Abstract
Objective:
Pharmacogenomic studies of antipsychotics have typically examined effects of individual polymorphisms; by contrast, polygenic risk scores (PRS) derived from genome-wide association studies (GWAS) can quantify the influence of thousands of common alleles of small effect in a single measure. We examined whether PRS for schizophrenia are predictive of antipsychotic efficacy in four independent cohorts of patients with first-episode psychosis (total n=510).
Method:
All subjects received initial treatment with antipsychotic medication for first-episode psychosis, and all were genotyped on standard SNP arrays imputed to the 1000 Genomes reference panel. PRS was computed based on the results of the large-scale schizophrenia GWAS reported by the Psychiatric Genomics Consortium. Symptoms were measured by total symptom rating scales at baseline and week 12 or at the last follow-up visit before drop-out.
Results:
In the discovery cohort, higher PRS significantly predicted higher symptom scores at 12-week follow-up (controlling for baseline symptoms, sex, age, and ethnicity). The PRS threshold of PT<0.01 gave the strongest result in the discovery cohort, and was used to replicate the findings in the other three cohorts. Higher PRS significantly predicted greater post-treatment symptoms in the combined replication analysis (p=.0095), and was individually significant in two of the three replication cohorts. Across the four cohorts, PRS was significantly predictive of adjusted 12-week symptom scores (pooled partial r=0.18, p=0.002, 3.24% of variance explained). Patients with low PRS were more likely to be treatment responders than those with high PRS (OR=1.91 in Caucasian samples, p<0.01).
Conclusion:
Patients with higher PRS for schizophrenia tended to have less improvement with antipsychotic drug treatment. PRS burden may have potential utility as a prognostic biomarker.
Introduction
Genetic susceptibility to schizophrenia is highly polygenic, including many associated loci of small effect(1, 2). While individual risk alleles may convey an odds ratio of 1.1 or lower, the combination of all such effects across the genome holds substantial explanatory power. For example, any individual can be characterized by a polygenic risk score (PRS), representing the total number of risk alleles he or she carries, weighted by the odds ratio associated with each allele as derived from previous genome-wide association study (GWAS) findings(3, 4). While a high PRS for schizophrenia is not deterministic, PRS derived from the Psychiatric Genomics Consortium(1) accounts for approximately 7% of variation in risk for schizophrenia (as measured on the liability scale(5), with about half of that variance accounted for by the top (genome-wide significant) loci. Additionally, individuals scoring in the top decile are ~15 times more likely to manifest the illness compared to those in the bottom decile(1).
Given the explanatory power of PRS for susceptibility to schizophrenia, it is reasonable to ask whether these scores can be informative regarding clinical heterogeneity within the disorder(2). For example, while antipsychotic drugs are the mainstay therapy for schizophrenia(6, 7), up to 30-40% of patients do not respond to antipsychotic treatment(8), and many patients discontinue their medications due to lack of efficacy(9). There is currently a paucity of clinically informative biomarkers, and pharmacogenomics is one approach to identifying predictors of treatment response(10). To date, candidate-gene studies and a small number of genome-wide association studies (GWAS) have had limited success in identifying genetic variants replicably associated with antipsychotic treatment response. So far, only two variants (at the DRD2 gene and the COMT gene) have demonstrated consistent effects across multiple cohorts as demonstrated by meta-analysis(11, 12). Although promising, their effect sizes are relatively small (OR’s = 1.54 and 1.37, respectively) and predictive power is limited(13).
Given previous findings suggesting that a family history of schizophrenia may be associated with poor clinical response(14, 15), patients with higher genetic burden of schizophrenia may have poorer clinical outcomes. Compared to candidate gene approaches, PRS methods may better capture the full genomic underpinnings of illness and improve clinical prediction, as has been recently demonstrated in prostate cancer, in which higher PRS was associated with more aggressive illness(16). One recent schizophrenia study utilized clinically assigned clozapine therapy as a proxy for treatment resistance (by comparing clozapine-treated patients to those who have never been prescribed clozapine) and found that PRS was significantly higher in clozapine patients than in non-clozapine patients(17), although another study failed to replicate the finding (18). However, both were cross-sectional studies that can be affected by ascertainment bias and inaccuracies of classification; for example, a similar cross-sectional study providing evidence for a pharmacogenetic role for the BDNF Val66Met variant(19) was not supported by subsequent longitudinal studies conducted in the context of clinical trials(20). Furthermore, PRS may have additional advantages in clinical prediction because it is a continuous variable that can have different cutoffs that maximize predictive power, whereas the candidate gene approach can compare only carriers to non-carriers. Moreover, its predictive power will increase as the discovery sample gets larger.
In the present study, we aimed to investigate whether PRS based on the large-scale GWAS conducted by the Psychiatric Genomics Consortium (PGC)(1) is predictive of antipsychotic efficacy in patients with first-episode non-affective psychosis. There are several advantages of studying first-episode psychosis, such as minimal or no prior drug exposure, increased effect size of genotype-phenotype association(21), and representation of the whole patient population compared to chronic patient samples which may be subject to ascertainment biases(22). While one prior study examined PRS in relation to clinical response to lurasidone in patients with chronic schizophrenia(23), this is the first study, to our knowledge, to longitudinally examine treatment response in first episode patients undergoing initial treatment with antipsychotics.
Methods
Participants
The discovery cohort consists of 77 patients from the Zucker Hillside Hospital First Episode schizophrenia trial (ZHH-FE)(24). They were treated with either risperidone or olanzapine for 16 weeks and psychotic symptoms were assessed by the Schedule for Affective Disorders and Schizophrenia Change Version with psychosis and disorganization items (SADS-C+PD) at baseline and weeks 1, 2, 3, 4, 6, 8, 10, 12, 14, and 16. To compute a total symptoms score, selected items from the SADS-C+PD were converted to corresponding items in the Brief Psychiatric Rating Scale (BPRS)(25) and a total score of the imputed BPRS was calculated(26). Last-observation-carry-forward (LOCF) method was used for missing data and the change score from baseline to week 12 was computed as the main phenotype of total symptom reduction after treatment. Week 12 data was chosen to be consistent with other cohorts. The patients were from several different continental ancestries including European, African, Asian, and mixed ancestry.
Three additional cohorts were used for replication of the findings from the discovery sample. 1) The European First Episode Schizophrenia Trial (EUFEST) cohort: patients were randomized to one of five antipsychotics: olanzapine, quetiapine, ziprasidone, amisulpride, and haloperidol, and treated for up to 12 months(27). Symptoms were assessed with the Positive and Negative Symptoms Scale (PANSS)(28). Genomic data was available for 150 patients. All 141 European-ancestry patients were included in the present study and 9 patients from other racial groups were excluded to make the sample more homogeneous. LOCF method was used for missing data at 3 months. 2) The Programa Asistencial Fases Iniciales de Psicosis (PAFIP) de Cantabria, Spain (29, 30). Patients were treated with aripiprazole, olanzapine, quetiapine, risperidone, and ziprasidone for 12 weeks. Data were available for 192 patients with DNA and BPRS ratings at baseline and 12 weeks. All subjects were of European ancestry. 3) The third cohort consists of 100 patients from the clinical trial as part of the Center for Intervention Development and Applied Research at ZHH (CIDAR) and they were treated with either risperidone or aripiprazole for 12 weeks, and symptoms were assessed by the BPRS(31). Again, LOCF method was used for missing data. Like the ZHH-FE sample, the CIDAR patients came from various ancestry groups. Table 1 shows the demographic data for the four cohorts.
Table 1.
Demographic and descriptive data for the four cohorts included in the study.
| Cohort | N | Age (M±SD) |
% Male | % White | Symptom Rating Scale |
# SNP’s included in PRS at PT<0.01 |
|---|---|---|---|---|---|---|
| ZHH-FE | 77 | 23.0±4.9 | 75% | 39.0% | Derived BPRS items from SADS-C+PD |
7736 |
| EUFEST | 141 | 25.6±5.2 | 60% | 100% | PANSS | 8903 |
| PAFIP | 192 | 31.8±10.2 | 52% | 100% | BPRS | 8634 |
| CIDAR | 100 | 21.5±5.1 | 75% | 35.4% | BPRS | 8110 |
Note: ZHH-FE: Zucker Hillside Hospital First Episode Schizophrenia trial; EUFEST: European First Episode Schizophrenia Trial; PAFIP: the Programa Asistencial Fases Iniciales de Psicosis de Cantabria, Spain; CIDAR: the clinical trial as part of the Center for Intervention Development and Applied Research at Zucker Hillside Hospital. BPRS: Brief Psychiatric Rating Scale; PANSS: Positive and Negative Symptoms Scale; SADS-C+PD: Schedule for Affective Disorders and Schizophrenia Change Version with psychosis and disorganization items. PRS: Polygenic Risk Score. PT: P-value threshold.
Genotyping
DNA was extracted from peripheral lymphocytes and genotyping was performed using the Illumina Omni-1 Quad (ZHH-FE and EUFEST samples) or Illumina Infinium HumanOmniExpressExome platform (CIDAR and PAFIP samples). Standard quality control procedures were performed to exclude SNPs with minor allele frequency (MAF) <2%, genotyping failure >5%, Hardy-Weinberg equilibrium p<10−6, mismatch between recorded and genotyped sex, as well as related individuals (the relative with the lower call rate was dropped). SNP imputation was conducted with IMPUTE2(32) against the full 1000 Genomes phase 3 reference panel(33). The imputed SNPs underwent another round of quality control and SNPs with missing data >5% and imputation information score <0.8 were excluded. The discovery cohort ended up with 6,143,400 high quality SNPs. The EUFEST, PAFIP, and CIDAR samples had 6,863,830, 7,302,869, and 7,302,858 SNPs, respectively, after quality control. Principle component analysis was conducted in each cohort and the top 3 principal component scores were saved for further analysis. All genomic data analysis was performed using the SVS software, version 8.7.0 (Golden Helix, Inc., Bozeman, MT, USA).
Polygenic Risk Scores (PRS)
PRS based on the PGC schizophrenia GWAS(1) represents a measure of genetic liability to schizophrenia. The higher an individual’s PRS, the higher his/her risk of schizophrenia. PRS was calculated for each participant in the sample as the weighted sum of the risk allele they carried, based on the summary statistics (effect alleles and odds ratios) derived from the clumped PGC GWAS results, which consists of 102,636 SNPs. The clumped PGC GWAS summary statistics file was downloaded from the LD Hub at the Broad Institute (available at http://ldsc.broadinstitute.org/ldhub/). The clumping parameters are as following: a SNP will be clumped to a more significant SNP with LD (r2 ≥ 0.10) within a 500kb window, with the MHC region represented by a single SNP. The calculation was carried out in the PRSice software(4) for the four cohorts separately. SNPs were selected to be included in the PRS calculation based on their p values in the original PGC GWAS. For the discovery cohort (ZHH-FE), the PRS was calculated at several p value thresholds (PT) based on the original PGC GWAS, in order to explore which one would maximize the signal of PRS-phenotype association. Specifically, PT’s ≤ 5×10−8, 0.001, 0.01, 0.05, 0.10, 0.20, and 0.50 were applied to compute seven sets of PRS for the discovery cohort. The PT with maximum prediction power for the outcome variable in the discovery cohort was then used for computing the PRS for the three replication cohorts. PRS data were approximately normally distributed, and were converted into z scores for easy interpretation.
Statistical Analysis
The primary phenotype was antipsychotic drug efficacy, defined by symptom reduction from baseline to 12 weeks or 3 months. Symptoms were measured using the total score of BPRS items (derived from the SADS-C+PD) for the ZHH-FE cohort, total BPRS scores for the PAFIP and CIDAR samples, or the total PANSS score for the EUFEST sample. The 12-week (or 3-month) scores, adjusted for the baseline scores, age, and sex, served as the primary dependent variable in a hierarchical linear regression; PRS was the predictor variable. The endpoint score adjusted for baseline value in a regression analysis is functionally equivalent to the simple change score from baseline to endpoint, but is statistically more powerful. Genomic principal component scores were also covaried to control for population stratification for the ZHH-FE and CIDAR cohorts because they consisted of subjects of various ancestries, while the EUFEST and PAFIP cohorts were entirely of European descent. Meta-analysis was performed to combine the effect sizes (partial correlation coefficients) from the three replication cohorts, as well as all four cohorts combined, because each cohort had a relatively small sample size. While it is not uncommon for replication tests to be reported with one-tailed p-values, we report two-tailed tests for all analyses for purposes of clarity and to remain conservative in reporting significant results. All statistical analyses were performed using SPSS version 24 (IBM, Armonk, NY, USA).
Results
Discovery Cohort
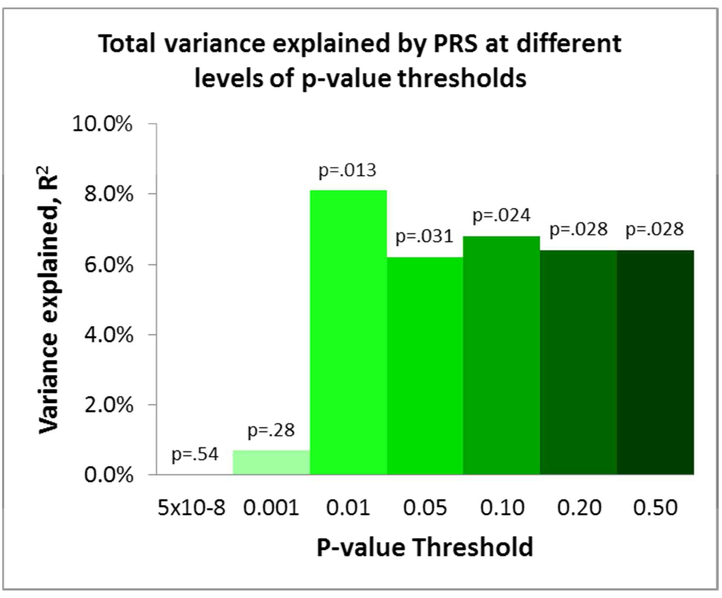
Among the 77 patients in the ZHH-FE cohort, higher PRS at the thresholds of PT<0.01, 0.05, 0.10, 0.20, and 0.50 significantly predicted worse response to treatment (i.e., higher symptom scores at 12-week follow-up), explaining between 6.4% and 8.1% of the total variance in outcome (all p’s<.05; Figure 1). PRS at the threshold PT < 5×10−8 and PT < 0.001 were not significant in predicting the symptom change scores (p=.54, p=.28, respectively). PRS at PT<0.01 gave the strongest result in the discovery sample, and therefore was used to replicate the findings in the other three cohorts.
Figure 1.
PRS at different levels of p-value thresholds explained percentages of total variance in 12-week symptom scores controlling for baseline symptoms and other covariates in the discovery sample.
Replication Cohorts and Meta-analysis
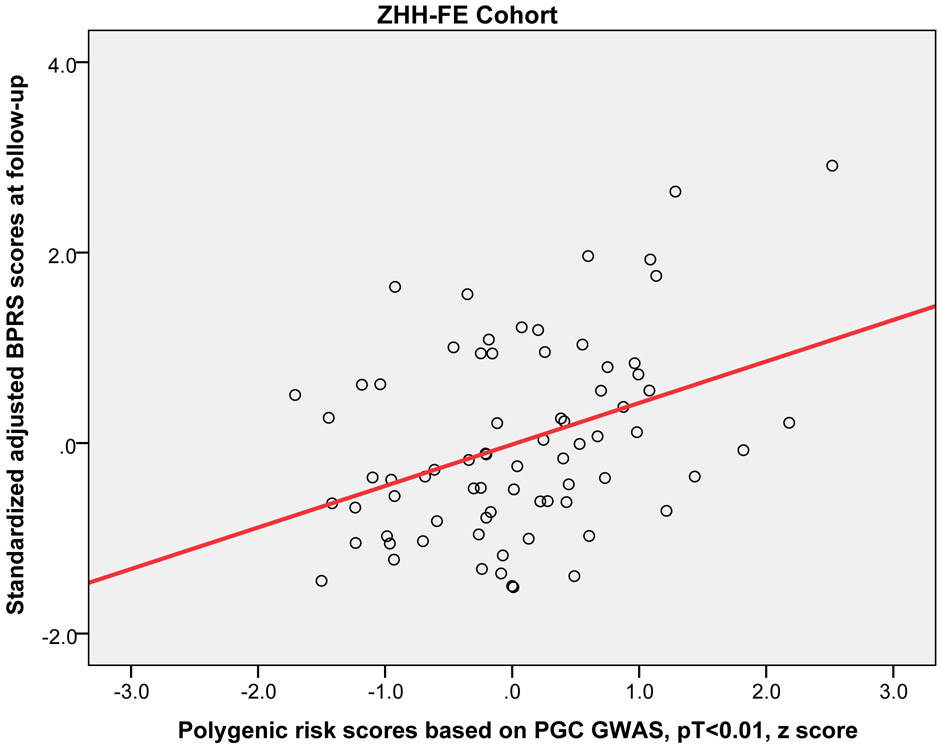
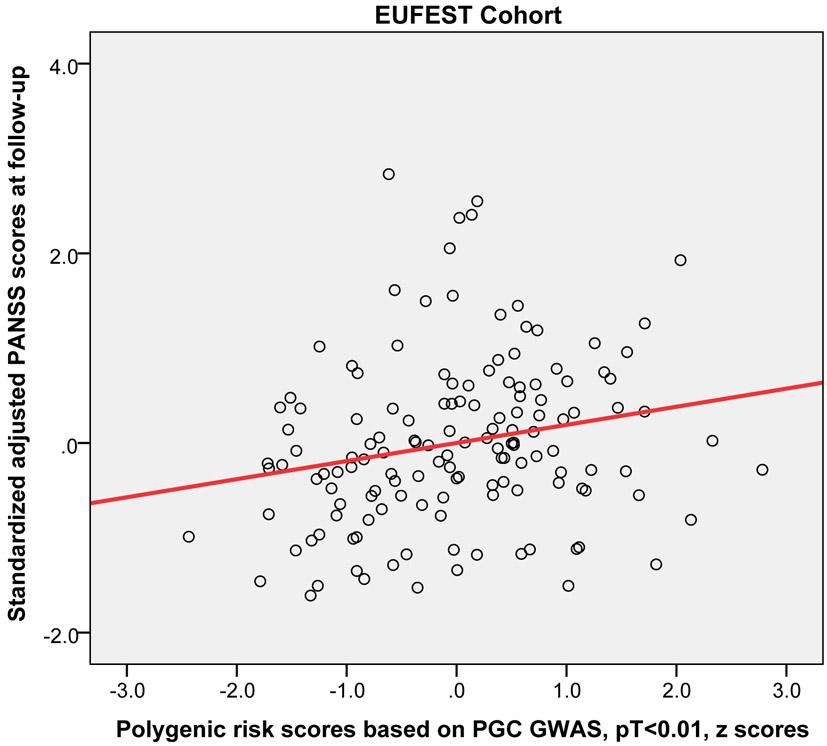
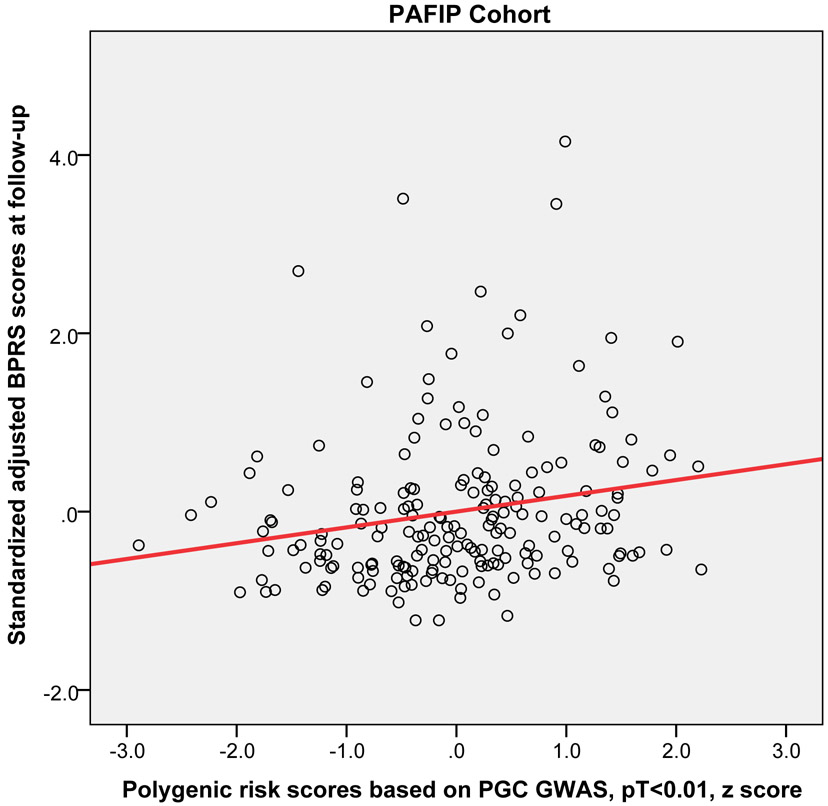
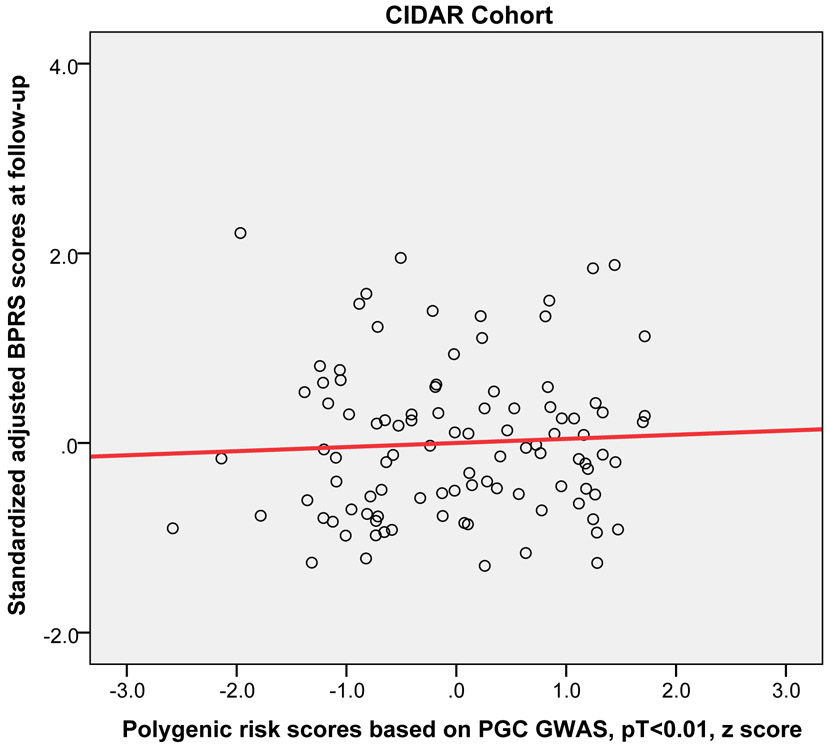
Higher PRS (at PT<0.01) significantly predicted worse outcome (i.e, higher symptoms at the 12-week or 3-month follow-up) across the three replication cohorts (pooled partial r = 0.15, p = 0.019). Moreover, this relationship was statistically significant in the EUFEST and PAFIP cohorts individually, explaining 3.5% and 3.7% of variance respectively (Table 2). Figure 2 shows the scatter plot and fitted regression line of PRS at PT<0.01 on adjusted symptom scores at the 12-week follow-up. Importantly, these results were not simply a function of PRS-related differences in baseline symptoms; PRS was not significantly correlated with baseline total symptoms in any of the four cohorts (p=.23, p=.52, p=.15, p=.43, respectively). As an exploratory analysis, PRS at other p-value thresholds were also used to predict antipsychotic efficacy in the same regression model (see Supplemental Table S1).
Table 2.
The results of hierarchical linear regression using PRS at PT <0.01 to predict symptom scores at the 12-week or 3-month follow-up while controlling for age, sex, baseline symptom score, and genomic principal components (in the ZHH-FE and CIDAR cohorts).
| N | Beta | Partial r | p (2-tailed) | R2 change | |
|---|---|---|---|---|---|
| ZHH-FE | 77 | .680 | .293 | .013 | 8.1% |
| EUFEST | 141 | .190 | .212 | .012 | 3.5% |
| PAFIP | 192 | .195 | .199 | .006 | 3.7% |
| CIDAR | 100 | −.013 | −.005 | NS | 0% |
Note: ZHH-FE: Zucker Hillside Hospital First Episode Schizophrenia trial; EUFEST: European First Episode Schizophrenia Trial; PAFIP: the Programa Asistencial Fases Iniciales de Psicosis de Cantabria, Spain; CIDAR: the clinical trial as part of the Center for Intervention Development and Applied Research at Zucker Hillside Hospital. PRS: Polygenic Risk Score. PT: P-value threshold.
Figure 2.
Scatter plots with linear regression line of polygenic risk scores (PT < 0.01) predicting standardized adjusted symptom scores at 12-week or 3-month follow-up controlling for age, sex, baseline symptom score, and genomic principal components (in the ZHH-FE and CIDAR cohorts). ZHH-FE: Zucker Hillside Hospital First Episode Schizophrenia trial; EUFEST: European First Episode Schizophrenia Trial; PAFIP: the Programa Asistencial Fases Iniciales de Psicosis de Cantabria, Spain; CIDAR: the clinical trial as part of the Center for Intervention Development and Applied Research at Zucker Hillside Hospital.
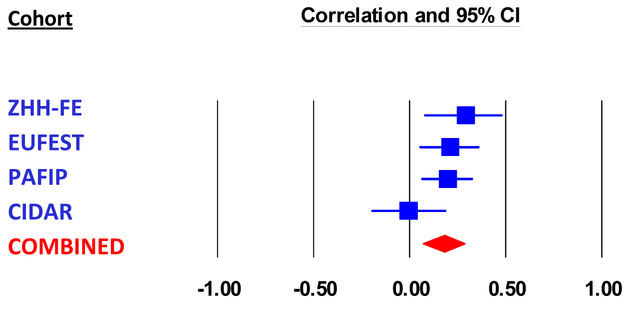
Combining the four cohorts in a meta-analysis with a random effects model, PRS (at PT<0.01) was significantly predictive of 12-week symptom scores (pooled partial correlation coefficient=0.18, p=0.002, total n = 510; Figure 3). Heterogeneity measures for the meta-analysis showed Q = 4.68, df=3, p=0.20, and I2 = 36%, indicating relatively homogeneous findings. The overall results remained significant when only European ancestry individuals were included in the meta-analysis (pooled partial r = 0.19, p<0.001, total n=387).
Figure 3.
Meta-analysis of the association between PRS at PT <0.01 and symptom scores at 12-week follow-up in four cohorts. Effect size was partial correlation coefficient after controlling for age, sex, baseline symptom score, and genomic principal components (in the ZHH-FE and CIDAR cohorts). ZHH-FE: Zucker Hillside Hospital First Episode Schizophrenia trial; EUFEST: European First Episode Schizophrenia Trial; PAFIP: the Programa Asistencial Fases Iniciales de Psicosis de Cantabria, Spain; CIDAR: the clinical trial as part of the Center for Intervention Development and Applied Research at Zucker Hillside Hospital.
To test the specificity of schizophrenia PRS predicting antipsychotic drug response, we repeated the same analysis using polygenic risk scores for type 2 diabetes based on the GWAS findings from the DIAGRAM (DIAbetes Genetics Replication And Meta-analysis) consortium(34), and polygenic risk scores for human height based on the GWAS findings from the GIANT consortium(35). Neither of these polygenic risk scores, at any PT threshold, significantly predicted symptom change in any of the four cohorts (all p>.05; mean p=0.69, median p=0.74).
To rule out the potential confounding effects of early drop-out, we ran the analysis for completers only in the ZHH-FE, EUFEST, and CIDAR cohorts excluding subjects who dropped out prior to the end of the study. (The PAFIP analysis was already completers-only based on the original design of that trial). The results were essentially unchanged.
Clinical Implications
To explore clinical significance of this finding, response rate was calculated in each cohort with the definition of treatment response as 50% or more reduction in total symptoms scores (either BPRS or PANSS) from baseline to 12-week follow-up. Each cohort was divided into high versus low PRS groups with a median split. Combining the four cohorts, the response rate was 60.9% (154 out of 253) in the low PRS group, compared to 52.1% (134 out of 257) in the high PRS group, χ2 = 3.95, df = 1, p = 0.047, odds ratio (OR) = 1.43. Because it is not possible to control for genomic principal components in this categorical analysis, we repeated the analysis for cohorts consisting of European ancestry only (i.e., EUFEST and PAFIP). Combining the two cohorts, the response rate in the low PRS group was 61.8% (102/165), while it was 45.8% (77/168) in the high PRS group (χ2 = 8.56, df = 1, p = 0.0034, OR = 1.91). Supplemental Table 2 provides the response rate in each cohort, separated by Caucasians and non-Caucasians.
Discussion
In multiple cohorts of first-episode patients with non-affective psychosis, we found that schizophrenia polygenic risk scores were significantly predictive of antipsychotic drug efficacy, with higher PRS associated with poorer treatment response. These results suggest that polygenic burden may impact severity of illness, in addition to reflecting risk for developing psychosis. To the best of our knowledge, this is the first study predicting antipsychotic efficacy based on schizophrenia PRS examining patients undergoing initial treatment for a first episode of illness and utilizing multiple cohorts for replication of effects.
Only a few prior studies have examined the relationship of PRS to treatment response. Consistent with our findings, a cross-sectional study reported significantly higher PRS in patients with treatment-resistant schizophrenia (as indexed by taking clozapine and who also had early, insidious onset and poor premorbid social function) as compared to patients who had never been prescribed clozapine.(17) However, in the same study, clozapine responders had higher schizophrenia polygene scores than non-responders to clozapine, suggesting that treatment with clozapine may be an important (and perhaps under-utilized) treatment option for patients with high PRS. A second cross-sectional study reported a similar trend, with clozapine initiation associated with elevated PRS(18). However, it should be noted that results fell just short of statistical significance (adjusted hazard ratio=1.23; 95% confidence interval=0.97-1.56), albeit with smaller sample size (n=105 clozapine patients) compared to the prior study (n=434 clozapine patients)(17). Notably, the association between PRS and clinical outcome was weaker (and nonsignificant) for more broadly defined treatment resistance based on chart history, indicating the importance of prospective studies(18). The only longitudinal study examining the relationship of PRS to treatment response demonstrated a paradoxical inverse relationship, such that higher scores were associated with greater reduction in symptoms after six weeks of treatment with lurasidone(23). It is possible that ascertainment criteria of this lurasidone clinical trial may have affected results, insofar as treatment-resistant patients were explicitly excluded, and patients with good clinical outcomes on standard treatments would not have enrolled in Phase III clinical trial of a novel antipsychotic.
Studies of patients in the first episode have the advantage of examining the full range of clinical trajectories of schizophrenia, before patients become lost to research due to either very good or very bad outcomes.(22) Only two reports have examined PRS in the context of first episode psychosis(36, 37). In contrast to the present study, both of these reports included patients with affective as well as non-affective psychosis, but results are largely consistent with the present findings. The first study revealed higher schizophrenia PRS in patients ultimately diagnosed with schizophrenia relative to those with affective psychoses(31). The second study, although longitudinal, did not directly report on treatment-related changes; nevertheless, higher PRS was significantly and positively correlated with PANSS total scores after one month of treatment(37).
Pharmacogenetic studies of antipsychotic drug response have typically focused on individual genes and SNPs in the candidate gene approach. Although a few genes have been reported to predict antipsychotic efficacy, such as DRD2(11, 38), HTR2A(39, 40), and genes in the glutamate system(41), most SNPs had small effect sizes, few have been convincingly replicated (10,11) and their clinical significance is questionable. Although dopamine D2 receptor antagonism is the common, and probably necessary, mechanism of action for antipsychotic drugs, these agents bind to many different receptors of various neurotransmitters(42), and it is very likely that some of these may involve in antipsychotic drug response(43). Perhaps more importantly, many of the drug effects may be from down-stream reactions within the dopamine signaling pathway. Therefore, the examination of multiple genes is important because this may help capture the potential down-stream effects from antipsychotic drugs. In addition, PRS represents the total genetic burden of liability to schizophrenia. Conceivably, higher genetic burden may implicate a broader range of etiopathophysiologic mechanisms, thereby rendering patients less responsive to drug treatment based primarily on a single mechanism of action (dopaminergic blockade). As such, the PRS approach may be useful in both practical and theoretical sense in predicting clinical treatment response.
There are several limitations in the present study. PRS is a weighted sum of risk alleles an individual carries. Many of the SNPs included in PRS may not be relevant to antipsychotic drug response, and inclusion of these could dilute the signal. We observed that statistical association was generally significant using PRS thresholds of PT≥0.01, suggesting that thousands of SNPs are required in order to saturate the relevant signal, while use of only SNPs attaining genome-wide significance in the PGC schizophrenia GWAS was insufficient to capture this variance. However, we currently do not have sufficient biological knowledge or statistical techniques to ascertain which SNPs are relevant and which are contributing noise. In addition, the four cohorts of patients were treated with various antipsychotic drugs which could increase the heterogeneity in outcomes, thereby decreasing our ability to detect significant signals. In the future, a very large sample of first-episode patients undergoing a single drug treatment would be required to discover which genetic variants are involved in antipsychotic drug response. Finally, PRS was not predictive of antipsychotic treatment response in the CIDAR cohort. If the true effect size is most accurately reflected in the meta-analytic result (r=.18), then a sample with n=100 (such as CIDAR) would only have power of .42 to detect a significant effect. Therefore, the failure to replicate in the CIDAR cohort was most likely due to chance variation, possibly exacerbated by the multi-ethnic nature of the sample. Notably, the overall pooled effect size is within the 95% confidence interval of the effect size in the CIDAR cohort, so this sample is not truly an outlier. At the same time, the effect size observed in the initial discovery cohort was substantially larger than the remaining cohorts, perhaps reflective of the winner’s curse. Given these variable results, it is noteworthy that the meta-analytic effect size (3.24%) is comparable to the effect sizes of the two largest (and most homogeneous studies (3.5-3.7%).
Future studies with larger samples may also result in the ability to identify a PRS cutoff with sufficient explanatory power to attain clinical utility. In the present study, we observed an odds ratio of nearly 2 for dichotomized treatment response in patients with low PRS compared to patients with high PRS. While this effect size is insufficient to guide clinical decision making, a recent large-scale study of PRS in bipolar disorder demonstrates how modest effect sizes may still allow clinical utility at the extremes(44). With a sample size of 2586 patients, the International Consortium on Lithium Genetics (ConLi+Gen) was able to divide the cohort into deciles on the basis of PRS, whereas the present study was limited to a median split due to relatively smaller sample size. In the ConLi+Gen study, bipolar patients in the lowest decile of schizophrenia PRS had a nearly 3.5-fold better response rate to lithium, compared to patients in the highest decile of PRS. Notably, a median split of the ConLi+Gen data would have provided an odds ratio of only 1.68, which is weaker than that observed in the present study. Given the linear relationships observed in the present study (Figure 2), it is reasonable to hypothesize that a larger sample size could provide an upper cutoff with strong prognostic ability. In this regard, PRS may ultimately be a more flexible and powerful biomarker than individual SNPs, which only permit 3 genotypic classifications. However, if greater schizophrenia polygenic burden is associated with poorer response to all conventional treatments, enhanced utilization of clozapine and/or novel therapeutic approaches(45) will be even more urgently needed for this subpopulation.
Supplementary Material
Disclosure
This work was supported in part by the National Institutes of Health (K23MH097108 to J.P.Z., R21MH099868 to T.L., P30MH090590 to J.M.K, and P50MH080173 to A.K.M) and by NARSAD Young Investigator Grants to J.P.Z. from the Brain & Behavior Research Foundation.
Dr. Zhang has received grant support from the National Institute of Mental Health, Brain & Behavioral Research Foundation, and Genomind, Inc.
Dr. Robinson has been a consultant to Asubio, Costello Medical Consulting, Innovative Science Solutions, Janssen, Neurocrine, Otsuka and Shire and he has received research support from Otsuka.
Mr. Yu and Dr. Gallego have nothing to disclose.
Dr. Kahn has received honoraria from Alkermes, Minerva neuroscience, Lundbeck, Gedeon Richter, and Janssen-Cilag.
Prof. Crespo-Facorro was supported by the Ministerio de Ciencia e Innovación (MICINN; https://sede.micinn.gob.es/facilita/), Madrid, in the coordinated project SAF2010-20840-C02-01/02 and by MINECO (SAF2013-46292-R). He has received in the last three years honoraria for his participation as a speaker at educational events from Otsuka, Lundbeck and Johnson & Johnson and has been a consultant and/or advisor to or has received honoraria from: Alkermes, Lundbeck, Otsuka, Casen Recordati and Teva.
Dr. Kane has been a consultant for or received honoraria from Alkermes, Eli Lilly, EnVivo Pharmaceuticals (Forum), Forest (Allergan), Genentech, H. Lundbeck. Intracellular Therapies, Janssen Pharmaceutica, Johnson and Johnson, Neurocrine, Otsuka, Pierre Fabre, Reviva, Roche, Sunovion, Takeda and Teva. He has received grant support from Otsuka and Janssen. He has participated in Advisory Boards for Alkermes, Intracellular Therapies, Lundbeck, Neurocrine, Otsuka, Pierre Fabre, Takeda, and Teva. Dr. Kane is a Shareholder in MedAvante, Inc., Vanguard Research Group and LB Pharmaceuticals, Inc.
Dr. Malhotra has received grant support from the National Institute of Mental Health. Dr. Malhotra is a consultant to Genomind, Inc and Concert Pharma, and is on the advisory board of InformedDNA.
Dr. Lencz has received grant support from the National Institute of Mental Health, Brain & Behavioral Research Foundation, and the US-Israel Binational Science Foundation.
References
- 1.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sullivan PF, Agrawal A, Bulik CM, Andreassen OA, Borglum AD, Breen G, Cichon S, Edenberg HJ, Faraone SV, Gelernter J, Mathews CA, Nievergelt CM, Smoller JW, O’Donovan MC, Psychiatric Genomics C. Psychiatric Genomics: An Update and an Agenda. Am J Psychiatry. 2017:appiajp201717030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Euesden J, Lewis CM, O’Reilly PF. PRSice: Polygenic Risk Score software. Bioinformatics. 2015;31:1466–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee SH, Goddard ME, Wray NR, Visscher PM. A better coefficient of determination for genetic profile analysis. Genetic epidemiology. 2012;36:214–224. [DOI] [PubMed] [Google Scholar]
- 6.Kahn RS, Sommer IE, Murray RM, Meyer-Lindenberg A, Weinberger DR, Cannon TD, O’Donovan M, Correll CU, Kane JM, van Os J, Insel TR. Schizophrenia. Nat Rev Dis Primers. 2015;1:15067. [DOI] [PubMed] [Google Scholar]
- 7.Zhang JP: The benefits of antipsychotic drugs: Symptom control and improved quality of life in Life Threatening Effects of Antipsychotic Drugs. Edited by Manu P, Flanagan RJ, Ronaldson KJ. London, United Kingdom, Elsevier; 2016. pp. 295–309. [Google Scholar]
- 8.Conley RR, Kelly DL. Management of treatment resistance in schizophrenia. Biol Psychiatry. 2001;50:898–911. [DOI] [PubMed] [Google Scholar]
- 9.Lieberman JA, Stroup TS, McEvoy JP, Swartz MS, Rosenheck RA, Perkins DO, Keefe RS, Davis SM, Davis CE, Lebowitz BD, Severe J, Hsiao JK. Effectiveness of antipsychotic drugs in patients with chronic schizophrenia. N Engl J Med 2005;353:1209–1223. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J-P, Malhotra AK. Pharmacogenetics and antipsychotics: therapeutic efficacy and side effects prediction. Expert Opinion on Drug Metabolism & Toxicology. 2011;7:9–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang JP, Lencz T, Malhotra AK. D2 receptor genetic variation and clinical response to antipsychotic drug treatment: a meta-analysis. Am J Psychiatry. 2010;167:763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang E, Zai CC, Lisoway A, Maciukiewicz M, Felsky D, Tiwari AK, Bishop JR, Ikeda M, Molero P, Ortuno F, Porcelli S, Samochowiec J, Mierzejewski P, Gao S, Crespo-Facorro B, Pelayo-Teran JM, Kaur H, Kukreti R, Meltzer HY, Lieberman JA, Potkin SG, Muller DJ, Kennedy JL. Catechol-O-Methyltransferase Val158Met Polymorphism and Clinical Response to Antipsychotic Treatment in Schizophrenia and Schizo-Affective Disorder Patients: a Meta-Analysis. Int J Neuropsychopharmacol 2016;19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J-P, Malhotra AK. Pharmacogenomics of Antipsychotic Drugs. Current Treatment Options in Psychiatry. 2017;4:127–138. [Google Scholar]
- 14.Crespo-Facorro B, de la Foz VO, Ayesa-Arriola R, Perez-Iglesias R, Mata I, Suarez-Pinilla P, Tabares-Seisdedos R, Vazquez-Barquero JL. Prediction of acute clinical response following a first episode of non affective psychosis: Results of a cohort of 375 patients from the Spanish PAFIP study. Prog Neuropsychopharmacol Biol Psychiatry. 2013;44:162–167. [DOI] [PubMed] [Google Scholar]
- 15.Hassan AN, De Luca V. The effect of lifetime adversities on resistance to antipsychotic treatment in schizophrenia patients. Schizophrenia research. 2015;161:496–500. [DOI] [PubMed] [Google Scholar]
- 16.Seibert TM, Fan CC, Wang Y, Zuber V, Karunamuni R, Parsons JK, Eeles RA, Easton DF, Kote-Jarai Z, Al Olama AA, Garcia SB, Muir K, Gronberg H, Wiklund F, Aly M, Schleutker J, Sipeky C, Tammela TL, Nordestgaard BG, Nielsen SF, Weischer M, Bisbjerg R, Roder MA, Iversen P, Key TJ, Travis RC, Neal DE, Donovan JL, Hamdy FC, Pharoah P, Pashayan N, Khaw KT, Maier C, Vogel W, Luedeke M, Herkommer K, Kibel AS, Cybulski C, Wokolorczyk D, Kluzniak W, Cannon-Albright L, Brenner H, Cuk K, Saum KU, Park JY, Sellers TA, Slavov C, Kaneva R, Mitev V, Batra J, Clements JA, Spurdle A, Teixeira MR, Paulo P, Maia S, Pandha H, Michael A, Kierzek A, Karow DS, Mills IG, Andreassen OA, Dale AM, Consortium* P. Polygenic hazard score to guide screening for aggressive prostate cancer: development and validation in large scale cohorts. BMJ. 2018;360:j5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frank J, Lang M, Witt SH, Strohmaier J, Rujescu D, Cichon S, Degenhardt F, Nothen MM, Collier DA, Ripke S, Naber D, Rietschel M. Identification of increased genetic risk scores for schizophrenia in treatment-resistant patients. Mol Psychiatry. 2015;20:150–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wimberley T, Gasse C, Meier SM, Agerbo E, MacCabe JH, Horsdal HT. Polygenic Risk Score for Schizophrenia and Treatment-Resistant Schizophrenia. Schizophrenia bulletin. 2017;43:1064–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang JP, Lencz T, Geisler S, DeRosse P, Bromet EJ, Malhotra AK. Genetic variation in BDNF is associated with antipsychotic treatment resistance in patients with schizophrenia. Schizophrenia research. 2013;146:285–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cargnin S, Massarotti A, Terrazzino S. BDNF Val66Met and clinical response to antipsychotic drugs: A systematic review and meta-analysis. European psychiatry : the journal of the Association of European Psychiatrists. 2016;33:45–53. [DOI] [PubMed] [Google Scholar]
- 21.Zhang JP, Lencz T, Zhang RX, Nitta M, Maayan L, John M, Robinson DG, Fleischhacker WW, Kahn RS, Ophoff RA, Kane JM, Malhotra AK, Correll CU. Pharmacogenetic Associations of Antipsychotic Drug-Related Weight Gain: A Systematic Review and Meta-analysis. Schizophrenia bulletin. 2016;42:1418–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malhotra AK, Zhang JP, Lencz T. Pharmacogenetics in psychiatry: translating research into clinical practice. Mol Psychiatry. 2012;17:760–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Yoshikawa A, Brennan MD, Ramsey TL, Meltzer HY. Genetic predictors of antipsychotic response to lurasidone identified in a genome wide association study and by schizophrenia risk genes. Schizophrenia research. 2017. [DOI] [PubMed] [Google Scholar]
- 24.Robinson DG, Woerner MG, Napolitano B, Patel RC, Sevy SM, Gunduz-Bruce H, Soto-Perello JM, Mendelowitz A, Khadivi A, Miller R, McCormack J, Lorell BS, Lesser ML, Schooler NR, Kane JM. Randomized comparison of olanzapine versus risperidone for the treatment of first-episode schizophrenia: 4-month outcomes. Am J Psychiatry. 2006;163:2096–2102. [DOI] [PubMed] [Google Scholar]
- 25.Overall JE, Gorham DR. The brief psychiatric rating scale. Psychol Rep 1962;10:799–812. [Google Scholar]
- 26.Gallego JA, Robinson DG, Sevy SM, Napolitano B, McCormack J, Lesser ML, Kane JM. Time to treatment response in first-episode schizophrenia: should acute treatment trials last several months? J Clin Psychiatry. 2011;72:1691–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kahn RS, Fleischhacker WW, Boter H, Davidson M, Vergouwe Y, Keet IP, Gheorghe MD, Rybakowski JK, Galderisi S, Libiger J, Hummer M, Dollfus S, Lopez-Ibor JJ, Hranov LG, Gaebel W, Peuskens J, Lindefors N, Riecher-Rossler A, Grobbee DE. Effectiveness of antipsychotic drugs in first-episode schizophrenia and schizophreniform disorder: an open randomised clinical trial. Lancet. 2008;371:1085–1097. [DOI] [PubMed] [Google Scholar]
- 28.<j/>Guy W: ECDEU Assessment Manual for Psychopharmacology. US Department of Health, Education, and Welfare publication ADM 76–338. Washington, DC, National Institute of Mental Health; 1976. pp. 217–222. [Google Scholar]
- 29.Crespo-Facorro B, Perez-Iglesias R, Ramirez-Bonilla M, Martinez-Garcia O, Llorca J, Luis Vazquez-Barquero J. A practical clinical trial comparing haloperidol, risperidone, and olanzapine for the acute treatment of first-episode nonaffective psychosis. J Clin Psychiatry. 2006;67:1511–1521. [DOI] [PubMed] [Google Scholar]
- 30.Crespo-Facorro B, Ortiz-Garcia de la Foz V, Mata I, Ayesa-Arriola R, Suarez-Pinilla P, Valdizan EM, Vazquez-Barquero JL, Perez-Iglesias R. Aripiprazole, Ziprasidone and Quetiapine in the treatment of first-episode nonaffective psychosis: A 12-week randomized, flexible-dose, open-label trial. Schizophrenia research. 2013;147:375–382. [DOI] [PubMed] [Google Scholar]
- 31.Robinson DG, Gallego JA, John M, Petrides G, Hassoun Y, Zhang JP, Lopez L, Braga RJ, Sevy SM, Addington J, Kellner CH, Tohen M, Naraine M, Bennett N, Greenberg J, Lencz T, Correll CU, Kane JM, Malhotra AK. A Randomized Comparison of Aripiprazole and Risperidone for the Acute Treatment of First-Episode Schizophrenia and Related Disorders: 3-Month Outcomes. Schizophrenia bulletin. 2015;41:1227–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics. 2009;5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genomes Project C, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, Prokopenko I, Kang HM, Dina C, Esko T, Fraser RM, Kanoni S, Kumar A, Lagou V, Langenberg C, Luan J, Lindgren CM, Muller-Nurasyid M, Pechlivanis S, Rayner NW, Scott LJ, Wiltshire S, Yengo L, Kinnunen L, Rossin EJ, Raychaudhuri S, Johnson AD, Dimas AS, Loos RJ, Vedantam S, Chen H, Florez JC, Fox C, Liu CT, Rybin D, Couper DJ, Kao WH, Li M, Cornelis MC, Kraft P, Sun Q, van Dam RM, Stringham HM, Chines PS, Fischer K, Fontanillas P, Holmen OL, Hunt SE, Jackson AU, Kong A, Lawrence R, Meyer J, Perry JR, Platou CG, Potter S, Rehnberg E, Robertson N, Sivapalaratnam S, Stancakova A, Stirrups K, Thorleifsson G, Tikkanen E, Wood AR, Almgren P, Atalay M, Benediktsson R, Bonnycastle LL, Burtt N, Carey J, Charpentier G, Crenshaw AT, Doney AS, Dorkhan M, Edkins S, Emilsson V, Eury E, Forsen T, Gertow K, Gigante B, Grant GB, Groves CJ, Guiducci C, Herder C, Hreidarsson AB, Hui J, James A, Jonsson A, Rathmann W, Klopp N, Kravic J, Krjutskov K, Langford C, Leander K, Lindholm E, Lobbens S, Mannisto S, Mirza G, Muhleisen TW, Musk B, Parkin M, Rallidis L, Saramies J, Sennblad B, Shah S, Sigurethsson G, Silveira A, Steinbach G, Thorand B, Trakalo J, Veglia F, Wennauer R, Winckler W, Zabaneh D, Campbell H, van Duijn C, Uitterlinden AG, Hofman A, Sijbrands E, Abecasis GR, Owen KR, Zeggini E, Trip MD, Forouhi NG, Syvanen AC, Eriksson JG, Peltonen L, Nothen MM, Balkau B, Palmer CN, Lyssenko V, Tuomi T, Isomaa B, Hunter DJ, Qi L, Wellcome Trust Case Control C, Meta-Analyses of G, Insulin-related traits Consortium I, Genetic Investigation of ATC, Asian Genetic Epidemiology Network-Type 2 Diabetes C, South Asian Type 2 Diabetes C, Shuldiner AR, Roden M, Barroso I, Wilsgaard T, Beilby J, Hovingh K, Price JF, Wilson JF, Rauramaa R, Lakka TA, Lind L, Dedoussis G, Njolstad I, Pedersen NL, Khaw KT, Wareham NJ, Keinanen-Kiukaanniemi SM, Saaristo TE, Korpi-Hyovalti E, Saltevo J, Laakso M, Kuusisto J, Metspalu A, Collins FS, Mohlke KL, Bergman RN, Tuomilehto J, Boehm BO, Gieger C, Hveem K, Cauchi S, Froguel P, Baldassarre D, Tremoli E, Humphries SE, Saleheen D, Danesh J, Ingelsson E, Ripatti S, Salomaa V, Erbel R, Jockel KH, Moebus S, Peters A, Illig T, de Faire U, Hamsten A, Morris AD, Donnelly PJ, Frayling TM, Hattersley AT, Boerwinkle E, Melander O, Kathiresan S, Nilsson PM, Deloukas P, Thorsteinsdottir U, Groop LC, Stefansson K, Hu F, Pankow JS, Dupuis J, Meigs JB, Altshuler D, Boehnke M, McCarthy MI, Replication DIG, Meta-analysis C. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 2012;44:981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, Ferreira T, Wood AR, Weyant RJ, Segre AV, Speliotes EK, Wheeler E, Soranzo N, Park JH, Yang J, Gudbjartsson D, Heard-Costa NL, Randall JC, Qi L, Vernon Smith A, Magi R, Pastinen T, Liang L, Heid IM, Luan J, Thorleifsson G, Winkler TW, Goddard ME, Sin Lo K, Palmer C, Workalemahu T, Aulchenko YS, Johansson A, Zillikens MC, Feitosa MF, Esko T, Johnson T, Ketkar S, Kraft P, Mangino M, Prokopenko I, Absher D, Albrecht E, Ernst F, Glazer NL, Hayward C, Hottenga JJ, Jacobs KB, Knowles JW, Kutalik Z, Monda KL, Polasek O, Preuss M, Rayner NW, Robertson NR, Steinthorsdottir V, Tyrer JP, Voight BF, Wiklund F, Xu J, Zhao JH, Nyholt DR, Pellikka N, Perola M, Perry JR, Surakka I, Tammesoo ML, Altmaier EL, Amin N, Aspelund T, Bhangale T, Boucher G, Chasman DI, Chen C, Coin L, Cooper MN, Dixon AL, Gibson Q, Grundberg E, Hao K, Juhani Junttila M, Kaplan LM, Kettunen J, Konig IR, Kwan T, Lawrence RW, Levinson DF, Lorentzon M, McKnight B, Morris AP, Muller M, Suh Ngwa J, Purcell S, Rafelt S, Salem RM, Salvi E, Sanna S, Shi J, Sovio U, Thompson JR, Turchin MC, Vandenput L, Verlaan DJ, Vitart V, White CC, Ziegler A, Almgren P, Balmforth AJ, Campbell H, Citterio L, De Grandi A, Dominiczak A, Duan J, Elliott P, Elosua R, Eriksson JG, Freimer NB, Geus EJ, Glorioso N, Haiqing S, Hartikainen AL, Havulinna AS, Hicks AA, Hui J, Igl W, Illig T, Jula A, Kajantie E, Kilpelainen TO, Koiranen M, Kolcic I, Koskinen S, Kovacs P, Laitinen J, Liu J, Lokki ML, Marusic A, Maschio A, Meitinger T, Mulas A, Pare G, Parker AN, Peden JF, Petersmann A, Pichler I, Pietilainen KH, Pouta A, Ridderstrale M, Rotter JI, Sambrook JG, Sanders AR, Schmidt CO, Sinisalo J, Smit JH, Stringham HM, Bragi Walters G, Widen E, Wild SH, Willemsen G, Zagato L, Zgaga L, Zitting P, Alavere H, Farrall M, McArdle WL, Nelis M, Peters MJ, Ripatti S, van Meurs JB, Aben KK, Ardlie KG, Beckmann JS, Beilby JP, Bergman RN, Bergmann S, Collins FS, Cusi D, den Heijer M, Eiriksdottir G, Gejman PV, Hall AS, Hamsten A, Huikuri HV, Iribarren C, Kahonen M, Kaprio J, Kathiresan S, Kiemeney L, Kocher T, Launer LJ, Lehtimaki T, Melander O, Mosley TH Jr., Musk AW, Nieminen MS, O’Donnell CJ, Ohlsson C, Oostra B, Palmer LJ, Raitakari O, Ridker PM, Rioux JD, Rissanen A, Rivolta C, Schunkert H, Shuldiner AR, Siscovick DS, Stumvoll M, Tonjes A, Tuomilehto J, van Ommen GJ, Viikari J, Heath AC, Martin NG, Montgomery GW, Province MA, Kayser M, Arnold AM, Atwood LD, Boerwinkle E, Chanock SJ, Deloukas P, Gieger C, Gronberg H, Hall P, Hattersley AT, Hengstenberg C, Hoffman W, Lathrop GM, Salomaa V, Schreiber S, Uda M, Waterworth D, Wright AF, Assimes TL, Barroso I, Hofman A, Mohlke KL, Boomsma DI, Caulfield MJ, Cupples LA, Erdmann J, Fox CS, Gudnason V, Gyllensten U, Harris TB, Hayes RB, Jarvelin MR, Mooser V, Munroe PB, Ouwehand WH, Penninx BW, Pramstaller PP, Quertermous T, Rudan I, Samani NJ, Spector TD, Volzke H, Watkins H, Wilson JF, Groop LC, Haritunians T, Hu FB, Kaplan RC, Metspalu A, North KE, Schlessinger D, Wareham NJ, Hunter DJ, O’Connell JR, Strachan DP, Wichmann HE, Borecki IB, van Duijn CM, Schadt EE, Thorsteinsdottir U, Peltonen L, Uitterlinden AG, Visscher PM, Chatterjee N, Loos RJ, Boehnke M, McCarthy MI, Ingelsson E, Lindgren CM, Abecasis GR, Stefansson K, Frayling TM, Hirschhorn JN. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vassos E, Di Forti M, Coleman J, Iyegbe C, Prata D, Euesden J, O’Reilly P, Curtis C, Kolliakou A, Patel H, Newhouse S, Traylor M, Ajnakina O, Mondelli V, Marques TR, Gardner-Sood P, Aitchison KJ, Powell J, Atakan Z, Greenwood KE, Smith S, Ismail K, Pariante C, Gaughran F, Dazzan P, Markus HS, David AS, Lewis CM, Murray RM, Breen G. An Examination of Polygenic Score Risk Prediction in Individuals With First-Episode Psychosis. Biol Psychiatry. 2017;81:470–477. [DOI] [PubMed] [Google Scholar]
- 37.Sengupta SM, MacDonald K, Fathalli F, Yim A, Lepage M, Iyer S, Malla A, Joober R. Polygenic Risk Score associated with specific symptom dimensions in first-episode psychosis. Schizophrenia research. 2017;184:116–121. [DOI] [PubMed] [Google Scholar]
- 38.Zhang JP, Robinson DG, Gallego JA, John M, Yu J, Addington J, Tohen M, Kane JM, Malhotra AK, Lencz T. Association of a Schizophrenia Risk Variant at the DRD2 Locus With Antipsychotic Treatment Response in First-Episode Psychosis. Schizophrenia bulletin. 2015;41:1248–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arranz MJ, Munro J, Sham P, Kirov G, Murray RM, Collier DA, Kerwin RW. Meta-analysis of studies on genetic variation in 5-HT2A receptors and clozapine response. Schizophrenia research. 1998;32:93–99. [DOI] [PubMed] [Google Scholar]
- 40.Chen SF, Shen YC, Chen CH. HTR2A A-1438G/T102C polymorphisms predict negative symptoms performance upon aripiprazole treatment in schizophrenic patients. Psychopharmacology (Berl). 2009;205:285–292. [DOI] [PubMed] [Google Scholar]
- 41.Stevenson JM, Reilly JL, Harris MS, Patel SR, Weiden PJ, Prasad KM, Badner JA, Nimgaonkar VL, Keshavan MS, Sweeney JA, Bishop JR. Antipsychotic pharmacogenomics in first episode psychosis: a role for glutamate genes. Translational psychiatry. 2016;6:e739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyamoto S, Duncan GE, Marx CE, Lieberman JA. Treatments for schizophrenia: a critical review of pharmacology and mechanisms of action of antipsychotic drugs. Mol Psychiatry. 2005;10:79–104. [DOI] [PubMed] [Google Scholar]
- 43.Ruderfer DM, Charney AW, Readhead B, Kidd BA, Kahler AK, Kenny PJ, Keiser MJ, Moran JL, Hultman CM, Scott SA, Sullivan PF, Purcell SM, Dudley JT, Sklar P. Polygenic overlap between schizophrenia risk and antipsychotic response: a genomic medicine approach. Lancet Psychiatry. 2016;3:350–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.International Consortium on Lithium G, Amare AT, Schubert KO, Hou L, Clark SR, Papiol S, Heilbronner U, Degenhardt F, Tekola-Ayele F, Hsu YH, Shekhtman T, Adli M, Akula N, Akiyama K, Ardau R, Arias B, Aubry JM, Backlund L, Bhattacharjee AK, Bellivier F, Benabarre A, Bengesser S, Biernacka JM, Birner A, Brichant-Petitjean C, Cervantes P, Chen HC, Chillotti C, Cichon S, Cruceanu C, Czerski PM, Dalkner N, Dayer A, Del Zompo M, DePaulo JR, Etain B, Falkai P, Forstner AJ, Frisen L, Frye MA, Fullerton JM, Gard S, Garnham JS, Goes FS, Grigoroiu-Serbanescu M, Grof P, Hashimoto R, Hauser J, Herms S, Hoffmann P, Hofmann A, Jamain S, Jimenez E, Kahn JP, Kassem L, Kuo PH, Kato T, Kelsoe J, Kittel-Schneider S, Kliwicki S, Konig B, Kusumi I, Laje G, Landen M, Lavebratt C, Leboyer M, Leckband SG, Tortorella A, Manchia M, Martinsson L, McCarthy MJ, McElroy S, Colom F, Mitjans M, Mondimore FM, Monteleone P, Nievergelt CM, Nothen MM, Novak T, O’Donovan C, Ozaki N, Osby U, Pfennig A, Potash JB, Reif A, Reininghaus E, Rouleau GA, Rybakowski JK, Schalling M, Schofield PR, Schweizer BW, Severino G, Shilling PD, Shimoda K, Simhandl C, Slaney CM, Squassina A, Stamm T, Stopkova P, Maj M, Turecki G, Vieta E, Volkert J, Witt S, Wright A, Zandi PP, Mitchell PB, Bauer M, Alda M, Rietschel M, McMahon FJ, Schulze TG, Baune BT. Association of Polygenic Score for Schizophrenia and HLA Antigen and Inflammation Genes With Response to Lithium in Bipolar Affective Disorder: A Genome-Wide Association Study. JAMA Psychiatry. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petrides G, Malur C, Braga RJ, Bailine SH, Schooler NR, Malhotra AK, Kane JM, Sanghani S, Goldberg TE, John M, Mendelowitz A. Electroconvulsive therapy augmentation in clozapine-resistant schizophrenia: a prospective, randomized study. Am J Psychiatry. 2015;172:52–58. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.