Summary
The derivation of human embryonic stem cells (hESCs) and the stunning discovery that somatic cells can be reprogrammed into human induced pluripotent stem cells (hiPSCs) holds the promise to revolutionize biomedical research and regenerative medicine. In this review, we focus on disorders of the central nervous system and explore how advances in human pluripotent stem cells (hPSCs) coincide with evolutions in genome engineering and genomic technologies to provide realistic opportunities to tackle some of the most devastating complex disorders.
Introduction
The first report of human pluripotent stem cells (hPSCs) remains a key discovery with the promise to fundamentally reshape regenerative medicine and the study of complex human diseases (Thomson, 1998). These cells termed human embryonic stem cells (hESCs) were derived directly from the human pre-implantation embryo and are generally considered the functional in vitro equivalent to the pluripotent cells of the blastocyst, thought to be the founder cells of the embryo proper (Reubinoff et al., 2000; Thomson, 1998). Once removed from the blastocyst, hESCs can be maintained in culture for an extended period and, comparable to their in vivo counterpart, have the developmental potential to differentiate into any somatic cell type. The derivation of hESCs has captured the imagination of biomedical researchers and the public likewise based on the promise to provide an essentially unlimited supply of human somatic cells for basic research and regenerative medicine.
In vitro differentiation of hESCs has already revolutionized our ability to study early human development and cell-type specification in a cell culture dish and provides suitable cells for cell replacement therapies. However, broad application of hESCs remains challenging due to technical difficulties like immune rejection after transplantation of non-autologous cells and ethical concerns associated with the use of human embryos for research. The discovery that transient expression of a few transcription factors (Oct4, Sox2, KLF4 and c-Myc) is sufficient to reprogram somatic cells to a pluripotent state (Maherali et al., 2007; Takahashi and Yamanaka, 2006; Wernig et al., 2007) allows the derivation of human induced pluripotent stem cells (hiPSCs) (Takahashi et al., 2007; Yu et al., 2007) and resolves many limitations associated with hESCs. The basic biological properties of hiPSCs are highly similar to hESCs, opening up the exciting opportunity for cell transplantation therapies using patient-derived autologous cells that should evade immune rejections. Clinical trials are already underway for a number of diseases, including macular degeneration (Mandai et al., 2017; Mehat et al., 2018; Schwartz et al., 2015; Song et al., 2015), ischemic heart disease (Menasché et al., 2018), diabetes (Viacyte; https://clinicaltrials.gov: NCT03163511) and spinal cord injury (Asterias Biotherapeutics; https://clinicaltrials.gov: NCT02302157). Despite these promising initial trials, the routine and safe application of cell replacement therapies for a broader spectrum of degenerative diseases is likely years away (Trounson and DeWitt, 2016).
Beside the enthusiasm for cell replacement therapies, hiPSC technology also provides the unique opportunity to establish cellular models of human diseases. Studying human diseases has been limited by the lack of relevant model systems that combine known genetic elements with disease-associated phenotypic readouts, especially for common medical conditions with no well-defined genetic etiology or Mendelian inheritance patterns, such as obesity, heart disease, diabetes and sporadic neurodegenerative disorders. The complex etiology of sporadic diseases, thought to result from the interaction between genetic and non-genetic (lifestyle and environmental) risk factors, has impeded understanding the molecular mechanisms of complex diseases (Lander, 2011; McClellan and King, 2010). Following in vitro differentiation, patient-derived hiPSCs provide somatic cells, which carry all the genetic elements implicated in disease development, allows the study of underlying genetic aberrations in human diseases-relevant cell types (Shi et al., 2016; Soldner and Jaenisch, 2012).
The development of hPSCs technology coincides with recent advances in genomic technologies and genome engineering. Genome-scale genetic and epigenetic information have provided valuable insights into the genetic basis of complex diseases and identified genetic and epigenetic variations associated with many human disorders. Unfortunately, most risk-associated variants have no established biological function or clinical relevance (McClellan and King, 2010). Therefore, translating these associations to mechanistic insights into disease development and progression remains a fundamental challenge. The remarkable progress of genome editing technologies, in particular the simplicity and versatility of the CRISPR/Cas system (Komor et al., 2017), has vastly increased our ability to modify the genome in human cells and enables us to begin to systematically dissect the functional effect of genetic variants.
In this review, we will summarize recent progress, limitations and potential solutions in using hPSCs technology to model complex human diseases focusing on neurological diseases, which pose unique challenges. We will highlight how integrating hPSC technology with CRISPR/Cas genome editing and genome-scale genetic and epigenetic information can systematically dissect the function of disease-associated risk alleles to provide a genetically controlled system to study sporadic diseases in culture. We will discuss the unique challenges to capture the full complexity of brain disorders including differentiation, maturation and aging of neuronal cell types and the analysis of non-cell autonomous interactions in complex organoids in vitro and interspecies chimeric in vivo systems. We will end by reviewing strategies to implement such systems for phenotypic genetic and compound screens for drug development.
HPSC-based models for monogenetic diseases
The identification of genes linked to familial forms of diseases such as cystic fibrosis, sickle cell anemia and diabetes fundamentally changed our understanding of many diseases by revealing insights into the molecular and cellular pathogenesis (Lander, 2011; McClellan and King, 2010). Detailed knowledge of disease-causing mutations allows us to develop effective model systems that combine known genetic elements with disease-associated phenotypic readouts. In the past, typical experimental systems were either based in vitro on primary human cells derived from patients with known mutations or in vivo using genetically engineered animal model systems. Unfortunately, studying diseases of the nervous system using primary cells has been challenging due to limited access to live brain tissue and the inability to reliably maintain and expand human neuronal cells such as post-mitotic neurons in culture. In contrast, genetically engineered animal models have provided remarkable insight into the development and function of the brain and the pathophysiology of neurological diseases; however, currently available rodent models for neurodevelopmental or neurodegenerative diseases recapitulate some but not all of the relevant features of the human disease (Bolker, 2017; Jucker, 2010), one of the major reasons why many therapeutic approaches show promise in preclinical disease models but fail in subsequent clinical trials.
hPSC technology now empowers the study of human brain physiology and neurological disease development in culture by providing a nearly unlimited access to all somatic cell types of the brain (Shi et al., 2016; Soldner and Jaenisch, 2012). The approach entails the derivation of hPSCs with known disease-associated mutations followed by in vitro differentiation into disease-relevant cell types. Differentiation into all major cell types of the CNS including a wide variety of specific neuronal subtypes, such as astrocytes, oligodendrocytes and more recently microglial cells (Muffat et al., 2016), can be induced with great precision (reviewed in (Tao and Zhang, 2016)). Using such in vitro generated cells in disease modeling may uncover alterations in molecular and cellular phenotypes associated with a specific disease, which can subsequently reveal detailed mechanistic insights and guide the development of novel therapeutics.
Initial hPSC models used hESC lines derived from blastocysts that carried mutations associated with monogenetic diseases or chromosomal abnormalities (Ilic and Ogilvie, 2016). Routine use of preimplantation genetic diagnosis (PGD), a procedure to detect genetic defects in embryos prior to transfer in utero, provides access to donated embryos with congenital defects, which would be otherwise discarded. To date, more than 100 hESC lines have been derived from genotyped embryos (Ilic and Ogilvie, 2016), including cell lines carrying mutations implicated in neurological disorders like spinal muscular atrophy, spinocerebellar ataxia, fragile X syndrome and Huntington disease. However, hESC based models remain challenging given the ethical and regulatory issues.
HiPSCs technology has resolved many of the ethical and technical issues in disease modeling and has rapidly evolved over the last decade. Following in vitro differentiation, patient-derived hiPSCs provide disease-relevant somatic cells, which carry all the genetic elements implicated in the disease reflecting the genetic spectrum of the patient population. While discussing all of these studies is beyond the scope of this work, excellent recent reviews summarize hiPSC diseases models for neurodevelopmental (Ardhanareeswaran et al., 2017), neuropsychiatric (Hoffman et al., 2018) and neurodegenerative diseases including Alzheimer’s disease (Sullivan and Young-Pearse, 2017) and Parkinson’s disease (Shi et al., 2016; Zhang et al., 2017)), motor neuron and Huntington’s disease. The spectrum of phenotypes, which can be assessed in hiPSC-based in vitro models include a wide range of molecular, metabolic, cellular and electrophysiological analysis. Such in vitro models may allow studying even complex disease-relevant phenotypes, such as pathologies associated processes in embryonic development, cellular maturation, non-cell autonomous effects between diverse neuronal cell populations, synaptic transmission and neuronal network function. HiPSC disease modeling has confirmed disease pathologies gained in other model systems and provided remarkable, novel molecular and cellular insights into many disorders.
Importantly, hiPSC-based drug discovery has identified novel compounds that can reverse disease-associated phenotypes in vitro and are currently under clinical investigation to treat diseases such as amyotrophic lateral sclerosis (McNeish et al., 2015), tauopathies in Alzheimer’s disease, progressive supranuclear palsy (Bright et al., 2015) and spinal muscular atrophy (Naryshkin et al., 2014). Two important observations emerge from this work. Robust and disease-relevant phenotypes in cell culture often occur (i) in neurodevelopmental or early age of onset disorders and (ii) in monogenetic disorders with highly penetrant mutations and severe, rapidly progressive well-defined disease-associated pathology and clinical manifestation. However, many of the most prevalent and devastating neurological disorders such as Alzheimer’s and Parkinson’s disease have no well-defined genetic etiology and manifest only late in life with slow progression over many years, a potentially serious complication for studying the complexity of late age of onset neurodegenerative disorders.
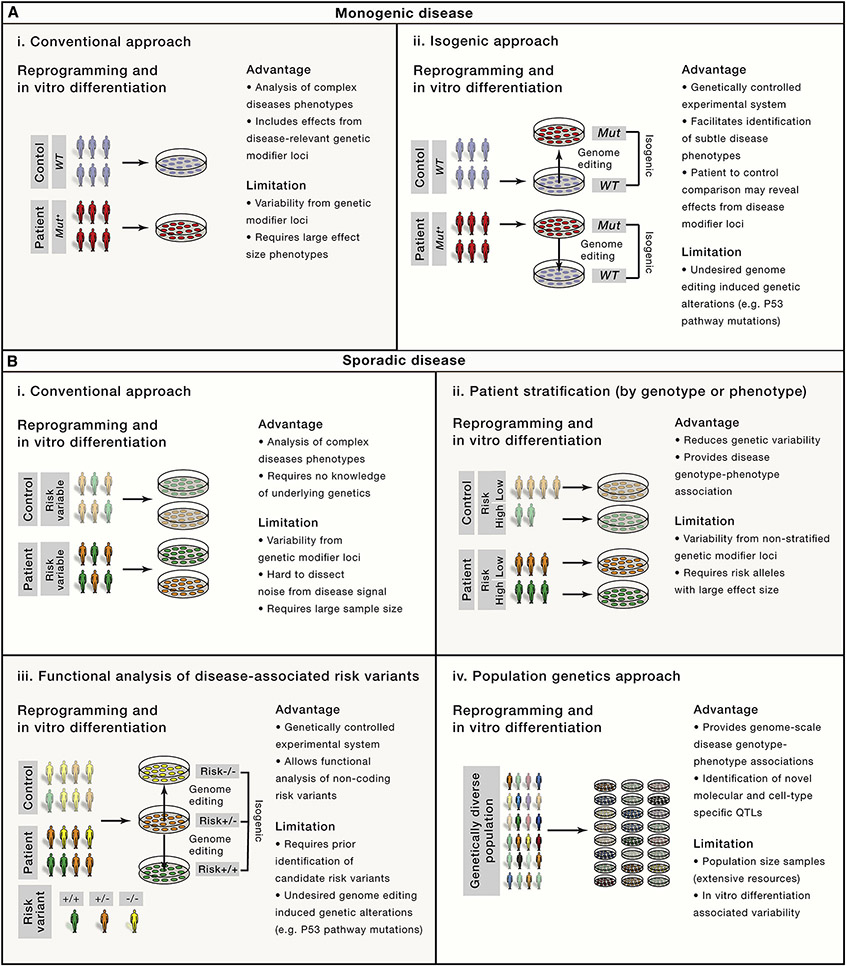
It is generally recognized that individual hiPSCs display a high degree of variability in their biological properties such as the propensity to differentiate into functional cell types, which is independent of genotype or disease status (Liang and Zhang, 2013; Soldner and Jaenisch, 2012). This critically limits the ability to identify robust disease-associated phenotypes by comparing patient-derived cells with unrelated controls (Figure 1). Such system-innate variability proves challenging for age-related disorders, as the disease-associated phenotypes typically progress slowly over many years. Due to the rather limited time in culture, in vitro phenotypes are expected to be rather mild and subtle. Reprogramming-associated genetic and epigenetic alterations, the lack of robust in vitro differentiation protocols and genetic background variations constitute the main sources of phenotypic cell to cell variation (Liang and Zhang, 2013; Soldner and Jaenisch, 2012) and remain a major limitation of the current hiPSC approach.
Figure 1: Comparison of hiPSC-based strategies to model complex diseases in cell culture.
A. HiPSC-based strategies to identify diseases-associated phenotypes for monogenetic diseases comparing (i) the conventional approach with (ii) an isogenic approach. Genome editing allows to generate an experimental system with controlled genetic background by either correcting a disease-associated mutation in patient-derived cells or inversely inserting the mutation into wild-type hPSCs.
B. HiPSC-based strategies to model sporadic diseases comparing (i) the conventional approach with (ii) a patient stratification approach based on known disease associated genotypes (risk score or specific risk genes) or clinical phenotypes and (iii) population genetics approaches to identify associations between specific genotypes and QTLs and (iv) the functional analysis of specific disease-disease associated risk variants. Genome editing allows to generate an allelic series of isogenic cell lines carrying distinct risk variants.
Genetic engineering of disease models
Even mutations that cause highly penetrant human diseases are influenced by genetic background and show significant phenotypic and clinical variation like disease penetrance or age of onset. Using concepts from genetically engineered mouse models, genetic engineering of hPSCs should provide an elegant solution to study the effect of disease-associated mutations in a genetically controlled context. However, classical gene targeting approaches using homologous recombination were surprisingly inefficient in human cells. Advances in genome editing technologies using engineered site-specific nucleases, such as meganucleases, zinc finger nucleases (ZFNs) (Hockemeyer et al., 2009; Urnov et al., 2005), transcription activator-like effector-based nucleases (TALEN) (Boch et al., 2009; Hockemeyer et al., 2011) and the CRISPR/Cas9 system (Cong et al., 2013; Jinek et al., 2012; 2013; Mali et al., 2013), have vastly increased our ability to modify the genome with unprecedented flexibility (Komor et al., 2017). In particular, the simplicity and versatility of the CRISPR/Cas9 system to efficiently modify the genome in human cells, even at multiple loci simultaneously, allows the engineering of genetically controlled hPSC based models, which differ only at known genetic disease-causing variants.
Testing specific disease-associated variants in an otherwise identical isogenic context should detect even subtle molecular or cellular phenotypes masked by variability in genetic background (Figure 1A). As a proof of principle, we used ZFNs to either seamlessly correct Parkinson’s disease-associated mutations in the SNCA gene in patient-derived hiPSCs or to insert similar variants into wild-type hESCs (Soldner et al., 2011). These isogenic hPSC lines provided an experimental system with a controlled genetic background (Figure 1A), which allowed us to identify mitochondrial dysfunction and nitrosative and oxidative stress as molecular events in the pathogenesis of Parkinson’s disease (Chung et al., 2013; Ryan et al., 2013). Engineering of isogenic hPSCs by either correcting the disease-associated mutation in patient-derived hiPSCs or insertion into wild-type hPSCs has become standard to distinguish disease-associated effects from background variation. Examples include Parkinson’s and Alzheimer’s disease (Paquet et al., 2016; Reinhardt et al., 2013; Schöndorf et al., 2014), hereditary motor and sensory neuropathy (Murakami et al., 2017), frontotemporal lobar degeneration (Imamura et al., 2016), Huntington disease (Xu et al., 2017), tetrahydrobiopterin metabolism disorder (Ishikawa et al., 2016), fragile X syndrom (Xie et al., 2016) and the functional analysis of neurorexin-1 in neuropsychiatric diseases (Pak et al., 2015). Limitations to this approach are off-target cleavage of site-specific nucleases and undesired genetic alterations in cell survival pathways such as P53 mutations as a consequence of genome engineering associated genotoxic stress and isolating single cell clones (Haapaniemi et al., 2018; Ihry et al., 2018; Merkle et al., 2017). Also, effects of genetic modifiers in a given genetic background may mask or alter relevant diseases-associate phenotypes.
HPSC models of complex diseases – genetic considerations
Monogenetic familial diseases are rare in neurological disorders. Most cases are sporadic with no well-defined genetic etiology and inheritance patterns. Epidemiology and population genetics suggest that sporadic diseases result from complex interactions between genetic and non-genetic risk factors including age, lifestyle, and environment (Lander, 2011; McClellan and King, 2010). This multi-faceted complexity of sporadic diseases complicates generating genetically defined disease-relevant cellular and animal models impeding our understanding of the underlying molecular mechanisms. While rare monogenetic forms may provide mechanistic insights into mechanisms, common pathological or clinical features between familial and sporadic forms may be a poor predictor for common underlying causes because of the highly variable genetic and phenotypic spectrum of sporadic diseases.
The evolution of sequencing and genotyping technologies allows dissecting the genetic basis of sporadic forms of common diseases in a systematic and cost effective manner (Lander, 2011). Genome-scale sequencing and large-scale genome-wide association studies (GWASs) have identified sequence variants such as single nucleotide polymorphisms (SNPs), deletions and insertions associated with a wide variety of neurological diseases including Parkinson’s disease (Chang et al., 2017), Alzheimer’s disease (Lambert et al., 2013), schizophrenia (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014), bipolar disorders (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013) and autism spectrum disorders (Gaugler et al., 2014). These studies suggest a fluent spectrum of genetic risk with disease reflecting the individual genetic architecture (Pihlstrøm et al., 2017). Prevailing models assume that disease susceptibility arises through a combination of multiple rare or common risk variants rather than determined by strict Mendelian inheritance (Lander, 2011; McClellan and King, 2010). While rare genetic alterations and de-novo mutations with large effects usually occur predominantly in neurodevelopmental and psychiatric diseases, common alleles with low effect size may be more significant for neurodegenerative disorders.
Risk variants pinpoint genomic loci and genes that may play key roles in disease pathogenesis, but functional interpretation of GWAS datasets remains challenging. Sequence variants merely are statistical associations with disease risk and have no established biological function (Lander, 2011; McClellan and King, 2010). Most GWAS loci contain numerous disease-associated risk variants in linkage disequilibrium (LD) spanning a wide genomic area with multiple target genes complicating the identification of relevant disease genes. Further, the majority of disease-associated genetic variants reside in the non-coding part of the genome, which largely prevents any functional analysis in non-human cellular or animal model systems given the limited sequence conservation of non-coding elements. Advancing from disease association to mechanistic insights into disease development and progression, however, is central to our understanding of complex diseases (Ward and Kellis, 2012).
HPSC models of complex diseases - conventional approach
hiPSCs have fundamentally changed our ability to investigate the pathogenesis of complex diseases. Without detailed knowledge of the underlying genetics, patient-derived hiPSCs after in vitro differentiation provide a nearly unlimited supply of disease-relevant somatic cells for in vitro modelling that carry all the genetic elements implicated in disease development. Comparing neurons derived from patients with schizophrenia or autism spectrum disorders to age-matched unaffected individuals (Figure 1B) identified disease-relevant phenotypic changes providing proof-of concept to modeling sporadic diseases (reviewed in (Ardhanareeswaran et al., 2017; Hoffman et al., 2018). While initially surprising, considering the patients’ genetic heterogeneity, such robust disease-associated phenotypes may result from diverse genetic variants that converge on common developmental pathways such as cortical development, synaptic function and epigenetic processes (Hoffman et al., 2018). To reduce variability and facilitate the identification of disease-associated phenotypes, large-scale hiPSC consortia have significantly increased the number of independent samples (HD iPSC Consortium, 2012; Soares et al., 2014).
Stringent patient selection based on genetic or clinical features may reduce heterogeneity in the patient population and significantly increase the power of hiPSC based studies (Figure 1B) (Hoekstra et al., 2017). One approach uses a polygenic risk score, that summarizes the number of GWAS identified risk alleles and the respective effect size for genetically informed patient stratification (Lewis and Vassos, 2017). Stratification of patients with schizophrenia by polygenic risk scores (Hoffman et al., 2018) or patients with autism spectrum disorders by common clinical features such as macrocephaly or microcephaly have facilitated the identification of robust diseases- associated molecular and cellular phenotypes (Marchetto et al., 2015; Mariani et al., 2017). A similar approach using genetic risk information has been successfully applied to neurodegenerative diseases to stratify patients, which carry rare, high-risk mutations in GBA (glucosylceramidase beta) associated with increased risk to develop Parkinson’s disease (Schöndorf et al., 2014) or specific APOE (Huang et al., 2017) or SORL1 (sortilin-related receptor, L(DLR class) A repeats containing) alleles (Young et al., 2015) associated with increased risk to develop Alzheimer’s disease.
HPSC models of complex diseases - population genetics approach
Common disease-associated risk alleles for late age of onset disorders typically have a small effect size, which often precludes observations in cellular models. One common strategy to dissect risk loci is to correlate disease-associated sequence variations with phenotypic or molecular quantitative traits. A quantitative trait locus (QTL) is a specific region in the genome where a particular sequence variant correlates with the variation in a quantitative trait. Although gene expression (eQTLs) is the most systematically analyzed trait (GTEx Consortium, 2015), a wide spectrum of molecular features can identify QTLs, such as the expression of microRNAs, noncoding RNAs, alternative splicing, protein expression, the abundance of metabolic products (mQTLs) or epigenetic modifications including chromatin accessibility, histone modifications, DNA methylation (meQTLs) and transcription factor binding (Ward and Kellis, 2012).
The promise of genome-scale eQTL mapping to gain insight into the functional effect of common risk variants initiated collaborative efforts to provide high-quality transcriptome datasets from genotyped individuals. The main databases that include brain samples are the Genotype-Tissue Expression (GTEx) project (GTEx Consortium, 2015), Braincloud (Colantuoni et al., 2011), CommonMind (Senthil et al., 2017), the PsychENCODE project (PsychENCODE Consortium, 2015) and the UK brain expression consortium (UKBEC; http://www.braineac.org). Although cis-acting effects of sequence variants on gene expression have been identified as major factors for phenotypic variation and disease susceptibility (GTEx Consortium, 2015; Lee and Young, 2013), this approach has many limitations in the CNS, in particular access to large numbers of high quality brain samples for sufficient statistical power to reliably identify eQTLs with small effect size. Cis-regulatory effects on gene expression are extremely cell-type specific, so resolving cellular heterogeneity within brain tissue (e.g. variable contribution from neurons, astrocytes, oligodendrocytes, microglia, epithelial cells and blood cells) represents a significant challenge to eQTL mapping. It remains to be seen if emerging single cell technologies, such as single nuclei FACS sorting from post mortem tissue and single-nuclei RNA-sequencing (snRNA-seq), will resolve some limitations (Habib et al., 2017).
An alternative approach to resolve cellular heterogeneity complements brain QTL analysis with in vitro hiPSC-derived datasets. Many collaborative initiatives established large hiPSC cell repositories from healthy subjects and patients with a wide spectrum of monogenic and complex diseases (Soares et al., 2014). A consortium supported by the National Heart, Lung, and Blood Institute’s Next Generation Association Studies (NextGen) and the Human Induced Pluripotent Stem Cell Initiative (HipSci) tested an idea from hiPSC-based QTL association studies in vitro (Figure 1B) (Warren et al., 2017a). Initial “population genetics in a dish” analysis combined more than 2000 hiPSCs and investigated sources of variation of gene expression in hiPSCs. Genetic background was the main source of variability allowing identification of numerous genetic variants that contribute to variation and cell-line specific regulation of transcription (DeBoever et al., 2017; Kilpinen et al., 2017). However, comparing transcriptional variation within and across individuals indicated a nearly equal amount of non-genetic variability, partly driven by Polycomb repression complex target genes, which may reflect reprogramming-associated variability (Carcamo-Orive et al., 2017).
The subsequent analysis of eQTLs and chromatin accessibility in hiPSC-derived sensory neurons (Schwartzentruber et al., 2017), and eQTLs and metabolic alterations (mQTLs) in hiPSC-derived hepatocyte-like cells (Pashos et al., 2017; Warren et al., 2017b) showed a significant overlap between primary tissues and in vitro identified QTLs that may complement in vivo datasets with novel associations not observed in primary tissues or patients. The unique advantage of this approach is that in vitro differentiated somatic cells not only serve as discovery platform for QTLs but at the same time provide an experimental system to perform functional cellular genomic experiments. Pashos and colleagues used CRISPR/Cas genome editing to confirm a functional effect of novel risk variants on gene expression (Pashos et al., 2017). Comparing in vitro derived sensory neurons with primary dorsal root ganglia however reveals experimental variation of in vitro derived cells as major limitation of this approach. Given that detecting effects of regulatory variants with moderately large effect sizes requires at least 20 to 80 hiPSC lines, this approach depends on simple and robust protocols to differentiate large cohorts of hPSCs (Schwartzentruber et al., 2017). It remains to be seen whether in vitro association studies can reliably detect novel associations not observed in primary tissues or patients.
HPSC models of complex diseases - Functional analysis of disease-associated risk variants
Correlating genomic variants with quantitative traits such as gene expression can isolate disease-relevant genes and pathways but does not identify causal risk variants or provide molecular insights into how sequence variation contributes to the pathogenesis of complex diseases (Ward and Kellis, 2012). Compelling evidence suggests that disease-association of non-coding risk variants is driven by sequence-specific alterations in the function of non-coding regulatory elements that affects transcription and disease susceptibility. Large-scale collaborative initiatives, such as the ENCODE (ENCODE Project Consortium, 2012), the PsychENCODE (PsychENCODE Consortium, 2015) and Roadmap Epigenomics project (Roadmap Epigenomics Consortium, 2015) are accelerating discovery of functional non-coding elements. These resources provide a comprehensive catalogue for cell and tissue-type specific regulatory sequences including the human brain.
Consistent with gene-regulatory variants impacting disease risk, GWAS disease associations are enriched in open chromatin regions with enhancer-associated histone modifications in tissues and cell-types affected by the respective disease. (Ernst et al., 2011; Hnisz et al., 2013; Maurano et al., 2012). Distal enhancer elements bound by transcription factors control gene expression in a tissue and cell-type specific manner and sequence-specific changes in transcription factor-binding in enhancers correlate with changes in chromatin state and cis-regulated gene expression (Karnuta and Scacheri, 2018; Lee and Young, 2013; Wamstad et al., 2014). This suggests a simple molecular mechanism of how individual sequence variants modify transcription factor binding, contribute to alterations in transcription and affect disease susceptibility.
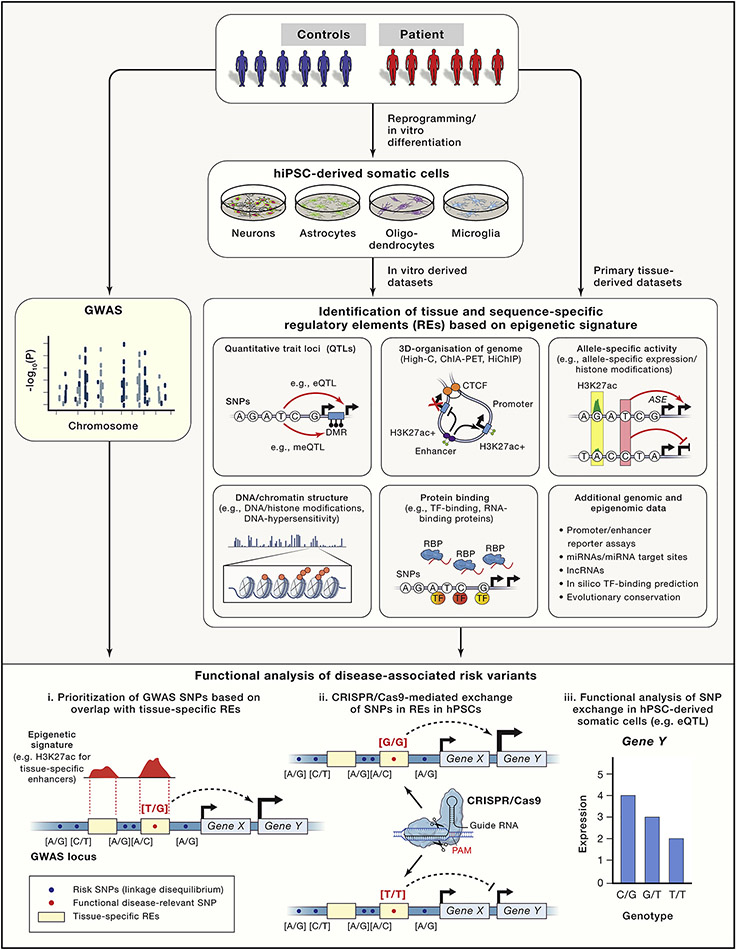
The integration of epigenetic signatures with GWAS datasets may facilitate the discovery of functional disease-associated risk variants in regulatory elements (Figure 2). Mapping disease-associated SNPs to transcription factor binding sites that regulate tissue-specific enhancer function and gene expression identified causal risk variants in metabolic (Claussnitzer et al., 2015) and cardiac diseases (Wang et al., 2016). To refine prioritizing plausible functional variants, current efforts focus on establishing a rigorous experimental and statistical framework integrating novel and more complex epigenetic data (Schaid et al., 2018; Ward and Kellis, 2012). Such datasets include the 3-dimensional organization of the genome to map interactions between enhancers and target genes (Hnisz et al., 2016), allele-specific epigenetic and gene expression information to facilitate the identification of cis-regulatory effects of sequence variants and mapping of RNA binding proteins to identify regulators of alternative splicing. Again, cellular heterogeneity and limited access to brain tissues restrict generation of high-quality datasets with sufficient sequencing coverage. One current strategy complements existing patient tissue-derived datasets with hiPSC-derived somatic cell populations, preferentially from the same donor with identical genetic background (Figure 2).
Figure 2: Identification of functional risk variants based on epigenetic signatures.
Candidate disease-associated risk variants (e.g.SNP) are prioritized based on integrating GWAS data with a variety of epigenetic datasets. Risk variants overlapping with tissue-specific regulatory elements are more likely to be functional compared to risk variants outside of regulatory elements. Epigenetic datasets can be generated either from primary tissue or from in vitro hPSC-derived somatic cell types. Schema at the bottom depicts (i) the identification of a hypothetical GWAS identified diseases-associated risk variant overlapping with a brain-specific distal enhancer (indicated by enrichment for histone H3 lysine 27 acetylation Chip-seq signal), (ii) the CRISPR/Cas9 mediated SNP exchange and (iii) the functional analysis of the SNP exchange indicated by a genotype-dependent cis-regulatory effect on expression of Gene Y (eQTL).
To advance from association to causal variant, we recently applied this approach to prioritize risk variants based on the epigenetic signature to sporadic Parkinson’s disease. After compiling a candidate list of probable causal risk variants in active brain enhancers by integrating epigenetic and GWAS datasets, we employed CRISPR/Cas genome editing to dissect the function of diseases-associated risk variants (Figure 2). We identified the sequence-specific binding of transcription factors in a non-coding risk variant in an intronic enhancer element to regulate the expression of SNCA, a key gene implicated in the pathogenesis of Parkinson’s disease (Soldner et al., 2016). This proof-of-concept illustrates that the integration of population genetic and genome-scale epigenetic data combined with hiPSC and genome editing technologies provide a platform to study complex disorders in a genetically controlled and systematic manner. A similar approach was used to dissect the genetic risk associated with 5 vascular diseases, including coronary artery disease, migraine, cervical artery dissection, fibro- muscular dysplasia and hypertension and identified a non-coding risk variant in an intronic enhancer of PHCTR1 to regulate the expression of endothelin 1, a gene with physiological function in vasculature (Gupta et al., 2017).
Current efforts focus on advancing from the “one risk variant at a time” approach to systematically evaluate risk variants on a genome scale. Targeted engineering of existing and newly identified species of Cas proteins has substantially broadened the versatility of the CRISPR/Cas toolbox to now target almost any genomic base (Komor et al., 2017). Several studies performed pooled genetic screens using tiling libraries of gRNAs to target and disrupt the function of thousands of endogenous non-coding elements for functional interrogation (reviewed in (Klann et al., 2018). Alternatively, targeting regulatory sequences with a nuclease deficient Cas9 protein (dCas9) fused to transcriptional activators (CRISPRa), suppressors (CRISPRi) (Gilbert et al., 2014) or active domains of epigenetic modifiers such as DNA methylation/demethylation or histone acetylation can change the activity of non-coding regulatory elements (Amabile et al., 2016; Kearns et al., 2015; Liu et al., 2016). An example of a potential candidate for therapeutic application of epigenetic editing is fragile X syndrome, which is caused by a GGC expansion leading to the silencing of FMR1. Targeting the repeat by a dCas9-Tet fusion protein caused demethylation of the repeat, restoration of FMR1 expression and reversal of electrophysiological abnormalities in fragile X patient-derived neurons (Liu et al., 2018). Pooled epigenome editing screens can systematically analyze the functional consequences of modulating the activity of non-coding elements and interrogate the function of GWAS identified risk variants in regulatory elements on a genome scale (Fulco et al., 2016; Hilton et al., 2015; Thakore et al., 2015). However, these experiments used tumor cell lines and examined large biological effects that enrich functional gRNAs. Whether small effects on gene expression as a molecular mechanism suggested for disease susceptibility are sufficient to identify functional gRNA targets and translate such approaches to human post- mitotic neurons remains an open question. A promising novel approach termed Pertrub-seq may combine single cell sequencing with CRISPR/Cas mediated modulation/disruption of non-coding regulatory elements to dissect the molecular effects on gene expression in individual cells (Adamson et al., 2016; Dixit et al., 2016; Jaitin et al., 2016).
HPSC models of complex phenotypes
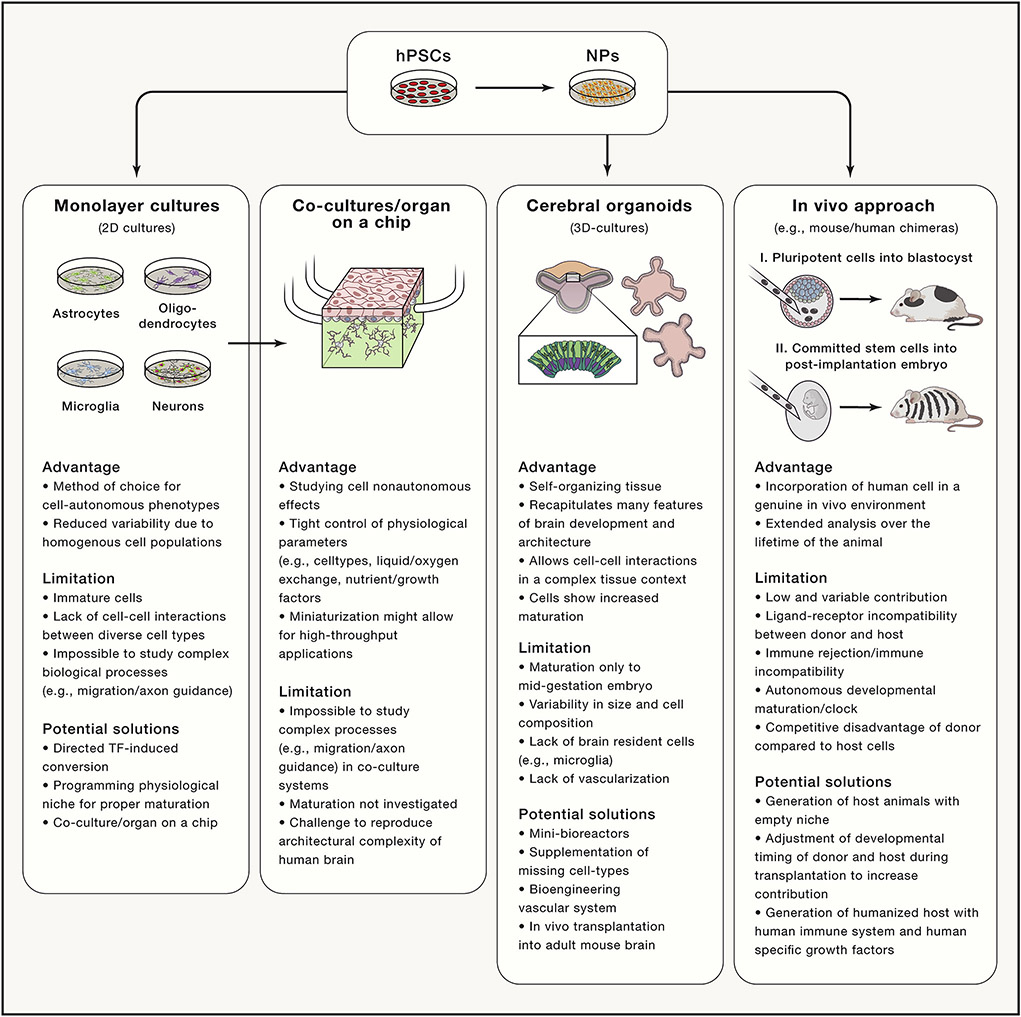
A major limitation of hiPSC approaches is the lack of robust differentiation protocols. The traditional protocols for in vitro differentiation recapitulate the order and timing of regional specification and neuronal commitment during embryonic development by supplying developmental cues like morphogens or modulating developmentally relevant pathways using small molecules (Keller, 1995). This approach can direct cell fates with remarkable precision to all major cell types of the nervous system including various distinct neuronal sub-types resembling dopaminergic, glutamatergic, GABAergic, motor, hypothalamic and striatal medium spiny neurons (Figure 3) (Tao and Zhang, 2016). However, differentiation outcomes may suffer from substantial clone-to-clone variability, adding a significant source of variability to cellular disease models (Soldner and Jaenisch, 2012). In addition, any disease-related genetic variant could also affect the differentiation process adding another layer of complexity to the interpretation of phenotypic variation between disease and control cultures.
Figure 3: Comparison of in vitro and chimeric in vivo approaches to model complex disease-associated phenotypes.
hPSCs, human pluripotent stem cells; NPs, neural progenitors; TF, transcription factor.
One strategy to robustly control in vitro differentiation is directed conversion of hPSCs to pure and functionally mature neurons driven by forced expression of cell fate specifying neuronal transcription factors such as NGN2 to induce cortical excitatory neurons (iNs) (Zhang et al., 2013) or NGN2, ISL1, LHX3, NEUROD1, BRN2, ASCL1 and MYT1L to induce motor neurons (iMNs) (Shi et al., 2018). This approach allowed dissection of the molecular effects of distinct ApoE isoforms, a strong risk factor for Alzheimer’s disease, amyloid-beta precursor protein (APP) expression and disease susceptibility (Huang et al., 2017). An alternate strategy selects specific cell types within a heterogeneous population by genetically inserting cell-type specific selection or fluorescent marker proteins. Using longitudinal tracking of the fluorescent motor neuron specific reporter Hb9-RFP in hiPSCs derived from patients with GGGGCC repeat expansion in C9ORF72, which underlies amyotrophic lateral sclerosis and frontotemporal dementia, Shi and colleagues observed a robust degeneration of patient-derived cells and identified disruption of vesicle trafficking as the pathogenic molecular mechanism (Shi et al., 2018).
HPSC models of complex phenotypes - Programming a physiological niche
Many in vitro differentiation protocols are thought to produce rather immature cells with fetal-stage like properties independent of the age of the initial donor (Studer et al., 2015). The failure to generate mature functional cells in vitro may result from the lack of a physiological environment to direct proper maturation. Using concepts from reprogramming of fibroblasts to hiPSCs or direct lineage conversion of fibroblast to induced neurons, it may be possible to convert cell identity from an immature to a more mature state by recreating the transcriptional program of mature cells. Access to transcriptional and epigenomic information from in vitro differentiated neuronal cell types and the in vivo counterparts derived from mature primary tissue will permit mapping epigenetic and transcriptional differences between immature and mature cell states. These differences may identify key transcriptional regulators that may circumvent the requirement for an appropriate exogenous niche and program the conversion of an immature to a mature cell identity.
Since aging is an important risk factor for many neurodegenerative diseases, current efforts attempt t o mimic age-related processes in cell culture. In vitro ectopic overexpression of Progerin, a truncated form of the mutated Lamin A gene associated with the premature aging syndrome Hutchinson-Gilford Progeria, in hPSC-derived neurons induce aging associated changes, such as genomic instability, epigenetic dysregulation and mitochondrial dysfunction. Overexpression of Progerin was sufficient to accelerate the appearance of disease-associated phenotypes in a hiPSC model of Parkinson’s disease (Miller et al., 2013). It remains to be seen whether this approach can be applied more broadly to age-related diseases and whether phenotypes observed in a premature aging context correlate with disease pathologies.
HPSC models of complex phenotypes - neuronal co-culture systems
Because neuronal cell loss is central to many neurological disorders, much current research focuses mainly on generating homogenous neuronal cultures. A pure cell population remains the gold standard to study cell-autonomous molecular and cellular phenotypes due to reduced confounding effects from cellular heterogeneity. However, non-neuronal cell types, including astrocytes and microglia, and non-cell autonomous interactions among these cells are essential for brain development and function, the proper maturation of in vitro cultured neuronal cells and play a key role in the pathogenesis of neurological diseases (Clarke and Barres, 2013; Salter and Stevens, 2017). Different strategies exist to recreate a physiological in vivo niche to allow for adequate differentiation, aging and interaction between in vitro derived cell types (Figure 3).
Initial hPSC-based disease modeling studies, using simple co-culture systems between astrocytes and neurons, successfully dissected the effects of SOD1 mutations in glial cells on motor neuron survival in amyotrophic lateral sclerosis (Di Giorgio et al., 2008). These advances spurred efforts to produce more sophisticated in vitro systems by combining hPSC technology with basic engineering principles using novel synthetic biomaterials, hydrogels, microfluidics and 3Dprinting with the goal to recreate some of the complexity of the human brain. These approaches allow tight control of physiological parameters such as cell-type composition, liquid and oxygen exchange or nutrients and growth factor delivery (Figure 3). Now, novel microdevices, commonly referred to “organs-on-a-chip”, are being developed to recapitulate the complex structure and functionality of a human organs (Ronaldson-Bouchard and Vunjak-Novakovic, 2018).
HPSC models of complex phenotypes - cerebral organoids
Although conventional 2-dimensional (2D) monolayer co-cultures can recapitulate many non-cell-autonomous interactions, such models are still limited to provide the complexity of 3-dimensional (3D) brain structures to study complex features, such as spatial organization of neuronal structures (e.g. neocortical layer formation), migration, axon guidance or establishment and response to morphogen gradients. These dysfunctional processes likely contribute to disease pathogenesis especially in neurodevelopmental disorders. Recently, 3D-culture models resembling human brain structures, commonly referred to as cerebral organoids, have facilitated the unprecedented opportunity to study complex aspects of human brain development and disease pathology in a cell culture dish (Kadoshima et al., 2013; Lancaster et al., 2013).
Exploiting the self-organizing capacity of neuroectodermal precursor cells, cerebral organoids recapitulate many features of human brain development and morphology, including ventricle and neural tube-like structures with apical-basal polarity, progenitor zone organization with outer radial glial stem cells, cortical plate development and electrophysiological mature neurons that participate in neuronal network activity (Kadoshima et al., 2013; Karzbrun et al., 2018; Lancaster et al., 2013; 2017; Li et al., 2017b; Paşca et al., 2015; Quadrato et al., 2017). Cerebral organoids have already provided novel insights into human-specific processes, which could not be studied in conventional 2D culture systems, such as brain gyrification (Karzbrun et al., 2018; Li et al., 2017b), cell migration and outer radial glial mitosis defects in Miller-Dieker syndrome (Bershteyn et al., 2017).
Organoid models are now a common tool to study neurodevelopmental disorders, including microcephaly (Lancaster et al., 2013; Li et al., 2017a), autism spectrum disease associated with macrocephaly (Mariani et al., 2017; Wang et al., 2017), lissencephally (Bershteyn et al., 2017; Karzbrun et al., 2018), Rett Syndrom (Mellios et al., 2017), Thymothy syndrome (Birey et al., 2017), lysosomal storage diseases such as Sandhoff disease (Allende et al., 2018), schizophrenia (Srikanth et al., 2018; Stachowiak et al., 2017) and Zika infection on neonatal brain development (reviewed in (Qian et al., 2017). Despite the early success with this approach for disease-modelling, challenges remain. The current organoid approach produces substantial variability between individual cerebral organoids regarding organoid size and cellular composition (Quadrato et al., 2017). More sophisticated bioengineering approaches such as the use of mini-bioreactors to grow individual organoids under controlled conditions (Qian et al., 2016) may reduce variability, which should allow studying a broader range of complex diseases.
Although neuronal cell types are abundant in cerebral organoids, the detailed analysis of cellular diversity reveals the lack of non-neuronal cell types such as endothelial cells and microglia. Since these cell types likely have a central role in brain development and disease pathology, one strategy is to externally provide the missing cell types during organoid formation. We demonstrated in vitro hPSC-derived microglia efficiently invade cerebral organoids, showing the utility of this strategy (Muffat et al., 2016).
One concern is whether neuronal cells in cerebral organoids mature past early events in brain development. In vitro derived organoids recapitulate gene expression and epigenetic programs of fetal cortical development. After extended periods in culture, organoids show a gene expression signature comparable to mid-gestation embryos between post-conception week 17-24 (Camp et al., 2015; Luo et al., 2016; Paşca et al., 2015; Quadrato et al., 2017). This may explain why most reported organoid studies describe the dysfunction of processes that occur during early brain development.
One major barrier for extended in vitro culture with further maturation is the lack of vascularization for nutrient and oxygen penetration, as late stage cerebral organoids commonly develop extensive necrosis and massive cell death. Current attempts to engineer a vascular system into organoids either exploit the self-organizing capacity of stem cells to construct a functional vascular system by providing appropriate vascular precursor populations during organoid development or using sophisticated bio and tissue-engineering tools to construct artificial vasculature like structures. One alternate strategy to overcome this limitation transplants cerebral organoids into the adult mouse brain. Transplanted organoids survive and mature for extended periods of time in the host brain. Surprisingly, mouse blood vessels invade the organoids to create a functional vascular system, resulting in significantly improved neuronal survival and the establishment of extensive functional-graft-to-host synaptic connection (Mansour et al., 2018).
It remains uncertain whether cerebral organoid models can be applied to study late age onset disorders. Yet, experimental evidence exists modelling Alzheimer’s disease like pathology in a 3D culture system with ectopic overexpression of familial Alzheimer’s diseases-associated mutations in beta-amyloid precursor protein and presenilin 1 (Choi et al., 2014; Park et al., 2018) and in cerebral organoids derived from Alzheimer’s patients (Raja et al., 2016), suggesting the feasibility to replicate disease-associated pathology not observed in Alzheimer’s disease mouse models.
HPSC models of complex phenotypes - interspecies chimeras
Generating interspecies chimeras using cells from more than one individual may provide the most physiologically relevant environment to study human disease in an in vivo context. Experimental chimeras derived from cells within the same species are routinely used for biomedical application including generating genetically engineered mouse models. The idea of interspecies chimeras between human cells and a model organism such as the mouse, rat, pig or monkey is particular tantalizing for late age of onset disorders, as it would allow studying patient-derived cells in a genuine in vivo environment for extended periods of time over the lifetime of the animal. Although transplantation of human cells into adult hosts animals provides an important research model to study the feasibility of cell-replacement therapies for regenerative applications, we will only focus here on approaches that may support the functional integration of host cells during normal embryonic development.
One approach generates interspecies chimeras by injecting hPSCs into the preimplantation embryo, so human cells would participate in normal development and functionally integrate throughout the entire host embryo. However, this approach currently seems rather inefficient, as the extent of functional integration into the host animal remains an intense debate (Gafni et al., 2013; Theunissen et al., 2016; Wu et al., 2016) A recent report described the robust contribution of human cells to the pre-implantation embryo but only limited contribution to the post-implantation pig embryo (Wu et al., 2017). Currently it is not clear whether the variable contributions result from differences in evolutionary distance, better matched developmental states or reduced ligand-receptor incompatibility between human donor cells and the host embryo.
An alternative approach generates chimeras by transplanting lineage-restricted multipotent stem cells into the developing post-gastrulation embryo. The donor cells, after transplanting into a developmental-stage matched host, should participate in normal embryonic development to generate functionally integrated human cells of the respective cell lineage. Using this approach, we showed that human neural crest stem cells transplanted into the gastrulating mouse embryo followed neural crest specific migration patterns and contributed to the pigment lineage in postnatal mice (Cohen et al., 2016). Importantly, genetic depletion of the host melanoblast lineage significantly increased integration of human cells, suggesting that an “empty host niche” gives donor cells a selective advantage to efficiently contribute to interspecies chimeras. Similarly, transplanted human neural precursor cells into E14.5 embryos or terminally differentiated cortical neurons into neonatal mice robustly functionally integrate into mouse brain (Espuny-Camacho et al., 2013; Nagashima et al., 2014).
One surprising difference between these two transplantation paradigms is that neural crest cells may acquire functional properties at the time of birth of the host animal, while transplanted human neuronal cells may functionally mature substantially slower than host neurons. This indicates, that human transplanted neural cells follow a developmental maturation pattern resembling human embryonic development rather than the host environment (Espuny-Camacho et al., 2013). Similarly, this cell autonomous developmental clock may inhibit or delay the appearance of mature cell types in vitro. In both experimental paradigms, the overall contribution of human cells is rather limited. This could be caused by technical challenges related to transplantation (Cohen et al., 2016; Nagashima et al., 2014), imperfect adjustment of developmental timing between donor and host cells at the time of transplantation (Cohen et al., 2018) or species-specific differences.
Inter-species immune-rejection and the lack of crucial signals from the host environment may contribute to limited survival. Using sophisticated immunodeficient mouse models or humanized hosts carrying fully functional human immune system or genetically engineered human specific growth factors could enhance the chimeric contribution of human cells (Zhang et al., 2004). Recent intraspecies chimera experiments suggest that matched developmental timing is crucial for successful contributions by transplanted cells (Cohen et al., 2018). It is still unclear, whether inhibition of cell death (e.g. overexpression of antiapoptotic factors such as Bcl2) is sufficient to relax this requirement (Cohen et al., 2018; Masaki et al., 2016).
Proof of principle examples demonstrate the unique promise of studying human cells in a physiologically relevant, in vivo environment. Human neurons transplanted into a mouse model of Alzheimer’s disease display signs of neuronal degeneration accompanied by cell loss and some pathological features observed in patients with Alzheimer’s disease, such as dystrophic presynaptic changes and hyper-phosphorylation of tau (Espuny-Camacho et al., 2017). The most stunning demonstration of interspecies chimera approach generated human glia chimeric mice by injecting bipotential astrocyte-oligodendrocyte precursors into the neonatal brain. The transplanted human cells did not only survive and integrate, but also replaced the entire glial population of the host brain. The engraftment of human glia in the mouse brain enhanced synaptic plasticity and learning in the transplanted mice (Han et al., 2013). Transplanting glial precursor cells derived from schizophrenia patients induced schizophrenia-like behaviors like excessive anxiety, antisocial traits and disrupted sleep in mice indicating a causal role of glial pathology in disease development (Windrem et al., 2017).
HPSC-based drug discovery
A main motivation behind hPSC-based disease modeling is to discover novel therapeutic targets and develop new drugs. Since human model system are anticipated to be more predictive for clinical efficacy of novel drugs, this approach may substantially reduce cost and time to translate potential therapies into the clinic. Initial proof-of concept experiments employed a candidate approach testing a small number of compounds to reverse disease-associated phenotypes in human disease-relevant cell types. Selected candidate small molecules included compounds known to target disease-relevant molecular pathways, compounds expected to be effective based on analysis in non-hiPSC disease models and compounds previously described as effective in related diseases to evaluate for repurposing. This strategy yielded numerous biologically active compounds that can reverse disease-associated phenotypes in vitro for a wide variety of neurodevelopmental, psychiatric and neurodegenerative disorders (reviewed in (Avior et al., 2016).
Initial compounds, which either used hiPSC-based disease models for pre-clinical drug development or were developed based on observations from hiPSC disease models, are already in clinical trials. These includes a novel class of splicing modifying compounds to treat spinal muscular atrophy (Naryshkin et al., 2014) (https://clinicaltrials.gov; NCT02913482), a humanized antibody targeting secreted eTau to treat tauopathies (Bright et al., 2015), including progressive supranuclear palsy (https://clinicaltrials.gov; NCT03068468) and early Alzheimer’s disease (https://clinicaltrials.gov; NCT03352557), and the Kv7.2/3 potassium channel agonist ezogabine, an approved antiepileptic medication to treat amyotrophic lateral sclerosis (Kiskinis et al., 2014; Wainger et al., 2014) (https://clinicaltrials.gov; NCT# 02450552).
A major promise of hiPSC technology is the potential to perform unbiased high-throughput compound (HTS) or genome-scale genetic screens. Drug development in the past predominantly employed target-based drug discovery (TDD) that requires identifying defined molecular targets relevant to disease pathogenesis (Moffat et al., 2017). However, TDD has been remarkably ineffective for neurological diseases given the complexity of pathological phenotypes and lack of reliable model systems to identify essential disease-relevant targets. As alternative, recent progress in hiPSC technology has revived phenotypic drug discovery (PDD). This approach does not rely on a priori knowledge of a specific drug target and its role in disease pathology. Instead, it screens for reversing complex disease-associated phenotypes, even with little insight into the underlying molecular mechanisms.
The critical requirement for phenotypical screens are robust, simple to analyze disease-relevant phenotypes, which can be easily scaled up for high-throughput applications. Despite the limitations, some unbiased medium-scale screens have already identified active disease-phenotype modifying compounds for neurological disorders, including familial dysautonomia (Ramirez et al., 2012), fragile X syndrome (Kaufmann et al., 2015; Kumari et al., 2015; Li et al., 2016), Alzheimer’s diseases (Brownjohn et al., 2017; Kondo et al., 2017; Xu et al., 2013), Parkinson’s disease (Ryan et al., 2013), motor neuron disease (Höing et al., 2012), amyotrophic lateral sclerosis (Burkhardt et al., 2013; Egawa et al., 2012; Shi et al., 2018) and Zika infection on fetal brain development (Zhou et al., 2017). Dictated by the underlying disease pathophysiology, the screening readouts can range from simple quantitative traits such as re-expression of the FMR1 gene for fragile-X-syndrome to complex higher order disease-associated traits such as cellular degeneration for amyotrophic lateral sclerosis (Shi et al., 2018).
Developing high-content screening platforms, which implement automatic digital microscopy combined with sophisticated computational image analysis to quantify complex traits, will become critical for successful phenotypical screening approaches (Finkbeiner et al., 2015). However, practical and technical challenges associated with the complexity of the hPSC platform currently limit scaling-up, required for a broader application of PDD. Efforts to automatize and miniaturize the hPSC-screening platforms using cutting edge robotics, microfluidics and nanotechnology may overcome this limitation. Current phenotypical screens only screen small molecule collections in the order of several hundred to several thousand compounds, not the million-compound libraries commonly used in the pharmaceutical industry. These screening libraries focus on well-curated compounds with known molecular targets or biological activity, or utilize small-molecule libraries that contain drugs already approved for clinical applications. Drug repurposing should substantially reduce cost and facilitate translation of novel therapeutics with known safety, pharmacodynamics and pharmacokinetics information.
An alternative to chemical compound-based drug discovery are genome-scale genetic screens. Genetic screens in various model systems like yeast, flies, nematodes and mice prove to be invaluable to investigate gene function, dissect complex biological processes and identify drug targets. In contrast to the enormous chemical space of pharmacological active molecules, the number of genes in the human genome constrains the scale of genetic compared to compound screens. Genetic screens resolve some limitations in chemical phenotypic screens like identifying molecular targets of compounds recovered in chemical screens as they directly pinpoint the gene and pathways implicated in the observed phenotypes. Importantly, genetic screens may provide targets for subsequent target-based drug discovery (TDD) efforts. The ease and efficiency of the CRISPR/Cas system to target any locus in the genome provides great flexibility to develop genome-wide knock-out, gene activation (CRISPRa), gene suppressor (CRISPRi) or epigenome modifier screens in human cells (Gilbert et al., 2014). Genome-scale CRISPR screens are already used to identify genes implicated in many biologically relevant phenotypes, such as gene essentiality (Wang et al., 2017), caner progression (Chen et al., 2015; Shalem et al., 2014; Shi et al., 2015), drug resistance (Shalem et al., 2014; Wang et al., 2014), viral infection, immune response and vulnerability to bacterial toxins (Zhou et al., 2014). While published genome-scale screens predominantly utilize fast proliferating tumor cell lines, genome-scale phenotypical genetic screens in post-mitotic neuronal cells may be already feasible and become an invaluable tool for drug discovery in neurological diseases.
Conclusion
Rapid advances in the hPSC approach combined with progress in genome editing technologies and the availability of genetic and epigenetic information now provide a realistic opportunity to study human neurological disorders in a disease-relevant and systematic manner. Most sporadic diseases arise through a complex interaction of multiple genetic and non-genetic risk factors suggesting that each person’s disease prognosis, including onset, progression and responsivity to treatment, is an individual trait. Personalized medicine will tailor therapies to a patient’s unique clinical, physiological and environmental information.
We described how hiPSCs could bridge this gap by generating disease relevant models from the patient’s own cells. This hPSC approach allows easily alternation between the in vitro disease platform and in vivo patient context. We can now generate novel disease hypotheses based on specific clinical observations and functionally test them with high molecular precision in disease-relevant cellular models from the same individual or reversely use insights gained from cellular models and directly confirm the relevance in a patient’s context. Combined with continuing advances in tissue engineering that can generate 3-dimensional organoid structures resembling entire organs, we envision that complex patient-derived human model systems will become a powerful research tool in our understanding of human physiology and disease development, as seen with other tractable model organisms. Working with human systems will overcome many limitations of non-human model organisms, which will more successfully translate newly identified molecular pathways or disease phenotype modifying chemical compounds into novel targets for therapeutic interventions. These advances remarkably occurred only 20 after the first report deriving a hESC line and just over 10 years after the stunning discovery that reprogrammed human somatic cells into hPSCs.
Now, the first clinical trials are already underway evaluating the efficacy of hPSC-derived therapeutics for transplantation of novel drugs developed in hPSC disease models. This stands in contrast to the initial uncertainty if cell culture models for late age of onset neurological disorders could identify any disease-relevant phenotype. The fundamental impact of hPSCs on disease modeling cannot be overstated. There remains no effective alternative approach to functionally dissect the molecular and cellular effects of non-coding variation and to develop reliable models for many devastating sporadic disorders. However, we acknowledge these initial steps will further the promise of hPSCs technology to transform biomedical research. As for every nascent field, unresolved issues of the hPSC approach remains, as discussed in this review. We are confident that the rapidly evolving field will overcome these barriers, so that hPSCs will increasingly yield basic biological and clinical translational insights for years to come.
Acknowledgements:
Work in our lab is supported by grants from the NIH. We would like to thank members of the Jaenisch lab as well as many other colleagues for stimulating discussions and Brandi Mattson for help with editing the manuscript. We apologize to many whose work we did not adequately describe or cite due to space constraints.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Adamson B, Norman TM, Jost M, Cho MY, Nuñez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, et al. (2016). A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell 167, 1867–1873.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allende ML, Cook EK, Larman BC, Nugent A, Brady JM, Golebiowski D, Sena-Esteves M, Tifft CJ, and Proia RL (2018). Cerebral organoids derived from Sandhoff disease-induced pluripotent stem cells exhibit impaired neurodifferentiation. J. Lipid Res. 59, 550–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amabile A, Migliara A, Capasso P, Biffi M, Cittaro D, Naldini L, and Lombardo A (2016). Inheritable Silencing of Endogenous Genes by Hit- and-Run Targeted Epigenetic Editing. Cell 167, 219–224.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardhanareeswaran K, Mariani J, Coppola G, Abyzov A, and Vaccarino FM (2017). Human induced pluripotent stem cellsfor modelling neurodevelopmentaldisorders. Nature Publishing Group 13, 265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avior Y, Sagi I, and Benvenisty N (2016). Pluripotent stem cells in diseasemodelling and drug discovery. Nat Rev Mol Cell Biol 1–13. [DOI] [PubMed] [Google Scholar]
- Bershteyn M, Nowakowski TJ, Pollen AA, Di Lullo E, Nene A, Wynshaw-Boris A, and Kriegstein AR (2017). Human iPSC-Derived Cerebral Organoids Model Cellular Features of Lissencephaly and Reveal Prolonged Mitosis of Outer Radial Glia. Cell Stem Cell 20, 435–449.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, Fan HC, Metzler KRC, Panagiotakos G, Thom N, et al. (2017). Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, and Bonas U (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512. [DOI] [PubMed] [Google Scholar]
- Bolker JA (2017). Animal Models in Translational Research: Rosetta Stone or Stumbling Block? BioEssays 39, 1700089. [DOI] [PubMed] [Google Scholar]
- Bright J, Hussain S, Dang V, Wright S, Cooper B, Byun T, Ramos C, Singh A, Parry G, Stagliano N, et al. (2015). Neurobiology of Aging. Neurobiology of Aging 36, 693–709. [DOI] [PubMed] [Google Scholar]
- Brownjohn PW, Smith J, Portelius E, Serneels L, Kvartsberg H, De Strooper B, Blennow K, Zetterberg H, and Livesey FJ (2017). Phenotypic screening identifies modulators of amyloid precursor protein processing in human stem cell models of Alzheimer’s disease. Stem Cell Reports 8, 870–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt MF, Martinez FJ, Wright S, Ramos C, Volfson D, Mason M, Garnes J, Dang V, Lievers J, Shoukat-Mumtaz U, et al. (2013). Molecular and Cellular Neuroscience. Molecular and Cellular Neuroscience 56, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, et al. (2015). Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proceedings of the National Academy of Sciences 201520760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcamo-Orive I, Hoffman GE, Cundiff P, Beckmann ND, D’Souza SL, Knowles JW, Patel A, Papatsenko D, Abbasi F, Reaven GM, et al. (2017). Analysis of Transcriptional Variability in a Large Human iPSC Library Reveals Genetic and Nongenetic Determinants of Heterogeneity. Cell Stem Cell 20, 518–531.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang D, Nalls MA, Hallgrímsdóttir IB, Hunkapiller J, van der Brug M, Cai F, International Parkinson's Disease Genomics Consortium, 23andMe Research Team, Kerchner GA, Ayalon G, et al. (2017). A meta-analysis of genome-wide association studies identifies 17 new Parkinson's disease risk loci. Nat Genet 2017 October; 49(10):1511–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, et al. (2015). Genome-wide CRISPR Screen in a Mouse Model of Tumor Growth and Metastasis. Cell 160, 1246–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D'Avanzo C, Chen H, Hooli B, Asselin C, Muffat J, et al. (2014). A three-dimensional human neural cell culture model of Alzheimer's disease. Nature 515, 274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CY, Khurana V, Auluck PK, Tardiff DF, Mazzulli JR, Soldner F, Baru V, Lou Y, Freyzon Y, Cho S, et al. (2013). Yeast Reveal a “Druggable” Rsp5/Nedd4 Network that Ameliorates α-Synuclein Toxicity in Neurons. Science 342, 979–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke LE, and Barres BA (2013). Emerging roles of astrocytes in neural circuit development. Nat Rev Neurosci 14, 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M, Dankel SN, Kim K-H, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V, et al. (2015). FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N Engl J Med 373, 895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MA, Markoulaki S, and Jaenisch R (2018). Matched developmental timing of donor cells with the host is crucial for chimera formation. Stem Cell Reports 10, 1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MA, Wert KJ, Goldmann J, Markoulaki S, Buganim Y, Fu D, and Jaenisch R (2016). Human neural crest cells contribute to coat pigmentation in interspecies chimeras after in utero injection into mouse embryos. Proceedings of the National Academy of Sciences 113, 1570–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, et al. (2011). Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 478, 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium (2013). Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 1371–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBoever C, Li H, Jakubosky D, Benaglio P, Reyna J, Olson KM, Huang H, Biggs W, Sandoval E, D’Antonio M, et al. (2017). Large-Scale Profiling Reveals the Influence of Genetic Variation on Gene Expression in Human Induced Pluripotent Stem Cells. Cell Stem Cell 20, 533–546.e537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio F, Boulting G, Bobrowicz S, and Eggan K (2008). Human Embryonic Stem Cell-Derived Motor Neurons Are Sensitive to the Toxic Effect of Glial Cells Carrying an ALS-Causing Mutation. Cell Stem Cell 3, 637–648. [DOI] [PubMed] [Google Scholar]
- Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, et al. (2016). Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 167, 1853–1857.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa N, Kitaoka S, Tsukita K, Naitoh M, Takahashi K, Yamamoto T, Adachi F, Kondo T, Okita K, Asaka I, et al. (2012). Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci Transl Med 4, 145ra104. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. (2011). Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espuny-Camacho I, Arranz AM, Fiers M, Snellinx A, Ando K, Munck S, Bonnefont J, Lambot L, Corthout N, Omodho L, et al. (2017). Hallmarks of Alzheimer’s Disease in Stem-Cell- Derived Human Neurons Transplanted into Mouse Brain. Neuron 93, 1066–1081.e1068. [DOI] [PubMed] [Google Scholar]
- Espuny-Camacho I, Michelsen KA, Gall D, Linaro D, Hasche A, Bonnefont J, Bali C, Orduz D, Bilheu A, Herpoel A, et al. (2013). Pyramidal Neurons Derived from Human Pluripotent Stem Cells Integrate Efficiently into Mouse Brain Circuits In Vivo. Neuron 77, 440–456. [DOI] [PubMed] [Google Scholar]
- Finkbeiner S, Frumkin M, and Kassner PD (2015). Cell-Based Screening: Extracting Meaning from Complex Data. Neuron 86, 160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco CP, Munschauer M, Anyoha R, Munson G, Grossman SR, Perez EM, Kane M, Cleary B, Lander ES, and Engreitz JM (2016). Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 354, 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, et al. (2013). Derivation of novel human ground state naive pluripotent stem cells. Nature 504, 282–286. [DOI] [PubMed] [Google Scholar]
- Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, Mahajan M, Manaa D, Pawitan Y, Reichert J, et al. (2014). Most genetic risk for autism resides with common variation. Nat Genet 46, 881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. (2014). Genome-Scale CRISPR-Mediated Controlof Gene Repression and Activation. Cell 159, 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium (2015). Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RM, Hadaya J, Trehan A, Zekavat SM, Roselli C, Klarin D, Emdin CA, Hilvering CRE, Bianchi V, Mueller C, et al. (2017). A Genetic Variant Associated with Five Vascular Diseases Is a Distal Regulator of Endothelin-1 Gene Expression. Cell 170, 522–526.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapaniemi E, Botla S, Persson J, Schmierer B, and Taipale J (2018). CRISPR-Cas9 genome editing induces a p53- mediated DNA damage response. Nat Med 1–10. [DOI] [PubMed] [Google Scholar]
- Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M, Choudhury SR, Aguet F, Gelfand E, Ardlie K, et al. (2017). Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods 14, 955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Chen M, Wang F, Windrem M, Wang S, Shanz S, Xu Q, Oberheim NA, Bekar L, Betstadt S, et al. (2013). Forebrain Engraftmentby Human Glial Progenitor Cells Enhances Synaptic Plasticity and Learning in Adult Mice. Cell Stem Cell 12, 342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HD iPSC Consortium (2012). Induced pluripotent stem cells from patients with Huntington's disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell 11, 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, D'Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, and Gersbach CA (2015). Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33, 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, and Young RA (2013). Super-Enhancers in the Control of Cell Identity and Disease. Cell 155, 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Day DS, and Young RA (2016). Insulated Neighborhoods: Structuraland Functional Units of Mammalian Gene Control. Cell 167, 1188–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, Dekelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. (2009). Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol 27, 851–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. (2011). Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol 29, 731–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra SD, Stringer S, Heine VM, and Posthuma D (2017). Genetically-Informed Patient Selection for iPSC Studies of Complex Diseases May Aid in Reducing Cellular Heterogeneity. Front. Cell. Neurosci. 11, 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman GE, Schrode N, Flaherty E, and Brennand KJ (2018). New considerations for hiPSC-based models of neuropsychiatric disorders. Mol. Psychiatry 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höing S, Rudhard Y, Reinhardt P, Glatza M, Stehling M, Wu G, Peiker C, Böcker A, Parga JA, Bunk E, et al. (2012). Discovery of Inhibitors of MicroglialNeurotoxicity Acting Through Multiple Mechanisms Using a Stem-Cell-Based Phenotypic Assay. Cell Stem Cell 11, 620–632. [DOI] [PubMed] [Google Scholar]
- Huang Y-WA, Zhou B, Wernig M, and Südhof TC (2017). ApoE2, ApoE3, and ApoE4 Differentially Stimulate APP Transcription and Ab Secretion. Cell 168, 427–441.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihry RJ, Worringer KA, Salick MR, Frias E, Ho D, Theriault K, Kommineni S, Chen J, Sondey M, Ye C, et al. (2018). p53 inhibits CRISPR–Cas9 engineering in human pluripotent stem cells. Nat Med 1–16. [DOI] [PubMed] [Google Scholar]
- Ilic D, and Ogilvie C (2016). Concise Review: Human Embryonic Stem Cells-What Have We Done? What Are We Doing? Where Are We Going? Stem Cells 35, 17–25. [DOI] [PubMed] [Google Scholar]
- Imamura K, Sahara N, Kanaan NM, Tsukita K, Kondo T, Kutoku Y, Ohsawa Y, Sunada Y, Kawakami K, Hotta A, et al. (2016). Calcium dysregulation contributesto neurodegeneration in FTLDpatient iPSC-derived neurons. Sci Rep 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Imamura K, Kondo T, Koshiba Y, Hara S, Ichinose H, Furujo M, Kinoshita M, Oeda T, Takahashi J, et al. (2016). Genetic and pharmacological correction of aberrant dopamine synthesis using patient iPSCs with BH4 metabolism disorders. Hum Mol Genet 347 Pt 1, ddw339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin DA, Weiner A, Yofe I, Lara-Astiaso D, Keren-Shaul H, David E, Salame TM, Tanay A, van Oudenaarden A, and Amit I (2016). Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA-Seq. Cell 167, 1883–1896.e15. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, and Charpentier E (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, and Doudna J (2013). RNA-programmed genome editing in human cells. Elife 2, e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jucker M (2010). The benefits and limitations of animal models for translational research in neurodegenerative diseases. Nat Med 16, 1210–1214. [DOI] [PubMed] [Google Scholar]
- Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, and Sasai Y (2013). Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proceedings of the National Academy of Sciences 110, 20284–20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnuta JM, and Scacheri PC (2018). Enhancers: bridging the gap between gene control and human disease. Hum Mol Genet 27, R219–R227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzbrun E, Kshirsagar A, Cohen SR, Hanna JH, and Reiner O (2018). Human brain organoids on a chip reveal the physics of folding. Nature Physics 14, 515–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann M, Schuffenhauer A, Fruh I, Klein J, Thiemeyer A, Rigo P, Gomez-Mancilla B, Heidinger-Millot V, Bouwmeester T, Schopfer U, et al. (2015). High-Throughput Screening Using iPSC-Derived Neuronal Progenitors to Identify Compounds Counteracting Epigenetic Gene Silencing in Fragile X Syndrome. J Biomol Screen 20, 1101–1111. [DOI] [PubMed] [Google Scholar]
- Kearns NA, Pham H, Tabak B, Genga RM, Silverstein NJ, Garber M, and Maehr R (2015). Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat Methods 12, 401–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller GM (1995). In vitro differentiation of embryonic stem cells. Current Opinion in Cell Biology 7, 862–869. [DOI] [PubMed] [Google Scholar]
- Kilpinen H, Goncalves A, Leha A, Afzal V, Alasoo K, Ashford S, Bala S, Bensaddek D, Casale FP, Culley OJ, et al. (2017). Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 546, 370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiskinis E, Sandoe J, Williams LA, Boulting GL, Moccia R, Wainger BJ, Han S, Peng T, Thams S, Mikkilineni S, et al. (2014). Pathways Disrupted in Human ALSMotor Neurons Identifiedthrough Genetic Correction of Mutant SOD1. Cell Stem Cell 14, 781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann TS, Black JB, and Gersbach CA (2018). ScienceDirectCRISPR-based methods for high-throughput annotation of regulatory DNA. Current Opinion in Biotechnology 52, 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komor AC, Badran AH, and Liu DR (2017). CRISPR-Based Technologiesfor the Manipulation of Eukaryotic Genomes. Cell 168, 20–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Imamura K, Funayama M, Tsukita K, Miyake M, Ohta A, Woltjen K, Nakagawa M, Asada T, Arai T, et al. (2017). iPSC-Based Compound Screening and In Vitro Trials Identify a Synergistic Anti-amyloid b Combination for Alzheimer’s Disease. Cell Rep 21, 2304–2312. [DOI] [PubMed] [Google Scholar]
- Kumari D, Swaroop M, Southall N, Huang W, Zheng W, and Usdin K (2015). High-Throughput Screening to Identify Compounds That Increase Fragile X Mental Retardation Protein Expression in Neural Stem Cells Differentiated From Fragile X Syndrome Patient-Derived Induced Pluripotent Stem Cells. STEM CELLS Translational Medicine 4, 800–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, et al. (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 45, 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Corsini NS, Wolfinger S, Gustafson EH, Phillips AW, Burkard TR, Otani T, Livesey FJ, and Knoblich JA (2017). Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol 35, 659–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, and Knoblich JA (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES (2011). Initial impact of the sequencing of the human genome. Nature 470, 187–197. [DOI] [PubMed] [Google Scholar]
- Lee TI, and Young RA (2013). Transcriptional Regulation and Its Misregulation in Disease. Cell 152, 1237–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CM, and Vassos E (2017). Prospects for using risk scores in polygenic medicine. 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhao H, Ananiev GE, Musser MT, Ness KH, Maglaque DL, Saha K, Bhattacharyya A, and Zhao X (2016). Establishment of Reporter Lines for Detecting Fragile X Mental Retardation (FMR1) Gene Reactivation in Human Neural Cells. Stem Cells 35, 158–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Le Sun, Fang A, Li P, Wu Q, and Wang X (2017a). Recapitulating cortical developmentwith organoid culture in vitro and modeling abnormal spindle-like (ASPM related primary) microcephaly disease. Protein & Cell 8, 823–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Muffat J, Omer A, Bosch I, Lancaster MA, Sur M, Gehrke L, Knoblich JA, and Jaenisch R (2017b). Induction of Expansion and Folding in Human Cerebral Organoids. Cell Stem Cell 20, 385–396.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, and Zhang Y (2013). Genetic and epigenetic variations in iPSCs: potential causes and implications for application. Cell Stem Cell 13, 149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, and Jaenisch R (2016). Editing DNA Methylation in the Mammalian Genome. Cell 167, 233–235.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Wu H, Krzisch M, Wu X, Graef J, Muffat J, Hnisz D, Li CH, Yuan B, Xu C, et al. (2018). Rescue of Fragile X Syndrome Neurons by DNA Methylation Editing of the FMR1 Gene. Cell 172, 979–991.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Lancaster MA, Castanon R, Nery JR, Knoblich JA, and Ecker JR (2016). Cerebral Organoids Recapitulate Epigenomic Signatures of the Human Fetal Brain. Cell Rep 17, 3369–3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. (2007). Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell 1, 55–70. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, and Church GM (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandai M, Watanabe A, Kurimoto Y, Hirami Y, Morinaga C, Daimon T, Fujihara M, Akimaru H, Sakai N, Shibata Y, et al. (2017). Autologous Induced Stem-Cell-Derived Retinal Cells for Macular Degeneration. N Engl J Med 376, 1038–1046. [DOI] [PubMed] [Google Scholar]
- Mansour AA, Gonçalves JT, Bloyd CW, Li H, Fernandes S, Quang D, Johnston S, Parylak SL, Jin X, and Gage FH (2018). An in vivo model of functional and vascularized human brain organoids. Nat Biotechnol 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Belinson H, Tian Y, Freitas BC, Fu C, Vadodaria KC, Beltrao-Braga PC, Trujillo CA, Mendes APD, Padmanabhan K, et al. (2015). FOXG1-Dependent Dysregulation of GABA/ Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell 162, 375–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, Amenduni M, Szekely A, Palejev D, Wilson M, et al. (2017). FOXG1-Dependent Dysregulation of GABA/ Glutamate Neuron Differentiation in Autism Spectrum Disorders. 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki H, Kato-Itoh M, Takahashi Y, Umino A, Sato H, Ito K, Yanagida A, Nishimura T, Yamaguchi T, Hirabayashi M, et al. (2016). Inhibition of Apoptosis Overcomes Stage-Related Compatibility Barriers to Chimera Formation in Mouse Embryos. Cell Stem Cell 19, 587–592. [DOI] [PubMed] [Google Scholar]
- Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et al. (2012). Systematic Localization of Common Disease- Associated Variation in Regulatory DNA. Science 337, 1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan J, and King M-C (2010). Genetic Heterogeneity in Human Disease. Cell 141, 210–217. [DOI] [PubMed] [Google Scholar]
- McNeish J, Gardner JP, Wainger BJ, Woolf CJ, and Eggan K (2015). From Dish to Bedside: Lessons Learned While Translating Findings from a Stem Cell Model of Disease to a Clinical Trial. Cell Stem Cell 17, 8–10. [DOI] [PubMed] [Google Scholar]
- Mehat MS, Sundaram V, Ripamonti C, Robson AG, Smith AJ, Borooah S, Robinson M, Rosenthal AN, Innes W, Weleber RG, et al. (2018). Transplantation of Human Embryonic Stem Cell-Derived Retinal Pigment Epithelial Cells in Macular Degeneration. Ophthalmology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellios N, Feldman DA, Sheridan SD, Ip JPK, Kwok S, Amoah SK, Rosen B, Rodriguez BA, Crawford B, Swaminathan R, et al. (2017). MeCP2-regulated miRNAs control early human neurogenesis through differential effects on ERK and AKT signaling. Mol. Psychiatry 23, 1051–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menasché P, Vanneaux V, Hagége A, Bel A, Cholley B, Parouchev A, Cacciapuoti I, Al-Daccak R, Benhamouda N, Blons H, et al. (2018). Transplantation of Human Embryonic Stem Cell-Derived Cardiovascular Progenitors for Severe Ischemic Left Ventricular Dysfunction. J. Am. Coll. Cardiol. 71, 429–438. [DOI] [PubMed] [Google Scholar]
- Merkle FT, Ghosh S, Kamitaki N, Mitchell J, Avior Y, Mello C, Kashin S, Mekhoubad S, Ilic D, Charlton M, et al. (2017). Human pluripotent stem cells recurrently acquire and expand dominant negative P53 mutations. Nature 545, 229–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JD, Ganat YM, Kishinevsky S, Bowman RL, Liu B, Tu EY, Mandal PK, Vera E, Shim J-W, Kriks S, et al. (2013). Human iPSC-Based Modeling of Late-Onset Disease via Progerin-Induced Aging. Cell Stem Cell 13, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat JG, Vincent F, Lee JA, Eder J, and Prunotto M (2017). Opportunities and challenges in phenotypic drug discovery: an industry perspective. Nat Rev Drug Discov 16, 531–543. [DOI] [PubMed] [Google Scholar]
- Muffat J, Li Y, Yuan B, Mitalipova M, Omer A, Corcoran S, Bakiasi G, Tsai L-H, Aubourg P, Ransohoff RM, et al. (2016). Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat Med 22, 1358–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami N, Imamura K, Izumi Y, Egawa N, Tsukita K, Enami T, Yamamoto T, Kawarai T, Kaji R, and Inoue H (2017). Proteasome impairment in neural cells derived from HMSN-P patient iPSCs. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima F, Suzuki IK, Shitamukai A, Sakaguchi H, Iwashita M, Kobayashi T, Tone S, Toida K, Vanderhaeghen P, and Kosodo Y (2014). Novel and Robust Transplantation Reveals the Acquisition of Polarized Processes by Cortical Cells Derived from Mouse and Human Pluripotent Stem Cells. Stem Cells and Development 23, 2129–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naryshkin NA, Weetall M, Dakka A, Narasimhan J, Zhao X, Feng Z, Ling KKY, Karp GM, Qi H, Woll MG, et al. (2014). Motor neuron disease. SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science 345, 688–693. [DOI] [PubMed] [Google Scholar]
- Pak C, Danko T, Zhang Y, Aoto J, Anderson G, Maxeiner S, Yi F, Wernig M, and Südhof TC (2015). Human Neuropsychiatric Disease Modeling using Conditional Deletion Reveals Synaptic Transmission Defects Caused by Heterozygous Mutations in NRXN1. Cell Stem Cell 17, 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, Olsen KM, Gregg A, Noggle S, and Tessier-Lavigne M (2016). Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533, 125–129. [DOI] [PubMed] [Google Scholar]
- Park J, Wetzel I, Marriott I, Dréau D, D'Avanzo C, Kim DY, Tanzi RE, and Cho H (2018). A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat Neurosci 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashos EE, Park Y, Wang X, Raghavan A, Yang W, Abbey D, Peters DT, Arbelaez J, Hernandez M, Kuperwasser N, et al. (2017). Large, Diverse Population Cohorts of hiPSCs and Derived Hepatocyte-like Cells Reveal Functional Genetic Variation at Blood Lipid-Associated Loci. Cell Stem Cell 20, 558–570.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paşca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park J-Y, O'Rourke NA, Nguyen KD, et al. (2015). Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods 12, 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihlstrøm L, Wiethoff S, and Houlden H (2017). Genetics of neurodegenerative diseases: an overview. Handb Clin Neurol 145, 309–323. [DOI] [PubMed] [Google Scholar]
- PsychENCODE Consortium, Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, Jaffe AE, Pinto D, Dracheva S, et al. (2015). The PsychENCODE project. Nat Neurosci 18, 1707–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Jacob F, Song H, and Ming G-L (2017). Using brain organoids to understand Zika virus-induced microcephaly. Development 144, 952–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, et al. (2016). Brain-Region-Specific Organoids Using Mini- bioreactors for Modeling ZIKV Exposure. Cell 165, 1238–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, Maria N, Scholvin J, Goldman M, Kinney JP, et al. (2017). Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545, 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja WK, Mungenast AE, Lin Y-T, Ko T, Abdurrob F, Seo J, and Tsai L-H (2016). Self-Organizing 3D Human Neural Tissue Derived from Induced Pluripotent Stem Cells Recapitulate Alzheimer’s Disease Phenotypes. PLoS ONE 11, e0161969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez CN, Kim H, Zeltner N, Liu B, Radu C, Bhinder B, Kim YJ, Choi IY, Mukherjee-Clavin B, Djaballah H, et al. (2012). large-scale screening using familial dysautonomia induced pluripotent stem cells identifies compounds that rescue IKBKAP expression. Nat Biotechnol 30, 1244–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt P, Schmid B, Burbulla LF, Schöndorf DC, Wagner L, Glatza M, Höing S, Hargus G, Heck SA, Dhingra A, et al. (2013). Genetic Correction of a LRRK2 Mutationin Human iPSCs Links Parkinsonian Neurodegeneration to ERK-Dependent Changes in Gene Expression. Cell Stem Cell 12, 354–367. [DOI] [PubMed] [Google Scholar]
- Reubinoff BE, Pera MF, Fong CY, Trounson A, and Bongso A (2000). Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol 18, 399–404. [DOI] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaldson-Bouchard K, and Vunjak-Novakovic G (2018). Organs-on-a-Chip: A Fast Track for Engineered Human Tissues in Drug Development. Cell Stem Cell 22, 310–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan SD, Dolatabadi N, Chan SF, Zhang X, Akhtar MW, Parker J, Soldner F, Sunico CR, Nagar S, Talantova M, et al. (2013). Isogenic Human iPSC Parkinson’s Model Shows Nitrosative Stress-InducedDysfunction in MEF2-PGC1a Transcription. Cell 155, 1351–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter MW, and Stevens B (2017). Microglia emerge as central players in brain disease. Nat Med 23, 1018–1027. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, Chen W, and Larson NB (2018). From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat Rev Genet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöndorf DC, Aureli M, McAllister FE, Hindley CJ, Mayer F, Schmid B, Sardi SP, Valsecchi M, Hoffmann S, Schwarz LK, et al. (2014). iPSC-derived neurons from GBA1-associated Parkinson's disease patients show autophagic defects and impaired calcium homeostasis. Nature Communications 5, 4028. [DOI] [PubMed] [Google Scholar]
- Schwartz SD, Regillo CD, Lam BL, Eliott D, Rosenfeld PJ, Gregori NZ, Hubschman J-P, Davis JL, Heilwell G, Spirn M, et al. (2015). Human embryonic stem cell-derived retinal pigment epithelium in patients with age-related macular degeneration and Stargardt's macular dystrophy: follow-up of two open-label phase 1/2 studies. Lancet 385, 509–516. [DOI] [PubMed] [Google Scholar]
- Schwartzentruber J, Foskolou S, Kilpinen H, Rodrigues J, Alasoo K, Knights AJ, Patel M, Goncalves A, Ferreira R, Benn CL, et al. (2017). Molecular and functional variation in iPSC-derived sensory neurons. Nat Genet 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil G, Dutka T, Bingaman L, and Lehner T (2017). Genomic resources for the study of neuropsychiatric disorders. Mol. Psychiatry 22, 1659–1663. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert L, Root DE, Doench JG, et al. (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, and Vakoc CR (2015). Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol 33, 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Inoue H, Wu JC, and Yamanaka S (2016). Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov 16, 115–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lin S, Staats KA, Li Y, Chang W-H, Hung S-T, Hendricks E, Linares GR, Wang Y, Son EY, et al. (2018). Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat Med 24, 313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares FAC, Sheldon M, Rao M, Mummery C, and Vallier L (2014). International coordination of large-scale human induced pluripotent stem cell initiatives: Wellcome Trust and ISSCR Workshops white paper. Stem Cell Reports 3, 931–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, and Jaenisch R (2012). Medicine. iPSC disease modeling. Science 338, 1155–1156. [DOI] [PubMed] [Google Scholar]
- Soldner F, Laganiére J, Cheng AW, Hockemeyer D, Gao Q, Alagappan R, Khurana V, Golbe LI, Myers RH, Lindquist S, et al. (2011). Generation of Isogenic Pluripotent Stem Cells Differing Exclusively at Two Early Onset Parkinson Point Mutations. Cell 146, 318–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Stelzer Y, Shivalila CS, Abraham BJ, Latourelle JC, Barrasa MI, Goldmann J, Myers RH, Young RA, and Jaenisch R (2016). Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature 533, 95–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WK, Park K-M, Kim H-J, Lee JH, Choi J, Chong SY, Shim SH, Del Priore LV, and Lanza R (2015). Treatment of macular degeneration using embryonic stem cell-derived retinal pigment epithelium: preliminary results in Asian patients. Stem Cell Reports 4, 860–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth P, Lagomarsino VN, Muratore CR, Ryu SC, He A, Taylor WM, Zhou C, Arellano M, and Young-Pearse TL (2018). Shared effects of DISC1 disruption and elevated WNT signaling in human cerebral organoids. Translational Psychiatry 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak EK, Benson CA, Narla ST, Dimitri A, Chuye LEB, Dhiman S, Harikrishnan K, Elahi S, Freedman D, Brennand KJ, et al. (2017). Cerebral organoids reveal early cortical maldevelopment in schizophrenia— computational anatomy and genomics, role of FGFR1. Translational Psychiatry 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer L, Vera E, and Cornacchia D (2015). Programming and Reprogramming Cellular Age in the Era of Induced Pluripotency. Cell Stem Cell 16, 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan SE, and Young-Pearse TL (2017). Induced pluripotent stem cells as a discovery tool for Alzheimer's disease. Brain Research 1656, 98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, and Yamanaka S (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, and Yamanaka S (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. [DOI] [PubMed] [Google Scholar]
- Tao Y, and Zhang S-C (2016). Neural Subtype Specificationfrom Human Pluripotent Stem Cells. Cell Stem Cell 19, 573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore PI, D'Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, Reddy TE, Crawford GE, and Gersbach CA (2015). Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Genome Biol 10, 1143–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen TW, Friedli M, He Y, Planet E, O’Neil RC, Markoulaki S, Pontis J, Wang H, Iouranova A, Imbeault M, et al. (2016). Molecular Criteria for Defining the Naive Human Pluripotent State. Cell Stem Cell 19, 502–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA (1998). Embryonic Stem Cell Lines Derived from Human Blastocysts. Science 282, 1145–1147. [DOI] [PubMed] [Google Scholar]
- Trounson A, and DeWitt ND (2016). Pluripotent stem cells progressing to the clinic. Nat Rev Mol Cell Biol 17, 194–200. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee Y-L, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, and Holmes MC (2005). Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435, 646–651. [DOI] [PubMed] [Google Scholar]
- Wainger BJ, Kiskinis E, Mellin C, Wiskow O, Han SSW, Sandoe J, Perez NP, Williams LA, Lee S, Boulting G, et al. (2014). Intrinsic Membrane Hyperexcitability of Amyotrophic Lateral Sclerosis Patient-Derived Motor Neurons. Cell Rep 7, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamstad JA, Wang X, Demuren OO, and Boyer LA (2014). Distal enhancers: new insights into heart development and disease. Trends in Cell Biology 24, 294–302. [DOI] [PubMed] [Google Scholar]
- Wang P, Mokhtari R, Pedrosa E, Kirschenbaum M, Bayrak C, Zheng D, and Lachman HM (2017). CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in cerebral organoids derived from iPS cells. 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, and Lander ES (2014). Genetic Screens in Human Cells Using the CRISPR-Cas9 System. Science 343, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tucker NR, Rizki G, Mills R, Krijger PH, de Wit E, Subramanian V, Bartell E, Nguyen X-X, Ye J, et al. (2016). Discovery and validation of sub-threshold genome-wide association study loci using epigenomic signatures. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward LD, and Kellis M (2012). Interpreting noncoding genetic variation in complex traits and human disease. Nat Biotechnol 30, 1095–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren CR, Jaquish CE, and Cowan CA (2017a). The NextGen Genetic Association Studies Consortium: A Forayinto In Vitro Population Genetics. Cell Stem Cell 20, 431–433. [DOI] [PubMed] [Google Scholar]
- Warren CR, O’Sullivan JF, Friesen M, Becker CE, Zhang X, Liu P, Wakabayashi Y, Morningstar JE, Shi X, Choi J, et al. (2017b). Induced Pluripotent Stem Cell Differentiation Enables Functional Validation of GWAS Variants in Metabolic Disease. Cell Stem Cell 20, 547–557.e547. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein B, and Jaenisch R (2007). In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature 448, 318–324. [DOI] [PubMed] [Google Scholar]
- Windrem MS, Osipovitch M, Liu Z, Bates J, Chandler-Militello D, Zou L, Munir J, Schanz S, McCoy K, Miller RH, et al. (2017). Human iPSC Glial Mouse Chimeras Reveal Glial Contributions to Schizophrenia. Cell Stem Cell 21, 195–208.e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Greely HT, Jaenisch R, Nakauchi H, Rossant J, and Belmonte JCI (2016). Stem cells and interspecies chimaeras. Nature 540, 51–59. [DOI] [PubMed] [Google Scholar]
- Wu J, Platero-Luengo A, Sakurai M, Sugawara A, Gil MA, Yamauchi T, Suzuki K, Bogliotti YS, Cuello C, Valencia MM, et al. (2017). Interspecies Chimerism with Mammalian Pluripotent Stem Cells. Cell 168, 473–486.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie N, Gong H, Suhl JA, Chopra P, Wang T, and Warren ST (2016). Reactivation of FMR1 by CRISPR/Cas9-Mediated Deletion of the Expanded CGG-Repeat of the Fragile X Chromosome. PLoS ONE 11, e0165499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Lei Y, Luo J, Wang J, Zhang S, Yang X-J, Sun M, Nuwaysir E, Fan G, Zhao J, et al. (2013). Prevention of β-amyloid induced toxicity in human iPS cell-derived neurons by inhibition of Cyclin-dependent kinases and associated cell cycle events. Stem Cell Research 10, 213–227. [DOI] [PubMed] [Google Scholar]
- Xu X, Tay Y, Sim B, Yoon S-I, Huang Y, Ooi J, Utami KH, Ziaei A, Ng B, Radulescu C, et al. (2017). Reversal of Phenotypic Abnormalities by CRISPR/Cas9-Mediated Gene Correction in Huntington Disease Patient-Derived Induced Pluripotent Stem Cells. Stem Cell Reports 8, 619–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JE, Boulanger-Weill J, Williams DA, Woodruff G, Buen F, Revilla AC, Herrera C, Israel MA, Yuan SH, Edland SD, et al. (2015). Elucidating Molecular Phenotypes Caused by the SORL1 Alzheimer’s Disease Genetic Risk Factor Using Human Induced Pluripotent Stem Cells. Cell Stem Cell 16, 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik M, Smuga-Otto K, Antosiewicz-Bourget J, Frane J, Tian S, Nie J, Jonsdottir G, Ruotti V, Stewart R, et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Chen W, Tan S, and Lin T (2017). Stem Cells for Modeling and Therapy of Parkinson's Disease. Human Gene Therapy 28, 85–98. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, et al. (2013). Rapid Single-Step Induction of Functional Neurons from Human Pluripotent Stem Cells. Neuron 78, 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-W, Su Y, Lanning N, Gustafson M, Shinomiya N, Zhao P, Cao B, Tsarfaty G, Wang L-M, Hay R, et al. (2004). Enhanced growth of human met-expressing xenografts in a new strain of immunocompromised mice transgenic for human hepatocyte growth factor/scatter factor. Oncogene 24, 101–106. [DOI] [PubMed] [Google Scholar]
- Zhou T, Tan L, Cederquist GY, Fan Y, Hartley BJ, Mukherjee S, Tomishima M, Brennand KJ, Zhang Q, Schwartz RE, et al. (2017). High-Content Screening in hPSC-Neural Progenitors Identifies Drug Candidates that Inhibit Zika Virus Infection in Fetal-like Organoids and Adult Brain. Cell Stem Cell 21, 274–283.e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, and Wei W (2014). High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 509, 487–491. [DOI] [PubMed] [Google Scholar]