Abstract
Exosomes are nano-sized vesicles composed of lipids, proteins, and nucleic acids. Their molecular landscape is diverse, and exosomes derived from different cell types have distinct biological activities. Since exosomes are now being utilized as delivery vehicles for exogenous therapeutic cargoes, their intrinsic properties and biological effects must be understood. We performed miRNA profiling and found substantial differences in the miRNA landscape of prostate cancer (PC3) and human embryonic kidney (HEK) 293 exosomes with little correlation in abundance of common miRNAs (R2=0.16). Using a systems-level bioinformatics approach, the most abundant miRNAs in PC3 exosomes but not HEK exosomes were predicted to significantly modulate integrin signaling, with integrin-β3 loss inducing macrophage M2 polarization. PC3 but not HEK exosomes downregulated integrin-β3 expression levels by 70%. There was a dose-dependent polarization of RAW 264.7 macrophages toward an M2 phenotype when treated with PC3-derived exosomes but not HEK-derived exosomes. Conversely, HEK exosomes, widely utilized as delivery vehicles were predicted to target cadherin signaling, with experimental validation showing a significant increase in the migratory potential of MCF7 breast cancer cells treated with HEK exosomes. Even widely utilized exosomes are unlikely to be inert, and their intrinsic activity ought to be assessed before therapeutic deployment.
Keywords: Exosomes, miRNA profiling, phenotypic effects, M2 polarization, cadherin signaling
Introduction
Exosomes are nano-sized vesicles secreted by many cells and are composed of a lipid bilayer, membrane proteins, and a hydrophilic core. They are highly versatile carriers of hydrophilic and hydrophobic molecules, proteins, and nucleic acids. While the phenotypic effects of exosomes can be modulated by loading with different therapeutic cargoes, the effects of their intrinsic properties, such as their protein, lipid or nucleic acid complement, are often overlooked. However, these properties and their effects are of critical importance for clinical translation of exosomes as therapeutics. Furthermore, the overall action of exosomes most likely results from the combined effects of all rather than just a single component (1).
An intriguing aspect of exosome biology is their ability to transfer genetic materials to recipient cells in the form of non-coding RNAs, especially microRNAs (miRNAs) (2). A single miRNA can modulate the expression of hundreds of genes; in total, 30–70% of human genes are predicted miRNA targets through base-pairing of their “seed” sequences (3). Exosomes used to be viewed as a ‘garbage disposal’ system of unwanted cellular proteins and RNAs, but the current view is that exosomes act as natural delivery vehicles for nucleic acids and bioactive proteins that cells use to achieve coherent inter-cellular communication with recipient cells (1, 4).
Exosomes seem to have evolved to deliver these bio-messages in vivo. They are relatively stable and protect their cargo from enzymatic degradation (5). Their surface lipids and integral membrane proteins can direct exosome uptake into specific cells (6, 7). For therapeutic purposes, exosomes have been supplemented with application-specific cargoes across many disciplines including cardiology, neurology, and oncology (1). Generally, RNA interference strategies such as siRNA or miRNAs are used (8–14), but exosomes have also been loaded with small molecules (15, 16). Additionally, exosomes have been genetically engineered to express specific targeting surface peptides to control their biodistribution and cellular uptake (8, 10, 14).
Current exosome manipulations do not completely alter their intrinsic properties. Given the heterogeneity of exosomes and extracellular vesicles and current limitations in characterizing subpopulations of released vesicles, total control over intrinsic exosome activity remains a challenge (1). This is an especially important consideration if the intrinsic biology of the exosomes is undesirable.
While it is now appreciated that exosomes display a wide range of biological effects, the underlying mechanisms are not fully understood. Cancer-derived exosomes can exert malignant phenotypes on recipient cells including promoting tumor cell proliferation, immune suppression or subversion, angiogenesis, and preparation of pre-metastatic niches (17–25). These effects are in stark contrast to the effects reported for exosomes from non-cancerous cells. Mesenchymal stem cell (MSC)-derived exosomes are mostly reported to offer tissue-regenerative effects in a host of settings ranging from protection after cerebral and myocardial ischemia to anti-fibrotic effects and restorative functions in damaged livers and kidneys (26–32). In the quest for scalable production of therapeutic exosomes, many researchers are utilizing easy-to-manipulate cells such as human embryonic kidney (HEK) 293 assuming that they represent a neutral phenotype (1). However, since HEK 293 represent a transformed cell line, it is not readily apparent if this assumption is valid (33).
The objective of this study was to determine how miRNA composition affects the biological functions of exosomes derived from different cellular sources using bioinformatics approaches to predict their biological effects and experiments to validate predictions. We compared the phenotypic effects of exosomes derived from two distinct cell types: the commonly utilized HEK 293 cells and PC3 prostate cancer cells. We first performed miRNA profiling and used bioinformatics analyses to predict which pathways are targeted by the miRNA cargoes of these two cell types. In vitro internalization assays were then used to determine the rate and extent of exosome uptake into various cell types before testing our in silico predictions in representative assays, including their effects on cell migration and macrophage polarization. Because the biological source of exosomes imparts them with intrinsic functional activities, this study emphasizes the importance of characterizing exosomes before clinical use.
Materials and Methods
Exosome isolation
Prostate cancer cells (PC3) and human embryonic kidney cells (HEK) cells were purchased from ATCC. 48 h before exosome isolation, PC3 and HEK cells were cultured in exosome-free medium. To isolate exosomes, cell culture supernatants were collected and centrifuged at 2000 × g for 30 min to remove cells and debris. Total exosome isolation reagent (Invitrogen, Carlsbad, CA) was added to the supernatant and incubated overnight at 4°C. To collect the exosomes, the mixture was centrifuged at 10,000 × g for 1 h at 4°C and the exosome pellet was re-suspended in phosphate buffered saline (PBS), pH 7.4.
Nanoparticle tracking analysis
Exosome sizing and enumeration was determined by nanoparticle tracking analysis (NTA). Exosome samples were serially diluted in PBS. Dilutions corresponding to 10–500 μl of culture medium were generally in the range of the Nanosight LM10 instrument (Malvern Instruments, Malvern, UK) of 0.5 and 20E8 particles per milliliter. Each sample was tracked for 30 s using a camera setting of 16. Analysis was performed with the software version 2.3 using a gain of 6.0 and a threshold of 11 for all samples.
Zeta Potential Analysis
Exosome samples with a concentration of 2 μg/ml in PBS were diluted 1:50 in filtered deionized water and analyzed on the NanoBrook Omni. Each HEK exosome and PC3 exosome sample was read a total of three times using the phase analysis light scattering method to obtain the exosome zeta potential. Samples were allowed to stabilize for 5 minutes and reads were set to 30 seconds. The analysis was performed in triplicate.
Western blot analysis
25 μg exosomes by protein were incubated with 30 μl anti-human CD63 beads (Thermo Fisher Scientific, Waltham, MA). Exosomes were bound to the beads overnight at 4°C using a combination of tumbling and orbital shaking (650 rpm) to prevent settling. Exosome-bound beads were cleaned by magnetic separation according to the manufacturer’s instructions. Exosome beads were re-suspended in 10 μl of RIPA buffer and sonicated for 10 min. Exosomes were held at 4°C for an additional 15 min in RIPA buffer for complete lysis before being combined with 4 μl of 4× LDS buffer. Samples were heated to 95°C for 5 min and then separated on a 4–12% Bis-Tris NuPage gel using SDS-MOPS buffer. The transfer onto PVDF membrane was performed at 100 V for 60 min. Isolates were then confirmed as CD63+ by western blotting using 1:1000 BD anti-human CD63 (556019; Becton Dickinson, Franklin Lakes, NJ) overnight at 4°C.
Flow cytometry
8 μg exosomes (quantified by protein) were combined with 30 μl of anti-human CD9 beads (Thermo Fisher Scientific). Exosomes were bound to the beads overnight at 4°C using a combination of tumbling and orbital shaking (650 rpm) to prevent settling. Exosome-bound beads were cleaned by magnetic separation according to the manufacturer’s instructions. Beads were then re-suspended to 100 μl in 1%-BSA-PBS and incubated with 1.5 μl of anti-human CD63-FITC (Miltenyi Biotech). Exosomes were stained for 1 h at room temperature with orbital shaking (450 rpm) in a glass test tube to prevent settling. Beads were analyzed on the MacsQuant® Analyzer 10 flow cytometer.
miRNA profiling
Exosomes from HEK and PC3 cells were collected and RNA isolated using Trizol reagent. miRNA profiling was performed using the NanoString platform (NanoString Technologies, Seattle, WA) as per the manufacturer’s instructions. nSolver was used to analyze the miRNA profiling data. miRNA reads were normalized to positive controls and the top 100 reads. Negative controls were used to determine the threshold for background. To assess reproducibility, duplicate biological samples were profiled.
miRNA pathway and network analyses
The top 28 miRNAs identified in HEK exosomes and the top 18 miRNAs identified in PC3 exosomes were present at >1% of the total read counts of all exosomal miRNAs and thus used for further analysis. Messenger RNA targets of the abundant miRNAs were identified with Diana (microT-CDS v5.0) using a filter threshold of greater than 0.7. Targeted pathways were analyzed by submitting the Diana predicted gene-targets to PANTHER v12.0 “statistical over-representation test” against the annotation data set “PANTHER pathway.” Statistically significantly targeted pathways were considered p<0.05.
Exosomal uptake and flow cytometry
To assess exosome uptake into HEK and PC3 cells, 50,000 MDA-MB-231, HUVEC, PC3, and RAW 264.7 cells plated in exosome-free media (DMEM supplemented with 10% exosome-free serum) were incubated with 2 μg of Bodipy-TR-labeled exosomes (Cat# D7540, Thermo Fisher Scientific). Stained exosomes were cleaned on Exosome Spin Columns (MW3000) according to the manufacturer’s instructions (Cat# 4484449, Thermo Fisher Scientific). Incubation periods of 0.5 h, 1 h, 2 h, and 4 h, were used. Cells were detached by trypsin digestion and thoroughly washed with citric acid buffer to remove surface-bound but non-internalized exosomes using the method described in (34). Cellular uptake was analyzed using the MACSQuant® Analyzer (Miltenyi Biotech).
Macrophage polarization study
RAW 264.7 macrophages (3 × 10 cells) were plated onto a T-25 flask with DMEM and 100 ng/mL LPS for 6 h at 37°C. Cells were then detached with TrypLE Express Enzyme (Cat#: 2605036, Gibco, Thermo Fisher Scientific). RAW 264.7 cells were plated at 1.6 × 105 cells/well of a 24-well plate with new medium. 4 μg,16 μg and 64 μg/ml HEK and PC3 exosomes were added. After 48 h, RNA was collected according to the TRIzol protocol and cDNA was obtained using the NEB ProtoScript II synthesis kit. A three-step qPCR protocol using PerfeCTa SYBR Green SuperMix (Cat#:95056–02K, Quantabio, Beverly, MA) was conducted utilizing GAPDH as a housekeeping gene. INOS (M1 marker), (FP 5’-TTTGCTTCCATGCTAATGCGAAAG-3’, RP: 5’-GCTCTGTTGAGGTCTAAAGGCTCCG-3’); TNF-α (M1 Marker), (FP: 5’- ACGGCATGGATCTCAAAGAC-3’, RP: 5’- AGATAGCAAATCGGCTGA-3’); arginase-1 (M2 marker): (FP: 5’-CAGAAGAATGGAAGAGTCAG-3’, RP: 5’-CAGATATGCAGGGAGTCACC-3’); MGL2 (M2 marker), (FP: 5’- AGCGGGAAGAGAAAAACCAG-3’, RP: 5’-AGATGACCACCAGTAGCAGGAG-3’); GAPDH, (FP: 5’-CAAGGTCATCCATGACAACTTTG-3’ RP: 5’-GTCCACCACCCTGTTGCTGTAG-3’).
Integrin β3 Targeting
To assess if macrophage polarization could be attributed to integrin-β3 targeting, as predicted, 2 × 105 macrophages were plated per well of a 24 well plate. Cells were treated in triplicate with 64.0 μg/ml of HEK or PC3 exosomes. As a positive control, 2 × 105 RAW 264.7 cells were electroporated in triplicate with 0.01 or 10 nM of Let-7b. After 48 h, RNA was collected according to the TRIzol protocol and cDNA was obtained using the NEB ProtoScript II synthesis kit. A three-step qPCR protocol using PerfeCTa SYBR Green SuperMix (Cat#:95056–02K, Quantabio, Beverly, MA) was conducted utilizing GAPDH as a housekeeping gene. ITGB3: (FP: 5’-TGACATCGAGCAGGTGAAAG-3’, RP: 5’- GAGTAGCAAGGCCAATGAGC-3’).
Cell viability assay
To assess the effect of HEK and PC3 exosomes on cellular proliferation or death, PC3 and MDA-MB-231 cells were plated onto 96-well plates at 3,000 cells/well and RAW 264.7 cells at 6,000 cells/well. 24 h after plating, cells were serum starved for 24 h in DMEM. Then medium was replaced with exosome-free medium and incubated with increasing exosome concentrations for 24h. After an additional 48 h, MTS substrate (Sigma Aldrich, St. Louis, MO) was added to the wells and incubated for 2 h at 37°C. The formazan dye produced by the viable cells was measured using a SpectraMax i3 plate reader at 490 nm.
Cell migration assay
MCF7 breast cancer cells were plated onto a 12 well plate at 300,000 cells per well with 1ml DMEM with 10% FBS. Cells were treated with 4.0, 16.0, or 64.0 μg/ml of HEK or PC3 exosomes. As a positive control one well was treated with 10nM of hsa-miR-374a-5p following the RNAiMAX protocol. After 24 h the media was removed and replaced with DMEM without FBS to serum starve the cells, additional doses of exosomes and miRNA were added and cells were incubated for an additional 24 h. After a total of 48 h under exosome treatment and 24 hours of serum starvation cells were detached and plated at 5,000 cells per well of a 5-μm 96-well migration assay plate (Millipore ECM512). Cells were plated apically in DMEM and attracted to the basolateral chamber by 10% FBS in DMEM. After 24 h total migrated cells were collected and lysed according to manufacturer’s protocol and detected by CyQuant GR dye. MCF7 cells treated with 64.0 μg/ml of HEK or PC3 exosomes over 48 h were also analyzed for expression of E-Cadherin, N-cadherin, and Vimentin. RNA was collected according to the TriZol protocol and cDNA was obtained using the NEB ProtoScript II synthesis kit. A three-step qPCR protocol using PerfeCTa SYBR Green SuperMix (Cat#:95056–02K, Quantabio, Beverly, MA) was conducted utilizing GAPDH as a housekeeping gene. The following primers were used: E-Cadherin, (FP: 5’- TGCCCAGAAAATGAAAAAGG-3’ RP: 5’- GTGTATGTGGCAATGCGTTC-3’) N-Cadherin, (FP: 5’- GACAATGCCCCTCAAGTGTT-3’ RP: 5’- CCATTAAGCCGAGTGATGGT-3’) Vimentin, (FP: 5’- GAGAACTTTGCCGTTGAAGC RP: 5’- GAGAACTTTGCCGTTGAAGC-3’) GAPDH, (FP: 5’-CAAGGTCATCCATGACAACTTTG-3’ RP: 5’-GTCCACCACCCTGTTGCTGTAG-3’).
Statistical analyses
Statistical analyses were performed using GraphPad Prism 7. Multiple comparisons were performed by one-way ANOVA followed by Tukey’s post hoc test. Statistically significant differences were marked with asterisks (*p<0.05, **p<0.01, ***p<0.001).
Results
Exosome characterization
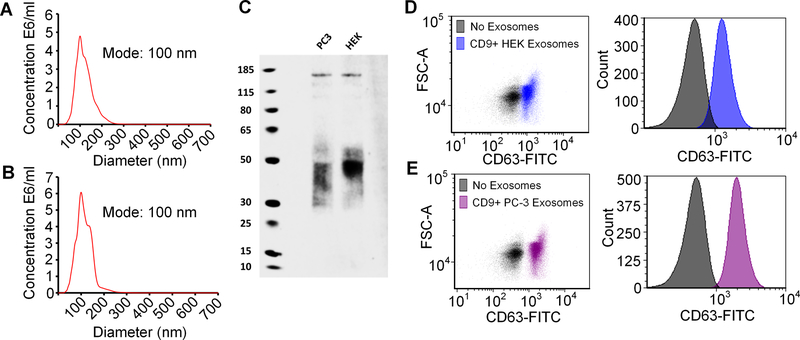
Exosome particle size was measured by nanoparticle tracking analysis (NTA) and the mode size was ~100 nm for both HEK and PC3 exosomes (Fig. 1A,B). HEK exosome samples had a mean diameter of 121 nm with an interquartile range of 93 to 141 nm and PC3 exosome samples had a mean diameter of 115 nm with an interquartile range of 109 to 134 nm. Both exosome samples displayed a moderate polydispersity as calculated by the standard deviation over the mean (%RSD) (Supplementary Figure 1). When isolated and analyzed by western blotting, both PC3 and HEK exosomes stained positive for CD63 (Fig. 1C). Although the molecular weight of CD63 is ~25kDa, it can be glycosylated at different sites, which can affect its migration during electrophoresis to produce a broad band between 30–60 kDa as seen here and as previously described (35). When analyzed by flow cytometry, HEK (Fig. 1D) and PC3 (Fig. 1E) exosomes were positive for CD9 and CD63. Dually CD9- and CD63-positive exosomes are considered bona fide exosomes (36). HEK and PC3 exosomes zeta potentials were also analyzed and in agreement with published data; both cell lines produced exosomes with a negative charge (37, 38). PC3 exosomes had a zeta potential of −11.84 ±4.39 mV and HEK exosomes had a zeta potential of −16.46 ±3.37 mV.
Figure 1.
Nanoparticle tracking analysis of (A) HEK exosomes and (B) PC3 exosomes. (C) CD63 western blot of PC3 and HEK exosomes. (D,E) HEK and PC3 exosomes are positive for both CD9 and CD63 as determined by staining CD9+ bead bound exosomes with CD63-FITC.
Profiling reveals disparate miRNA landscapes of HEK and PC3 exosomes
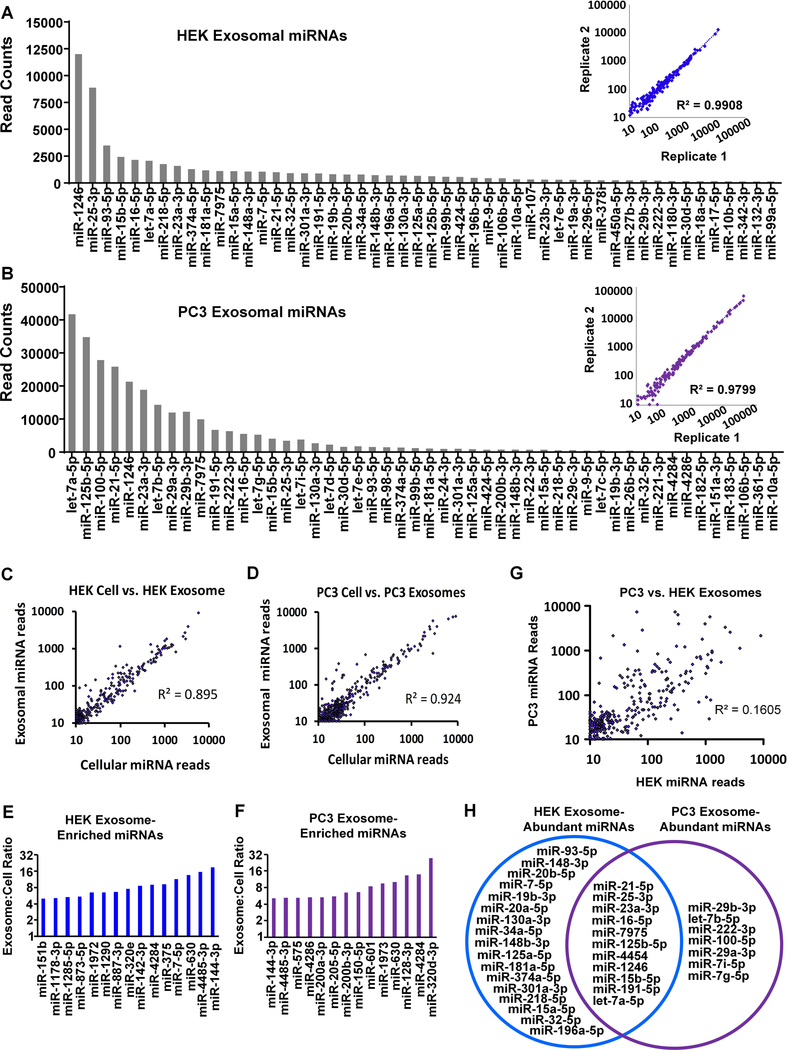
miRNA profiling was next performed to predict the potential phenotypic effects of HEK and PC3 exosomes, and miRNAs were rank ordered based on read counts (Fig. 2A,B). Biological replicates demonstrated a strong correlation between two different samples of the same cell line (R2=0.99 and R2=0.98 for HEK and PC3, respectively). We also observed a strong correlation between exosomal and cellular miRNA reads for each cell line indicating that, in general, the abundance of exosomal miRNAs reflects abundant miRNAs in the producing cell. The correlation between HEK cell and exosome miRNA reads was R2=0.895 (Fig. 2C), and the correlation between PC3 cell and exosome reads was R2=0.924 (Fig. 2D). Although exosomal miRNAs generally represent the miRNA landscape of the producing cell, both HEK and PC3 cells selectively packaged subsets of miRNAs into exosomes. Fifteen miRNAs were present in HEK exosomes at read counts at least four-fold greater than in HEK cells (Fig. 2E), and fourteen miRNAs were selectively packaged in PC3 cells (Fig. 2F). The selectively packaged miRNAs were mostly dissimilar between HEK and PC3 cells; however, miR-4284, miR-630, miR-4485–3p, and miR-144–3p were exosomally enriched by both cell types. When the miRNAs in PC3 exosomes were plotted against the miRNAs in HEK-293 exosomes, the correlation was low (R2=0.16) (Fig. 2G) and therefore substantially different.
Figure 2.
The top 50 exosomal miRNAs by number of reads are shown for HEK (A) and PC3 (B). Cellular versus exosomal read counts show high correlation for HEK (C) and PC3 (D), but a subset of miRNAs were enriched above four-fold in HEK cells (E) and PC3 cells (F). (G) Conversely, of the miRNAs found in both HEK and PC3 exosomes, a poor correlation in abundance was observed (R2=0.16). Abundant exosome miRNAs were considered to be present at >1% of the total mapped reads for all miRNAs. For HEK these 28 miRNAs represented 75% of all reads and for PC3 the 18 abundant miRNAs represented 80% of all reads. (H) Comparing the abundant miRNAs in HEK and PC3 exosomes demonstrated that some abundant miRNAs are shared between the HEK and PC3 cells (11), while others were unique to HEK (17) or PC3 (7) exosomes.
We next considered that the most abundant miRNAs were likely mediators of the miRNA effects of HEK or PC3 exosomes. We defined high-abundance miRNAs as those representing greater than 1% of the total reads of all mapped miRNAs. High-abundance miRNAs present at greater than 1% of the total reads represented ~75% of miRNAs in HEK exosomes (28 total miRNAs) and ~80% of miRNAs in PC3 exosomes (18 total miRNAs). Several of the miRNAs found at high read counts in PC3 exosomes but not HEK exosomes or were present at very low numbers. For example, let-7b/i/g-5p, miR-100–5p, miR-222–3p and miR-29a-3p were present at high reads in PC3 but not HEK exosomes. Conversely, the second most abundant miRNA in HEK exosomes, miR-25–3p, was only found at low read counts in PC3 exosomes (Fig. 2H).
In silico analysis of HEK and PC3 exosome miRNA landscapes
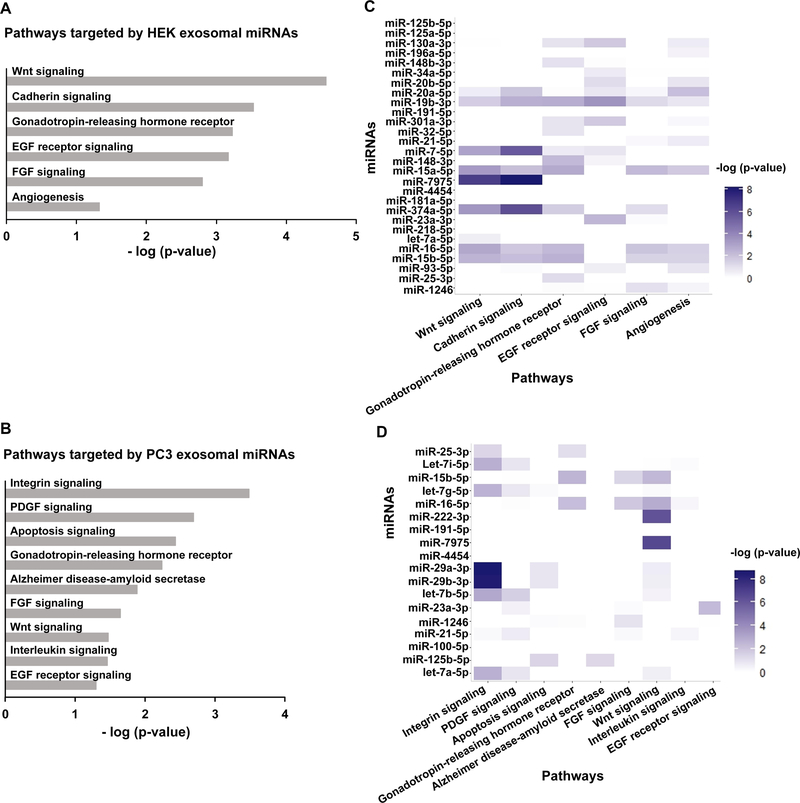
After observing that HEK and PC3 exosomes packaged different miRNA into exosomes with poorly correlated abundance, we next sought to understand which pathways were most targeted by these unique miRNA landscapes. The top 28 HEK miRNAs and the top 18 PC3 miRNAs were analyzed with Diana to ascertain the mRNA targets of the abundant exosomal miRNAs. After obtaining the gene targets of all abundant miRNAs, these were collectively analyzed by PANTHER to predict which pathways were most significantly targeted. The 28 abundant miRNAs in HEK exosomes were most significantly predicted to target Wnt signaling, cadherin signaling, and the gonadotropin-releasing hormone receptor pathways (Fig. 3A). The 18 abundant miRNAs in PC3 exosomes were most significantly predicted to target three unique pathways: integrin signaling, PDGF signaling, and apoptosis signaling (Fig. 3B). Since 11 miRNAs were common to the abundant miRNAs in HEK and PC3 exosomes, we assessed the targets of each miRNA separately to obtain a more granular view of which pathways were targeted by specific miRNAs. The statistical significance of targeted pathways of the abundant HEK and PC3 exosomal miRNAs showed that pathways that were significantly targeted by the miRNA landscape were not necessarily highly targeted by numerous individual miRNAs. This highlights the importance of considering the combined effect of all abundant miRNAs in predicting miRNA-mediated phenotypic effects of exosomes (Fig. 3C, D).
Figure 3.
The gene targets of abundant miRNAs in HEK and PC3 exosomes were obtained from Diana using a cut-off threshold of 0.7. To ascertain which pathways are most targeted, the gene lists were analyzed with PANTHER (HEK (A), PC3 (B)). Analyzing the gene-target list for each miRNA individually with PANTHER shows which of the significant pathways are targeted by specific miRNAs (HEK (C), PC3 (D)).
Cell-specific uptake of exosomes into cells
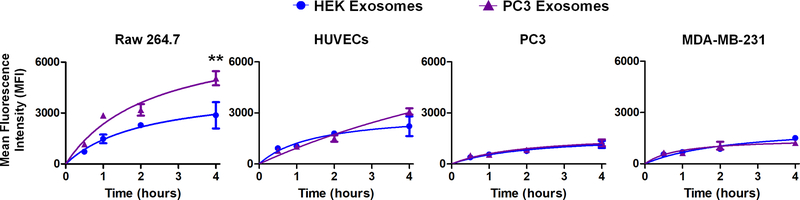
To assess how cellular origin affects exosome uptake, we incubated four different cell lines (RAW 264.7 macrophages, HUVECs, PC3, and MDA-MB-231 cells) with PC3 and HEK exosomes for 4 h. Cellular uptake was quantified by flow cytometry after removing non-internalized surface-bound exosomes. As shown in Figure 4, HEK and PC3 exosomes were taken up into HUVECs, PC3, and MDA-MB-231 cells to a similar degree. Uptake was lowest into MDA-MB-231 and PC3 cancer cells at ~1200–1300 mean fluorescence intensity (MFI). Almost twice as many exosomes were taken up into HUVECs. Significant differences in PC3 and HEK exosomal uptake were observed for RAW 264.7 cells. Not only was the uptake significantly higher in RAW 264.7 cells, in line with previous literature (34), but PC3 exosomes showed almost twice as much uptake compared to HEK exosomes (5051 MFI vs. 2871 MFI, p<0.01) into the macrophages.
Figure 4.
Uptake of PC3 and HEK exosomes into RAW 264.7 macrophages, HUVECs, PC3, and MDA-MB-231 cells. Cells were incubated with 2μg of BODIPY-labeled exosomes and internalization was assessed at 0.5, 1.0, 2.0, and 4.0 h by digesting cells in trypsin and performing a stringent citric acid buffer wash, followed by flow cytometric analysis (n=3 for each time point).
Effects of exosomes on cell viability
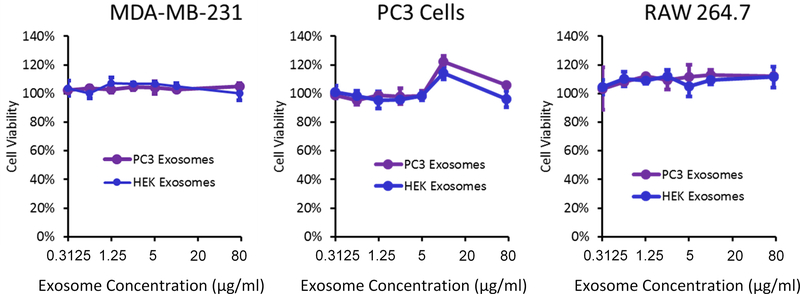
To assess the effects of these different exosomes on cell viability, PC3 cells, RAW 264.7 cells, and MDA-MB-231 cells were incubated with increasing concentrations of PC3 and HEK exosomes for 24h at concentrations of between 0.3 μg/mL to 75 μg/mL. No significant changes in viability were observed at all exosome concentrations tested (Fig. 5). Cell viability at very large doses (75 μg/mL) was not significantly reduced, highlighting the overall low toxicity of exosomes (Fig. 5).
Figure 5.
PC3, MDA-MB-231, and RAW 264.7 cells were incubated with PC3 or HEK exosomes. None of the tested cell lines demonstrated a significant decrease in viability at any of the tested doses of either HEK or PC3 exosomes.
Exosomes from different cellular sources differentially modulate macrophage polarization
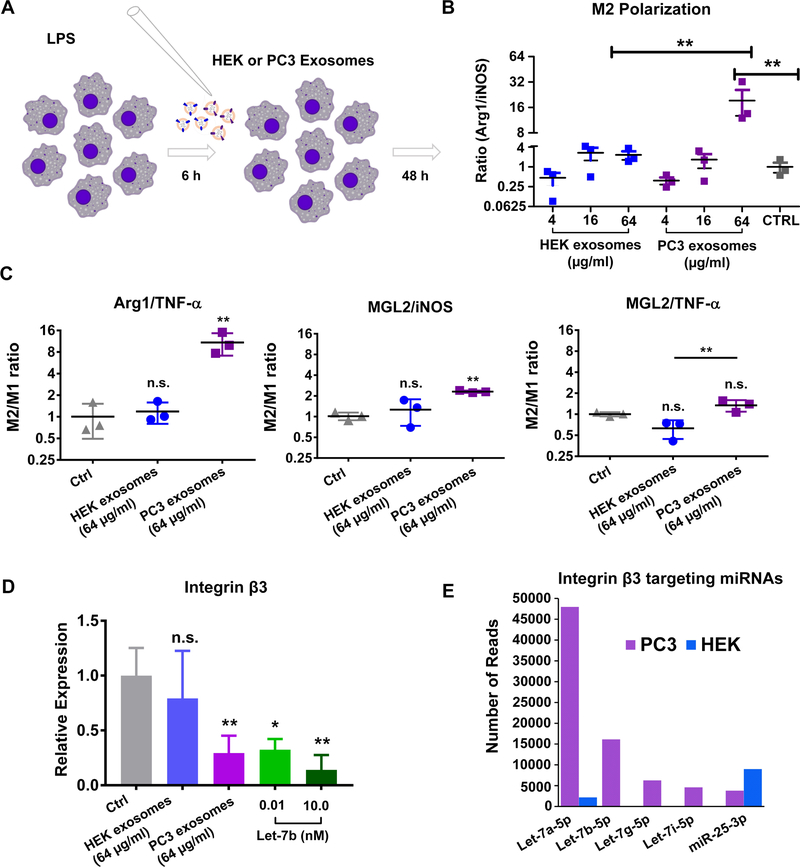
Since integrin signaling can affect macrophage polarization and our analysis indicated that PC3 exosomal miRNAs, but not HEK exosomal miRNAs, would target integrin signaling, we tested the hypothesis that PC3 exosomes would polarize RAW 264.7 macrophages to an M2 phenotype. When RAW 264.7 cells were first treated with LPS and then treated with exosomes (Fig. 6A), we observed that PC3 but not HEK exosomes polarized macrophages back to an M2 phenotype. The effect of PC3 exosomes was dose-dependent as assessed by the ratio of arginase (an M2 marker) expression to iNOS (an M1 marker) expression (Fig. 6B). A similar trend in the M2/M1 macrophage ratio was observed when MGL2 was used as an alternative M2 marker and TNF-α was used as an alternative M1 marker (Figure 6C) (39, 40). While there was a trend toward a greater M2/M1 ratio with increasing doses of HEK exosomes, these were not statistically significant compared to non-exosome-treated macrophages. At 32μg of HEK exosomes, the Arg1/iNOS ratio was 2.29-fold greater than control cells. At 32 μg of PC3 exosomes, there was a 19.2-fold increase in the Arg1/iNOS ratio, which was significantly greater than control cells and the 32 μg dose of HEK exosomes (p<0.01). In line with our predictions, only PC3 but not HEK exosomes downregulated integrin-β3, which when reduced causes M2 polarization (Fig. 6C). Numerous exosomally-abundant PC3 miRNAs were predicted to target integrin-β3 including the most abundant miRNA (Let-7a-5p) and the other Let-7 miRNAs (Let7-b, -g, & -i). Indeed, we show that PC3 exosomes downregulate integrin-β3 in RAW 264.7 macrophages ~70% (p < 0.01), while HEK exosomes did not significantly alter the expression of integrin-β3 at this dose. We confirmed that Let-7b-5p, which was only present in PC3 and not HEK exosomes, down-regulated integrin-β3 in RAW 264.7 macrophages. When Raw 264.7 cells were electroporated with Let-7b we observed a comparable level of integrin-β3 knockdown as when PC3 exosomes were given to RAW macrophages at 64 μg/ml (Fig. 6D). Of the predicted integrin-β3-targeting miRNAs in PC3 exosomes, hsa-miR-25–3p was the only one expressed to a similar extent in HEK exosomes, the rest were abundant only in PC3 exosomes (Fig. 6E). These observations agree with strong evidence that the Let-7 miRNA family controls macrophage polarization (41).
Figure 6.
(A) RAW 264.7 cells were pre-treated with LPS prior to addition of HEK or PC3 exosomes. (B) M2 polarization was assessed by comparing the ratio of Arg1 to iNOS in cells treated with HEK and PC-3 exosomes. (C) Additional M2 marker MGL2 and M1 marker TNF-α were quantified and the ratios indicated that a high dose of PC3 exosomes had an M2 polarizing effect. (D) Expression of integrin β3, which affects macrophage polarization, was reduced in RAW 264.7 cells treated with the high dose of PC3 exosomes (p<0.01). Let-7b which was present in PC3 exosomes but not HEK exosomes induced a similar level of integrin β3 knockdown when electroporated into RAW 264.7. (E) This is in agreement with the observation that integrin β3 targeting miRNAs are more numerous and abundant in PC3 exosomes.
Exosomes from HEK cells and PC-3 cells modulate cellular migration
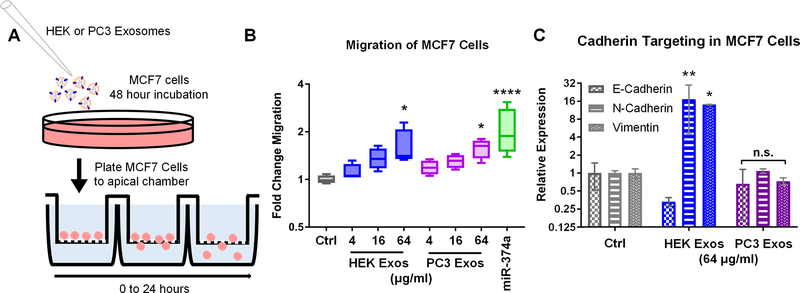
While PC3 exosomes were predicted and shown to exert malignant phenotypic effects such as M2 polarization through targeting of integrin signaling, HEK exosomes were also predicted to modulate specific pathways not targeted by PC3 exosomes. HEK, but not PC3 exosomes, were predicted to target cadherin signaling. Several types of cancer, especially breast cancer acquire metastatic potential through transitioning from an epithelial to mesenchymal phenotype. Expression of N-cadherin, specifically, is correlated with breast cancer motility, and is not typically highly expressed by poorly malignant breast cancer cells such as MCF7 (42). To test the hypothesis that HEK exosomes would exert a greater effect on the migratory potential of MCF7 cells via regulation of cadherin genes, we treated MCF7 cells with HEK and PC3 exosomes and assessed cell motility in a transwell migration assay (Fig. 7A). Since HEK exosomes but not PC3 exosomes contained high amounts of miRNA-374a, which is shown in the literature to induce migration of MCF7 cells via cadherin targeting, we included a positive control of transfected miRNA-374a (43). MCF7 cells treated with HEK and PC3 exosomes showed dose-dependent increases in the number of cells migrating across a 5 μm membrane. At the highest dose both HEK and PC3 exosomes had a significant effect with ~1.5- to 2-fold increases in migrating cells over non-treated MCF7 cells (p<0.05) (Fig. 7B). Gene expression analyses showed that PC3-exosome effects on MCF7 migration was not due to alterations in E-cadherin, N-Cadherin, or Vimentin expression levels (Fig. 7C). As predicted, HEK exosomes demonstrated dramatic effects on cadherin gene expression. The epithelial cadherin (E-cadherin) was down-regulated to ~33% while the mesenchymal markers (N-cadherin and Vimentin) were both upregulated over 10-fold (p < 0.05, p < 0.001 respectively) (Fig. 7C).
Figure 7.
(A) MCF7 cells were treated with PC3 or HEK exosomes or transfected with 10 nM of hsa-miR-374a-5p. (B) Cellular migration was assessed using a transwell migration assay. The highest dose of PC3 and HEK exosomes induced statistically significant increases in MCF7 migration (p<0.05) as did the positive control miR-374a (p<0.0001). (C) To assess if the increased migration of MCF7 cells upon exposure to HEK exosomes was related to changes in the expression of cadherin genes as predicted, qPCR was performed. HEK exosomes but not PC3 exosomes significantly upregulated the mesenchymal gene markers N-Cadherin and Vimentin (p<0.05, p<0.01) while also displaying a trend toward the reduction of the epithelial marker E-cadherin.
Discussion
Using exosomes as a delivery vehicle for exogenous therapeutic cargoes represents an exciting new area of pharmaceutical research. Before exosomes can be translated into the clinic, however, their innate biological effects must be understood. Since miRNAs mediate many of the epigenetic “reprogramming” effects of exosomes on recipient cells, we performed miRNA profiling of two distinct cell types: a commonly used exosome-producing cell type (HEK 293 human embryonic kidney cells) versus a cancer cell type (PC3 prostate cancer cells). We then used in silico analyses to predict the biological effects of the most abundant miRNAs packaged in HEK 293 versus PC3 exosomes. To assess the predictability of our bioinformatics analysis, the most significant biological pathways for each type of exosomes was assessed experimentally. Our analyses indicated that PC3 exosomes contained an miRNA landscape that would most significantly target integrin signaling (p<0.0001). Integrin signaling is known to affect macrophage polarization (44). HEK exosomes were not predicted to target integrin signaling, but were instead predicted to uniquely target cadherin-signaling, which plays a role in epithelial to mesenchymal transition and cellular migration.
With regard to integrin signaling, loss of integrin-β3 in macrophages promotes M2 polarization (45, 46). Not only was integrin signaling the most significantly predicted pathway in PC3 exosomes but the most abundant miRNA in PC3 exosomes, let-7a-5p, targets and down-regulates integrin-β3 expression (47). This prompted us to assess the effect of PC3 exosomes versus HEK exosomes in affecting M2 macrophage polarization. Confirming our in silico predictions, PC3 exosomes downregulated integrin-β3 to 30% of control. Additionally, PC3 but not HEK exosomes showed a dose-dependent ability to polarize macrophages to an M2 phenotype as determined by the expression patterns of several M2 and M1 markers. Commonly, the metabolic state of macrophages is used to understand if the cells are in an inflammatory M1-state, indicated by the high expression of iNOS and NO metabolism, or a reparative, immune-suppressive M2 state, indicated by the high expression of Arginase-1 and arginine metabolism (48). PC3 exosomes shifted RAW 264.7 macrophages towards an M2 phenotype with an 8.4-fold increase in the Arg1/iNOS ratio compared to HEK exosomes. PC3 exosomes polarized macrophages towards an M2 phenotype over HEK exosomes when additional M1 (TNF-α) and M2 (MGL2) markers were analyzed. Generally, we observed a higher M2/M1 ratio with Arginase-1 than with MGL2. This is in line with a recently published study that showed that Arginase-1 is the highest upregulated gene when macrophages are polarized towards the M2 phenotype (39).
This observation provides not only an important phenotypic characterization of prostate cancer exosomes, but also offers a biological explanation of how prostate cancer may mediate macrophage polarization in vivo. Macrophage polarization toward an M2 phenotype is a prominent and deleterious hallmark of many cancers including prostate cancer. Tumor-associated macrophages (TAMs) that adopt an M2 phenotype promote tumorigenesis through many mechanisms including increased angiogenesis, cancer cell proliferation via growth factor secretion, degradation of the extracellular matrix and release of chemokines, (which promote metastasis), and suppression of an immune response against the tumor (49). Conditioned medium from PC3 cells was previously shown to polarize RAW 264.7 macrophages to M2 (50), and it has also been shown that highly apoptotic RM-1 prostate cancer cells (treated with CoCl2 to achieve >60% apoptosis) caused M2 polarization of bone marrow macrophages through MFG-E8 (51). The exact mechanisms of M2 polarization via non-apoptotic prostate cancer cells is, however, largely unknown.
Our findings are consistent with recent reports that posit a role for exosomes in polarizing macrophages via their miRNA cargoes. In pancreatic cancer, exosomes have been shown to mediate macrophage M2 polarization, and this polarization was modulated by the exosome miRNA content (52). Further, Saha et al. directly demonstrated that the miRNA cargo of exosomes from alcohol-treated monocytes polarized naive monocytes to M2 macrophages (53). Through miRNA-profiling and phenotyping of prostate cancer exosomes, we now suggest a miRNA-mediated effect of exosomes on macrophage polarization via down-regulation of integrin-β3 in prostate cancer.
While it has been shown that cancer-cell derived exosomes exert malignant phenotypes and should probably be avoided as delivery vehicles, it is less clear how the miRNA landscapes of other cell lines impacts their biological activity and use as delivery vehicles. HEK cells, for example, are widely used as producer cells of therapeutic exosomes (1). While researchers are modifying and engineering HEK exosomes for drug delivery, it is challenging to completely remove their miRNA cargo. Thus, the intrinsic biological effects of HEK exosomes mediated by miRNAs would remain and could potentially contribute to undesired side-effects.
The miRNA landscape of HEK exosomes indicated that they would target cadherin signaling, a pathway not represented in the predicted targets of PC3 exosome miRNAs. When MCF7 breast cancer cells were treated with HEK or PC3 exosomes, HEK exosomes significantly increased the expression of N-cadherin and Vimentin, two genes which are implicated in the metastatic potential of breast cancer (54, 55). Furthermore, there was a trend toward reduction of E-cadherin in MCF7 cells treated with HEK exosomes; loss of E-cadherin is also associated with increases in breast cancer motility (56). Not surprisingly from these effects on cadherin genes, HEK exosomes did in fact exert a dose-dependent increase in the migratory potential of MCF7 cells, up to 2-fold over the control MCF7 cells. It appears that in some settings, such as macrophage polarization, HEK exosomes are relatively benign compared to PC3 exosomes, while in other settings HEK exosomes may cause undesired adverse effects. Conversely, although PC3 exosomes did not alter the expression of the assessed cadherin genes in MCF7, these cancer exosomes did promote the migration of MCF7 cells. PC3 exosomes were predicted to modulate integrin signaling, a pathway that is known to play a major role in the migratory potential of breast cancers (57). Additionally, PC3 exosomes were predicted to target PDGF signaling, which could also increase MCF7 motility (58). Further, PC3 exosomes contain hsa-miR-29b in high abundance, a miRNA that is present at very low read counts in HEK exosomes. In a study specific to MCF7 cells, transfection with hsa-miR-29b resulted in significant increases in the migratory ability of the cells (59).
In this study, exosomes were isolated using a precipitation-based method. Studies have shown that different routes of exosome isolation and RNA extraction can result in differential enrichment of different RNA species. While the composition of different RNA species, such as rRNAs and tRNAs, can vary dramatically when using different processing strategies, Tang et al. have shown that there is a high correlation between miRNA composition isolated by ultracentrifugation and precipitation-based exosome isolation methods (R2 ≥ 0.85) (60). In both commonly used methods, ultracentrifugation-based and precipitation-based isolation, exosomes are treated as bulk isolates. Future studies are warranted to assess if and how RNA composition differ between subpopulations of exosomes.
While many of the phenotypic effects of exosomes may be mediated by miRNAs, not all exosomal effects or behaviors are related to miRNAs. Other molecular characterizations such as proteomic and lipidomic studies should also be considered within a complete pipeline of exosome characterization (61). Also, the influence of other non-coding RNAs besides miRNAs, which are also present in exosomes, need further study (1). The ability of PC3 exosomes to target macrophages twice as efficiently as HEK exosomes could be due to differences in protein and lipid composition. For example, it has been shown that exosomes derived from different cells have different membrane proteins that promote specific uptake and internalization into cells (62). Further studies to understand how PC3 exosomes are taken up by macrophages could reveal strategies for engineering macrophage-targeted exosomes or synthetic delivery vehicles.
As exosomes are explored as delivery vehicles, their intrinsic biological activities cannot be ignored. Our data show that PC3 exosomes should likely not be used as a delivery vehicle in anti-neoplastic therapies where M2 macrophage polarization is known to exacerbate cancer progression. The use of HEK exosomes for drug delivery applications should also be critically evaluated, as HEK exosomes significantly modulated the expression of cadherin genes away from an epithelial phenotype and toward a more migratory mesenchymal phenotype.
Conclusion
Here we show that exosomes derived from different cellular sources show different intrinsic biological effects that can at least in part be explained by their miRNA composition. Our data suggest that prostate cancer exosomes modulate macrophage polarization and that HEK exosomes increase the migration of MCF7 cancer cells. These findings highlight that before exosomes are clinically employed as delivery vehicles, it is critical to understand their intrinsic biological activities without assuming inertness.
Supplementary Material
Acknowledgements
We acknowledge support by the NIH through awards R01EB023262 and R21EB021454. We thank Dr. Prashant Singh for performing the miRNA profiling for HEK and PC3 cells and exosomes. The authors declare no competing financial interests.
References
- 1.Ferguson SW, Nguyen J. Exosomes as therapeutics: The implications of molecular composition and exosomal heterogeneity. J Control Release. 2016;228:179–90. [DOI] [PubMed] [Google Scholar]
- 2.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–U72. [DOI] [PubMed] [Google Scholar]
- 3.Lu J, Clark AG. Impact of microRNA regulation on variation in human gene expression. Genome Res. 2012;22(7):1243–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quesenberry PJ, Aliotta J, Deregibus MC, Camussi G. Role of extracellular RNA-carrying vesicles in cell differentiation and reprogramming. Stem Cell Res Ther. 2015;6:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallo A, Tandon M, Alevizos I, Illei GG. The Majority of MicroRNAs Detectable in Serum and Saliva Is Concentrated in Exosomes. Plos One. 2012;7(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mulcahy LA, Pink RC, Carter DR. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franzen CA, Simms PE, Van Huis AF, Foreman KE, Kuo PC, Gupta GN. Characterization of uptake and internalization of exosomes by bladder cancer cells. Biomed Res Int. 2014;2014:619829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29(4):341–5. [DOI] [PubMed] [Google Scholar]
- 9.Chen L, Charrier A, Zhou Y, Chen R, Yu B, Agarwal K, et al. Epigenetic regulation of connective tissue growth factor by MicroRNA-214 delivery in exosomes from mouse or human hepatic stellate cells. Hepatology. 2014;59(3):1118–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooper JM, Wiklander PB, Nordin JZ, Al-Shawi R, Wood MJ, Vithlani M, et al. Systemic exosomal siRNA delivery reduced alpha-synuclein aggregates in brains of transgenic mice. Mov Disord. 2014;29(12):1476–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katakowski M, Buller B, Zheng X, Lu Y, Rogers T, Osobamiro O, et al. Exosomes from marrow stromal cells expressing miR-146b inhibit glioma growth. Cancer Lett. 2013;335(1):201–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Li D, Liu Z, Zhou Y, Chu D, Li X, et al. Targeted exosome-mediated delivery of opioid receptor Mu siRNA for the treatment of morphine relapse. Sci Rep. 2015;5:17543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munoz JL, Bliss SA, Greco SJ, Ramkissoon SH, Ligon KL, Rameshwar P. Delivery of Functional Anti-miR-9 by Mesenchymal Stem Cell-derived Exosomes to Glioblastoma Multiforme Cells Conferred Chemosensitivity. Mol Ther Nucleic Acids. 2013;2:e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21(1):185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim MS, Haney MJ, Zhao Y, Mahajan V, Deygen I, Klyachko NL, et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine. 2016;12(3):655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Batrakova EV, Kim MS. Development and regulation of exosome-based therapy products. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2016;8(5):744–57. [DOI] [PubMed] [Google Scholar]
- 17.Harris DA, Patel SH, Gucek M, Hendrix A, Westbroek W, Taraska JW. Exosomes released from breast cancer carcinomas stimulate cell movement. Plos One. 2015;10(3):e0117495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y, Gu Y, Han Y, Zhang Q, Jiang Z, Zhang X, et al. Tumor Exosomal RNAs Promote Lung Pre-metastatic Niche Formation by Activating Alveolar Epithelial TLR3 to Recruit Neutrophils. Cancer Cell. 2016;30(2):243–56. [DOI] [PubMed] [Google Scholar]
- 19.Ye SB, Li ZL, Luo DH, Huang BJ, Chen YS, Zhang XS, et al. Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget. 2014;5(14):5439–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhuang G, Wu X, Jiang Z, Kasman I, Yao J, Guan Y, et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012;31(17):3513–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tadokoro H, Umezu T, Ohyashiki K, Hirano T, Ohyashiki JH. Exosomes derived from hypoxic leukemia cells enhance tube formation in endothelial cells. J Biol Chem. 2013;288(48):34343–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fong MY, Zhou W, Liu L, Alontaga AY, Chandra M, Ashby J, et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. 2015;17(2):183–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou W, Fong MY, Min Y, Somlo G, Liu L, Palomares MR, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25(4):501–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32(3–4):623–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, Yuan X, Shi H, Wu L, Qian H, Xu W. Exosomes in cancer: small particle, big player. J Hematol Oncol. 2015;8:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Camussi G, Cantaluppi V, Deregibus MC, Gatti E, Tetta C. Role of microvesicles in acute kidney injury. Contrib Nephrol. 2011;174:191–9. [DOI] [PubMed] [Google Scholar]
- 27.Barile L, Lionetti V, Cervio E, Matteucci M, Gherghiceanu M, Popescu LM, et al. Extracellular vesicles from human cardiac progenitor cells inhibit cardiomyocyte apoptosis and improve cardiac function after myocardial infarction. Cardiovasc Res. 2014;103(4):530–41. [DOI] [PubMed] [Google Scholar]
- 28.Lai RC, Arslan F, Lee MM, Sze NS, Choo A, Chen TS, et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010;4(3):214–22. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Z, Yang J, Yan W, Li Y, Shen Z, Asahara T. Pretreatment of Cardiac Stem Cells With Exosomes Derived From Mesenchymal Stem Cells Enhances Myocardial Repair. J Am Heart Assoc. 2016;5(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Y, Sun X, Cao W, Ma J, Sun L, Qian H, et al. Exosomes Derived from Human Umbilical Cord Mesenchymal Stem Cells Relieve Acute Myocardial Ischemic Injury. Stem Cells Int. 2015;2015:761643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katsuda T, Ochiya T. Molecular signatures of mesenchymal stem cell-derived extracellular vesicle-mediated tissue repair. Stem Cell Res Ther. 2015;6:212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferguson SW, Wang J, Lee CJ, Liu M, Neelamegham S, Canty JM, et al. The microRNA regulatory landscape of MSC-derived exosomes: a systems view. Sci Rep. 2018;8(1):1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36(1):59–74. [DOI] [PubMed] [Google Scholar]
- 34.Feng D, Zhao WL, Ye YY, Bai XC, Liu RQ, Chang LF, et al. Cellular Internalization of Exosomes Occurs Through Phagocytosis. Traffic. 2010;11(5):675–87. [DOI] [PubMed] [Google Scholar]
- 35.Tominaga N, Hagiwara K, Kosaka N, Honma K, Nakagama H, Ochiya T. RPN2-mediated glycosylation of tetraspanin CD63 regulates breast cancer cell malignancy. Mol Cancer. 2014;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proceedings of the National Academy of Sciences. 2016;113(8):E968–E77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sokolova V, Ludwig AK, Hornung S, Rotan O, Horn PA, Epple M, et al. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids Surf B Biointerfaces. 2011;87(1):146–50. [DOI] [PubMed] [Google Scholar]
- 38.Ohshima H Encyclopedia of biocolloid and biointerface science. Hoboken, New Jersey: John Wiley & Sons, Inc.; 2016. volumes cm p. [Google Scholar]
- 39.Jablonski KA, Amici SA, Webb LM, Ruiz-Rosado Jde D, Popovich PG, Partida-Sanchez S, et al. Novel Markers to Delineate Murine M1 and M2 Macrophages. Plos One. 2015;10(12):e0145342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li C, Levin M, Kaplan DL. Bioelectric modulation of macrophage polarization. Sci Rep. 2016;6:21044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Banerjee S, Xie N, Cui H, Tan Z, Yang S, Icyuz M, et al. MicroRNA let-7c regulates macrophage polarization. J Immunol. 2013;190(12):6542–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hazan RB, Phillips GR, Qiao RF, Norton L, Aaronson SA. Exogenous expression of N-cadherin in breast cancer cells induces cell migration, invasion, and metastasis. J Cell Biol. 2000;148(4):779–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai J, Guan H, Fang L, Yang Y, Zhu X, Yuan J, et al. MicroRNA-374a activates Wnt/beta-catenin signaling to promote breast cancer metastasis. J Clin Invest. 2013;123(2):566–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cha BH, Shin SR, Leijten J, Li YC, Singh S, Liu JC, et al. Integrin-Mediated Interactions Control Macrophage Polarization in 3D Hydrogels. Adv Healthc Mater. 2017;6(21). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Su XM, Esser AK, Amend SR, Xiang JY, Xu YL, Ross MH, et al. Antagonizing Integrin beta 3 Increases Immunosuppression in Cancer. Cancer Res. 2016;76(12):3484–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang LP, Dong YJ, Dong YL, Cheng JZ, Du J. Role of Integrin-beta 3 Protein in Macrophage Polarization and Regeneration of Injured Muscle. J Biol Chem. 2012;287(9):6177–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muller DW, Bosserhoff AK. Integrin beta(3) expression is regulated by let-7a miRNA in malignant melanoma. Oncogene. 2008;27(52):6698–706. [DOI] [PubMed] [Google Scholar]
- 48.Rath M, Muller I, Kropf P, Closs EI, Munder M. Metabolism via Arginase or Nitric Oxide Synthase: Two Competing Arginine Pathways in Macrophages. Front Immunol. 2014;5:532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mantovani A, Sozzani S, Locati M, Allavena P, Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23(11):549–55. [DOI] [PubMed] [Google Scholar]
- 50.Soki FN, Koh AJ, Jones JD, Kim YW, Dai JL, Keller ET, et al. Polarization of Prostate Cancer-associated Macrophages Is Induced by Milk Fat Globule-EGF Factor 8 (MFG-E8)-mediated Efferocytosis. J Biol Chem. 2014;289(35):24560–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen PC, Cheng HC, Wang J, Wang SW, Tai HC, Lin CW, et al. Prostate cancer-derived CCN3 induces M2 macrophage infiltration and contributes to angiogenesis in prostate cancer microenvironment. Oncotarget. 2014;5(6):1595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su MJ, Aldawsari H, Amiji M. Pancreatic Cancer Cell Exosome-Mediated Macrophage Reprogramming and the Role of MicroRNAs 155 and 125b2 Transfection using Nanoparticle Delivery Systems. Sci Rep-Uk. 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saha B, Momen-Heravi F, Kodys K, Szabo G. MicroRNA cargo of extracellular vesicles from alcohol-exposed monocytes signals naive monocytes to differentiate into M2 macrophages with increased phagocytic activity. Eur J Clin Invest. 2016;46:82–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu CY, Lin HH, Tang MJ, Wang YK. Vimentin contributes to epithelial-mesenchymal transition cancer cell mechanics by mediating cytoskeletal organization and focal adhesion maturation. Oncotarget. 2015;6(18):15966–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nieman MT, Prudoff RS, Johnson KR, Wheelock MJ. N-cadherin promotes motility in human breast cancer cells regardless of their E-cadherin expression. J Cell Biol. 1999;147(3):631–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hiraguri S, Godfrey T, Nakamura H, Graff J, Collins C, Shayesteh L, et al. Mechanisms of inactivation of E-cadherin in breast cancer cell lines. Cancer Res. 1998;58(9):1972–7. [PubMed] [Google Scholar]
- 57.Taherian A, Li X, Liu Y, Haas TA. Differences in integrin expression and signaling within human breast cancer cells. BMC Cancer. 2011;11:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hurst NJ Jr., Najy AJ, Ustach CV, Movilla L, Kim HR. Platelet-derived growth factor-C (PDGF-C) activation by serine proteases: implications for breast cancer progression. Biochem J. 2012;441(3):909–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang C, Bian Z, Wei D, Zhang JG. miR-29b regulates migration of human breast cancer cells. Mol Cell Biochem. 2011;352(1–2):197–207. [DOI] [PubMed] [Google Scholar]
- 60.Tang YT, Huang YY, Zheng L, Qin SH, Xu XP, An TX, et al. Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. Int J Mol Med. 2017;40(3):834–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haraszti RA, Didiot MC, Sapp E, Leszyk J, Shaffer SA, Rockwell HE, et al. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J Extracell Vesicles. 2016;5:32570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.