Abstract
Chronic granulomatous disease (CGD) is a primary immunodeficiency characterized by impaired antimicrobial activity in phagocytic cells. As a monogenic disease affecting the hematopoietic system, CGD is amenable to gene therapy. Indeed in a phase I/II clinical trial, we demonstrated a transient resolution of bacterial and fungal infections. However, the therapeutic benefit was compromised by the occurrence of clonal dominance and malignant transformation demanding alternative vectors with equal efficacy but safety-improved features. In this work we have developed and tested a self-inactivating (SIN) gammaretroviral vector (SINfes.gp91s) containing a codon-optimized transgene (gp91phox) under the transcriptional control of a myeloid promoter for the gene therapy of the X-linked form of CGD (X-CGD). Gene-corrected cells protected X-CGD mice from Aspergillus fumigatus challenge at low vector copy numbers. Moreover, the SINfes.gp91s vector generates substantial amounts of superoxide in human cells transplanted into immunodeficient mice. In vitro genotoxicity assays and longitudinal high-throughput integration site analysis in transplanted mice comprising primary and secondary animals for 11 months revealed a safe integration site profile with no signs of clonal dominance.
Stein and colleagues report on the comprehensive preclinical and safety studies that they conducted in order to transition a self-inactivating gammaretroviral vector into the clinic for use in subjects with X-linked chronic granulomatous disease. Their studies included assessment of dosage requirements, therapeutic efficacy, and resistance to transgene silencing and genotoxic potential.
Introduction
Gene therapy of inherited diseases has provided convincing evidence of therapeutic benefits for many treated patients. In particular, treatment of primary severe congenital immunodeficiencies by gene transfer into hematopoietic stem cells has proven in some cases to be as beneficial as allogeneic stem cell transplantation, the treatment of choice for these diseases if human leukocyte antigen-matched donors are available [reviewed in (Booth et al., 2011; Fischer et al., 2011; Naldini, 2011; Aiuti et al., 2012; Rivat et al., 2012; Seymour and Thrasher, 2012)]. However, sustained therapeutic success has been hampered by serious adverse events occurring in a few of the treated patients as a consequence of vector integration and activation of proto-oncogenes leading to the development of myeloproliferative diseases and leukemias (Hacein-Bey-Abina et al., 2008; Howe et al., 2008; Stein et al., 2010; Aiuti et al., 2012; Cavazzana-Calvo et al., 2012). Despite these adverse effects, most patients responded to chemotherapy treatment, are currently in complete remission, and still do contain gene-marked cells that have contributed to the recovery of immune functions. Three patients (two with the X-linked form of chronic granulomatous disease [X-CGD] and one with SCID-X1) succumbed as a consequence of insertion-mutagenesis-driven leukemia (Rivat et al., 2012).
Previously, we conducted a phase I clinical trial aimed at the correction of X-CGD, a rare inherited immunodeficiency characterized by severe and life-threatening bacterial and fungal infections as well as widespread tissue granuloma formation. Phagocytic cells of CGD patients fail to kill ingested microbes because of a defect in the nicotinamide dinucleotide phosphate (NADPH) oxidase complex resulting in compromised antimicrobial activity (Segal, 1996; Segal et al., 2000; Holland, 2010). In this clinical trial, we used a gammaretroviral vector with strong enhancer–promoter sequences in the long terminal repeats (LTRs) to genetically modify CD34+ cells in two X-CGD patients. After successful reconstitution of phagocytic functions, both patients experienced a clonal outgrowth of gene-marked cells caused by vector-mediated insertional activation of the EVI1, PRDM16, and SETBP1 proto-oncogenes leading to the development of myeloid malignancies in both patients (Ott et al., 2006; Stein et al., 2010). Moreover, functional correction of gene-transduced cells decreased with time because of epigenetic inactivation of the vector promoter within the LTR, resulting in the accumulation of nonfunctional gene-transduced cells (Stein et al., 2010). A similar expansion of gene-transduced cells with activated EVI1 was observed in two patients treated at the Children's Hospital in Zurich. Both patients underwent allogeneic bone marrow (BM) transplantation before or at the onset of myelodysplasia and are currently in complete remission (R. Seger and J. Reichenbach, personal communication).
The understanding of the molecular basis of insertional mutagenesis after gene therapy has motivated the development of advanced integrating vectors with equal therapeutic potency but reduced genotoxicity (Baum et al., 2004; Naldini, 2011). In our clinical study, insertional oncogenesis was initiated either by enhancer-mediated activation of proto-oncogene promoters or by aberrant splicing of transcripts initiated at the LTR and spliced out into exons of proto-oncogenes (Ott et al., 2006; Stein et al., 2010). Similarly, LTR-driven activation of LMO2 was identified as the major cause of lymphoid leukemia after gene therapy of X-linked severe combined immunodeficiency syndrome (X1-SCID (Hacein-Bey-Abina et al., 2008; Howe et al., 2008). Therefore, the redesign of vector architecture has been a matter of intense work during the past years (Corrigan-Curay et al., 2012). In particular, the deletion of the enhancer elements within the viral LTR unique 3′ (U3) regions has significantly contributed to the reduction of genotoxic effects associated with LTR-driven gammaretroviral vectors. Despite their integration preference in proximity to cellular promoter regions, these U3-deleted self-inactivating (SIN) vectors have shown an improved safety profile in vitro and in vivo (Modlich et al., 2006; Montini et al., 2006, 2009). SIN vector safety can be further improved by the careful selection of internal promoters to drive transgene expression (Zychlinski et al., 2008). Accordingly, the use of tissue-specific promoters, which are inactive in stem/progenitor cells but active in terminally differentiated cells, should further increase the safety level of SIN vectors (Modlich et al., 2009). Indeed, SIN gammaretroviral and lentiviral vectors have shown promising results in terms of efficacy and safety in several preclinical studies (Thornhill et al., 2008; Zhou et al., 2010; Avedillo Diez et al., 2011; Huston et al., 2011; Tolar et al., 2012; Scaramuzza et al., 2013) and ongoing clinical trials, although the follow-up time of treated patients is still too short to make conclusive statements at present (Cartier et al., 2009; Cavazzana-Calvo et al., 2010; Biffi et al., 2011). On the basis of the aforementioned advancements, we developed a SIN gammaretroviral vector for the safe and effective gene therapy of X-linked CGD (SINfes.gp91s). We combined the SIN configuration with an internal promoter, with preferential expression in myeloid cells. However, the introduction of a new vector into the clinic demands a series of sophisticated preclinical studies that are quite challenging in particular within an academic environment (Will et al., 2007). Here, we report on the comprehensive and thorough preclinical efficacy and safety testing of the SINfes.gp91s retroviral vector assessing dosage requirements, therapeutic efficacy, resistance to transgene silencing, and genotoxic potential.
Results and Discussion
A SIN gammaretroviral vector for X-CGD gene therapy
We designed a new gammaretroviral vector for the gene therapy of X-CGD based on an advanced SIN vector platform with the following important features: (1) all retroviral coding sequences are deleted, (2) a primer-binding-site sequence is used that escapes targeting by epigenetic silencers recognizing conventional murine gammaretroviral sequences, and (3) modifications of the 5′ promoter of the retroviral vector plasmid in combination with a safety-modified version of the woodchuck hepatitis virus post-transcriptional regulatory element support production at high titers (Schambach et al., 2006, 2009). The backbone used for the construction of SINfes.gp91s was derived from the SERS11 vector backbone described by Schambach et al. (2006) containing an internal therapeutic cassette consisting of a 507 bp DNA fragment derived from the promoter region of the human c-FES gene driving the expression of a synthetic, codon-optimized gp91phox cDNA (Moreno-Carranza et al., 2009). Similar vectors containing either the elongation factor short promoter or the migration inhibitory factor-related protein 8 promoter were constructed in parallel and tested for gp91phox expression and reconstitution of superoxide activity in vitro and in vivo. Although all vectors performed similarly, the SINfes.gp91s vector was selected for preclinical studies based on slightly higher levels of gp91phox expression and reconstituted NADPH oxidase activity per vector copy number (VCN) in granulocytes derived from transplanted X-CGD mice (Supplementary Fig. S1; Supplementary Data are available online at www.liebertonline.com/humc).
Pharmacodynamic studies: correction of the X-CGD phenotype by low VCNs in vitro and in vivo
To determine the gene dosage required for an effective therapeutic effect, we first assessed reconstitution of superoxide activity in gene-corrected X-CGD granulocytes in vitro and in vivo. We tested the performance of SINfes.gp91s in vivo using male/female or CD45.2/CD45.1 animals to distinguish donor from recipient cells. Therefore, we transplanted SINfes.gp91s-modifed X-CGD cells differing in the mean VCN per cell in X-CGD mice, resulting in two cohorts with low and high VCNs, respectively. In the low-VCN group, a total of 8 primary and 10 secondary animals (mean VCN 0.42) were analyzed, whereas the high-VCN group consisted of 9 primary and 10 secondary transplanted animals (mean VCN 2.9). Engraftment of donor cells was similar in both groups as assessed by quantitative polymerase chain reaction (qPCR) using specific primers for the murine SRY gene (transplantation of male X-CGD cells into female X-CGD recipients) or by fluorescence-activated cell sorting analysis of CD45.2 X-CGD cells engrafted into CD45.1 recipients. Independently of the setting used, donor cell engraftment ranged between 80% and 100% in all animals.
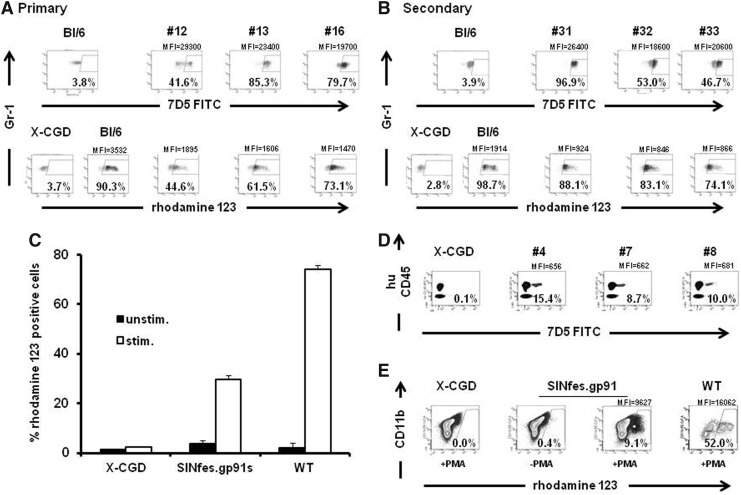
Gp91phox expression was measured by flow cytometry 18–19 weeks after transplantation in the peripheral blood cells of treated mice. Gp91phox expression was found on average in 27.1%±7.7% of the peripheral blood cells in primary and 25.2%±16.7% in secondary animals in the low-VCN group, whereas in peripheral blood cells from the high-VCN group gp91phox expression was on average 64.7%±23.2% and 56.7%±23.3% in primary and secondary animals, respectively (Fig. 1A and B, and Supplementary Fig. S2A and B, p<0.003 between low- and high-VCN groups). Rescue of the CGD phenotype was tested by assessing superoxide production in gene-modified granulocytes using the dihydrorhodamine-123 (DHR) assay (Bohler et al., 1986). Superoxide production in granulocytes of primary animals ranged between 17.1% and 70.3% of wild-type (wt) levels (low-VCN animals) and between 44.6% and 97.8% in the high-VCN animals (Fig. 1A and Supplementary Fig. S2C and D). In secondary animals, rhodamine-123-positive granulocytes ranged between 15.9% and 41.8% in the low-VCN group and between 27.0% and 93.5% in the high-VCN animals, respectively (Fig. 1B and Supplementary Fig. S2C, p<0.0001, between low- and high-VCN groups). Thus, even at low VCNs (mean 0.42) gp91phox expression and reconstitution of superoxide activity at therapeutic relevant levels were achieved in primary and secondary animals, respectively.
FIG. 1.
Gp91phox expression and functional reconstitution of superoxide production by SINfes.gp91s-transduced cells in vivo. Representative examples of gp91phox expression (A and B, upper panels) and superoxide production (A and B, lower panels) in granulocytes of primary (A) and secondary (B) animals transplanted with SINfes.gp91s-transduced lin− cells. Transgene expression analysis was performed 18 (primary) and 16 (secondary) weeks after transplantation. Intracellular staining using a monoclonal antibody (7D5) was used to detect gp91phox expression. NADPH oxidase activity (A and B, lower panels) was assessed in the same samples as above by the oxidative burst assay (DHR). The percentage of gp91phox expressing Gr-1+ cells and superoxide production in CD11b+ Gr-1+ donor cells as well as the MFI of rhodamine 123 is shown. (C) Reconstitution of oxidase activity in the progeny of primary human CD34+ cells from X-CGD patients transduced with SINfes.gp91s after in vitro differentiation. NADPH oxidase activity was measured in CD11b+ cells by the DHR assay after in vitro differentiation of nontransduced (X-CGD) or transduced cells. In vitro differentiated CD34+ cells from a healthy donor served as positive control (WT). (D) Reconstitution of gp91phox expression in primary human hematopoietic cells in vivo. X-CGD CD34+ cells were transduced with SINfes.gp91s or left untreated (X-CGD). About 107 cells were transplanted into sublethally irradiated (2.5 Gy) immunodeficient mice. Three weeks after transplantation, blood was obtained from the tail vein of the animals and gp91phox expression was analyzed in human cells (huCD45) by fluorescence-activated cell sorting. (E) Reconstitution of oxidase activity in vivo. Bone marrow cells obtained from NOD/scid/IL-2Rγ null mice transplanted either with nontransduced (X-CGD) or SINfes.gp91s-transduced X-CGD CD34+ cells or cells from a healthy donor (WT) were differentiated ex vivo for 2 weeks in the presence of G-CSF. NADPH oxidase activity was measured after phorbol-12-myristate-13-acetate stimulation in the huCD45+ cell fraction by the DHR assay. Cells were costained for CD11b expression. The percentage of superoxide production in huCD45+ CD11b+ cells and the MFI of rhodamine-123-positive cells are indicated. DHR, dihydrorhodamine-123; MFI, mean fluorescence intensity; NADPH, nicotinamide dinucleotide phosphate; WT, wild type; X-CGD, X-linked form of chronic granulomatous disease.
The performance of the SINfes.gp91s vector was also assessed in human hematopoietic cells from X-CGD patients. For these studies, a pre-GMP vector preparation was used (see Supplementary Materials and Methods for details). Transduction efficiency into X-CGD CD34+ cells was 28.7%±8.5% as estimated from gp91phox expression at a mean VCN of 0.58±0.11. After transduction, X-CGD CD34+ cells were differentiated in vitro in the presence of G-CSF to measure reconstitution of oxidase activity in mature granulocytes. The percentage of superoxide-producing CD11b-positive cells (29.7%±10.7%) correlated well with the percentage of gp91phox-expressing cells and corresponded to 40% of the values detected in CD11b+ cells derived from healthy CD34+ cells (Fig. 1C). Transduced X-CGD CD34+ cells were also injected in sublethally irradiated immunodeficient mice. Gp91phox expression and superoxide production were estimated 3 weeks after transplantation in the peripheral blood of recipient mice and in vitro differentiated BM cells from transplanted animals, respectively. Gp91phox expression was found in 11.4%±3.6% of the huCD45+ human peripheral blood cells (Fig. 1D), whereas 9.1% of the cells were found to produce superoxide after G-CSF-induced myeloid differentiation and phorbol-12-myristate-13-acetate stimulation (Fig. 1E). Notably, the mean levels of superoxide production per cell were 60% of wt levels, as estimated from the mean fluorescence intensity (MFI; Fig. 1E). These values are therapeutically relevant and likely to be sufficient to improve health conditions in CGD patients, as residual superoxide production level is a predictor of survival in CGD patients. Indeed, in a recent study comprising a total of 287 CGD patients (67.9% X-CGD patients), a strong association between long-term survival and residual superoxide production was observed (Kuhns et al., 2010). According to this study, a survival benefit is expected for CGD patients with as low as 1% of normal superoxide production levels. From this analysis, we anticipate a significant benefit for X-CGD patients receiving an autologous transplant containing SINfes.gp91s gene-modified cells.
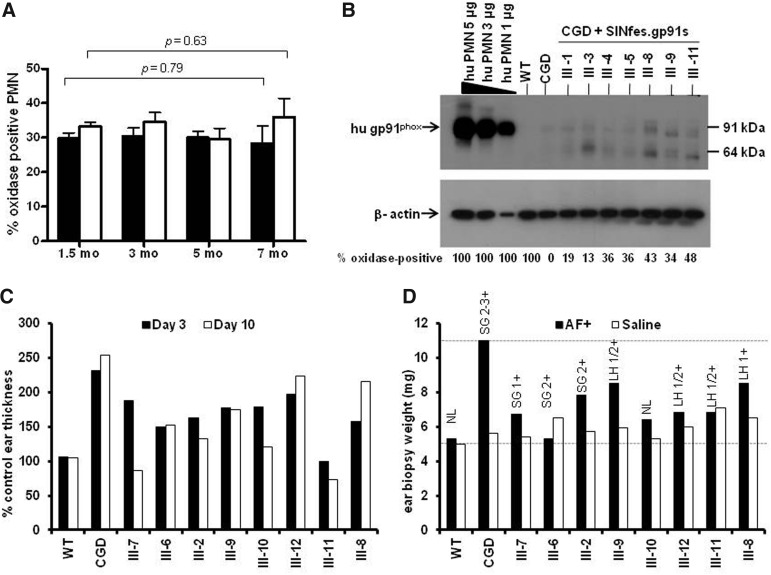
Decisive for the successful treatment of CGD patients is the production of sufficient superoxide levels in transduced cells to alleviate typical CGD symptoms. Aspergillus fumigatus (AF) is an opportunistic fungal pathogen that can cause serious infectious and inflammatory complications in patients with CGD. As measures of therapeutic efficacy in X-CGD animals that were transplanted with retrovirally transduced syngeneic hematopoietic cells, we evaluated whether these animals were protected from hyperinflammatory responses caused by the intradermal injection of sterile AF hyphae (Petersen et al., 2002; Goebel et al., 2005) and from pneumonia after respiratory challenge with live AF conidia (Bjorgvinsdottir et al., 1997; Dinauer et al., 2001). In these experiments, busulfan-conditioned CGD mice were transplanted with SINfes.gp91s-transduced cells (see Supplementary Materials and Methods). Oxidase activity in peripheral blood polymorphonuclear leukocytes (PMNs) (mostly neutrophils) was sustained during the first 10 months after transplantation and ranged between 12% and 48% with a MFI of about 1/10 of wt levels (Fig. 2A and Supplementary Table S1). The mean VCN in donor-derived cells obtained from the BM of selected animals (86–90% chimerism) 7 months after transplantation was 0.17 vector copies per cell (range 0.12–0.22; Supplementary Table S1), with oxidase activity ranging between 13% and 36% in the PMNs obtained from of these animals.
FIG. 2.
Suppression of hyperinflammatory response in X-CGD mice transplanted with SINfes.gp91s-transduced cells. (A) Oxidase activity in peripheral blood PMNs over time after transplantation of either 1×106 (solid bars) or 2×106 (open bars) SINfes.gp91s-transduced lin− cells into X-CGD animals. Mean values±SEM are given. p-Values according to two-tailed t-test. (B) Human gp91phox expression in peripheral blood PMNs of animals transplanted with SINfes.gp91s-transduced cells. PMNs were obtained from the indicated animals 7 months after transplantation. Gp91phox was detected by Western blotting using the CL5 antibody (Sadat et al., 2009). Cell extracts from human PMN were used at different concentrations as control. No cross-reaction with murine gp91phox was observed as indicated by the lack of hybridization in cell extracts obtained from wild-type mice (WT). The percentage of oxidase-positive cells is indicated below. (C and D) X-CGD animals transplanted with SINfes.gp91s-transduced cells are protected from Aspergillus fumigatus (AF) challenge. Seven months after transplantation, sterile AF hyphae were injected intradermally into one of the ears of the animals. As a control, saline was injected into the contra lateral ear. The degree of inflammation at the AF injection site was estimated at 3 and 10 days after injection from the ear thickness at the injection site (C) or the weight of a 5 mm ear punch biopsy taken 30 days after injection (D). Black bars, AF injection; white bars, saline injection. Values between 1 and 3 denote the severity of the inflammation, with 1 being low and 3 high. LH, lymphocytic/histiocytic (macrophages) infiltration; NL, normal; PMNs, polymorphonuclear leukocytes; SEM, standard error of the mean; SG, suppurative/granulomatous.
After intradermal challenge with sterile AF hyphae, X-CGD mice develop a persistent inflammatory response that is characterized by an acute infiltration of neutrophils at the site of injection (1–3 days) followed by a chronic inflammatory reaction 30 days postinjection because of infiltrating neutrophils, mononuclear cells, and giant cells (Petersen et al., 2002; Goebel et al., 2005). Seven months after transplantation, eight animals were challenged by intradermal injection of 2.5 μg sterilized AF hyphae into one ear, with the contralateral ear injected with saline only. No significant changes in oxidase activity were observed at this time point compared with the initial values (p>0.05; Fig. 2A). Indeed, PMNs of transplanted animals expressed substantial amounts of glycosylated (91 kDa) and nonglycosylated (64 kDa) gp91phox protein at this time point as demonstrated in Western blots (Fig. 2B). The degree of inflammation was measured by the thickness of the ear at the site of injection 3 and 10 days after injection (Fig. 2C). While wt animals showed a reduced inflammatory response at day 3, which was not enhanced at day 10, X-CGD mice suffered from a severe inflammatory response that was not controlled at day 10 postinjection. In contrast, most of the animals transplanted with SINfes.gp91s-transduced cells showed limited inflammation at the site of injection, which decreased at day 10, demonstrating effective control of inflammation by gene-modified cells. At 30 days postinjection, the mice were sacrificed and a 5 mm punch biopsy was then taken from each ear. Biopsy specimens were evaluated for the presence or absence of inflammatory cells, and were graded on a 0–3+scale, with a score of 0 indicating no inflammation (e.g., as seen in ears injected with phosphate buffered saline), and a score of 3+ denoting maximum inflammation as observed in X-CGD controls. X-CGD mice ears showed a severe (grade 2–3+) inflammatory response, characterized by a massive infiltration of neutrophils and macrophages (Fig. 2D). In contrast, the ear challenge data showed that treated CGD mice had no severe suppuration (grade 3+) as observed in untreated X-CGD mice, although many of the mice still had some thickening. In the treated group, six mice showed lymphocytic/histiocytic (macrophages) chronic infiltrates graded 1–3+, whereas three mice were found to have grade 1–2+ suppurative granulocytic infiltration.
As a second measure of therapeutic efficacy, CGD mice transplanted with SINfes.gp91s-transduced marrow were challenged with live AF conidia administered by intratracheal instillation. In addition to wt and CGD controls, we also studied mice that were mixed chimeras of wt and CGD leukocytes, generated by transplanting lethally irradiated mice with mixtures of wt and CGD marrow. Four to seven months after transplantation, 200 conidia, a dose previously shown to establish chronic pneumonia in 100% of CGD mice (Dinauer et al., 2001), was instilled into the lungs. Fourteen days later, mice were sacrificed and lung histology was scored (Table 1). All X-CGD mice had multifocal granulomas with microabscesses or diffuse inflammation, with scores ranging from 1 to 4 (mean score 2.2). The wt mice had no evidence of lung disease, as expected. Almost all of the WT/X-CGD chimeric mice and SINfes.gp91S-transplanted mice also had no evidence of lung disease, even for mice with as few as 10–20% NADPH-oxidase-positive neutrophils. In the one mouse in each of these categories that had detectable lung disease, only small focal granulomas (with a score of 1) were detected. Thus, the SINfes.gp91s vector was able to protect animals from AF-induced inflammatory reactions and gave CGD mice a high degree of protection from pneumonia induced by AF at very low VCNs. In addition, our analysis indicates that both expression of gp91phox and NADPH oxidase activity were sustained for 7 months in vivo.
Table 1.
Pulmonary Disease in Mice After Challenge with Aspergillus fumigatus
| Group | n | % Oxidase-positive PMNs | A. fumigatus lung disease |
|---|---|---|---|
| WT mice | 8 | >95 | 0/8 |
| X-CGD mice | 9 | 0 | 9/9 |
| WT/X-CGD transplantation chimeras | 4 | 13–15 | 1/4 |
| 5 | 32–54 | 0/5 | |
| SINfes.gp91S-transplanted X-CGD mice | 4 | 10–19 | 1/4 |
| 3 | 26–29 | 0/3 | |
| 4 | 34–37 | 0/4 |
Oxidase-positive PMNs were determined by dihydrorhodamine-123 assay before intratracheal challenge with 200 A. fumigatus conidia. Mice were sacrificed 14 days after challenge. Lung histology was scored in a blinded manner, following a scoring system correlating with histopathologic severity. Three cross sections were scored in three categories: focal granulomas and/or microabscesses, multifocal granulomas and/or microabscesses, or diffuse inflammation covering the lung lobe. All X-CGD mice had multifocal granulomas with microabscesses or diffuse inflammation, with scores ranging from 1 to 4 (mean score 2.2). The WT/X-CGD chimera and SINfes.gp91S-transplanted mice with detectable lung disease had small focal granulomas only. PMNs, polymorphonuclear leukocytes; WT, wild type; X-CGD, X-linked form of chronic granulomatous disease.
The c-FES promoter is less prone to CpG methylation in hematopoietic cells
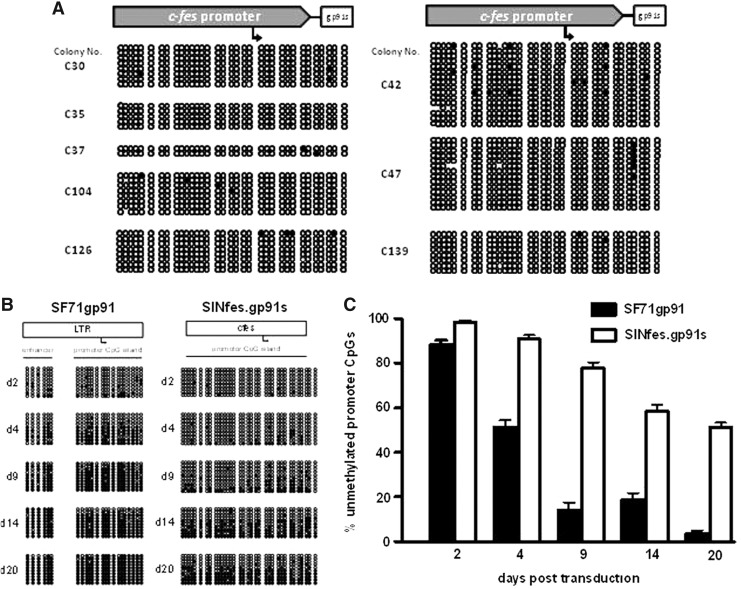
As transcriptional silencing of transgene expression was one of the unwanted effects observed in our previous phase I clinical trial conducted with the SF71gp91phox vector (Ott et al., 2006; Stein et al., 2010), we investigated the level of CpG methylation at the c-FES promoter in SINfes.gp91s-transduced hematopoietic cells. BM cells obtained from transplanted animals (low-VCN group) at week 30 were plated in methylcellulose and tested for the presence of vector sequences by PCR and superoxide production by the nitroblue tetrazolium assay. Superoxide production was detected in 38% of the colonies (WT 55.7%), which is similar to the percentage of superoxide-positive granulocytes detected in peripheral blood of these animals (20–40%, Supplementary Fig. S2C). Isolated vector-positive BM colonies but negative for superoxide production were tested for CpG methylation at the c-FES promoter by bisulfite sequencing. Only sporadic CpG methylation was detected within the fes promoter in the samples analyzed (Fig. 3A), suggesting that the lack of superoxide activity was not caused by inactivation of the c-FES promoter by methylation, but rather by chromosomal positional effects or by partial myeloid differentiation of BM colonies. Alternatively, codon optimization may favor CpG methylation because of increased GC content. Indeed Di et al. (2012) have shown that codon optimization may lead to CpG methylation and transgene silencing. However, this phenomenon was only seen when the spleen focus-forming virus (SFFV) promoter was placed in front of the codon-optimized transgene. The inclusion of a cellular promoter with low CpG content did not lead to CpG methylation of the transgene or to transgene silencing (Di et al., 2012). Therefore, the most plausible explanation for the lack of gp91phox expression in BM-derived colonies is position effect or partial myeloid differentiation. To confirm these results, a more stringent methylation assay using the murine embryonic carcinoma cell line P19 was performed. Methylation-dependent repression of gene expression in P19 cells occurs shortly after DNA transfer into the cells and recapitulates the usually slow process of promoter silencing observed in somatic cells. Thus, P19 cells provide a sensitive system to test DNA methylation-dependent inactivation of gene expression. We transduced P19 cells with the SINfes.gp91s vector. As a control we used the SF71gp91phox vector, in which gp91phox expression is under the transcriptional control of the SFFV LTR. DNA was extracted from transduced cells at different time points and analyzed for CpG methylation by bisulfite modification followed by sequencing. As expected, a rapid methylation at CpG dinucleotides occurred within the SFFV enhancer/promoter, with more than 80% of the CpG sites methylated already at day 9 post-transduction. In contrast, the c-FES promoter was at least three- to sixfold less sensitive to CpG methylation than the SFFV promoter, with 60% of the CpGs remaining unmethylated even at day 20 after transduction, a time point at which the SFFV LTR was almost fully methylated (Fig. 3B and C). These results suggest that the c-FES promoter is less prone to methylation-mediated silencing during hematopoiesis and is capable of maintaining therapeutic gene expression even in an unfavorable environment. This observation correlates well with the long-term and stable reconstitution of superoxide activity observed in transplanted CGD animals (Fig. 2).
FIG. 3.
The c-FES promoter is less prone to CpG methylation in hematopoietic cells. (A) Assessment of CpG methylation at the c-FES promoter in CFC. DNA was extracted from individual vector-positive but oxidase-negative colonies derived from the BM of transplanted X-CGD mice 30 weeks after transplantation and subjected to bisulfite conversion and sequencing. (B) The c-FES promoter is less sensitive than the SFFV promoter to CpG methylation. P19 cells were transduced with SF71gp91phox or SINfes.gp91s. DNA was extracted at different time points and analyzed for CpG methylation by bisulfite sequencing. (C) Schematic diagram summarizing the data presented in (B). Mean values±SEM are shown. BM, bone marrow; CFC, colony-forming cells; SFFV, spleen focus forming virus.
SINfes.gp91s exhibits a low genotoxic potential in vitro and in vivo
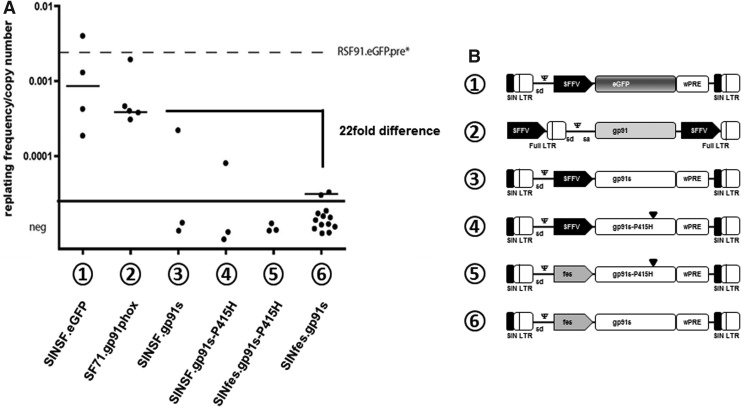
Former clinical gene therapy trials using LTR-driven vectors for modification of hematopoietic stem cells have shown that genotoxicity and subsequent development of hematological malignancies can be a major threat for the patients. Therefore, an essential component of preclinical assessment of new vectors is toxicology, including evaluation of genotoxicity in vitro and in vivo. Initially, we used the in vitro immortalization (IVIM) assay to estimate vector genotoxicity, as this assay relies on the induction of a survival advantage by insertional activation of cellular proto-oncogenes, which becomes apparent when primary murine hematopoietic cells are cultured under differentiating cytokine conditions and plated in limiting dilution (Modlich et al., 2006). This assay is frequently being used as a surrogate assay for in vivo experiments as it faithfully predicts malignant events, which may occur in gene therapy trials, at least those affecting the myeloid compartment (Baum et al., 2011). Therefore, genotoxicity of SINfes.gp91s was first investigated in the IVIM assay, using the LTR-driven SF71gp91phox and the SIN vector SINSF.eGFP vector as positive controls to validate the assay (Modlich et al., 2006). In parallel, we tested a SINSF.gp91s vector as well as a vector containing a catalytic inactive mutant of gp91phox (gp91s-P415H) (Dinauer et al., 1989) to exclude any potential toxicity of gp91phox that could have affected the readout of the assay. Lastly, in most experiments we used murine lineage negative (lin−) cells obtained from X-CGD animals to preserve a disease-specific cell background. No differences in the frequency of immortalized clones were found neither when lin− cells derived from C57Bl/6 or X-CGD animals, nor when gp91phox or its catalytic inactive mutant were used (Fig. 4). In line with previous studies of SIN vectors (Modlich et al., 2006), SINfes.gp91s displayed a significantly lower frequency of immortalization compared with its LTR-driven counterpart (Fig. 4). The mean replating frequency observed with the SINfes.gp91s vector was 3.13E-05±0.198E-05 (n=13), a value 46.6-fold lower than that observed with the SINSF.eGFP vector and 22-fold lower than that of SF71gp91phox, the vector used in our previous clinical trial (Fig. 4). Besides providing a quantitative estimation on the incidence of malignant transformation, the IVIM assay also allows to estimate the fitness of transformed cells, which is calculated from the replating frequency of transformed cells as a function of vector dose and vector design (Modlich et al., 2006, 2009; Zychlinski et al., 2008). The SINfes.gp91s vector was tested 10 times in gp91phox knockout cells in five independent assays and 3 times in wt cells (Supplementary Table S2). In two assays, one clone with replating capacity could be recovered; all others gave negative results. In eight assays, the LTR-driven SF71gp91phox vector was tested side by side and gave rise to numerous clones in all assays. Therefore, the incidence of replating clones was significantly lower for the SINfes.gp91s vector (p=0.0002; Fisher's exact test). Moreover, the two clones arising from SINfes.gp91s-transduced cells grew poorly in vitro and could not be expanded to mass cultures for extensive molecular analysis, which is an indication for low fitness of these clones. Despite this, we attempted to determine the genomic vector location in these clones by ligation-mediated PCR. A single integration site (IS) was detected for each clone. None of these could be assigned to Evi1 or Prdm16, the most common events usually found in this assay (Modlich et al., 2009). The IS in one clone mapped 790 bp upstream of the vinculin gene on chromosome 14 in forward orientation. Unfortunately, the sequence retrieved from the second clone was too short (16 bp) for a definite alignment on the mouse genome.
FIG. 4.
Frequency of immortalization of lin− cells by SINfes.gp91s in vitro. (A) lin− cells from X-CGD mice were transduced on two consecutive days at an MOI of 10 per day with different gp91phox or eGFP coding vectors, expanded in bulk for 14 days as described in the Supplementary Materials and Methods and plated at a density of 100 cells per well in 96-well plates. After 14 days, plates were screened for colony growth, and the frequency of replating was calculated based on Poisson statistics using L-Calc software. The graphic shows the frequency of replating cells corrected for the average vector copy number as measured by quantitative PCR on day 4 after transduction. The limit of detection of the assay is indicated as a thick black line. The dotted line reflects the immortalization frequency of RSF91.eGFPpre*, an LTR-driven gammaretroviral vector expressing eGFP. (B) Schematic diagram of the vectors used in this study. lin−, lineage negative; LTR, long terminal repeat; MOI, multiplicity of infection; PCR, polymerase chain reaction.
Finally, we determined the genotoxic potential of the SINfes.gp91s vector in a long-term in vivo study. Primary lin− murine BM cells isolated from male/female C57BL/6 X-CGD (CD45.2) mice were transduced with SINfes.gp91s at a total multiplicity of infection (MOI) of 6.6 and with the control vector SF91eGFP at an MOI of 5.2 to ensure high VCNs per cell, thus enhancing the probability of insertional mutagenesis. The LTR-driven SF91eGFP vector was chosen as a positive control for assay validation, as this vector is known to induce clonal dominance and leukemia in vivo (Modlich et al., 2008, 2009). Transduction efficiency on lin− cells was measured on day 10 by qPCR on vector sequences. A mean VCN of 2.8 per cell was determined for SINfes.gp91s-transduced cells, whereas this value was 1.6 vector copies per cell for SF91eGFP-transduced cells. After transduction, cells were expanded in a serum-free medium supplemented with a cytokine cocktail previously shown to expand long-term repopulating cells (Zhang et al., 2006). Expanded cells were transplanted into lethally irradiated recipients (C57BL/6 CD45.1) together with 100.000 fresh BM-derived mononuclear cells (CD45.1) to ensure efficient repopulation of transplanted animals. Animals were monitored for a total of 30 and 16 weeks for primary and secondary transplanted recipients, respectively, resulting in a total observation time of 11 months (Supplementary Fig. S3).
Engraftment of donor X-CGD CD45.2 cells into CD45.1 animals was assessed by fluorescence-activated cell sorting. Nearly all animals were reconstituted to levels above 90% with donor cells. VCNs were determined in DNA samples obtained from peripheral blood, BM, and spleen of transplanted animals with mean values ranging between 2.5 and 3.4 in primary and between 2.6 and 3.7 in secondary animals for SF91eGFP- and SINfes.gp91s-transduced cells, respectively (Fig. 5A and B). Despite high engraftment and VCN in BM, spleen, and peripheral blood, no vector-related malignant abnormalities in hematologic organs were observed in primary or secondary transplanted animals at any time point. The percentage of lymphocytes (∼70%), monocytes (∼10%), and granulocytes (∼20–25%) in the peripheral blood of the SINfes.gp91s animals were not different from those observed in control C57BL/6 animals. Similarly, the body weight of animals in the test group remained constant in a range between 20 and 30 g, and was thus not different from the control groups. Occasionally, we found CGD-related symptoms, such as granulomas, in the lungs of animals transplanted with SF91eGFP-transduced X-CGD cells.
FIG. 5.
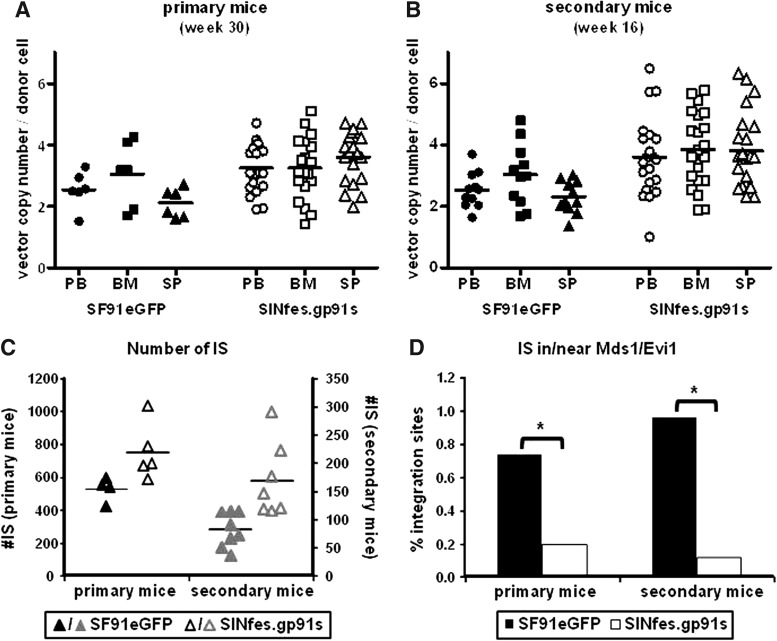
Vector copy numbers and total number of ISs per mouse in SINfes.gp91s- and SF91eGFP-transplanted animals. Vector copy numbers in DNA samples obtained from PB, BM, and SP of transplanted primary (A) and secondary (B) animals. (C) Total number of ISs per mouse. ISs were obtained from serially transplanted SF91eGFP and SINfes.gp91s mice by ligation-mediated/linear-amplification-mediated PCR and subsequent 454 sequencing. (D) Percentage of ISs found in or close to the Mds1/Evi1 locus for primary and secondary transplanted mice. With SINfes.gp91s, significantly less ISs were detected in primary and in secondary animals, respectively. *p=0.077 and 0.026 for primary and secondary mice, respectively, as determined by χ2-test. BM, bone marrow; ISs, integration sites; PB, peripheral blood; SP, spleen.
Because of the lack of overt malignancies in transplanted animals, we decided to perform a clonal distribution analysis by ligation-mediated/linear-amplification-mediated PCR in combination with deep sequencing. The identification of unique vector integration site (IS) as molecular markers for each individual cell and its clonal progeny provides information on clonal dynamics, degree of in vivo clonal skewing, and frequency of targeting worrisome genomic regions, that is, proto-oncogenes, and allows the assessment of the genotoxic potential of the vectors used (Schmidt et al., 2007). We randomly selected four SF91eGFP and five SINfes.gp91s primary animals and their respective progeny for this analysis.
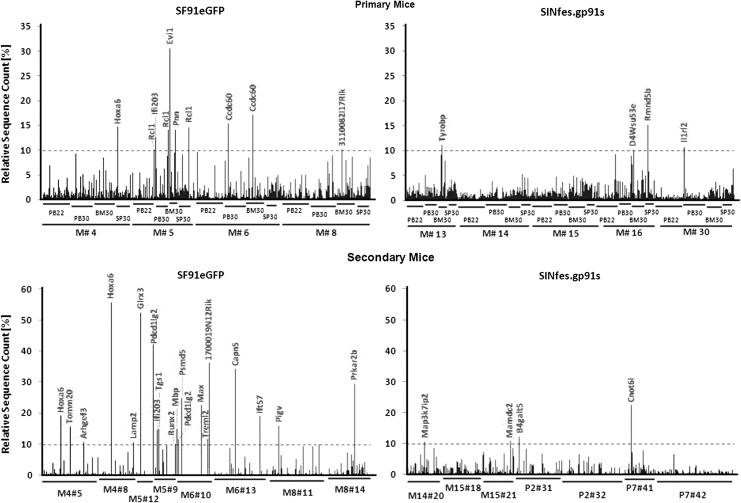
We found a mean of 482 and 714 unique IS per mice in primary SF91eGFP and SINfes.gp91s animals, respectively (Fig. 5C and Supplementary Fig. S4). A remarkably high retrieval frequency of IS sequence reads for particular genes targeted repeatedly by the SF91eGFP vector was evident in most primary animals, with Hoxa6 (15% retrieval frequency in M#4), Evi1 (31% retrieval frequency in M#5), and Ccdc60 (15–17% retrieval frequency in M#6) being among the most prominent integrations found (Fig. 6 and Supplementary Fig. S4). In contrast, in SINfes.gp91s-transplanted animals, we did not detect multiple independent vector integrations (>4) in the same genomic region (common insertion sites) with an increased retrieval frequency above 10%. Only 4 clones containing single integrations into Il1rl2 (10.1%), D4Wsu53e (10.7%), Tyrobp (11%), or Rmnd5b (15%) showed retrieval frequencies above 10%, the threshold arbitrarily set for defining dominant clones. Notably, measuring clonal contributions using the retrieval frequency of individual IS sequences gives an estimate on the presence of the respective cell clone(s) in the analyzed tissue/cell fraction and allows a reliable longitudinal monitoring of these clones over time, although the method used here does not allow a precise determination of clone size in a mixed population.
FIG. 6.
Retrieval frequency of individual ISs in SINfes.gp91s- and SF91eGFP-transplanted animals. Retrieval frequency of individual sequences in PB, BM, and SP of SF91eGFP- and SINfes.gp91s-transplanted primary (upper diagram) and secondary animals (lower diagram) at different time points after transplantation. DNA was extracted from the indicated samples, and retroviral vector integrations sites were retrieved by ligation-mediated/linear-amplification-mediated PCR and subsequent 454 high-throughput sequencing. The relative sequence count gives a semiquantitative estimation for the contribution of clones carrying a unique IS to total gene-marked hematopoiesis. A threshold of 10% was arbitrarily set to define a significant contribution of a clone to hematopoiesis. All ISs with a sequence retrieval frequency above 10% are marked.
Three of the predominant clones found in SINfes.gp91s primary animals (Tyrobp, Rmnd5b, and D4Wsu53e) were found in secondary animals, but with declining clonal contributions, most likely because of the lack of repopulating capabilities of these particular clones (Supplementary Fig. S4). Similarly, the dominant Evi1 clone detected in the BM of SF91eGFP mouse M#5 was barely detectable in secondary animals, with a clonal contribution of less than 0.04% (Supplementary Table S1). In contrast, the predominant Hoxa6 clone was found at a retrieval frequency of 19.3% and 55.8% in animals transplanted with BM cells derived from M#4, indicating a dominant clone with engraftment potential (Fig. 6). Further predominant clones in secondary animals transplanted with SF91eGFP-transduced cells contained integrations into Glrx3 (retrieval frequency 52%), Pdcd1lg2 (42%), 1700019N12Rik (36%), Capn5 (34%), and Prkar2b (30%) (Fig. 6). All of these clones were also detected in the corresponding primary animals. On the basis of the sequence reads, however, the contribution to gene marked hematopoiesis was below 5%. Most likely, clonal dominance correlated with increased transcription at the targeted locus, as we detected increased transcription levels for Evi1, Hoxa6, and Glrx3 in primary and secondary animals, respectively (Supplementary Fig. S5).
Three clones with clonal contributions slightly above the threshold of 10% were found in secondary SINfes.gp91s animals (Map3k7ip2, 10.7%; Mamdc2, 10.9%; B4galt5, 12.2%). However, two of these were not detected in a second recipient of the same BM cells (Mamdc2) or its contribution was well below the threshold of 10% (B4galt5). For the third one (Map3k7ip2), we had only one secondary mouse, as the second animal transplanted with the same BM cells died at week two post-transplantation because of engraftment failure. One clone with a dominance above 20% was observed in an animal (P7#41) transplanted with a pool of BM cells obtained from M#29 and M#30 (Fig. 6). This particular clone contained a proviral integration upstream of the Cnot6I gene (CCR4-NOT transcription complex; NM_144910) and was found at a frequency of 22.3%. However, attempts to detect increased transcripts in BM cells from this animal failed (Supplementary Fig. S5). Indeed, the transcription levels for Cnot6I in P7#41 were not significantly different from those found in animal P7#42 (p>0.05), in which the contribution of the Cnot6I clone to total gene marked hematopoiesis was less than 7% (Supplementary Fig. S4). This is reminiscent of a previous study in which clonal dominance was observed upon transplantation of hematopoietic cells transduced with a SIN vector lacking an internal enhancer–promoter sequence, suggesting that clonal restriction is a physiological phenomenon in this transplantation setting (Cornils et al., 2009, 2012).
In view of the relevance of integrations into or close to the Mds1/Evi1 locus, we analyzed the frequency of integrations at this locus in SINfes.gp91s- and SF91eGFP-transplanted animals independently of their contribution to gene-marked hematopoiesis. From a total of 1759 (primary mice)/418 (secondary mice) and 3324 (primary mice)/836 (secondary mice) unique IS for SF91eGFP and SINfes.gp91s, respectively, the percentage of IS that were identified in or close to the Mds1/Evi1 locus in primary and secondary animals was significantly less in SINfes.gp91s- than in SF91eGFP-transplanted animals (p=0.077 and 0.026, respectively, determined by χ2-test) (Fig. 5D). On the basis of this observation, the risk of insertional activation of Evi1 by the SINfes.gp91s vector is substantially reduced compared with SF91eGFP.
In addition, we applied the Shannon and the Simpson diversity indexes for a more precise assessment of the heterogeneity of hematopoietic repopulation in the treated animals. The Shannon index is used to determine the number of unique repopulating clones (richness of a population), whereas the Simpson index measures the degree of clonal dominance with a high index indicating low clonal dominance (Shannon, 1948; Simpson, 1949). We observed that in both cohorts the average indices were substantially increased for SINfes.gp91s-treated animals compared with the SF91 controls (Supplementary Fig. S6), thus confirming the reduced probability of this vector to affect the transduced cell and to cause unwanted clonal outgrowth. These data, together with the low frequency of in vitro immortalized clones found in SINfes.gp91s-transduced lin− cells, even at >15 vector copies per cell (Fig. 4), demonstrate a low genotoxicity profile for this vector and reinforce the relevance of vector design for the safety of future clinical studies (Modlich et al., 2006; Montini et al., 2009; Schambach et al., 2009). In addition, our studies support the use of clonal dominance as a surrogate assay to define the genotoxicity profile of integrating vectors. The use of new technologies, that is, barcoding, to mark gene-transduced cells (Gerrits et al., 2010; Glimm et al., 2011; Lu et al., 2011) will certainly facilitate the analysis of clonal dominance in the future.
Conclusions
Gene therapy has proven to be effective in correcting X-CGD, at least temporarily. A series of phaseI/II studies using a variety of different gammaretroviral vectors have shown partial correction of the disease shortly after reinfusion of gene-transduced cells [reviewed in (Grez et al., 2011)]. However, long-term correction of the disease failed in most cases. Probably, the low-intensity conditioning used in these trials accounts for treatment failure, as in the absence of in vivo selection autologous transplantation of either gammaretrovirus- or lentivirus-transduced hematopoietic stem cells in nonhuman primates treated with nonmyeloablative regimens have generally resulted in low levels (1% or less) of long-term marking (Huhn et al., 1999; Rosenzweig et al., 1999; Brenner et al., 2006; Kahl et al., 2006; Kang et al., 2006). Thus, for future trials, intensified conditioning may be necessary, as gene-corrected cells are not expected to have a selection advantage in CGD. However, the application of LTR-driven vectors, as used in previous trials, would pose a serious safety concern under these conditions as more stem and primitive progenitor cells are supposed to engraft after myeloablative conditioning. These considerations prompted us to develop an optimized SIN gammaretrovirus-based vector (SINfes.gp91s) for the clinical application in X-CGD. The SINfes.gp91s vector restores superoxide production in X-CGD cells to therapeutically relevant levels even at low VCNs and has a significantly improved genotoxicity profile without signs of transformation in genotoxicity assays, making this vector a promising candidate for safe future clinical application. Indeed, a stable packaging cell line for vector production was established and clinical vector lots have been produced (Loew et al., 2010). Moreover, the SINfes.gp91s vector was recently approved for the modification of human hematopoietic cells in the context of a clinical phase I/II study (Paul Ehrlich Institute reference number A.11492.01.1).
Supplementary Material
Acknowledgments
This work was supported by grants from the Bundesministerium für Bildung und Forschung (Grant 01GU0811, TP2b, to M.G. and IFB-Tx [01EO0802] to C.Ba.) and iGENE to M.G., C.Ba., M.S., and C.v.K.); the Chronic Granulomatous Disorder Research Trust, London (Grant J4G/04B/GT to M.G.); the German Academic Exchange Service (DAAD, 0315187), the Research Priority Program 1230 from the Deutsche Forschungsgemeinschaft (to M.G., C.Ba., M.S., and C.v.K.); the LOEWE Center for Cell and Gene Therapy Frankfurt funded by the Hessische Ministerium für Wissenschaft und Kunst (HMWK; funding reference number: III L 4–518/17.004 [2010]), by the Deutsche Forschungsgemeinschaft (DFG) Graduate Program GK1172-Biologicals (to C.Br. and K.B.K.); and National Institutes of Health Grants P01 HL53586 and the Riley Children's Foundation (to M.C.D.). The Georg-Speyer-Haus is supported by the Bundesministerium für Gesundheit and the Hessisches Ministerium für Wissenschaft und Kunst. We also thank Natalie Stull and Sutha John for assistance with Aspergillus challenge models.
Author Disclosure Statement
The authors declare that they have no conflicts of interest.
References
- Aiuti A. Bacchetta R. Seger R., et al. Gene therapy for primary immunodeficiencies: part 2. Curr. Opin. Immunol. 2012;24:585–591. doi: 10.1016/j.coi.2012.07.012. [DOI] [PubMed] [Google Scholar]
- Avedillo Diez I. Zychlinski D. Coci E.G., et al. Development of novel efficient SIN vectors with improved safety features for Wiskott-Aldrich Syndrome stem cell based gene therapy. Mol. Pharmaceutics. 2011;8:1525–1537. doi: 10.1021/mp200132u. [DOI] [PubMed] [Google Scholar]
- Baum C. Von Kalle C. Staal F.J., et al. Chance or necessity? Insertional mutagenesis in gene therapy and its consequences. Mol. Ther. 2004;9:5–13. doi: 10.1016/j.ymthe.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Baum C. Modlich U. Gohring G., et al. Concise review: managing genotoxicity in the therapeutic modification of stem cells. Stem Cells. 2011;29:1479–1484. doi: 10.1002/stem.716. [DOI] [PubMed] [Google Scholar]
- Biffi A. Bartolomae C.C. Cesana D., et al. Lentiviral vector common integration sites in preclinical models and a clinical trial reflect a benign integration bias and not oncogenic selection. Blood. 2011;117:5332–5339. doi: 10.1182/blood-2010-09-306761. [DOI] [PubMed] [Google Scholar]
- Bjorgvinsdottir H. Ding C. Pech N., et al. Retroviral-mediated gene transfer of gp91phox into bone marrow cells rescues defect in host defense against Aspergillus fumigatus in murine X-linked chronic granulomatous disease. Blood. 1997;89:41–48. [PubMed] [Google Scholar]
- Bohler M.C. Seger R.A. Mouy R., et al. A study of 25 patients with chronic granulomatous disease: a new classification by correlating respiratory burst, cytochrome b, and flavoprotein. J. Clin. Immunol. 1986;6:136–145. doi: 10.1007/BF00918746. [DOI] [PubMed] [Google Scholar]
- Booth C. Gaspar H.B. Thrasher A.J. Gene therapy for primary immunodeficiency. Curr. Opin. Pediatr. 2011;23:659–666. doi: 10.1097/MOP.0b013e32834cd67a. [DOI] [PubMed] [Google Scholar]
- Brenner S. Ryser M.F. Choi U., et al. Polyclonal long-term MFGS-gp91phox marking in rhesus macaques after nonmyeloablative transplantation with transduced autologous peripheral blood progenitor cells. Mol. Ther. 2006;14:202–211. doi: 10.1016/j.ymthe.2006.01.015. [DOI] [PubMed] [Google Scholar]
- Cartier N. Hacein-Bey-Abina S. Bartholomae C.C., et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- Cavazzana-Calvo M. Payen E. Negre O., et al. Transfusion independence and HMGA2 activation after gene therapy of human β-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavazzana-Calvo M. Fischer A. Hacein-Bey-Abina S., et al. Gene therapy for primary immunodeficiencies: part 1. Curr. Opin. Immunol. 2012;24:580–584. doi: 10.1016/j.coi.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Cornils K. Lange C. Schambach A., et al. Stem cell marking with promoter-deprived self-inactivating retroviral vectors does not lead to induced clonal imbalance. Mol. Ther. 2009;17:131–143. doi: 10.1038/mt.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornils K. Bartholomae C.C. Thielecke L., et al. Comparative clonal analysis of reconstitution kinetics after transplantation of hematopoietic stem cells gene marked with a lentiviral SIN or a gamma-retroviral LTR vector. Exp. Hematol. 2012;41:28–38.e3. doi: 10.1016/j.exphem.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Corrigan-Curay J. Cohen-Haguenauer O. O'reilly M., et al. Challenges in vector and trial design using retroviral vectors for long-term gene correction in hematopoietic stem cell gene therapy. Mol. Ther. 2012;20:1084–1094. doi: 10.1038/mt.2012.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di W.L. Semenova E. Larcher F., et al. Human involucrin promoter mediates repression-resistant and compartment-specific LEKTI expression. Hum. Gene Ther. 2012;23:83–90. doi: 10.1089/hum.2011.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinauer M.C. Curnutte J.T. Rosen H., et al. A missense mutation in the neutrophil cytochrome b heavy chain in cytochrome-positive X-linked chronic granulomatous disease. J. Clin. Invest. 1989;84:2012–2016. doi: 10.1172/JCI114393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinauer M.C. Gifford M.A. Pech N., et al. Variable correction of host defense following gene transfer and bone marrow transplantation in murine X-linked chronic granulomatous disease. Blood. 2001;97:3738–3745. doi: 10.1182/blood.v97.12.3738. [DOI] [PubMed] [Google Scholar]
- Fischer A. Hacein-Bey-Abina S. Cavazzana-Calvo M. Gene therapy for primary immunodeficiencies. Hematol. Oncol. Clin. North. Am. 2011;25:89–100. doi: 10.1016/j.hoc.2010.11.007. [DOI] [PubMed] [Google Scholar]
- Gerrits A. Dykstra B. Kalmykowa O.J., et al. Cellular barcoding tool for clonal analysis in the hematopoietic system. Blood. 2010;115:2610–2618. doi: 10.1182/blood-2009-06-229757. [DOI] [PubMed] [Google Scholar]
- Glimm H. Ball C.R. Von Kalle C. You can count on this: barcoded hematopoietic stem cells. Cell Stem Cell. 2011;9:390–392. doi: 10.1016/j.stem.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Goebel W.S. Mark L.A. Billings S.D., et al. Gene correction reduces cutaneous inflammation and granuloma formation in murine X-linked chronic granulomatous disease. J. Invest. Dermatol. 2005;125:705–710. doi: 10.1111/j.0022-202X.2005.23908.x. [DOI] [PubMed] [Google Scholar]
- Grez M. Reichenbach J. Schwäble J., et al. Gene therapy of chronic granulomatous disease: the engraftment dilemma. Mol. Ther. 2011;19:28–35. doi: 10.1038/mt.2010.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. Garrigue A. Wang G.P., et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland S.M. Chronic granulomatous disease. Clin. Rev. Allergy Immunol. 2010;38:3–10. doi: 10.1007/s12016-009-8136-z. [DOI] [PubMed] [Google Scholar]
- Howe S.J. Mansour M.R. Schwarzwaelder K., et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhn R.D. Tisdale J.F. Agricola B., et al. Retroviral marking and transplantation of rhesus hematopoietic cells by nonmyeloablative conditioning. Hum. Gene Ther. 1999;10:1783–1790. doi: 10.1089/10430349950017464. [DOI] [PubMed] [Google Scholar]
- Huston M.W. Van Til N.P. Visser T.P., et al. Correction of murine SCID-X1 by lentiviral gene therapy using a codon-optimized IL2RG gene and minimal pretransplant conditioning. Mol. Ther. 2011;19:1867–1877. doi: 10.1038/mt.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl C.A. Tarantal A.F. Lee C.I., et al. Effects of busulfan dose escalation on engraftment of infant rhesus monkey hematopoietic stem cells after gene marking by a lentiviral vector. Exp. Hematol. 2006;34:369–381. doi: 10.1016/j.exphem.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Kang E.M. Hsieh M.M. Metzger M., et al. Busulfan pharmacokinetics, toxicity, and low-dose conditioning for autologous transplantation of genetically modified hematopoietic stem cells in the rhesus macaque model. Exp. Hematol. 2006;34:132–139. doi: 10.1016/j.exphem.2005.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhns D.B. Alvord W.G. Heller T., et al. Residual NADPH oxidase and survival in chronic granulomatous disease. NEJM. 2010;363:2600–2610. doi: 10.1056/NEJMoa1007097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loew R. Meyer Y. Kuehlcke K., et al. A new PG13-based packaging cell line for stable production of clinical-grade self-inactivating gamma-retroviral vectors using targeted integration. Gene Ther. 2010;17:272–280. doi: 10.1038/gt.2009.134. [DOI] [PubMed] [Google Scholar]
- Lu R. Neff N.F. Quake S.R., et al. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat. Biotechnol. 2011;29:928–933. doi: 10.1038/nbt.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlich U. Bohne J. Schmidt M., et al. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108:2545–2553. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlich U. Schambach A. Brugman M.H., et al. Leukemia induction after a single retroviral vector insertion in Evi1 or Prdm16. Leukemia. 2008;22:1519–1528. doi: 10.1038/leu.2008.118. [DOI] [PubMed] [Google Scholar]
- Modlich U. Navarro S. Zychlinski D., et al. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol. Ther. 2009;17:1919–1928. doi: 10.1038/mt.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montini E. Cesana D. Schmidt M., et al. Hematopoietic stem cell gene transfer in a tumor-prone mouse model uncovers low genotoxicity of lentiviral vector integration. Nat. Biotechnol. 2006;24:687–696. doi: 10.1038/nbt1216. [DOI] [PubMed] [Google Scholar]
- Montini E. Cesana D. Schmidt M., et al. The genotoxic potential of retroviral vectors is strongly modulated by vector design and integration site selection in a mouse model of HSC gene therapy. J. Clin. Invest. 2009;119:964–975. doi: 10.1172/JCI37630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Carranza B. Gentsch M. Stein S., et al. Transgene optimization significantly improves SIN vector titers, gp91phox expression and reconstitution of superoxide production in X-CGD cells. Gene Ther. 2009;16:111–118. doi: 10.1038/gt.2008.143. [DOI] [PubMed] [Google Scholar]
- Naldini L. Ex vivo gene transfer and correction for cell-based therapies. Nat. Rev. Genet. 2011;12:301–315. doi: 10.1038/nrg2985. [DOI] [PubMed] [Google Scholar]
- Ott M.G. Schmidt M. Schwarzwaelder K., et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Petersen J.E. Hiran T.S. Goebel W.S., et al. Enhanced cutaneous inflammatory reactions to Aspergillus fumigatus in a murine model of chronic granulomatous disease. J. Invest. Dermatol. 2002;118:424–429. doi: 10.1046/j.0022-202x.2001.01691.x. [DOI] [PubMed] [Google Scholar]
- Rivat C. Santilli G. Gaspar H.B., et al. Gene therapy for primary immunodeficiencies. Hum. Gene Ther. 2012;23:668–675. doi: 10.1089/hum.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig M. Macvittie T.J. Harper D., et al. Efficient and durable gene marking of hematopoietic progenitor cells in nonhuman primates after nonablative conditioning. Blood. 1999;94:2271–2286. [PubMed] [Google Scholar]
- Sadat M.A. Dirscherl S. Sastry L., et al. Retroviral vector integration in post-transplant hematopoiesis in mice conditioned with either submyeloablative or ablative irradiation. Gene Ther. 2009;16:1452–1464. doi: 10.1038/gt.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaramuzza S. Biasco L. Ripamonti A., et al. Preclinical safety and efficacy of human CD34(+) cells transduced with lentiviral vector for the treatment of Wiskott-Aldrich syndrome. Mol. Ther. 2013;21:175–184. doi: 10.1038/mt.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schambach A. Bohne J. Chandra S., et al. Equal potency of gammaretroviral and lentiviral SIN vectors for expression of O6-methylguanine-DNA methyltransferase in hematopoietic cells. Mol. Ther. 2006;13:391–400. doi: 10.1016/j.ymthe.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Schambach A. Swaney W.P. Van Der Loo J.C. Design and production of retro- and lentiviral vectors for gene expression in hematopoietic cells. Methods Mol. Biol. 2009;506:191–205. doi: 10.1007/978-1-59745-409-4_14. [DOI] [PubMed] [Google Scholar]
- Schmidt M. Schwarzwaelder K. Bartholomae C., et al. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR) Nat. Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- Segal A.W. The NADPH oxidase and chronic granulomatous disease. Mol. Med. Today. 1996;2:129–135. doi: 10.1016/1357-4310(96)88723-5. [DOI] [PubMed] [Google Scholar]
- Segal B.H. Leto T.L. Gallin J.I., et al. Genetic, biochemical, and clinical features of chronic granulomatous disease. Medicine (Baltimore) 2000;79:170–200. doi: 10.1097/00005792-200005000-00004. [DOI] [PubMed] [Google Scholar]
- Seymour L.W. Thrasher A.J. Gene therapy matures in the clinic. Nat. Biotechnol. 2012;30:588–593. doi: 10.1038/nbt.2290. [DOI] [PubMed] [Google Scholar]
- Shannon C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948;27:379–423. and 623–656. [Google Scholar]
- Simpson E.H. Measurement of diversity. Nature. 1949;163:688. [Google Scholar]
- Stein S. Ott M.G. Schultze-Strasser S., et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat. Med. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- Thornhill S.I. Schambach A. Howe S.J., et al. Self-inactivating gammaretroviral vectors for gene therapy of X-linked severe combined immunodeficiency. Mol. Ther. 2008;16:590–598. doi: 10.1038/sj.mt.6300393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolar J. Becker P.S. Clapp D.W., et al. Gene therapy for fanconi anemia: one step closer to the clinic. Hum. Gene Ther. 2012;23:141–144. doi: 10.1089/hum.2011.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will E. Bailey J. Schuesler T., et al. Importance of murine study design for testing toxicity of retroviral vectors in support of phase I trials. Mol. Ther. 2007;15:782–791. doi: 10.1038/sj.mt.6300083. [DOI] [PubMed] [Google Scholar]
- Zhang C.C. Kaba M. Ge G., et al. Angiopoietin-like proteins stimulate ex vivo expansion of hematopoietic stem cells. Nat. Med. 2006;12:240–245. doi: 10.1038/nm1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S. Mody D. Deravin S.S., et al. A self-inactivating lentiviral vector for SCID-X1 gene therapy that does not activate LMO2 expression in human T cells. Blood. 2010;116:900–908. doi: 10.1182/blood-2009-10-250209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zychlinski D. Schambach A. Modlich U., et al. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol. Ther. 2008;16:718–725. doi: 10.1038/mt.2008.5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.