Abstract
Background
Molecular size determination of circulating free fetal DNA in maternal plasma is an important detection method for noninvasive prenatal testing (NIPT). The fetal DNA molecule is the primary factor determining the overall performance of NIPT and its clinical interpretation. The proportion of cell-free fetal DNA molecules is expressed as the fetal DNA fraction in the plasma of pregnant women.
Methods
We proposed an effective method to deduce fetal chromosomal aneuploidy based on the proportion of a certain range of DNA fragment lengths from maternal plasma. We gradually narrowed the range of the upper and lower boundary via a traversing algorithm.
Results
We explored the optimal range of the upper and lower boundary by using size-based DNA fragment length. Using this range, the accuracy of the sensitivity and specificity could be improved by up to 100% for detecting the three most common autosomal aneuploidies, namely trisomy 13, trisomy 18, trisomy 21 in the sample set.
Conclusions
Numerical experiments demonstrate that our method is effective and efficient. The program is available upon request.
Introduction
Noninvasive prenatal testing (NIPT) is now widely used in clinical practices worldwide and does not require traumatic sampling, with high accuracy, high sensitivity and specificity. For most pregnant women, the invasive sampling of fetal genetic material through amniocentesis or chorionic villus sampling is gradually replaced by noninvasive prenatal testing [1,2]. NIPT can screen for fetal chromosome aneuploidy and certain copy number variations [2]. In 1997, Dennis Lo found that cell-free fetal DNA (cffDNA) was present in the maternal plasma and it increased stably with the gestational weeks and disappeared rapidly with the delivery of pregnant women, which can be used as an ideal material for NIPT [3]. The cffDNA was discovered in plasma of pregnant women in 1997 which developed a new technique to analyze and measure accurately fetal DNA in maternal plasma [3].
Detection of the three most common autosomal aneuploidies is an important indication for noninvasive prenatal diagnosis [4]. The three aneuploidies are trisomy 21 syndrome (Down syndrome), trisomy 13 syndrome (Patau syndrome) and trisomy 18 syndrome (Edwards syndrome) [5]. In recent years, high accuracy has been achieved in prenatal screening for Down syndrome. However, identification of trisomy 13 and trisomy 18 remains a significant secondary goal [6]. Down syndrome occurs in 1 in 800 live births, whereas about 1 in 10,000 newborns is estimated to carry trisomy 13, and the incidence of trisomy 18 is about 1 out of every 6,000 live births [6,7]. Due to Down syndrome and Patau syndrome, as well as Edwards syndrome with high rates of spontaneous loss, live-born infants are less likely to survive beyond the early stage [8]. Only 5–10% of live-born infants with the three aneuploidies can survive more than one year [9,10].
A new generation of DNA sequencing technology is used to sequence the cell-free DNA fragments in the maternal plasma. The sequencing results are subjected to bioinformatics diagnostic algorithms for obtaining the fetal genetic information and detecting the presence of the three most common autosomal aneuploidies [11–15]. With the availability of massively parallel sequencing technologies, some applied data can be obtained, such as the genomic identities and quantities of millions of DNA molecules in biological samples originating from the plasma of pregnant women [4]. To date, a series of new avenues of noninvasive diagnostic applications have developed; for example, chromosomal aneuploidy detection, fetal sex determination, and detection of monogenic diseases [16–18]. The cffDNA contained in the maternal plasma accounts for about 5%-20% of the total cell-free DNA fragments [19–21]. The fractional fetal DNA concentration is a paramount factor for determining the overall performance of NIPT based on the analysis of DNA in maternal plasma.
In previous studies, the detection of fetal chromosomal aneuploidy has been discussed, but its processing is cumbersome and complicated. Most studies take advantage of the fetal DNA fraction to analyze sequencing data of maternal plasma DNA though various methods and instruments. In this study, we proposed a simple and effective method, which employs a range of cell-free DNA segment lengths to determine fetal chromosomal aneuploidy. A chromosomal abnormality would lead to an increased or reduced representation of the chromosome in the contents of cell-free DNA fragments in the maternal plasma [22,23]. If a woman is pregnant with a trisomic fetus, the amounts of its corresponding chromosome fragments should be increased [4,22]. Because of the extra chromosome, the proportion of corresponding cell-free DNA fragments would also be elevated in the maternal plasma when compared to a pregnancy with a euploid fetus. Based on this, the study discussed the suitable range of cell-free DNA fragment lengths derived from maternal plasma for the detection of fetal chromosomal aneuploidy.
Materials
One sample set was applied which had been used in previous studies [2,4–5]. There were 144 maternal plasma sample cases, which were divided into four parts in the sample set. These included 21 cases each with a trisomy 13 fetus, 27 cases each with a trisomy 18 fetus, 36 cases each with a trisomy 21 fetus, and 60 cases each with a euploid fetus. None of the pregnancy samples collected were tested for any invasive sampling of fetal genetic material [5]. The sample set we used came from “Size-based molecular diagnostics using plasma DNA for noninvasive prenatal testing”, contributed by Y. M. Dennis Lo, April 2, 2014, PNAS. USA. The data information is shown in S1 Dataset.
Methods and results
Calculating the proportion of chromosomes
First, we calculated the percentage of each chromosome contained in the sample set separately. The sample set contained the size distribution of cell-free DNA fragments originating from maternal plasma. The following formula was used to calculate the proportion of every corresponding chromosome, and the result was shown in S1 Appendix.
Where PchrN denotes the proportion of target chromosomes fragments in all chromosomes fragments originating from maternal plasma samples; SumchrN denotes the sum of target chromosomes fragments originating from maternal plasma samples; and Sumall denotes the sum of all chromosomes fragments originating from maternal plasma samples.
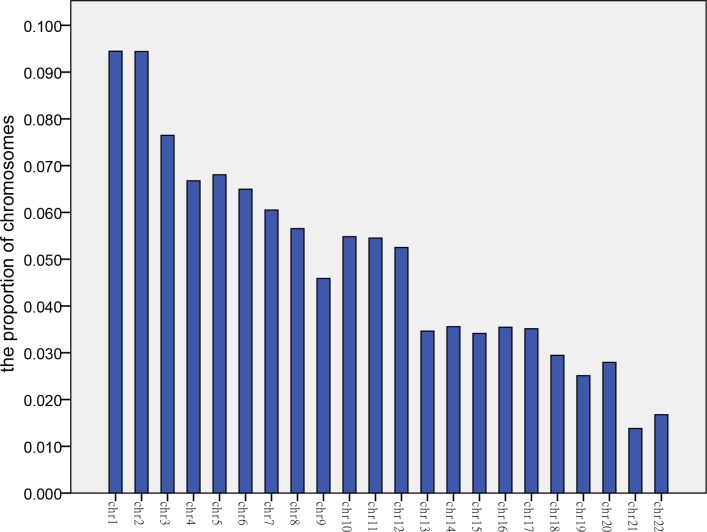
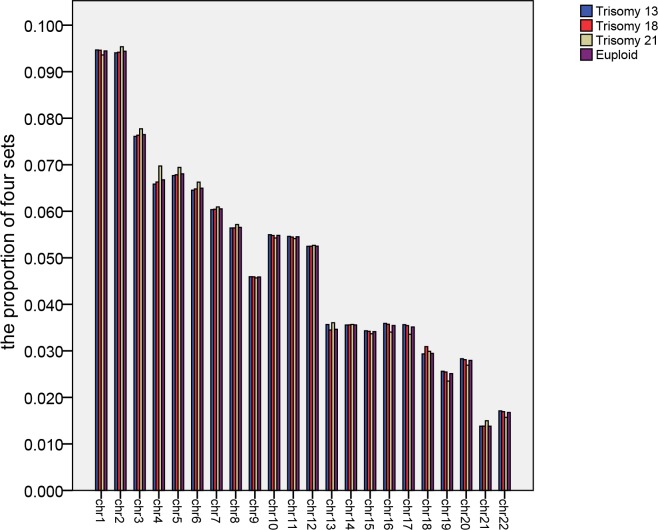
Next, we calculated the average value of each chromosome for 60 euploid samples originating from maternal plasma DNA (Fig 1). Similarly, we calculated the average value of each chromosome for 21 cases with a trisomy 13 fetus, 27 cases with a trisomy 18 fetus, and 36 cases with a trisomy 21 fetus. We compared the proportions of these four records represented them graphically (Fig 2).
Fig 1. The proportion of autosomes.
This bar chart shows the average proportion of autosomes in maternal plasma for 60 euploid samples.
Fig 2. Comparison of four sets of data.
Blue, red, yellow, and purple strips indicate trisomy 13, trisomy 18, Down syndrome, and euploid pregnancies, respectively.
Fig 1 indicates that the proportion of DNA fragments from each corresponding autosome ranged from about 2% to 9%. We found out that the proportion of 9 chromosomes from chromosome 7 to chromosome 15 was almost the same, except for chromosome 13 in Fig 2. The three ratios corresponding to chromosomes 13, 18 and 21 were of particular interest. Among the four groups of values corresponding to chromosome 13, the average of samples with a 13 trisomy and a 21 trisomy was higher. Among the four groups of values corresponding to chromosome 18, the average of 27 cases with a trisomy 18 fetus was highest. The average of 36 cases with a trisomy 21 fetus had a larger value than others among the four groups of values corresponding to chromosome 21.
We computed the average of 144 samples for each chromosome (all autosomes except chromosomes 13, 18, 21). We were not certain of the sex of all samples from maternal plasma DNA. The number of Y chromosome fragments in male fetuses was generally larger than that in female fetuses. If we determined the average value directly, it would lead to a large error in the calculation result. The sex chromosome calculation data should thus be discarded. The method we used could also detect the sex of the fetus in this study. We noted the average value and standard deviation of each chromosome in S2 Appendix.
Exploring the most suitable range
The primary research question of the study was to explore the most suitable range of cell-free DNA fragment lengths for detecting the three fetal chromosomal aneuploidies with a high accuracy.
We gradually narrowed the range of the interval via a traversing algorithm, and finally settled on a range of DNA fragment lengths of 80 to 155 bp (details of sample collection and processing available in supplementary files). We first calculated the value of the lower boundary from 30 to 130 in 10 bp steps and the value of the upper boundary from 150 to 170 in 5 bp steps. We obtained the most suitable range in the 114 sample cases through the calculated results. The lower limit range started from 75 to 85 bp and the upper limit range started from 150 to 155 bp; the resulting accuracy was relatively high throughout the range.
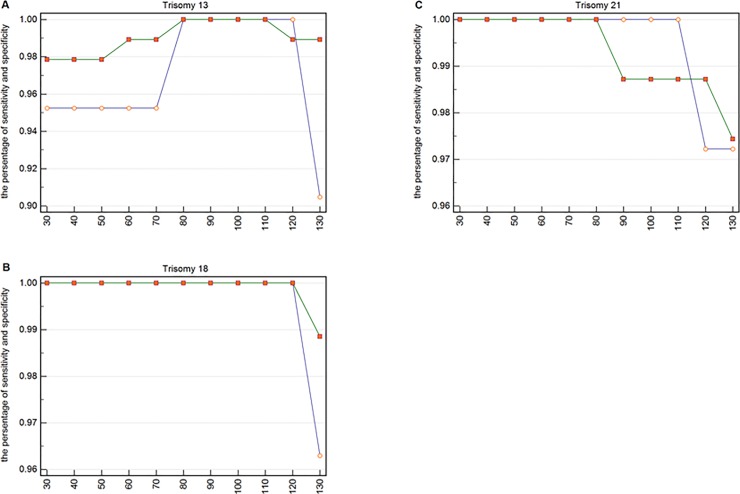
We could infer the lower boundary by analyzing the specificity and sensitivity of these three chromosomal diseases shown in Fig 3. The results of the identification of Edwards syndrome using cell-free DNA fragments ranging from 30 to 120 bp had little change.
Fig 3. The specificity and sensitivity of each lower boundary for trisomy 13, 18, and 21.
(A) The specificity and sensitivity for 21 pregnancies with trisomy 13 and for 93 pregnancies without trisomy 13. The small circles represent sensitivity and the small squares represent specificity. (B) The specificity and sensitivity for 27 pregnancies with trisomy 18 and for 87 pregnancies without trisomy 18. The small circles represent sensitivity and the small squares represent specificity. (C) The specificity and sensitivity for 36 pregnancies with trisomy 21 and for 78 pregnancies without trisomy 21. The small circles represent sensitivity and the small squares represent specificity.
In Fig 3, the abscissa represents the value of the lower bound of each interval, and the upper bound was 150 bp. The two points of ordinate represents the sensitivity and specificity, respectively. For instance, the two points corresponding to the abscissa 30 represent the value of the sensitivity and specificity, respectively, when the range is 30 to 150 bp. We calculated the value of the lower boundary from 30 to 130 in 10 bp steps and observed the best intervals to be 80 to 110 bp for Trisomy 13, 30 to 120 bp for Trisomy 18, and 30 to 80 bp for Trisomy 21. The most suitable range we obtained was 80 to 150 bp, through the “AND” algorithm, and this was obtained with 150 bp as the upper bound.
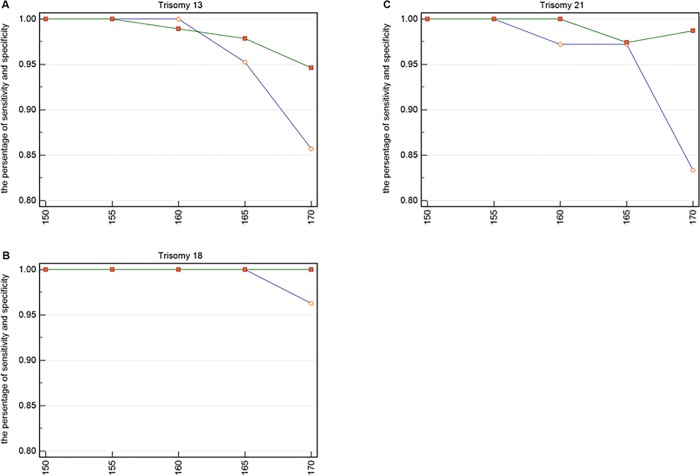
Similarly, we could obtain the upper boundary by analyzing the specificity and sensitivity of these three chromosomal diseases shown in Fig 4.
Fig 4. The specificity and sensitivity of each upper boundary for trisomy 13, 18, and 21.
(A) The specificity and sensitivity for 21 pregnancies with trisomy 13 and for 93 pregnancies without trisomy 13. The small circles represent sensitivity and the small squares represent specificity. (B) The specificity and sensitivity for 27 pregnancies with trisomy 18 and for 87 pregnancies without trisomy 18. (C) The specificity and sensitivity for 36 pregnancies with trisomy 21 and for 78 pregnancies without trisomy 21.
In Fig 4, the abscissa represents the value of the upper bound of each interval, and the lower bound is 80 bp. The two points of ordinate represent the sensitivity and specificity, respectively. For example, the two points corresponding to the abscissa 150 represent the value of the sensitivity and specificity, respectively, when the range is 80 to 150 bp. We calculated the value of the upper boundary from 150 to 170 in 5 bp increments and observed the best interval to be 150 to 155 bp for Trisomy 13, 150 to 165 bp for Trisomy 18, and 150 to 155 bp for Trisomy 21. The most suitable range we obtained was 80 to 155 bp through the “AND” algorithm, and this was obtained using 80 bp as the lower bound.
From this, we obtained the values of the upper and lower bounds when determining the three most common autosomal aneuploidies. The accuracy and sensitivity of the DNA fragments between 80 bp and 155 bp was relatively high. Through the above calculations, the suitable range of DNA fragments length was found to be 80 to 155 bp for the sample set we used.
Detection of fetal trisomies 18 and 21 based on the total range of fragment lengths
We randomly selected 30 sample cases with a euploid fetus originating from the sample set as training samples. We calculated a z-score for the target chromosome 18 and 21 for test samples using the following equation:
Where PchrN is the proportion of the corresponding target chromosome; the mean PchrN_training is the average value of 30 training samples; the SDchrN_training is the standard deviation of 30 training samples.
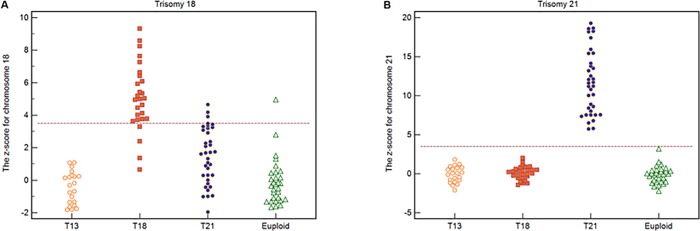
Using a z-score cutoff value of >3.5, all 36 cases each with a trisomy 21 fetus could be correctly distinguished and 78 non-trisomy 21 cases could be correctly identified (Fig 5). The sensitivity and specificity both were 100%. Twenty-three out of 27 cases each with a trisomy 18 fetus could be correctly distinguished, and 83 out of 87 non-trisomy 18 cases could be correctly identified (Fig 5). The sensitivity was 85.2% and the specificity was 95.4%.
Fig 5. The z-scores for chromosomes 18 and 21.
(A) The z-score of chromosome 18 for 114 test samples which included 21 trisomy 13, 27 trisomy 18, 36 trisomy 21, and 30 euploid cases. Dotted line denotes the z-score cutoff value of 3.5. (B) The z-score of chromosome 21 for 114 test samples which included 21 trisomy 13, 27 trisomy 18, 36 trisomy 21, and 30 euploid cases. Dotted line denotes the z-score cutoff value of 3.5.
Detection of fetal trisomies 13, 18 and 21 by the certain range of fragment lengths
In the previous method, the accuracy we obtained for the total range of fragment lengths was not very high. However, the specificity and sensitivity of the DNA fragments between 80 and 155 bp was relatively high. We would use the range, which was obtained through previous calculation to detect the three most common autosomal aneuploidies. If a fetus suffers from an autosomal aneuploidy, it would have one chromosome more than a euploid fetus. The number of fetal DNA fragments from the extra chromosome would thus increase.
We used the ratio to represent the relationship between the target chromosome (chromosomes 13, 18 and 21) and the other chromosomes (all autosomes except chromosomes 13, 18 and 21), denoted by RchrN, using the following equation:
Where P(80−155)chrN denotes the proportion of DNA fragments originating from the goal chromosome with sizes ranging from 80 bp to 155 bp and P(80−155)chrother denotes the proportion of DNA fragments originating from the other chromosomes with sizes range from 80 bp to 155 bp.
Then, we randomly selected 30 sample cases with a euploid fetus originating from the sample set as training samples. We used an improved z-score to distinguish the fetus sample cases, defining a new formula as follows:
Where RchrN_Test is the RchrN for the test sample, meanRchrN_Training is the mean RchrN of the training samples, and SDRchrN_Training is the SD of the RchrN of the training samples.
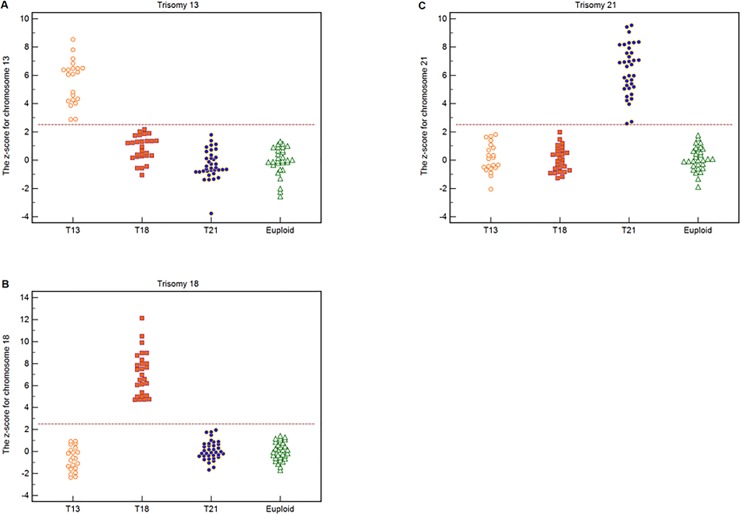
Using a z-score cutoff value of >2.5, all 21 cases each with a trisomy 13 fetus could be correctly distinguished and 93 non-trisomy 13 cases could be correctly identified (Fig 6A). The sensitivity and specificity both were 100%. All 27 cases each with a trisomy 18 fetus could be correctly distinguished and 87 non-trisomy 18 cases could be correctly identified (Fig 6B). There was an outlier replaced by the average value. The sensitivity and specificity were both 100%. All 36 cases each with a trisomy 21 fetus could be correctly distinguished and 78 non-trisomy 21 cases could be correctly identified (Fig 6C). The sensitivity and specificity both were 100%.
Fig 6. The z-scores for chromosomes 13, 18 and 21.
(A) The z-score of chromosome 13 for 114 test samples, which include 21 trisomy 13, 27 trisomy 18, 36 trisomy 21, and 30 euploid cases. Dotted line denotes the z-score cutoff value of 2.5. (B) The z-score of chromosome 18 for 114 test samples, which include 21 trisomy 13, 27 trisomy 18, 36 trisomy 21, and 30 euploid cases. (C) The z-score of chromosome 21 for 114 test samples, which include 21 trisomy 13, 27 trisomy 18, 36 trisomy 21, and 30 euploid cases.
Discussion
In the study, we calculated the proportions of all 24 chromosomes by observation and comparison. In general, the ratio of each chromosome from sequencing samples tends to be consistent. When a fetus had autosome aneuploidy, its corresponding chromosome ratio would increase. We know that if there are extra or missing chromosomes, the number of corresponding fetal DNA fragments will increase or decrease [24–26]. For instance, the proportion of chromosome 21 in a sample from a pregnant woman carrying a trisomy 21 fetus is higher than in a normal maternal plasma sample [27]. We can use this principle to predict multiple chromosome diseases. This method we used is simple and effective, and can be used as a first-tier screening test to determine multiple types of fetal autosomal aneuploidies.
We explored the values of the upper and lower bounds when determining the three most common autosomal aneuploidies and finally settled on a size range from 80 bp to 155 bp. We suspected that cell-free fetal DNA fragments are mainly concentrated in this size range through this processing. With the accumulation of data for all 24 human chromosomes, all genetic problems are expected to become detectable, allowing for accurate screening during pregnancy to ultimately reduce the incidence of fetal birth defects [28–30].
Noninvasive prenatal testing is now widely used in the medical field. The basic principle of NIPT is the need to extract cell-free DNA from the plasma of pregnant women to perform high-throughput sequencing [31,32]. Combined with economic and health data, NIPT can be used as a sequential screening program for traditional detection techniques. At present, the clinical application of NIPT can generally make a clear analysis of the three most common autosomal aneuploidies [31–33].
Our analysis showed that this method improves the accuracy of detecting the three most common autosomal aneuploidies to some extent. Our method requires a full range of genome sequencing, and massively parallel sequencing is currently expensive. The applicability of the method that is used as a first tier screening test requires a formal investigation of its diagnostic performance and cost-effectiveness. Furthermore, the amount of cell-free fetal DNA in maternal plasma is extremely small and the length of the pregnancy may affect the accuracy of diagnosis. The method is based on the length of the fragment and the corresponding number. Therefore, it is ensured that the cell-free fetal DNA in the plasma of pregnant women reaches a certain concentration. It is worthy of recognition that the results of this study provide the support for existing clinical data. In order to make this method apply to clinical diagnosis, we will apply our approach to the latest clinical data for improve the feasibility and make it be put into clinical diagnosis as soon as possible.
Conclusions
In conclusion, numerical experiments demonstrate that our method is effective and efficient. Research into the size range of cell-free fetal DNA fragments is important for the performance of NIPT and its clinical assessment. This study suggests that most of the cell-free fetal DNA fragments are between 80 and 155 bp in length, which may serve as a valuable reference point for future research. The results of sample collection and processing are supplemented S1 Files.
Supporting information
The appendix is the proportion of every corresponding chromosome for 144 samples.
(XLSX)
The appendix is the average value and standard deviation of chromosomes for 144 samples.
(XLSX)
The sample set included 21 cases each with a trisomy 13 fetus, 27 cases each with a trisomy 18 fetus, 36 cases each with a trisomy 21 fetus, and 60 cases each with a euploid fetus.
(XLSX)
The supplementary files include the result of sample collection and processing.
(ZIP)
Acknowledgments
Thanks to the provider of the data used in this article.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This project was supported by the National Key Research and Development Program of China 2016YFC1000307. The sub-project of National Key Research and Development Program of China 2016YFC1000307-10,the Program of National Research Institute for Family Planning(2017GJM04, 2018 CNV).
References
- 1.Gregg AR, Skotko BG, Benkendorf JL, Monaghan KG, Bajaj K, Best RG, et al. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: a position statement of the American College of Medical Genetics and Genomics. Genetics In Medicine. 2016; 18(10): 1056–1065. 10.1038/gim.2016.97 [DOI] [PubMed] [Google Scholar]
- 2.Chiu RW, Akolekar R, Zheng YW, Leung TY, Sun H, Chan KC, et al. Noninvasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: Large scale validity study. BMJ. 2011; 342: c7401 10.1136/bmj.c7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CWG, et al. Presence of fetal DNA in maternal plasma and serum. The Lancet. 1997; 350(9076): 485–487. [DOI] [PubMed] [Google Scholar]
- 4.Chen EZ, Chiu RW, Sun H, Akolekar R, Chan KC,Leung TY, et al. Noninvasive prenatal diagnosis of fetal trisomy 18 and trisomy 13 by maternal plasma DNA sequencing. PLoS One. 2011; 6(7): e21791 10.1371/journal.pone.0021791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu SC, Chan KC, Zheng YW, Jiang P, Liao GJ, Sun H, et al. Size-based molecular diagnostics using plasma DNA for noninvasive prenatal testing. Proceedings of the National Academy of Science of the United States of America. 2014; 111(23): 8583–8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palomaki GE, Deciu C, Kloza EM, Lambertmesserlian GM, Haddow JE, Neveux LM, et al. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: an international collaborative study. Genetics in Medicine. 2012; 14(3): 296–305. 10.1038/gim.2011.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Driscoll DA, Gross S. Clinical practice. Prenatal screening for aneuploidy. The New England journal of medicine. 2009; 360(24): 2556–2562. 10.1056/NEJMcp0900134 [DOI] [PubMed] [Google Scholar]
- 8.Morris JK, Savva GM. The risk of fetal loss following a prenatal diagnosis of trisomy 13 or trisomy 18. Am J Med Genet A. 2008; 146A(7): 827–832. 10.1002/ajmg.a.32220 [DOI] [PubMed] [Google Scholar]
- 9.Nembhard WN, Waller DK, Sever LE, Canfield MA. Patterns of first-year survival among infants with selected congenital anomalies in Texas, 1995–1997. Teratology. 2001; 64(5): 267–275. 10.1002/tera.1073 [DOI] [PubMed] [Google Scholar]
- 10.Rasmussen SA, Wong LY, Yang Q, May KM, Friedman JM. Population-based analyses of mortality in trisomy 13 and trisomy 18. Pediatrics. 2003; 111(4 Pt 1): 777–784. [DOI] [PubMed] [Google Scholar]
- 11.Lun FM, Tsui NB, Chan KC, Leung TY, Lau TK, Charoenkwan P, et al. Noninvasive prenatal diagnosis of monogenic diseases by digital size selection and relative mutation dosage on DNA in maternal plasma. Proceedings of the National Academy of Sciences of the United States of America. 2008; 105(50): 19920–19925. 10.1073/pnas.0810373105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nygren AO, Dean J, Jensen TJ, Kruse S, Kwong W, Boom D, et al. Quantification of fetal DNA by use of methylation-based DNA discrimination. Clin Chem. 2010; 56(10): 1627–1635. 10.1373/clinchem.2010.146290 [DOI] [PubMed] [Google Scholar]
- 13.Gregg AR, Skotko BG, Benkendorf JL, Monaghan KG, Bajaj K, Best RG, et al. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: a position statement of the American College of Medical Genetics and Genomics. Genetics In Medicine. 2016; 18(10): 1056–1065. 10.1038/gim.2016.97 [DOI] [PubMed] [Google Scholar]
- 14.Lam KW., Jiang P, Liao GJ, Chan KC, Leung TY, Chiu RW, et al. Noninvasive prenatal diagnosis of monogenic diseases by targeted massively parallel sequencing of maternal plasma: Application to β-thalassemia. Clin Chem. 2012; 58(10): 1467–1475. 10.1373/clinchem.2012.189589 [DOI] [PubMed] [Google Scholar]
- 15.Chan KC, Jiang PY, Sun K, Cheng KY, Tong YK, Cheng H et al. Second generation noninvasive fetal genome analysis reveals demnovo mutations, single-base parental inheritance, and preferred DNA ends. Proceedings of the National Academy of Sciences. 2016; 113(50): E8159–E8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lun FM, Chiu RW, Chan KC, Leung TY, Lau TK, Lo YM. Microfluidics digital PCR reveals a higher than expected fraction of fetal DNA in maternal plasma. Clin Chem. 2008; 54(10): 1664–1672. 10.1373/clinchem.2008.111385 [DOI] [PubMed] [Google Scholar]
- 17.Tsui NB, Kadir RA, Chan KC, Chi C, Mellars G, Tuddenham EG, et al. Noninvasive prenatal diagnosis of hemophilia by microfluidics digital PCR analysis of maternal plasma DNA. Blood. 2011; 117(13): 3684–3691. 10.1182/blood-2010-10-310789 [DOI] [PubMed] [Google Scholar]
- 18.Chiu RW, Chan KC, Gao Y, Lau VY, Zheng W, Leung TY, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proceedings of the National Academy of Science of the United States of America. 2008; 105(51): 20458–20463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo YM, Zhang J, Leung N, Lau K, Chang MZ, Hjelm NM. Rapid Clearance of Fetal DNA from Maternal Plasma. American journal of human Genetics. 1999; 64(1): 218–224 10.1086/302205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo YM, Lun FM, Chan KC, Tsui NB, Chong KC, Lau TK, et al. Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proceedings of the National Academy of Sciences of the United States of America. 2007; 104(32): 13116–13121. 10.1073/pnas.0705765104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sparks AB, Struble CA, Wang ET, Song K, Oliphant A. Noninvasive prenatal detection and selective analysis of cell-free DNA obtained from maternal blood: evaluation for trisomy 21 and trisomy 18. American Journal of Obstetrics and Gynecology. 2012; 206(4): 319e1–319e9. [DOI] [PubMed] [Google Scholar]
- 22.Lo YM, Chan KC, Hao S, Chen Z, Jiang PY, Lun MF, et al. Maternal Plasma DNA Sequencing Reveals the Genome-Wide Genetic and Mutational Profile of the Fetus. Science Translational Medicine. 2010; 2(61): 61–91. [DOI] [PubMed] [Google Scholar]
- 23.Sparks AB, Struble CA, Wang ET, Song K, Oliphant A. Noninvasive prenatal detection and selective analysis of cell-free DNA obtained from maternal blood: Evaluation for trisomy 21 and trisomy 18. American Journal of Obstetrics and Gynecology. 2012; 206(4): 319.e1–319.e9. [DOI] [PubMed] [Google Scholar]
- 24.Chu T, Bunce K, Hogge WA, Peters DG. Statistical model for whole genome sequencing and its application to minimally invasive diagnosis of fetal genetic disease. Bioinformatics 2009; 25(10): 1244–1250. 10.1093/bioinformatics/btp156 [DOI] [PubMed] [Google Scholar]
- 25.Nicolaides KH, Syngelaki A, Ashoor G, Birdir C, Touzet G. Noninvasive prenatal testing for fetal trisomies in a routinely screened first-trimester population. American journal of obstetrics and gynecology. 2012; 207(5): 374.e371–374.e376. [DOI] [PubMed] [Google Scholar]
- 26.Hui WW, Jiang P, Tong YK, Lee WS, Cheng YK, New MI, et al. Universal haplotype-based noninvasive prenatal testing for single gene diseases. Clinical Chemistry. 2017; 63(2): 513–524. 10.1373/clinchem.2016.268375 [DOI] [PubMed] [Google Scholar]
- 27.Ehrich M, Deciu C, Zwiefelhofer T, Tynan JA, Cagasan L, Tim R, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: A study in a clinical setting. Am. J. Obstet. Gynecol. 2011; 204(3): 205.e201–205.e211. [DOI] [PubMed] [Google Scholar]
- 28.Bianchi DW, Platt LD, Goldberg JD, Abuhamad AZ, Sehnert AJ, Rava RP. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012; 119(5): 890–901. 10.1097/AOG.0b013e31824fb482 [DOI] [PubMed] [Google Scholar]
- 29.Palomaki GE, Deciu C, Kloza EM, Lambert-Messerlian GM, Haddow JE, Neveux LM, et al. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as down syndrome: An international collaborative study. Genet Med. 2012; 14(3): 296–305. 10.1038/gim.2011.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norton ME, Wapner RJ. Cell-free DNA analysis for noninvasive examination of trisomy. The New England journal of medicine. 2015; 373(26): 2582 10.1056/NEJMc1509344 [DOI] [PubMed] [Google Scholar]
- 31.Meng M, Li X, Ge H, Chen F, Han M, Zhang Y, et al. Noninvasive prenatal testing for autosomal recessive conditions by maternal plasma sequencing in a case of congenital deafness. Genet Med. 2014; 16(12): 972–976. 10.1038/gim.2014.51 [DOI] [PubMed] [Google Scholar]
- 32.New MI, Tong YK, Yuen T, Jiang P, Pina C, Chan KC, et al. Noninvasive prenatal diagnosis of congenital adrenal hyperplasia using cell-free fetal DNA in maternal plasma. The Journal of clinical endocrinology. 2014; 99(6): E1022–E1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu Y, Li X, Ge HJ, Xiao B, Zhang YY, Ying XM, et al. Haplotype-based approach for noninvasive prenatal tests of duchenne muscular dystrophy using cell-free fetal DNA in maternal plasma. Genet Med. 2015; 17(11): 889–896. 10.1038/gim.2014.207 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The appendix is the proportion of every corresponding chromosome for 144 samples.
(XLSX)
The appendix is the average value and standard deviation of chromosomes for 144 samples.
(XLSX)
The sample set included 21 cases each with a trisomy 13 fetus, 27 cases each with a trisomy 18 fetus, 36 cases each with a trisomy 21 fetus, and 60 cases each with a euploid fetus.
(XLSX)
The supplementary files include the result of sample collection and processing.
(ZIP)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.