Abstract
Background
Intestinal barrier defects are common in patients with inflammatory bowel disease (IBD). To identify which components could underlie these changes, we performed an in-depth analysis of epithelial barrier genes in IBD.
Methods
A set of 128 intestinal barrier genes was selected. Polygenic risk scores were generated based on selected barrier gene variants that were associated with Crohn’s disease (CD) or ulcerative colitis (UC) in our study. Gene expression was analysed using microarray and quantitative reverse transcription PCR. Influence of barrier gene variants on expression was studied by cis-expression quantitative trait loci mapping and comparing patients with low and high risk scores.
Results
Barrier risk scores were significantly higher in IBD patients than controls. At single-gene level, the associated barrier SNPs were most significantly enriched in PTGER4 for CD, and HNF4A for UC. As a group, the regulating proteins were most enriched for CD and UC. Expression analysis showed that many epithelial barrier genes were significantly dysregulated in active CD and UC, with overrepresentation of mucus layer genes. In uninflamed CD ileum and IBD colon, most barrier gene levels restored to normal, except for MUC1 and MUC4 that remained persistently increased compared to controls. Expression levels did not depend on cis-regulatory variants, nor combined genetic risk.
Conclusions
We found genetic and transcriptomic dysregulations of key epithelial barrier genes and components in IBD. Of these, we believe mucus genes, in particular MUC1 and MUC4, play an essential role in the pathogenesis of IBD, and could represent interesting targets for treatment.
Keywords: intestinal barrier, genetic analysis, inflammatory bowel disease, mucosal gene expression
Introduction
Inflammatory bowel diseases (IBDs), including Crohn’s disease (CD) and ulcerative colitis (UC), are a group of chronic, relapsing inflammatory disorders of the gut that affect an increasing number of individuals around the world (1). The current hypothesis on the pathogenesis of IBD is that the disease results from complex interactions between the host genome, exposome, gut microbiome and mucosal immune system (2, 3). In this regard, also a dysfunctional intestinal barrier has long been recognized as a key pathogenic factor in IBD (4). The intestinal barrier is located at the interface between the external luminal environment and the internal immune system, and has the complex task to defend against potentially harmful molecules and microorganisms, while being permeable to essential nutrients and solutes (5). It is thought that intestinal barrier defects in IBD patients result in an increased uptake of luminal antigens across the intestinal epithelium, which in turn would trigger the immune system and the development of mucosal inflammation. However, whether mucosal barrier alterations represent a primary dysfunction in the aetiology of IBD, or develop as consequence of ongoing inflammatory processes in IBD patients, is not entirely clear (6). Observations of increased intestinal permeability in a proportion of healthy first-degree relatives of IBD patients suggest that intestinal barrier dysfunction might be genetically determined, and not only due to the impact of inflammatory mediators (7–15). Genome-wide association studies (GWAS) have also implicated the intestinal epithelial barrier as one of the key pathways in the pathogenesis of IBD (16–18).
The general structure of the intestinal barrier is based on several components contributing to its function as a physical barrier between the luminal and internal environment, together with elements from the mucosal immune system that create an immunological defence barrier (6, 19). The mucus layer provides the most apical line of defence against the luminal environment, and forms a sieve-like gel structure that prevents large particles and bacteria from contacting the underlying intestinal epithelium (20). Besides the predominant enterocytes, the epithelium is composed of other specialised cell types with a wide array of functions, including goblet cells that produce the gel-like mucus; paneth cells that secrete antimicrobial peptides reinforcing the immune barrier; and microfold cells that support transport of large luminal antigens and microbiota to immune cells in the lamina propria (5, 21). The intestinal epithelial cells themselves constitute by far the strongest determinants of the physical intestinal barrier through the establishment of an almost impermeable polarised monolayer along the gut wall in the absence of specific transporters. The intercellular space is furthermore sealed by junctional protein complexes, of which the tight junctions are located at the most apical pole of the epithelial cells. Tight junctions are the main gatekeepers of the paracellular space and can mediate permeability of ions and small molecules up to 20 kDa. Adherens junctions and desmosomes, in contrast, form strong adhesive bonds and are primarily responsible of maintaining tissue cohesion and integrity (22, 23). Both tight junctions and adherens junctions are dependent on scaffolding proteins for their formation, and may interact with the cytoskeleton and a broad range of signalling molecules for their regulation (24). At the basal side of the epithelium, hemidesmosomes take care of the firm attachment of the cells to the basement membrane and the extracellular matrix, which in turn also control intestinal functions (25). Given the complex organisation and regulation of the intestinal mucosal barrier, there is a need to identify which elements are most critical for the pathophysiology of IBD.
In the present study, we performed an in-depth characterisation of intestinal epithelial barrier genes in IBD patients, and combined genetic and transcriptomic approaches to get a better view on disease-relevant genes and components of the intestinal epithelial barrier. We first evaluated genetic risk scores based on variants in barrier genes, and searched for genes and barrier components that were most enriched at genetic level. Second, we investigated expression levels of barrier genes using intestinal mucosal tissue from IBD patients. Finally, we also analysed whether the barrier gene variants regulated the mucosal gene expression levels in our study cohort.
Materials and Methods
Ethical statement
Subjects were recruited at the outpatient IBD clinic of the University Hospitals Leuven, Belgium. The study was approved by the ethics committee of the UZ/KU Leuven (S53684/B322201213950), with written informed consent from all individuals prior to sample collection.
Selection of intestinal barrier genes
A literature search was performed in PubMed to select genes involved in intestinal epithelial barrier function. Different combinations of the following search terms were used: “inflammatory bowel disease”, “Crohn’s disease”, “ulcerative colitis”, “intestinal barrier function”, “intestinal integrity”, “intestinal epithelium”, “gut barrier”, “mucosal permeability” and “barrier genes”. Importantly, also genes without previous evidence for their significance in IBD, but essential for the structure of the intestinal barrier were included. For gene selection, we focused on the intestinal epithelium as physical barrier, and excluded genes involved in immunological barrier function. Subdivision of the genes into barrier components/categories was performed at the end of the selection.
Genetic risk study
Genotyping of 1696 CD patients, 884 UC patients and 849 unrelated controls from our center was performed before via Immunochip (Table 1) (17, 18). For this study, we first extracted all SNPs in the selected barrier genes, including markers located within 50 kb up- or downstream of the transcription start/end site of the genes (n=3220). All these SNPs passed quality control according to the criteria as described before (17, 18). Highly correlated SNPs (SNPs in high linkage disequilibrium, r2>0.7) were subsequently excluded, leaving 1317 barrier SNPs for association. Comparative analysis between cases (CD or UC) and controls was performed using logistic regression in PLINK (v1.07). A CD or UC polygenic barrier risk score was defined for each individual by counting the total number of risk alleles for the nominally significant disease-associated SNPs (defined as uncorrected p<0.05) in the CD or UC versus controls analyses respectively. Comparison of the combined barrier risk scores between cases (CD or UC) and controls was done using Mann-Whitney U tests. Quartile analysis was done using the Chi-squared test in R 3.2.5.
Table 1. Overview of the number of samples.
| Active UC | Inactive UC | Active CD | Inactive CD | Controls | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Colon | Ileum | Colon | Ileum | Colon | Ileum | Colon | Ileum | Colon | Ileum | |
| Genetic risk | 884 | 1696 | 849 | |||||||
| Microarray | 74 | - | 23 | - | 8 | 51 | - | 16 | 11 | 11 |
| qRT-PCR | 72 | - | 22 | - | 8 | 51 | 26 | 14 | 11 | 11 |
| eQTL* | 56 | - | 20 | - | 3 | 34 | - | 12 | 0 | 0 |
| Q1/Q4-expressiona* | 32 | - | 12 | - | 2 | 15 | - | 4 | 0 | 0 |
UC, ulcerative colitis; CD, Crohn’s disease; qRT-PCR, quantitative reverse transcription PCR; eQTL, expression quantitative trait loci; Q1/Q4-expression, expression analysis of the lowest genetic risk scores (quartile 1, Q1) and highest genetic risk scores (quartile 4, Q4).
eQTL and risk comparisons were performed for the individuals with both genetic and microarray data.
Number of samples within Q1 and Q4 of the CD or UC genetic risk scores.
To evaluate if specific genes or barrier components were enriched in independently associated SNPs, we compared the number of (non-)associated variants in a given gene to the number of (non-)associated variants in the other genes for gene-level enrichment; and the number of (non-)significant genes (significant defined as having at least one associated SNP) in a given barrier component with those in the other components for component-level enrichment. Comparisons were done using the Fisher Exact test in R 3.2.5 for 2x2 contingency tables, taking into account the total number of variants in each gene or barrier component. P<0.05 was considered as enriched.
Mucosal gene expression study
Patients and biopsies
Endoscopic mucosal biopsies were obtained from 198 IBD patients and 22 controls for microarray and/or quantitative reverse transcription PCR (qRT-PCR) analysis. The biopsy specimens included colon from 97 UC patients (74 with active disease, 23 with inactive disease), 34 CD patients (eight with active colonic disease, 26 with inactive disease), and 11 controls; and terminal ileum from 67 CD patients (51 with active ileal disease, 16 with inactive disease) and 11 controls (Table1). The uninflamed colon biopsies from CD patients were solely used for qRT-PCR analysis. Baseline characteristics from the individuals are presented in Supplementary Table 1. All biopsies were taken from different patients (no paired samples). Disease activity of the patients was based on endoscopic findings, with active disease defined as Mayo endoscopic subscore ≥2 for UC, and the presence of ulcers for CD patients. The control group, who underwent endoscopy for polyp screening, had normal mucosa at endoscopic level. The biopsies were immediately snap-frozen in liquid nitrogen upon extraction, and stored at -80°C until RNA isolation.
RNA isolation and microarray analysis
Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Venlo, the Netherlands). Assessment of RNA integrity and quantity was performed by the 2100 Bioanalyzer (Agilent, Waldbronn, Germany) and the Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, US), respectively. The isolated RNA was analysed with Affymetrix GeneChip Human Gene 1.0 ST Arrays (Affymetrix, Santa Clara, CA, USA) (GSE75214), and as previously described by Vanhove et al. (26). Comparative analyses between the studied groups were performed using the R/Bioconductor LIMMA (linear models for microarray data) package (27). Differential expression was calculated based on moderated t-statistics with correction for multiple testing according to the Benjamini-Hochberg false discovery rate (FDR) (28). Because the main focus of this study was to evaluate the intestinal epithelial barrier, we filtered the results from the genome-wide comparative analyses for the gene probe sets representing the selected barrier genes. Gene probe sets with a >2-fold change (FC) and FDR<0.05, showing multiple testing correction for the entire array, were considered biologically significant. The gene probe set of IL8 (inflammatory marker) was included to evaluate inflammation. Enrichment of genes in the specified categories of the intestinal barrier was evaluated using the Fisher-Exact test in R 3.2.5. Correlations with IL8 were studied with the Spearman’s Rank Correlation test in IBM SPSS statistics 22. The microarray log2 expression values were used for the correlation analyses, and the colon (n=116) and ileum (n=78) samples were studied separately. P<0.05 was considered significant.
In order to evaluate the relation between the barrier gene expression levels and genetic barrier risk, pairwise comparisons in LIMMA (as above) were performed for patients with low and high genetic barrier risk. Low and high risk was defined as a genetic barrier risk score below the 25th percentile value (quartile (Q) 1 = Q1), or above the 75th percentile value (Q4) respectively (Table 1).
Quantitative reverse transcription PCR
Based on the significance levels in the comparisons and/or their relevance for both tissue types (colon and ileum), the following genes were selected for validation by qRT-PCR: MUC1, MUC4, TFF1, CLDN1, CLDN8, OCLN, DSG3 and MAGI1. Beta-actin was used as endogenous reference gene. The primer and probe sequences (Sigma-Aldrich, Diegem, Belgium) for the genes were custom-designed using OligoAnalyzer 3.1 software (see Supplementary Table 2). The RevertAid H Minus First Strand cDNA synthesis kit (Fermentas, St. Leon-Rot, Germany) was used to synthesize cDNA from 0.5 µg total RNA, according to the manufacturer’s protocol. Five samples were excluded, because insufficient cDNA was available. The qRT-PCR experiments were performed in duplicate using the SensiFast Probe No-ROX Kit (GC Biotech, Alphen aan den Rijn, The Netherlands) in a final reaction volume of 20 µl, on a Rotor-Gene 3000 instrument (Corbett Research, Mortlake, Australia). Cycle threshold values for each gene were determined by the Rotor-Gene 6 software package. The relative mRNA expression values of the barrier genes were calculated as ratio relative to the endogenous reference gene beta-actin (Pfaffl method) (29). Statistical analysis of the results was performed using two-tailed Mann-Whitney U tests for unpaired samples (IBM SPSS statistics 22), and a significance level of 0.05 was used.
Expression quantitative trait loci (eQTL) mapping
The genotype profiles and gene expression data were combined by performing cis-eQTL mapping on the available set of samples in our cohort with both genetic marker and microarray expression information: inflamed (n=56) and normal (n=20) colon from UC patients, inflamed colon (n=3) from CD patients, and inflamed (n=34) and normal (n=12) ileum from CD patients (Table 1). The maximal distance between each gene and SNP was limited to one mega-base (Mb). Only SNPs with a minor allele frequency (MAF)>0.05, and low linkage disequilibrium (r2<0.1) were selected for analysis (n=17.108, of which 2329 in cis of the barrier genes). Direct pairwise regressions were performed using the Matrix eQTL package in R 3.2.5 (30). Each location (colon, ileum) and disease activity status (active, inactive) was analysed separately, because the microarray results pointed towards distinct profiles for these groups. Within each subgroup, we again filtered for MAF<0.05 during the eQTL analysis to avoid false positive results. Correction for multiple testing was performed using the Benjamini-Hochberg procedure implemented in Matrix eQTL.
Results
Epithelial barrier gene selection and classification
We selected 128 genes related to physical intestinal barrier function. Of these, 25 were classified as part of the mucus layer, 34 as tight junctions, five as adherens junctions, 14 as desmosomes, four as hemidesmosomes, 17 as cytoskeleton, nine as extracellular matrix, and 20 as regulating proteins (see Supplementary Table 3).
Genetics of epithelial barrier genes
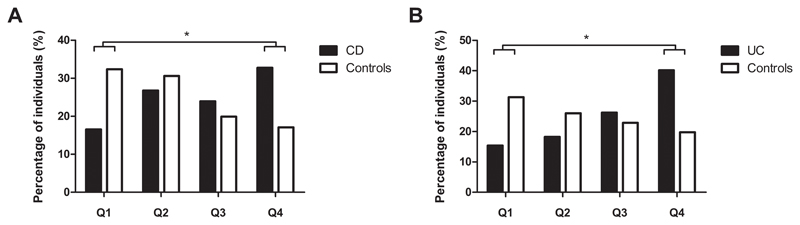
Eighty-two SNPs were nominally significant for association with CD, and 69 SNPs with UC (see Supplementary Table 4). None of these remained significant after correction for multiple testing (<3.8x10-5 for 1317 SNPs). When considering the total number of risk alleles for the nominally significant SNPs per individual, CD patients had significantly higher CD barrier risk scores compared to controls (median 82 [interquartile range (IQR): 77-87] versus 78 [IQR: 73-83], p<2.2x10-16) (see Supplementary Figure 1A for the distribution of the CD barrier risk scores). The median number of UC barrier risk alleles also was significantly higher in UC patients than in controls (68 [IQR: 64-73] versus 64 [IQR: 60-69], p<2.2x10-16) (see Supplementary Figure 1B for the distribution of the UC barrier risk scores). Quartile analysis of the barrier risk scores showed that a higher proportion of CD patients had CD barrier risk scores in Q4 versus controls (32.8% versus 17.1%), with proportionally less patients in Q1 versus controls (16.5% versus 32.4%) (p<2.2x10-16) (Figure 1A). Similar findings were seen for the UC barrier risk scores: more UC patients in Q4 (40.2% versus 19.8%), while less patients were found in Q1 compared to controls (15.4% versus 31.3%) (p<2.2x10-16) (Figure 1B).
Figure 1. Quartile analysis of the barrier risk scores in patients and controls.
The percentage of individuals in the quartiles (Q1-Q4) of the CD barrier risk scores (A) and UC barrier risk scores (B). Comparisons of the number of individuals in Q1 and Q4 was done with Chi-squared testing. *Statistically significant (p<2.2x10-16 for A and B).
CD, Crohn’s disease; UC, ulcerative colitis; Q, quartile.
In addition to the combined risk of the genetic barrier variants, we evaluated if the nominally associated SNPs were overrepresented in specific genes or components of the intestinal barrier. Comparison of the numbers of associated variants in the selected barrier genes showed enrichment in MUC19, MUC22 and TFF1 [mucus layer], and PTGER4 [regulating proteins] for CD (p=4.30x10-2, 9.41x10-3, 1.12x10-2 and 8.94x10-4, respectively), whereas for UC most enrichment was seen in MUC21 and MUC22 [mucus layer], and GNA12 and HNF4A [regulating proteins] (p=4.87x10-2, 2.47x10-2, 7.85x10-3 and 5.67x10-3, respectively) (see Supplementary Table 5). The barrier component with most genes associated with CD and UC was the group of regulating proteins, although enrichment of this component was only significant for UC (p=2.18x10-3) (see Supplementary Table 6 and Figure 2).
Figure 2. Enrichment analysis of associated barrier genes (≥1 SNP) per component.
Bar plots representing the percentage of significant genes in each of the barrier components for CD (left) and UC (right). Only the set of regulating proteins was significantly enriched for UC in associated barrier genes compared to the other barrier components using Fisher Exact testing (*p<0.05).
CD, Crohn’s disease; UC, ulcerative colitis.
Mucosal barrier gene expression
Microarray analysis
Of the 128 selected genes, 125 were represented on the Human Gene 1.0 ST arrays by 132 different gene probe sets. To correlate the barrier gene mRNA expression levels with inflammation, we included the expression profile of IL8, represented by one extra gene probe set. In agreement with endoscopic disease activity, IL8 expression was significantly higher in active IBD (UC and/or CD) compared to controls, whereas no differences were detected for IL8 in uninflamed biopsies of IBD patients versus controls. Results of all comparisons are given in Supplementary Table 7. A heat map of the colonic and ileal expression values per gene probe set and individual is provided as Supplementary Figure 2 and 3.
Colonic expression of the epithelial barrier genes did not differ between UC and CD patients with active disease. As compared to controls, however, the expression of many barrier genes was dysregulated in the colon of active IBD (UC and/or CD) patients. The mRNA expression levels of MUC1, MUC5B, EMCN, MCAM and TFF1 [mucus layer], CLDN1 and JAM2 [tight junctions], DSG3 [desmosomes], LAMA4 and LAMC1 [extracellular matrix], and TCF4 and F2RL2 [regulating proteins] were >2-fold significantly upregulated in the inflamed colon of IBD patients, while the mRNA expression levels of RETNLB [mucus layer], CLDN8 and OCLN [tight junctions], and MAGI1 and MEP1A [regulating proteins] were >2-fold significantly downregulated in active IBD patients when compared to the colon of controls (Table 2). Of the different barrier components, the mucus layer was most enriched in differentially expressed genes (p=4.97x10-2) (see Supplementary Table 8). None of the barrier genes remained significantly dysregulated in the colon of UC patients with inactive disease as compared to their expression levels in controls (Table 2). All colonic dysregulated genes showed a highly significant correlation with IL8, confirming the direct impact of inflammation on epithelial barrier gene expression (see Supplementary Table 9 and Supplementary Figure 4 for the highest correlated ones).
Table 2. Significant barrier genes in the colon of UC and CD patients versus controls.
| Gene symbol | Gene probeset ID | Colonic expression | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Active UC vs controls | Inactive UC vs controls | Active CD vs controls | Active IBD vs controls | ||||||
| FC | P | FC | P | FC | P | FC | P | ||
| MUC1 | 7920642 | 2.30 | 9.39E-06 | 1.65* | 4.17E-03 | 2.52 | 6.05E-05 | 2.32 | 2.83E-06 |
| MUC5B | 7937612 | 2.66 | 1.41E-05 | 1.84* | 2.62E-02 | 2.53 | 6.76E-03 | 2.65 | 1.13E-05 |
| EMCN | 8101957 | 2.08 | 4.83E-04 | 1.29 | 2.70E-01 | 1.69* | 2.49E-02 | 2.04 | 5.27E-04 |
| MCAM | 7952205 | 2.94 | 2.87E-08 | 1.48* | 3.59E-02 | 2.66 | 2.92E-04 | 2.91 | 2.64E-08 |
| TFF1 | 8070579 | 3.23 | 1.04E-07 | 1.31 | 4.54E-01 | 1.95* | 2.20E-02 | 3.08 | 3.01E-07 |
| RETNLB | 8089394 | 0.40 | 8.10E-03 | 1.73 | 6.48E-02 | 1.19 | 7.01E-01 | 0.45 | 2.31E-02 |
| CLDN1 | 8092726 | 4.69 | 8.74E-12 | 1.31 | 4.29E-01 | 2.82 | 1.52E-03 | 4.46 | 5.45E-11 |
| CLDN8 | 8069795 | 0.07 | 2.95E-15 | 1.25 | 8.17E-01 | 0.15 | 2.42E-02 | 0.07 | 1.82E-13 |
| OCLN | 8105908 | 0.45 | 4.67E-04 | 0.74* | 3.73E-03 | 0.55* | 4.48E-05 | 0.46 | 3.46E-04 |
| JAM2 | 8068024 | 2.10 | 5.56E-06 | 1.78* | 3.11E-03 | 2.08 | 1.70E-03 | 2.10 | 2.96E-06 |
| DSG3 | 8020762 | 4.74 | 1.04E-05 | 1.09 | 8.35E-01 | 1.17 | 6.48E-01 | 4.13 | 1.25E-04 |
| LAMA4 | 8128991 | 2.37 | 8.08E-07 | 1.47 | 1.29E-01 | 2.26 | 5.18E-03 | 2.36 | 1.07E-06 |
| LAMC1 | 7908041 | 2.94 | 1.12E-09 | 1.51* | 4.17E-02 | 2.50 | 1.70E-03 | 2.89 | 1.96E-09 |
| MAGI1 | 8088602 | 0.48 | 5.78E-18 | 0.78* | 9.60E-04 | 0.56* | 5.03E-05 | 0.49 | 2.59E-17 |
| HNF4A | 8062823 | 0.49 | 1.00E-05 | 0.93 | 3.69E-01 | 0.64* | 1.46E-03 | 0.51* | 1.43E-05 |
| TCF4 | 8023415 | 3.05 | 1.79E-08 | 1.59 | 1.13E-01 | 2.36 | 1.70E-02 | 2.98 | 3.59E-08 |
| MEP1A | 8120088 | 0.21 | 6.11E-07 | 0.58* | 1.89E-04 | 0.37 | 1.65E-04 | 0.22 | 8.80E-07 |
| F2RL2 | 8112731 | 3.18 | 9.99E-10 | 1.31 | 2.49E-01 | 1.99* | 6.63E-03 | 3.04 | 7.46E-09 |
Fold changes and FDR-corrected p for the barrier genes that were significantly upregulated (bold) or downregulated (bold and underlined) in the colon of IBD patients versus controls. Fold changes indicated with an asterisk represent genes with significant p, but less than 2-fold differential expression.
UC, ulcerative colitis; CD, Crohn’s disease; IBD, inflammatory bowel disease; FC, fold change; vs, versus.
In addition to the colonic mRNA expression levels, differences in barrier gene expression in the terminal ileum of CD patients with active and inactive disease, and controls were evaluated. Eight genes (MUC1, MUC4, MUC5B, MUC6 and TFF1 [mucus layer], CLDN1 and CLDN18 [tight junctions] and F2RL2 [regulating proteins]) showed a >2-fold significantly increased expression in the inflamed ileal mucosa of CD patients compared to uninflamed tissue of controls, while the expression of CLDN8 [tight junctions] was significantly downregulated (Table 3). The barrier component most enriched in genes with differential expression in the ileum of CD patients with active disease versus controls also was the mucus layer (p=8.54x10-3) (see Supplementary Table 8). Interestingly, the ileal expression of MUC1, MUC4 [mucus layer] and CLDN8 [tight junctions] remained dysregulated in the ileum of CD patients with inactive disease in comparison to controls. The mRNA expression of MUC1 and MUC4 was >2-fold significantly upregulated in inactive CD patients, while CLDN8 was >2-fold significantly downregulated in patients compared to controls (Table 3). Again, significant correlations were found between the ileal mRNA levels of the dysregulated genes and IL8 (see Supplementary Table 9 and Supplementary Figure 4 for the highest correlated ones).
Table 3. Significant barrier genes in the ileum of CD patients versus controls.
| Gene symbol | Gene probeset ID | Ileal expression | |||
|---|---|---|---|---|---|
| Active CD vs controls | Inactive CD vs controls | ||||
| FC | P | FC | P | ||
| MUC1 | 7920642 | 8.47 | 8.19E-11 | 4.17 | 2.42E-03 |
| MUC4 | 8092978 | 4.64 | 3.02E-06 | 2.81 | 2.35E-02 |
| MUC5B | 7937612 | 2.53 | 4.45E-02 | 1.13 | 8.52E-01 |
| MUC6 | 7945595 | 3.88 | 1.88E-02 | 1.50 | 3.12E-01 |
| TFF1 | 8070579 | 2.54 | 9.86E-04 | 1.57 | 9.68E-02 |
| CLDN1 | 8092726 | 2.95 | 8.78E-05 | 1.68 | 1.02E-01 |
| CLDN8 | 8069795 | 0.39 | 2.84E-06 | 0.47 | 3.44E-02 |
| CLDN18 | 8082928 | 3.15 | 2.27E-03 | 1.83* | 5.68E-03 |
| F2RL2 | 8112731 | 2.06 | 6.61E-03 | 1.23 | 3.62E-01 |
Fold changes and FDR-corrected p for the barrier genes that were significantly upregulated (bold) or downregulated (bold and underlined) in the ileum of CD patients versus controls. Fold changes indicated with an asterisk represent genes with significant p, but less than 2-fold differential expression.
UC, ulcerative colitis; CD, Crohn’s disease; IBD, inflammatory bowel disease; FC, fold change; vs, versus.
Validation by qRT-PCR
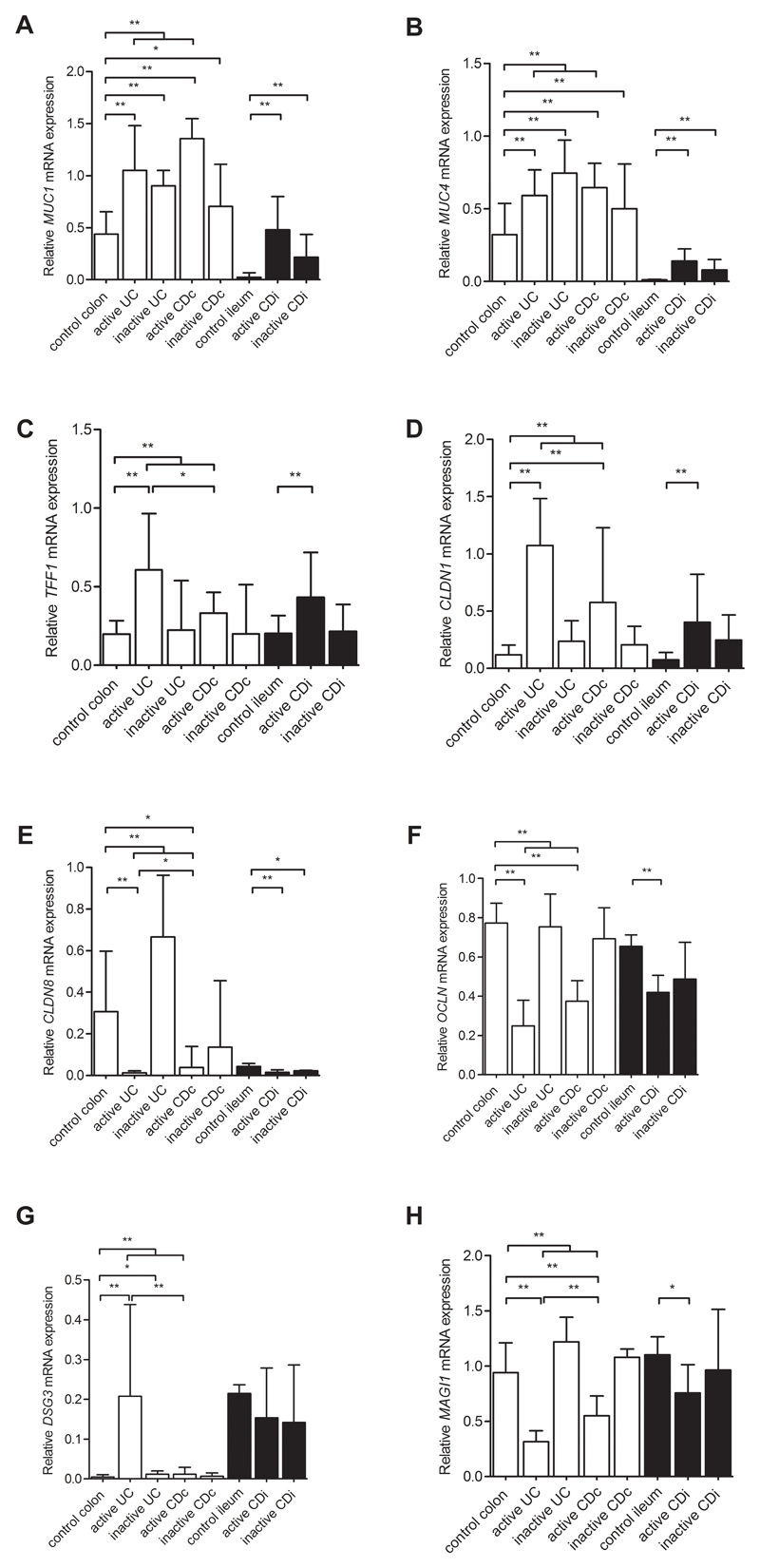
The differential barrier gene expression levels of MUC1, MUC4 and TFF1 [mucus layer], CLDN1, CLDN8 and OCLN [tight junctions], DSG3 [desmosomes], and MAGI1 [regulating proteins] from the microarray analysis were confirmed by qRT-PCR (Figure 3).
Figure 3. Relative expression levels of eight barrier genes with quantitative reverse transcription PCR.
Bar plots representing the relative expression levels of eight barrier genes (A-H) measured by qRT-PCR in colon (white bars) from controls (n=11), active UC (n=72), inactive UC (n=22), active CD patients (n=8) and inactive CD patients (n=26); and ileum (black bars) from controls (n=11), active CD (n=51) and inactive CD patients (n=14). The expression levels are normalised to beta-actin. Data are expressed as medians with interquartile range. Comparisons between the subgroups were performed with Mann-Whitney U testing. Significant differences as described in the main text are indicated (*p<0.05, **p<0.01).
UC, ulcerative colitis; CDc, colon of Crohn’s disease patients; CDi, ileum of CD patients.
As compared to the normal colon of controls, we found that the mRNA levels of MUC1, TFF1, CLDN1 and DSG3 were significantly upregulated in the inflamed colon of UC and/or CD patients, while the colonic expression levels of CLDN8, OCLN and MAGI1 were significantly decreased in active IBD patients compared to controls. The more sensitive qRT-PCR results also showed increased mRNA expression levels of MUC4 for these comparisons, while OCLN and MAGI1 levels were significantly decreased in active CD patients. In addition, while no significant alterations were previously found for the colonic expression of the barrier genes between UC and CD patients with active disease, qRT-PCR analysis did identify significantly different levels of CLDN8, DSG3, TFF1 and MAGI1 in the colon of active UC patients when compared to active CD patients. Finally, qRT-PCR showed significantly increased expression levels of MUC1, MUC4 and DSG3 in the colon of UC patients with inactive disease versus controls. Evaluation of the genes in an additional cohort of 26 inactive CD patients demonstrated that MUC1 and MUC4 also were significantly upregulated in uninflamed colon samples from CD patients compared to healthy controls (p=0.043 and 0.009 respectively) (Figure 3).
In the ileum, we confirmed the differential expression of MUC1, MUC4, TFF1, CLDN1 and CLDN8 in active CD patients when compared to controls. The ileal expression of MUC1, MUC4 and CLDN8 also remained dysregulated in CD patients with inactive disease as seen in the microarray analysis. Additional differences were observed for OCLN and MAGI1, having significantly decreased levels in the inflamed ileum of CD patients when compared to the ileum of controls.
Influence of genetics on mucosal barrier gene expression
In order to evaluate if there were any cis-acting genetic variants affecting the barrier gene expression levels, we performed cis-eQTL mapping in each of the patient sample groups (inflamed colon, normal colon, inflamed ileum, normal ileum). No significant cis-eQTL signals were found after correction for multiple testing in any of the groups. We also did not find significant differences in the barrier gene expression levels between CD and UC patients with the lowest (<75 for CD, <62 for UC) and highest genetic barrier risk scores (>86 for CD, >70 for UC).
Discussion
This study represents a comprehensive report in which the different components of the intestinal epithelial barrier were analysed at genetic and transcriptomic level in the context of IBD, taking into account disease type (CD and UC), biopsy location (colon and ileum) and activity status (inflamed and uninflamed).
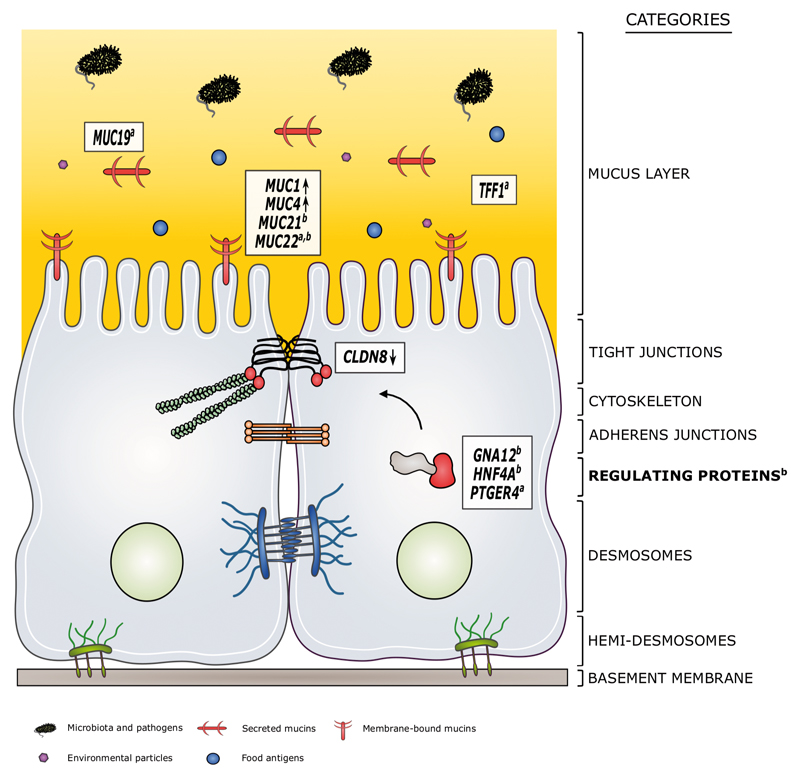
We found that the total number of risk alleles in epithelial barrier genes was significantly higher in CD and UC patients compared to controls, validating the known impact of the intestinal barrier for the pathogenesis of IBD. Further analysis of barrier gene variants highlighted the potential role of MUC19, MUC22, TFF1 and PTGER4 for CD, and MUC21, MUC22, GNA12 and HNF4A for UC. At component-level, genes with associated variants were most enriched in the group of regulating proteins for both CD and UC. The mucosal gene expression study showed that the mRNA expression of many epithelial barrier genes was dysregulated in the inflamed colon and ileum of IBD patients, with a significant over-representation of mucus layer genes in both. During inactive disease, the expression of MUC1 and MUC4 remained commonly disturbed in intestinal samples of CD and UC patients, suggesting that these genes act as crucial players in IBD. In CD ileum, CLDN8 also remained significantly lower expressed compared to controls as evaluated by both microarray and qRT-PCR. Analysis of the link between the genetic variants in the barrier genes and their expression alterations, however, did not show significant findings, which might indicate that both levels are not necessarily directly related and influenced by many other disease-specific factors. A schematic overview of the most interesting results is shown in Figure 4.
Figure 4. Schematic overview of the main results.
The genetic and transcriptomic approaches in this study identified the potential role of particular epithelial barrier genes and components in the context of IBD.
We found that disease-associated variants were significantly enriched in MUC19 (secreted mucin), MUC22 (membrane-bound mucin), TFF1 (stabilizing mucus layer protein) and PTGER4 (regulating protein) for CD, and MUC21 (membrane-bound mucin), MUC22 (membrane-bound mucin), GNA12 (regulating protein) and HNF4A (regulating protein) for UC. The most enriched barrier component was the set of regulating proteins for both CD and UC. At mRNA level, persistent changes in ileal and colonic MUC1 (membrane-bound mucin) and MUC4 (membrane-bound mucin) expression were found during inactive disease for CD and UC, pinpointing to a possible central role of these genes in IBD onset/relapse. In uninflamed CD ileum, also CLDN8 expression remained strongly dysregulated.
Genetic predispositions and expression changes may together induce barrier dysfunction of the intestinal epithelium in IBD patients, and result in an enhanced uptake of harmful luminal antigens and initiation of inflammation.
Symbols within the mucus layer are explained below the figure. Structures within the epithelial cells are defined by the category names. Arrows indicate persistent up- or downregulation of the genes during inactive disease. asignificant enrichment of variants associated with CD; bsignificant enrichment of variants associated with UC.
GWAS have previously identified multiple individual SNPs that are associated with the risk to IBD. Although the functional relevance of many of these SNPs is not known yet, the observed higher genetic barrier risk scores in IBD patients compared to controls suggest that patients also more commonly have a combination of disease-associated variants in intestinal barrier genes which could cause an intensification of the small effects from the individual risk variants. Part of the IBD patients may thus have a distinct genetic predisposition to have intestinal barrier defects, and respond differently – most likely in combination with other predisposing factors - to common environmental stimuli triggering disease onset or relapse. Enrichment analysis with associated barrier SNPs for CD showed that the most significantly enriched gene in this study was PTGER4. The PTGER4 locus has already been identified in several other studies as associated with CD (17, 31). The gene encodes the prostaglandin receptor EP4, of which activation has been suggested to result in redistribution of junctional proteins and the cytoskeleton, with an increase in epithelial barrier disruption (32). The most significant enriched gene for UC was HNF4A, a transcription factor known for its essential role in the development and regulation of intestinal epithelial cells, and previously associated in a number of GWAS with UC (17, 33, 34). Ahn et al. showed that mice with a conditional knock-out of Hnf4a in intestinal epithelial cells had a markedly increased intestinal permeability and susceptibility to acute DSS colitis (35). Amongst the other enriched genes, MUC19 (secreted gel-forming mucin) and GNA12 (TJ regulator) are also extensively described based on their association in large GWAS and meta-analyses, whereas reports on genetic evidence for MUC21, MUC22 and TFF1 in IBD are rather limited (17, 18, 34). MUC21 encodes a recently identified transmembrane mucin protein, in which one particular SNP has been associated to UC by Achkar et al. who looked into the major histocompatibility complex on chromosome 6p (36). In the context of lung diseases, both MUC21 and MUC22, another membrane-bound mucin at epithelial surfaces, have been proposed as candidates for association with asthma, although it could not be excluded that other genes in close proximity including HLA regions are responsible for these signals (37). Changes in the integrity of the bronchial epithelium are thought to play a central role in the sensitisation to allergens and the development of asthma, a chronic inflammatory disease of the airways which represent a similar defence barrier as in the gut (38). The family of trefoil factors, including TFF1, has received considerable attention in a number of animal and intestinal expression studies, but has so far not been associated with the risk to IBD or other immune-related disorders. While its precise physiological function and regulation in the gut is not clear, TFF1 is thought to act in mucosal repair and reinforcement of the mucus layer by interaction with mucin molecules (39).
In addition to the enrichment analysis at single gene-level, which searched for multiple risk signals within the same genomic location, a component-level analysis was performed where we evaluated which barrier components had the highest number of genes with at least one associated SNP. We showed that the regulating proteins were most overrepresented, with multiple significant genes for both disease types, although only significant at p<0.05 for UC. We could assume that IBD barrier defects partly originate from effects of variants within different regulating barrier genes, together with some strong signals from individual genes of other barrier components like mucus layer factors that showed high enrichment at single-gene level. Of note, the group of regulating proteins involved a broad mixture of scaffolding proteins, transcription factors and previously associated regulatory genes, possibly creating a selection bias towards association. We also are aware that the genetic analysis had limited power to detect genome-wide significant findings. Given our current sample size and a significance level of 3.8x10-5, we only had 57% and 40% power to identify variants with an effect size of 1.5 and allele frequency of 0.1 for CD and UC respectively. Still, some of the most significant signals that we found, were already described in larger studies, as were the genes enriched in independent significant variants (e.g. MUC19, HNF4A).
The results from the gene expression study showed that IBD patients with active disease had major gene expression changes at different levels of the intestinal epithelial barrier validating many previous reports. Interestingly, there was a considerable overlap between genes dysregulated in the colon and ileum of both CD and UC patients during active disease (e.g. MUC1, MUC5B, TFF1, CLDN1, CLDN8 and F2RL2). This suggests that these barrier molecules are affected in a similar way and represent largely the same barrier defects at both tissue sites under the influence of inflammatory mediators. The most aberrant changes during inflammation were found for MUC1 in the ileum of CD patients, and CLDN8 in the colon of UC and CD patients. MUC1 is synthesized by goblet and absorptive cells from the intestinal epithelium, and is expressed as a membrane-bound glycoprotein in the mucus layer (40). Consistent with our results, different studies have previously implicated increased MUC1 gene and protein expression during inflammation (41–44). It was suggested by Kadayakkara et al. that an increase in MUC1 gene expression may initially serve to protect the gut epithelium by strengthening the function of the mucus layer, while repetitive cycles of inflammation can induce an increased expression of an abnormal hypo-glycosylated protein form of MUC1 which attracts innate inflammatory cells and promotes the development of chronic inflammation and oncogenesis (45). CLDN8 was the most downregulated gene in active IBD patients, as also frequently described in previous studies (46–49). CLDN8 belongs to the “sealing” proteins of the claudin family which restrict paracellular flux and decrease intestinal permeability, in contrast to pore-forming claudins such as CLDN2 which increase permeability of the intestinal barrier (50). In a study of Zeissig and colleagues, downregulation of CLDN8 was accompanied by CLDN2 upregulation at the tight junctions, intensely enhancing tight junction permeability in active CD patients (47). In our study, CLDN2 gene expression was increased in the colon of active IBD patients compared to controls, but not more than 2-fold different. When comparing the number of differentially expressed genes for IBD patients with active disease versus controls, the mucus layer genes were most enriched. Taken these results together with the findings from the genetic study, we suggest a key role for the mucus layer component in the pathogenesis of IBD. Future studies should look at protein levels of mucus layer genes to dissect their biological working mechanism and functional relevance for IBD.
Remarkably, the common barrier genes that remained dysregulated during inactive disease were MUC1 and MUC4 in CD and UC colon (qRT-PCR) and CD ileum (microarray and qRT-PCR). In inactive ileum of CD patients, also CLDN8 expression remained strongly dysregulated according to both microarray and qRT-PCR analysis. Like MUC1, MUC4 is a membrane-bound mucin protein at the apical side of the intestinal epithelial cells, and forms the glycocalyx which is situated just below the gel-like mucus layer (51). As opposed to studies of barrier gene expression changes during inflammation, less information is available on these barrier gene levels in quiescent disease (52, 53). A recent study of Peloquin et al. investigated a selection of 678 genes within previously identified IBD risk loci, and found that uninflamed samples of CD patients exhibited perturbed expression levels of particular genes with increased variances compared to healthy controls. They suggested that these genes are normally held under tight regulatory control, which is lost in the setting of CD (54). It could thus be that MUC1 and MUC4 are in a continuously, dysregulated state (primary or due to subclinical molecular inflammation) which can trigger disease onset and relapse in predisposed patients – and worsen with active inflammation. We should then suppose that high MUC1 and MUC4 levels have a detrimental effect on the intestinal barrier by expression of an aberrant form as suggested earlier for MUC1, or by causing a general imbalance in mucins which affects the mucus composition and function. An alternative hypothesis on persistent increases in MUC1 and MUC4 expression during inactive disease could be that they represent a secondary defence or repair mechanism to protect the gut and account for the damage of previous inflammation. Since CLDN8 encodes a pore-sealing protein, it is acceptable that its expression has not returned to normal levels in controls when secondary to inflammation, and thus is less dynamic than other barrier genes, again promoting chronic reactivation of the disease. Although not significant in our genetic study, Franke et al. showed that the MUC1 locus was genetically associated with CD, which would be in favour for the hypothesis of a primary role for this gene (55). For MUC4 and CLDN8, no association reports are available in current literature.
In previous studies, our group has evaluated the effect of infliximab therapy on the mucosal expression of several genes involved in IBD (56–58). Albeit the primary goals of these studies were different, the majority of the dysregulated genes during active disease in the current study were also significantly dysregulated in the inflamed mucosa of IBD patients before their first infusion of infliximab. Strikingly, MUC1 and MUC4 also remained significantly upregulated in the colon of CD responders after infliximab treatment compared to controls, and the same was found for MUC1 in the ileum of CD responders versus controls (see Supplementary Methods for a description of this cohort, and Supplementary Table 10 for all comparisons before and after infliximab treatment). Both genes did not show significantly altered levels in UC responders compared to controls, which is similar to our microarray findings in uninflamed colon from UC patients. Analysis with qRT-PCR, in contrast, did show persistent higher levels in the latter samples in our study, which might be explained by the higher sensitivity of qRT-PCR as opposed to microarray, or a more pronounced effect in the phenotype of CD versus UC.
When combining the results from the genetic and mucosal gene expression study, there was no direct link between variants in the epithelial barrier genes and differences in their expression. Neither single cis-acting variants in the barrier genes, nor the combined barrier risk scores were associated with the expression of the barrier genes at an FDR level of 5%. These results could imply that other mechanisms are primarily involved for the genetic barrier risk and expression changes seen in IBD patients in this cohort. For the MUC19 risk locus, for example, it has been suggested that associated SNPs in the gene region probably exert their effect by inducing changes in mRNA conformation, translational efficiency or subcellular localisation rather than gene expression (59). Gene expression alterations of the barrier genes could also be regulated by SNPs further away from the genes, but because of the limited sample size of the overlapping cohort, we only examined eQTLs acting in cis (including a strict window of 1 Mb). Trans-eQTLs (>1 Mb from the barrier gene start/end sites) were not described here as it was shown that small effects of trans-variants are harder to detect and much more sensitive to statistical power (60). Unmistakeably, interesting signals could be missed in that way, indicating the need for larger sample sizes. Some recent studies have investigated genome-wide eQTLs in primary tissue cell types for IBD, and their overlap with the known IBD susceptibility loci (61–65). None of the top signals from these studies correspond with one of our selected barrier genes, confirming that we should search for other regulating mechanisms in these regions.
Taken together, the data in this study allowed us to get a better view on which genes and components from the intestinal epithelial barrier pathway are most critical for IBD, based on their genetic and transcriptomic significance. Identification of the most critical molecules could be necessary to enhance the development of novel barrier-restoring therapeutics. Today, several agents that modify intestinal barrier integrity have been proposed, but their clinical application is still limited - mostly due to shortcomings in the mechanistic and functional understanding of the intestinal barrier. One of the most promising agents for UC currently includes phosphatidylcholine, a major class of phospholipids in the colonic mucus layer. The delayed release of phosphatidylcholine in the gut is thought to reinforce the mucus layer. Our data also support the intestinal mucus layer as a key therapeutic target within the intestinal barrier. The compound has been shown to be an effective and safe therapeutic option for UC patients in phase II clinical trials, but more research is needed to understand its exact working mechanism, and its lack of efficacy for CD (66–68).
In conclusion, we provided an in-depth view on the genetic and transcriptomic basis of intestinal epithelial barrier defects in IBD. By using three different approaches, we identified a selection of barrier genes (e.g. MUC1, MUC4, MUC22) and components (e.g. mucus layer, regulating proteins) that may be plausible candidates for the onset or perpetuation of chronic gastrointestinal inflammation in IBD (Figure 4). Future studies focussing on the functional working mechanism of these genes and categories are required to uncover their precise role in the disease pathogenesis, and therapeutic potential.
Supplementary Material
Acknowledgement
The authors would like to thank Karolien Claes, Nooshin Ardeshir Davani, Sophie Organe and Willem-Jan Wollants for the technical support; and Vera Ballet for the patient inclusion and database management. We also thank Leentje Van Lommel from the Gene Expression Unit for technical assistance with the microarray and quantitative reverse transcription PCR experiments.
Source of Funding and Conflicts of Interest:
This work was supported by the Research Foundation Flanders (FWO) [G.0440.06, G.0479.10] and the European Crohn’s and Colitis Organisation [ECCO Grant 2013]. T.V., M.F., G.V.A. and Sé.V. are senior clinical investigators of the FWO. This work was also supported by an Advanced European Research Council [ERC] Grant [ERC-2015-AdG].
T.V. receives lecture fees from Will Pharma and consulting fees from Shire. M.F. reports financial support for research from Takeda; lecture fees from Abbvie, Boehringer-Ingelheim, Chiesi, Falk, Ferring, Janssen, Mitsubishi Tanabe, MSD, Takeda, Tillotts and Zeria; and consulting fees from Abbvie, Boehringer-Ingelheim, Ferring, Janssen and MSD. G.V.A. receives support for research from Abbvie and MSD; lecture fees from Abbvie, MSD, Ferring, Janssen and Takeda; and consulting fees from Abbvie, MSD and Takeda. P.R. receives research support and lecture fees from Abbvie, Centocor and Merck; and consulting fees from Abbvie, Centocor, Merck, UCB, Takeda, Genentech/Hoffman-LaRoche, Serono, Bristol Myers Squibb, Robarts, Tillotts, Pfizer and Falk Pharma. Sé.V. reports grant support from Abbvie, MSD and Takeda; lecture fees from Abbvie, MSD, Takeda, Ferring, Falk Pharma, Hospira and Tillotts; and consulting fees from Abbvie, MSD, Takeda, Ferring, Genentech/Roche, Shire, Pfizer, Galapagos, Mundipharma, Hospira, Celgene, Second Genome and Janssen. For the remaining authors none were declared.
References
- 1.Molodecky NA, Soon IS, Rabi DM, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54 e42. doi: 10.1053/j.gastro.2011.10.001. quiz e30. [DOI] [PubMed] [Google Scholar]
- 2.Bouma G, Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat Rev Immunol. 2003;3:521–533. doi: 10.1038/nri1132. [DOI] [PubMed] [Google Scholar]
- 3.Rampton D, Shanahan F. Fast facts inflammatory bowel disease. 3. ed. Abingdon, Oxford: Health Press Ltd; 2008. [Google Scholar]
- 4.McGuckin MA, Eri R, Simms LA, et al. Intestinal barrier dysfunction in inflammatory bowel diseases. Inflamm Bowel Dis. 2009;15:100–113. doi: 10.1002/ibd.20539. [DOI] [PubMed] [Google Scholar]
- 5.Pastorelli L, De Salvo C, Mercado JR, et al. Central role of the gut epithelial barrier in the pathogenesis of chronic intestinal inflammation: lessons learned from animal models and human genetics. Front Immunol. 2013;4:280. doi: 10.3389/fimmu.2013.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salim SY, Soderholm JD. Importance of disrupted intestinal barrier in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:362–381. doi: 10.1002/ibd.21403. [DOI] [PubMed] [Google Scholar]
- 7.Hollander D, Vadheim CM, Brettholz E, et al. Increased intestinal permeability in patients with Crohn's disease and their relatives. A possible etiologic factor. Ann Intern Med. 1986;105:883–885. doi: 10.7326/0003-4819-105-6-883. [DOI] [PubMed] [Google Scholar]
- 8.Ainsworth M, Eriksen J, Rasmussen JW, et al. Intestinal permeability of 51Cr-labelled ethylenediaminetetraacetic acid in patients with Crohn's disease and their healthy relatives. Scand J Gastroenterol. 1989;24:993–998. doi: 10.3109/00365528909089246. [DOI] [PubMed] [Google Scholar]
- 9.Teahon K, Smethurst P, Levi AJ, et al. Intestinal permeability in patients with Crohn's disease and their first degree relatives. Gut. 1992;33:320–323. doi: 10.1136/gut.33.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bjarnason I, O'Morain C, Levi AJ, et al. Absorption of 51chromium-labeled ethylenediaminetetraacetate in inflammatory bowel disease. Gastroenterology. 1983;85:318–322. [PubMed] [Google Scholar]
- 11.Jenkins RT, Ramage JK, Jones DB, et al. Small bowel and colonic permeability to 51Cr-EDTA in patients with active inflammatory bowel disease. Clin Invest Med. 1988;11:151–155. [PubMed] [Google Scholar]
- 12.Ukabam SO, Clamp JR, Cooper BT. Abnormal small intestinal permeability to sugars in patients with Crohn's disease of the terminal ileum and colon. Digestion. 1983;27:70–74. doi: 10.1159/000198932. [DOI] [PubMed] [Google Scholar]
- 13.Marin ML, Geller SA, Greenstein AJ, et al. Ultrastructural pathology of Crohn's disease: correlated transmission electron microscopy, scanning electron microscopy, and freeze fracture studies. Am J Gastroenterol. 1983;78:355–364. [PubMed] [Google Scholar]
- 14.Marin ML, Greenstein AJ, Geller SA, et al. A freeze fracture study of Crohn's disease of the terminal ileum: changes in epithelial tight junction organization. Am J Gastroenterol. 1983;78:537–547. [PubMed] [Google Scholar]
- 15.Pearson AD, Eastham EJ, Laker MF, et al. Intestinal permeability in children with Crohn's disease and coeliac disease. Br Med J (Clin Res Ed) 1982;285:20–21. doi: 10.1136/bmj.285.6334.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellinghaus D, Jostins L, Spain SL, et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet. 2016;48:510–518. doi: 10.1038/ng.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu JZ, van Sommeren S, Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 20.Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 21.Coskun M. Intestinal epithelium in inflammatory bowel disease. Front Med (Lausanne) 2014;1:24. doi: 10.3389/fmed.2014.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groschwitz KR, Hogan SP. Intestinal barrier function: molecular regulation and disease pathogenesis. J Allergy Clin Immunol. 2009;124:3–20. doi: 10.1016/j.jaci.2009.05.038. quiz 21–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matter K, Balda MS. Signalling to and from tight junctions. Nat Rev Mol Cell Biol. 2003;4:225–236. doi: 10.1038/nrm1055. [DOI] [PubMed] [Google Scholar]
- 24.Niessen CM. Tight junctions/adherens junctions: basic structure and function. J Invest Dermatol. 2007;127:2525–2532. doi: 10.1038/sj.jid.5700865. [DOI] [PubMed] [Google Scholar]
- 25.Walko G, Castanon MJ, Wiche G. Molecular architecture and function of the hemidesmosome. Cell Tissue Res. 2015;360:529–544. doi: 10.1007/s00441-015-2216-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vanhove W, Peeters PM, Staelens D, et al. Strong Upregulation of AIM2 and IFI16 Inflammasomes in the Mucosa of Patients with Active Inflammatory Bowel Disease. Inflamm Bowel Dis. 2015;21:2673–2682. doi: 10.1097/MIB.0000000000000535. [DOI] [PubMed] [Google Scholar]
- 27.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B. 1995:289–300. [Google Scholar]
- 29.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Libioulle C, Louis E, Hansoul S, et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 2007;3:e58. doi: 10.1371/journal.pgen.0030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez-Lagunas MJ, Martin-Venegas R, Moreno JJ, et al. PGE2 promotes Ca2+-mediated epithelial barrier disruption through EP1 and EP4 receptors in Caco-2 cell monolayers. Am J Physiol Cell Physiol. 2010;299:C324–334. doi: 10.1152/ajpcell.00397.2009. [DOI] [PubMed] [Google Scholar]
- 33.Garrison WD, Battle MA, Yang C, et al. Hepatocyte nuclear factor 4alpha is essential for embryonic development of the mouse colon. Gastroenterology. 2006;130:1207–1220. doi: 10.1053/j.gastro.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCole DF. IBD candidate genes and intestinal barrier regulation. Inflamm Bowel Dis. 2014;20:1829–1849. doi: 10.1097/MIB.0000000000000090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahn SH, Shah YM, Inoue J, et al. Hepatocyte nuclear factor 4alpha in the intestinal epithelial cells protects against inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:908–920. doi: 10.1002/ibd.20413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Achkar JP, Klei L, de Bakker PI, et al. Amino acid position 11 of HLA-DRbeta1 is a major determinant of chromosome 6p association with ulcerative colitis. Genes Immun. 2012;13:245–252. doi: 10.1038/gene.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galanter JM, Gignoux CR, Torgerson DG, et al. Genome-wide association study and admixture mapping identify different asthma-associated loci in Latinos: the Genes-environments & Admixture in Latino Americans study. J Allergy Clin Immunol. 2014;134:295–305. doi: 10.1016/j.jaci.2013.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heijink IH, Nawijn MC, Hackett TL. Airway epithelial barrier function regulates the pathogenesis of allergic asthma. Clin Exp Allergy. 2014;44:620–630. doi: 10.1111/cea.12296. [DOI] [PubMed] [Google Scholar]
- 39.Aamann L, Vestergaard EM, Gronbaek H. Trefoil factors in inflammatory bowel disease. World J Gastroenterol. 2014;20:3223–3230. doi: 10.3748/wjg.v20.i12.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim YS, Ho SB. Intestinal goblet cells and mucins in health and disease: recent insights and progress. Curr Gastroenterol Rep. 2010;12:319–330. doi: 10.1007/s11894-010-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beatty PL, Plevy SE, Sepulveda AR, et al. Cutting edge: transgenic expression of human MUC1 in IL-10-/- mice accelerates inflammatory bowel disease and progression to colon cancer. J Immunol. 2007;179:735–739. doi: 10.4049/jimmunol.179.2.735. [DOI] [PubMed] [Google Scholar]
- 42.Longman RJ, Poulsom R, Corfield AP, et al. Alterations in the composition of the supramucosal defense barrier in relation to disease severity of ulcerative colitis. J Histochem Cytochem. 2006;54:1335–1348. doi: 10.1369/jhc.5A6904.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheng YH, Triyana S, Wang R, et al. MUC1 and MUC13 differentially regulate epithelial inflammation in response to inflammatory and infectious stimuli. Mucosal Immunol. 2013;6:557–568. doi: 10.1038/mi.2012.98. [DOI] [PubMed] [Google Scholar]
- 44.Furr AE, Ranganathan S, Finn OJ. Aberrant expression of MUC1 mucin in pediatric inflammatory bowel disease. Pediatr Dev Pathol. 2010;13:24–31. doi: 10.2350/08-06-0479.1. [DOI] [PubMed] [Google Scholar]
- 45.Kadayakkara DK, Beatty PL, Turner MS, et al. Inflammation driven by overexpression of the hypoglycosylated abnormal mucin 1 (MUC1) links inflammatory bowel disease and pancreatitis. Pancreas. 2010;39:510–515. doi: 10.1097/MPA.0b013e3181bd6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oshima T, Miwa H, Joh T. Changes in the expression of claudins in active ulcerative colitis. J Gastroenterol Hepatol. 2008;23(Suppl 2):S146–150. doi: 10.1111/j.1440-1746.2008.05405.x. [DOI] [PubMed] [Google Scholar]
- 47.Zeissig S, Burgel N, Gunzel D, et al. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn's disease. Gut. 2007;56:61–72. doi: 10.1136/gut.2006.094375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Clark PM, Dawany N, Dampier W, et al. Bioinformatics analysis reveals transcriptome and microRNA signatures and drug repositioning targets for IBD and other autoimmune diseases. Inflamm Bowel Dis. 2012;18:2315–2333. doi: 10.1002/ibd.22958. [DOI] [PubMed] [Google Scholar]
- 49.Toedter G, Li K, Sague S, et al. Genes associated with intestinal permeability in ulcerative colitis: changes in expression following infliximab therapy. Inflamm Bowel Dis. 2012;18:1399–1410. doi: 10.1002/ibd.22853. [DOI] [PubMed] [Google Scholar]
- 50.Koval M. Differential pathways of claudin oligomerization and integration into tight junctions. Tissue Barriers. 2013;1:e24518. doi: 10.4161/tisb.24518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andrianifahanana M, Moniaux N, Batra SK. Regulation of mucin expression: mechanistic aspects and implications for cancer and inflammatory diseases. Biochim Biophys Acta. 2006;1765:189–222. doi: 10.1016/j.bbcan.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 52.Hensel KO, Boland V, Postberg J, et al. Differential expression of mucosal trefoil factors and mucins in pediatric inflammatory bowel diseases. Sci Rep. 2014;4:7343. doi: 10.1038/srep07343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hamm CM, Reimers MA, McCullough CK, et al. NOD2 status and human ileal gene expression. Inflamm Bowel Dis. 2010;16:1649–1657. doi: 10.1002/ibd.21208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peloquin JM, Goel G, Kong L, et al. Characterization of candidate genes in inflammatory bowel disease-associated risk loci. JCI Insight. 2016;1:e87899. doi: 10.1172/jci.insight.87899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Bruyn M, Machiels K, Vandooren J, et al. Infliximab restores the dysfunctional matrix remodeling protein and growth factor gene expression in patients with inflammatory bowel disease. Inflamm Bowel Dis. 2014;20:339–352. doi: 10.1097/01.MIB.0000438430.15553.90. [DOI] [PubMed] [Google Scholar]
- 57.Arijs I, De Hertogh G, Lemaire K, et al. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PloS one. 2009;4:e7984. doi: 10.1371/journal.pone.0007984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arijs I, De Hertogh G, Machiels K, et al. Mucosal gene expression of cell adhesion molecules, chemokines, and chemokine receptors in patients with inflammatory bowel disease before and after infliximab treatment. Am J Gastroenterol. 2011;106:748–761. doi: 10.1038/ajg.2011.27. [DOI] [PubMed] [Google Scholar]
- 59.Shastry BS. SNPs: impact on gene function and phenotype. Methods Mol Biol. 2009;578:3–22. doi: 10.1007/978-1-60327-411-1_1. [DOI] [PubMed] [Google Scholar]
- 60.Pinpointing expression differences. Nat Genet. 2007;39:1175. doi: 10.1038/ng1007-1175. [DOI] [PubMed] [Google Scholar]
- 61.Di Narzo AF, Peters LA, Argmann C, et al. Blood and Intestine eQTLs from an Anti-TNF-Resistant Crohn's Disease Cohort Inform IBD Genetic Association Loci. Clin Transl Gastroenterol. 2016;7:e177. doi: 10.1038/ctg.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peters JE, Lyons PA, Lee JC, et al. Insight into Genotype-Phenotype Associations through eQTL Mapping in Multiple Cell Types in Health and Immune-Mediated Disease. PLoS Genet. 2016;12:e1005908. doi: 10.1371/journal.pgen.1005908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hulur I, Gamazon ER, Skol AD, et al. Enrichment of inflammatory bowel disease and colorectal cancer risk variants in colon expression quantitative trait loci. BMC Genomics. 2015;16:138. doi: 10.1186/s12864-015-1292-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kabakchiev B, Silverberg MS. Expression quantitative trait loci analysis identifies associations between genotype and gene expression in human intestine. Gastroenterology. 2013;144:1488–1496. doi: 10.1053/j.gastro.2013.03.001. 1496 e1481–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Singh T, Levine AP, Smith PJ, et al. Characterization of expression quantitative trait loci in the human colon. Inflamm Bowel Dis. 2015;21:251–256. doi: 10.1097/MIB.0000000000000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Karner M, Kocjan A, Stein J, et al. First multicenter study of modified release phosphatidylcholine "LT-02" in ulcerative colitis: a randomized, placebo-controlled trial in mesalazine-refractory courses. Am J Gastroenterol. 2014;109:1041–1051. doi: 10.1038/ajg.2014.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stremmel W, Merle U, Zahn A, et al. Retarded release phosphatidylcholine benefits patients with chronic active ulcerative colitis. Gut. 2005;54:966–971. doi: 10.1136/gut.2004.052316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gibson PR, Muir JG. Reinforcing the mucus: a new therapeutic approach for ulcerative colitis? Gut. 2005;54:900–903. doi: 10.1136/gut.2004.058453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.