Abstract
Luminal breast cancers are typically estrogen receptor positive and generally have the best prognosis. However, a subset of luminal tumors, namely luminal B cancers, frequently metastasize and recur. Unfortunately, the causal events that drive their progression are unknown and therefore it is difficult to identify individuals who are likely to relapse and should receive escalated treatment. Here we identify a bi-functional RasGAP tumor suppressor that is lost in almost 50% of luminal B tumors. Moreover, we show that two RasGAP genes are concomitantly inactivated in the most aggressive luminal malignancies. Importantly, these genes cooperatively regulate two major oncogenic pathways, Ras and NF-kappa B through distinct domains, and when inactivated drive the metastasis of luminal tumors in vivo. Finally, while the cooperative effects on Ras drive invasion, NF-κB activation triggers EMT and is required for metastasis. Collectively, these studies reveal important mechanistic insight into the pathogenesis of luminal B tumors and provide functionally relevant prognostic biomarkers that may guide treatment decisions.
Keywords: Ras, RasGAPs, NK-κB, Breast Cancer, Luminal B
Introduction
Breast cancer is one of the most common malignancies in women worldwide and the second leading cause of cancer deaths in women in the United States (1). In the clinic breast tumors are primarily defined by hormone receptor and HER2 status (2). However, transcriptional profiling studies demonstrate that breast tumors can be further classified into subtypes that include: luminal cancers (A and B), basal-like, claudin-low, HER2/ERBB2 overexpressing, and normal-like (3–5). Luminal tumors are typically ER+ and represent the majority of breast cancers. Notably, while the Luminal A subtype of tumors respond well to endocrine therapy, luminal B tumors exhibit a higher proliferative index, are less responsive, and are more likely to become metastatic (6,7). In fact, the relapse risk rate for luminal B tumors is similar to that of basal and HER2/ERBB2-amplified tumors (8). Genomic efforts have identified many genetic, epigenetic, and transcriptional differences between these two luminal subtypes, however little is known about the functional events that underlie the aggressive behavior of luminal B tumors (9–11). A better mechanistic understanding of the signals that drive their progression would not only help identify individuals who could benefit from upfront additional adjuvant treatment, but might also provide insight into new therapeutic strategies.
The Ras pathway is one of the most frequently deregulated pathways in human cancer. Activating mutations in RAS genes occur in nearly 30% of all human tumors (12); however, RAS mutations are conspicuously rare in breast cancer and current TCGA data indicates that RAS is mutated in only 1% of all breast lesions (13,14). Nevertheless, the Ras pathway is aberrantly activated in more than 50% of human breast tumors and has been suggested to contribute to progression and recurrence (15–17). These observations raise the question of how the Ras pathway becomes hyperactivated in this tumor type.
Ras GTPase Activating Proteins promote the hydrolysis of Ras-GTP to Ras-GDP, thereby turning off Ras (18). Accordingly, the inactivation or loss of a RasGAP leads to enhanced Ras-GTP levels, thus providing an alternative means of activating Ras. The NF1 gene is an example of a RasGAP tumor suppressor that is inactivated in a variety of sporadic cancers (reviewed in (19)). Here we show that a unique bi-functional RasGAP tumor suppressor, one that serves as a signaling scaffold to coordinately suppress both Ras and NF-κB is selectively and frequently lost in luminal B tumors (20). However, we also find that the most aggressive luminal cancers lack two distinct RasGAP genes, which cooperatively trigger the activation of multiple Ras isoforms as well as NF-κB. Collectively, the studies presented here reveal the mechanism by which two major oncogenic pathways are activated and cooperate to drive the progression of luminal B breast cancers, a finding that has important prognostic and therapeutic implications.
Results
The RasGAP Gene DAB2IP is Selectively Lost in Luminal B Breast Cancers
Despite the low incidence of oncogenic RAS mutations, the Ras/ERK pathway is hyperactivated in more than half of human breast cancers (16). Interestingly, the RasGAP gene, RASAL2, has been shown to function as a tumor suppressor and is lost in a subset of breast tumors (21). However, given that there are fourteen RasGAPs in the human genome, we speculated that other RasGAP genes might also contribute to breast cancer development. Because RasGAPs are frequently inactivated by epigenetic mechanisms in cancer (19), we first evaluated mRNA expression levels of RasGAP genes across breast cancer subtypes. Strikingly, we found that DAB2IP, a RasGAP tumor suppressor that drives prostate cancer progression (20), was selectively suppressed in luminal B breast tumors (Fig. 1A and B). Importantly, a CpG island in the DAB2IP promoter was frequently hypermethylated in the luminal B subtype (Fig. 1C, chr9: 123,331,678) and methylation at this site inversely correlated with mRNA expression levels (Fig. 1D). Moreover, we found that low DAB2IP expression was associated with a significant decrease in relapse-free survival in patients with luminal B tumors (Fig. 1E, log rank p=0.0028), raising the possibility that DAB2IP loss might contribute to the progression of this breast cancer subtype.
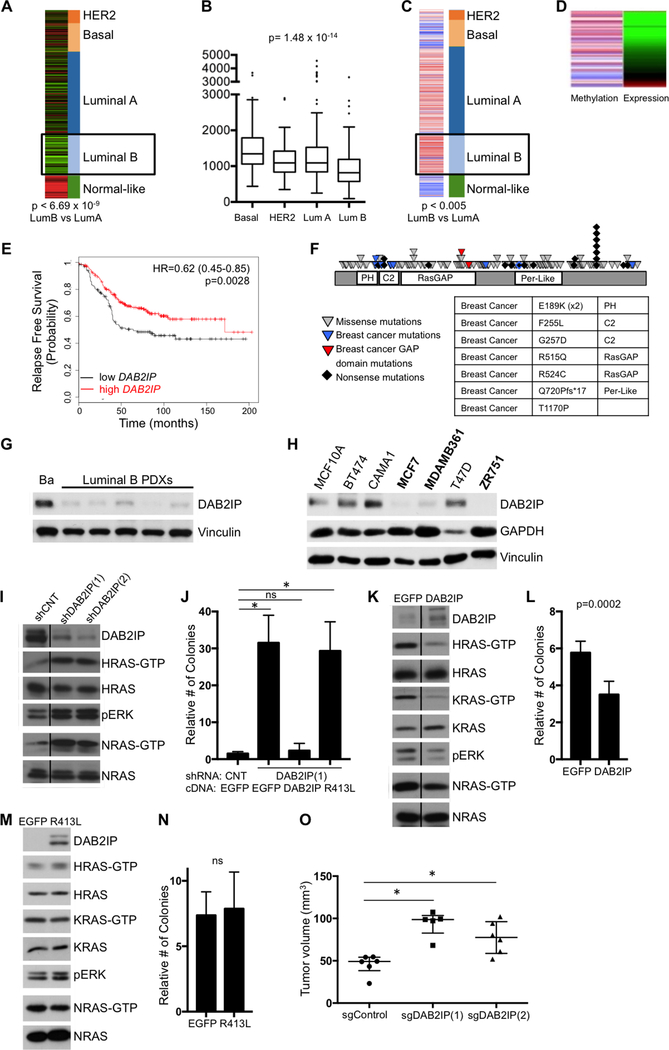
Figure 1. DAB2IP is selectively lost in luminal B breast cancers and functions as a tumor suppressor by affecting multiple Ras Isoforms.
A, DAB2IP mRNA expression across the different subtypes of human breast cancer. DAB2IP is specifically low in the luminal B subtype (p< 6.69×10−9, t-test comparing LumA vs LumB). B, Tukey boxblot showing the median DAB2IP expression per subtype (p=1.5×10−14, ANOVA). C, Methylation of the CpG island in the DAB2IP promoter (chr9: 123,331,678) across subtypes. The DAB2IP promoter is specifically methylated in the Luminal B subtype (p< 0.005, t-test comparing LumA vs LumB). D, Methylation at the CpG island in the DAB2IP promoter inversely correlates with expression. E, Kaplan-Meier curve showing relapse-free survival of luminal B tumors with high or low DAB2IP expression (log rank p=0.0028). F, DAB2IP mutations in human tumor samples. Each triangle represents a nonsynonymous mutation and each diamond represents a nonsense mutation. Blue triangles indicate breast cancer mutations and red triangles indicate breast cancer mutations in the catalytic RasGAP domain. The table shows DAB2IP mutations in human breast tumors and the domain in which they occur. G, DAB2IP expression in lysates of basal (Ba, HCI-004) and luminal B (HCI-011, −013, −017, −003, −005) PDX tumors. H, DAB2IP expression in a panel of ER+ human breast cancer cell lines and normal immortalized mammary epithelial cells (MCF10A). Cell lines with minimal or no DAB2IP are indicated in bold. I, Western blot showing an increase in HRas-GTP, NRas-GTP, and pERK levels in CAMA1 cells following shRNA-mediated inactivation of DAB2IP or control shRNA. Samples were run on the same gels, but a lane was cut out as indicated by the line. Lysates were run in duplicate. DAB2IP, HRAS/HRAS-GTP and pERK were probed for on the same gel, while NRAS/NRAS-GTP were probed for on a separate gel. J, Soft agar colony formation of CAMA1 cells infected with a 3’-UTR shRNA targeting DAB2IP or nontargeting control, followed by expression of EGFP control vector, wild-type DAB2IP, or DAB2IP GAP point mutant (R413L). Data show relative number of colonies +/− SD. There was a significant increase in soft agar colony formation upon DAB2IP suppression (p=0<0.0001, t-test comparing EGFP/shCNT vs EGFP/shDAB2IP(1)). Reconstitution with wild-type DAB2IP rescued colony formation, however, the R413L GAP mutant failed to rescue (p<0.0001, t-test comparing EGFP/shCNT vs R413L/shDAB2IP(1)). K, Western blot showing a reduction of HRas-GTP, KRas-GTP, and pERK levels in MCF7 cells upon expression of DAB2IP compared to EGFP. Samples were run on the same gel, but a lane was cut out as indicated by the line. Lysates were run in duplicate. DAB2IP, HRAS/HRAS-GTP, KRAS/KRAS-GTP and pERK were probed for on the same gel, while NRAS/NRAS-GTP were probed for on a separate gel. L, Soft agar colony formation of MCF7 cells expressing DAB2IP or EGFP. Data show relative number of colonies +/− SD. There was a statistically significant decrease in soft agar colony formation upon DAB2IP-reconstitution (p=0.0002, t-test). M, Western blot showing that expression of the R413L DAB2IP-GAP mutant fails to suppress HRas-GTP, KRas-GTP, NRas-GTP, and pERK levels in MCF7 cells. Lysates were run in duplicate. DAB2IP, HRAS/HRAS-GTP, KRAS/KRAS-GTP and pERK were probed for on the same gel, while NRAS/NRAS-GTP were probed for on a separate gel. N, Soft agar colony formation of MCF7 cells expressing the R413L GAP mutant or EGFP. Data show relative number of colonies +/− SD. There was no statistically significant decrease in anchorage-independent growth upon ectopic expression of the R413L mutant. O, Xenograft tumor formation of CAMA1 cells infected with DAB2IP-targeting CRISPR guide RNA or empty control vector. Cells were injected subcutaneously into female NOD/SCID mice. Horizontal bars indicate median tumor volume and the error bars indicate +/− interquartile range. There was a statistically significant increase in tumor volume upon CRISPR-mediated DAB2IP knock-out (p=0.0043 for sgDAB2IP(1); p=0.0087 for sgDAB2IP(2), Mann-Whitney).
To complement expression analyses, the mutational status of DAB2IP was also examined. Numerous mutations predicted to be deleterious were identified in DAB2IP across several tumor types, suggesting that DAB2IP might function as a broad human tumor suppressor (Fig. 1F). A subset of these alterations were present in breast cancers, including damaging mutations in the catalytic RasGAP domain, and the period-like domain, a region shown to be important for NF-κB signaling (to be discussed). However, similar to RASAL2, the frequency of DAB2IP mutations in breast cancer was relatively low, consistent with the observation that RasGAPs are more typically silenced by epigenetic events in cancer (19).
To confirm that DAB2IP protein levels were indeed suppressed in human luminal B tumors, we assessed DAB2IP protein expression in a panel of patient-derived xenografts (PDXs). Notably, DAB2IP protein was minimally expressed in all five luminal B tumors examined (Fig. 1G), as compared to a basal PDX control, a sub-type in which the DAB2IP gene is infrequently methylated and suppressed (Fig. 1A-C). Immunohistochemical analysis on a larger set of primary human tumors will be described below.
DAB2IP Controls the Activation of Multiple Ras Proteins and Functions as a Tumor Suppressor in Luminal Breast Cancers
To investigate the function of DAB2IP in luminal tumor models, DAB2IP expression was assessed in a panel of ER+ breast cancer cell lines. While primary luminal A and B tumors can readily be distinguished by transcriptional profiling analysis, there have been discrepancies in classifying luminal cell lines into A and B subtypes. Some suggest that cell lines cannot be molecularly distinguished as luminal A or B and/or that luminal A tumors are not represented by existing cell lines (22,23), while others classify cell lines by different criteria (24,25) (Various annotations of cell lines examined are shown in Supplementary Table S1). Therefore, to select cell lines for functional studies we merely sought to identity luminal cell lines that either lacked DAB2IP expression (for gain-of-function analysis), or expressed DAB2IP (for loss-of-function studies). In contrast to normal mammary epithelial cells (MCF10As), DAB2IP was absent or minimally expressed in 3 out of 6 luminal lines (Fig. 1H and Supplementary Fig. S1A). Importantly, we found that the DAB2IP promoter was differentially methylated in breast cancer cell lines with low DAB2IP expression as compared to DAB2IP-expressing cell lines (Supplementary Fig. S1B) and DAB2IP mRNA levels directly correlated with protein expression across a broad panel of human breast cancer cell lines (Supplementary Fig. S1C and S1D, p<0.0001), further supporting the conclusion that promoter methylation regulates DAB2IP silencing in breast cancer.
The biochemical and biological consequences of either ablating or reconstituting DAB2IP were evaluated in two luminal breast cancer cell lines: CAMA1 and MCF7 cells, respectively. Notably, DAB2IP ablation led to potent Ras pathway activation in CAMA1 cells (Fig. 1I). Specifically, DAB2IP knock-down using two different shRNAs, activated H-Ras, N-Ras and ERK, although it should be noted that the K-Ras protein was not detected in this particular cell line (data not shown). While shRNA-mediated DAB2IP suppression had no effect on CAMA1 cell proliferation in two-dimensional (2D) cultures (Supplementary Fig. S1E), it dramatically enhanced anchorage-independent colony formation (1J). Moreover, anchorage-independent growth was suppressed by reconstitution with wild-type DAB2IP, but was not inhibited by a catalytically inactive RasGAP mutant (DAB2IPR413L), demonstrating that the RasGAP domain is required for the tumor suppressor function of DAB2IP (Fig. 1J and Supplementary Fig.S1F).
DAB2IP was also reconstituted in MCF7 cells, which normally lack DAB2IP (Figure 1K). Only low levels of DAB2IP expression were tolerated in these cells, potentially due to an evolved dependence on its suppression. Nevertheless, reconstitution potently suppressed H- and K-Ras-GTP levels, had more modest effects on N-Ras, and suppressed phospho-ERK (Figure 1K). Consistent with ablation studies, DAB2IP reconstitution did not affect proliferation under 2D-culture conditions (Supplementary Fig. S1G), but reduced colony formation (Figure 1L), whereas the inactivating mutation in the catalytic RasGAP domain failed to inhibit Ras and ERK activation (Fig. 1M), and had no effect on colony formation, (Fig. 1N). Finally, CRISPR-mediated ablation of DAB2IP using two different guide RNAs promoted primary tumor growth in a xenograft model in vivo (Figure 1O). Collectively, these studies demonstrate that 1) DAB2IP is frequently lost in luminal B breast cancers and its suppression promotes the growth of primary tumors, 2) DAB2IP can regulate multiple Ras isoforms, and 3) its RasGAP activity is required for its tumor suppressor function.
Concomitant Loss of DAB2IP and RASAL2 Occurs in the Most Aggressive Luminal B Tumors
Because DAB2IP and RASAL2 both regulate Ras (Figure 1 and (21)), we expected that their loss might be mutually exclusive and that tumors would exhibit either the loss of one or the other RasGAP gene. However, immunoblot analysis demonstrated that DAB2IP and RASAL2 proteins were concomitantly suppressed in some luminal cell lines (Fig. 2A). In addition, DAB2IP and RASAL2 were both minimally expressed or undetectable in 3/5 of the luminal B PDX tumors (Fig. 2B). Like DAB2IP, we found that the RASAL2 promoter was hypermethylated in cells that expressed low levels of RASAL2 (Supplementary Fig. S2A, chr1: 176,334,344), and mRNA levels precisely correlated with protein levels (Supplementary Fig. S2B and S2C). To more accurately determine the relative frequency of DAB2IP and/or RASAL2 loss in luminal B tumors, mRNA expression levels were analyzed in primary luminal B tumors from TCGA datasets. DAB2IP was suppressed in a striking 46% percent of luminal B malignancies and nearly half of these tumors had also lost RASAL2 (Fig. 2C). Notably, ER+ tumors with low levels of both RASAL2 and DAB2IP more frequently presented as Stage III/IV lesions as compared to those with high levels (30% versus 17%; p<0.05 Fisher Exact Test), which more frequently presented as Stage I lesions. Moreover, in the context of luminal B disease alone, low levels of both genes together were associated with a significant decrease in relapse-free survival (Fig. 2D, log rank p=3.1e-08).
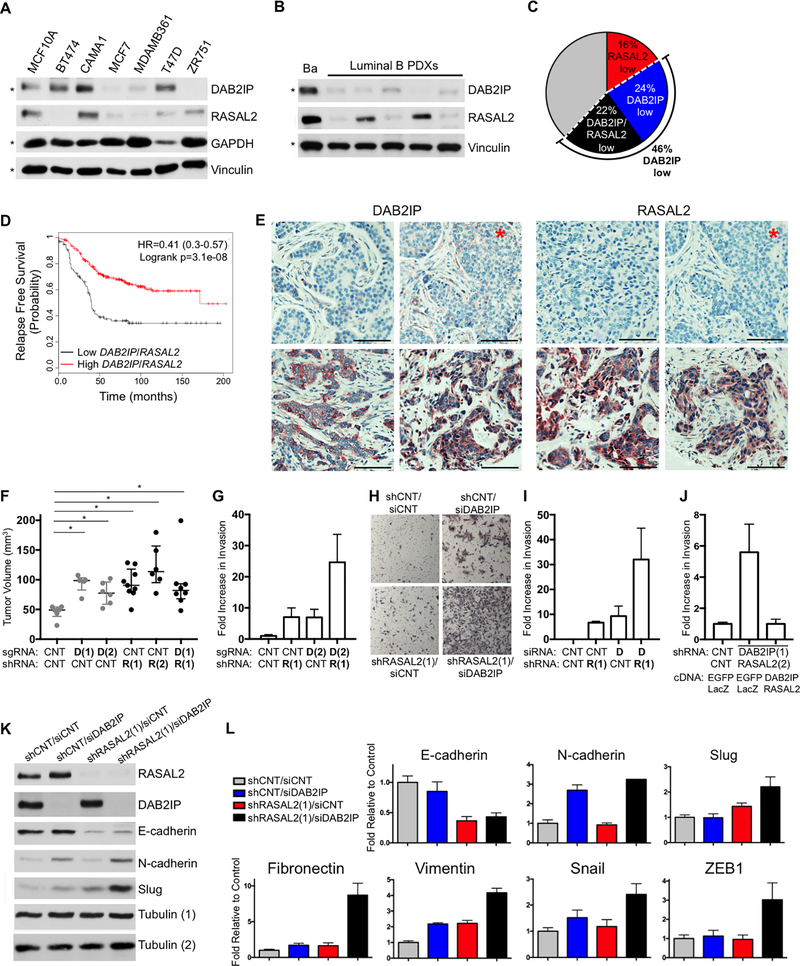
Figure 2. Concomitant loss of DAB2IP and RASAL2 occur in the most aggressive luminal B tumors and specifically enhance invasion and EMT.
A, RASAL2 and DAB2IP protein expression in a panel of ER+ human breast cancer cell lines and normal immortalized mammary epithelial cells (MCF10A). The blots marked by an asterisk are a duplicate from Figure 1H for comparison. B, DAB2IP and RASAL2 expression in lysates of basal (Ba, HCI-004) and luminal B (HCI-011, −013, −017, −003, −005) PDX tumors. The blots marked by an asterisk are a duplicate from Figure 1G for comparison. C, Pie chart of human luminal B tumors. 16% of luminal B tumors have low levels of RASAL2 (red), 24% have low levels of DAB2IP (blue), and 22% have low levels of both, RASAL2 and DAB2IP (black). D, Kaplan-Meier curve showing relapse-free survival of luminal B tumors with high RASAL2 and DAB2IP expression or low RASAL2 and DAB2IP expression (log rank p=3.1e-08). E, Representative IHC images of human luminal B tumors with low (top) and high (bottom) DAB2IP and RASAL2 expression. The images marked by the red asterisk are images of the same section of the same tumor. The scale bars correspond to 100μm. F, Xenograft tumor formation of CAMA1 cells infected with nontargeting control vectors, DAB2IP-targeting CRISPR guide RNAs (p=0.0043 for sgDAB2IP(1); p=0.0087 for sgDAB2IP(2), Mann-Whitney), RASAL2-targeting shRNAs (p=0.0016 for shRASAL2(1); p=0.0022 for shRASAL2(2), Mann-Whitney), or both sgDAB2IP(1)/shRASAL2(1)-targeting constructs (p=0.008, Mann-Whitney). Cells were injected subcutaneously into female NOD/SCID mice. Horizontal bars indicate median tumor volume and the error bars indicated +/− interquartile range. The data indicated in grey is a duplicate from Figure 1O. There was no statistically significant increase in tumor volume upon combined inactivation of DAB2IP and RASAL2. G, Transwell invasion of MCF10A cells infected with control shRNA, an shRNA targeting RASAL2 (R(1)) and/or a sgRNA targeting DAB2IP (D(2)). Invasion was measured after 24 hours and reported as average +/− SD. H, Transwell invasion of MCF10A cells infected with control shRNA, an shRNA targeting RASAL2 (R(1)) and/or an siRNA targeting DAB2IP (D). Invasion was measured after 24 hours and representative images are shown. I, Transwell invasion of CAMA1 cells infected with control shRNA, an shRNA targeting RASAL2 (R(1)) and/or an siRNA targeting DAB2IP (D). Invasion was measured after 24 hours and reported as average +/− SD. J, Transwell invasion of MCF10A cells infected with control shRNA or 3’-UTR shRNAs targeting RASAL2 and DAB2IP, followed by expression of EGFP/LacZ control vectors or wild-type DAB2IP/RASAL2. Invasion was measured after 24 hours and reported as average +/− SD. K, Western blot showing the expression of molecular markers of an EMT, E-cadherin, N-cadherin, and Slug in MCF10A cells infected with an shRNA targeting RASAL2 and/or an siRNA targeting DAB2IP. Lysates were run in duplicate. DAB2IP and E-cadherin were probed for on the same gel with the loading control Tubulin (1), while RASAL2, N-cadherin and Slug were probed for separately with the loading control Tubulin (2). L, Real-time PCR quantification of E-cadherin, N-cadherin, Slug, Fibronectin, Vimentin, Snail, and ZEB1 in MCF10A cells in response to RASAL2 and/or DAB2IP suppression. Data show average relative amount of mRNA +/− SD.
To confirm and quantify DAB2IP and RASAL2 protein loss in primary human tumors, immunohistochemical analysis was performed. Antibodies were first validated using human xenografts that expressed or lacked DAB2IP and RASAL2 (Supplementary Fig. S2D and S2E). DAB2IP and RASAL2 expression were then examined in 63 primary human luminal B tumors (as described in (26)). Consistent with mRNA analysis, DAB2IP expression was uniformly absent or minimally detected in 43% of human luminal B tumors and 21% of luminal B tumors exhibited a loss of both DAB2IP and RASAL2 (Fig. 2E). While outcome data is not yet available, low levels of DAB2IP and RASAL2 in this luminal B cohort were also associated with more advanced stage at the time of diagnosis. Specifically, 90% of tumors that lacked DAB2IP and RASAL2 were categorized as Stage II-IV, while only 10% were categorized as Stage I lesions. In contrast 45% of luminal B tumors that expressed DAB2IP and RASAL2 were categorized as Stage I, p<0.05 Fisher Exact Test). Taken altogether these observations suggest that DAB2IP and RASAL2 are frequently co-inactivated in luminal B tumors and raise the possibility that they may functionally cooperate in the development and/or progression of these cancers.
Concomitant Loss of DAB2IP and RASAL2 Cooperatively Drives Invasion, EMT, and Metastasis
To elucidate the precise biological consequences of concomitant DAB2IP and RASAL2 loss, we first investigated whether the suppression of both genes might enhance tumor formation in vivo. While DAB2IP and RASAL2 inactivation individually promoted the growth of human breast cancer xenografts, there was no additional increase in primary tumor growth when both genes were ablated (Fig. 2F). However, because the concomitant loss of DAB2IP and RASAL2 was associated with advanced disease and recurrence in humans, we evaluated the effects of suppression on invasiveness and epithelial-to-mesenchymal transition (EMT): cellular properties associated with tumor progression and metastasis (27). While the individual ablation of DAB2IP and RASAL2 modestly increased invasiveness, their concomitant loss dramatically enhanced the invasion of human mammary epithelial cells (Fig. 2G and H) and luminal breast cancer cells (Fig. 2I). Results were confirmed using multiple shRNA, siRNA, and sgRNA sequences (Figure 2G-I and Supplementary Figure S2F) and this enhanced invasiveness was rescued by reconstitution with RASAL2 and DAB2IP cDNA (Fig. 2J).
Interestingly, the combined loss of DAB2IP and RASAL2 also cooperatively enhanced EMT. For example, EMT is characterized by a decrease in E-cadherin and an increase in N-cadherin levels. Only the combined ablation of RASAL2 and DAB2IP resulted in both the effective suppression of E-cadherin and upregulation of N-cadherin (Fig. 2K). Immunoblot and/or q-PCR analysis also revealed a dramatic cooperative increase in the expression of the EMT transcriptional regulators, Slug, Snail, and Zeb1 and other EMT markers such as Fibronectin and Vimentin (Figure 2K and L). Again, the induction of EMT was rescued by ectopic expression of RASAL2 and DAB2IP cDNA (Supplementary Fig. S2G). Together these findings suggest that while the loss of one RasGAP gene is sufficient to promote primary tumor development, the concomitant loss of DAB2IP and RASAL2 together potently promote invasion and EMT, consistent with a potential role for these genes in tumor progression.
To determine whether DAB2IP and RASAL2 regulate metastasis in vivo, both gain- and loss-of-function studies were performed. First, DAB2IP and RASAL2 were reconstituted individually or together in MDA-MB-361 breast cancer cells that lack endogenous DAB2IP and RASAL2 and were engineered to express luciferase. Following intra-cardiac injection, metastatic lesions were monitored by Xenogen imaging. Parental MDA-MB-361 cells are metastatic (Fig. 3A and B). Notably, while reconstitution with DAB2IP or RASAL2 alone reduced the number of metastases, only combined reconstitution prevented metastasis formation altogether (Fig. 3A and B). This complete suppression of metastasis was maintained for four months when the experiment was terminated (Fig. 3C, p=0.004 Mann-Whitney). Importantly, DAB2IP and RASAL2 reconstitution had no effect on the proliferation of these cells, indicating that the absence of metastases was not merely a consequence of impaired proliferation (Fig. 3D and E). To complement these studies DAB2IP and RASAL2 were also ablated in a luminal mouse cancer cell line using shRNA sequences, and tumor cells were injected into tail veins of syngeneic mice as previously described (Fig. 3F and G) (28). Metastases in the lungs were visualized and quantified by counting individual tumors from multiple adjacent histological sections. Importantly, co-inactivation of DAB2IP and RASAL2 dramatically enhanced metastasis (Fig. 3G and3H, p=0.0019, Mann-Whitney).
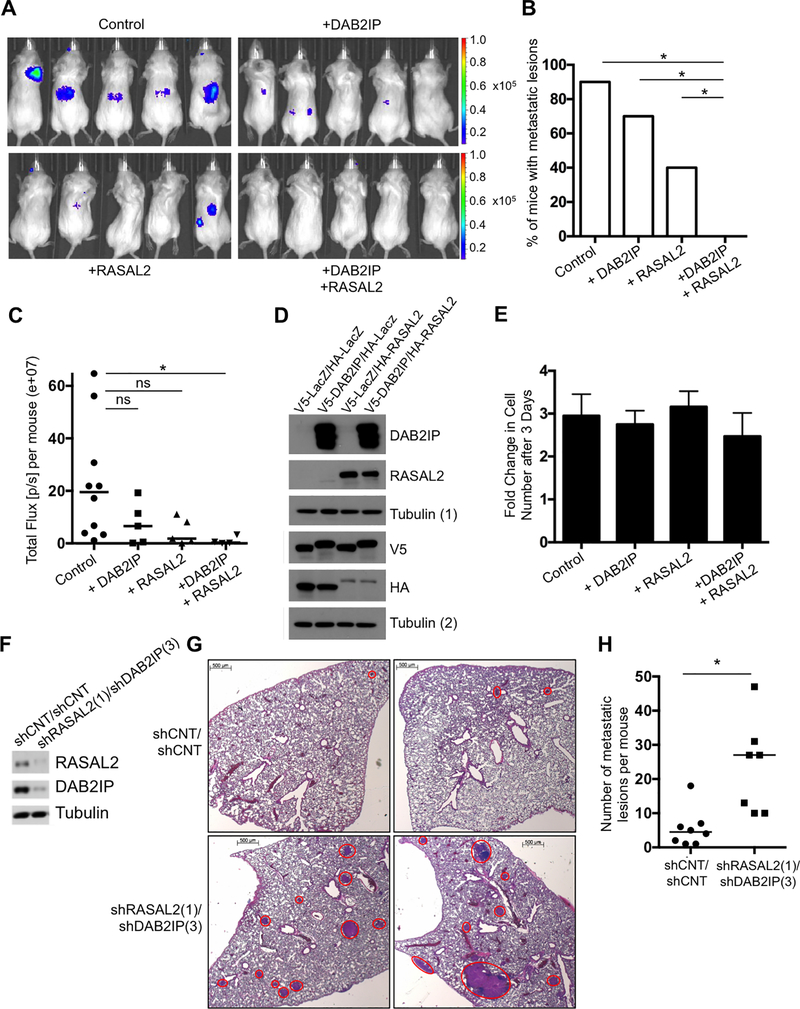
Figure 3. DAB2IP and RASAL2 cooperatively regulate metastasis in vivo.
A, Representative bioluminescence images of luciferase-expressing MDA-MB-361 cells that were reconstituted with LacZ/LacZ, LacZ/RASAL2, DAB2IP/LacZ or DAB2IP/RASAL2 and injected intracardially into NOD/SCID mice. B, Quantification of the number of mice with metastatic lesions as detected by bioluminescence imaging two months post injection of MDA-MB-361 cells. Expression of RASAL2 and DAB2IP together significantly suppressed metastasis formation compared to expression of RASAL2 alone (p=0.043, Fisher Exact), DAB2IP alone (p= 0.002, Fisher Exact), or LacZ control (p=6×10−5, Fisher Exact). C, Total Flux [p/s] (e+07) per mouse as determined by bioluminescence imaging four months post inter-cardiac injection. Only expression of RASAL2 and DAB2IP together significantly suppressed bioluminescence signal compared to the LacZ control (p=0.004, Mann-Whitney). D, Western blot confirming ectopic expression of DAB2IP and RASAL2. Lysates were run in duplicate. DAB2IP and RASAL2 were probed for on the same gel with the loading control Tubulin (1), while the tags, V5 and HA, were probed for separately with the loading control Tubulin (2). E, Fold growth at day three of MDA-MB-361 cells expressing LacZ/LacZ, LacZ/RASAL2, DAB2IP/LacZ or DAB2IP/RASAL2 +/− SD. F, Western blot confirming RASAL2 and DAB2IP suppression in McNeu cells, a luminal mouse cancer cell line that was infected with shRNAs targeting RASAL2 and DAB2IP or non-targeting controls. G, Representative H&E images of lungs, with McNeu metastases marked by red circles. McNeu cells were infected with shRNAs targeting RASAL2 and DAB2IP or non-targeting controls and were injected into tail veins of syngeneic mice. Lungs were harvested three weeks post injection and the number of metastases per mouse was quantified. Scale bars correspond to 500μm. H, Suppression of RASAL2 and DAB2IP significantly enhanced metastasis formation (p=0.0019, Mann-Whitney). Horizontal bars indicate the median number of metastatic lesions per mouse.
Interestingly, we previously crossed the autochthonous genetically engineered version of this luminal model to a Rasal2 knock-out mouse and found that the frequency of metastasis was enhanced (21). Retrospective analysis of DAB2IP expression revealed that nearly 60% of primary tumors from this model had spontaneously lost DAB2IP (Supplementary Fig. S3A), and that animals with primary tumors that lost both RasGAPs developed significantly more metastases (Supplementary Fig. S3B, p=0.004, Mann-Whitney). Moreover, in 2/2 instances examined, in which DAB2IP was still expressed in the primary Rasal2−/− tumors, it was selectively lost in metastatic lesions from these same animals (Supplementary Fig. S3C). Together, these gain and loss-of-function studies demonstrate that DAB2IP and RASAL2 cooperatively regulate the metastasis of luminal cancers. Expression analysis in both human tumors and mouse models confirm that these two tumor suppressors are in fact frequently co-inactivated in aggressive luminal malignancies.
Ras Pathway Activation Drives Invasion, while NF-κB Activation is Required for EMT and Metastasis
To deconstruct the specific signals that drive invasion, EMT and metastasis, we first evaluated the combined effects of DAB2IP and RASAL2 loss on Ras signaling. While the suppression of each RasGAP activated K-Ras, H-Ras, ERK and AKT, the concomitant ablation of both RasGAPs dramatically enhanced the activation of all of these components, which overall appeared to be greater than an additive effect (Fig. 4A and B). We then assessed the contribution of the Ras pathway to invasion and EMT triggered by DAB2IP/RASAL2 loss. As shown in Fig. 2, MCF10A cells are not normally invasive, but become potently invasive when DAB2IP and RASAL2 are suppressed. Both MEK and PI3K inhibitors suppressed invasion triggered by combined DAB2IP and RASAL2 loss, underscoring the importance of both of these effector pathways in the process of invasion (Fig. 4C) (29–31). Moreover, while reconstitution with wild-type DAB2IP and RASAL2 suppressed Ras pathway activation (Supplementary Fig. S4) and reversed the invasive phenotype (Fig. 2J and 4D), a catalytically inactive RasGAP point mutant (20,21) failed to do so (Fig. 4D and Supplementary Fig. S4). In direct contrast, PI3K and MEK inhibitors had no effect on EMT triggered by DAB2IP/RASAL2 suppression (Figure 4E), suggesting that aberrant Ras pathway activation primarily serves to drive invasion in DAB2IP/RASAL2-deficient cells.
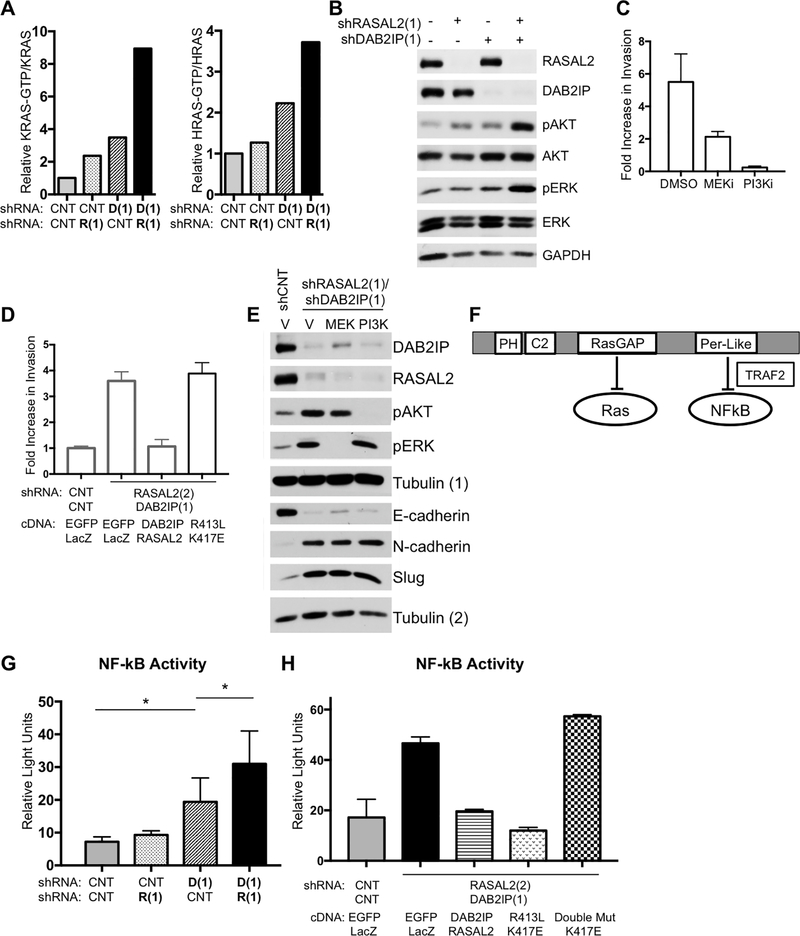
Figure 4. RASAL2 and DAB2IP loss cooperates to activate the Ras and the NFκB pathways.
A, Quantification of KRas- and HRas-GTP levels in MCF10A cells upon shRNA-mediated suppression of RASAL2 (R(1)) and/or DAB2IP (D(1)). The Ras-GTP/Ras ratio of each sample was calculated and normalized to the non-targeting shRNA control. B, Western blot showing downstream Ras pathway activation (pAKT and pERK) following RASAL2 and/or DAB2IP knock down in MCF10A cells. C, Transwell invasion of MCF10A cells infected with shRNAs targeting RASAL2 and DAB2IP. Invasion was measured after 24 hours in the presence of vehicle (DMSO), MEK inhibitor (PD-0325901 at 500nM) and PI3K inhibitor (GDC-0941 at 750nM). The number of cells that invaded was normalized to the non-targeting shRNA control and reported as average +/− SD. D, Transwell invasion of MCF10A cells infected with control shRNA or 3’-UTR shRNAs targeting RASAL2 and DAB2IP, followed by expression of EGFP and LacZ control vectors, wild-type DAB2IP and RASAL2, or the GAP mutants (DAB2IP413L and RASAL2K417E). Invasion was measured after 24 hours and reported as average +/− SD. The data indicated in grey is a duplicate from Figure 2J for comparison. E, Western blot showing the expression of molecular markers of an EMT in MCF10A cells. Cells were infected with non-targeting shRNAs or shRNAs targeting RASAL2 and DAB2IP. Addition of MEK or PI3K inhibitors had no effect on the induction of an EMT. Lysates were run in duplicate. DAB2IP, RASAL2, pAKT and pERK were probed for on the same gel with the loading control Tubulin (1), while E-cadherin, N-cadherin and Slug were probed for separately with the loading control Tubulin (2). F, Schematic of DAB2IP’s functional domains. The RasGAP domain allows DAB2IP to negatively regulate Ras while the Period-like (Per-like) domain allows DAB2IP to negatively regulate NF-κB through a direct interaction with TRAF2. G, NF-κB activity reported as relative light units +/− SD. MCF10A cells were infected with non-targeting control shRNAs (CNT) or shRNAs targeting RASAL2 (R(1)) and DAB2IP (D(1)) alone or together and NF-κB activity was measured using a reporter assay. While suppression of DAB2IP alone significantly induced NF-κB activity compared to the control (p=0.0025, t-test), suppression of both RASAL2 and DAB2IP even further induced NF-κB activity (p=0.04, t-test shCNT/shDAB2IP vs shRASAL2/shDAB2IP). H, NF-κB activity reported as relative light units +/− SD. MCF10A cells were infected with non-targeting control shRNAs or shRNAs targeting RASAL2/DAB2IP. Subsequently cells were infected with control cDNA (EGFP and LacZ), wild-type RASAL2/DAB2IP cDNA, GAP point mutant cDNA (DAB2IPR413L/RASAL2K417E) and DAB2IP double mutant/RASAL2 GAP point mutant cDNA (DAB2IPR413L/S728A/RASAL2K417E). Data was reported as average +/− SD. While expression of wild-type RASAL2/DAB2IP and expression of the GAP mutants rescued NF-κB activation, the DAB2IP double mutant failed to suppress NF-κB activity.
In addition to its catalytic RasGAP domain, DAB2IP is unique among RasGAPs in that it also possesses a period-like domain, which has been shown to directly bind TRAF-2 and suppress NF-κB signaling (Fig. 4F, (20,32)). Therefore the effect of DAB2IP and/or RASAL2 loss on NF-κB activation in mammary epithelial cells was examined using an NF-κB transcriptional reporter assay. While RASAL2 ablation did not affect NF-κB activation, DAB2IP suppression significantly stimulated NF-κB activity (Fig. 4G). Interestingly, the concomitant loss of RASAL2 further enhanced NF-κB activation (Fig. 4G). This finding is consistent with the observation that the Ras pathway can converge with and enhance NF-κB activation in other cell types ((33,34)). Nevertheless, in this setting NF-κB activation first requires DAB2IP loss, which appears to function as a dominant suppressive signal. NF-κB activity can only then be further enhanced by the loss of a second RasGAP, RASAL2.
While the period-like domain of DAB2IP has been shown to be critical for NF-κB regulation in prostate cancer (20), we also assessed its importance in this setting. Notably, reconstitution with either wild-type RASAL2 and DAB2IP, or catalytically inactive RasGAP point mutants (DAB2IPR413L and RASAL2K417E) effectively restored NF-κB activity to baseline levels in DAB2IP/RASAL2-depleted cells (Fig. 4H), supporting the conclusion that Ras pathway activation is not sufficient to induce NF-κB in these cells. However, when a deleterious point mutation in the period-like domain of DAB2IP (DAB2IPR413L/S728A) was also included (32), NF-κB activity remained elevated (Fig. 4H). Taken together, these data demonstrate that the concomitant loss of DAB2IP and RASAL2 triggers a potent activation of both Ras and NF-κB in breast cancers, and illustrate the importance of the RasGAP and period-like domains in regulating these pathways, respectively.
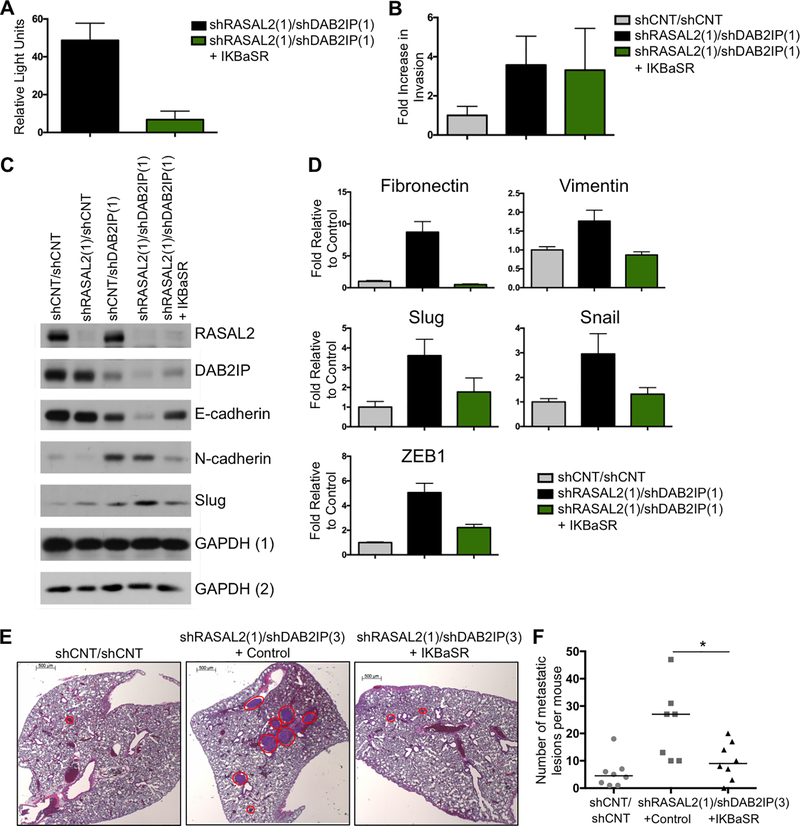
To investigate the contribution of NF-κB activation to invasion, EMT, and metastasis triggered by DAB2IP and RASAL2 loss, we utilized an IKBα super-repressor (IKBα-SR), which effectively suppresses NF-κB activation in DAB2IP/RASAL2-deficient cells (Fig. 5A) (35). In contrast to Ras pathway inhibitors, IKBα suppression had no effect on the invasive properties of DAB2IP/RASAL2-deficient cells (Fig. 5B). However it potently blocked the suppression of E-cadherin and the induction of N-cadherin (Fig. 5C) and dramatically inhibited the upregulation of all other EMT markers, triggered by DAB2IP and RASAL2 loss (Fig. 5D). Finally, to determine whether NF-κB activation was required for metastasis in vivo, we evaluated its effects in the luminal tumor model shown in Figure 3G. While the combined inactivation of DAB2IP and RASAL2 triggered metastasis in this model, co-expression of the IKBα-SR significantly suppressed the development of metastatic lesions (Fig. 5E and F, p=0.037 Mann-Whitney). Taken together these data suggest that DAB2IP and RASAL2 inactivation drive the metastasis of luminal breast cancers by potently activating the Ras and NF-κB pathways. These data further suggest that while the Ras pathway primarily regulates invasion, NF-κB drives EMT and is required for metastasis, thus providing a unique mechanism of cooperativity between these two tumor suppressors (Fig. 6).
Figure 5. NF-κB activation is required for EMT and metastasis.
A, NF-κB activity, reported as relative light units +/− SD, from MCF10A cells infected with shRNAs targeting RASAL2 and DAB2IP and co-infected with either vector control or the IKBα−SR. Co-infection with the IKBα−SR completely ablates NF-κB activity. B, Transwell invasion of MCF10A cells infected with control shRNAs or shRNAs targeting RASAL2 and DAB2IP. Invasion was measured after 24 hours and reported as average +/− SD Suppression of NF-κB activity (co-infection of the IKBα−SR) had no effect on invasion. C, Western blot showing the expression of the EMT markers E-cadherin and N-cadherin in MCF10A cells that were infected with control shRNAs or shRNAs targeting RASAL2 and/or DAB2IP in the absence or presence of the IKBα−SR. Lysates were run in duplicate. RASAL2, N-cadherin and Slug were probed for on the same gel with the loading control GAPDH (1), while DAB2IP and E-cadherin were probed for separately with the loading control GAPDH (2). D, Real-time PCR quantification of Fibronectin, Vimentin, Slug, Snail, and ZEB1 in MCF10A cells in response to RASAL2 and DAB2IP suppression with and without the IKBα−SR. Data show average relative amount of mRNA +/− SD. E, Representative H&E images of lungs, with McNeu metastases marked by red circles. McNeu cells were infected with non-targeting control shRNAs or shRNAs targeting RASAL2 and DAB2IP with and without the IKBα−SR. Cells were injected into tail veins of syngeneic mice, lungs were harvested three weeks post injection and the number of metastases per mouse was quantified. Scale bars correspond to 500μm. F, Co-infection of the IKBα−SR significantly suppressed metastasis formation (p=0.037, Mann-Whitney). Horizontal bars indicate the median number of metastatic lesions per mouse. Note: The data indicated in grey is a duplicate from Figure 3G.
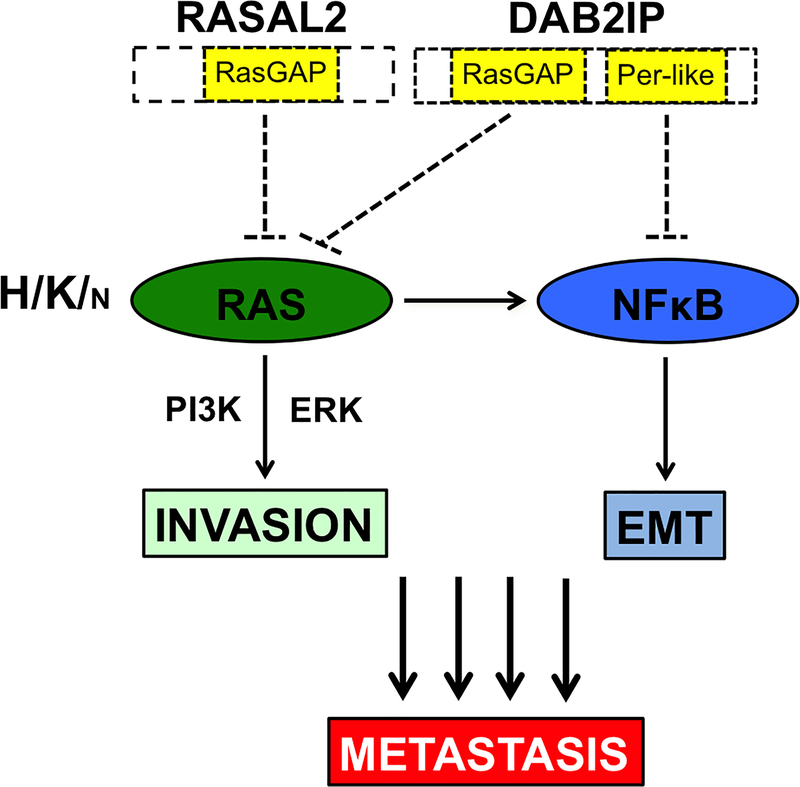
Figure 6. DAB2IP and RASAL2 Cooperate to Drive Distinct Aspects of Metastasis Through Ras and NF-κB.
Cartoon depicting the mechanism by which RASAL2 and DAB2IP regulate invasion, EMT, and metastasis in breast cancer. DAB2IP and RASAL2 both possess catalytic RasGAP domains. Accordingly, loss of RASAL2 and DAB2IP together potently activate all three major Ras isoforms and downstream effectors (although when all Ras isoforms are expressed they appear to exert more potent effects on K- and H-Ras). Loss of RASAL2 and DAB2IP also potently activates NF-κB. NF-κB activation requires the loss of DAB2IP, which directly affects the NF-κB pathway through its period-like domain, however NF-κB activity is further enhanced by Ras pathway activation. Our studies further suggest that while Ras activation drives invasion, NF-κB is required for EMT and metastasis. We hypothesize that it is the combined and potent activation of these two important signaling pathways that underlies the aggressive and metastatic nature of luminal B breast cancers that have lost both DAB2IP and RASAL2.
Discussion
Luminal tumors comprise approximately 75% of all breast cancers and many patients are cured by surgery and/or anti-estrogen therapies (1). However, a subset of luminal cancers, namely luminal B tumors, are highly proliferative, metastatic and frequently recur. Transcriptional, genomic, and epigenetic profiling studies demonstrate that the more aggressive luminal B tumors are distinct from the less aggressive luminal A tumors (3–5). However, the genetic or epigenetic drivers of this aggressive behavior are still largely unknown (9). This lack of insight impedes both treatment decisions and therapeutic development.
In this study we show that two RasGAP tumor suppressors, DAB2IP and RASAL2, play an important role in the pathogenesis of luminal B tumors. Strikingly, up to 62% of luminal B cancers have lost at least one of these genes. However, we find that tumors that have lost both genes frequently present as advanced disease and are more likely to recur. Importantly, we report that DAB2IP and RASAL2 can individually function as tumor suppressors in breast cancer, and when suppressed promote primary tumor growth in animal models. However, the loss of both genes triggers the potent activation of multiple Ras isoforms, the Ras effectors ERK and AKT, as well as NF-κB. We show that NF-κB is primarily activated by DAB2IP loss, which directly regulates the NF-κB pathway through its period-like domain; however, this signal is further enhanced by the dramatic activation of Ras, caused by the inactivation of both RasGAPs. Our data further suggest that both the Ras and NF-κB pathways play important and distinct roles in invasion, EMT, and metastasis. Taken together these studies reveal a mechanism by which these two major oncogenic pathways are activated in luminal B breast cancers and cooperate to drive their progression, which has been a longstanding unanswered question in the field (9).
In addition to providing insight into the pathogenesis of luminal B cancers, this finding may also have important clinical implications. Currently, breast cancers are not functionally classified as luminal A or luminal B tumors in the clinic and all ER+ luminal tumors are often treated as ER-responsive cancers (9). While clinical tests have been developed to assess the risk of recurrence, better prognostic markers are urgently needed to identify patients who are at the highest risk, in order to more accurately determine who should receive (or be spared) from escalated treatments such as adjuvant chemotherapy (2). Given that DAB2IP and RASAL2 play a direct causal role in driving metastasis and are concurrently lost in almost a quarter of luminal B tumors, our data suggest that DAB2IP and RASAL2 loss should be evaluated as potential predictive biomarkers. The antibodies used and validated in this study may be valuable tools in this context. Moreover, analysis could be coupled with the assessment of downstream effectors such as NF-κB, AKT/mTOR, or ERK, all of which have been implicated in disease progression (9,17,36). If successful, these efforts could complement or improve current tests, which could have a significant impact on treatment decisions.
Finally, the mechanism of epigenetically silencing two related tumor suppressors in cancer may have interesting teleological implications. Notably, the potent activation of Ras through the acquisition of an activating point mutation in a RAS gene directly requires a single genetic event. This may be advantageous for some tumor types in which a burst of Ras activity provides a growth or survival advantage. However despite the frequent activation of the Ras pathway in advanced tumors, RAS mutations are notably rare in breast cancer, raising the intriguing possibility that there may be selection against such an event. This could be due to enhanced sensitivity to a strong oncogenic signal, which might result in replicative stress, senescence, or some other growth inhibitory response. Thus, the sequential epigenetic silencing of two genes might provide an alternative mechanism of Ras activation, perhaps resulting in a more gradual, progressive activation of the Ras pathway during tumor evolution. The direct convergence of the Ras and NF-κB pathways via DAB2IP, also provides a means of further expanding and coupling these important oncogenic signals. Regardless, the studies presented here provide important mechanistic insight into this poorly understood tumor type and provide a framework for investigating this gene family, and more broadly this mechanism of inactivation, in other cancers.
Methods
Cell Culture and DNA Constructs
MCF7 cells were purchased from ATCC in 2013. BT549, MDA-MB-231, MDA-MB-453, SKBR3, and T47D cells were obtained from Dr. William Hahn (Dana-Farber Cancer Institute) in 2011. BT474, SUM149PT, SUM159PT, and ZR-75–1 cells were obtained from Dr. Frank McCormick in 2011 (University of California San Francisco). CAMA1 cells were obtained from Dr. Marcia Haigis (Harvard Medical School) in 2010. MDA-MB-361 cells were provided by Dr. Charlotte Kupperwasser in 2012 (Tufts Medical Center) and MCF10A cells were provided by Dr. Jayanta Debnath (University of California San Francisco) in 2013. All human breast cancer cell lines were authenticated in August 2016 by the Molecular Diagnostics Laboratory at the Dana-Farber Cancer Institute according to an American National Standards Institute (ANSI) recommended protocol. Briefly, cell lines were authenticated using the Promega GenePrint® 10 system, amplified DNA fragments were resolved on the Applied Biosystems 3130xl Genetic Analyzer and were analyzed with Applied Biosystems GeneMapper v 4.0 software. MCNeu cells were obtained from Dr. Sandra McAllister in 2015 (Brigham and Women’s Hospital) and were originally established by Dr. Laura Esserman (University of California San Francisco, (28)).
shRNAs were purchased from the RNAi Consortium (Broad Institute, MIT) with the following sequences: RASAL2 shRNA1 (5’-CCCTCGTGTTCTTGCTGATAT-3’), RASAL2 shRNA2 (3’-UTR shRNA) (5’-ATGGAGTGCAATAGGACATTG-3’), DAB2IP shRNA1 (3’-UTR shRNA) (5’- GTAATGTAACTATCTCACCTA −3’), DAB2IP shRNA2 (5’- AGGGATAGGCTAAGGAGTAAG −3’), DAB2IP shRNA3 (5’- GCACATCACTAACCACTACCT-3’). A control shRNA was purchased from Addgene (5’-CCTAAGGTTAAGTCGCCCTCG-3’). siControl and siDAB2IP were purchased from Dharmacon (ON-TARGETplus Non-targeting Control Pool, cat. # D-001810–10, and SMARTpool HUMAN siGENOME DAB2IP, cat. # M-008249–01-0005: 5’-CGCAGUUGUUAGAAGACGA-3’ / 5’-GGCUAAGGAGUAAGGACGA-3’ / 5’-GGACCAACAUGCAGCGCUU-3’ / 5’-GAUAGAUUUCACCCGGUUA-3’). The CRISPR guides (DAB2IP guide-1 (5’- TGGGATGCACCGCTCGCCGGagg-3’) and DAB2IP guide-2 (5’- GATGGAGAACCTCCGGCGAGcgg-3’)) were cloned into the lentiviral Cas9-containing pXPR-BRD001 construct obtained from Dr. William Hahn (Dana-Farber Cancer Institute). The Control pXBR-BRD001 vector contained no guideRNA.
The RASAL2 and RASAL2K417E (GAP point mutant) cDNA in the pHAGE-N-Flag-HA lentiviral expression vector was previously described (21). The wild-type, R413L GAP point mutant, and R413L/S728A double mutant (GAP point mutant and Per-domain point mutant) DAB2IP cDNA, previously described in Minn et al. (20), was cloned into the pLX-C-V5 lentiviral expression vector (obtained from Dr. William Hahn (Dana-Farber Cancer Institute)) and the pFB retroviral expression vector (generously provided by Pablo Viciana-Rodriguez). The pBABE IKBα−SR construct was obtained from Dr. William Hahn (Dana-Farber Cancer Institute).
Infections and Transfections
ShRNA or cDNA constructs were prepared and virus was harvested as previously described (21). Virus was incubated on target cells for 6–12 hours at a 1:2–1:10 dilution with 8μg/ml polybrene. Infected cells were selected in 0.5–2.0μg/ml puromycin, 400–800 μg/mL neomycin or 1–2μg/mL blasticidin, depending on the construct and optimized for each cell line. For transfections, cells were transfected for six hours with 10μM siRNA constructs using a 1:400 dilution of Lipofectamine RNAiMAX (Invitrogen, cat. # 13778–075) in antibiotic-free media.
Proliferation and Soft Agar Assays
For proliferation assays 150,000 cells were plated in triplicate in 6-well dishes. The following day cells were counted for day 0, and then triplicate sets of cells were counted on day 3. Cells for soft agar assays were plated in six biological replicates in 6-well dishes using the appropriate growth media, cell number and agarose concentration as previously described (21).
Protein Lysates and Western Blot Analyses
Protein extracts were isolated from cells in 1% SDS boiling lysis buffer. Ras-GTP levels were determined using a Ras Activation Assay kit (EMD Millipore cat. # 17–218). The following antibodies were used for immunoblots: phospho-AKT (Ser473, Cell Signaling cat. # 4060), AKT (Cell Signaling cat. # 9272), DAB2IP (Abcam cat. # 87811), E-cadherin (BD Transduction Laboratories cat. # 610181) phospho-ERK (Thr202/Thr204, Cell Signaling cat. # 4370), ERK (Cell Signaling cat. # 9102), GAPDH (Cell Signaling cat. # 2118), HA (Covance cat. # MMS-101P), N-cadherin (BD Transduction Laboratories cat. # 610920), HRas (Santa Cruz cat. # SC-520), KRas (Santa Cruz cat. # SC-30), NRas (Santa Cruz cat. # SC-519), RASAL2 (GeneTex cat. # 120989), Slug (Cell Signaling cat. # 9585), α-Tubulin (Sigma cat. # T5168), β-Tubulin (Sigma cat. # T4026), V-5 (Life Technologies cat. # 46–0705), Vinculin (Cell Signaling cat. # 4650). For Ras-GTP assays and analysis of EMT markers, cells were typically serum-starved in 1% serum for 36 hours. Ras-GTP levels were assessed by Western Blot and quantified using ImageJ software. For inhibitor experiments, drugs were added to the cells (PD-0325901 (MEK inhibitor) was used at 500nM and GDC-0941 (PI3K inhibitor) was used at 750nM) and lysates were harvested 48 hours post drug addition.
For the DAB2IP/RASAL2 protein/mRNA correlation analaysis, mRNA levels were assessed as described in the ‘Quantitative real-time PCR’ section and samples were normalized to the internal control glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Protein levels were assessed by Western Blot, quantified using ImageJ software and samples were normalized to GAPDH. Following normalization to GAPDH, mRNA and protein levels were normalized to DAB2IP/RASAL2 levels in normal mammary epithelial cells (MCF10As). The correlation analysis was performed in PRISM.
Patient-derived Xenograft Analysis
Patient-derived xenografts were produced as previously described by DeRose et al. (HCI-003–005, and HCI-011) (37) and Sikora et al. (HCI-013) (38). Tumors were harvested and flash-frozen. The frozen tissue was homogenized and protein extracts were isolated in 1% SDS boiling lysis buffer.
Molecular Subtype Association, Mutational, Survival, and Expression Analysis
Human expression (RNAseq, IlluminaHiSeq) and DNA methylation (HumanMethylation 450K) TCGA breast invasive carcinoma data was analyzed using the UCSC TCGA genome browser (39). For statistical analyses, the expression data was downloaded and statistical test were performed in PRISM. Mutational data was downloaded from the cBio portal (13,14). All survival curves were generated using the Kaplan-Meier Plotter browser (40) using the following probes: DAB2IP-225020_at; RASAL2–217201_at).
The frequency of DAB2IP and RASAL2 suppression in human breast cancers was determined using the TCGA breast invasive carcinoma human expression data set (RNAseq, IlluminaHiSeq) and defining the lowest 25% expressing samples across all subtypes as DAB2IP/RASAL2 ‘low’. Conversely, ‘high’ expression was defined as the highest 25% expressing samples across all subtypes. Based on this threshold, luminal B tumors were analyzed to assess the percentage of luminal B tumors that had low RASAL2 and/or DAB2IP expression. The TCGA clinical classifications of the same data set were analyzed and stage annotations of ER+ tumors with ‘co-low’ versus ‘co-high’ expression (‘low’ and ‘high’ defined as described above) were compared (Fisher Exact test).
Immunohistochemistry and Tissue Microarrays
Human breast tissue samples were acquired and tissue microarrays were constructed as previously described (26). Institutional review board (IRB) approval for the study was originally obtained through the Dana-Farber/ Harvard Cancer Center IRB; the Newton Wellesley Hospital IRB; the University of Colorado, Colorado Multiple Institutional Review Board; the Sunnybrook Health Sciences Center research ethics board; and the Mayo Clinic IRB. The study was conducted in accordance with the Declaration of Helsinki and every patient participating in the study provided written informed consent. Luminal B tumors were classified as previously described (26). TMAs were sectioned onto ProbeOn Plus microscope slides (Fisher Scientific) for immunohistochemistry. Briefly, tissues were dewaxed and washed in 100% EtOH. Antigen retrieval was performed with Citrate buffer pH6 for 6 minutes full power and 6 minutes medium power in a microwave. Samples were then treated with Redusol (Biomeda Corp., Foster City, CA) for 2 minutes at 40o C, incubated with fresh H2O2/MeOH (1% vol/vol) for 2 minutes at 40o C, blocked with appropriate serum for 5 minutes at 40o C, incubated with appropriate antibodies at 4o C o/n, washed, and then incubated with secondary antibodies for 15 minutes at 40o C. Slides were treated by washing with peroxidase enhancer and incubated with AEC chromogen (Biomeda) for 2 minutes at 40o C. All washes were performed with Automation Buffer (Biomeda). Cell nuclei were counterstained with aqueous hematoxylin QS (Vector Laboratories, Burlingame, CA). Slides were mounted with Faramount (Dako Corp.). Antibodies used for staining tissue sections were: DAB2IP (1:200; abcam87811), and RASAL2 (1:200; GTX120989). Vectastain Elite ABC system kits was used (Vector Laboratories). Images were captured under indicated magnification with identical exposure and gain for any given experiment using a Nikon Eclipse 90i microscope. We evaluated immunostaining blinded to clinicopathological information and scored the stains as negative or positive. We used the average value from replicate cores for correlation with clinical outcome.
In vitro Invasion Assays
BioCoat Matrigel Invasion Chambers (BD cat. # 354480) were prepared according to manufacturer’s instructions. 50,000 MCF10A cells were plated in triplicate in assay media (described by Debnath et al., (41)) in the top chamber of the invasion inserts. Assay media with 20ng/mL EGF was added to the bottom and cells were incubated over night. After incubation, cells were fixed and stained using the Diff Stain Kit (IMEB Inc cat. # K7128) according to the manufacturer’s instructions. The number of cells that had invaded through the matrigel was quantified by counting four random distinct fields using a light microscope. For CAMA1 cells, 100,000 cells were plated per insert in serum-free media and full serum (10% FBS) was added to the bottom as the chemoattractant. For inhibitor experiments, drugs were added to the top and bottom of the invasion inserts. PD-0325901 (MEK inhibitor) was used at 500nM and GDC-0941 (PI3K inhibitor) was used at 750nM.
Xenograft Assays
All animal procedures were approved by the Center for Animal and Comparative Medicine at Harvard Medical School in accordance with the NIH Guildelines for the Care and Use of Laboratory Animals and the Animal Welfare Act.
Female NOD/SCID mice were purchased from Charles River Laboratories (cat. # 394) and CAMA1 cells (3×106 cells, 50% matrigel from BD Biosciences cat. # 354234) were injected subcutaneously. Tumor size was measured by caliper and tumor volume was calculated using the formula volume = (length × width2) × π/6.
For the MDA-MB-316 metastasis experiment, the pLenti PGK V5 Luc vector (Addgene #21471) was introduced into MDA-MB-316 cells and cells were selected with G418. 1×106 cells were injected intracardically into NOD/SCID mice (Jackson Laboratories cat. # 001303) using an ultrasound guidance device (VEVO 200). Mice were injected with 150 mg of luciferin intra-peritoneally and imaged monthly using a Perkin Elmer IVIS Spectrum CT Biophotonic Imager. Bioluminescence was measured as total flux in photons per second and a common scale was used for all images across the study.
For the MCNeu metastasis experiment, transgenic FVB/N-Tg (MMTVneu) mice were purchased from Jackson Laboratory (cat. # 002376) and 3×105 cells (establizhed previously (28)) were injected into the tail vein. Lungs were harvested three weeks post injection, fixed in buffered formalin, stored in 70% ethanol, paraffin embedded, and sectioned. Sections were stained with hematoxylin and eosin and metastases were quantified by counting all metastases per lung in each animal using a light microscope.
Nuclear factor-κB reporter assays.
2 × 104 cells were seeded in triplicate in 24-well plates. After 18 hours, cells were transfected with 100ng of pNF-κB–luciferase (BD Bioscience) and 25ng Renilla luciferase–encoding plasmid (pRL-TK) with X-tremeGENE 9 DNA Transfection Reagent (Roche cat. # 06365809001). 36 hours after transfection, luciferase activity was measured followed by Renilla luciferase activity using the Dual Luciferase Reporter Assay Kit (Promega cat. # E-1910). All data were normalized as relative luciferase light units / Renilla luciferase light units.
Quantitative real-time PCR
Total RNA was isolated using Trizol (Invitrogen) and was subsequently Dnase-treated (Roche). cDNA was synthesized using the Quanta qScript kit (VWR cat. # 101414–098). Standard curves were run with each primer set and sample reactions were run in triplicate using PerfeCTA SYBR Green Supermix (Quanta Script cat. # 95071–250). Samples were normalized to the internal control glyceraldehyde 3-phosphate dehydrogenase. The primer sequences used were as follows:
Ecadherin
5’ TGCCCAGAAAATGAAAAAGG
3’ GTGTATGTGGCAATGCGTTC
Fibronectin
5’ CAGTGGGAGACCTCGAGAAG
3’ TCCCTCGGAACATCAGAAAC
GAPDH
5’ CATGTTCGTCATGGGTGTGAACCA
3’ ATGGCATGGACTGTGGTCATGAGT
Ncadherin
5’ ACAGTGGCCACCTACAAAGG
3’ CCGAGATGGGGTTGATAATG
Slug
5’ GGGGAGAAGCCTTTTTCTTG
3’ TCCTCATGTTTGTGCAGGAG
Snail
5’ CTGGGTGCCCTCAAGATGCA
3’ CCGGACATGGCCTTGTAGCA
Vim
3’ GCTTCCTGTAGGTGGCAATC
5’ GAGAACTTTGCCGTTGAAGC
ZEB1
3’ TGGTGATGCTGAAAGAGACG
5’ TGCACTGAGTGTGGAAAAGC
RASAL2
3’ ACCCTCTTTGCTCGTACAAC
5’ GGGATGTTGACTAGCCCTAC
DAB2IP
3’ CGCGCTACCAAACCATCAC
5’ ATCATCAGGTCTGTCAGGAAGT
Bisulfite Sequencing
Genomic DNA was isolated using a Qiagen kit (cat. # 69504) and the bisulfite conversion reaction was performed using a New England BioLabs kit (cat. # E3318S). The primers used for amplifying up the CpG region of interest were designed using the MethPrimer browser ((42)) and were as follows:
DAB2IP
5’ AGAATTCGGGATAGGTTTAAAGTTGTAGT
3’ AGAATTCCCTCTTAACCCCAAATAACTAT
RASAL2
5’ AGAATTCTGTTTTTTAGTAAAAGAATGGGATTG
3’ AGAATTCACAAACTCTTACCTAAAATCTACAT
The amplification PCR reaction was carried out using a kit from New England Biolabs (cat. # M0490) and the reaction was set up and run according to the manufacturer’s instructions. Following amplification, PCR products were send out for sequencing.
Supplementary Material
Statement of Significance.
The lack of insight into mechanisms that underlie the aggressive behavior of luminal B breast cancers impairs treatment decisions and therapeutic advances. Here we show that two RasGAP tumor suppressors are concomitantly lost in aggressive luminal B tumors and demonstrate that they drive metastasis by activating Ras and NF-κB.
Acknowledgements
We thank Ayana Henderson for technical assistance.
Financial Support: This work was supported by grants from the Ludwig Center at DF/HCC (K.C.) and NCI (R01 1R01CA188659, K.C.)
Footnotes
There are no conflicts of interest to disclose.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: A Cancer Journal for Clinicians. 2016;66:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Redig AJ, McAllister SS. Breast cancer as a systemic disease: a view of metastasis. J Intern Med. 2013;274:113–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours Nature. Nature Publishing Group; 2000;406:747–52. [DOI] [PubMed] [Google Scholar]
- 4.Sotiriou C, Neo S-Y, McShane LM, Korn EL, Long PM, Jazaeri A, et al. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003;100:10393–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, et al. A Comparison of PAM50 Intrinsic Subtyping with Immunohistochemistry and Clinical Prognostic Factors in Tamoxifen-Treated Estrogen Receptor-Positive Breast Cancer. Clin Cancer Res. 2010;16:5222–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran B, Bedard PL. Luminal-B breast cancer and novel therapeutic targets. Breast Cancer Res. 2011;13:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis MJ, Perou CM. The genomic landscape of breast cancer as a therapeutic roadmap. Cancer Discovery. 2013;3:27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koboldt DC, Fulton RS, McLellan MD, Schmidt H, Kalicki-Veizer J, McMichael JF, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J, Aksoy BA, Dogrusoz U, Dresdner G. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;269:pl1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discovery. 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sivaraman VS, Wang H, Nuovo GJ, Malbon CC. Hyperexpression of mitogen-activated protein kinase in human breast cancer. J Clin Invest. 1997;99:1478–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lintig von FC, Dreilinger AD, Varki NM, Wallace AM, Casteel DE, Boss GR. Ras activation in human breast cancer. Breast Cancer Res Treat. 2000;62:51–62. [DOI] [PubMed] [Google Scholar]
- 17.Mueller H, Flury N, Eppenberger-Castori S, Kueng W, David F, Eppenberger U. Potential prognostic value of mitogen-activated protein kinase activity for disease-free survival of primary breast cancer patients. Int J Cancer. 2000;89:384–8. [DOI] [PubMed] [Google Scholar]
- 18.Bernards A GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim Biophys Acta. 2003;1603:47–82. [DOI] [PubMed] [Google Scholar]
- 19.Maertens O, Cichowski K. An expanding role for RAS GTPase activating proteins (RAS GAPs) in cancer Advances in Biological Regulation. Elsevier Ltd; 2014;55:1–14. [DOI] [PubMed] [Google Scholar]
- 20.Min J, Zaslavsky A, Fedele G, McLaughlin SK, Reczek EE, De Raedt T, et al. An oncogene–tumor suppressor cascade drives metastatic prostate cancer by coordinately activating Ras and nuclear factor-κB. Nat Med. 2010;16:286–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLaughlin SK, Olsen SN, Dake B, De Raedt T, Lim E, Bronson RT, et al. The RasGAP gene, RASAL2, is a tumor and metastasis suppressor. Cancer Cell. 2013;24:365–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prat A, Karginova O, Parker JS, Fan C, He X, Bixby L, et al. Characterization of cell lines derived from breast cancers and normal mammary tissues for the study of the intrinsic molecular subtypes. Breast Cancer Res Treat. 2013;142:237–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subik K, Lee JF, Baxter L, Strzepek T. The expression patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by immunohistochemical analysis in breast cancer cell lines. Breast cancer (Auckl.). 2010;4:35–41 [PMC free article] [PubMed] [Google Scholar]
- 25.Holliday DL, Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011;13:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins LC, Gelber S, Marotti JD, White S, Ruddy K, Brachtel EF, et al. Molecular Phenotype of Breast Cancer According to Time Since Last Pregnancy in a Large Cohort of Young Women. The Oncologist. 2015;20:713–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaffer CL, Weinberg RA. A Perspective on Cancer Cell Metastasis. Science. 2011;331:1559–64. [DOI] [PubMed] [Google Scholar]
- 28.Campbell MJ, Wollish WS, Lobo M, Esserman LJ. Epithelial and fibroblast cell lines derived from a spontaneous mammary carcinoma in a MMTV/neu transgenic mouse. In Vitro Cell Dev Biol Anim. 2002;38:326–33. [DOI] [PubMed] [Google Scholar]
- 29.Shin S, Dimitri CA, Yoon S-O, Dowdle W, Blenis J. ERK2 but Not ERK1 Induces Epithelial-to-Mesenchymal Transformation via DEF Motif-Dependent Signaling Events. Molecular Cell. 2010;38:114–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Virtakoivu R, Pellinen T, Rantala JK, Perälä M, Ivaska J. Distinct roles of AKT isoforms in regulating β1-integrin activity, migration, and invasion in prostate cancer. Mol Biol Cell. 2012;23:3357–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chin YR, Toker A. Akt isoform-specific signaling in breast cancer. Cell Adhesion & Migration. 2014;5:211–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H AIP1/DAB2IP, a Novel Member of the Ras-GAP Family, Transduces TRAF2-induced ASK1-JNK Activation. Journal of Biological Chemistry. 2004;279:44955–65. [DOI] [PubMed] [Google Scholar]
- 33.Hutti JE, Pfefferle AD, Russell SC, Sircar M, Perou CM, Baldwin AS. Oncogenic PI3K Mutations Lead to NF- B-Dependent Cytokine Expression following Growth Factor Deprivation. Cancer Research. 2012;72:3260–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dhawan P, Richmond A. A Novel NF- B-inducing Kinase-MAPK Signaling Pathway Up-regulates NF- B Activity in Melanoma Cells. Journal of Biological Chemistry. 2002;277:7920–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, et al. Integrative Genomic Approaches Identify IKBKE as a Breast Cancer Oncogene. Cell. 2007;129:1065–79. [DOI] [PubMed] [Google Scholar]
- 36.Oida K, Matsuda A, Jung K, Xia Y, Jang H, Amagai Y, et al. Nuclear factor-ĸB plays a critical role in both intrinsic and acquired resistance against endocrine therapy in human breast cancer cells. Sci Rep. 2014;4:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.<b>DeRose YS, Wang G, Lin Y-C, Bernard PS, Buys SS, Ebbert MTW, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes Nat Med. Nature Publishing Group; 2011;17:1514–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sikora MJ, Cooper KL, Bahreini A, Luthra S, Wang G, Chandran UR, et al. Invasive lobular carcinoma cell lines are characterized by unique estrogen-mediated gene expression patterns and altered tamoxifen response. Cancer Res. 2014;74:1463–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu J, Sanborn JZ, Benz SC, Craft B, Szeto C. The UCSC Cancer Genomics Browser. Nat Methods. 2009;6:239–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2009;123:725–31. [DOI] [PubMed] [Google Scholar]
- 41.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–68. [DOI] [PubMed] [Google Scholar]
- 42.Li L-C, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.