Abstract
Background
Human tuberculosis (TB) is caused by seven phylogenetic lineages of the Mycobacterium tuberculosis complex (MTBC), Lineage 1–7. Recent advances in rapid genotyping of MTBC based on single nucleotide polymorphisms (SNP), allow for phylogenetically robust strain classification, paving the way for defining genotype-phenotype relationships in clinical settings. Such studies have revealed that, in addition to host and environmental factors, strain variation in the MTBC influences the outcome of TB infection and disease. In Tanzania, such molecular epidemiological studies of TB however are scarce in spite of a high TB burden.
Methods and findings
Here we used SNP-typing to characterize a nationwide collection of 2,039 MTBC clinical isolates representative of 1.6% of all new and retreatment TB cases notified in Tanzania during 2012 and 2013. Four lineages, namely Lineage 1–4 were identified within the study population. The distribution and frequency of these lineages varied across regions but overall, Lineage 4 was the most frequent (n = 866, 42.5%), followed by Lineage 3 (n = 681, 33.4%) and 1 (n = 336, 16.5%), with Lineage 2 being the least frequent (n = 92, 4.5%). We found Lineage 2 to be independently associated with female sex (adjusted odds ratio [aOR] 2.14; 95% confidence interval [95% CI] 1.31 – 3.50, p = 0.002) and retreatment cases (aOR 1.67; 95% CI 0.95 – 2.84, p = 0. 065) in the study population. We found no associations between MTBC lineage and patient age or HIV status. Our sublineage typing based on spacer oligotyping on a subset of Lineage 1, 3 and 4 strains revealed the presence of mainly EAI, CAS and LAM families. Finally, we detected low levels of multidrug resistant isolates among a subset of 144 retreatment cases.
Conclusions
This study provides novel insights into the MTBC lineages and the possible influence of pathogen–related factors on the TB epidemic in Tanzania.
Introduction
Tuberculosis (TB) is the leading cause of mortality due to an infectious disease [1]. In 2017, an estimated 10 million people developed TB globally, with 1.6 million dying of the disease. Tanzania is among the thirty high burden countries, with a national average TB notification rate of 129 cases per 100,000; however, some regions show higher notification rates [2]. Like in most sub-Saharan African countries, the HIV epidemic contributes substantially to the high TB incidence in Tanzania, where a-third of the TB patients are co-infected with HIV [2]. Contrarily, drug-resistant TB is still low in this setting [3]. Other risk factors such as poverty also influence the epidemiology of TB in Tanzania [4].
Transmission of TB occurs via infectious aerosols, where upon exposure individuals can either clear the infection, develop active disease or remain latently infected [5]. The complex dynamics of TB infection and disease are determined by the environment, the host and the pathogen [6]. Seven main phylogenetic lineages of the Mycobacterium tuberculosis complex (MTBC) (Lineage 1–7) cause TB in humans [7]. These lineages are phylogeographically distributed, partially reflecting human migration histories [8–10]. Genomic differences among the MTBC strains translate into relevant biological and epidemiological phenotypes [11]. Epidemiologically speaking, these phenotypes are demonstrated by indicators such as transmission potential, disease severity and progression rates from infection to disease [12–15]. In general, strains of the widely distributed lineages, Lineage 2 and 4 or “generalists”, appear to be more virulent than those of the geographically restricted lineages, Lineage 5 and 6 or “specialists” [7,11].
Studying genotype-phenotype relationships requires understanding the genetic diversity of MTBC clinical strains in a given clinical setting. In Tanzania, few studies have described the genetic diversity of the MTBC [16–19]. These previous works used conventional genotyping tools such as the spacer oligonucleotide typing (spoligotyping) technique and revealed the presence of mainly the East African Indian (EAI), Central Asian (CAS) and Latin American Mediterranean (LAM) spoligo families, and the Beijing family reported only at the lowest frequencies. Based on phylogenetically robust techniques, which include single nucleotide polymorphisms (SNPs), the spoligo families correspond to Lineage 1, 3, 4 and 2, respectively. These previous studies from Tanzania are limited as they only focused on few specific geographical locations on the country and only one study profiled MTBC on a countrywide scale albeit with low sampling coverage [18]. Moreover, despite the invaluable contribution of techniques like spoligotyping in the molecular epidemiology field, such techniques are suboptimal for phylogenetically robust strain classification due to high rates of convergent evolution [20,21].
In this study, we applied for the first time a robust SNP typing method to classify the largest so far nationwide representative collection of clinical isolates to gain insights into unknown patterns of MTBC diversity in different regions of Tanzania. Given that only few studies have assessed and identified lineage-specific differences in clinical settings, we then looked for potential associations between the MTBC lineages and the available clinical and epidemiological characteristics of the patients in the study population.
Material and methods
Ethics statement
The study was approved by the National Tuberculosis and Leprosy Programme and the ethical clearance was provided by the National Institute for Medical Research of Tanzania (Dar es Salaam, Tanzania). The data in this study were analyzed anonymously.
The National Tuberculosis and Leprosy Program routine drug surveillance system
Our study was based on a nationwide convenience sample of sputum smear positive new and retreatment TB cases diagnosed between 2012 and 2013 in Tanzania. The collection was obtained via a platform established for routine TB drug resistance surveillance by the National Tuberculosis Leprosy Program (NTLP) of Tanzania, covering health facilities in all geographical regions of the country. Briefly, smear positive sputa specimens from approximately 25% of new TB cases and from all retreatment cases were obtained for the drug resistance surveillance. To obtain 25% sputa from new cases each region was allocated four months per annum, where the respective health facilities in the region submitted sputa samples to zonal reference laboratories for culture. The zonal laboratories include the Central Tuberculosis Reference Laboratory (CTRL) in Dar es Salaam, Bugando Medical Center (BMC) in Mwanza and Kilimanjaro Christian Medical Center (KCMC) in Kilimanjaro, which serve the Coastal and Southern zone, the Lake zone, and the Northern zone, respectively. Isolates from the two zonal laboratories, BMC and KCMC were then sent to the CTRL for drug susceptibility testing (DST). For this study we included all the isolates we could retrieve from the culture archives at the CTRL.
Study population and data collection
We included a total of 2,039 unique (single patient) culture-confirmed TB cases, each of whom we could retrieve the respective culture isolate from the CTRL culture archives. This study population represents 1.6% of all the estimated TB notified cases in the country between 2012 and 2013 (S1 Fig). We also obtained corresponding socio-demographic and clinical information collected during patients’ consultation at the respective health facilities. The demographic data collected included age, sex and geographical location of the patients, whereas clinical data included HIV status and disease category (i.e., new case and retreatment case).
Processing of culture isolates
The smear positive sputa samples were cultured on Löwenstein Jensen (LJ) growth medium according to laboratory protocols. For this study, we included MTBC clinical isolates retrieved from archived LJ media. We then prepared heat inactivated samples for the retrieved clinical isolates by suspending MTBC colonies into 1ml sterile water and heat inactivate at 95°C for one hour.
Molecular genotyping
We then classified the MTBC clinical isolates into main phylogenetic lineages by TaqMan real-time PCR according to standard protocols (Applied Biosystems, Carlsbad, USA) and as previously described [22]. Briefly, the TaqMan PCR uses fluorescently labeled allele-specific probes for singleplex SNP-typing that are specific for each MTBC lineage. For comparisons, we also performed 43-spacer spoligotyping on a membrane for a subset of representative MTBC clinical strains following standard protocols [23], since spoligotyping is still widely used as a gold standard for genotyping in similar settings. We randomly selected 107 samples out of the 2,039 representative of three lineages, Lineage 1, 3 and 4. We excluded Lineage 2 strains for this analysis given that such strains almost exclusively belong to the Beijing family. The clinical strains were assigned to spoligotype families using the online database SITVITWEB [24].
Drug resistance genotyping
We selected a subset of 144 clinical isolates from the 321 retreatment cases to perform molecular drug resistance testing. We used a previously described multiplex polymerase chain reaction (PCR) to target the rifampicin resistance determining region of rpoB gene [25]. The PCR assay targets both the tuberculous and non-tuberculous Mycobacteria (MTBC and NTMs, respectively) rpoB gene, so we could also rule out the presence of non-tuberculous isolates in our study sample using the assay. The amplified rpoB gene product was confirmed by electrophoresis on a 2% agarose gel and sent for Sanger sequencing. We analyzed the resulting sequences by Staden software package [26] and using MTBC H37Rv rpoB gene as reference sequence.
Statistical analysis
For statistical analysis, we applied descriptive statistics to delineate patients’ characteristics. We used Chi-square or Fisher’s exact tests for assessment of differences between groups in categorical variables, whenever applicable. We used univariate and multivariate logistic regression models to assess for the association between MTBC lineages and patients’ clinical and demographic characteristics. The associations were assessed for Lineage 2 compared to all other lineages (Lineages 1, 3 and 4), adjusting for age, sex, disease category and HIV status. All statistical analyses were performed in R 3.5.0 [27].
Results
Patients’ demographic and clinical characteristics
The patients’ demographic and clinical information in our study included age, sex, geographical location, HIV and disease category (new or retreatment case). Table 1 describes patients’ characteristics of the study population. The proportions of the observed and missing data for the study population are summarized in S2 Fig.
Table 1. Clinical and demographic characteristics of the TB cases.
| Characteristics | Valid Proportion % |
Total (%) n = 2,039 |
|---|---|---|
| Age, median (IQR) | ||
| 35 (27–44) | ||
| Age groups (years) | ||
| Child age (< 15) | 9.9 | 20 (1.0) |
| Young age (15–24) | 29.7 | 312 (15.3) |
| Early adult (25–44) | 48.0 | 1170 (57.4) |
| Late adult (45–64) | 10.0 | 379 (18.6) |
| Old age (> 65) | 2.5 | 73 (3.6) |
| Not available | 85 (4.2) | |
| total n = 1,954 | ||
| Sex | ||
| Female | 32.4 | 645 (31.6) |
| Male | 67.6 | 1346 (66.0) |
| Not available | 48 (2.4) | |
| total n = 1,991 | ||
| HIV status | ||
| Negative | 67.7 | 1086 (53.3) |
| Positive | 32.2 | 517 (25.4) |
| Indeterminate | 0.06 | 1 (0.1) |
| Not available | 435 (21.3) | |
| total n = 1,604 | ||
| Patient category | ||
| New case | 84.0 | 1,679 (82.3) |
| Retreatment | 16.1 | 321 (15.7) |
| Not available | 39 (1.9) | |
| total n = 2,000 | ||
| Geographical zone | ||
| Central | 1.1 | 22 (1.1) |
| Coastal | 51.6 | 1,029 (50.5) |
| Lake | 17.9 | 358 (17.6) |
| Northern | 20.2 | 403 (19.8) |
| S. Highlands | 8.1 | 162 (8.0) |
| Western | 0.5 | 10 (0.5) |
| Zanzibar | 0.6 | 12 (0.6) |
| Not available | 43 (2.1) | |
| total n = 1,996 |
IQR, interquartile range; valid proportion, proportion excluding missing values; Total n, all values including NA (not available).
Our study population consisted of TB patients ranging between the age of 2 and 89 years with a median age of 28 years (interquartile range [IQR] 27–44). To further probe the age distribution in the study population, we categorized the TB patients into five different age groups (Table 1). We detected approximately three-quarters of the TB cases to occur among the “young age” and “early adult” age groups. Further, our findings show that about 1.0% of the TB cases were pediatric cases (< 15 years).
Similar to other settings [1], we identified a higher proportion of male TB cases compared to female TB cases. However, the male-to-female ratio observed in our study population was higher than the national estimates for the two years of sampling (2.2:1 vs., 1.4:1). The striking gender imbalance among TB cases seems to peak at adolescence onwards and is less pronounced among pediatric TB cases (S1 Table). Additionally, a-third (32.2%, 517/1604) of the TB cases with available HIV status were HIV co-infected. In contrast, TB/HIV co-infected cases were more likely to be female (44.5%, CI 38.3–50.7% vs., 25.8%, 95% CI 20.6–31.0%) which is consistent with HIV being generally more prevalent in females than males in Tanzania [28]. We found that our study population comprised 16.1% (321/2000) of TB retreatment cases, which was four-fold higher than the overall countrywide notifications [29]. Finally, more than half (51.6%, 1029/1996) of the TB patients in our study population were diagnosed in the Coastal zone of Tanzania and about 40% were either diagnosed in the Lake and Northern zones. In addition to higher TB notification rates, the three former mentioned geographical zones contain the country’s zonal TB reference laboratories. The remaining 10% of the patients were diagnosed in any of the remaining four geographical zones of Tanzania.
Main MTBC lineages in Tanzania
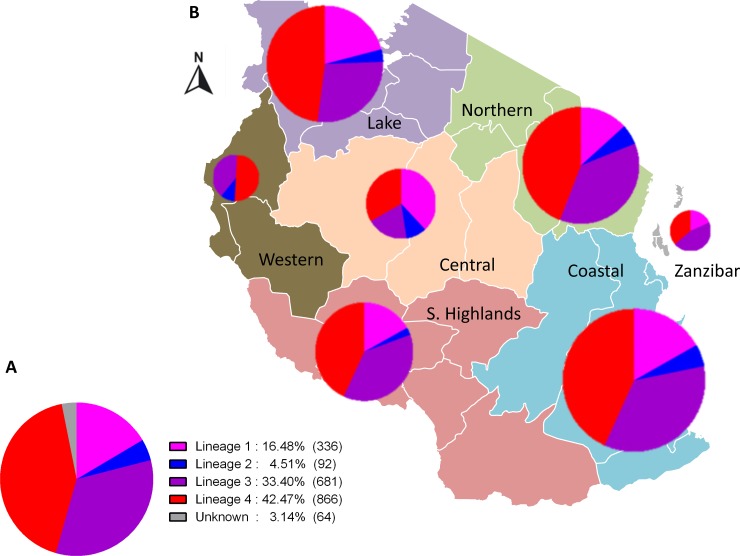
Using SNP-typing, we detected four of the seven known MTBC lineages (Fig 1), albeit at varying proportions. In our study population, Lineage 4 and Lineage 3 were the most frequent (866, 42.5% and 681, 33.4%, respectively) followed by Lineage 1 (336, 16.5%). Lineage 2 was the least frequent (92, 4.5%). The remaining 64 clinical isolates (3.1%) could not be assigned to any of the MTBC lineages possibly because there was insufficient amount of DNA in the samples (below the detection limit). Of the seven geographical zones, four (Coastal, Northern, Lake and Southern Highlands) were highly represented with more than 100 clinical strains each (Table 2). The distribution of the MTBC lineages varied within the geographical zones (Fig 1 and S3 Fig). Our findings reveal that Lineage 1 strains were more frequent in the Lake zone compared to the overall average frequency (20.9% vs. 16.8%), whereas the frequency of Lineage 3 in this zone was lower (27.6% vs. 34.3%) compared to other geographical zones. By contrast, Lineage 4 was the most predominant in all geographical zones and showed relatively similar frequencies across the zones.
Fig 1. MTBC lineages in Tanzania.
A. MTBC lineage classification of 2,039 nationwide clinical strains. B. MTBC lineage frequencies and geographical distribution in Tanzania.
Table 2. MTBC lineage distribution across geographical regions in Tanzania.
| Geographical Zone | Lineage | Total | |||
|---|---|---|---|---|---|
| L1 (%) | L2 (%) | L3 (%) | L4 (%) | ||
| Central | 8 (38.1 ) | 2 (9.5 ) | 4 (19 ) | 7 (33.3 ) | 21 |
| Coastal | 168 (16.8 ) | 50 (5 ) | 350 (35 ) | 432 (43.2 ) | 1,000 |
| Lake | 72 (20.9 ) | 12 (3.5 ) | 95 (27.6 ) | 165 (48 ) | 344 |
| Northern | 52 (13.3 ) | 22 (5.6 ) | 145 (37 ) | 173 (44.1 ) | 392 |
| S. Highlands | 27 (16.9 ) | 4 (2.5 ) | 60 (37.5 ) | 69 (43.1 ) | 160 |
| Western | 0 (0 ) | 1 (10 ) | 4 (40 ) | 5 (50 ) | 10 |
| Zanzibar | 2 (18.2 ) | 0 (0 ) | 5 (45.5 ) | 4 (36.4 ) | 11 |
| Total | 329 (17 ) | 91 (4.7 ) | 663 (34.2 ) | 855 (44.1 ) | 1,938 |
L1, Lineage 1; L2, Lineage 2; L3, Lineage 3; L4, Lineage 4
Sublineage classification
After we detected four main MTBC lineages, we next explored the respective subfamilies on a subset of Lineage 1, 3 and 4 strains using spoligotyping. Lineage 2 strains were excluded from this analysis since the global strains almost exclusively belong to one spoligotype family, Beijing with almost identical fingerprint pattern. We identified 24 spoligotypes (SITs; Spoligotype International Type) among the 107 clinical strains analyzed (S2 Table). Twenty six (24.3%) of the spoligo patterns had not been previously reported in the SITVITWEB database and therefore we assigned them as orphan spoligotypes. Several spoligotypes were identified within each of the three lineages. EAI5 was the common spoligotype among the Lineage 1 and CAS1_Kili spoligotype among the Lineage 3 strains. Within Lineage 4 strains, LAM, T, and H families were detected and the LAM subfamily, particularly LAM_ZWE was the most frequent.
Association between lineage and patients’ characteristics
Having described the circulating main lineages of the MTBC we then assessed the relationship between these lineages and patients’ characteristics in the study population (Table 3). We detected a higher proportion of female sex among TB patients infected with Lineage 2 (52.1%) compared to those infected with the other three lineages (range from 31% to 34.5%, p = 0.009). Moreover, we observed that retreatment cases were frequently infected with Lineage 2 strains (26.8%), which was twofold higher compared to Lineage 1 and 4 strains (p < 0.001). We found no evidence for association between lineages and patients’ characteristics such as age and HIV status (Table 3).
Table 3. Frequency distribution of MTBC main lineages across patients’ characteristic groups.
| Patient characteristics | Lineage | |||
|---|---|---|---|---|
| L1, n (%) | L2, n (%) | L3, n (%) | L4, n (%) | |
| Age group | ||||
| Child age (< 15) | 2 (0.7) | 0 (0) | 7 (1.3) | 8 (1.2) |
| Young age (15–24) | 44 (16.2) | 10 (13.9) | 79 (14.9) | 112 (16.9) |
| Early adult (25–44) | 156 (57.4) | 44 (61.1) | 336 (63.5) | 378 (57.1) |
| Late adult (45–64) | 57 (21.0) | 15 (20.8) | 85 (16.1) | 140 (21.1) |
| Old age (> 65) | 13 (4.8) | 3 (4.2) | 22 (4.2) | 24 (3.6) |
| Sex | ||||
| Female | 85 (31.3) | 37 (51.4) | 184 (34.8) | 223 (33.7) |
| Male | 187 (68.8) | 35 (48.6) | 345 (65.2) | 439 (66.3) |
| HIV status | ||||
| Negative | 184 (67.6) | 45 (62.5) | 349 (66.0) | 459 (69.3) |
| Positive | 88 (32.4) | 27 (37.5) | 180 (34.0) | 203 (30.7) |
| Patient category | ||||
| New case | 235 (86.4) | 53 (76.6) | 405 (76.6) | 560 (84.6) |
| Retreatment | 37 (13.6) | 19 (26.4) | 124 (23.4) | 102 (15.4) |
| Total | 272 (17.7) | 72 (4.7) | 529 (34.4) | 662 (43.2) |
L1, Lineage 1; L2, Lineage 2; L3, Lineage 3; L4, Lineage 4
Lineage 2 has previously been associated with retreatment cases, drug resistance and lately also with female sex [15,25]. We therefore investigated if similar associations exist in our study population using a subset of TB cases with complete clinical and demographic information (n = 1,535). To assess these associations we performed logistic regression analyses comparing Lineage 2 to all other lineages pooled together (Table 4). Our analyses revealed Lineage 2 to be independently associated with female sex (adjusted odds ratio [aOR] 2.14; 95% confidence interval [95% CI] 1.31 – 3.50, p = 0.002) and retreatment cases (aOR 1.67; 95% CI 0.95 – 2.84, p = 0.065). We did not detect any association between the lineages and patients’ age and the HIV status.
Table 4. Associations of patients’ clinical and demographic characteristics with MTBC Lineage 2 (n = 72) compared to all other lineages (n = 1,463).
| Patient characteristics | Lineage 2 | Unadjusted | Adjusted | ||
|---|---|---|---|---|---|
| n (%) | OR (95% CI) | p value | OR (95% CI) | p value | |
| Age, median (IQR) | 35 (28–44) | 0.99 (0.97 – 1.01) | 0.329 | ||
| Female sex | 37 (51.4) | 2.09 (1.30–3.36) | 0.002 | 2.14 (1.31 – 3.50) | 0.002 |
| Retreatment case | 19 (26.4) | 1.64 (0.93–2.76) | 0.075 | 1.67 (0.95 – 2.84) | 0.065 |
| HIV positive | 27 (37.5) | 0.79 (0.49–1.31) | 0.349 | 0.90 (0.55–1.51) | 0.91 |
| Observations | 1,535 | 1,535 |
IQR, Interquartile range; OR, Odds ratio; 95% CI, 95% confidence interval.
Mutations within rpoB gene in retreatment cases
To investigate whether drug resistance was linked to a particular lineage, we included in total 144 out of 321 retreatment cases for drug resistance genotyping of the rpoB gene that confers resistance to rifampicin. Out of these, 112 (77.8%) had no mutations compared to the H37Rv reference gene and 15 (10.4%) contained at least one mutation, either synonymous (3/15) or non-synonymous (12/15) (S4 Fig and S3 Table). We could not determine mutation status in the rpoB gene of 17 (11.8%) retreatment cases due to PCR and sequencing failure. Among the 12 strains detected with non-synonymous rpoB mutations, five belonged to Lineage 2, four to Lineage 4, and three to Lineage 3 (S4 Table). Table 5 summarizes the non-synonymous rpoB mutations detected.
Table 5. Non-synonymous mutations detected on the rpoB gene among retreatment cases.
Discussion
In this study, we classified the countrywide collection of 2,039 MTBC isolates representing 1.6% of all smear positive new and retreatment TB cases notified during 2012 and 2013 in Tanzania. Our findings show that the MTBC strains among the study population are diverse, comprising four main phylogenetic lineages (Lineage 1–4) which occur throughout the country. Specifically, we found that Lineage 4 was the most frequent, followed by Lineage 3 and 1. Despite Lineage 2’s recent global dissemination [13], it was the least frequent in our study population. Finally, our analysis on the relationship between MTBC lineages and patients’ characteristics revealed associations of Lineage 2 with female sex and retreatment TB cases included in the study population.
Among the 7 human–adapted MTBC lineages, Lineage 4 is the most broadly distributed and occurs at high frequencies in Europe, the Americas and Africa [24,34]. In our study, we observe that TB epidemics in Tanzania are also predominated by Lineage 4, which is regarded as the most successful of MTBC lineages [34]. In general, the wide geographical range of Lineage 4 is postulated to be driven by a combination of its enhanced virulence, high rates of human migration linked to its spread and ultimately its ability to infect different human population backgrounds [34,35]. In contrast, Lineage 1 and 3 are known to be mainly confined to the rim of the Indian Ocean [7], which is consistent with our observation that nearly 50% of the MTBC strains included in the study belong to these two lineages. This high frequency of Lineage 1 and 3 likely reflects the long-term migrations between Eastern Africa and the Indian subcontinent [36]. In addition, the distribution and frequency of Lineage 1 and 3 in the mainland subset did not vary from that of the coastal region, suggesting spread via internal migrations. Lineage 1 was proposed to have evolved in East Africa prior disseminating out of the continent [10]. Based on this, one might expect higher frequencies of Lineage 1 in the region. Instead, the so called “modern” (TbD1−) lineages (4 and 3 in this case) could be dominating in Tanzania despite presumably being introduced into the African continent only after the first European contact [34,37]. This perhaps illustrates the ability of “modern” lineages to thrive in co-existence with the pre-existing “ancient” (TbD1+) lineages such as Lineage 1 in our case, perhaps because of the comparably higher virulence [14,38]. The neighboring countries of Tanzania on the other hand show comparable MTBC lineage composition to our study [39,40], suggesting common demographic histories and ongoing exchanges that resulted into distinct MTBC populations. Our findings would suggest the frequency of Lineage 2–Beijing in Tanzania, like in most parts of the continent except for South Africa [39,40] to be relatively low, despite the long-standing African-Asian contacts [40]. Evidence from recent studies show that Lineage 2–Beijing was only recently introduced into Africa [13,41].
The burden of TB disease is generally higher in males [1,42], rendering male sex as a potential risk factor for TB. Furthermore, the male bias among TB patients is also observed in settings with no obvious sex-based differences in health-seeking behavior [43]. Whilst we show similar trends in this study, our findings reveal that the proportion of females was higher among TB patients infected with Lineage 2. This finding is consistent with several other previous studies conducted in different settings [15,25,44]. Social and physiological factors predisposing males to higher risk of TB have been indicated [45]. On the one hand, these include risk behaviors such as substance abuse (alcoholism, tobacco smoking) and gender specific roles such as risk occupations (e.g., mining) that are male dominated and known to increase the risk for TB. On the other hand, genetic makeup and sex hormones might contribute to the differences in TB susceptibility among females and males, as epidemiological and experimental studies have suggested female sex hormones to be protective [45]. These observations would propose that the sex imbalance in TB emerges after the onset of puberty. Of note, we observe less sex imbalance in “child” age group (<15 years) which also corroborates the national notification rates [29]. However, this observation can be confounded by BCG vaccination which might be most effective in this age group. Despite the high prevalence of HIV among young females in sub-Saharan Africa [28] and HIV being the strongest risk factor for TB, TB burden remains higher in males. While social and physiological aspects play an important role, findings from this study and others previously conducted in Nepal and Vietnam [15,25] suggest that bacterial factors could disrupt the trends towards male bias in TB, a finding which warrants further investigation. Our hypothesis is that because of higher virulence, Lineage 2 strains are able to overcome the resistance poised by female sex which could explain the less pronounced sex imbalanced.
In addition to its association with female sex, we found that retreatment TB cases were more likely infected with Lineage 2. A retreatment case in our study population represented recurrent TB case either due to relapse or reinfection. We hypothesized that this observation was possibly linked to drug resistance, given the previous reported association between Lineage 2 and drug resistance [46]. However, we detected only 8.3% (12/144) of strains among the retreatment subset tested to contain mutations conferring resistance to rifampicin, five of which belonged to Lineage 2. These findings would suggest that retreatment cases included in this study are mainly driven by reinfection as opposed to treatment failure or relapse.
Finally, based on the age distribution of TB cases in our study, recent or ongoing transmission in high burden countries is implicated as the main contributor to the TB burden rather than disease reactivation (following longer latency periods) [47]. Additionally, an association with young age has been used as an epidemiological proxy for highly transmissible strains and faster rates of disease progression [48,49]. In this study, we did not detect any differences in median age of TB patients infected with different lineages (S5 Fig), an observation that could speak for high ongoing transmission rates in general, irrespective of lineage.
Our study is limited by focusing on a convenient collection of MTBC clinical isolates that could be retrieved from the culture archives, representing 1.6% of all TB cases notified in 2012 and 2013. Given that our findings are based on a limited number of TB cases, the results particularly those related to associations between lineages and patients’ characteristics should be taken with caution as the strength or lack of such associations could likely be affected by the sampling. In addition, most of the geographical zones were underrepresented which could in turn underestimate the respective regional lineage composition and the overall countrywide distribution. Unfortunately, data on drug susceptibility based on other methods such as Xpert MTB/RIF, phenotypic DST and Line Probe Assay (LPA) were unavailable, which could have complemented the drug resistance genotyping performed on a limited subset of the retreatment cases. Systematic sampling would allow for better resolution on the distribution patterns, the frequencies and on epidemiological features of MTBC lineages, which might partially determine the regional specific epidemics.
In conclusion, this study addresses for the first time the countrywide MTBC population structure based on robust SNP-typing. We show that MTBC population in Tanzania is diverse with four of the seven known lineages detected. This study sets the stage for further in depth investigations on epidemiological impact of MTBC lineages in Tanzania.
Supporting information
(PDF)
Proportion of observed and missing data for the variables included in the study.
(TIF)
Distribution of MTBC lineages across different regions of Tanzania. Size of the circle is proportional to the number of isolates analyzed from the regions.
(TIF)
A subset of MTBC strains from retreatment cases included for rpoB drug resistance genotyping.
(PDF)
The age distributions of TB patients grouped by infecting MTBC lineage.
(TIFF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We would like to thank the National Tuberculosis Leprosy Programme (NTLP) through the Central Reference Laboratory (CTRL) for permission to use the MTBC isolate collection for this study.
Data Availability
The data used are held in a public repository under the given link: https://github.com/SwissTPH/Mtb_diversity_TZ.git.
Funding Statement
This work was supported by the Swiss National Science Foundation (grants 310030_166687, IZRJZ3_164171, IZLSZ3_170834 and CRSII5_177163 to SG; http://www.snf.ch), the European Research Council (309540-EVODRTB to SG; https://erc.europa.eu) and SystemsX.ch (http://www.systemsx.ch to SG). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO. Global tuberculosis report Geneva: World Health Organization; 2017. [Google Scholar]
- 2.NTLP. Annual report for 2016. Dar es Salaam. 2016;1–48.
- 3.Nagu TJ, Aboud S, Mwiru R, Matee M, Fawzi W, Mugusi F. Multi Drug and Other Forms of Drug Resistant Tuberculosis Are Uncommon among Treatment Naïve Tuberculosis Patients in Tanzania. Surolia A, editor. PLoS One. 2015;10(4):e0118601 10.1371/journal.pone.0118601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MoHSW. First tuberculosis prevalence survey in the United Republic of Tanzania. 2013; [Google Scholar]
- 5.Rieder HL. Epidemiologic Basis of Tuberculosis Control First edition 1999. 1999. [Google Scholar]
- 6.Comas I, Gagneux S. The Past and Future of Tuberculosis Research. Manchester M, editor. PLoS Pathog. 2009;5(10):e1000600 10.1371/journal.ppat.1000600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gagneux S. Ecology and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol. 2018;16(4):202–13. 10.1038/nrmicro.2018.8 [DOI] [PubMed] [Google Scholar]
- 8.Gagneux S, DeRiemer K, Van T, Kato-Maeda M, de Jong BC, Narayanan S, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci. 2006;103(8):2869–73. 10.1073/pnas.0511240103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenner L, Egger M, Bodmer T, Furrer H, Ballif M, Battegay M, et al. HIV Infection Disrupts the Sympatric Host–Pathogen Relationship in Human Tuberculosis. Gibson G, editor. PLoS Genet. 2013;9(3):e1003318 10.1371/journal.pgen.1003318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comas I, Coscolla M, Luo T, Borrell S, Holt KE, Kato-Maeda M, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 2013;45(10):1176–82. 10.1038/ng.2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coscolla M. Biological and Epidemiological Consequences of MTBC Diversity In: Strain variation in the Mycobacterium tuberculosis complex:Its role in biology, epidemiology and control. Springer, Cham; 2017. p. 95–116. [DOI] [PubMed] [Google Scholar]
- 12.Hanekom M, van der Spuy GD, Streicher E, Ndabambi SL, McEvoy CRE, Kidd M, et al. A Recently Evolved Sublineage of the Mycobacterium tuberculosis Beijing Strain Family Is Associated with an Increased Ability to Spread and Cause Disease. J Clin Microbiol. 2007;45(5):1483–90. 10.1128/JCM.02191-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowley D, Govender D, February B, Wolfe M, Steyn L, Evans J, et al. Recent and Rapid Emergence of W‐Beijing Strains of Mycobacterium tuberculosis in Cape Town, South Africa. Clin Infect Dis. 2008;47(10):1252–9. 10.1086/592575 [DOI] [PubMed] [Google Scholar]
- 14.Stavrum R, PrayGod G, Range N, Faurholt-Jepsen D, Jeremiah K, Faurholt-Jepsen M, et al. Increased level of acute phase reactants in patients infected with modern Mycobacterium tuberculosis genotypes in Mwanza, Tanzania. BMC Infect Dis. 2014;14(1):309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holt KE, McAdam P, Thai PVK, Thuong NTT, Ha DTM, Lan NN, et al. Frequent transmission of the Mycobacterium tuberculosis Beijing lineage and positive selection for the EsxW Beijing variant in Vietnam. Nat Genet. 2018;50(6):849–56. 10.1038/s41588-018-0117-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eldholm V, Matee M, Mfinanga SGM, Heun M, Dahle UR. A first insight into the genetic diversity of Mycobacterium tuberculosis in Dar es Salaam, Tanzania, assessed by spoligotyping. BMC Microbiol. 2006;6(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kibiki GS, Mulder B, Dolmans WM, de Beer JL, Boeree M, Sam N, et al. M. tuberculosis genotypic diversity and drug susceptibility pattern in HIV- infected and non-HIV-infected patients in northern Tanzania. BMC Microbiol. 2007;7(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mfinanga SGM, Warren RM, Kazwala R, Kahwa A, Kazimoto T, Kimaro G, et al. Genetic profile of Mycobacterium tuberculosis and treatment outcomes in human pulmonary tuberculosis in Tanzania. Tanzan J Health Res. 2014;16(2):58–69. [DOI] [PubMed] [Google Scholar]
- 19.Mbugi E V., Katale BZ, Siame KK, Keyyu JD, Kendall SL, Dockrell HM, et al. Genetic diversity of Mycobacterium tuberculosis isolated from tuberculosis patients in the Serengeti ecosystem in Tanzania. Tuberculosis. 2015;95(2):170–8. 10.1016/j.tube.2014.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Comas I, Homolka S, Niemann S, Gagneux S. Genotyping of Genetically Monomorphic Bacteria: DNA Sequencing in Mycobacterium tuberculosis Highlights the Limitations of Current Methodologies. Litvintseva AP, editor. PLoS One. 2009;4(11):e7815 10.1371/journal.pone.0007815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fenner L, Malla B, Ninet B, Dubuis O, Stucki D, Borrell S, et al. “Pseudo-Beijing”: Evidence for Convergent Evolution in the Direct Repeat Region of Mycobacterium tuberculosis. Sechi LA, editor. PLoS One. 2011;6(9):e24737 10.1371/journal.pone.0024737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stucki D, Malla B, Hostettler S, Huna T, Feldmann J, Yeboah-Manu D, et al. Two New Rapid SNP-Typing Methods for Classifying Mycobacterium tuberculosis Complex into the Main Phylogenetic Lineages. Mokrousov I, editor. PLoS One. 2012;7(7):e41253 10.1371/journal.pone.0041253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35(4):907–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demay C, Liens B, Burguière T, Hill V, Couvin D, Millet J, et al. SITVITWEB–A publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect Genet Evol. 2012;12(4):755–66. 10.1016/j.meegid.2012.02.004 [DOI] [PubMed] [Google Scholar]
- 25.Malla B, Stucki D, Borrell S, Feldmann J, Maharjan B, Shrestha B, et al. First Insights into the Phylogenetic Diversity of Mycobacterium tuberculosis in Nepal. Sola C, editor. PLoS One. 2012;7(12):e52297 10.1371/journal.pone.0052297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staden R. The Staden sequence analysis package. Mol Biotechnol. 1996;5(3):233–41. [DOI] [PubMed] [Google Scholar]
- 27.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna; 2018. [Google Scholar]
- 28.Hegdahl HK, Fylkesnes KM, Sandøy IF. Sex Differences in HIV Prevalence Persist over Time: Evidence from 18 Countries in Sub-Saharan Africa. Faragher EB, editor. PLoS One. 2016;11(2):e0148502 10.1371/journal.pone.0148502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.National Tuberculosis and Leprosy Programme (NTLP). Annual report for 2013. Dar es Salaam. 2013.
- 30.Phelan J, Coll F, McNerney R, Ascher DB, Pires DE V, Furnham N, et al. Mycobacterium tuberculosis whole genome sequencing and protein structure modelling provides insights into anti-tuberculosis drug resistance. BMC Med. 2016;14(1):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker TM, Kohl TA, Omar S V, Hedge J, Del C, Elias O, et al. Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect Dis. 2015;15:1193–202. 10.1016/S1473-3099(15)00062-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miotto P, Cabibbe AM, Borroni E, Degano M, Cirillo DM. Role of Disputed Mutations in the rpoB Gene in Interpretation of Automated Liquid MGIT Culture Results for Rifampin Susceptibility Testing of Mycobacterium tuberculosis. J Clin Microbiol. 2018;56(5):e01599–17. 10.1128/JCM.01599-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heyckendorf J, Andres S, Köser CU, Olaru ID, Schön T, Sturegård E, et al. What Is Resistance? Impact of Phenotypic versus Molecular Drug Resistance Testing on Therapy for Multi- and Extensively Drug-Resistant Tuberculosis. Antimicrob Agents Chemother. 2018;62(2):1550–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stucki D, Brites D, Jeljeli L, Coscolla M, Liu Q, Trauner A, et al. Mycobacterium tuberculosis lineage 4 comprises globally distributed and geographically restricted sublineages. Nat Genet. 2016;48(12):1535–43. 10.1038/ng.3704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coscolla M, Gagneux S. Consequences of genomic diversity in Mycobacterium tuberculosis. Semin Immunol. 2014;26(6):431–44. 10.1016/j.smim.2014.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Neill MB, Shockey AC, Zarley A, Aylward W, Eldholm V, Kitchen A, et al. Lineage specific histories of Mycobacterium tuberculosis dispersal in Africa and Eurasia. bioRxiv. 2018;210161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Comas I, Hailu E, Kiros T, Bekele S, Mekonnen W, Gumi B, et al. Population Genomics of Mycobacterium tuberculosis in Ethiopia Contradicts the Virgin Soil Hypothesis for Human Tuberculosis in Sub-Saharan Africa. Curr Biol. 2015;25(24):3260–6. 10.1016/j.cub.2015.10.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Portevin D, Gagneux S, Comas I, Young D. Human Macrophage Responses to Clinical Isolates from the Mycobacterium tuberculosis Complex Discriminate between Ancient and Modern Lineages. Bessen DE, editor. PLoS Pathog. 2011;7(3):e1001307 10.1371/journal.ppat.1001307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mbugi E V., Katale BZ, Streicher EM, Keyyu JD, Kendall SL, Dockrell HM, et al. Mapping of Mycobacterium tuberculosis Complex Genetic Diversity Profiles in Tanzania and Other African Countries. Sreevatsan S, editor. PLoS One. 2016;11(5):e0154571 10.1371/journal.pone.0154571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chihota VN, Niehaus A, Streicher EM, Wang X, Sampson SL, Mason P, et al. Geospatial distribution of Mycobacterium tuberculosis genotypes in Africa. Arez AP, editor. PLoS One. 2018;13(8):e0200632 10.1371/journal.pone.0200632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rutaihwa LK, Menardo F, Stucki D, Gygli SM, Ley SD, Malla B, et al. Multiple introductions of the Mycobacterium tuberculosis Lineage 2 Beijing into Africa over centuries. bioRxiv. 2018;413039. [Google Scholar]
- 42.Guerra-Silveira F, Abad-Franch F. Sex Bias in Infectious Disease Epidemiology: Patterns and Processes. Nishiura H, editor. PLoS One. 2013;8(4):e62390 10.1371/journal.pone.0062390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhines AS. The role of sex differences in the prevalence and transmission of tuberculosis. Tuberculosis. 2013;93(1):104–7. 10.1016/j.tube.2012.10.012 [DOI] [PubMed] [Google Scholar]
- 44.Buu TN, Huyen MN, Lan NTN, Quy HT, Hen N V, Zignol M, et al. The Beijing genotype is associated with young age and multidrug-resistant tuberculosis in rural Vietnam. Int J Tuberc Lung Dis. 2009;13(7):900–6. [PubMed] [Google Scholar]
- 45.Nhamoyebonde S, Leslie A. Biological Differences Between the Sexes and Susceptibility to Tuberculosis. J Infect Dis. 2014;209(suppl 3):S100–6. [DOI] [PubMed] [Google Scholar]
- 46.Borrell S, Gagneux S. Infectiousness, reproductive fitness and evolution of drug-resistant Mycobacterium tuberculosis. Int J Tuberc Lung Dis. 2009;13(12):1456–66. [PubMed] [Google Scholar]
- 47.Yates TA, Khan PY, Knight GM, Taylor JG, McHugh TD, Lipman M, et al. The transmission of Mycobacterium tuberculosis in high burden settings. Lancet Infect Dis. 2016;16(2):227–38. 10.1016/S1473-3099(15)00499-5 [DOI] [PubMed] [Google Scholar]
- 48.de Jong BC, Hill PC, Aiken A, Awine T, Antonio M, Adetifa IM, et al. Progression to Active Tuberculosis, but Not Transmission, Varies by Mycobacterium tuberculosis Lineage in The Gambia. J Infect Dis. 2008;198(7):1037–43. 10.1086/591504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borgdorff MW, van Soolingen D. The re-emergence of tuberculosis: what have we learnt from molecular epidemiology? Clin Microbiol Infect. 2013;19(10):889–901. 10.1111/1469-0691.12253 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Proportion of observed and missing data for the variables included in the study.
(TIF)
Distribution of MTBC lineages across different regions of Tanzania. Size of the circle is proportional to the number of isolates analyzed from the regions.
(TIF)
A subset of MTBC strains from retreatment cases included for rpoB drug resistance genotyping.
(PDF)
The age distributions of TB patients grouped by infecting MTBC lineage.
(TIFF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
The data used are held in a public repository under the given link: https://github.com/SwissTPH/Mtb_diversity_TZ.git.