin L, Pook CE, and Walker DE (2008). A nesting of vipers: Phylogeny and historical biogeography of the Viperidae (Squamata: Serpentes). Mol. Phylogenet. Evol 49, 445–459. [DOI] [PubMed] [Google Scholar]
- 7.Hendry CR, Guiher TJ, and Pyron RA (2014). Ecological divergence and sexual selection drive sexual size dimorphism in New World pitvipers (Serpentes: Viperidae). J. Evol. Biol 27, 760–771. [DOI] [PubMed] [Google Scholar]
- 8.French WJ, Hayes WK, Bush SP, Cardwell MD, Bader JO, and Rael ED (2004). Mojave toxin in venom of Crotalus helleri (Southern Pacific Rattlesnake): molecular and geographic characterization. Toxicon 44, 781–791. [DOI] [PubMed] [Google Scholar]
- 9.Sunagar K, Undheim EA, Scheib H, Gren EC, Cochran C, Person CE, Koludarov I, Kelln W, Hayes WK, King GF, et al. (2014). Intraspecific venom variation in the medically significant Southern Pacific Rattlesnake (Crotalus oreganus helleri): biodiscovery, clinical and evolutionary implications. J. Proteomics 99, 68–83. [DOI] [PubMed] [Google Scholar]
- 10.Glenn JL, Straight RC, Wolfe MC, and Hardy DL (1983). Geographical variation in Crotalus scutulatus scutulatus (Mojave rattlesnake) venom properties. Toxicon 21, 119–130. [DOI] [PubMed] [Google Scholar]
- 11.Glenn JL, and Straight RC (1989). Intergradation of two different venom populations of the Mojave rattlesnake (Crotalus scutulatus scutulatus) in Arizona. Toxicon 27, 411–418. [DOI] [PubMed] [Google Scholar]
- 12.Borja M, Neri-Castro E, Castarñeda-Gaytan G, Strickland JL, Parkinson CL, Castañeda-Gaytan J, Ponce-López R, Lomonte B, Olvera-Rodríguez A, Alagón A, and Pérez-Morales R (2018). Biological and Proteolytic Variation in the Venom of Crotalus scutulatus scutulatus from Mexico. Toxins (Basel) 10, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glenn JL, Straight RC, and Wolt TB (1994). Regional variation in the presence of canebrake toxin in Crotalus horridus venom. Comp Biochem Physiol Pharmacol Toxicol Endocrinol 107, 337–346. [DOI] [PubMed] [Google Scholar]
- 14.Davis MA, Douglas MR, Collyer ML, and Douglas ME (2016). Deconstructing a Species-Complex: Geometric Morphometric and Molecular Analyses Define Species in the Western Rattlesnake (Crotalus viridis). PLoS ONE 11, e0146166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massey DJ, Calvete JJ, Sánchez EE, Sanz L, Richards K, Curtis R, and Boesen K (2012). Venom variability and envenoming severity outcomes of the Crotalus scutulatus scutulatus (Mojave rattlesnake) from Southern Arizona. J. Proteomics 75, 2576–2587. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez EE, Galán JA, Powell RL, Reyes SR, Soto JG, Russell WK, Russell DH, and Pérez JC (2005). Disintegrin, hemorrhagic, and proteolytic activities of Mohave rattlesnake, Crotalus scutulatus scutulatus venoms lacking Mojave toxin. Comp. Biochem. Physiol. C Toxicol. Pharmacol 141, 124–132. [DOI] [PubMed] [Google Scholar]
- 17.Calvete JJ, Fasoli E, Sanz L, Boschetti E, and Righetti PG (2009). Exploring the venom proteome of the western diamondback rattlesnake, Crotalus atrox, via snake venomics and combinatorial peptide ligand library approaches. J. Proteome Res 8, 3055–3067. [DOI] [PubMed] [Google Scholar]
- 18.Fox JW, Ma L, Nelson K, Sherman NE, and Serrano SMT (2006). Comparison of indirect and direct approaches using ion-trap and Fourier transform ion cyclotron resonance mass spectrometry for exploring viperid venom proteomes. Toxicon 47, 700–714. [DOI] [PubMed] [Google Scholar]
- 19.Mackessy SP (2010). Evolutionary trends in venom composition in the western rattlesnakes (Crotalus viridis sensu lato): toxicity vs. tenderizers. Toxicon 55, 1463–1474. [DOI] [PubMed] [Google Scholar]
- 20.Calvete JJ, Sanz L, Cid P, de la Torre P, Flores-Díaz M, Dos Santos MC, Borges A, Bremo A, Angulo Y, Lomonte B, et al. (2010). Snake venomics of the Central American rattlesnake Crotalus simus and the South American Crotalus durissus complex points to neurotoxicity as an adaptive paedomorphic trend along Crotalus dispersal in South America. J. Proteome Res 9, 528–544. [DOI] [PubMed] [Google Scholar]
- 21.Sanz L, Gibbs HL, Mackessy SP, and Calvete JJ (2006). Venom proteomes of closely related Sistrurus rattlesnakes with divergent diets. J. Proteome Res 5, 2098–2112. [DOI] [PubMed] [Google Scholar]
- 22.Gibbs HL, Sanz L, and Calvete JJ (2009). Snake population venomics: proteomics-based analyses of individual variation reveals significant gene regulation effects on venom protein expression in Sistrurus rattlesnakes. J. Mol. Evol 68, 113–125. [DOI] [PubMed] [Google Scholar]
- 23.Durban J, Pérez A, Sanz L, Gomez A, Bonilla F, Rodríguez S, Chacon D, Sasa M, Angulo Y, Gutiérrez JM, and Calvete JJ (2013). Integrated “omics” profiling indicates that miRNAs are modulators of the ontogenetic venom composition shift in the Central American rattlesnake, Crotalus simus simus. BMC Genomics 14, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wooldridge BJ, Pineda G, Banuelas-Ornelas JJ, Dagda RK, Gasanov SE, Rael ED, and Lieb CS (2001). Mojave rattlesnakes (Crotalus scutulatus scutulatus) lacking the acidic subunit DNA sequence lack Mojave toxin in their venom. Comp. Biochem. Physiol. B Biochem. Mol. Biol 130, 169–179. [DOI] [PubMed] [Google Scholar]
- 25.Zancolli G, Baker TG, Barlow A, Bradley RK, Calvete JJ, Carter KC, de Jager K, Owens JB, Price JF, Sanz L, et al. (2016). Is hybridization a source of adaptive venom variation in rattlesnakes? A test, using a crotalus scutulatus × viridis hybrid zone in Southwestern New Mexico. Toxins (Basel) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rokyta DR, Wray KP, McGivern JJ, and Margres MJ (2015). The transcriptomic and proteomic basis for the evolution of a novel venom phenotype within the Timber Rattlesnake (Crotalus horridus). Toxicon 98, 34–48. [DOI] [PubMed] [Google Scholar]
- 27.Dagda RK, Gasanov S, De La Oiii Y, Rael ED, and Lieb CS (2013). Genetic basis for variation of metalloproteinase-associated biochemical activity in venom of the mojave rattlesnake (crotalus scutulatus scutulatus). Biochem. Res. Int 2013, 251474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dowell NL, Giorgianni MW, Kassner VA, Selegue JE, Sanchez EE, and Carroll SB (2016). The Deep Origin and Recent Loss of Venom Toxin Genes in Rattlesnakes. Curr. Biol 26, 2434–2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murakami T, Kariu T, Takazaki S, Hattori S, Chijiwa T, Ohno M, and Oda-Ueda N (2009). Island specific expression of a novel [Lys(49)]phospholipase A(2) (BPIII) in Protobothrops flavoviridis venom in Amami-Oshima, Japan. Toxicon 54, 399–407. [DOI] [PubMed] [Google Scholar]
- 30.Tsai IH, Chen YH, Wang YM, Tu MC, and Tu AT (2001). Purification, sequencing, and phylogenetic analyses of novel Lys-49 phospholipases A(2) from the venoms of rattlesnakes and other pit vipers. Arch. Biochem. Biophys. 394, 236–244. [DOI] [PubMed] [Google Scholar]
- 31.Reyes-Velasco J, Meik JM, Smith EN, and Castoe TA (2013). Phylogenetic relationships of the enigmatic longtailed rattlesnakes (Crotalus ericsmithi, C. lannomi, and C. stejnegeri). Mol. Phylogenet. Evol 69, 524–534. [DOI] [PubMed] [Google Scholar]
- 32.Lomonte B, and Rangel J (2012). Snake venom Lys49 myotoxins: From phospholipases A(2) to non-enzymatic membrane disruptors. Toxicon 60, 520–530. [DOI] [PubMed] [Google Scholar]
- 33.Scarborough RM, Rose JW, Naughton MA, Phillips DR, Nannizzi L, Arfsten A, Campbell AM, and Charo IF (1993). Characterization of the integrin specificities of disintegrins isolated from American pit viper venoms. J. Biol. Chem 268, 1058–1065. [PubMed] [Google Scholar]
- 34.Masuda S, Maeda H, Miao JY, Hayashi H, and Araki S (2007). cDNA cloning and some additional peptide characterization of a single-chain vascular apoptosis-inducing protein, VAP2. Endothelium 14, 89–96. [DOI] [PubMed] [Google Scholar]
- 35.Markland FS Jr., and Swenson S (2013). Snake venom metalloproteinases. Toxicon 62, 3–18. [DOI] [PubMed] [Google Scholar]
- 36.Bjarnason JB, and Fox JW (1994). Hemorrhagic metalloproteinases from snake venoms. Pharmacol. Ther 62, 325–372. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y-M, Parmelee J, Guo Y-W, and Tsai I-H (2010). Absence of phospholipase A(2) in most Crotalus horridus venom due to translation blockage: comparison with Crotalus horridus atricaudatus venom. Toxicon 56, 93–100. [DOI] [PubMed] [Google Scholar]
- 38.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N,Li H, Zhai W, Fritz MHY,et al. (2010). A Draft Sequence of the Neandertal Genome. Science 328, 710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Durand EY, Patterson N, Reich D, and Slatkin M (2011). Testing for ancient admixture between closely related populations. Mol. Biol. Evol 28, 2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith CF, and Mackessy SP (2016). The effects of hybridization on divergent venom phenotypes: Characterization of venom from Crotalus scutulatus scutulatus × Crotalus oreganus helleri hybrids. Toxicon 120, 110–123. [DOI] [PubMed] [Google Scholar]
- 41.Aird SD, Thirkhill LJ, Seebart CS, and Kaiser II (1989). Venoms and Morphology of Western Diamondback/Mojave Rattlesnake Hybrids. J. Herpetol 23, 131–141. [Google Scholar]
- 42.Murphy RW, and Ben Crabtree C (1988). Genetic Identification of a Natural Hybrid Rattlesnake: Crotalus scutulatus scutulatus × C. viridis viridis. Herpetologica 44, 119–123. [Google Scholar]
- 43.Pease JB, Haak DC, Hahn MW, and Moyle LC (2016). Phylogenomics Reveals Three Sources of Adaptive Variation during a Rapid Radiation. PLoS Biol. 14, e1002379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suh A, Smeds L, and Ellegren H (2015). The dynamics of incomplete lineage sorting across the ancient adaptive radiation of neoavian birds. PLoS Biol. 13, e1002224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Joly S, McLenachan PA, and Lockhart PJ (2009). A statistical approach for distinguishing hybridization and incomplete lineage sorting. Am. Nat 174, E54–E70. [DOI] [PubMed] [Google Scholar]
- 46.Martin SH, Davey JW, and Jiggins CD (2015). Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol. Biol. Evol 32, 244–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parham P, and Moffett A (2013). Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat. Rev. Immunol 13, 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gendzekhadze K, Norman PJ, Abi-Rached L, Graef T, Moesta AK, Layrisse Z, and Parham P (2009). Co-evolution of KIR2DL3 with HLA-C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc. Natl. Acad. Sci. USA 106, 18692–18697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gibbs HL, and Mackessy SP (2009). Functional basis of a molecular adaptation: prey-specific toxic effects of venom from Sistrurus rattlesnakes. Toxicon 53, 672–679. [DOI] [PubMed] [Google Scholar]
- 50.Martinez RR, Pérez JC, Sánchez EE, and Campos R (1999). The antihemorrhagic factor of the Mexican ground squirrel, (Spermophilus mexicanus). Toxicon 37, 949–954. [DOI] [PubMed] [Google Scholar]
- 51.Biardi JE, and Coss RG (2011). Rock squirrel (Spermophilus variegatus) blood sera affects proteolytic and hemolytic activities of rattlesnake venoms. Toxicon 57, 323–331. [DOI] [PubMed] [Google Scholar]
- 52.Neves-Ferreira AGC, Cardinale N, Rocha SLG, Perales J, and Domont GB (2000). Isolation and characterization of DM40 and DM43, two snake venom metalloproteinase inhibitors from Didelphis marsupialis serum. Biochim. Biophys. Acta 1474, 309–320. [DOI] [PubMed] [Google Scholar]
- 53.Hittinger CT, Gonçalves P, Sampaio JP, Dover J, Johnston M, and Rokas A (2010). Remarkably ancient balanced polymorphisms in a multilocus gene network. Nature 464, 54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Myers EW, Sutton GG, Delcher AL, Dew IM, Fasulo DP, Flanigan MJ, Kravitz SA, Mobarry CM, Reinert KH, Remington KA, et al. (2000).Awhole-genome assembly of Drosophila. Science 287,2196–204. [DOI] [PubMed] [Google Scholar]
- 55.Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, Ganapathy G, Wang Z, Rasko DA, McCombie WR, and Jarvis ED; Adam M Phillippy (2012). Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat. Biotechnol 30, 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berlin K, Koren S, Chin C-S, Drake JP, Landolin JM, and Phillippy AM (2015). Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol 33, 623–630. [DOI] [PubMed] [Google Scholar]
- 57.Chin C-S, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. [DOI] [PubMed] [Google Scholar]
- 58.Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ (1990). Basic local alignment search tool. J. Mol. Biol 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 59.Thompson JD, Higgins DG, and Gibson TJ (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kielbasa SM, Wan R, Sato K, Horton P, and Frith MC (2011). Adaptive seeds tame genomic sequence comparison. Genome Res. 21, 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, SanchezAlvarado A, and Yandell M (2008). MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18, 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, and Gascuel O (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol 59, 307–321. [DOI] [PubMed] [Google Scholar]
- 68.Gouy M, Guindon S, and Gascuel O (2010). SeaViewversion 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol 27, 221–224. [DOI] [PubMed] [Google Scholar]
- 69.Fox JW, and Serrano SMT (2008). Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. FEBSJ. 275, 3016–3030. [DOI] [PubMed] [Google Scholar]
- 70.Vu GT, Caligari PD, and Wilkinson MJ (2010). A simple, high throughput method to locate single copy sequences from Bacterial Artificial Chromosome (BAC) libraries using High Resolution Melt analysis. BMC Genomics 11, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for clone 125O12 reads reported in this paper is SRA: SRR5858077. The accession number for clone 133D14 reads reported in this paper is SRA: SRR5858076. The accession number for clone 85A17 reads reported in this paper is SRA: SRR5858079. The accession number for clone 46A6 reads reported in this paper is SRA: SRR5858078. The accession number for clone 136F17 reads reported in this paper is SRA: SRR5858071. The accession number for clone 190H17 reads reported in this paper is SRA: SRR5858070. The accession number for clone 27I3 reads reported in this paper is SRA: SRR5858069. The accession number for clone 145P9 reads reported in this paper is SRA: SRR5858068. The accession number for clone 132M5 reads reported in this paper is SRA: SRR5858067. The accession number for C. scutulatus 927 type A RNA-seq reads reported in this paper is SRA: SRR5858073; The accession number for C. scutulatus 993 type A RNA-seq reads reported in this paper is SRA: SRR5858072. The accession number for C. scutulatus 928 type B RNA-seq reads reported in this paper is SRA: SRR5858075. The accession number for C. scutulatus 983 type B RNA-seq reads reported in this paper is SRA: SRR5858074. See Table S2. See Table S2. The assembled BAC clones and genomic sequences are available on FigShare:
SVMP genomic sequence files: https://figshare.com/articles/SVMP_genomic_sequences/
PLA2 genomic sequence files: https://figshare.com/articles/SVMP_genomic_sequences/
SVMP BAC clone sequence files: https://figshare.com/articles/SVMP_genomic_sequences/
PLA2 BAC clone sequence files: https://figshare.com/articles/SVMP_genomic_sequences/