Abstract
Rationale:
ER stress dysregulates ER proteostasis, which activates the transcription factor, ATF6, an inducer of genes that enhance protein folding and restore proteostasis. Due to increased protein synthesis, it is possible that protein folding and, thus, ER proteostasis are challenged during cardiac myocyte growth. However, it is not known whether ATF6 is activated, and if so, what its function is during hypertrophic growth of cardiac myocytes.
Objective:
To examine the activity and function of ATF6 during cardiac hypertrophy.
Methods and Results:
We found that ATF6 was activated and ATF6-target genes were induced in mice subjected to an acute model of trans-aortic constriction (TAC), or to free-wheel exercise, which promote adaptive cardiac myocyte hypertrophy with preserved cardiac function. Cardiac myocyte-specific deletion of Atf6 (ATF6 cKO) blunted TAC- and exercise-induced cardiac myocyte hypertrophy and impaired cardiac function, demonstrating a role for ATF6 in compensatory myocyte growth. Transcript profiling and chromatin immunoprecipitation identified RHEB as an ATF6-target gene in the heart. RHEB is an activator of mTORC1, a major inducer of protein synthesis and subsequent cell growth. Both TAC and exercise upregulated RHEB, activated mTORC1, and induced cardiac hypertrophy in WT mouse hearts, but not in ATF6 cKO hearts. Mechanistically, knockdown of ATF6 in neonatal rat ventricular myocytes blocked phenylephrine (PE)-, and insulin-like growth factor 1 (IGF1)-mediated Rheb induction, mTORC1 activation, and myocyte growth, all of which were restored by ectopic RHEB expression. Moreover, AAV9-RHEB restored cardiac growth to ATF6 cKO mice subjected to TAC. Finally, ATF6 induced RHEB in response to growth factors, but not in response to other activators of ATF6 that do not induce growth, indicating that ATF6 target gene induction is stress-specific.
Conclusions:
Compensatory cardiac hypertrophy activates ATF6, which induces Rheb and activates mTORC1. Thus, ATF6 is a previously unrecognized link between growth stimuli and mTORC1-mediated cardiac growth.
Keywords: Myocytes, cardiac protein folding, proteostasis, cardiac hypertrophy, ATF6, Rheb, mTORC1, ER stress, Basic Science Research
INTRODUCTION
Protein homeostasis, or proteostasis involves the coordination of protein synthesis and folding to ensure proteome integrity and vital cell function1. In cardiac myocytes the endoplasmic reticulum (ER) is a major site of synthesis of proteins that are critical for proper function of the heart, including many calcium-handling proteins, receptors, and secreted proteins, such as hormones, stem cell homing factors, and growth factors2, 3. Therefore, ER proteostasis maintains the integrity of the cardiac myocyte proteome and, thus, cardiac contractility. Increased protein synthesis in growing cardiac myocytes must be balanced by increased protein-folding to avoid the accumulation of toxic misfolded proteins; thus growth poses a potential challenge to cardiac myocyte proteostasis4. However, the molecular mechanisms underlying the maintenance of ER proteostasis during cardiac myocyte growth are not well understood.
ER proteostasis is controlled in all mammalian cells by several ER-transmembrane sensors of protein misfolding, including the adaptive transcription factor, ATF65. When protein synthesis surpasses the capacity of the protein-folding machinery, increases in misfolded proteins cause the translocation of the ER-transmembrane, 670-amino acid, 90 kD form of ATF6, to the Golgi, where it is clipped, liberating an N-terminal fragment that serves as a transcription factor. This 50 kD active form of ATF6 regulates a gene program that is responsible for the expression of numerous proteins that enhance ER protein folding, which adaptively restores the balance between protein synthesis and folding6, 7. Thus, nodal proteostasis regulators, such as ATF6, that sense and maintain this balance could play important roles in optimizing cardiac myocyte growth; however, neither the activation nor the function of ATF6 in the setting of hypertrophic cardiac growth has been examined. Accordingly, here we studied the effects of Atf6 deletion in mouse hearts and in cultured cardiac myocytes during physiologically-relevant maneuvers known to promote compensatory cardiac hypertrophy in either a concentric (pressure overload)8 or eccentric (exercise)9 manner, positing that the absence of ATF6 would imbalance proteostasis, which would be maladaptive.
Both growth maneuvers activated ATF6, and Atf6 deletion was maladaptive, as evidenced by impaired cardiac function. However, surprisingly, cardiac myocyte growth was also impaired upon Atf6 deletion. This was unexpected, since Atf6 is not known to be required for cardiac myocyte growth. Further mechanistic studies showed that ATF6 is activated as a result of the increased demands placed on the ER protein folding machinery during growth-related increases in protein synthesis. Moreover we found that Atf6 serves a previously unrecognized role as a molecular link between growth stimuli and activation of mammalian/mechanistic target of rapamycin complex 1 (mTORC1), a major promoter of protein synthesis and consequent growth of cardiac myocytes10–15. Two conditions need to be met for mTORC1 to be activated; 1) in response to a growth stimulus, mTORC1 needs to translocate to organelles, such as lysosomes16, where it encounters the small GTPase activator of mTORC1, Rheb17, and 2) Rheb must be active and present in sufficient quantities18. In terms of Rheb activation, it is known that growth stimuli lead to the phosphorylation and, thus, inhibition of the Rheb GTPase-activating protein (GAP), TSC1/TSC219, which increases the GTP-loading state and, thus, the activity of Rheb. However, the molecular mechanisms underlying the Rheb gene expression are less well understood. Here, we showed, for the first time, that ATF6 is an inducer of Rheb, and in this way, ATF6 coordinates protein synthesis and protein folding, ensuring the adaptive maintenance of proteostasis in growing cardiac myocytes. Thus, ATF6 is a newly identified and essential member of mTORC1 growth signaling in cardiac myocytes in the heart.
METHODS
All supporting data are available within the article. Further details on the Methods can be found in the Online Supplement.
Laboratory animals.
The research reported in this paper has been reviewed and approved by the SDSU Institutional Animal Care and Use Committee and it conforms to the Guide for the Care and Use of Laboratory Animals published by the National Research Council.
ATF6 floxed mice.
Atf6fl/fl mice used in this study were generated by Dr. Gokhan S. Hotamisligil20. All of the mice used in this study were 10 week-old males.
Statistics.
Unless otherwise stated, values shown are mean ± SEM and statistical treatments are either a t-test or a one-way ANOVA followed by Newman-Keuls post hoc analysis.
RESULTS
ATF6 is required for cardiac myocyte hypertrophy in response to pressure overload.
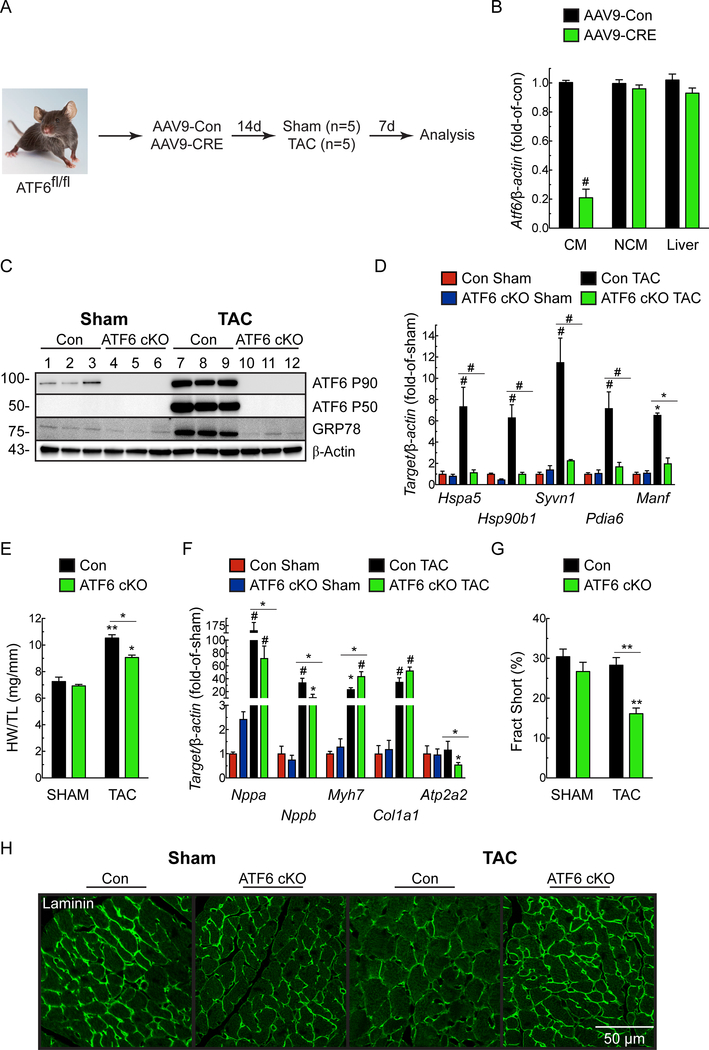
To examine the role of Atf6 in cardiac myocytes on heart growth, we generated an Atf6 conditional knockout mouse (ATF6 cKO) by injecting Atf6fl/fl mice with a recombinant AAV9 that encodes Cre under the control of the cardiac troponin T promoter (Fig. 1A). Compared to Atf6fl/fl injected with AAV9-Con, injection with AAV9-CRE effectively reduced Atf6 mRNA from cardiac myocytes isolated from Atf6fl/fl mice, but not non-cardiac myocytes, or liver (Fig. 1B). Atf6fl/fl mice injected with AAV9-Con (Con) or AAV9-CRE (ATF6 cKO), were subjected to TAC and examined 7d later, when hypertrophic growth is maximal21 and structural remodeling is compensatory22, 23. TAC activated ATF6 in Con mouse hearts, as evidenced by increased levels of the active, 50 kD form of ATF6 (Fig. 1C). This was unexpected, since ATF6 is not known to be activated in cardiac myocytes by any growth stimulus. Coordinate with ATF6 activation, TAC increased expression of numerous canonical ATF6 target genes (Fig. 1C-D; Online Fig. IA). As expected, ATF6 was undetectable in ATF6 cKO mouse hearts (Fig. 1C-D). TAC increased Con mouse heart weights, but, surprisingly, this growth effect was significantly blunted in ATF6 cKO mouse hearts (Fig. 1E). TAC increased Nppa and Nppb expression to similar extents in both Con and ATF6 cKO mice, while the induction of Myh7 and Col1a1 was slightly greater in the ATF6 cKO mice (Fig. 1F). This, coupled with the decrease in Atp2a2 i.e. SERCA2a in ATF6 cKO mice, suggests a blunted compensatory response in the absence of ATF6. In Con mouse hearts, cardiac function, including fractional shortening was preserved, while chamber dimensions were unchanged after TAC (Fig. 1G; Online Table I) and cardiac myocyte size was increased (Fig. 1H), consistent with the compensatory nature of cardiac hypertrophy in mice at this time after pressure overload24. However, in contrast to Con, in ATF6 cKO mice subjected to TAC myocyte size was decreased compared to Con (Fig. 1H) and fractional shortening was impaired (Fig. 1G) with increased chamber dimensions, such as LVEDV and LVESV, despite high frequency Doppler measurements between right and left carotid arteries demonstrating consistent and identical pressure overload in TAC-operated Con and ATF6 cKO mice (Online Table I). Along with increased plasma levels of cTnI (Online Fig. IB), these results are consistent with the initial stages of chamber dilation, as well as myocardial damage and decompensation in the ATF6 cKO mice. Thus, ATF6 is activated by pressure overload and is required for hypertrophy.
Figure 1. Effect of cardiac myocyte-specific ATF6 gene deletion in hearts of mice subjected to TAC.
A, Protocol for AAV9 administration to ATF6fl/fl mice and TAC. B, ATF6 mRNA levels determined by qRT-PCR on isolated cardiac myocytes (CM), non-cardiac myocytes (NCM), and liver extracts from ATF6fl/fl mice injected with AAV9-Con (Con) or AAV9-CRE (i.e. ATF6 cKO) n=3. C, Immunoblot of LV extracts from Con or ATF6 cKO mice. D, mRNA for ATF6 target genes determined by qRT-PCR. E, Heart weight/tibia lengths (HW/TL). F, mRNA levels for fetal genes determined by qRT-PCR. Nppa, natriuretic peptide A); Nppb, natriuretic peptide B); Myh7, β-myosin heavy chain; Col1a1, Collagen 1A1; Atp2a2, Serca2a. G, Fractional shortening (%), determined by echocardiography, see Online Table I. H, Confocal immunocytofluorescence microscopy (ICF) analysis of mouse heart sections for laminin (green). Data are mean ± SEM. *P≤0.05, **P≤0.01, #P≤0.001 different from Con Sham, or from the value shown by the bar.
ATF6 is required for cardiac myocyte hypertrophy in response to exercise.
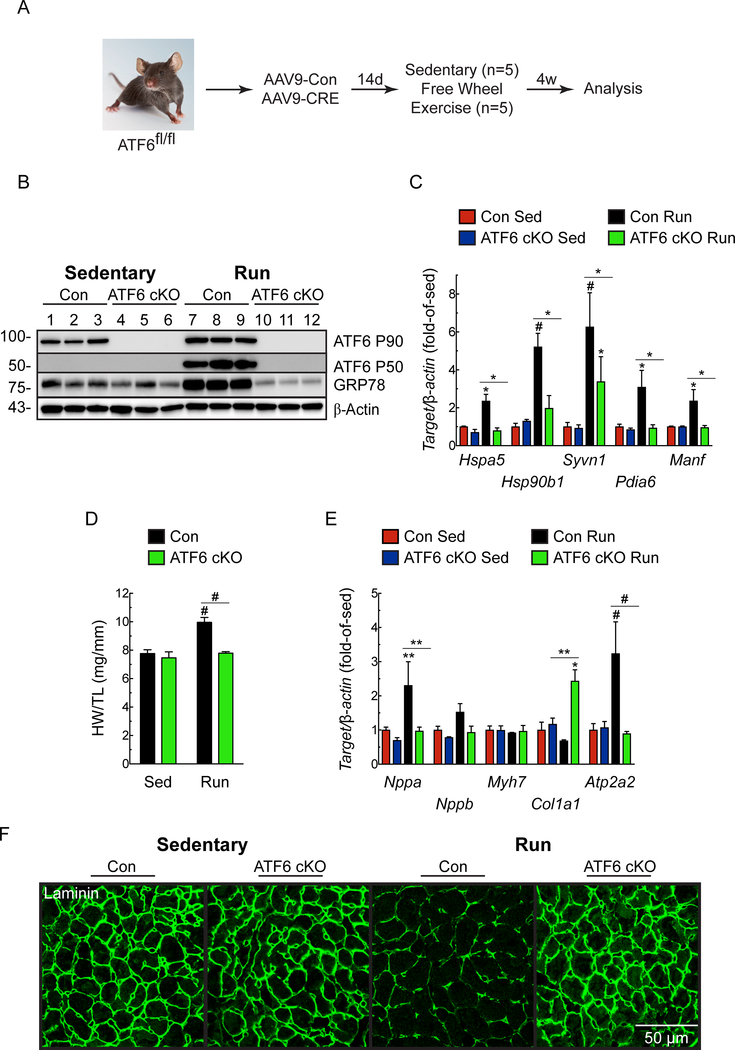
To assess the breadth of the impact of ATF6 on heart growth, we examined the effects of cardiac myocyte-specific ATF6 deletion in mice subjected to free-wheel exercise25, 26 (Fig. 2A). Similar to TAC, exercise surprisingly activated ATF6 and induced ATF6 target genes in Con, but not in ATF6 cKO mice (Fig. 2B-C). As expected, compared to Con sedentary mice, Con mice subjected to exercise exhibited increased heart weights and LV wall thickness, as well as myocyte size (Fig. 2D, 2F; Online Table II). While Nppa and Nppb were mildly increased, Atp2a2 was robustly increased by exercise in Con mouse hearts, and there was no change in Myh7 or Col1a1 (Fig. 2E); this gene profile is typical of adaptive cardiac hypertrophy in exercising mice24, 27. In contrast to Con, in ATF6 cKO mice subjected to exercise there was no change in heart weights or LV wall thickness (Fig. 2D; Online Table II), reduced increases in myocyte size (Fig. 2F), and reduced induction of ATF6 target genes (Fig. 2C). Compared to exercised Con mice, exercised ATF6 cKO mice showed no increase in Nppa, and neither Con nor ATF6 cKO mice showed significant changes in Nppb or Myh7. Importantly, while Con mice exhibited decreased Col1a1 and increased Atp2a2 after exercise, which are beneficial genetic changes typical of this regime, the ATF6 cKO mice failed to adapt and had increased Col1a1 and no change in Atp2a2 (Fig. 2E). Thus, ATF6 is activated by exercise and is required for compensatory hypertrophy in this exercise model.
Figure 2. Effect of cardiac myocyte-specific ATF6 gene deletion in hearts of mice subjected to free wheel exercise.
A, Protocol for AAV9 administration to ATF6fl/fl mice and free wheel exercise. B, Immunoblot of LV extracts from Con or ATF6 cKO mice. C, mRNA levels for ATF6 target genes determined by qRT-PCR. D, Heart weights/tibia lengths (HW/TL). E, mRNA levels for fetal genes determined by qRT-PCR. F, ICF analysis of mouse heart sections for laminin (green). Data are mean ± SEM. *P≤0.05, **P≤0.01, #P≤0.001. Echocardiography details are in Online Table II.
Rheb is an ATF6-inducible gene in the heart.
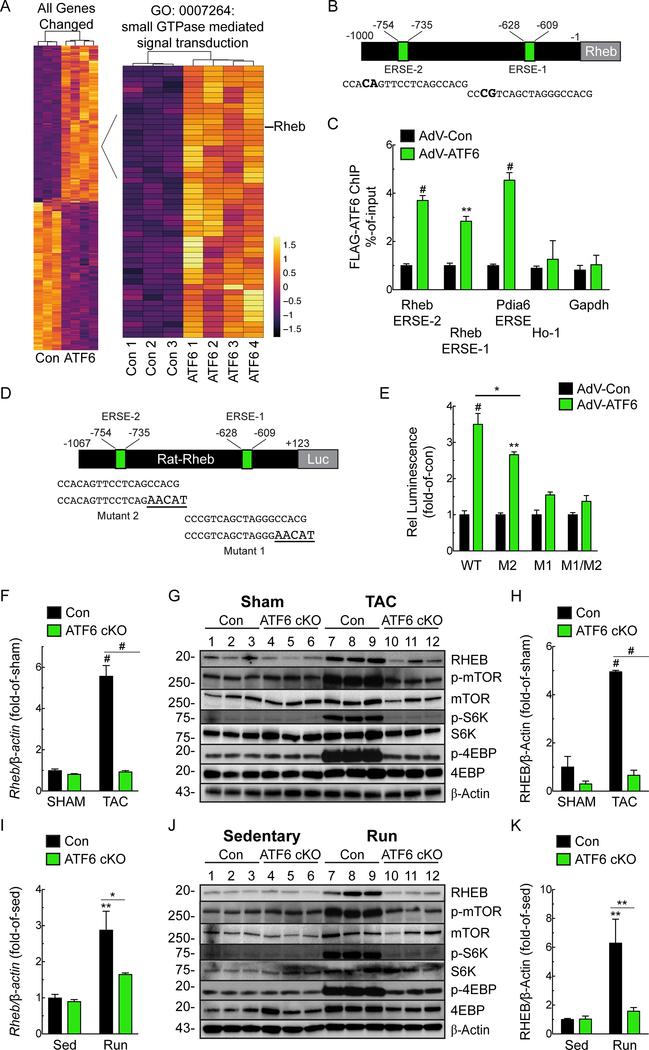
Since there are no known Atf6-inducible genes that are required for cardiac myocyte growth, we turned to transcript profiling for clues to the identities of such genes. RNA sequencing of the hearts of our previously published transgenic mice that express activated ATF628 (Online Table III) revealed that ATF6 induced 51 genes in the gene ontology category, small GTPase mediated signal transduction; this category includes the ras-related small GTPase, Ras homologue enriched in brain (RHEB) (Fig. 3A; Online Fig. IIA). Rheb is required for activation of mTORC1, however, only in the presence of a growth stimulus. Accordingly, we focused on Rheb as a candidate gene through which ATF6 might contribute to cardiac hypertrophy, pursuing the hypothesis that increased Rheb gene expression and subsequent mTORC1 activation under growth conditions are Atf6-dependent. The upregulation of RHEB by ATF6 in mouse hearts observed by RNA sequencing was confirmed by qRT-PCR (Online Fig. IIB). Consistent with ATF6 as a possible transcriptional inducer of Rheb was our finding that the Rheb promoter has two potential ATF6 binding sites, which we call ER stress response elements (ERSEs)-1 and −2 (Fig. 3B). Chromatin immunoprecipitation (ChIP) showed that ATF6 binds to both sites in the RHEB gene in neonatal rat ventricular myocytes (NRVM) (Fig. 3C). The progressive decline in RHEB promoter activity in plasmids that encode 5’-truncation deletions of the rat RHEB promoter driving luciferase demonstrated the importance of these putative ERSEs in ATF6-mediated RHEB promoter activation (Online Fig. IIC). To mechanistically investigate the functional involvement of these ERSEs further, we mutated either or both ERSE (Fig. 3D). Mutating either ERSE decreased ATF6 RHEB promoter activation by ATF6, however, the promoter-proximal site, i.e. ERSE-1 appeared to have the largest effect (Fig. 3E). To determine whether ATF6 is sufficient to induce Rheb in the heart, in vivo, mice were injected with a recombinant AAV9 that encodes activated ATF6, i.e. ATF6(1–373). qRT-PCR and immunoblotting demonstrated that activated ATF6 increased RHEB mRNA and protein in the heart (Online Fig. IID-F). These results are the first demonstration in any cell type that ATF6 induces RHEB, implicating ATF6 as a critical link between growth stimuli and mTORC1 activation.
Figure 3. Regulation of Rheb Expression by ATF6.
A, Heat map of transcript profiling showing z-score-transformed RPKM values (Reads Per Kilobase per Million mapped reads) with hierarchical clustering of transcripts of control and ATF6 transgenic mouse hearts. Differentially expressed genes with p values and FDR ≤0.05 and a subset of genes annotated with term GO:0007264 are shown. All the genes increased or decreased by ATF6 are in Online Table III. B, Locations of consensus ATF6-binding motifs, i.e. ER stress response elements 1 and 2 (ERSE-1 and 2) and their sequences in the RHEB gene 5’-flanking region. Nucleotide differences from canonical ERSE elements are bold. C, Neonatal rat ventricular myocytes (NRVM) were infected with AdV encoding control or FLAG-ATF6(1–373) [active form], and then ATF6 binding to endogenous ERSE-1 or ERSE-2, as well as to the endogenous PDIA6 ERSE, used here as a positive control, and the negative control targets heme oxygenase 1 (ho-1) and gapdh were examined by chromatin immunoprecipitation (ChIP) (n=3). D, Locations of ERSE-1 and 2 in the RHEB 5’-flanking region, their sequences (lower case), and the mutations that were made (bold and upper case). E, NRVM were transfected with rat-rheb(−1067/+123)-Luc WT, M2, M1 or M1/M2 then infected with AdV FLAG-ATF6(1–373),. Then, 48h later, luciferase activity was measured in extracts (n=6). F-H, mRNA for RHEB determined by qRT-PCR (F) and Rheb protein and mTOR pathway components measured by immunoblots (G) and quantified by densitometry (H) from Con or ATF6 cKO mouse heart extracts after 7 days of Sham or TAC. I-K, mRNA for RHEB determined by qRT-PCR (I) and Rheb protein and mTOR pathway components immunoblots (J) and quantified by densitometry (K) from Con or ATF6 cKO mouse heart extracts after 4 weeks of sedentary or free wheel exercise (Run). Data are mean ± SEM. *P≤0.05, **P≤0.01, #P≤0.001.
RHEB induction during pressure-overload and exercise requires ATF6.
We found that RHEB was strongly induced in Con mice after either TAC or exercise, but not in ATF6 cKO mouse hearts (Fig. 3F-K). Thus, ATF6 is necessary for the upregulation of RHEB in these models of cardiac hypertrophy, in vivo. Since RHEB is required for mTORC1 activation in response to a growth stimulus, we assessed mTORC1 pathway activation. As expected, pressure-overload and exercise both activated mTORC1, as shown by increased phosphorylation of mTORC1 (Ser2448), p70 ribosomal S6 kinase (S6K; Thr389), and eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1; Thr37/46); however, mTORC1 activation was blunted in ATF6 cKO mouse hearts (Fig. 3G, J), consistent with the key role for ATF6 in mTORC1 activation by growth stimuli. To examine whether ATF6 might affect other signaling pathways leading to mTORC1 activation, we assessed the phosphorylation of Akt on Ser308 and the phosphorylation of TSC2, both of which lie upstream of Rheb in the mTORC1 signaling pathway. We found that pressure overload increased phosphorylation of Akt (Thr308) and TSC2 (Thr1462), as expected; however, in contrast to Rheb expression, neither of these phosphorylation events were affected by ATF6 deletion (Online Fig. IIIA). Thus, the deficit in mTORC1 activation in ATF6 cKO mice must reside downstream of Akt and TSC2, i.e. Rheb. We also examined whether ATF6 deletion affected other well known canonical hypertrophy signaling pathways, but found that neither phosphorylation of Akt on Ser473, Erk phosphorylation (Online Fig. IIIA) or calcineurin activation (Online Fig. IIIB) were affected by ATF6 deletion. These results pinpoint the growth deficit in the ATF6 cKO mouse hearts to the inability to upregulate Rheb.
RHEB is required for PE- and IGF1-induced cardiac myocyte growth.
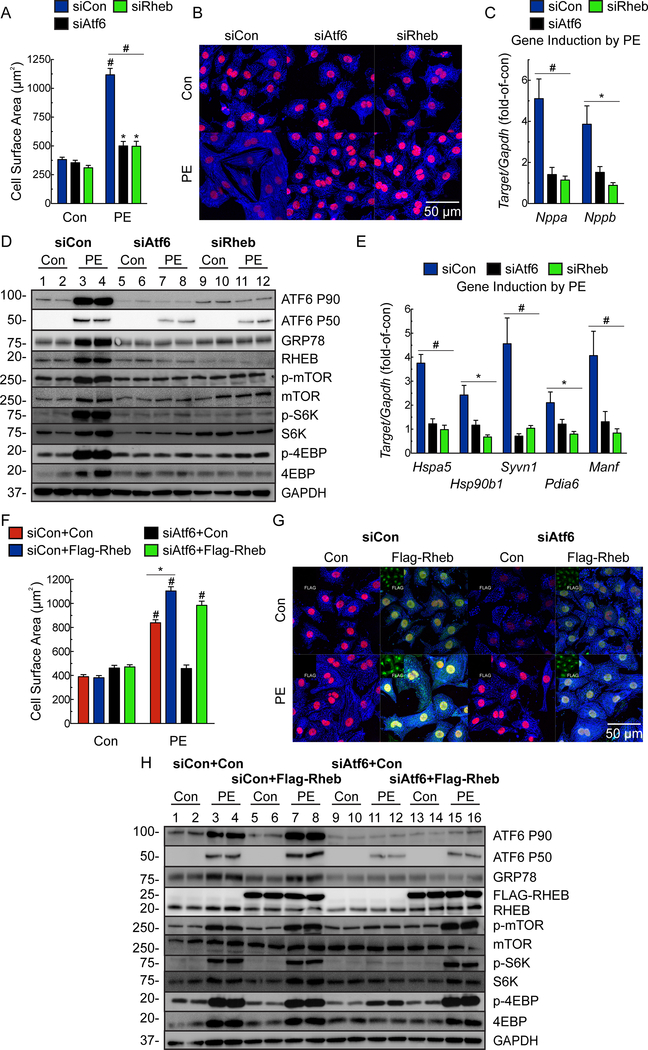
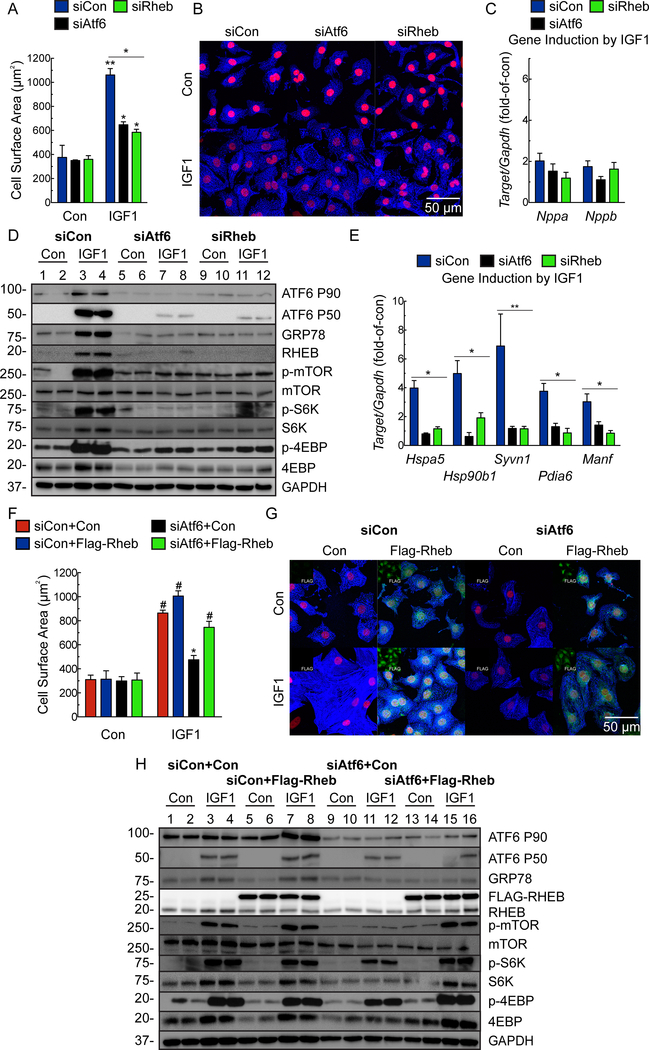
To explore the mechanistic relationship between ATF6 and RHEB we used RHEB and ATF6 loss-of-function approaches in NRVM treated with the α1-adrenergenic receptor agonist, phenylephrine (PE) or insulin-like growth factor 1 (IGF1), which recapitulate much of the intracellular signaling during pressure-overload or exercise-induced hypertrophy, respecitvely29. Knocking down either ATF6 or RHEB abrogated the effects of PE or IGF1 on cardiac myocyte hypertrophy, fetal gene induction, ATF6 target gene induction and mTORC1 signaling (Fig. 4A-E; Online Fig. IVA, IVC; Fig. 5A-E; Online Fig. IVB), but had no effect on mTORC2 signaling, as assessed by phosphorylation of Akt on Ser-473 (Online Fig. IVD-E). To further substantiate the results with Rheb siRNA, we used a different Rheb loss-of-function approach involving the Rheb inhibitor, Lonafarnib30. Lonafarnib mimicked the effects of Rheb siRNA on PE- and IGF1-mediated ATF6 activation, mTORC1 signaling, ATF6 gene induction and growth in NRVM (Online Fig. V).
Figure 4. Effects of ATF6- and RHEB knockdown and ectopic Rheb expression on phenylephrine-induced hypertrophy in cultured cardiac myocytes.
A-E, NRVM were transfected with a nontargeted siRNA (siCon) or with siRNAs targeted to either rat ATF6 (siAtf6) or RHEB (siRheb), and then treated ± phenylephrine (PE; 50μM) for 48 hours. A, Cell surface area was determined by photomicroscopy and morphometry (n=6). B, ICF of NRVM for α-actinin (blue) and TOPRO-3 (red). Bar = 50μm. C, qRT-PCR examination of Nppa and Nppb. Values are expressed as fold-of-control cardiac myocytes in the absence of PE (n=6). D, Immunoblot of NRVM. E, mRNA for ATF6 target genes determined by qRT-PCR. Values are expressed as fold-of-control myocytes in the absence of PE (n=3). F-H, NRVM were transfected with a control plasmid or a plasmid encoding Flag-Rheb and either siCon or siAtf6, followed by treatment ± PE for 48 hours. Cell surface area (F) was determined by morphometry after ICF (G). NRVM were stained for FLAG (green; isolated channel displayed in inset), α-actinin (blue), and TOPRO-3 (red). Bar = 50μm. Only FLAG-positive cells were used for cell surface area analysis (n=3). H, Immunoblot of NRVM. Data are mean ± SEM. *P≤0.05, **P≤0.01, #P≤0.001.
Figure 5. Effects of ATF6- and RHEB knockdown and ectopic Rheb expression on insulin like growth factor 1-induced hypertrophy in cultured cardiac myocytes.
A-E, NRVM were transfected with siCon, siAtf6 or siRheb, then treated ± IGF1 (100ng/ml) for 48 hours. A, Cell surface area was determined by was determined by morphometry after ICF (n=6). B, ICF of NRVM for α-actinin (blue) and TOPRO-3 (red). Bar = 50μm. C, qRT-PCR for Nppa and Nppb. Values are fold-of-control myocytes in the absence of IGF1 (n=6). D, Immunoblot of NRVM. E, mRNA levels of ATF6 target genes determined by qRT-PCR. Values are fold-of-control myocytes in the absence of IGF1 (n=3). F-H, NRVM were transfected with a control plasmid or a plasmid encoding Flag-Rheb and then either siCon or siAtf6, followed by treatment ± IGF1 for 48 hours. Cell surface area (F) was determined by morphometry after ICF (G). NRVM were stained for FLAG (green; isolated channel displayed in inset), α-actinin (blue) and TOPRO-3 (red). Bar = 50μm. Only FLAG-positive cells were used for cell surface area analysis (n=3). H, Immunoblot of NRVM. Data are mean ± SEM. *P≤0.05, **P≤0.01, #P≤0.001.
To complement ATF6 loss-of-function approach, we used a gain-of-function approach, examining the effects of ectopic expression of ATF6 and RHEB. In the absence of a growth stimulus, ectopic expression of ATF6 did not increase myocyte growth, as expected, due to the absence of mTORC1 activation under these conditions (Online Fig. VIA Con). Either PE or IGF1 increased myocyte growth, which was completely blocked by the mTORC1 inhibitor, rapamycin, as expected (Online Fig. VIA, PE and IGF1, red vs blue). Ectopic ATF6 augmented the growth-promoting effects of PE and IGF1, which were also completely blocked by rapamycin (Online Fig. VIA, PE and IGF1, black and green). Moreover, ectopic ATF6 slightly augmented PE- and IGF1-stimulated NRVM growth, however, it was not able to restore growth in cells treated with either RHEB siRNA or Lonafarnib (Online Fig. VIB-C). As expected, ectopic expression of RHEB had no effect in the absence of a growth stimulus; however, upon a growth stimulus, the loss of growth and mTORC1 activation seen with ATF6 siRNA were completely restored by ectopically expressed RHEB (Fig. 4F-H; Fig. 5F-H).
Mechanistic relationship between growth signaling and the UPR.
The unfolded protein response (UPR), which in addition to ATF6, is mediated by PERK and IRE15, is activated by the misfolding of proteins induced by a variety of chemical and pathophysiological treatments, most of these do not promote growth. In fact, the UPR is not widely considered to be growth-promoting. Accordingly, since we found here that ATF6 can be activated during growth, we assessed how growth affected the other arms of the UPR. We found that PE and IGF1 activated all three arms of the UPR in a rapamycin-sensitive manner (Online Fig. VIIA), indicating that mTORC1 activation is required for UPR activation during growth. We then individually knocked down ATF6, PERK and IRE1, and found that only ATF6 knockdown blunted growth (Online Fig. VIIB-C). To ensure that the effects of ATF6 on growth are dependent on the transcriptional effects of ATF6, we showed that NRVM infected with AdV-ATF6(1–373) [active] exhibited increased growth in response to PE, especially when endogenous ATF6 was knocked down, however AdV-ATF6(94–373) [trancriptionally inactive] did not (Online Fig. VIID).
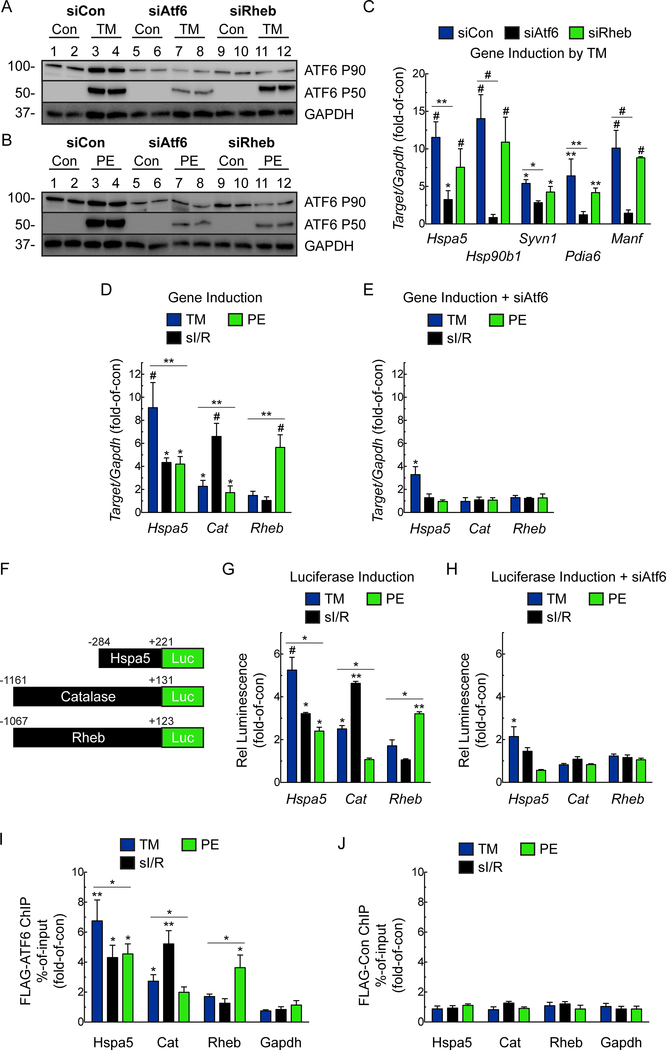
Next, we examined the effect on mTORC1 signaling of other UPR stimulators that do not affect growth, such as tunicamycin (TM), which increases ER protein misfolding by inhibiting protein glycosylation in the ER. In contrast to PE, activation of ATF6 by TM was not dependent on RHEB (Fig. 6A-B). Additionally, while RHEB knockdown blocked PE- and IGF1-mediated induction of ATF6 target genes, (Fig. 4E, 5E), it had no effect on TM-mediated induction of ATF6 target genes (Fig. 6C). Thus, there are RHEB/growth-dependent and RHEB/growth-independent pathways that lead to ATF6 activation and induction of ATF6 target genes.
Figure 6. Examination of Rheb Requirement for Growth-dependent but not Growth-independent Activation of the ATF6.
A-B, NRVM were transfected with siCon, siAtf6 or siRheb then treated ± tunicamycin (TM; 10μg/mL) (A) or PE (50μM) (B) for 24 hours, then analyzed for ATF6 activation by immunoblotting. C, mRNA levels for ATF6 target genes determined by qRT-PCR. Values are fold-of-control, i.e. not treated with TM (n=6). D, E, NRVM were transfected with siCon (D) or siAtf6 (E), then treated ± TM (10μg/mL), or PE (50μM) for 24 hours, or subjected to simulated ischemia/reperfusion (sI/R; 8 hours of sI, followed by 24 hours of reperfusion) and mRNA for ATF6 target genes determined by qRT-PCR (n=6). F, Diagram of constructs that encode luciferase driven by the grp78, catalase, and rheb 5’-flanking region. G, H, NRVM were transfected with human-grp78(−284/+221)-Luc WT, rat-catalase(−1161/+131)-Luc WT, or rat-rheb(−1067/+123)-Luc WT and then transfected with siCon (G) or siAtf6 (H), then treated ± TM (10μg/mL), or PE (50μM) for 24 hours, or subjected to sI/R and luciferase activity measured in extracts (n=6). I, J, NRVM infected with AdV FLAG-ATF6(1–670) (I) or control (J), and then ATF6 binding to the endogenous grp78, catalase, or rheb genes, as well as to the negative control gene, gapdh, examined by ChIP under the same experimental conditions described above (n=3). Data are mean ± SEM. *P≤0.05, **P≤0.01, #P≤0.001.
Stimulus-dependent differential induction of ATF6 target genes.
We dived deeper into the mechanism of RHEB/growth-dependent and RHEB/growth-independent pathways of ATF6 activation. We previously showed that ATF6 induces some proteins targeted to the ER, where they enhance protein folding (e.g. HSPA5/GRP78), and others located outside the ER, where they serve other functions. One example of the latter is our finding that I/R activates ATF6-dependent induction of catalase (CAT), which resides in peroxisomes and neutralizes damaging ROS. Here, we provide an additional example of an ATF6-inducible gene, RHEB, that encodes a protein that resides outside the ER. Because of the differences in the locations and functions of Hspa5, Cat, and Rheb, we posited that they might be differentially induced by treatments that cause ER protein misfolding (TM), or oxidative stress (I/R) but do not induce growth, or to a treatment that induces growth (PE). While, for the most part, the mRNA levels for all three genes were increased by all the treatments, the quantitative nature of induction differed depending on the treatments, such that TM, sI/R, and PE had the greatest effects on induction of Hspa5, Cat, and Rheb, respectively (Fig. 6D). Notably, CAT induction was highly selective, showing an approximate 6-fold induction by sI/R, and much less induction by either TM or PE (Fig. 6D, Cat). Remarkably, RHEB induction was also highly selective, showing the least induction by TM or sI/R, while being induced by over 5-fold by PE (Fig. 6D, Rheb). Importantly, all of these effects depended on ATF6 (Fig. 6E).
To dissect this stimulus-dependent differential gene induction further, we showed that promoter/luciferase reporter constructs for Hspa5, Cat, and Rheb (Fig. 6F) were also differentially induced by TM, sI/R and PE, mimicking mRNA induction (Fig. 6G). Importantly, as with the mRNA, all of these effects depended upon ATF6 (Fig. 6H).
These stimulus-specific effects of ATF6 on Hspa5, Cat, and Rheb could be due to the stimulus-dependent binding of ATF6 to the ERSEs in these genes. To test this, we developed a new method for measuring ATF6 binding to the HSPA5, CAT, and RHEB promoters in cells treated with TM, sI/R or PE. To this end we generated a recombinant AdV FLAG full-length p90 ATF6, i.e. ATF6(1–670), which remains in the ER in the absence of ER stress, and, therefore, can not bind to ERSEs. NRVM expressing FLAG-ATF6(1–670) were treated with TM, sI/R or PE, each of which induce the formation of the FLAG-tagged N-terminal, active p50 form of ATF6, so it can bind to ERSEs. ChIP demonstrated that the binding of ATF6 to these genes differed, depending on the stimulus, mimicking the mRNA induction and promoter activation (Fig. 6I). These effects were not seen with AdV encoding only FLAG, verifying ATF6-specificity (Fig. 6J). This shows, for the first time in any cell type, that ATF6 can be activated by a broad spectrum of conditions that affect proteostasis in a variety of ways, yet the relative induction of ATF6 targets differs in a condition-dependent manner.
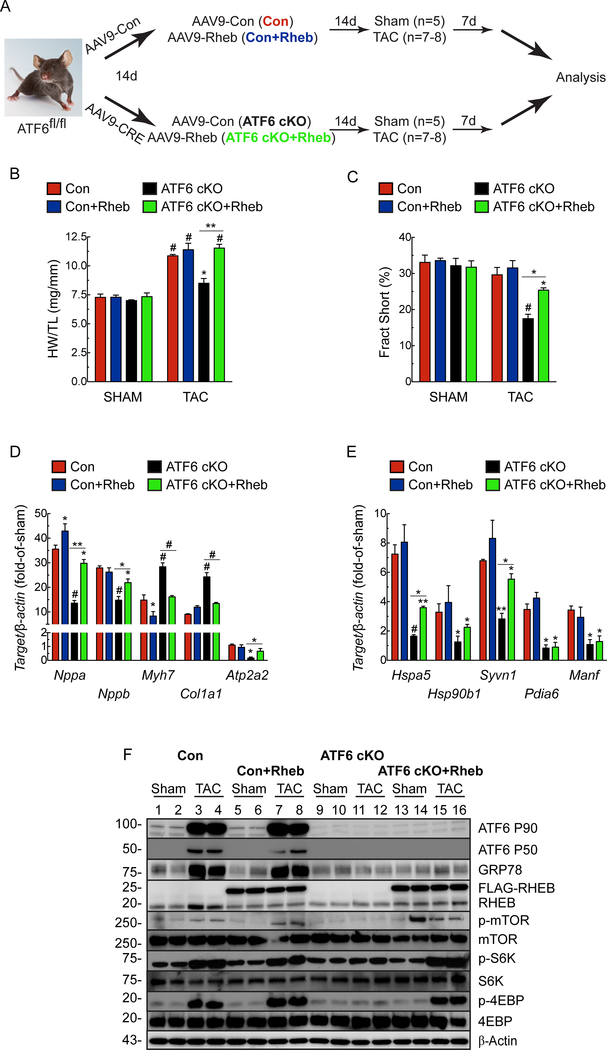
Ectopic expression of RHEB restores cardiac growth to ATF6 cKO mouse hearts.
Next, we assessed the effects of ectopic expression of RHEB in the heart, in vivo using a new recombinant AAV9-RHEB (Fig. 7A). In ATF6 cKO mice, AAV9-RHEB effectively restored the loss of mTORC1 signaling, hypertrophic growth and cardiac function, as well as the hypertrophic and ATF6 gene programs in response to TAC (Fig. 7B-F; Online Table IV). Thus, it is by increasing RHEB that ATF6 influences mTORC1 signaling and subsequent cardiac myocyte growth, fetal gene expression and ATF6-target gene expression.
Figure 7. Effect of cardiac myocyte-specific ectopic Rheb expression in ATF6 gene deleted mouse hearts subjected to TAC.
A, Experimental protocol for AAV9 administration to ATF6fl/fl mice and TAC. B, Heart weights/tibia lengths (HW/TL). C, Fractional shortening (%), as determined by echocardiography, see Online Table IV. D, mRNA for fetal genes determined by qRT-PCR. E, mRNA for ATF6 target genes determined by qRT-PCR. F, Immunoblots of LV extracts. Data are mean ± SEM. *P≤0.05, **P≤0.01, #P≤0.001.
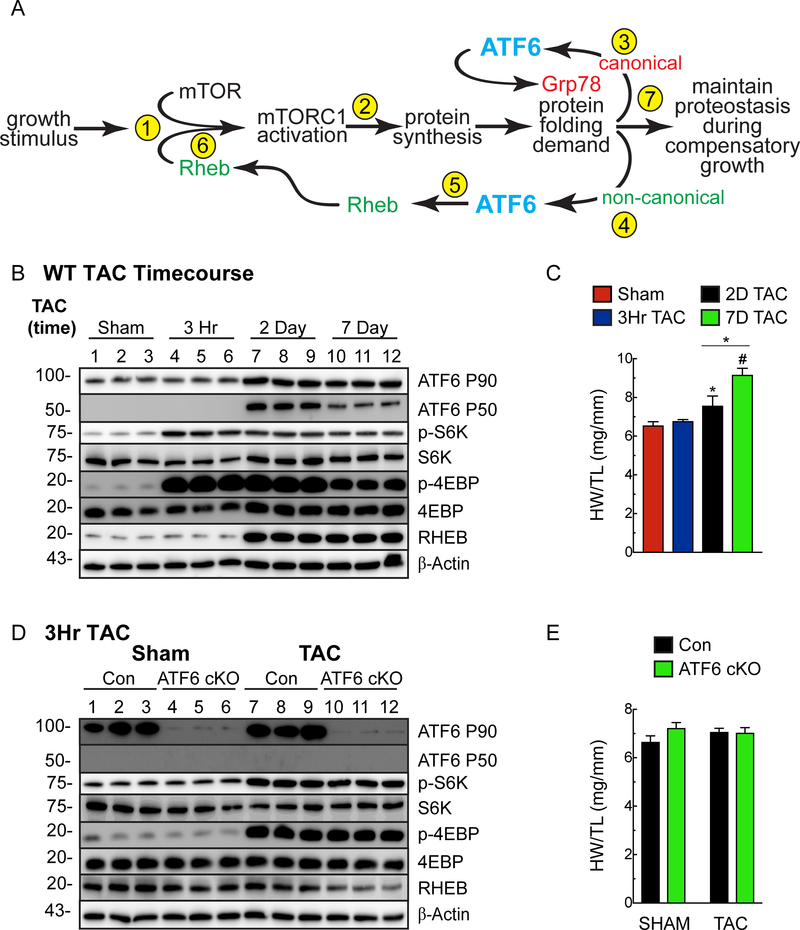
ATF6 activation in response to growth requires mTORC1 activation, protein synthesis and protein misfolding.
To this point, mTORC1 and ATF6 activation were shown to be dependent on each other under the growth conditions examined. To account for this interdependence, we posited a temporal sequence of events, wherein the initial event is mTORC1 activation, which depends on basal levels of Rheb (Fig. 8A, Step 1). This initial mTORC1 activation precedes, but drives initial increases in protein synthesis that place demands on the protein-folding machinery (Fig. 8A, Step 2), which activates ATF6. Then, ATF6 serves canonical- and non-canonical roles (Fig. 8A, Steps 3, 4), the latter of which includes RHEB induction (Fig. 8A, Step 5), which is necessary to sustain mTORC1 activation (Fig. 8A, Step 6) and the continued increases in protein synthesis that required for growth and cardiac myocyte hypertrophy (Fig. 8A, Step 7). To examine this hypothesis, a TAC time course was carried out. At 3h of TAC, a time when mTORC1 is activated, but protein synthesis has not yet increased, mTORC1 signaling was activated, but ATF6 was not activated and RHEB was not induced (Fig. 8B, 3h). However, at both 2 and 7d of TAC, when protein synthesis is increased, mTORC1 signaling and ATF6 were activated, and RHEB was induced (Fig. 8B, 2d and 7d). As expected, heart weights increased as a function of TAC time from 3h to 7d (Fig. 8C; Online Table V). Thus, mTORC1 activation occurred soon after TAC and preceded ATF6 activation. Further supporting our hypothesis that initially, mTORC1 activation precedes ATF6 activation were results of a 3h TAC experiment in ATF6 cKO mice, where, in contrast to longer times of TAC (i.e. 7d - Fig. 3G), the deletion of ATF6 did not affect mTORC1 activation (Fig. 8D). As expected, heart weights did not change under these conditions (Fig. 8E; Online Table VI).
Figure 8. Mechanism whereby ATF6 acts as a nodal regulator of both protein synthesis and protein folding during cardiac hypertrophy.
A, Shown are the temporal sequence of steps involved in mediating the initial (Steps 1–4) and sustained (Steps 5–7) aspects of growth and the interdependent roles of mTORC1 and ATF6. B, C, Immunoblot of LV extracts (B) and heart weights/tibia lengths (HW/TL) (C) from WT mice subjected to TAC for 3 hours, 2 days, or 7 days. Echocardiography details in Online Table V. D, E, Immunoblot of LV extracts (D) and heart weights/tibia lengths (HW/TL) (E) from Con or ATF6 cKO mice subjected to 3 hours of TAC. Echocardiography details in Online Table VI. Data are mean ± SEM. *P≤0.05, #P≤0.001.
Consistent with these results, when examining the effect of PE and IGF1 at the earliest time points, just prior to when protein synthesis is greatest in NRVM, knocking down ATF6 did not affect mTORC1 activation (Online Fig. VIIIA), but again, ATF6 activation was rapamycin-dependent (Online Fig. VIIIB). Moreover, inhibiting protein synthesis with cycloheximide had no effect on mTORC1 activation at these short times of PE or IGF1 treatment, but impaired ATF6 activation and RHEB induction, indicating that protein synthesis is required for ATF6 activation and subsequent RHEB induction (Online Fig. VIIIC). Finally, in NRVM treated with the chemical chaperone, 4PBA, PE and IGF1 activated mTORC1 however, ATF6 was not activated and RHEB was not induced (Online Fig. VIIID), indicating the increase in protein folding demand driven by increases in protein synthesis are responsible for activating ATF6.
DISCUSSION
ATF6 is required for growth of the heart.
While previous studies reported increased expression of a few ER stress genes in mouse models of pressure overload, implicating ER protein misfolding31–34, prior to our study here, neither the activation nor the roles for ATF6 in cardiac myocytes during cardiac growth had been examined. Here, we showed, for the first time that ATF6, a major mediator of the UPR, is activated by diverse growth stimuli and that ATF6 is required for growth of the heart in response to these stimuli. We determined that the mechanism of this effect involves ATF6-mediated induction of RHEB (Fig. 8A). It was surprising to find that ATF6 is required for heart growth, considering the UPR is not widely known to be involved in growth processes. However, this non-canonical role for ATF6 complements its canonical role as a sensor of misfolded proteins in the ER and, as such, a sensor of increases in protein folding demand, which occur during growth. Thus, ATF6 maintains proteostasis and proteome integrity when the heart is stimulated to grow in a compensatory manner.
ATF6 exhibits stress-specific transcriptional gene induction.
We also found that, depending on the stimulus, ATF6 target genes are differentially expressed due to the unique effects that the stresses have on ATF6 binding to, and thus, transcriptional activation of ATF6 target genes. Such differential ATF6 target gene induction by treatments that all activate ATF6 suggests that there are yet-to-be-described regulatory layers that fine-tune the ATF6 gene program to best adapt to the conditions. Some possible mechanisms that could contribute to this differential expression are beginning to emerge, as it has been shown that ATF6 can interact with other transcription factors, such as Nrf1, PGC1α and β, and ERRγ,35–37 which changes the transcriptional programming in ways that fine-tune ATF6 target gene induction. Additionally, it was recently shown that ATF6 can be differentially activated by lipids and proteotoxic stress at least partly by virtue of separate activation domains on ATF6 for these two different stresses38.
Rheb in the heart.
Rheb was originally documented as an mTORC1 activator in the brain39, this role has been demonstrated in numerous other tissues and organs17, 40, 41. Global deletion of Rheb is embryonic lethal, in part due to cardiac defects42, demonstrating the importance of Rheb-mediated mTORC1 activation in heart growth and development. The growth-promoting effect of Rheb gain-of-function was demonstrated in adult rat ventricular myocytes transfected with adenovirus encoding Rheb43. However, overexpression of Rheb in transgenic mice increased infarct size, in part because Rheb inappropriately decreased autophagy, which is adaptive in this disease setting44. Pharmacological inhibition of Rheb in mice subjected to TAC for three weeks was cardioprotective14. These findings differ from our study, perhaps because different times after TAC were studied, or different approaches to decreasing Rheb. It is also possible that Rheb induction and mTORC1 activation have different roles in a severe afterload-induced hypertrophy model, such that acute activation works in a compensatory manner, but chronic activation drives decompensation. The αMHC-CRE-dependent conditional deletion of Rheb from mouse cardiac myocytes resulted in atrophic hearts, heart failure, and death within 1–2 weeks after birth, a timeframe that aligns with the time of αMHC expression after birth12, 45. Although there have been no studies prior to ours mechanistically connecting Atf6 with Rheb induction, one study in tumor cells46, and another in the setting of Huntington’s disease47, have implicated such a connection and, therefore, support the findings reported here.
Feedback regulation of ATF6-mediated growth.
Our study describes a mechanism whereby ATF6 matches protein synthesis with folding in times of increased growth; since this constitutes a positive feedback mechanism, we reason that there must also be mechanisms that interrupt this feedback, thereby limiting the rate of growth driven by the ATF6-Rheb-mTORC1 axis. One such mechanism might involve Rheb itself, which has been shown to activate PERK48. Mechanisms such as this underscore the complexities of proteostasis, raising questions about how Rheb switches from protein synthesis activator to inhibitor.
Conclusions.
The results of our study firmly place ATF6 in a critical position as a determinant of cardiac growth (Fig. 8A). Moreover, since ATF6 is ubiquitously expressed, our findings underscore the widespread importance of the ATF6-Rheb-mTORC1-growth signaling axis described here in non-cardiac cells and tissues in addition to the heart.
Supplementary Material
NOVELTY AND SIGNFICANCE
What Is Known?
Cardiac myocytes in the heart exhibit adaptive hypertrophic growth in response to pathological and physiological conditions, both of which activate mTORC1 signaling and increase protein synthesis, critical drivers of cardiac growth.
Increased protein synthesis places demands on the protein-folding machinery in cardiac myocytes, leading to some protein misfolding, which activates the transcription factor, ATF6, which regulates a gene program that fortifies cellular protein-folding capacity.
What new information does this article contribute?
Selective deletion of ATF6 in cardiac myocytes of mice impaired mTORC1 signaling, protein synthesis and cardiac hypertrophy, demonstrating that ATF6 is required for heart growth.
Selective activation of ATF6 in mouse hearts induced RHEB, a required activator of mTORC1 and protein synthesis, identifying ATF6 as a previously unrecognized link between pathological and physiological growth stimuli and mTOR-mediated increases in protein synthesis, protein folding and cardiac growth.
ER stress impairs ER protein folding, which activates the transcription factor, ATF6, an inducer of genes that restores ER protein folding. We hypothesized that increased protein synthesis during cardiac hypertrophy might challenge protein folding by ER, leading toactivation of ATF6. We found that ATF6 was activated in mice subjected to pathological and physiological growth conditions that promote adaptive cardiac myocyte hypertrophy. Cardiac myocyte-specific deletion of ATF6 blunted adaptive cardiac myocyte hypertrophy, while ectopic expression of ATF6 enhanced the hypertrophic response. Here, ATF6 was found to promote the induction of RHEB, an obligate activator of mTORC1, a known regulator of protein synthesis and cellular growth. Ectopic expression of RHEB restored cardiac growth to ATF6 cKO mice. ATF6 induced RHEB in response to growth factors, but not in response to other activators of ATF6 that do not induce growth, indicating that ATF6 target gene induction is stress-specific. Thus, ATF6 is a newly identified, essential member of mTORC1-mediated growth signaling in the heart. Moreover, since ATF6 is ubiquitously expressed, our findings underscore the potential widespread importance of this new ATF6-Rheb-mTORC1-growth signaling axis in non-cardiac cells and tissues in addition to the heart.
ACKNOWLEDGMENTS
We thank Dr. Gokhan S. Hotamisligil (Harvard T.H. Chan School of Public Health, Boston, MA) for the ATF6fl/fl mice.
SOURCES OF FUNDING
EAB American Heart Association (17PRE33670796), the Rees-Stealy Research Foundation and SDSU Heart Institute, the Inamori Foundation, and the ARCS Foundation, Inc., San Diego Chapter.
CH Boehringer Ingelheim Fonds Travel Grant, the DZHK Mobilitätsprogramm and the Deutsche Herzstiftung
OJM the DZHK and by the BMBF (German Ministry of Education and Research)
SD was supported by an Excellence Grant from the German Centre for Cardiovascular Research (DZHK) and Department of Cardiology, Angiology, and Pneumology, University Hospital Heidelberg.
CCG by (NIH) grants R01 HL75573, R01 HL104535, and P01 HL085577.
Nonstandard Abbreviations and Acronyms:
- AAV
adeno-associated virus
- AdV
adenovirus
- ANOVA
analysis of variance
- ATF6
activating transcription factor 6 alpha
- ATF6 cKO
ATF6 alpha conditional knockou
- Cat
Catalase
- ER
endoplasmic reticulum
- Grp78
78 kilodalton glucose-regulated protein, Hapa5
- HR
heart rate
- HW
heart weight
- ICF
immunocytofluorescence
- LV
left ventricle
- LVEDV
left ventricular end diastolic volume
- LVESV
left ventricular end systolic volume
- LVIDD
left ventricular inner diameter in diastole
- LVIDS
left ventricular inner diameter in systole
- PWTD
left ventricular posterior wall thickness in diastole
- PWTS
left ventricular posterior wall thickness in systole
- Rheb
Ras homologue enriched in brain
- SR
sarcoplasmic reticulum
- TAC
transverse aortic constriction
- TL
tibia length
- TM
tunicamycin
- UPR
unfolded protein response
Footnotes
DISCLOSURES
None.
REFERENCES
- 1.Balch WE, Morimoto RI, Dillin A and Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–9. [DOI] [PubMed] [Google Scholar]
- 2.Glembotski CC. Roles for the sarco-/endoplasmic reticulum in cardiac myocyte contraction, protein synthesis, and protein quality control. Physiology (Bethesda). 2012;27:343–50. [DOI] [PubMed] [Google Scholar]
- 3.Gidalevitz T, Stevens F and Argon Y. Orchestration of secretory protein folding by ER chaperones. Biochim Biophys Acta. 2013;1833:2410–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glembotski CC. Roles for ATF6 and the sarco/endoplasmic reticulum protein quality control system in the heart. Journal of Molecular and Cellular Cardiology. 2014;71:11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arrieta A, Blackwood EA and Glembotski CC. ER Protein Quality Control and the Unfolded Protein Response in the Heart. Curr Top Microbiol Immunol. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu C, Johansen FE and Prywes R. Interaction of ATF6 and serum response factor. Mol Cell Biol. 1997;17:4957–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haze K, Yoshida H, Yanagi H, Yura T and Mori K. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol Biol Cell. 1999;10:3787–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luckey SW, Walker LA, Smyth T, Mansoori J, Messmer-Kratzsch A, Rosenzweig A, Olson EN and Leinwand LA. The role of Akt/GSK-3beta signaling in familial hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2009;46:739–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernardo BC and McMullen JR. Molecular Aspects of Exercise-induced Cardiac Remodeling. Cardiol Clin. 2016;34:515–530. [DOI] [PubMed] [Google Scholar]
- 10.Zhang D, Contu R, Latronico MV, Zhang J, Rizzi R, Catalucci D, Miyamoto S, Huang K, Ceci M, Gu Y, Dalton ND, Peterson KL, Guan KL, Brown JH, Chen J, Sonenberg N and Condorelli G. MTORC1 regulates cardiac function and myocyte survival through 4E-BP1 inhibition in mice. J Clin Invest. 2010;120:2805–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shende P, Plaisance I, Morandi C, Pellieux C, Berthonneche C, Zorzato F, Krishnan J, Lerch R, Hall MN, Ruegg MA, Pedrazzini T and Brink M. Cardiac raptor ablation impairs adaptive hypertrophy, alters metabolic gene expression, and causes heart failure in mice. Circulation. 2011;123:1073–82. [DOI] [PubMed] [Google Scholar]
- 12.Tamai T, Yamaguchi O, Hikoso S, Takeda T, Taneike M, Oka T, Oyabu J, Murakawa T, Nakayama H, Uno Y, Horie K, Nishida K, Sonenberg N, Shah AM, Takeda J, Komuro I and Otsu K. Rheb (Ras homologue enriched in brain)-dependent mammalian target of rapamycin complex 1 (mTORC1) activation becomes indispensable for cardiac hypertrophic growth after early postnatal period. J Biol Chem. 2013;288:10176–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volkers M, Toko H, Doroudgar S, Din S, Quijada P, Joyo AY, Ornelas L, Joyo E, Thuerauf DJ, Konstandin MH, Gude N, Glembotski CC and Sussman MA. Pathological hypertrophy amelioration by PRAS40-mediated inhibition of mTORC1. Proc Natl Acad Sci U S A. 2013;110:12661–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu X, Cao Y, Nie J, Liu H, Lu S, Hu X, Zhu J, Zhao X, Chen J, Chen X, Yang Z and Li X. Genetic and pharmacological inhibition of Rheb1-mTORC1 signaling exerts cardioprotection against adverse cardiac remodeling in mice. Am J Pathol. 2013;182:2005–14. [DOI] [PubMed] [Google Scholar]
- 15.Sciarretta S, Forte M, Frati G and Sadoshima J. New Insights Into the Role of mTOR Signaling in the Cardiovascular System. Circ Res. 2018;122:489–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L and Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duran RV and Hall MN. Regulation of TOR by small GTPases. EMBO Rep. 2012;13:121–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Betz C and Hall MN. Where is mTOR and what is it doing there? J Cell Biol. 2013;203:563–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tee AR, Manning BD, Roux PP, Cantley LC and Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr Biol. 2003;13:1259–68. [DOI] [PubMed] [Google Scholar]
- 20.Engin F, Yermalovich A, Nguyen T, Hummasti S, Fu W, Eizirik DL, Mathis D and Hotamisligil GS. Restoration of the unfolded protein response in pancreatic beta cells protects mice against type 1 diabetes. Sci Transl Med. 2013;5:211–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Zhang Y, Ding G, May HI, Xu J, Gillette TG, Wang H and Wang ZV. Temporal dynamics of cardiac hypertrophic growth in response to pressure overload. Am J Physiol Heart Circ Physiol. 2017;313:H1119–H1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamura A, Rokosh DG, Paccanaro M, Yee RR, Simpson PC, Grossman W and Foster E. LV systolic performance improves with development of hypertrophy after transverse aortic constriction in mice. Am J Physiol Heart Circ Physiol. 2001;281:H1104–12. [DOI] [PubMed] [Google Scholar]
- 23.Takaoka H, Esposito G, Mao L, Suga H and Rockman HA. Heart size-independent analysis of myocardial function in murine pressure overload hypertrophy. Am J Physiol Heart Circ Physiol. 2002;282:H2190–7. [DOI] [PubMed] [Google Scholar]
- 24.Harvey PA and Leinwand LA. The cell biology of disease: cellular mechanisms of cardiomyopathy. J Cell Biol. 2011;194:355–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen DL, Harrison BC, Maass A, Bell ML, Byrnes WC and Leinwand LA. Cardiac and skeletal muscle adaptations to voluntary wheel running in the mouse. J Appl Physiol (1985) 2001;90:1900–8. [DOI] [PubMed] [Google Scholar]
- 26.Chung E, Heimiller J and Leinwand LA. Distinct cardiac transcriptional profiles defining pregnancy and exercise. PLoS One. 2012;7:e42297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernardo BC, Weeks KL, Pretorius L and McMullen JR. Molecular distinction between physiological and pathological cardiac hypertrophy: experimental findings and therapeutic strategies. Pharmacol Ther. 2010;128:191–227. [DOI] [PubMed] [Google Scholar]
- 28.Martindale JJ, Fernandez R, Thuerauf D, Whittaker R, Gude N, Sussman MA and Glembotski CC. Endoplasmic reticulum stress gene induction and protection from ischemia/reperfusion injury in the hearts of transgenic mice with a tamoxifen-regulated form of ATF6. Circ Res. 2006;98:1186–93. [DOI] [PubMed] [Google Scholar]
- 29.Simpson P Stimulation of hypertrophy of cultured neonatal rat heart cells through an alpha 1-adrenergic receptor and induction of beating through an alpha 1- and beta 1-adrenergic receptor interaction. Evidence for independent regulation of growth and beating. Circ Res. 1985;56:884–94. [DOI] [PubMed] [Google Scholar]
- 30.Basso AD, Mirza A, Liu G, Long BJ, Bishop WR and Kirschmeier P. The farnesyl transferase inhibitor (FTI) SCH66336 (lonafarnib) inhibits Rheb farnesylation and mTOR signaling. Role in FTI enhancement of taxane and tamoxifen anti-tumor activity. J Biol Chem. 2005;280:31101–8. [DOI] [PubMed] [Google Scholar]
- 31.Okada K, Minamino T, Tsukamoto Y, Liao Y, Tsukamoto O, Takashima S, Hirata A, Fujita M, Nagamachi Y, Nakatani T, Yutani C, Ozawa K, Ogawa S, Tomoike H, Hori M and Kitakaze M. Prolonged endoplasmic reticulum stress in hypertrophic and failing heart after aortic constriction: possible contribution of endoplasmic reticulum stress to cardiac myocyte apoptosis. Circulation. 2004;110:705–12. [DOI] [PubMed] [Google Scholar]
- 32.Sari FR, Widyantoro B, Thandavarayan RA, Harima M, Lakshmanan AP, Zhang S, Muslin AJ, Suzuki K, Kodama M and Watanabe K. Attenuation of CHOP-mediated myocardial apoptosis in pressure-overloaded dominant negative p38alpha mitogen-activated protein kinase mice. Cell Physiol Biochem. 2011;27:487–96. [DOI] [PubMed] [Google Scholar]
- 33.Park CS, Cha H, Kwon EJ, Sreenivasaiah PK and Kim DH. The chemical chaperone 4-phenylbutyric acid attenuates pressure-overload cardiac hypertrophy by alleviating endoplasmic reticulum stress. Biochem Biophys Res Commun. 2012;421:578–84. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Kwak D, Lu Z, Xu X, Fassett J, Wang H, Wei Y, Cavener DR, Hu X, Hall J, Bache RJ and Chen Y. Endoplasmic reticulum stress sensor protein kinase R-like endoplasmic reticulum kinase (PERK) protects against pressure overload-induced heart failure and lung remodeling. Hypertension. 2014;64:738–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vekich JA, Belmont PJ, Thuerauf DJ and Glembotski CC. Protein disulfide isomerase-associated 6 is an ATF6-inducible ER stress response protein that protects cardiac myocytes from ischemia/reperfusion-mediated cell death. J Mol Cell Cardiol. 2012;53:259–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu J, Ruas JL, Estall JL, Rasbach KA, Choi JH, Ye L, Bostrom P, Tyra HM, Crawford RW, Campbell KP, Rutkowski DT, Kaufman RJ and Spiegelman BM. The unfolded protein response mediates adaptation to exercise in skeletal muscle through a PGC-1alpha/ATF6alpha complex. Cell Metab. 2011;13:160–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Misra J, Kim DK, Choi W, Koo SH, Lee CH, Back SH, Kaufman RJ and Choi HS. Transcriptional cross talk between orphan nuclear receptor ERRgamma and transmembrane transcription factor ATF6alpha coordinates endoplasmic reticulum stress response. Nucleic Acids Res. 2013;41:6960–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tam AB, Roberts LS, Chandra V, Rivera IG, Nomura DK, Forbes DJ and Niwa M. The UPR Activator ATF6 Responds to Proteotoxic and Lipotoxic Stress by Distinct Mechanisms. Dev Cell. 2018;46:327–343 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamagata K, Sanders LK, Kaufmann WE, Yee W, Barnes CA, Nathans D and Worley PF. rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J Biol Chem. 1994;269:16333–9. [PubMed] [Google Scholar]
- 40.Potheraveedu VN, Schopel M, Stoll R and Heumann R. Rheb in neuronal degeneration, regeneration, and connectivity. Biol Chem. 2017;398:589–606. [DOI] [PubMed] [Google Scholar]
- 41.Heard JJ, Fong V, Bathaie SZ and Tamanoi F. Recent progress in the study of the Rheb family GTPases. Cell Signal. 2014;26:1950–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goorden SM, Hoogeveen-Westerveld M, Cheng C, van Woerden GM, Mozaffari M, Post L, Duckers HJ, Nellist M and Elgersma Y. Rheb is essential for murine development. Mol Cell Biol. 2011;31:1672–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Huang BP, Luciani DS, Wang X, Johnson JD and Proud CG. Rheb activates protein synthesis and growth in adult rat ventricular cardiomyocytes. J Mol Cell Cardiol. 2008;45:812–20. [DOI] [PubMed] [Google Scholar]
- 44.Sciarretta S, Zhai P, Shao D, Maejima Y, Robbins J, Volpe M, Condorelli G and Sadoshima J. Rheb is a critical regulator of autophagy during myocardial ischemia: pathophysiological implications in obesity and metabolic syndrome. Circulation. 2012;125:1134–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao Y, Tao L, Shen S, Xiao J, Wu H, Li B, Wu X, Luo W, Xiao Q, Hu X, Liu H, Nie J, Lu S, Yuan B, Han Z, Xiao B, Yang Z and Li X. Cardiac ablation of Rheb1 induces impaired heart growth, endoplasmic reticulum-associated apoptosis and heart failure in infant mice. Int J Mol Sci. 2013;14:24380–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schewe DM and Aguirre-Ghiso JA. ATF6alpha-Rheb-mTOR signaling promotes survival of dormant tumor cells in vivo. Proc Natl Acad Sci U S A. 2008;105:10519–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernandez-Fernandez MR, Ferrer I and Lucas JJ. Impaired ATF6alpha processing, decreased Rheb and neuronal cell cycle re-entry in Huntington’s disease. Neurobiol Dis. 2011;41:23–32. [DOI] [PubMed] [Google Scholar]
- 48.Tyagi R, Shahani N, Gorgen L, Ferretti M, Pryor W, Chen PY, Swarnkar S, Worley PF, Karbstein K, Snyder SH and Subramaniam S. Rheb Inhibits Protein Synthesis by Activating the PERK-eIF2alpha Signaling Cascade. Cell Rep. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.