Abstract
DNA origami, a method for constructing nanoscale objects, relies on a long single strand of DNA to act as the ‘scaffold’ to template assembly of numerous short DNA oligonucleotide ‘staples’. The ability to generate custom scaffold sequences can greatly benefit DNA origami design processes. Custom scaffold sequences can provide better control of the overall size of the final object and better control of low-level structural details, such as locations of specific base pairs within an object. Filamentous bacteriophages and related phagemids can work well as sources of custom scaffold DNA. However, scaffolds derived from phages require inclusion of multi-kilobase DNA sequences in order to grow in host bacteria, and those sequences cannot be altered or removed. These fixed-sequence regions constrain the design possibilities of DNA origami. Here, we report the construction of a novel phagemid, pScaf, to produce scaffolds that have a custom sequence with a much smaller fixed region of 393 bases. We used pScaf to generate new scaffolds ranging in size from 1512 to 10 080 bases and demonstrated their use in various DNA origami shapes and assemblies. We anticipate our pScaf phagemid will enhance development of the DNA origami method and its future applications.
Keywords: DNA origami, phagemid, DNA nanotechnology
1. Introduction
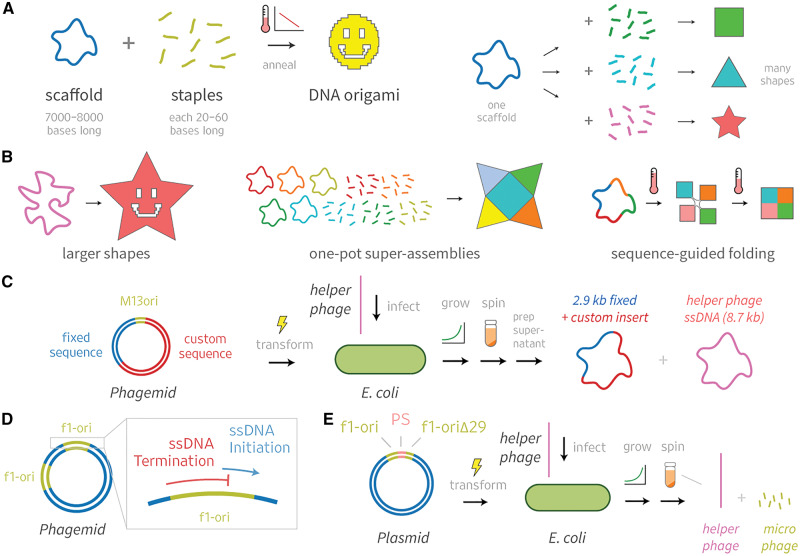
DNA origami builds tiny shapes using long single-stranded DNA (ssDNA) scaffolds and short ssDNA staples (1). A key strength of this method is that one scaffold sequence can be reused with different staples to create many different shapes (Figure 1A). However, although shapes generated from a single scaffold sequence can vary in nanometer-scale geometry, it is difficult to engineer sub-nanometer-scale details of shapes using only a single scaffold, or to create structures that vary in size from a single scaffold precursor (4). Thus, it is important to generate new scaffolds to expand the space of possible DNA origami designs. For example, new scaffolds could help create larger structures, or shapes with novel multimeric assemblies, or could help elucidate the principles of DNA origami self-assembly (Figure 1B). Here, our goal was to create new scaffolds of almost arbitrary custom sequence, up to 10 kilobases (kb) long, which are suitable for production in milligram (mg) quantities.
Figure 1.
DNA origami design would benefit from custom scaffolds. (A) Many DNA origami shapes can be folded from a single scaffold. (B) New scaffolds will expand the space of possible designs. (C) Phagemids are excellent sources of scaffolds, but have multi-kilobase sequence constraints. Previous studies offer hints of how these constraints could be circumvented. (D) Dotto et al. (2) used phagemids with modified origins to show that f1-ori ssDNA initiation and termination functions overlap, but can be inactivated separately by modifying distinct sequences. (E) Specthrie et al. (3) produced phage-like particles with ssDNA as short as 292 bases using a truncated f1-ori that acts as a terminator (f1-oriΔ29).
Many methods for producing scaffolds of custom sequence have been reported previously (5–19). Typically, ssDNA scaffolds are derived from double-stranded DNA (dsDNA) sources via combinations of selective amplification, isolation or degradation of one of the two strands. While reported methods offer excellent sequence customizability, they can be difficult to scale up in sequence length and production yield due to limitations in various in vitro enzymatic processing steps. We set out to develop a general approach for easily generating new scaffolds that would be scalable in both length and production yield, thus overcoming a significant hurdle in expanding their usability.
We selected filamentous bacteriophages as a platform that offers mg-scale yields in shake flasks, and whose yield can be boosted with bioreactors if needed (20). Filamentous phages, such as M13, are bacteria-specific viruses that package and export their single-stranded genomes into rod-like particles that have a protein coat. The phages can be recovered from the culture media and the ssDNA purified by standard molecular biology techniques (21). Custom sequences up to 2.5 kb can be inserted reliably into the M13 genome (22), though schemes to create much longer scaffolds have been reported (11). However, sequence customizability in the M13 phage is limited because most of its genome (>6 kb) consists of protein-coding and regulatory sequences that cannot be easily modified without disrupting phage growth.
Phagemids can be used to create scaffolds that have improved sequence customizability compared to M13 (9, 17, 23). These plasmids typically contain a host origin of replication (ori) sequence, a phage ori from M13 or relative such as f1, and an antibiotic resistance gene. Phagemid ssDNA can be exported in phage-like particles if the host cell is co-infected with a ‘helper phage’ or transformed with a ‘helper plasmid’ to express the necessary viral proteins (24). Phagemids can accommodate custom inserts several kb in size, but include a 2–3 kb fixed region that limits their usefulness in producing custom origami scaffolds (Figure 1C).
To increase scaffold sequence customizability, we sought to create a phagemid that could be produced using established preparation methods but would be able to package and export custom ssDNA sequences that have a relatively small fixed region. We were inspired by two papers that reported making use of modified f1-ori sequences to manipulate ssDNA synthesis (Figure 1D and E). In 1982, Dotto et al. (2) used phagemids with modified origins to show that f1-ori ssDNA synthesis initiation and termination functions overlap, but can be inactivated separately by modifying distinct sequences. In 1992, Specthrie et al. (3) packaged ssDNA as short as 292 bases into phage-like particles they called microphages. They were able to build small ssDNA strands using a phagemid that included an f1-ori, a packaging sequence (PS) and a truncated f1-ori that acts as a terminator (f1-oriΔ29). The terminator interrupts ssDNA synthesis of the full phagemid sequence, leading to packaging and export of only the region flanked by the ori and terminator.
We constructed a new phagemid, pScaf, for making custom DNA origami ssDNA scaffolds. After optimizing the pScaf ssDNA synthesis terminator, we used an iterative restriction-enzyme digestion and ligation scheme to clone several sequence inserts to produce scaffolds ranging from 1512 to 10080 bases in total length, each including an identical 393-base fixed region. We designed and folded several DNA origami shapes using the custom scaffolds, including a one-pot assembly of three scaffolds into a six-helix-bundle trimer, and analyzed the folding quality by gel and negative stain transmission electron microscopy (TEM).
2. Materials and methods
2.1 Construction of the pScaf vector
We initially converted the pUC18 vector into a phagemid by cloning the M13 origin from M13mp18 at the NdeI and KpnI restriction sites. Digested fragments were ligated with T4 DNA ligase (New England Biolabs (NEB)) and transformed into XL1-Blue MR competent cells (Agilent). We grew the pUC18-M13ori phagemid and recovered the DNA with a miniprep. Next, we used M13mp18 as a template to polymerase chain reaction (PCR)-amplify the ssDNA synthesis terminator, based on the Δ29 design from Specthrie et al. (3) The terminator was then inserted into the BamHI and EcoRI sites of the pUC18-M13ori. All subsequent variants of the terminator (Figure 1) were assembled by PCR and cloned into the same BamHI and EcoRI sites, and verified for correctness using DNA sequencing. We used the variant with three thymine bases (TTT) as the final pScaf vector in all subsequent experiments in this work.
2.2 Cloning scheme
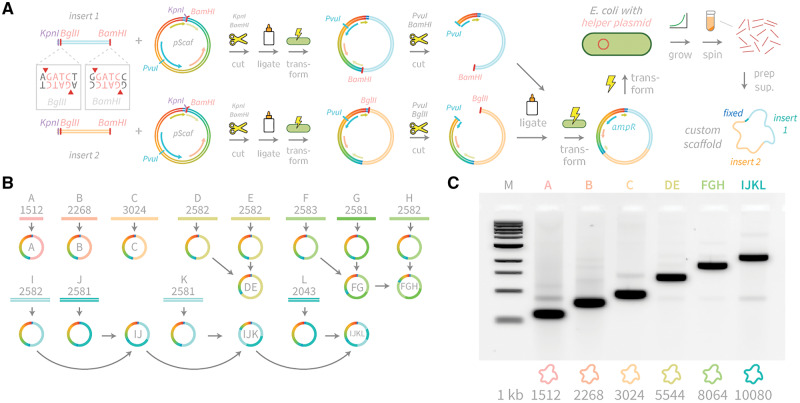
We used KpnI and BamHI restriction sites between the ssDNA synthesis initiator and terminator regions to clone inserts into the pScaf vector (Figure 3). In one step, we cloned short PCR-amplified insert sequences (A, B, C) directly into pScaf vectors that then generated the three smallest scaffolds (1512, 2268 and 3024). To create longer custom scaffolds, we adapted a multi-step cloning scheme previously used for cloning repeat protein cDNA constructs (25). We amplified custom sequence inserts in a PCR using forward and reverse primers designed to incorporate into the product a 5′ KpnI and BglII site, and a 3′ BamHI site, respectively. We cloned PCR-amplified sequence inserts of lengths up to 2.5 kb into separate pScaf vectors (D–L). To combine two inserts to create one larger insert, we digested the pScaf vector containing the intended 5′ region with PvuI and BamHI, and digested the vector containing the intended 3′ region with PvuI and BglII. BamHI and BglII digestions create compatible sticky-end overhangs that can be used to ligate the two inserts. The bases adjacent to the compatible sticky ends are different, so the ligation product does not reconstitute an internal BamHI or BglII site. However, the ligated fragment maintains a 5′ BglII site and a 3′ BamHI site for subsequent cloning rounds. We digest each plasmid at the PvuI restriction site to split the ampicillin-resistance gene bla into two parts that are only reconstituted by successful ligation of the full-length product. Thus, we generated three vectors with tandem inserts (DE, FG and IJ), and then combined FG with H to create FGH. We combined IJ with K to create IJK, which was subsequently combined with L to create IJKL. The pScaf vectors containing inserts DE, FGH and IJKL were then ready to produce the larger scaffolds (5544, 8064 and 10080).
Figure 3.
Cloning scheme and gel analysis of new scaffolds. (A) Custom sequence inserts are PCR-amplified with a forward primer containing KpnI and BglII sites and reverse primer containing a BamHI site. Inserts up to 3 kb in length were directly transformed into E. coli bearing helper plasmid. Larger scaffolds can be assembled by iterative PvuI+BamHI digestion of the vector containing the 5′ fragment, and PvuI+BglII digestion of the vector containing the 3′ fragment, followed by ligation, transformation and miniprep. (B) Twelve inserts (A–L) were cloned into pScaf vector at the KpnI-BamHI site. Inserts A, B and C were used to produce scaffolds with lengths of 1512, 2268 and 3024 bases. Larger scaffolds were assembled in multiple rounds as shown. (C) All scaffolds were grown in XL1-Blue cells containing helper plasmid M13cp, recovered and analyzed by agarose gel electrophoresis to determine purities ranging from 46% to 83%.
2.3 Scaffold amplification and purification
We transformed XL1-Blue-MRF’ using the M13cp helper plasmid (24) and then made it chemically competent with transformation and storage solution (10% polyethylene glycol (PEG)-8000, 30 mM MgCl2, 5% dimethyl sulfoxide, in 2xYT, pH 6, filtered) in order to produce a competent XL1-Blue-Helper strain. XL1-Blue-Helper was transformed with the desired pScaf construct to create a custom-phage-producing strain. We selected and grew a single colony for 18 h in an incubator-shaker (30°C, 225 rpm) in 3-mL 2xYT media (1.6% tryptone, 1% yeast extract, 0.25% NaCl) containing kanamycin, carbenicillin and chloramphenicol. Subsequently, the culture was transferred to a shake flask containing 100 mL 2xYT, 10 mL phosphate buffer (7% potassium phosphate dibasic, 3% sodium phosphate monobasic, pH 7, autoclaved), 1 mL 50% glucose, 0.5 mL 1 M MgCl2 and the appropriate antibiotics. The flask was then grown for 24 h on a shaker (30°C, 225 rpm). The flask was harvested by transferring the culture to a 500-mL ultracentrifuge bottle, stored on ice for 30 min and then centrifuged at 7000 g for 15 min at 4°C, pelleting the bacteria while allowing the phage to remain in the supernatant. The phage-containing supernatant was transferred to a clean 500-mL ultracentrifuge bottle, where 4 g PEG-8000 and 3 g NaCl were added. The bottle was then incubated on ice for 30 min prior to centrifugation at 9000 g for 15 min at 4°C, allowing the phage to form a pellet. The supernatant waste was decanted, and the phage pellet was resuspended in 3 mL Tris EDTA (TE) buffer. The bottle was then centrifuged at 15 000 g for 15 min at 4°C to pellet any residual bacteria. The supernatant containing the concentrated phage was then transferred to a 50-mL ultracentrifuge tube. Six microliter lysis buffer (0.2 M NaOH, 1% sodium dodecyl sulfate) was added, and the tube was mixed. Next, 4.5 mL neutralization buffer (3 M KOAc, pH 5.5) was added, and the tube was again mixed. The tube was incubated on ice for 15 min and then centrifuged at 15 000 g for 15 min at 4°C. The resulting supernatant was then decanted into a new 50-mL ultracentrifuge tube and 27 mL pure ethanol was added to it. The tube was capped and mixed by inversion prior to incubation at −20°C for 18 h. After incubation, the tube was centrifuged at 16 000 g for 15 min at 4°C to pellet the DNA. The supernatant waste was decanted leaving the DNA pellet at the bottom of the tube. The pellet was washed with 9 mL ice-cold 70% ethanol and centrifuged at 16 000 g for 5 min at 4°C. The supernatant waste was again decanted. The pellet was subsequently dried by gently blowing air into the tube, and finally the dried pellet was resuspended in up to 1 mL TE, depending on the desired concentration.
2.4 DNA origami preparation
Custom scaffold and staples were mixed at final concentrations of 20 nM and 200 nM, respectively, in a buffer containing 5 mM Tris, 1 mM ethylenediaminetetraacetic acid (EDTA) and 13 mM MgCl2. The solution was then subject to the following temperature ramp: denaturation at 65°C for 15 min, followed by cooling from 60°C to 38°C with a decrease of 1°C per 50 min.
2.5 Agarose gel analysis
Purified scaffolds and folded origami products were analyzed using 2% agarose gel electrophoresis in Tris-borate-EDTA (45 mM tris-borate and 1 mM EDTA) supplemented with 11 mM MgCl2 and SYBR Safe. Upon sample-loading, gels were run for 2 h at 80 V and subsequently scanned using a Typhoon FLA imager. DNA origami products folded from 1512-, 2268- and 3024-base scaffolds were subsequently purified by extracting the desired gel band using a razor blade, muddled to break down the agarose and then centrifuged through a Freeze ‘N Squeeze column (Bio-Rad).
2.6 PEG purification
DNA origami products folded from scaffolds of 5544 bases, and larger were purified using PEG precipitation. Assembly products were mixed with an equal volume of PEG precipitation buffer (15% PEG-8000, 10 mM Tris, 20 mM MgCl2 and 500 mM NaCl) and immediately centrifuged at 16 000 g at 20°C for 25 min. The supernatant was removed, and the pellet (which is often invisible) was resuspended in 1× folding buffer with magnesium (5 mM Tris, 1 mM EDTA and 13 mM MgCl2).
2.7 Transmission electron microscopy
Purified origami structures were diluted to approximately 25 ng/μL prior to imaging. 5 μL of the diluted origami was applied to glow-discharged, carbon-coated, 400-mesh formvar grids (Ted Pella) for 1.5 min. The grid was then blotted dry on filter paper (Whatman). Washing and staining was performed by preparing a pierce of parafilm with two 15-μL droplets of 1× folding buffer and two 15-μL droplets of 2% aqueous uranyl formate (electron microscopy sciences (EMS)) stain solution. The grid was dipped onto the first buffer droplet, blotted dry, dipped onto the next buffer droplet, blotted dry, dipped onto the first stain droplet, blotted dry, and then held onto the final stain droplet for 45 s before being blotted dry. Grids were then allowed to air-dry for 10 min prior to imaging. Electron micrographs were collected using an FEI TECNAI T12 transmission electron microscope using a using a 4k × 4k charge-coupled device camera (UltraScan 4000, Gatan) at 26 000× and 52 000× magnifications. Class averages were obtained using EMAN2 software.
3. Results
We constructed a phagemid, pScaf that can be used to create custom DNA origami scaffolds (Figure 2). We converted pUC18 into a phagemid for custom ssDNA production by adding four components: a full-length M13 origin for ssDNA initiation, KpnI and BamHI restriction sites for insert cloning, the M13 PS for phage particle export, and a modified M13 origin to serve as the ssDNA synthesis terminator (Supplementary Figure S1).
Figure 2.
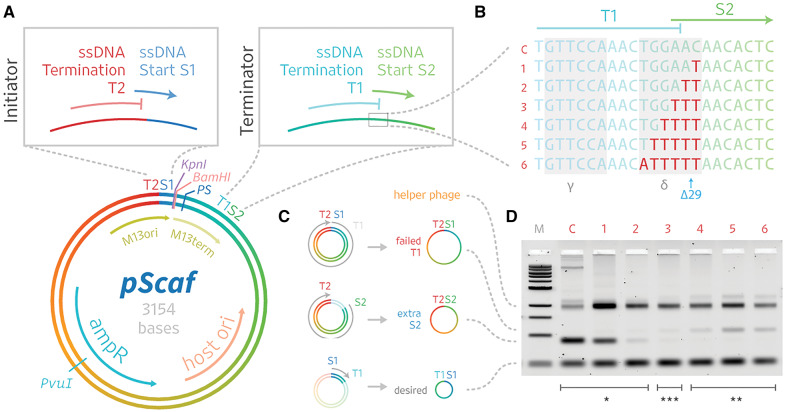
Construction and optimization of the pScaf phagemid. (A) The pScaf phagemid is derived from the pUC18 plasmid and M13mp18 phage vectors. The phagemid includes an M13 origin of replication (M13ori) that contains ssDNA start site S1 and ssDNA termination site T2, a KpnI and BamHI cloning site, a packaging sequence (PS), and a terminator of ssDNA synthesis (M13term) containing ssDNA termination site T1 and ssDNA start site S2. The M13term sequence was adapted from the M13ori sequence by selectively deactivating the S2 ssDNA initiation function. (B) The ssDNA synthesis initiation and termination functions of the origin overlap near the γ and δ regions that form an inverted repeat (highlighted in gray). Therefore, we used a mutational screen of the δ region to optimize the M13term performance. (C) Three primary species were observed: S1T2, representing failed termination at terminator T1, S2T2, representing spurious initiation at initiator S2, and S1T1, the desired product. (D) We analyzed the variants by agarose gel. Substituting 0–2 thymines yielded the S2T2 species (*). Substituting 4–6 bases yielded the S1T2 species (**). Substituting three thymines yielded the best balance between spurious S2 initiation and failed T1 termination, producing the most pure S1T1 species (***).
The works by Dotto and Specthrie informed the design and optimization of pScaf. We reasoned that if a terminator sequence can be used to generate microphages, a similar approach could be used to package larger custom sequences. We started with a construct that was similar to the Specthire microphage, except we added a restriction site and custom sequence inserts upstream of the terminator. When we tested the ‘Δ29’ terminator used by Specthrie et al. (3), we were able to produce our desired custom ssDNA scaffolds. However, our phage cultures also yielded an off-target ssDNA species consistent in size with residual packaging of the region downstream of our custom insert. We suspected that the Δ29 terminator might not fully abolish initiation of ssDNA synthesis, resulting in multiple ssDNA species produced from a single phagemid similar to constructs reported by Dotto et al. (2). Thus, we sought to optimize the terminator to reduce spurious initiation and produce a more pure ssDNA scaffold product (Figure 2).
Our goal was to optimize the phagemid so it would initiate ssDNA scaffold synthesis at only one start site (see S1 in Figure 2A, top left), continue synthesis through any custom inserted sequence, followed by the packaging site (PS) and finally end ssDNA synthesis at the termination site (T1 in Figure 2A, top-right). To minimize the synthesis of a second ssDNA fragment, we needed to reduce the activity of the second start site (S2) immediately following T1, which would produce an extra fragment that ends at the second terminator T2.
3.1 Terminator optimization
We started by truncating the last 50 bases of the 381-base M13 ori sequence so it included the core origin of plus-strand replication (α,β, γ and δ regions), but omitted the downstream initiation enhancer region. In his 1997 review, Horiuchi noted the enhancer can be removed without interfering with termination (26). Our goal was to focus on optimizing the terminator region while minimizing any off-target initiation, so we reasoned that the shortened fragment would serve as a good starting point. We then focused on optimizing the T1-S2 region (Figure 2B) to reduce unwanted initiation at S2, while maintaining T1 termination function. In the filamentous phage propagation, the plus-strand (i.e. phage-exported strand) ssDNA synthesis occurs by rolling-circle amplification starting at a single-stranded break created by gpII, a multi-function phage protein which also mediates unwinding, replication and termination by cleaving and circularizing the amplified strand (26). We focused on the γ and δ regions (Figure 2B, highlighted in grey) which are known binding sites for gpII where the termination and initiation functions overlap. We performed a mutational screen starting at the 3′ end of the δ region, near the site of the Specthrie et al. Δ29 truncation (Figure 2B, blue label). We cloned variants of the phagemid with up to six δ-region bases substituted with thymines (T), except for one thymine base which we replaced with adenine (Figure 2B). We tested ssDNA production of each variant in Escherichia coli culture co-infected with helper phage. Three ssDNA species were observed: the target species S1T1, and off-target species S1T2 and S2T2 (Figure 2C). We observed that 0-, 1-, or 2-base substitutions permitted ssDNA initiation, leading to synthesis of an S2T2 off-target species of ssDNA. Meanwhile, 4-, 5- or 6-base substitutions interfered with ssDNA termination, leading to an alternate S1T2 off-target ssDNA species (Figure 2D). We achieved the best balance between target (S1T1) and off-target (S1T2 and S2T2) ssDNA species using the 3-base substitution. When we analyzed the gel with ImageJ, we found the 3-base substitution produced off-target S1T2 and S2T2 bands with 3% and 1% the integrated density of the target S1T1 band, respectively. In comparison, the 2-base substitution produced a S2T2 species with 7% the integrated density of the S1T1 band, while the 4-base substitution produced a S1T2 band with 8% of the integrated density of the S1T1 band. We subsequently used the 3-base substitution in all pScaf constructs.
3.2 Custom scaffolds and test origami
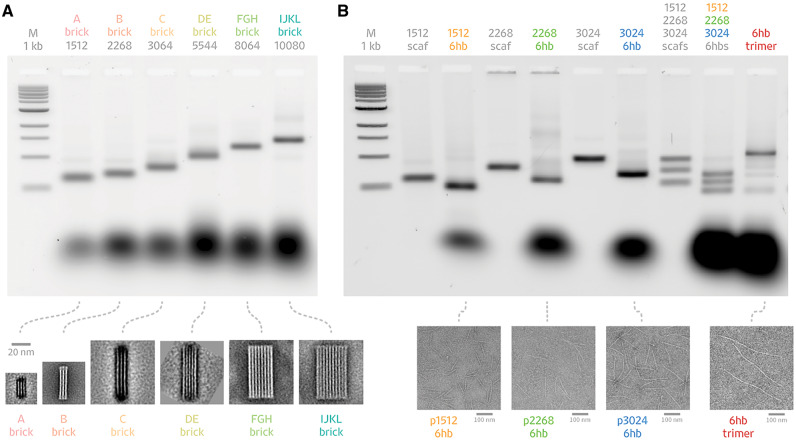
We created custom scaffolds with lengths of 1512, 2268, 3024, 5544, 8064 and 10080 bases (Figure 3, Supplementary Figure S2). Each scaffold was verified by sequencing and analyzed by agarose gel electrophoresis. We used ImageJ to estimate that the purity of correct-length scaffold products ranged from 46% to 83% of total integrated intensity of the sample lane (Figure 3C). We used Cadnano (27) to design a series of DNA origami shapes that approximate rectangular cuboids or brick-like structures. We folded the resulting DNA origami shapes and visualized them using agarose gel electrophoresis and negative stain TEM (Figure 4 and Supplementary Data S4–S10). When analyzed for purity with ImageJ, the leading gel bands representing the folded structures ranged from 68% to 83% of the total integrated intensity of the sample lane (Figure 4A). We assessed the folding quality of the shapes by manual counting of particles in the TEM micrographs. We considered a particle to be well folded if it appeared intact with no obvious defects, kinks, missing helices or aggregation. We estimated that 76% to 90% of all shapes were well folded.
Figure 4.
Folding custom scaffolds into DNA origami shapes and assemblies. Six scaffolds of varying sizes were folded into DNA origami shapes and assemblies. Folding reactions were analyzed by gel electrophoresis, and origami analyzed negative stain transmission electron microscopy. (A) Brick-like DNA origami shapes were folded from scaffolds of lengths 1512, 2268, 3024, 5544, 8064 and 10080 bases. Each brick was PEG-purified and imaged by negative stain TEM. Class averages for each brick are shown, along with a 20-nm scale bar. (B) Scaffolds of lengths 1512, 2268 and 3024 were folded separately into six-helix-bundle (6hb) nanotubes. All three scaffolds and 6hb-staple sets were combined into one-pot reaction mixtures. When connector staples are included, the three scaffolds assemble into a 6hb trimer. TEM micrographs of the individual 6hb nanotubes and combined trimer are shown with 100-nm scale bars.
3.3 One-pot folding of multi-scaffold assemblies
The ability to synthesize custom scaffolds with relatively little sequence overlap affords the possibility of folding DNA origami from multiple scaffolds in a one-pot reaction. To validate this concept, we designed a six-helix-bundle nanotube comprised of three scaffolds with lengths of 1512, 2268 and 3024 bases. These scaffold sequences are unique and derived from the same plasmid (see Supplementary Figure S2). The scaffolds were folded in a one-pot reaction both with and without connector staples to join them into a single nanotube.
4. Discussion
Our approach has significant advantages over pre-existing methods for making custom scaffolds to build DNA origami nanostructures. First, we have demonstrated that custom scaffolds can be generated with lengths of up to 10 kb. Thus, the method is compatible with nearly all origami shapes that have been published to date and will enable the creation of additional large shapes. Second, although cloning large (>3 kb) custom insert sequences into phagemids presents difficult challenges, the cloning scheme we report here circumvents some of them. We found the insertion of long sequences is much more reliable with a multi-step assembly approach that cuts the antibiotic resistance gene during each round and includes a positive selection for full-length constructs that reconstitute the gene. Earlier, when we attempted to clone several 7–8 kb PCR-amplified inserts in a single round without splitting the antibiotic resistance gene, we typically needed to screen 36 colonies at a time to observe 5–8% transformation efficiency. Once we implemented the multi-step scheme, our transformation efficiency improved to 80–100% when screening five colonies per round. Third, because we rely on phagemids to grow custom scaffolds in E. coli, the method is compatible with techniques that can be scaled to produce milligram-scale quantities of custom scaffolds using flasks and a shaker-incubator. Moreover, bacterial production offers a potentially reliable and feasible method for scaling up production to much larger quantities using bioreactors, as has been demonstrated for other scaffolds (20, 28).
We PCR-amplified 12 unique custom insert sequences from two different plasmids (Supplementary Figure S2) and incorporated them into the pScaf vector. We used multiple rounds of restriction and ligation to concatenate nine of these sequences into larger inserts as shown in Figure 3B, producing an additional six inserts. We did not observe any particular constraints on the sequences that can be incorporated into custom scaffolds using our method, with the caveat that we have only tested a limited number of sequence inserts from plasmid sources. Like any approach that requires cloning, our method is likely to encounter challenges with highly repetitive or low-complexity sequence inserts.
With our method, it will now be possible to rapidly generate large DNA origami shapes with highly customized scaffold sequences. We recognized the need for easier methods to make custom scaffolds when making DNA origami shapes that fold from limited sets of reusable staple sequences (29). For convenience, we incorporated sequences from pre-existing sources that we already had on hand [pFastBac-green fluorescent protein (GFP)-dynein-2(D1091–Q4307) plasmid and the bacteriophage Lambda genome, see Supplementary Figure S2]. Additional scaffolds of custom length can be generated quickly using a similar approach. Sequences also might be ordered from gene synthesis vendors and combined with our approach to generate scaffolds tailored for answering specific questions about DNA origami design principles, or to incorporate protein-binding sites.
Our scaffold lengths were chosen for convenience of designing DNA origami shapes using the honeycomb lattice architecture, which assumes B-form DNA with 34.3° mean twist per base, or 21 base pairs every two turns (6). In practice, circular scaffolds whose lengths are divisible by 42 are useful for creating shapes without any unused bases, and so we designed scaffolds as multiples of 42, i.e. 36 × 42 = 1512, 72 × 42 = 3024, 132 × 42 = 5044, 192 × 42 = 8064, 240 × 42 = 10080.
Custom scaffolds will be useful for studying the DNA origami method itself. For example, new scaffolds can be used to examine sequence-level determinants of folding yield. Sequences of particular base compositions (high-guanine-cytosine, high-adenine-thymine, 3-letter alphabets) with well-defined repetition or self-complementarity can be constructed and used to test hypotheses explaining the role of melting temperature or secondary structure in origami folding. Additionally, custom scaffolds can have practical benefits for routine origami design. We have often desired to create large DNA origami shapes with specific custom sequences at multiple non-contiguous locations within the scaffold. Generating these custom scaffolds is now much easier compared to cloning large inserts directly into M13mp18 in a single step.
It is also useful to be able to produce multiple unique scaffolds with very little sequence overlap. Sets of multiple unique custom sequences can be used for facile one-pot assembly of multimeric asymmetric structures. We demonstrated the one-pot assembly of small six-helix-bundle nanotube shapes and expect our approach can extend to reliably making larger shapes. Multi-scaffold DNA origami will enable to creation of assemblies with larger uniquely addressable surface areas when compared to single scaffold DNA origami. Larger assemblies will increase the maximum distances between unique modifications such as protein or nanoparticle attachments. While future studies will be needed to determine how much sequence overlap can be tolerated in multi-scaffold assemblies of DNA origami, we expect it should now be possible to design three custom 10-kb scaffolds to assemble into an asymmetric 30 kb origami in which nearly the entire structure is uniquely addressable, which could prove beneficial for certain applications.
Before we settled on the pScaf design, we attempted to generate custom scaffolds using a phagemid with a terminator based on the Δ29 truncation used by Specthrie et al. (3). Our observations of an off-target ssDNA species led us to attempt to optimize the terminator sequence. To guide our approach and select the site for our mutational screen, we reviewed previous studies of the initiation and termination of filamentous phage plus-strand synthesis (2, 26, 30). We were fortunate to find a sequence that seems to reduce the initiation function of gpII while retaining its termination function, though we lack a mechanistic understanding of why our variant gave better results. We also do not know the sources of trace amounts of off-target ssDNA that is produced in addition to our scaffolds (i.e. the faint extra bands and smearing in Figure 3C). A better understanding of the fundamental biology of filamentous phages could lead to further improvements in custom scaffold production with phagemids derived from pScaf. For example, a better understanding of how DNA sequence and structure in the vicinity of the termination site affect production of off-target ssDNA products can help us engineer an improved pScaf system with no off-target species.
Although the TTT terminator variant greatly improved the scaffold purity, we were not able to completely eliminate incomplete termination or spurious initiation in our scaffold preps. We also observed some prep-to-prep variation in total scaffold yield and purity and production of off-target species when using the helper plasmid (Supplementary Figure S3). We typically screened 4 to 5 colonies of XL1-Blue-Helper transformed with each pScaf variant and retained the transformant with the best purity. Our maximum purity is similar to commercially available M13-derived scaffold (31). Our DNA origami gel analysis and TEM images show that we obtained a relatively homogeneous population of folded origami structures, so the current levels of purity will be suitable for many applications.
The sequence insertion locus used KpnI and BamHI restriction sites, which would need modification if those sites are to be incorporated into a custom scaffold. A multiple cloning site would provide greater flexibility for cloning sequence inserts. We did not attempt to reduce the size of the terminator region that is incorporated into the scaffold ssDNA, so a further reduction in size of the fixed region of 393 bases may be possible with additional optimization. While we produced scaffolds up to 10 kb in length, we note that this length does not represent an upper limit. We have not yet attempted to make longer scaffolds, but it should be possible following our scheme.
In summary, we have developed and validated a novel phagemid, pScaf, for creating highly customized ssDNA DNA origami scaffolds of 10-kb lengths that can be produced with techniques compatible with milligram-scale yields. Our approach removes a longstanding constraint that has held back progress in the use of large DNA origami scaffolds, which is that researchers have had to rely on the genome of M13 phage which cannot be easily customized. The ability to generate many long, unique custom scaffolds will enable researchers to resolve many unanswered questions about what the optimal methods are to create and design DNA origami sequences, how to easily incorporate functional sequences, such as protein-binding sites, into multiple sites within each shape, and how to better adapt these methods in future applications.
Availability
Plasmids have been deposited at AddGene (https://www.addgene.org/). SnapGene files, custom scaffold sequences, Cadnano source files and DNA origami staple sequences available at https://github.com/douglaslab/pScaf.
Accession Numbers
Nucleotide sequences for the pScaf vector and custom sequence inserts used to make the reported scaffolds have been deposited in GenBank under the accession numbers: MH319458, MH319459, MH319460, MH319461, MH319462, MH319468 and MH319473.
Supplementary Material
Acknowledgements
We thank P. Rothemund and L. Bienen for comments on the manuscript. We are grateful to A. Bradbury for providing the M13cp helper plasmid.
Funding
Army Research Office [W911NF-14-1-0507]; Del E. Webb Foundation p13-2-28]; and National Science Foundation [CCF-1317640]; T.A. holds a Ruth L. Kirschstein NRSA Postdoctoral Fellowship (F32GM119322); S.M.D. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund (1010247). Funding for open access charge: Army Research Office.
Conflict of interest statement. None declared.
References
- 1. Rothemund P.W.K. (2006) Folding DNA to create nanoscale shapes and patterns. Nature, 440, 297–302. [DOI] [PubMed] [Google Scholar]
- 2. Dotto G.P., Horiuchi K., Zinder N.D. (1982) Initiation and termination of phage f1 plus-strand synthesis. Proc. Natl. Acad. Sci. U. S. A., 79, 7122–7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Specthrie L., Bullitt E., Horiuchi K., Model P., Russel M., Makowski L. (1992) Construction of a microphage variant of filamentous bacteriophage. J. Mol. Biol., 228, 720–724. [DOI] [PubMed] [Google Scholar]
- 4. Pinheiro A.V., Han D., Shih W.M., Yan H. (2011) Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol., 6, 763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pound E., Ashton J.R., Becerril H.A., Woolley A.T. (2009) Polymerase chain reaction based scaffold preparation for the production of thin, branched DNA origami nanostructures of arbitrary sizes. Nano Lett., 9, 4302–4305. [DOI] [PubMed] [Google Scholar]
- 6. Douglas S.M., Dietz H., Liedl T., Högberg B., Graf F., Shih W.M. (2009) Self-assembly of DNA into nanoscale three-dimensional shapes. Nature, 459, 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang H., Chao J., Pan D., Liu H., Huang Q., Fan C. (2012) Folding super-sized DNA origami with scaffold strands from long-range PCR. Chem. Commun., doi: 10.1039/c2cc32204h. [DOI] [PubMed] [Google Scholar]
- 8. Yang Y., Han D., Nangreave J., Liu Y., Yan H. (2012) DNA origami with double-stranded DNA as a unified scaffold. ACS Nano, 6, 8209–8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zadegan R.M., Jepsen M.D.E., Thomsen K.E., Okholm A.H., Schaffert D.H., Andersen E.S., Birkedal V., Kjems J. (2012) Construction of a 4 zeptoliters switchable 3D DNA box origami. ACS Nano, 6, 10050–10053. [DOI] [PubMed] [Google Scholar]
- 10. Ducani C., Kaul C., Moche M., Shih W.M., Högberg B. (2013) Enzymatic production of ‘monoclonal stoichiometric’ single-stranded DNA oligonucleotides. Nat. Methods, 10, 647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marchi A.N., Saaem I., Tian J., LaBean T.H. (2013) One-pot assembly of a hetero-dimeric DNA origami from chip-derived staples and double-stranded scaffold. ACS Nano, 7, 903–910. [DOI] [PubMed] [Google Scholar]
- 12. Said H., Schüller V.J., Eber F.J., Wege C., Liedl T., Richert C. (2013) M1.3—a small scaffold for DNA origami. Nanoscale, 5, 284–290. [DOI] [PubMed] [Google Scholar]
- 13. Li W., Yang Y., Jiang S., Yan H., Liu Y. (2014) Controlled nucleation and growth of DNA tile arrays within prescribed DNA origami frames and their dynamics. J. Am. Chem. Soc., 136, 3724–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nickels P.C., Ke Y., Jungmann R., Smith D.M., Leichsenring M., Shih W.M., Liedl T., Högberg B. (2014) DNA origami structures directly assembled from intact bacteriophages. Small, 10, 1765–1769. [DOI] [PubMed] [Google Scholar]
- 15. Marchi A.N., Saaem I., Vogen B.N., Brown S., LaBean T.H. (2014) Toward larger DNA origami. Nano Lett., 14, 5740–5747. [DOI] [PubMed] [Google Scholar]
- 16. Erkelenz M., Bauer D.M., Meyer R., Gatsogiannis C., Raunser S., Saccà B., Niemeyer C.M. (2014) A Facile Method for Preparation of Tailored Scaffolds for DNA-Origami. Small, 10, 73–77. [DOI] [PubMed] [Google Scholar]
- 17. Brown S., Majikes J., Martínez A., Girón T.M., Fennell H., Samano E.C., LaBean T.H. (2015) An easy-to-prepare mini-scaffold for DNA origami. Nanoscale, 7, 16621–16624. [DOI] [PubMed] [Google Scholar]
- 18. Veneziano R, Ratanalert S., Zhang K, Zhang F., Yan H, Chiu W., Bathe M. (2016) Designer nanoscale DNA assemblies programmed from the top down. Science, doi: 10.1126/science.aaf4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krieg E., Shih W.M. (2018) Selective nascent polymer catch-and-release enables scalable isolation of multi-kilobase single-stranded DNA. Angew. Chem. Int. Ed. Engl., 57, 714–718. [DOI] [PubMed] [Google Scholar]
- 20. Kick B., Praetorius F., Dietz H., Weuster-Botz D. (2015) Efficient production of single-stranded phage DNA as scaffolds for DNA origami. Nano Lett., 15, 4672–4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sambrook J, Russell D.W, (2001) Molecular Cloning: A Laboratory Manual, 3rd edn.Cold Spring Harbor Laboratory Press, New York. [Google Scholar]
- 22. Messing J. (1983) New M13 vectors for cloning. Methods Enzymol., 101, 20–78. [DOI] [PubMed] [Google Scholar]
- 23. Shih W.M., Quispe J.D., Joyce G.F. (2004) A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature, 427, 618–621. [DOI] [PubMed] [Google Scholar]
- 24. Chasteen L., Ayriss J., Pavlik P., Bradbury A.R.M. (2006) Eliminating helper phage from phage display. Nucleic Acids Res., 34, e145.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aksel T., Majumdar A., Barrick D. (2011) The contribution of entropy, enthalpy, and hydrophobic desolvation to cooperativity in repeat-protein folding. Structure, 19, 349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Horiuchi K. (1997) Initiation mechanisms in replication of filamentous phage DNA. Genes Cells, 2, 425–432. [DOI] [PubMed] [Google Scholar]
- 27. Douglas S.M., Marblestone A.H., Teerapittayanon S., Vazquez A., Church G.M., Shih W.M. (2009) Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res., 37, 5001–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Praetorius F., Kick B., Behler K.L., Honemann M.N., Weuster-Botz D., Dietz H. (2017) Biotechnological mass production of DNA origami. Nature, 552, 84. [DOI] [PubMed] [Google Scholar]
- 29. Niekamp S., Blumer K., Nafisi P.M., Tsui K., Garbutt J., Douglas S.M. (2016) Folding complex DNA nanostructures from limited sets of reusable sequences. Nucleic Acids Res., 44, e102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dotto G.P., Horiuchi K. (1981) Replication of a plasmid containing two origins of bacteriophage f1. J. Mol. Biol., 153, 169–176. [DOI] [PubMed] [Google Scholar]
- 31. Kuzyk A., Yang Y., Duan X., Stoll S., Govorov A.O., Sugiyama H., Endo M., Liu N. (2016) A light-driven three-dimensional plasmonic nanosystem that translates molecular motion into reversible chiroptical function. Nat. Commun., 7, 10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.