Abstract
Fuchs' endothelial corneal dystrophy (FECD) leads to vision loss and is one of the most common inherited eye diseases. Corneal transplants are the only curative treatment available, and there is a major unmet need for treatments that are less invasive and independent of donor tissue. Most cases of FECD are associated with an expanded CUG repeat within the intronic region of TCF4 and the mutant RNA has been implicated as the cause of the disease. We previously presented preliminary data suggesting that single-stranded antisense oligonucleotides (ASOs) can inhibit CUG RNA foci in patient-derived cells and tissue. We now show that duplex RNAs and single-stranded silencing RNAs (ss-siRNAs) reduce the number of cells with foci and the number of foci per cells. Potencies are similar to those that are achieved with chemically modified ASOs designed to block foci. These data widen the potential for synthetic nucleic acids to be used to treat a widely prevalent and debilitating disease.
Keywords: RNAi, Fuchs' dystrophy, ophthalmology, ss-siRNA, siRNA
Introduction
The eye is a tempting target for nucleic acid therapeutics [1]. There is a substantial unmet need for better treatments for many eye diseases and the patient population that might benefit from such treatments is large. The volume of the eye, approximately 6 mL, is small, reducing the cost of treatment. Modified oligonucleotides might be delivered by convenient topical administration and intraocular injection is a well-established mode of delivery [2–5].
These advantages have contributed to the approval of two nucleic acid drugs. Vitravene (fomivirsen) was an antisense first generation phosphorothioate oligonucleotide that was approved in 1998 for the treatment of cytomegaloviral retinitis [6]. Macugen (pegaptanib) was an aptamer approved in 2003 for the treatment of macular degeneration [7]. Both were delivered by intraocular injection. While neither drug is currently widely used, they demonstrated that it was possible to successfully introduce beneficial therapeutic oligonucleotides to the eye.
Fuchs' Endothelial Corneal Dystrophy (FECD) is an age-related disorder that affects 4% of the population over the age of 40 [8]. FECD is characterized by degeneration of corneal endothelium, leading to progressive loss of vision. Currently, the only curative therapy is corneal transplantation. While usually effective, corneal transplantation is limited by the availability of donor corneas and access to treatment centers capable of performing the surgery [9]. Patients require a 4–6 week recovery period and may be subjected to a range of potential complications that vary depending on surgical center location. FECD patients may lack access to treatment or may suffer for many years before the disease becomes advanced enough to require surgery. If a safe and effective nonsurgical treatment was available, it would be beneficial to hundreds of thousands of patients.
The causes of FECD are not fully understood, but 70% of FECD cases are linked to an expansion of the trinucleotide CUG within intron 2 of the TCF4 gene [10–13]. Patients with myotonic dystrophy, a disease caused by CUG repeats within the DMPK gene, are also at increased risk for FECD [14–16]. The finding that CUG repeats within noncoding regions of two different genes, both cause FECD, supports the belief that expanded CUG repeat RNA is a major contributor to disease.
We have previously shown that antisense oligonucleotides (ASOs) can block foci in FECD patient-derived cells [17]. These ASOs were designed to be complementary to the CUG repeat and contained locked nucleic acid (LNA) modifications. Because the LNA modifications were spread throughout the strand and the ASOs lack the central DNA region of gapmer ASOs that allow recruitment of RNAse H, the ASOs function through a “steric block” mechanism [18]. Davidson and colleagues achieved similar effects using a 2′-O-methyl RNA-modified ASO [19].
To maximize the range of synthetic oligonucleotide designs available for blocking or eliminating CUG foci, we now report the testing of silencing agents that function through the RNAi pathway [18]. These included duplex RNAs [20] and single-stranded silencing RNAs (ss-siRNAs) [21–23]. We compare these data to inhibition of foci by heavily modified steric block ASOs. We find that all strategies can produce active compounds, providing a broad base for developing effective delivery strategies and for pursuing compounds that will possess optimal in vivo potencies.
Materials and Methods
Human corneal endothelial cell culture
The F35T corneal endothelial cell line derived from FECD patient expressing TCF4 transcript with approximately 1500 CUG repeats was a generous gift of Dr. Albert Jun (Johns Hopkins). The F45 corneal endothelial cell was a primary culture from FECD patient expressing TCF4 transcript with approximately 1500 CUG repeats and subsequently used to generate immortalized F45SV corneal endothelial cell line (SV 40-mediated immortalization by ALSTEM, Richmond, CA). The HCEN19 corneal endothelial cell line was a primary culture from a healthy donor cornea from eye bank.
The dissected Descemet's membrane monolayer was incubated with 2 mg/mL collagenase A (Roche) in culture media at 37°C for 4 h to dissociate the cells, spun down at 800 g for 5 min, and plated on culture dish precoated with fibronectin (FNC) (Athena Environmental Sciences). Cells were grown in modified Eagle's minimal essential media (OptiMEM) (ThermoFisher) supplemented with 8% fetal bovine serum, 5 ng/mL human epidermal growth factor (ThermoFisher), 20 ng/mL nerve growth factor (Fisher Scientific), 100 μg/mL bovine pituitary extract (ThermoFisher), 20 μg/mL ascorbic acid (Sigma-Aldrich), 200 mg/L calcium chloride (Sigma-Aldrich), 0.08% chondroitin sulfate (Sigma-Aldrich), 50 μg/mL gentamicin (ThermoFisher), and antibiotic/antimycotic solution (diluted 1/100) (Sigma-Aldrich). Cultures were incubated at 37°C in 5% CO2 and passaged when confluent.
Synthesis and transfection of oligonucleotides
Duplex RNAs were purchased from IDT (Newark, NJ). ss-siRNAs and ASOs were synthesized by Ionis Pharmaceutics (Carlsbad, CA) using standard solid phase coupling method as previously described [21]. Oligonucleotides were characterized by LCMS. Duplex RNAs or ss-siRNAs were transfected into cells with lipid RNAiMAX (Life Technologies) as previously described [24]. Cells were plated at a density of 300K per well of a six-well plate and transfection was performed at the same time. Cells were typically harvested 4 days after transfection for RNA fluorescence in situ hybridization (FISH) assay.
Melting temperature determination
Thermal denaturation analysis of oligonucleotides to determine melting temperature (Tm) values was carried out using a CARY Varian 100 Bio UV-Vis spectrophotometer. Duplex RNAs, or single-strand siRNA/ASO, with their complementary DNA strand were added into 0.1 M NaH2PO4 buffer (1 μM final concentration, 400 μL total volume) in a 1 cm quartz cuvette, and was annealed and melted from 15°C to 95°C at a ramp rate of 2°C/min. The UV absorbance was monitored at 260 nm. The melting temperature was calculated and averaged from at least seven technical replicates.
FISH assay
Cornea endothelial cells post oligonucleotide transfection were harvested by trypsin and replated on glass slides using a cytospin 4 centrifuge (ThermoFisher). Cells were fixed with 4% formaldehyde in 1× phosphate-buffered saline (PBS) and permeabilized with 0.2%Triton100 in 2× saline sodium citrate buffer (SCC) at 4°C for 10 min. Cells were washed with 2× SSC and wash buffer (10% formamide in 2× SSC), and then incubated with prehybridization buffer (40% formamide in 2× SSC) at 45°C for 20 min. (CAG)6CA-5′ Texas red-labeled 2′-O-methyl RNA 20-mers probe in hybridization buffer (100 mg/mL dextran sulfate and 40% formamide in 2× SSC) was added. The slides were placed in a humidified chamber and incubated in the dark at 37°C overnight. On the next day, cells were washed twice with wash buffer at 37°C for 15 min, and then stained with mounting media with DAPI (H-1500; Vector Labs).
Cells were imaged at 60× magnification using a Widefield DeltaVision microscope. Images were processed by blind deconvolution with AutoQuant X3. Visualization of RNA foci were made using ImageJ with Bio-Formats plugins. For quantification, at least 20 pictures were taken from randomly chosen microscopic fields, containing 100–300 cells for each treatment. Counting of foci was performed manually by different investigators. Statistical analysis was performed by Student's t-test, with a homoscedastic two-tailed distribution [25].
Results
Experimental design
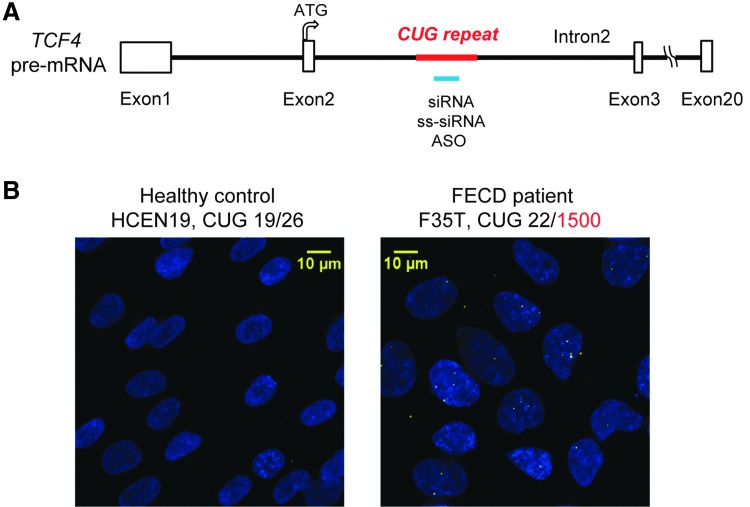
The expanded CUG repeat is within intron 2 of the TCF4 gene (Fig. 1A). The mutant RNA containing the expanded CUG repeat can be characterized by FISH and microscopy as foci that are counted to determine the average number of foci per cell (Fig. 1B). Each focus consists of a single RNA transcript, and each cell contains only a handful of RNA molecules [17].
FIG. 1.
Experimental design. (A) Scheme of TCF4 pre-mRNA intron 2 region. The colored bar shows the target region for the various nucleic acids at the CUG repeat. (B) Representative FISH images of healthy control and FECD patient-derived endothelial cell lines obtained by fluorescent microscopy. A (CAG)6CA RNA probe was used for detecting the CUGexp RNA foci. FECD, Fuchs' endothelial corneal dystrophy.
We transfected duplex RNAs, ss-siRNAs, or ASOs into FECD patient-derived corneal endothelial cells using cationic lipid. Over 100 cells were evaluated after FISH assay for each experimental condition and all experiments were done in duplicate or triplicate. Experiments were performed in F35T corneal endothelial cells, F45 primary corneal endothelial cells, or F45SV, an immortalized line derived from F45 cells. We used the F45SV immortalized cells for some experiments because primary F45 cells were difficult to culture and transfect.
Inhibition of CUGexp foci by duplex RNAs
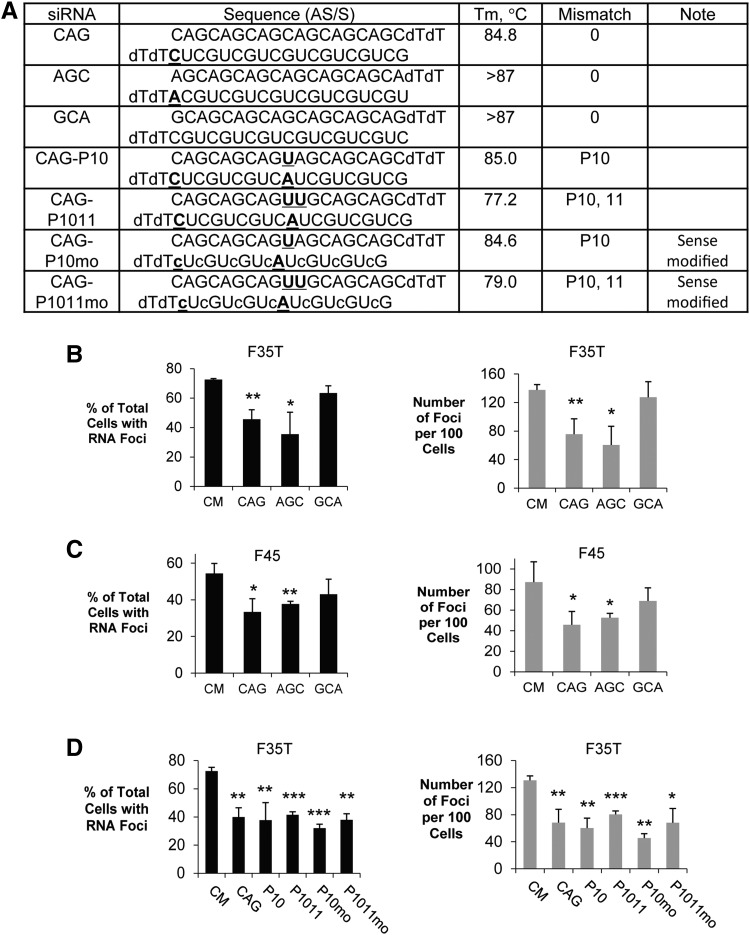
We used duplex RNAs to test the hypothesis that mutant TCF4 intronic RNA could be blocked by synthetic oligonucleotides acting through RNAi. Nineteen base (not including two terminal dT nucleotides) duplex RNAs were designed to be complementary to the expanded CUG repeat in each of three registers with sense strands beginning with C, U, or G (Fig. 2A). The antisense strand of the three duplex RNAs (CAG, AGC, and GCA) were fully complementary to the repeat region. One mismatched base was introduced into the duplex to make the 5’end of antisense strand unstable and increase its loading into the RISC complex [26].
FIG. 2.
Inhibition of CUGexp RNA foci by repeat-targeting duplex RNAs in F35T (CUG 22/1500) or F45 (CUG16/1500) endothelial cell line evaluated by fluorescent microscopy. (A) List of duplex RNAs. Mismatched base is shown in bold and underlined. 2′-O-methyl modified RNA base is in lowercase. dT, DNA base T. (B) Effect of duplex RNAs on inhibition of CUGexp RNA foci in F35T cells. (C) Effect of duplex RNAs on inhibition of CUGexp RNA foci in F45 cells. (D) Duplex RNAs with central mismatches inhibit CUGexp RNA foci in F35T cell line. Error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001 compared with noncomplementary negative control siRNA CM by Student's t-test.
Four other duplex RNAs (CAG-P10, CAG-1011, CAG-P10mo, and CAG-P1011mo) contained one or two central mismatched base within the antisense strands. The mismatch relative to the CUG repeat target was included because central mismatches prevent argonaute 2 (AGO2)-mediated cleavage of target RNA [27] without reducing the potential for recognition of expanded trinucleotide repeats [24].
This strategy of using centrally mismatched duplex RNAs has been used successfully for other expanded repeat targets, including the CAG repeat, within huntingtin mRNA [24] and the GGGGCC repeat within c9orf72 intronic RNA [28]. Previous studies have demonstrated requirement for argonaute 2 (AGO2), supporting believe that action is through RNAi [23,29,30]. Since our proposed mechanism of action requires binding but not cleavage, it avoids any potential for siRNA-induced cleavage of complementary CUG RNAs and may have the advantage of reducing off-target effects. Duplex RNAs CAG-P10mo and CAG-P1011mo have their sense strands modified with 2′-O-methyl bases to enhance potency and reduce off-target effects. Effects on foci were evaluated relative to an RNA duplex (CM) that lacked complementarity to the CUG repeat.
We observed that fully complementary duplex RNAs CAG and AGC significantly reduced the number of F35T cells with observable RNA foci and the number of foci per 100 cells (Fig. 2B). Duplex RNA GCA had no significant effect. A similar outcome was observed when duplex RNAs CAG, AGC, and GCA were introduced into F45 cells (Fig. 2C). All four mismatch-containing duplex RNAs, CAG-P10, CAG-P1011, CAG-P10mo, and CAG-P1011mo produced significant reductions in the total number of cells with foci and the number of foci per 100 cells (Fig. 2D). These data establish that duplex RNAs can block mutant TCF4 foci and that blocking does not require perfect complementarity to target.
Inhibition of CUGexp foci by ss-siRNAs
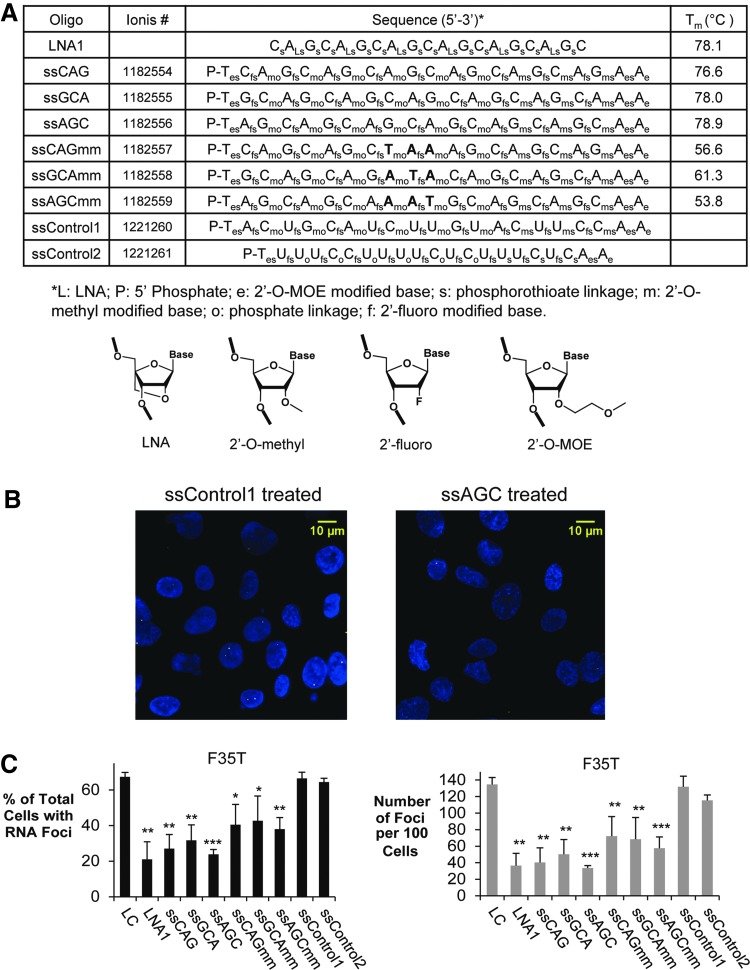
We designed ss-siRNAs ss-CAG, ss-GCA, and ss-ACG to be fully complementary to the expanded repeat in three different registers (Fig. 3A). Three other ss-siRNAs, ss-CAGmm, ss-GCAmm, and ss-ACGmm, were designed to possess three central mismatches relative to the CUG repeat target. As noted above, we had previously demonstrated that centrally located mismatches within duplex RNAs do not reduce recognition of repetitive sequences. The ss-siRNAs were composed of a mixture of 2′-O-methyl, 2′-fluoro, and 2′-O-methoxyethyl nucleotides. In other systems, we have observed that the introduction of mismatched bases into ss-siRNAs does not affect the potency ss-siRNAs [23].
FIG. 3.
ss-siRNAs inhibit RNA foci in F35T endothelial cell line. (A) Table of fully complementary or centrally mismatched ss-siRNAs targeting the CUG repeat. Mismatched bases are in bold face. Structures of the chemical modifications are below. (B) Representative FISH images showing effect of ss-siRNAs on inhibition of RNA foci in F35T cells. (C) Effect of ss-siRNAs on inhibition of RNAi foci in F35T cells. Error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001 relative to noncomplementary LC by Student's t-test. LNA1 is a CUG-targeting LNA that had been shown to inhibit foci in our previous work [17]. LC, LNA control.
As a positive control, we also included a LNA-modified ASO that was complementary to the expanded CUG repeat (LNA1) previously shown by our group to block foci and reduce the number of foci detected per cell [17]. Noncomplementary negative controls included one single-stranded LNA (LC) and two ss-siRNAs (ssControl1 and ssControl2).
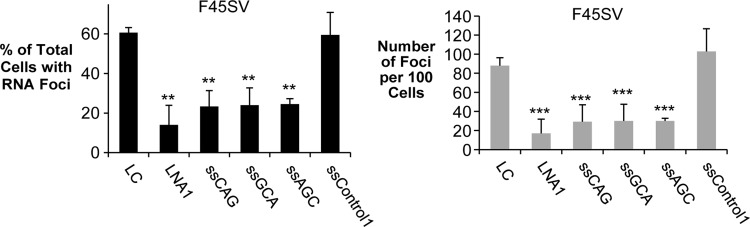
We observed that all six CUG-targeted ss-siRNAs caused significant decreases in the number of cells with foci and the number of foci per one hundred cells (Fig. 3B, C), comparable in potency to our previously reported LNA ASO LNA1. The fully complementary ss-siRNAs appeared to have higher efficacies than the ss-siRNAs that contained three mismatched bases. This difference may reflect the lower melting temperature values for hybridization to complementary targets (Fig. 3A). We observed a similar reduction in foci for a second corneal endothelial cell line (F45SV) (Fig. 4).
FIG. 4.
ss-siRNAs inhibit RNA foci in F45-SV40 endothelial cell line. **P < 0.01; ***P < 0.001 relative to noncomplementary LC by Student's t-test.
Inhibition of CUGexp foci by steric block ASOs
To provide context for evaluating duplex RNAs and ss-siRNAs, we also evaluated 18 or 16 base single-stranded ASOs that are extensively modified with 2′-O-methoxyethyl or 2′, 4′ constrained ethyl (cET)-modified nucleotides [31] (Figs. 5A and 6A). The cET modification reduces the entropic penalty paid upon binding and results in substantial increase in binding affinity for complementary targets, a phenomenon reflected in the measure Tm values for ASOs that contained cET. Single-stranded oligonucleotides that contain heavy cET and MOE modifications have not been observed to function through RNAi and do not recruit RNAi protein factors.
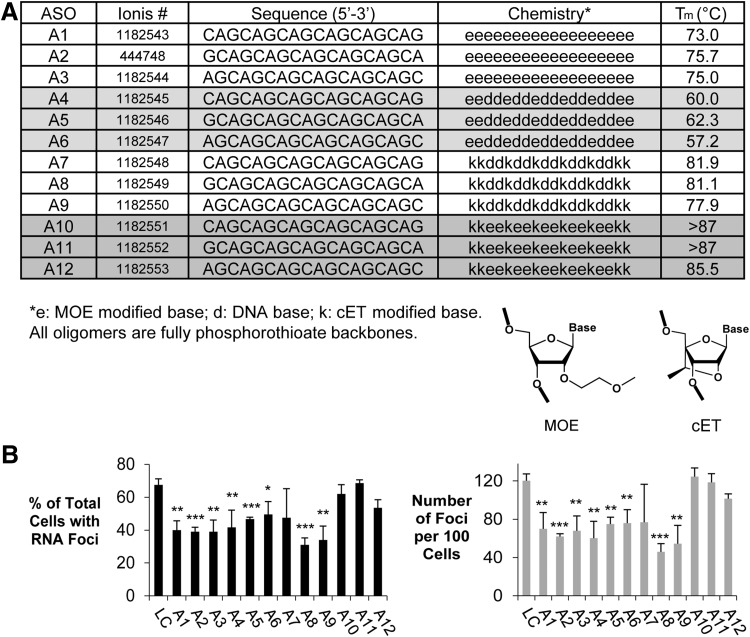
FIG. 5.
cET and MOE-modified ASOs targeting CUG repeat reduced RNA foci in F35T endothelial cells. (A). Table of 18-mer ASOs for CUG repeat. (B) Effect of ASOs on inhibition of RNA foci in F35T cells. Error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001 relative to noncomplementary LC by Student's t-test.
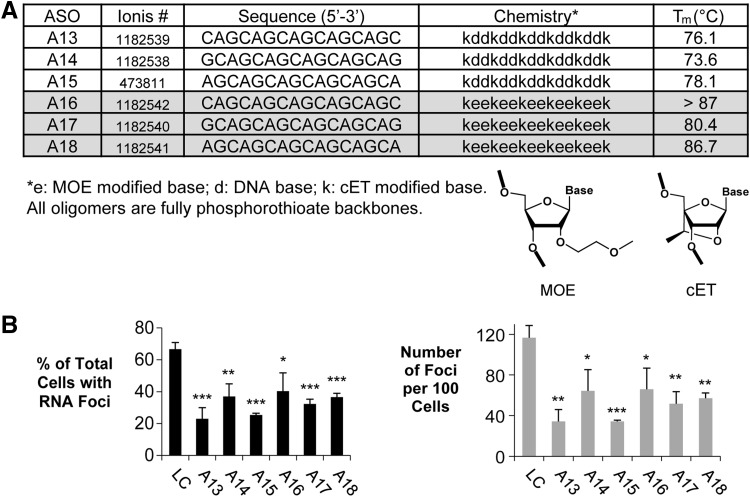
FIG. 6.
Shorter (16-mer) cET or MOE-modified ASOs reduced RNA foci in F35T endothelial cells. (A). Table of 16-mer ASOs for CUG repeat. (B) Effect of 16-mer ASOs on inhibition of RNA foci in F35T cells. Error bars represent SEM. *P < 0.05; **P < 0.01; ***P < 0.001 relative to noncomplementary LC by Student's t-test.
Most of the 18-base ASOs were active but showed efficacies that were below the three best ss-siRNAs (ss-CAG, ss-GCA, and ss-AGC) (Figs. 3C and 5B). Active compounds were entirely composed of 2′-O-methoxyethyl nucleotides, a mixture of DNA and 2′-O-methoxyethyl nucleotides, or a mixture of DNA and cET nucleotides. For the 18-base ASOs, the use of 2′-O-methoxyethyl nucleotides mixed with cET bases yielded inactive compounds.
The shorter 16-base ASOs possessed better efficacies than the 18-base ASOs (Fig. 6). Unlike the 18-base compounds, the ASOs were active regardless of whether they were mixtures of DNA and cET nucleotides or 2′-O-methoxylethyl and cET nucleotides. Lower Tm values may explain the better performance of 16-base 2′-O-methoxyethyl/cET ASOs relative to the inactive 18-base 2′-O-methoxyethyl/cET ASOs. Our data may define a window for optimal Tm values. When Tm values are too high, internal structure or off-target binding to related sequences may prevent efficient recognition of the CUG repeat. When Tm values are too low, binding to the repeat may be inefficient.
Discussion
FECD is a potentially important disease target for oligonucleotide therapeutics
Several factors support the conclusion that FECD may be a good target for oligonucleotide therapeutics. FECD is caused by mutant RNA containing an expanded CUG repeat [10–13]. FECD patients can possess the repeat within the TCF4 gene or the DMPK gene [14–16], demonstrating that the mutant RNA is the most likely trigger for the disease. Because of the central importance of the mutant RNA, synthetic oligonucleotides that block the RNA should be excellent candidates for controlling disease progression.
From a clinical perspective, the mutation within the TCF4 gene associated with FECD occurs in 2.5% of the Caucasian population, making it one of the most common disease-associated mutations [10–13]. Studies are also revealing relatively high occurrences in other ethnic groups [12]. While disease progression is slow, it eventually causes a debilitating loss of vision and curative treatments would benefit many people over the age of 60.
Currently, the only current curative option for FECD is corneal transplantation. While this option alleviates the disease for many patients, a small fraction of surgeries is partially or completely unsuccessful [32,33]. The fact that surgery is the only option discourages some patients and often leads to delays in treatment until vision loss is severe. A safe, effective, nonsurgical treatment would greatly expand the number of patients who would benefit from alleviating the course of the disease.
Comprehensive genetic testing and analysis is becoming more common. It is easy to imagine a future in which millions of people know whether they have the expanded repeat mutation within the TCF4 gene. Early signs of FECD can be detected during a standard visit to an ophthalmologist, and patients who know that they have a genetic propensity would be good candidates for early diagnosis. A well-tolerated and convenient drug would allow these patients to be treated early before loss of vision becomes noticeable.
Oligonucleotides have already been used in the clinic for treating eye disease [1–7]. Intraocular injection of oligonucleotides is well tolerated and a potential route for administration. ASOs and duplex RNAs have demonstrated remarkable long-active half-lives in clinical trials when used in the liver or central nervous system [18]. For chronic disease, administration as infrequently as three to four times per year can be effective. Such longevity makes it more reasonable to believe that intraocular injection would be tolerated by patients. We note that intraocular injection in the ophthalmologist's office would have the added benefit of ensuring patient compliance over the months or years needed for drug administration.
Topical delivery may also be possible. The affected corneal endothelial tissue is only one half millimeter away from the surface of the eye and previous results have suggested that topical delivery can be effective [3–5,17]. If delivery of ASOs or duplex RNAs in eye drop formulation was possible, the convenience would be a major advantage.
Path forward for anti-FECD oligonucleotide therapeutics
We have now shown that steric block ASOs, duplex RNAs, and ss-siRNAs can block the RNA foci formed by mutant TCF4 intronic RNA. In our previous study, we demonstrated reversal of the splicing defects in patient tissue associated with FECD [17] and Davidson and colleagues observed similar results using a steric-block 2′-O-methyl ASO [19]. While we did not test whether the RNAi protein machinery was necessary for activity in this study, we have previously shown that argonaute 2 protein was necessary when using duplex RNAs or ss-siRNAs to successfully target other expanded trinucleotide repeat genes [23,29,30].
Duplex RNAs, ss-siRNAs, and steric blocking ASOs function by different mechanisms and provide different options for the development of therapeutics [18]. Duplex RNAs and ss-siRNAs function through RNAi and taking advantage of the natural gene silencing machinery may lead to improved potency. Both ss-siRNAs and ASOs consist of just one strand, possibly improving cellular uptake and reducing cost. A definitive balancing of advantages and disadvantages of these approaches will require additional testing. At the current stage of characterization, all appear promising.
In the future, there are several objectives that should be achieved before clinical development of anti-CUG compounds. There is considerable evidence that ASOs can be active in the eye. We have observed that soaking the corneal endothelium is sufficient for ASO uptake [17]. In vivo, however, the efficiency of uptake is unknown. It is also unknown whether intraocular injection will be the most practical mode of delivery or whether topical delivery can be an option. Regardless of which mode of administration is possible, it is likely that delivery will need to be repeated multiple times, possibly for a lifetime. A high level of safety and tolerability will need to be demonstrated.
Many other genes have CUG repeats that are not expanded, but are long enough to permit binding of at least one anti-CUG repeat oligonucleotide. Previous studies in a mouse model of myotonic dystrophy revealed that expression of these other CUG-repeat genes was largely unaffected [34]. The fact that “Off-target” CUG repeats are much shorter than the expanded repeat within the TCF4 gene may also make binding less likely. That said, the potential for binding to other human CUG repeat genes will require study. However, our ability to tailor compounds through chemical modification and the introduction of strategically placed base alterations that introduce mismatches relative to the target may also help increase selectivity if problems are encountered.
The design of clinical trials will face challenges. The slow progression of FECD will require extended clinical trials. It will be beneficial for these trials to be designed to be as brief as possible and achieving that goal will require a finer understanding of the disease course of FECD and an ability to monitor the progression of findings and symptoms over months rather than years. While research will be needed to achieve that understanding, the cornea can be readily examined by ophthalmologists and changes to the cornea become visible long before disease symptoms become severe. Monitoring biomarkers may be a viable option for following the course of early disease and relatively rapid evaluation of new treatments.
In summary, FECD is a major cause of vision loss that affects hundreds of thousands of people. There are no nonsurgical curative treatments. The disease is caused by a mutant RNA, and the selectivity of ASOs and duplex RNAs makes them ideal starting points for therapeutic development. Specific to compounds that function through the RNAi pathway, we have identified several lead compounds that can block mutant RNA in patient-derived cells.
Acknowledgments
We thank Albert Jun for generously sharing F35T cell line. This study was supported by grants R01EY022161 (VVM), P30EY020799 (VVM), and R35GM118103 (DRC) from the National Institutes of Health, Bethesda, MD, an unrestricted grant from Research to Prevent Blindness, New York (VVM), the Alfred and Kathy Gilman Special Opportunities in Pharmacology Fund (DRC), and the Robert A. Welch Foundation I-1244 (DRC). VVM is the Paul T. Stoffel/Centex Professor in Clinical Care. DRC is the Rusty Kelley Professor of Biomedical Science.
Author Disclosure Statement
T.P. Prakash and F. Rigo are employees of Ionis Pharmaceuticals. V.V.M. and D.R.C. have filed a patent related to this research. J.H. declares no conflicts of interest.
References
- 1. Fattal E. and Bochot A. (2006). Ocular delivery of nucleic acids: antisense oligonucleotides, aptamers, and siRNA. Adv Drug Deliv Rev 58:1203–1223 [DOI] [PubMed] [Google Scholar]
- 2. Shen WY, Garrett KL, Wang CG, Zhang K, Ma ZZ, Constable IJ. and Rakoczy PE. (2002). Preclinical evaluation of a phosphorothioate oligonucleotide in the retina of rhesus monkey. Lab Invest 82:167–182 [DOI] [PubMed] [Google Scholar]
- 3. Cursiefen C, Viaud E, Bock F, Geudelin B, Ferry A, Kadlecova P, Levy M, Al S. Mahmood S. Colin, et al. (2014). Aganisrsen antisense oligonucleotide eye drops inhibit keratitis-induced corneal neovascularization and reduce the need for transplantation: the I-CAN study. Ophthalmology 121:1683–1692 [DOI] [PubMed] [Google Scholar]
- 4. Martínez T, González MV, Roehl I, Wright N, Pañeda C. and Jiménez AI. (2014). In Vitro and In vivo efficacy of SYL040012, a novel siRNA compound for treatment of glaucoma. Mol Ther 22:81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Benitez-Del-Castillo JM, Moreno-Montanes J, Jimenez AI, Munoz-Negrete J, Turman K, Palumaa K, Sadaba B, Gonzalez MV, Ruz V, et al. (2016). Safety and efficacy trials for SYL1001, a novel short interfering RNA for the treatment of dry eye disease. Cornea 57:6447–6454 [DOI] [PubMed] [Google Scholar]
- 6. Geary RS, Henry SP. and Grillone LR. (2002). Fomivirsen: clinical pharmacology and potential drug interactions. Clin Pharmacokinet 41:255–260 [DOI] [PubMed] [Google Scholar]
- 7. Nimjee SM, White RR, Becker RC. and Sullenger BA. (2017). Aptamers as Therapeutics. Annu Rev Pharmacol Toxicol 57:61–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lorenzetti DW, Uotila MH, Parikh N. and Kaufman HE. (1967). Central cornea guttata. Incidence in the general population. Am J Ophthalmol 64:1155–1158 [PubMed] [Google Scholar]
- 9. Deng SX, Lee WB, Hammersmith KM, Kuo AN, Li JY, Shen JF, Weikert MP. and Shtein RM. (2018). Descemet Membrane Endothelial Keratoplasty: safety and Outcomes: a Report by the American Academy of Ophthalmology. Ophthalmology 125:295–310 [DOI] [PubMed] [Google Scholar]
- 10. Wieben ED, Aleff RA, Tosakulwong N, Butz ML, Highsmith WE, Edwards AO. and Baratz KH. (2012). A common trinucleotide repeat expansion within the transcription factor 4 (TCF4, E2-2) gene predicts Fuchs corneal dystrophy. PLoS One 7:e49083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mootha VV, XGong, HC Ku. and C Xing. (2014). Association and familial segregation of CTG18.1 trinucleotide repeat expansion of TCF4 gene in Fuchs' endothelial corneal dystrophy. Invest Ophthalmol Vis Sci 55:33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xing C, Gong X, Hussain I, Khor CC, Tan DT, Aung T, Mehta JS, Vithana EN. and Mootha VV. (2014). Transethnic Replication of Association of CTG18.1 Repeat Expansion of TCF4 Gene With Fuchs' Corneal Dystrophy in Chinese Implies Common Causal Variant. Invest Ophthalmol Vis Sci 55:7073–7078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Soliman AZ, Xing C, Radwan SH, Gong X. and Mootha VV. (2015). Correlation of severity of fuchs endothelial corneal dystrophy with triplet repeat expansion in TCF4. JAMA Ophthalmol 133:1386–1391 [DOI] [PubMed] [Google Scholar]
- 14. Gattey D, Zhu AY, Stagner A, Terry MA. and Jun AS. (2014). Fuchs endothelial corneal dystrophy in patients with myotonic dystrophy: a case series. Cornea 33:96–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mootha VV, Hansen B, Rong Z, Mammen PP, Zhou Z, Xing C. and Gong X. (2017). Fuchs' Endothelial Corneal Dystrophy and RNA Foci in Patients with myotonic dystrophy. Invest Ophthalmol Vis Sci 58:4579–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Winkler NS, Milone M, Martinez-Thompson JM, Raja H, Aleff RA, Patel SV, Wieben ED. and Baratz KH. (2018). Fuch's corneal endothelial dystrophy in patients with myotonic dystrophy, type 1. Invest Opthamol Vis Sci 59:3053–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hu J, Rong Z, Gong X, Zhou Z, Sharma VK, Xing C, Watts JK, Corey DR. and Mootha VV. (2018). Oligonucleotides targeting TCF4 triplet repeat expansion inhibit RNA foci and mis-splicing in Fuchs' dystrophy. Hum Mol Genet 27:1015–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shen X. and Corey DR. (2017). Chemistry, mechanism, and clincial status of antisense oligonucleotides and duplex RNAs. Nucl Acids Res 46:1584–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zarouchlioti C, Sanchez-Pintado B, Hafford Tear NJ, Klein P, Liskova P, Dulla K, Semo M, Vugler AA, Muthusamy K, et al. (2018). Antisense Therapy for a Common Corneal Dystrophy Ameliorates TCF4 Repeat Expansion-Mediated Toxicity. Am J Hum Genet 102:528–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K. and Tuschl T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494–498 [DOI] [PubMed] [Google Scholar]
- 21. Lima WF, Prakash TP, Murray HM, Kinberger GA, Li W, Chappell AE, Li CS, Murray SF, Gaus H, et al. (2012). Single-stranded siRNAs activate RNAi in animals. Cell 150:883–894 [DOI] [PubMed] [Google Scholar]
- 22. Chorn G, Klein-McDowell M, Zhao L, Saunders MA, Flanagan WM, Willingham AT. and Lim LP. (2012). Single-stranded microRNA mimics. RNA 18:1796–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu D, Pendergraff H, Liu J, Kordasiewicz HB, Cleveland DW, Swayze EE, Lima WF, Crooke ST, Prakash TP. and Corey DR. (2012). Single-stranded RNAs that function through RNAi are potent and allele-selective inhibitors of huntingtin expression. Cell 150:895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu J, Liu J. and Corey DR. (2010). Allele-selectivity by switching to an miRNA-like RNAi mechanism. Chem Biol 17:1183–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zar JH. (1999). Biostatistical Analysis. Pearson Education, India [Google Scholar]
- 26. Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N. and Zamore PD. (2003). Asymmetry in the assembly of the RNAi enzyme complex. Cell 115:199–208 [DOI] [PubMed] [Google Scholar]
- 27. Wang Y, Juranek S, Li H, Sheng G, Tuschl T. and Patel DJ. (2008). Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456:921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu J, Liu J, Liande L, Gagnon KT. and Corey DR. (2015). Targeted recognition of GGGGCC/CCCCGG repeats at the C9orf72 locus by duplex RNA. Chem Biol 22:1505–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu J, Liu J, Chu Y, Yu D. and Corey DR. (2012). Mechanism of allele-selective inhibition of huntingtin expression by duplex RNAs that target CAG repeats. Nucl Acids Res 40:11270–11280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu J, Yu D, Aiba Y, Pendergraff H, Swayze EE, Lima WF, Prakash TP. and Corey DR. (2013). ss-siRNAs allele-selectively inhibit ataxin-3 expression: multiple mechanisms for an alternative gene silencing strategy. Nucl Acids Res 41:9570–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seth PP, Siwkowski A, Allerson CR, Vasquez G, Lee S, Prakash TP, Wancewicz EV, Witchell D. and Swayze EE. (2009). Short antisense oligonucleotides with novel 2’-4’ conformationally restricted nucleoside analogues show improved potency without increased toxicity in animals. J Med Chem 52:10–13 [DOI] [PubMed] [Google Scholar]
- 32. Price MO, Fairchild KM, Price DA. and Price FW. (2011). Descemet's stripping endothelial keratoplasty five-year graft survival and endothelial cell loss. Ophthalmology 118:725–729 [DOI] [PubMed] [Google Scholar]
- 33. Ang M, Soh Y, Htoon HM, Mehta JS. and Tan D. (2016). Five-year graft survival comparing descemet stripping automated endothelial keratoplasty and penetrating keratoplasty. Ophthalmology 123:1646–1652 [DOI] [PubMed] [Google Scholar]
- 34. Sobczak K, Wheeler TM, Wang W. and Thornton CA. (2013). RNA interference targeting CUG repeats in a mouse model of myotonic dystrophy. Mol Ther 21:380–387 [DOI] [PMC free article] [PubMed] [Google Scholar]