Abstract
Protein arginine methyltransferases (PRMTs) are found in a wide variety of eukaryotic organisms and can regulate gene expression, DNA repair, RNA splicing, and stem cell biology. In mammalian cells, nine genes encode a family of sequence-related enzymes; six of these PRMTs catalyze the formation of ω-asymmetric dimethyl derivatives, two catalyze ω-symmetric dimethyl derivatives, and only one (PRMT7) solely catalyzes ω-monomethylarginine formation. Purified recombinant PRMT7 displays a number of unique enzymatic properties including a substrate preference for arginine residues in R-X-R motifs with additional flanking basic amino acid residues and a temperature optimum well below 37 °C. Evidence has been presented for crosstalk between PRMT7 and PRMT5, where methylation of a histone H4 peptide at R17, a PRMT7 substrate, may activate PRMT5 for methylation of R3. Defects in muscle stem cells (satellite cells) and immune cells are found in mouse Prmt7 homozygous knockouts, while humans lacking PRMT7 are characterized by significant intellectual developmental delays, hypotonia, and facial dysmorphisms. The overexpression of the PRMT7 gene has been correlated with cancer metastasis in humans. Current research challenges include identifying cellular factors that control PRMT7 expression and activity, identifying the physiological substrates of PRMT7, and determining the effect of methylation on these substrates.
Keywords: PRMT7, monomethylarginine, protein arginine methylation, pluripotency, epigenetics, cancer
1. Introduction
Protein function is often dependent upon the posttranslational derivatization of the twenty amino acid residues on polypeptides that emerge from the ribosome [1]. A prominent site of such modification in eukaryotic cells is the arginine residue that can be methylated at its terminal and bridging nitrogen atoms [2–6]. Methylation of arginine residues can change the local chemical environment in a protein by introducing a degree of hydrophobicity and extra electron density, potentially shielding the positive charge of the guanidino group [3]. Additionally, Methyl groups can also change the pattern of hydrogen bonding. Although it had been thought that methylation decreases the number of potential H-bonds which can be formed by replacing the guanidino protons [3], computational and structural studies have shown the emergence of a new type of H-bond (at least for biochemists): the CH⋯O bond [7–9]. CHO bonds are weaker than the conventional H-bond, but in the case of arginine methylation, the addition of a single methyl group may actually increase the number of possible H-bonds. The specific effects of methylation on the hydrogen bonding patterns of arginine residues still largely remain to be determined.
In mammals, a family of nine sequence-related protein arginine methyltransferases (PRMT1-9) modifies a wide variety of proteins using the cofactor S-adenosylmethionine (AdoMet) to generate terminal monomethylated (MMA) and terminal symmetric (SDMA) and asymmetric (ADMA) dimethylated residues [2–6]. Much is already understood about the structure and function of the predominant ADMA generating (type I) PRMT1 enzyme and the predominant SDMA-generating (type II) enzyme, PRMT5 [3,4,10–13]. For example, methylation of histones by these two enzymes has been shown to regulate transcription and epigenetics [1,3–5,14]. Other type I ADMA-forming enzymes include PRMT2, 3, 4 (also known as CARM1), 6, and 8 [2–4,10,15–17]; PRMT9 is the other type II SDMA-forming enzyme in the family [18,19]. However, the only enzyme that is specific for only monomethylating the terminal nitrogen on arginine residues is the type III enzyme PRMT7 (Figure 1). In this review, we will focus on the unique features of PRMT7 enzymology and substrate specificity, as well as recent studies that have pointed to the role of this enzyme in gene regulation, stem cell biology, and cancer pathologies.
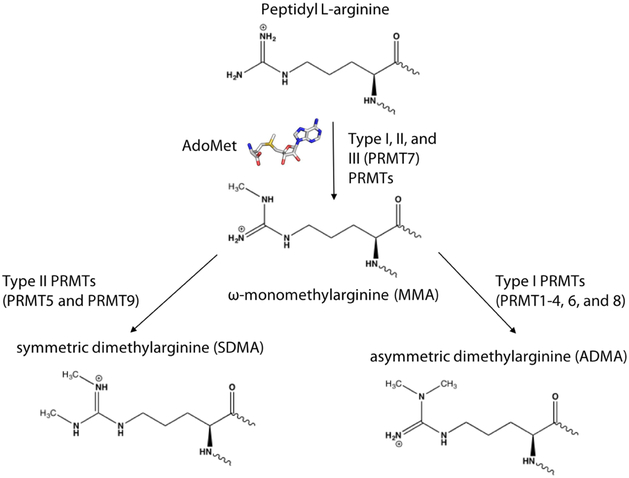
Figure 1. Scheme of arginine methylation carried out by PRMTs.
ADMA and MMA produced by type I (PRMT1-4, 6, and 8), SDMA and MMA produced by type II (PRMT5 and 9), and only MMA produced by type III (PRMT7) enzymes.
2. PRMT7 as the only strictly monomethylating, type III, PRMT
The gene for PRMT7 was first identified in Chinese hamster ovary (CHO) cells [20,21]. Since then, enzymological studies have been conducted on this enzyme from the protozoan Trypanosoma brucei (Tb) [22–25], the nematode worm Caenorhabditis elegans (Ce) [26–28], and humans [29–32]. First biochemically identified as an arginine monomethyltransferase by Miranda et al. in 2004 [29], PRMT7’s product specificity has been the subject of some controversy. In 2005 Lee et al. [33] reported that FLAG-tagged PRMT7 catalyzes SDMA formation. However, earlier work from Nishioka and Reinberg in 2003 demonstrated that the anti-FLAG tag antibodies used to purify FLAG-PRMT7 by Lee et al. also co-purify the major SDMA-producing enzyme PRMT5 [34]. The presence of contaminating PRMT5 in the FLAG-tagged PRMT7 enzyme preparation from Lee et al. likely led to PRMT7's incorrect identification as a dimethylating enzyme. Subsequent in vitro studies have thoroughly characterized PRMT7 as a solely monomethylating, type III enzyme [22,30–32]. The specific mechanisms by which PRMT7’s product specificity is determined are discussed in detail in sections 4 and 5 below.
3. Substrate recognition by PRMT7 in mammals and trypanosomes
Major PRMTs, such as PRMT1 and PRMT5, primarily recognize glycine- and arginine-rich regions (GAR or RGG/RG motifs) of polypeptides for methylation [2–5,10]. TbPRMT7 exhibits a similar sequence specificity in recognizing GAR regions [22–25]. Specifically, reactions with TbPRMT7 and histone H4 N-terminal peptides show that this enzyme prefers methylating arginine-3, a residue flanked by glycine residues [24,25]. However, the metazoan PRMT7 orthologs from C. elegans, M. musculus, and H. sapiens show significantly different recognition of substrate arginine residues [28,31,32]. First shown with mouse and human PRMT7, this enzyme has a strong preference for RXR motifs surrounded by basic amino acids [31,32]. In vitro studies from Feng et al. showed that the major sites of methylation on the N-terminal tail of histone H2B are the arginine residues 29, 31, and 33 in the context of lysine residues at positions 27, 28, 30, and 34 [31,32]. Feng et al. further showed that when mammalian PRMT7 was incubated with the same histone H4 N-terminal peptide used in the TbPRMT7 experiments, the mammalian ortholog only methylated residues R17 and R19 (adjacent to lysine residues) and not residue R3 [31].
Recent work on PRMT7 methylation of the mammalian translational initiation factor 2α (eIF2α) at residues 52-56 (RRRIR) has confirmed the importance of the RXR substrate recognition motif [35]. In this study mutation of the central R54 residue to a lysine residue, resulting in the loss of the RXR motif, eliminated methylation of the eIF2α protein [35]. However, it has also been recently reported that mammalian heat shock proteins are substrates for PRMT7 at an arginine residue not in an RXR motif [36]. Here, evidence was presented for methylation at R469 for the HSP70 protein HSPA8, where the residues adjacent to R469 (PAPRGVP) include two residues on the N-terminal side that are identical to the ones on the known PRMT4/CARM1 methylation sites on histone H3 R17 (KAPRKQL) and PABP1 R459 (AAPRPPF) [36]. Indeed, Gao et al. [37] have presented evidence suggesting that CARM1/PRMT4 may specifically methylate R469 in HSP70. Further studies will be necessary to resolve this controversy. If HSP70 R469 is in fact a methylation site for PRMT7, this enzyme would appear to recognize more than the RXR motif described in the studies above.
A recent characterization study of Ce PRMT-7 reveals a similar, though not identical, substrate specificity; while the Ce ortholog does indeed prefer RXR motifs for substrates, this enzyme is not as specific for such motifs as its mammalian counterparts [28]. Such differences may be accounted for by the minor sequence changes between each enzyme’s substrate binding motif—the double E loop (Figure 2); the importance of the double E loop residues will be further discussed in sections 4 and 5. Since the mammalian PRMT7 enzymes appear to recognize distinctly different substrate motifs than their dimethylating cousins, this suggests that PRMT7 is not simply a redundant monomethylating enzyme, but rather a writer of unique monomethylarginine posttranslational modifications (PTMs) [32,38].
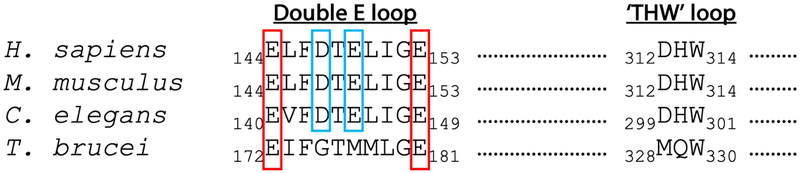
Figure 2. Sequence alignment of PRMT7 Double E and THW loops.
Sequences from humans, mice, nematode worms, and trypanosome PRMT7 have been aligned. Critical glutamate residues that flank the Double E loop are highlighted in red boxes. Acidic residues that direct substrate specificity in human, mouse, and worm PRMT7 are highlighted in cyan boxes. The sequences for substrate stabilizing ‘THW’ loops are also represented.
4. Structural biology studies of PRMT7 from protozoans to metazoans
To date, the structures of PRMT7 from T. brucei, C. elegans, and M. musculus have been solved [23,24,27,39,40] (Figure 3). Metazoan PRMT7 is distinct from the major dimethylating PRMTs, PRMT1, PRMT3, PRMT4/CARM1, PRMT5, and PRMT6, as well as TbPRMT7, because it does not appear to form dimers or other higher-ordered oligomeric structures [3,10,27,39,40]. However, the metazoan PRMT7 gene contains a gene duplication which results in two tandem methyltransferase domains that fold together to resemble the tertiary configuration of a homodimer [40] (Figure 3B and C). It should be noted here that PRMT9, a dimethylating enzyme, also has a similar gene duplication, though little is known about its structure [18]. Mutational studies show the C-terminal domain of PRMT7 to be catalytically inactive but crucial for overall activity of the enzyme [29,40]; S-adenosylhomocysteine (AdoHcy), the unmethylated analogue of AdoMet, has only been found to be bound to the N-terminal domain of the mouse [40] and C. elegans [27] structures. Many of the important residues involved in AdoMet and methyl-accepting substrate binding have been substituted in the C-terminal domain, including the second glutamate residue in the substrate arginine binding double E loop described below where a proline residue is found in both the mouse and C. elegans structures.
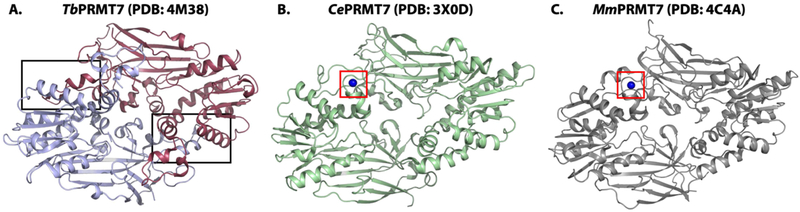
Figure 3. Overall Crystal structures of PRMT7 from a protist (Trypanosoma brucei, TbPRMT7), a nematode worm (Caenorhabditis elegans, CePRMT7), and a mammal (Mus musculus, MmPRMT7).
A) Crystal structure of TbPRMT7 (PDB: 4M38) as a homodimer; one monomer is colored raspberry and the other light blue. The dimerization arms are indicated in the black boxes. B) Crystal structure of CePRMT7 (PDB: 3X0D) in pale green; the Zn-finger like domain is boxed in red and the Zn2+ ion is shown as a blue sphere. C) Crystal structure of MmPRMT7 (PDB: 4C4A) in gray; the Zn-finger like domain is boxed in red and the Zn2+ ion is shown as a blue sphere.
The catalytic seven-β strand fold of the PRMTs contains both AdoMet-binding motifs and a substrate binding loop referred to as the “double E loop” due to the glutamates flanking a series of well-conserved residues [5,41,42] (Figures 2 and 4). These glutamate residues are important in attracting and sustaining substrate-enzyme binding interactions; specifically, crystal structures indicate that the glutamate residues closely coordinate the guanidine groups of substrate arginine side chains. Recent studies have shown the second of these glutamate residues to be a crucial determinant of type I (asymmetric dimethylation) versus type III (monomethylation) product specificity as discussed in section 5 below [24]. Additionally, the “THW” loop, composed of a threonine-histidine-tryptophan segment in type I PRMTs, and with a conserved tryptophan residue in the other PRMTs, aids in the stabilization of the positively charged arginine guanidino group through cation-π and hydrogen-bonding interactions (Figure 2). The “THW” loop has been shown to contribute in the type II SDMA producing activity of PRMTs; when this loop was mutated in a type III PRMT to mimic the sterics of a type II enzyme, the mutated enzyme was able to produce SDMA as described below in section 5 [25].
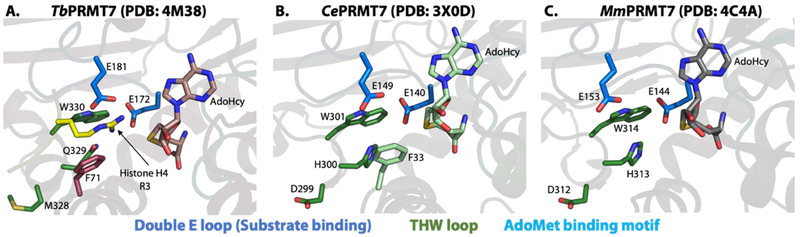
Figure 4. Structure of PRMT7 active sites from T. brucei, C. elegans, and M. musculus.
The Double E loops are represented in marine, the THW loop is represented in forest green, and the AdoMet binding motif is represented in cyan. A) Active site of TbPRMT7; the enzyme was co-crystallized with an N-terminal histone H4 peptide (shown in yellow and blue). B) Active site of CePRMT7. C) Active site of MmPRMT7.
5. Site-directed mutagenesis may help explain the specificity of PRMT7 for monomethylation
To understand the reasons behind PRMT7’s unique monomethylating ability, the product specificity of the enzyme has been studied by analyzing the products of active site point mutants. Most notably, these mutagenesis studies have been conducted on the T. brucei ortholog of PRMT7 to highlight the importance of active site residues such as E172 and E181 (the glutamates of the double E loop), Q329 (part of the THW loop), and F71 (part of helix αY) (Figure 2 and Figure 4A) [23–25,43]. In light of structural and enzyme activity data, Wang et al. concluded that TbPRMT7 may contain a narrow active site which causes the enzyme to be restricted to monomethylation [23]. Following this study, a comprehensive set of active site TbPRMT7 mutants were tested by Debler et al. and Jain et al. to show that there are distinct architectural features in the active site of this enzyme which regulate its ability to methylate arginine residues [24,25]. By comparing the makeup of active sites from type I, II, and III PRMT structures, it was shown that a more “open” region around the double E loop and a “crowded” region around the THW loop are characteristic of type I PRMTs, while the opposite configuration is characteristic of type II enzymes. All current PRMT7 structures show that both of these regions are much more constricted than type I or II PRMTs and contain constraining H-bonds and cation-π interactions which appear to prevent PRMT7 from accepting a previously monomethylated arginine residue for dimethylation (Figure 4) [25,43]. These results not only highlight active site configurations which are necessary for imparting product specificity to the entire PRMT family, but also shows that the architecture/volume of the active site, whether it is mediated by steric factors alone or by additional protein-protein interactions, plays a vital role in making PRMT7 a uniquely monomethylating enzyme.
6. Crosstalk between human PRMT5 and PRMT7—how PRMT7 activity may indirectly affect cellular SDMA levels
A number of studies have shown that changing the expression level of PRMT7 in cells can affect the level of SDMA in spite of the fact that PRMT7 can only form MMA. Downregulation of PRMT7 lowers SDMA and upregulation increases it, specifically at Arg-3 on histone H4, leading some authors to conclude that PRMT7 methylates this residue [4,44–49]. Interestingly, a study by Jelinic and coworkers also links PRMT7 and methylation of Arg-3 on histone H4; in this case however, the methylation is enhanced when PRMT7 associates with the CTCF-like (CTCFL) protein [50]. However, it is clear that PRMT5 and PRMT7 methylate different arginine residues on histone H4 - Arg-3 by PRMT5 and Arg-17 and Arg-19 by PRMT7 [31,32]. Given that mammalian PRMT7 methylates distinctly different residues on the same histone tails that PRMT5 methylates, experiments were conducted to understand how these enzymes could potentially engage in indirect regulation through their methylation activity [38]. It was shown that monomethylation of residue R17 on histone H4’s N-terminal tail not only drastically increased PRMT5-mediated methylation of residue R3, but also converted PRMT5’s low-activity conformation to the high-activity form at low substrate concentrations through allosteric regulation. The R17 monomethylation mark—a simulated product of PRMT7-mediated methylation—thus seems to function as an allosteric activator of PRMT5 function [38]. Intriguingly, as with histone H4, SDMA levels in human Sm proteins are also affected by PRMT7 expression [51]. Knowing that Sm proteins are readily methylated by PRMT5 and this methylation is crucial for the formation of Sm rings [51–54], crosstalk between PRMT5 and PRMT7 may be a new and general feature of gene regulation.
PRMT1 can also exhibit positive cooperativity with respect to histone H4 (1-21) peptides [38]. However, monomethylation of residue R17 had a smaller effect on PRMT1 activity than it did with PRMT5 [38]. Having shown that these major dimethylating PRMTs may function through positive cooperativity and that PRMT5’s methylation of histone H4 R3 may be significantly affected by PRMT7-mediated methylation of histone H4 R17, it is of interest to ask whether other members of the PRMT family participate in similar kinetic and regulatory behavior. Substantial enzymatic analysis on some of the PRMTs has been done [12,15–18,31,32,55–58], but at relatively high substrate and enzyme concentrations, suggesting that these studies may have missed any evidence of cooperativity and allosteric behavior exhibited by these enzymes. If other PRMTs do indeed display positive cooperativity, this will be a new avenue to target this class of enzymes for therapeutics. Allostery is a hallmark of many oligomeric proteins [59], and since most PRMTs functions as homo-oligomers [11,12,15–17,57,58], it is also important to explore the role of oligomerization in affecting potential cooperativity. That PRMTs can influence the activity of other PRMTs by indirect methylation of the same protein thus presents an opportunity to study the regulation of arginine methyltransferase family.
7. What are the physiological substrates for mammalian PRMT7?
The fact that histone proteins and peptides can be good substrates for PRMT7 in in vitro experiments has suggested that arginine residues in these sequences may also be in vivo substrates [28–32]. In vitro experiments identify histone H2B to be a robust substrate for PRMT7 at arginine residues 29, 31, and 33 [31,32]. These residues appear to be directly bound to the DNA in at least one structure of nucleosomes [60]. It is intriguing that a major age-dependent spontaneous modification results in the formation of a D-isoaspartyl residue at Asp-25, highly enriched in active chromatin, just upstream from the three sites of methylation on histone H2B by PRMT7 in vitro [61]. These results suggest that the formation of the modified aspartyl residue at position 25 can alter the availability of the downstream arginine residues for methylation by PRMT7 and gene activation.
However, there is no evidence yet from proteomic analyses for the methylation of histone H2B in vitro sites methylated by PRMT7. Nevertheless, as described above, one proteomic study provides evidence that histone H4 R17, an in vitro methylation site for PRMT7 [31], is monomethylated in mouse testes [62]. The difficulty in identifying PRMT7 substrates in cells may be largely because of PRMT7’s low activity [32] relative to the major dimethylating enzymes, PRMT1 and PRMT5 [38]. It has been shown that even a 5% change in histone content can lead significant physiological changes such as the onset of diseases like chondroblastoma [63]. These results suggest that while PRMT7-mediated methylation might be an uncommon event in cells, it may still occur at levels that can affect epigenetic pathways involved in chromatin regulation. In fact, with some 19,000 human genes, the modification of histones by PRMT7 in association with chromatin at only a few gene loci may be physiologically important while still resulting in the overall methylation of less than one histone in a thousand. An additional complication is that analytical methods such as mass spectrometry, which readily detect modifications at a stoichiometric level, may not be sensitive enough to detect methylation catalyzed by PRMT7 at only a small fraction of protein sites. Finally, as discussed below, common sample preparation methods for mass spectrometric analyses can preclude the identification of methylated sites in highly basic regions such as the RXR substrate motifs for PRMT7.
There is also evidence that PRMT7 may have non-histone substrates. The methylation state of proteins such as Sm proteins and elongation factors have been shown to be affected by the expression of PRMT7 [31,51,64]. Evidence associating PRMT7 in the regulation of the function of skeletal muscles [47], stem cells [45,49], and immune cells [48] also suggests the possibility of non-histone PRMT7 substrates, although it is possible that some of these phenotypes may be indirectly due to histone methylation.
Two specific cytosolic proteins have been recently proposed to be physiological substrates of PRMT7 - HSP70 [36] and eIF2α [35]. In a number of mouse and human cell lines, evidence has been presented for PRMT7-dependent methylation of HSP70 proteins at R469, affecting survival after heat shock or proteasomal stress [36]. Immunoprecipitation of a PRMT7 fusion construct with green fluorescent protein (PRMT7-GFP) in the MCF7 human breast cancer cell line resulted in the identification of ribosomal proteins and translational initiation factors, including eIF2α, as interaction partners [35]. PRMT7 was found to comigrate with monoribosomes and their subunits in the human embryonic kidney HEK293T cell line and with eIF2α [35]. As described above, eIF2α is an effective in vitro substrate for PRMT7 and it appears that its methylation may play an important role in phosphorylation-dependent eIF2α stress granule formation [35]. Here methylation of arginine residues in the eIF2α RRRIR sequence can affect the phosphorylation of the preceding serine residue [35].
Studies of the cellular localization of PRMT7-GFP in HEK293T [41] and breast cancer (MCF7) [35] cell lines have suggested that the bulk of PRMT7 is cytoplasmic with only weak fluorescent staining in the nucleus and no staining detected in nucleoli. Assuming that the GFP fusion protein has a similar distribution as native PRMT7, these results may suggest that the major physiological substrates would be also cytosolic. However, it has been shown that the localization of PRMT1 and PRMT4 GFP fusion proteins can be mostly cytoplasmic in some cell lines and mostly nuclear in others [41]. Szewczyk et al. also provide evidence that the PRMT7 antigen and the PRMT7-FLAG-tagged protein are almost entirely localized to the cytoplasmic fraction in mouse embryo fibroblasts and a variety of mouse and human cell lines [36]. If PRMT7 does in fact have roles in the nucleus, they may be catalyzed by small amounts of enzyme or enzyme that is transported into the nucleus only under specific conditions.
Finally, typical bottom-up mass spectrometry techniques utilizing trypsin and other enzymes that cleave adjacent to arginine and lysine residues are of limited utility in identifying posttranslational modifications on sites in highly basic amino acid content such as histone N-terminal tails and putative PRMT7 substrates that contain multiple arginine (and lysine) residues. At these sites, these proteases can digest peptides into individual amino acids, dipeptides or other species too small for typical mass spectrometric analyses. These problems may have greatly limited our understanding of PTMs catalyzed by enzymes such as PRMT7. The inability of trypsin to cleave adjacent to asymmetric dimethylated arginine residues has allowed for the identification of peptides containing ADMA [65] but it is unclear how the activity of trypsin is affected by the MMA residues formed by PRMT7. Peptides containing C-terminal MMA residues have been detected in tryptic digests [66]; if these identifications are correct, it would suggest that trypsin can cleave these bonds, perhaps at a lesser rate. The bottom line is that such cleavages contribute to the generation of methylated species too small for effective immunoprecipitation and mass spectroscopic identification. In fact, it was reported in a study of the mammalian proteome that digestion with chymotrypsin resulted in the identification of a “sizable” number of peptides containing methylated arginine residues that were not found when trypsin was used [67]. In future work, weaning away from trypsin may allow for the better identification of PRMT7 substrates, particularly those with RXR motifs adjacent to multiple lysine and arginine residues.
In the last few years, with the emergence of middle-down (and potentially top-down) proteomic techniques, the ability to leverage MS techniques to probe non-abundantly occurring PTMs, particularly those occurring in basic regions, has improved [68]. Using a combination of biochemical and mass spectrometric approaches, it may then be possible to elucidate the direct substrates of mammalian PRMT7 so that this enzyme’s exact role in the epigenetic landscape may be more clearly understood. At present, however, there is still a significant lack of knowledge of the direct substrates of PRMT7 in the cell.
8. Unique sub-physiological temperature and salt dependence of mammalian PRMT7 activity
Two unusual features have been found in studies of the in vitro activity of PRMT7. In the first place, both the bacterially-expressed human GST-PRMT7 and the insect expressed mouse PRMT7 display significantly reduced activity at 37 °C than at lower temperatures [28,32]. Most mammalian enzymes optimally function in a narrow range around 37 °C [69], but human and mouse PRMT7 are most active from 10 °C to 25 °C with less than 10% of the optimal activity at 37 °C in vitro [28,32]. The discovery of PRMT7’s preference for sub-physiological temperatures is quite unexpected since the other members of the PRMT family are active at 37 °C [12,15–18,56–58]. It should be mentioned, however, that we are aware of no studies that have specifically characterized the temperature dependence of PRMTs other than PRMT7. Cura et al. demonstrate that human PRMT7’s thermal denaturation temperature is around 55 °C so the lack of activity at 37 °C does not appear to be a result of the unfolding of the protein [39]. Furthermore, biophysical experiments such as CD spectroscopy and analytical ultracentrifugation have revealed that the secondary and tertiary structure PRMT7 are not significantly affected by varying temperatures from 4 °C to 37 °C [70]. Taken together, these studies indicate that at 37 °C, PRMT7’s relative inactivity may not be a result of structural denaturation.
It is thus possible that PRMT7 activity is physiologically controlled by temperature. Human cells are not always at 37 °C; the temperature at the extremities and in the lung can be significantly lower in a cold environment. It is clear that mammalian cells do respond to lowered temperatures with the induction of at least two "cold shock" proteins in humans - CIRP and RBM3 [71]. Interestingly as least one of these proteins appears to migrate from the nucleus to cytoplasmic stress granules after methylation by PRMT1 [72]. Significantly, it has also been shown that low temperatures can preserve the sternness of neural stem cells, potentially linking PRMT7 to stem cell maintenance (see sections 10 and 14) [73].
An additional link of PRMT7 with the response of mammals to cold temperatures comes from a genomic study suggesting that PRMT7 was one of five genes that differentiated modern African and Asian elephants and their ancient cold-adapted relatives including two species of woolly mammoths [74]. Given the various studies which implicate PRMT7 as an important player in the regulation of vital cellular processes such as transcription and DNA damage repair [44], as well as the regulation of diseases such as cancer [45,75,76], it will be important to understand how this enzyme’s activity in cells is affected by temperature.
Secondly, PRMT7 does not appear to be fully active under typical cellular salt concentrations [32]. Sodium chloride strongly inhibits the activity of the human GST-PRMT7 enzyme; half maximal activity is seen at about 25 mM with a peptide substrate based on histone H2B and about 200 mM for the GST-GAR protein substrate [32]. Further studies here will be needed to assess the generality of such inhibition.
9. Changes in expression of PRMT7 in cancer
Recent cancer research has implicated PRMTs and their posttranslational methylation of proteins in the genesis and proliferation of tumors [4,42]. A number of studies have shown that PRMTs are overexpressed in various cancer tissues [42,75,77,78] 79]. Specifically, overexpression of PRMT5 and PRMT7 has been associated with cancer metastasis [75,76,80], further highlighting the importance of regulatory interplay between the two enzymes. PRMT7 has primarily been linked to breast cancer progression [75,76]. In these studies, PRMT7 overexpression was shown to induce the epithelial to mesenchymal transition (EMT) associated with metastasis [76] and to contribute to invasiveness of tumor cells in breast cancer [75]. Additionally, in a study by Geng et al., PRMT7 automethylation at residue R531 was reported to enhance breast cancer metastasis [81]. In this work, automethylation was not found to affect PRMT7 methylation activity but could enhance the recruitment of PRMT7 to the E-cadherin promoter by the YY1 transcription factor [81]. Though a large sector of disease-related PRMT7 studies has focused on its role in breast cancer, some work also suggests that this enzyme may be a key regulator in leukemia [45]. In particular, protein lysine methyltransferase MLL4 (mixed lineage leukemia 4)-mediated methylation of histone H3 K4, an important gene activating mark in cancer regulation, is trans-activated by symmetric demethylation of histone H4 R3 [45]—a mark placed by PRMT5 and regulated by PRMT7 [38]. PRMT7 also appears to be involved in cell-proliferating processes such as maintaining pluripotency [82] and response to DNA damage [44] as discussed in sections 13 and 14 below. A possible mechanistic link between PRMTs and cancer is through histone methylation which is a PTM that has already been strongly associated with cancer [3,10]. In fact, it is becoming clearer that mammalian PRMT7’s methylating activity may be crucial in the regulation of various types of cancers as well [45,46,75,76].
10. Effects of knocking out the PRMT7 gene in mice - defects in skeletal muscle and immune systems
The goal of many studies involving gene perturbations in mice is to discover the normal function of the gene by examining the phenotype(s) of the homozygous mutant animals. However, in recent years it has become clear that such efforts can be complicated and compromised by redundant and compensatory pathways [83]. Indeed, genetic robustness appears to be strong enough so that up to 35% of knockout mouse ES cell lines in a large study appeared to be completely normal [84]. With this caveat in mind, what is the situation in mice with homozygous knockouts for the PRMT7 gene? Given the family of nine sequence-related mammalian PRMTs [5], it might be expected that little or no effect may be seen due to functional redundancies. However, while six of these enzymes catalyze asymmetric dimethylation reactions and two catalyze symmetric dimethylation reactions, PRMT7 is the only member of the family that is restricted to forming monomethylarginine residues and, as described above, has a rather unique specificity for the sequences in which arginine residues are modified.
There have now been four studies that report on significant physiological effects from the loss of the Prmt7 gene in mice [47,48,85,86]. In one study, homozygous global knockout mice died shortly after birth [48]; however, in the three other studies at least some of the global knockout mice were viable to adulthood [47,85,86]. Interestingly, distinct (and largely non-overlapping) phenotypes were seen as described below in each of these three studies.
An initial study on the effects of the loss of the Prmt7 gene in mice focused on the immune system [48]. In preliminary studies, the authors found that homozygous Prmt7 knockout mice were born from heterozygote parents at the expected Mendelian frequency of 25% (indicating normal survival in fetal development) but died within 10 days of birth. This result suggested that PRMT7 is essential for adult life, although the survival of other Prmt7−/− mice strains may indicate that other mutations were responsible for the early deaths in these mice. The authors then focused their work on the immune system by creating mice with a conditional Prmt7 knockout in B cells. These mice survived into adulthood but the loss of PRMT7 reportedly resulted in a redistribution of two types of mature B cells; a reduction of marginal zone B cells was found along with an increase in native follicular B cells. The latter cells can proliferate to form a germinal center where recombination and mutation occur to generate and select the differentiated plasma cells that produce antibodies with high affinities for their antigens. The authors showed that immunization of Prmt7−/− mice resulted in a hyperplasia of the germinal center compared to normal response in wild type mice. They suggested that PRMT7 mediates increases in the expression of the Bcl6 gene that encodes a transcriptional regulator that would normally limit germinal center formation. In the absence of PRMT7, unregulated cell proliferation then would occur in the germinal center. Additional experiments using chromatin immunoprecipitation provided evidence that PRMT7 could increase the SDMA levels of histone H4 R3 at the promoter of the Bcl6 gene. It is possible that such an increase may be due to the activating effect on PRMT5 by prior methylation of histone H4 at arginine-17 and/or arginine-19 residues by PRMT7 [38]. In a second study, homozygous reporter-tagged gene-trap Prmt7−/− mice were found to have slightly increased fat mass, slightly reduced body length, and, in females only, slightly reduced bone mineral content [85]. Just less than half of the expected knockout animals were found at fourteen days after birth, suggesting some lethality in utero or postnatally.
In a third study, mice with homozygous knockout of Prmt7 in all tissues were shown to be viable to adulthood but had significant defects in their muscles [47]. These mutant mice were found to have reduced muscle regeneration upon injury concomitant with loss of function of muscle satellite cells, the stem cells that can differentiate into myofibrils. The authors showed a substantial (~600-fold) induction of PRMT7 mRNA upon activation of satellite cells. In the absence of PRMT7, satellite cells went into cell cycle arrest and were subject to premature senescence. It was proposed that PRMT7 exerted these effects by regulating the expression of the de novo DNA methyltransferase DNMT3b. The authors detected decreased levels SDMA of histone H4 R3 at the Dnmt3b gene promoter in the Prmt7−/− mice, suggesting a similar mechanism of action as described above for the Bcl6 gene. These results point to a major function of PRMT7 in the generation and preservation of muscle stem cells.
In a fourth study of PrmtT7−/− mice, animals were also found to survive into adulthood, but alterations were reported in muscle structure with a shift from glycolysis-dependent type I fast-twitch fibers to mitochondrial-dependent type II slow-twitch fibers [86]. Clear evidence was presented, for example, of the replacement of fast-twitch fibers expressing myosin Myh-IIb for slow-twitch fibers expressing myosin Myh-IIa in the transverse abdominal muscle of PrmtT7−/− mice. Importantly, the authors found that the expression of the gene encoding the transcriptional coactivator PGC-1α is markedly reduced in muscles of PrmtT7−/− mice. PGC-1α stimulates mitochondrial biogenesis and is a hallmark of the formation of slow-twitch muscle fibers that depend on oxidative metabolism [86]. The authors proposed that PRMT7 may function in muscle stimulating the p38MAPK cascade that leads to the transcription of the PGC-1α gene, possibly by directly methylating p38MAPK. Further work will be necessary to test this hypothesis. It is certainly possible that PRMT7 can affect PGC-1α production by activating PRMT5-dependent histone H4 R3 symmetric dimethylation as described above in the studies of Ying et al. and Blanc et al. [47,48].
The distinct phenotypes observed in the four Prmt7 knockout studies suggest that we may only be at the tip of the iceberg in recognizing the physiological functions dependent upon PRMT7 activity. It is possible that there is a wide range of phenotypes that correspond to the loss of the PRMT7 gene and that each study above focused on only a subset of the phenotypes. Although PRMT7-dependent methylation of histone H4 is a common possible mechanism in these studies, PRMT7 also recognizes histone H2B as well as many additional proteins in vitro [31,32].
Finally, we should stress a major caveat in examining the phenotypes of knockout mice generated from genetically-modified embryonic stem cells. In most cases, multiple rounds of breeding can reduce, but not totally eliminate, what have been described as "passenger mutations" that can occur in nearby genes and be resistant to elimination by recombination [87]. These mutations may explain the apparent lethality Prmt7 homozygous knockout in mice seen by Ying et al. and Akawi et al. [48,85]. The fact that different adult phenotypes were seen in the four studies described above make it hard to rule out the presence of distinct passenger mutations in each of the knockout mice strains developed. In future studies, it may be possible to do gene rescue assays to show that the addition of a transgenic wild type PRMT7 gene reverses the effects seen in the PrmtT7−/− knockout animals.
11. Effects of mutations in the human PRMT7 gene—defects in development
Twelve individuals from eight families have now been found with defects in both the maternal and paternal Prmt7 genes [85,88–91]. In general, these mutations were found by global exomic sequencing of children that presented with developmental delays and intellectual disability. In each case, mutations in both of the parental alleles were found; heterozygote siblings and parents were unaffected. Ten distinct mutations have been identified including single nucleotide changes resulting in Arg32Thr, Arg387Gly, Trp494Arg, and Arg497Gln, two single nucleotide changes in intronic regions adjacent to splice junctions, four changes resulting in premature stop codons, and one 15,309 base pair deletion that spans exons 1 and 2. The mutation resulting in Arg32Thr also occurs adjacent to a splice junction, and it has been proposed that defective splicing may occur here as well [85]. Akawi et al. analyzed possible changes in hydrogen bonding at residues 32, 387, and 494 that may affect PRMT7 activity [85]. These authors point out that Arg387 hydrogen bonds to Glu478; mutating the latter residue to Gln has previously been found to result in a loss of 99.9% of the enzyme activity [40]. It is not yet clear how much these mutations result in a loss of PRMT7 activity itself or an overall loss of PRMT7 protein due to effects of the mutations on PRMT7 mRNA synthesis or degradation, mRNA translation, or proteolytic processing of the protein product. It will be interesting to assess the catalytic function of recombinantly-produced PRMT7 with the mutations observed in the affected individuals. Thus, it is unclear in these individuals how much, if any, residual PRMT7 activity remains in their tissues. For the individual with the homozygous deletion of exon 1 and exon 2, no protein was detected in an immunoblot [88]. It might be expected that the individual with the homozygous mutation resulting in an early stop codon [89] and the individuals with the homozygous frameshift mutations [90,91] would also completely lack PRMT7 activity. However, it is possible that some activity remains for the other affected individuals.
There is a range of clinical features found in the twelve individuals identified so far with mutations in both alleles of Prmt7. However, all of these individuals have abnormal facial features, short stature, brachydactyly (short fingers) and a global developmental and intellectual delay (reviewed in Valenzuela et al. [90] and Birnbaum et al. [91]). Eleven of the individuals have been diagnosed with hypotonia (low muscle tone and reduced muscle strength). In one additional case, a homozygous frameshift in PRMT7 was detected in the autopsy of a fetus that displayed significant fetal growth restriction [91]. The common phenotypes have now been described as "Short Stature, Brachydactyly, Intellectual Developmental Disability, and Seizures" or "SBIDDS" in Phenotype 617157 of the Online Mendelian Inheritance in Man (OMIM) database.
How much similarity is there between the effects of PRMT7 loss in humans and mice? One point of comparison is in muscle function where children deficient in PRMT7 have a lack of tonus at birth (reflecting relaxed muscles) and have a significant developmental delay in walking. However, no defects have been noted to date in human immune function, fat mass, or bone mineral content— all phenotypes seen in Prmt7 knockout mice [85]. As observed in mice previously, the effects of human mutations in PRMT7 may be compensated for, thereby masking the full effect of the loss of this methyltransferase.
12. Polymorphisms and known mutations in the human PRMT7 gene
As of November 2018, the Genome Aggregation Database (gnomAD; gnomad.broadinstitute.org) has presented sequences from large-scale studies including 15,708 genomes and 125,748 exomes from unrelated humans. From these studies, there appear to be no common polymorphisms in Prmt7 (defined as alterations of a gene that occur in at least 1% of the alleles) that result in amino acid changes; the most common missense mutation (Ser to Asn at position 573) occurs at a frequency of ~ 0.2%, resulting in only six homozygotes in this population. But there were changes in the DNA sequence seen at about half of the codons in the Prmt7 gene that would result in amino acid changes, including frameshifts, deletions, and the introduction of stop codons. Most of these changes were seen in a single allele, and the possibility remains that some or even many of these may reflect sequencing errors rather than actual mutations of the Prmt7 gene. However, fifty-two of the changes (some at the same codon) were seen at least eighteen times, suggesting that they represent a true variability in the human gene sequence. Six of the mutations were present at frequencies sufficient that one to six homozygotes were found, including the six at the Ser573Asn site.
The gnomAD does include 14 polymorphisms in Prmt7 that occur in intronic regions near splice junctions and in codons that do not change the amino acid sequence. These polymorphisms could change splicing patterns and translational and transcriptional efficiency. To date, there is no evidence that alternative splicing plays a role in PRMT7 function. Several possible isoforms have been identified on the UniProtKB database (uniprot).org from computational analyses but none of these have been experimentally verified.
The frequency with which PRMT7 mutations are seen in humans that result in disease might suggest that there is some physiological advantage associated with heterozygotic mutants, where one would expect to see about half of the enzyme activity. However, the mutations described for human patients in the PRMT7 gene do not appear to be common in the gnomAD of exomic and genomic sequencing studies. Of the eleven mutations described above that result in physiological defects in humans, eight are found in gnomAD in 1 to 19 alleles, representing frequencies ranging from 4.0 × 10−6 to 6.7 × 10−5. The additive mutation frequency for all of these mutations is 2 × 10−4, suggesting that in the population of the United States there may be some 65,000 heterozygotes and some 13 homozygotes.
13. DNA repair and PRMT7
One of the earliest biological studies done concerning PRMT7 after its discovery in CHO cells [20] reported that down-regulation of PRMT7 led to cellular resistance to DNA damaging agents [21]. A more recent study from Karkhanis and coworkers further showed that in human cell lines PRMT7 expression and subsequent methylation of histone H4 R3 leads to repression of DNA damage repair genes such as APEX2, POLD1, and POLD2 [44], highlighting PRMT7’s role in regulating the cell’s DNA repair machinery. Contrastingly, a study by Verbiest et al. found PRMT7-mediated DNA damage repair in CHO cells was impaired and the cells became susceptible to camptothecins—topoisomerase inhibitors—when PRMT7 was downregulated [92]. Taken together, there still seems to be some debate on how exactly PRMT7 is acting on DNA damage repair mechanisms, but it seems to be clear that the enzyme is a critical player in the regulation of processes involved in repairing DNA damage.
14. PRMT7 and stem cells
PRMT7 is expressed in both embryonic stem and germ cells (ESCs and EGCs, respectively) [93]. Buhr and colleagues hypothesized that PRMT7’s methylation of chromatin-associated proteins such as histones may indicate its involvement in stem cell pluripotency. In support of this theory, they showed that PRMT7 is selectively expressed in undifferentiated ESCs and EGCs, but not in differentiated ones [93]. A recent study more specifically showed that PRMT7 was indeed a pluripotent factor in mouse ESCs and that it regulated stemness through upregulation of symmetric demethylation of H4 R3 [94]. Furthermore, a proteomic study revealed PRMT7 to be a key factor in the induction of pluripotency and replacing the Yamanaka factor, SOX2 [95]. Certain micro-RNAs have also been identified as inhibitors of PRMT7-mediated stemness and pluripotency [96]. Outside of ESCs, PRMT7 has been implicated in muscle stem cell renewal [47]. Specifically, Blanc and Richard found that PRMT7 appears to regulate DNMT3b activity in order to maintain muscle tissue regeneration capacity.
15. Perspectives and conclusions
PRMT7 has become the subject of much study over the last two decades. From being initially discovered in CHO cells and sequentially classified as a putative arginine methyltransferase to being thoroughly biochemically characterized as a uniquely monomethylating PRMT, this enzyme has emerged as a vital player in the regulation of such processes as DNA damage repair, tumorigenesis, and stem cell pluripotency. Specifically, now that the field has established PRMT7 to be a non-redundant and distinct type of PRMT, mechanisms by which it engages with other enzymes like PRMT5 to regulate epigenetic control of transcription are becoming clearer. Furthermore, genetic defects in PRMT7 have been shown to present themselves in a range of phenotypes from physical to mental abnormalities in both mice and humans. As compelling as the literature on PRMT7 thus far may be, however, there are still many aspects of this enzyme that must be further explored and analyzed. For one, although vast biochemical in vitro data and indirect sequencing methods such as ChIP-seq have indicated that PRMT7 methylates proteins such as histones and Sm proteins, few direct biological links to PRMT7 substrates have been made. Biochemical evidence has also uncovered this enzyme’s preference for sub-physiological temperatures in vitro. Whether this is due to the lack of external factors such as binding partners or if this feature of PRMT7 activity is physiologically relevant remains to be seen. Additionally, given this enzyme’s medical relevance in cancer regulation and genetic syndromes, there is an apparent absence for chemical probes and small molecule inhibitors specific to PRMT7, while many have been described for enzymes such as PRMT1 and PRMT5 [97]. However, the recent description of a potent (nM IC50 values) and relatively specific PRMT7 inhibitor SGC8158 and its cell-permeable pro-inhibitor SGC3027 may mark a new effective approach [36]. Taken together, the various studies reviewed and summarized in this study exemplify PRMT7 to be a unique member of the PRMT family and highlight the need for further biological and biochemical analysis in light of its emergence as a vital player in cellular regulatory pathways.
Acknowledgements
Work in the author's laboratories was supported by National Science Foundation Grant MCB-1714569 (to S.G.C.), by a predoctoral Ruth L. Kirschstein National Service Award GM007185 (to K.J.) and by a Faculty Research Grant from the UCLA Academic Senate (to S.G.C.). Additionally, K.J. is supported by a postdoctoral Ruth L. Kirschstein National Service Award T32CA217824. We thank our colleagues for their helpful comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Walsh G, Jefferis R, Post-translational modifications in the context of therapeutic proteins., Nat. Biotechnol 24 (2006) 1241–52. doi: 10.1038/nbt1252. [DOI] [PubMed] [Google Scholar]
- [2].Morales Y, Cáceres T, May K, Hevel JM, Biochemistry and regulation of the protein arginine methyltransferases (PRMTs)., Arch. Biochem. Biophys 590 (2016) 138–52. doi: 10.1016/j.abb.2015.11.030. [DOI] [PubMed] [Google Scholar]
- [3].Fuhrmann J, Clancy KW, Thompson PR, Chemical biology of protein arginine modifications in epigenetic regulation, Chem. Rev 115 (2015) 5413–5461. doi: 10.1021/acs.chemrev.5b00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Blanc RS, Richard S, Arginine methylation: the coming of age, Mol. Cell 65 (2017) 8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- [5].Bedford MT, Clarke SG, Protein arginine methylation in mammals: who, what, and why, Mol. Cell 33 (2009) 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Clarke SG, The ribosome: A hot spot for the identification of new types of protein methyltransferases, J. Biol. Chem 293 (2018) 10438–10446. doi : 10.1074/jbc.AW118.003235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yesselman JD, Horowitz S, Brooks CL, Trievel RC, Frequent side chain methyl carbon-oxygen hydrogen bonding in proteins revealed by computational and stereochemical analysis of neutron structures., Proteins. 83 (2015) 403–10. doi: 10.1002/prot.24724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Desiraju GR, C-H⋯O and other weak hydrogen bonds. From crystal engineering to virtual screening., Chem. Commun. (Camb). (2005) 2995–3001. doi: 10.1039/b504372g. [DOI] [PubMed] [Google Scholar]
- [9].Cannizzaro CE, Houk KN, Magnitudes and Chemical Consequences of R 3 N + ─C─H⋯OC Hydrogen Bonding, J. Am. Chem. Soc 124 (2002) 7163–7169. doi: 10.1021/ja012417q. [DOI] [PubMed] [Google Scholar]
- [10].Fuhrmann J, Thompson PR, Protein arginine methylation and citrullination in epigenetic regulation., ACS Chem. Biol 11 (2016) 654–68. doi: 10.1021/acschembio.5b00942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang X, Cheng X, Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides, Structure. 11 (2003) 509–520. doi: 10.1016/S0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sun L, Wang M, Lv Z, Yang N, Liu Y, Bao S, Gong W, Xu R-M, Structural insights into protein arginine symmetric dimethylation by PRMT5, Proc. Natl. Acad. Sci 108 (2011) 20538–20543. doi: 10.1073/pnas.1106946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Antonysamy S, Bonday Z, Campbell RM, Doyle B, Druzina Z, Gheyi T, Han B, Jungheim LN, Qian Y, Rauch C, Russell M, Sauder JM, Wasserman SR, Weichert K, Willard FS, Zhang A, Emtage S, Crystal structure of the human PRMT5:MEP50 complex, Proc. Natl. Acad. Sci 109 (2012) 17960–17965. doi: 10.1073/pnas.1209814109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Peng C, Wong CC, The story of protein arginine methylation: characterization, regulation, and function, Expert Rev. Proteomics 14 (2017) 157–170. doi: 10.1080/14789450.2017.1275573. [DOI] [PubMed] [Google Scholar]
- [15].Frankel A, Clarke S, PRMT3 is a distinct member of the protein arginine N-methyltransferase family. Conferral of substrate specificity by a zinc-finger domain., J. Biol. Chem 275 (2000) 32974–82. doi: 10.1074/jbc.M006445200. [DOI] [PubMed] [Google Scholar]
- [16].Lakowski TM, Frankel A, A kinetic study of human protein arginine N-methyltransferase 6 reveals a distributive mechanism., J. Biol. Chem 283 (2008) 10015–25. doi: 10.1074/jbc.M710176200. [DOI] [PubMed] [Google Scholar]
- [17].Pak ML, Lakowski TM, Thomas D, Vhuiyan MI, Hüsecken K, Frankel A, A Protein Arginine N -Methyltransferase 1 (PRMT1) and 2 Heteromeric Interaction Increases PRMT1 Enzymatic Activity, Biochemistry. 50 (2011) 8226–8240. doi: 10.1021/bi200644c. [DOI] [PubMed] [Google Scholar]
- [18].Hadjikyriacou A, Yang Y, Espejo A, Bedford MT, Clarke SG, Unique features of human protein arginine methyltransferase 9 (PRMT9) and its substrate RNA splicing factor SF3B2., J. Biol. Chem 290 (2015) 16723–16743. doi: 10.1074/jbc.M115.659433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yang Y, Hadjikyriacou A, Xia Z, Gayatri S, Kim D, Zurita-Lopez C, Kelly R, Guo A, Li W, Clarke SG, Bedford MT, PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145., Nat. Commun 6 (2015) 6428. doi: 10.1038/ncomms7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Gros L, Delaporte C, Frey S, Decesse J, de Saint-Vincent BR, Cavarec L, Dubart A, V Gudkov A, Jacquemin-Sablon A, Identification of new drug sensitivity genes using genetic suppressor elements: protein arginine N-methyltransferase mediates cell sensitivity to DNA-damaging agents., Cancer Res 63 (2003) 164–71. http://www.ncbi.nlm.nih.gov/pubmed/12517794 (accessed October 25, 2018). [PubMed] [Google Scholar]
- [21].Gros L, Renodon-Cornière A, de Saint Vincent BR, Feder M, Bujnicki JM, Jacquemin-Sablon A, Characterization of prmt7alpha and beta isozymes from Chinese hamster cells sensitive and resistant to topoisomerase II inhibitors., Biochim. Biophys. Acta 1760 (2006) 1646–56. doi: 10.1016/j.bbagen.2006.08.026. [DOI] [PubMed] [Google Scholar]
- [22].Fisk JC, Sayegh J, Zurita-Lopez C, Menon S, Presnyak V, Clarke SG, Read LK, A type III protein arginine methyltransferase from the protozoan parasite Trypanosoma brucei., J. Biol. Chem 284 (2009) 11590–11600. doi: 10.1074/jbc.M807279200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang C, Zhu Y, Caceres TB, Liu L, Peng J, Wang J, Chen J, Chen X, Zhang Z, Zuo X, Gong Q, Teng M, Hevel JM, Wu J, Shi Y, Structural determinants for the strict monomethylation activity by trypanosoma brucei protein arginine methyltransferase 7., Structure. 22 (2014) 756–68. doi: 10.1016/j.str.2014.03.003. [DOI] [PubMed] [Google Scholar]
- [24].Debler EW, Jain K, Warmack RA, Feng Y, Clarke SG, Blobel G, Stavropoulos P, A glutamate/aspartate switch controls product specificity in a protein arginine methyltransferase., Proc. Natl. Acad. Sci. U. S. A 113 (2016) 2068–2073. doi: 10.1073/pnas.1525783113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Jain K, Warmack RA, Debler EW, Hadjikyriacou A, Stavropoulos P, Clarke SG, Protein arginine methyltransferase product specificity Is mediated by distinct active-site architectures, J. Biol. Chem 291 (2016) 18299–18308. doi: 10.1074/jbc.M116.740399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Takahashi Y, Daitoku H, Yokoyama A, Nakayama K, Kim J-D, Fukamizu A, The C. elegans PRMT-3 possesses a type III protein arginine methyltransferase activity., J. Recept. Signal Transduct. Res 31 (2011) 168–72. doi: 10.3109/10799893.2011.555768. [DOI] [PubMed] [Google Scholar]
- [27].Hasegawa M, Toma-Fukai S, Kim J-DD, Fukamizu A, Shimizu T, Protein arginine methyltransferase 7 has a novel homodimer-like structure formed by tandem repeats, FEBS Lett 588 (2014) 1942–1948. doi: 10.1016/j.febslet.2014.03.053. [DOI] [PubMed] [Google Scholar]
- [28].Hadjikyriacou A, Clarke SG, Caenorhabditis elegans PRMT-7 and PRMT-9 Are Evolutionarily Conserved Protein Arginine Methyltransferases with Distinct Substrate Specificities, Biochemistry. 56 (2017) 2612–2626. doi: 10.1021/acs.biochem.7b00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Miranda TB, Miranda M, Frankel A, Clarke S, PRMT7 is a member of the protein arginine methyltransferase family with a distinct substrate specificity, J. Biol. Chem 279 (2004) 22902–22907. doi: 10.1074/jbc.M312904200. [DOI] [PubMed] [Google Scholar]
- [30].Zurita-Lopez CI, Sandberg T, Kelly R, Clarke SG, Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming omega-NG-monomethylated arginine residues, J. Biol. Chem 287 (2012) 7859–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Feng Y, Maity R, Whitelegge JP, Hadjikyriacou A, Li Z, Zurita-Lopez C, Al-Hadid Q, Clark AT, Bedford MT, Masson JY, Clarke SG, Mammalian protein arginine methyltransferase 7 (PRMT7) specifically targets RXR sites in lysine- and arginine-rich regions, J. Biol. Chem 288 (2013) 37010–37025. doi: 10.1074/jbc.M113.525345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Feng Y, Hadjikyriacou A, Clarke SG, Substrate specificity of human protein arginine methyltransferase 7 (PRMT7): the importance of acidic residues in the double E loop., J. Biol. Chem 289 (2014) 32604–16. doi: 10.1074/jbc.M114.609271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lee J-H, Cook JR, Yang Z-H, Mirochnitchenko O, Gunderson SI, Felix AM, Herth N, Hoffmann R, Pestka S, PRMT7, a new protein arginine methyltransferase that synthesizes symmetric dimethylarginine., J. Biol. Chem 280 (2005) 3656–64. doi: 10.1074/jbc.M405295200. [DOI] [PubMed] [Google Scholar]
- [34].Nishioka K, Reinberg D, Methods and tips for the purification of human histone methyltransferases., Methods. 31 (2003) 49–58. http://www.ncbi.nlm.nih.gov/pubmed/12893173 (accessed April 28, 2017). [DOI] [PubMed] [Google Scholar]
- [35].Haghandish N, Baldwin RM, Morettin A, Dawit HT, Adhikary H, Masson J-Y, Mazroui R, Trinkle-Mulcahy L, Côté J, PRMT7 Methylates Eukaryotic Translation Initiation Factor 2α and Regulates its Role in Stress Granule Formation, Mol. Biol. Cell (2019) mbc.E18-05-0330. doi: 10.1091/mbc.E18-05-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Szewczyk MM, Ishikawa Y, Organ S, Sakai N, Li F, Ackloo S, Eram MS, Dilworth D, Fukushi H, Harding R, dela Sena CC, Sugo T, Hayashi K, Macleod D, Zepeda C, Takagi S, Al-Awar R, Richard S, Takizawa M, Arrowsmith CH, Vedadi M, Brown PJ, Nara H, Barsyte-Lovejoy D, Pharmacological inhibition of PRMT7 links arginine monomethylation to the cellular stress response, BioRxiv. (2018) 503136. doi: 10.1101/503136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gao W, Xiao R, Peng B, Xu H, Shen H, Huang M, Shi T, Yi J, Zhang W, Wu X, Gao X, Lin X, Dorrestein PC, Rosenfeld MG, Liu W, Arginine methylation of HSP70 regulates retinoid acid-mediated RARβ2 gene activation, Proc. Natl. Acad. Sci 112 (2015) E3327–E3336. doi: 10.1073/pnas.1509658112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jain K, Jin CY, Clarke SG, Epigenetic control via allosteric regulation of mammalian protein arginine methyltransferases., Proc. Natl. Acad. Sci. U. S. A 114 (2017) 10101–10106. doi: 10.1073/pnas.1706978114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Cura V, Troffer-Charlier N, Lambert M-A, Bonnefond L, Cavarelli J, Cloning, expression, purification and preliminary X-ray crystallographic analysis of mouse protein arginine methyltransferase 7., Acta Crystallogr. Sect. F, Struct. Biol. Commun 70 (2014) 80–6. doi: 10.1107/S2053230X13032871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Cura V, Troffer-Charlier N, Wurtz JM, Bonnefond L, Cavarelli J, Structural insight into arginine methylation by the mouse protein arginine methyltransferase 7: a zinc finger freezes the mimic of the dimeric state into a single active site., Acta Crystallogr. D. Biol. Crystallogr 70 (2014) 2401–12. doi: 10.1107/S1399004714014278. [DOI] [PubMed] [Google Scholar]
- [41].Herrmann F, Pably P, Eckerich C, Bedford MT, Fackelmayer FO, Human protein arginine methyltransferases in vivo--distinct properties of eight canonical members of the PRMT family., J. Cell Sci 122 (2009) 667–77. doi: 10.1242/jcs.039933. [DOI] [PubMed] [Google Scholar]
- [42].Yang Y, Bedford MT, Protein arginine methyltransferases and cancer., Nat. Rev. Cancer 13 (2013) 37–50. doi: 10.1038/nrc3409. [DOI] [PubMed] [Google Scholar]
- [43].Cáceres TB, Thakur A, Price OM, Ippolito N, Li J, Qu J, Acevedo O, Hevel JM, Phe71 in Type III Trypanosomal Protein Arginine Methyltransferase 7 (TbPRMT7) Restricts the Enzyme to Monomethylation., Biochemistry. 57 (2018) 1349–1359. doi: 10.1021/acs.biochem.7b01265. [DOI] [PubMed] [Google Scholar]
- [44].Karkhanis V, Wang L, Tae S, Hu YJ, Imbalzano AN, Sif S, Protein arginine methyltransferase 7 regulates cellular response to DNA damage by methylating promoter histones H2A and H4 of the polymerase δ catalytic subunit gene, POLD1, J. Biol. Chem 287 (2012) 29801–29814. doi: 10.1074/jbc.M112.378281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Dhar SS, Lee S-H, Kan P-Y, Voigt P, Ma L, Shi X, Reinberg D, Lee MG, Trans-tail regulation of MLL4-catalyzed H3K4 methylation by H4R3 symmetric dimethylation is mediated by a tandem PHD of MLL4., Genes Dev 26 (2012) 2749–62. doi: 10.1101/gad.203356.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Migliori V, Müller J, Phalke S, Low D, Bezzi M, Mok WC, Sahu SK, Gunaratne J, Capasso P, Bassi C, Cecatiello V, De Marco A, Blackstock W, Kuznetsov V, Amati B, Mapelli M, Guccione E, Symmetric dimethylation of H3R2 is a newly identified histone mark that supports euchromatin maintenance, Nat. Struct. Mol. Biol 19 (2012) 136–144. doi: 10.1038/nsmb.2209. [DOI] [PubMed] [Google Scholar]
- [47].Blanc RS, Vogel G, Chen T, Crist C, Richard S, PRMT7 preserves satellite cell regenerative capacity, Cell Rep 14 (2016) 1528–1539. doi: 10.1016/j.celrep.2016.01.022. [DOI] [PubMed] [Google Scholar]
- [48].Ying Z, Mei M, Zhang P, Liu C, He H, Gao F, Bao S, Histone arginine methylation by PRMT7 controls germinal center formation via regulating Bcl6 transcription, J. Immunol 195 (2015) 1538–1547. doi: 10.4049/jimmunol.1500224. [DOI] [PubMed] [Google Scholar]
- [49].Blanc RS, Richard S, Regenerating muscle with arginine methylation, Transcription. (2017) e1291083. doi: 10.1080/21541264.2017.1291083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Jelinic P, Stehle J-C, Shaw P, The Testis-Specific Factor CTCFL Cooperates with the Protein Methyltransferase PRMT7 in H19 Imprinting Control Region Methylation, PLoS Biol 4 (2006) e355. doi: 10.1371/journal.pbio.0040355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Gonsalvez GB, Tian L, Ospina JK, Boisvert F-M, Lamond AI, Matera AG, Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins, J. Cell Biol 178 (2007) 733–740. doi: 10.1083/jcb.200702147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Pesiridis GS, Diamond E, Van Duyne GD, Role of pICLn in methylation of Sm proteins by PRMT5, J. Biol. Chem 284 (2009) 21347–21359. doi: 10.1074/jbc.M109.015578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Avasarala S, Van Scoyk M, Karuppusamy Rathinam MK, Zerayesus S, Zhao X, Zhang W, Pergande MR, Borgia JA, DeGregori J, Port JD, Winn RA, Bikkavilli RK, PRMT1 is a Novel Regulator of Epithelial-Mesenchymal-Transition in Non-Small Cell Lung Cancer., J. Biol. Chem 290 (2015) 13479–89. doi: 10.1074/jbc.M114.636050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Azzouz TN, Pillai RS, Däpp C, Chari A, Meister G, Kambach C, Fischer U, Schümperli D, Toward an Assembly Line for U7 snRNPs, J. Biol. Chem 280 (2005) 34435–34440. doi: 10.1074/jbc.M505077200. [DOI] [PubMed] [Google Scholar]
- [55].Frankel A, Brown JI, Evaluation of kinetic data: What the numbers tell us about PRMTs, Biochim. Biophys. Acta - Proteins Proteomics (2018). doi: 10.1016/j.bbapap.2018.10.010. [DOI] [PubMed] [Google Scholar]
- [56].Feng Y, Xie N, Jin M, Stahley MR, Stivers JT, Zheng YG, A transient kinetic analysis of PRMT1 catalysis., Biochemistry. 50 (2011) 7033–44. doi: 10.1021/bi200456u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson BL, Henschen-Edman A, Mackay DR, Stallcup MR, Aswad DW, Methylation of histone H3 by coactivator-associated arginine methyltransferase 1, Biochemistry. 40 (2001) 5747–5756. doi: 10.1021/bi002631b. [DOI] [PubMed] [Google Scholar]
- [58].Dillon MBC, Rust HL, Thompson PR, Mowen KA, Automethylation of protein arginine methyltransferase 8 (PRMT8) regulates activity by impeding S-adenosylmethionine sensitivity., J. Biol. Chem 288 (2013) 27872–80. doi: 10.1074/jbc.M113.491092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Monod J, Wyman J, Changeux J-P, On the nature of allosteric transitions: A plausible model, J. Mol. Biol 12 (1965) 88–118. doi: 10.1016/S0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- [60].Davey CA, Sargent DF, Luger K, Maeder AW and Richmond TJ, Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 A resolution, J. Mol. Biol 319, 2002, 1097–1113, 10.1016/S0022-2836(02)00386-8 [DOI] [PubMed] [Google Scholar]
- [61].Qin Z, Zhu JX, Aswad DW, The D-isoAsp-25 variant of histone H2B is highly enriched in active chromatin: potential role in the regulation of gene expression?, Amino Acids. 48 (2016) 599–603. doi: 10.1007/s00726-015-2140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Luense LJ, Wang X, Schon SB, Weller AH, Lin Shiao E, Bryant JM, Bartolomei MS, Coutifaris C, Garcia BA, Berger SL, Comprehensive analysis of histone post-translational modifications in mouse and human male germ cells, Epigenetics Chromatin. 9 (2016) 24. doi: 10.1186/s13072-016-0072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Jayaram H, Hoelper D, Jain SU, Cantone N, Lundgren SM, Poy F, Allis CD, Cummings R, Bellon S, Lewis PW, S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3., Proc. Natl. Acad. Sci. U. S. A 113 (2016) 6182–7. doi: 10.1073/pnas.1605523113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Jung GA, Shin BS, Jang YS, Sohn JB, Woo SR, Kim JE, Choi G, Lee KHKM, Min BH, Lee KHKM, Park GH, Methylation of eukaryotic elongation factor 2 induced by basic fibroblast growth factor via mitogen-activated protein kinase., Exp. Mol. Med 43 (2011) 550–60. doi: 10.3858/emm.2011.43.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Merrill BM, Lopresti MB, Stone KL, Williams KR, Amino acid sequence of UP1, an hnRNP-derived single-stranded nucleic acid binding protein from calf thymus., Int. J. Pept. Protein Res 29 (1987) 21–39. http://www.ncbi.nlm.nih.gov/pubmed/3032834 (accessed February 5, 2019). [DOI] [PubMed] [Google Scholar]
- [66].Pang CNI, Gasteiger E, Wilkins MR, Identification of arginine- and lysine-methylation in the proteome of Saccharomyces cerevisiae and its functional implications., BMC Genomics. 11 (2010) 92. doi: 10.1186/1471-2164-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Uhlmann T, Geoghegan VL, Thomas B, Ridlova G, Trudgian DC, Acuto O, A Method for Large-scale Identification of Protein Arginine Methylation, Mol. Cell. Proteomics. 11 (2012) 1489–1499. doi: 10.1074/mcp.M112.020743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Sidoli S, Garcia BA, Middle-down proteomics: a still unexploited resource for chromatin biology, Expert Rev. Proteomics 14 (2017) 617–626. doi: 10.1080/14789450.2017.1345632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Held C, Sadowski G, Thermodynamics of Bioreactions, Annu. Rev. Chem. Biomol. Eng 7 (2016) 395–414. doi: 10.1146/annurev-chembioeng-080615-034704. [DOI] [PubMed] [Google Scholar]
- [70].Jain K, Protein Arginine Methyltransferases: Catalytic Mechanisms and Crosstalk in Epigenetics, University of California, Los Angeles, 2018. 90–112. https://escholarship.org/uc/item/4ft92012. [Google Scholar]
- [71].LLeonart ME, A new generation of proto-oncogenes: Cold-inducible RNA binding proteins, Biochim. Biophys. Acta - Rev. Cancer 1805 (2010) 43–52. doi: 10.1016/j.bbcan.2009.11.001. [DOI] [PubMed] [Google Scholar]
- [72].De Leeuw F, Zhang T, Wauquier C, Huez G, Kruys V, Gueydan C, The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor., Exp. Cell Res 313 (2007) 4130–44. doi: 10.1016/j.yexcr.2007.09.017. [DOI] [PubMed] [Google Scholar]
- [73].Saito K, Fukuda N, Matsumoto T, Iribe Y, Tsunemi A, Kazama T, Yoshida-Noro C, Hayashi N, Moderate low temperature preserves the stemness of neural stem cells and suppresses apoptosis of the cells via activation of the cold-inducible RNA binding protein., Brain Res 1358 (2010) 20–9. doi: 10.1016/j.brainres.2010.08.048. [DOI] [PubMed] [Google Scholar]
- [74].Lynch VJ, Bedoya-Reina OC, Ratan A, Sulak M, Drautz-Moses DI, Perry GH, Miller W, Schuster SC, Elephantid Genomes Reveal the Molecular Bases of Woolly Mammoth Adaptations to the Arctic., Cell Rep 12 (2015) 217–28. doi: 10.1016/j.celrep.2015.06.027. [DOI] [PubMed] [Google Scholar]
- [75].Baldwin RM, Haghandish N, Daneshmand M, Amin S, Paris G, Falls TJ, Bell JC, Islam S, Côté J, Protein arginine methyltransferase 7 promotes breast cancer cell invasion through the induction of MMP9 expression., Oncotarget 6 (2015) 3013–32. http://www.ncbi.nlm.nih.gov/pubmed/25605249 (accessed March 20, 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Yao R, Jiang H, Ma Y, Wang L, Wang L, Du J, Hou P, Gao Y, Zhao L, Wang G, Zhang Y, Liu D-X, Huang B, Lu J, PRMT7 induces epithelial-to-mesenchymal transition and promotes metastasis in breast cancer., Cancer Res 74 (2014) 5656–67. doi: 10.1158/0008-5472.CAN-14-0800. [DOI] [PubMed] [Google Scholar]
- [77].Koh CM, Bezzi M, Low DHP, Ang WX, Teo SX, Gay FPH, Al-Haddawi M, Tan SY, Osato M, Sabò A, Amati B, Wee KB, Guccione E, MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis., Nature. 523 (2015) 96–100. doi: 10.1038/nature14351. [DOI] [PubMed] [Google Scholar]
- [78].Hu D, Gur M, Zhou Z, Gamper A, Hung M-C, Fujita N, Lan L, Bahar I, Wan Y, Interplay between arginine methylation and ubiquitylation regulates KLF4-mediated genome stability and carcinogenesis, Nat. Commun 6 (2015) 8419. doi: 10.1038/ncomms9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Poulard C, Corbo L, Le Romancer M, Protein arginine methylation/demethylation and cancer., Oncotarget 7 (2016) 67532–67550. doi: 10.18632/oncotarget.11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Bao X, Zhao S, Liu T, Liu YY, Liu YY, Yang X, Overexpression of PRMT5 promotes tumor cell growth and is associated with poor disease prognosis in epithelial ovarian cancer., J. Histochem. Cytochem 61 (2013) 206–17. doi: 10.1369/0022155413475452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Geng P, Zhang Y, Liu XX, Zhang N, Liu Y, Liu XX, Lin C, Yan X, Li Z, Wang G, Li Y, Tan J, Liu D-X, Huang B, Lu J, Automethylation of protein arginine methyltransferase 7 and its impact on breast cancer progression., FASEB J. 31 (2017) 2287–2300. doi: 10.1096/fj.201601196R. [DOI] [PubMed] [Google Scholar]
- [82].Wang Y-C, Peterson SE, Loring JF, Protein post-translational modifications and regulation of pluripotency in human stem cells., Cell Res 24 (2014) 143–60. doi: 10.1038/cr.2013.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].El-Brolosy MA, Stainier DYR, Genetic compensation: A phenomenon in search of mechanisms, PLOS Genet 13 (2017) e1006780. doi: 10.1371/journal.pgen.1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].White JK, Gerdin A-K, Karp NA, Ryder E, Buljan M, Bussell JN, Salisbury J, Clare S, Ingham NJ, Podrini C, Houghton R, Estabel J, Bottomley JR, Melvin DG, Sunter D, Adams NC, Institute Mouse Genetics Project, Sanger D, Tannahill D, Logan DW, Macarthur DG, Flint J, Mahajan VB, Tsang SH, Smyth I, Watt FM, Skarnes WC, Dougan G, Adams DJ, Ramirez-Solis R, Bradley A, Steel KP, Barnes C, Beveridge R, Cambridge E, Carragher D, Chana P, Clarke K, Hooks Y, Igosheva N, Ismail O, Jackson H, Kane L, Lacey R, Lafont DT, Lucas M, Maguire S, McGill K, McIntyre RE, Messager S, Mottram L, Mulderrig L, Pearson S, Protheroe HJ, Roberson L-A, Salsbury G, Sanderson M, Sanger D, Shannon C, Thompson PC, Tuck E, Vancollie VE, Brackenbury L, Bushell W, Cook R, Dalvi P, Gleeson D, Habib B, Hardy M, Liakath-Ali K, Miklejewska E, Price S, Sethi D, Trenchard E, von Schiller D, Vyas S, West AP, Woodward J, Wynn E, Evans A, Gannon D, Griffiths M, Holroyd S, Iyer V, Kipp C, Lewis M, Li W, Oakley D, Richardson D, Smedley D, Agu C, Bryant J, Delaney L, Gueorguieva NI, Tharagonnet H, Townsend AJ, Biggs D, Brown E, Collinson A, Dumeau C-E, Grau E, Harrison S, Harrison J, Ingle CE, Kundi H, Madich A, Mayhew D, Metcalf T, Newman S, Pass J, Pearson L, Reynolds H, Sinclair A, Wardle-Jones H, Woods M, Alexander L, Brown T, Flack F, Frost C, Griggs N, Hrnciarova S, Kirton A, McDermott J, Rogerson C, White G, Zielezinski P, DiTommaso T, Edwards A, Heath E, Mahajan MA, Yalcin B, Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes., Cell. 154 (2013) 452–64. doi: 10.1016/j.cell.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Akawi N, McRae J, Ansari M, Balasubramanian M, Blyth M, Brady AF, Clayton S, Cole T, Deshpande C, Fitzgerald TW, Foulds N, Francis R, Gabriel G, Gerety SS, Goodship J, Hobson E, Jones WD, Joss S, King D, Klena N, Kumar A, Lees M, Lelliott C, Lord J, McMullan D, O’Regan M, Osio D, Piombo V, Prigmore E, Rajan D, Rosser E, Sifrim A, Smith A, Swaminathan GJ, Turnpenny P, Whitworth J, Wright CF, V Firth H, Barrett JC, Lo CW, FitzPatrick DR, Hurles ME, DDD study, Discovery of four recessive developmental disorders using probabilistic genotype and phenotype matching among 4,125 families., Nat. Genet 47 (2015) 1363–9. doi: 10.1038/ng.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Jeong H-J, Lee H-J, Vuong TA, Choi K-S, Choi D, Koo S-H, Cho SC, Cho H, Kang J-S, Prmt7 Deficiency Causes Reduced Skeletal Muscle Oxidative Metabolism and Age-Related Obesity., Diabetes. 65 (2016) 1868–82. doi: 10.2337/db15-1500. [DOI] [PubMed] [Google Scholar]
- [87].Vanden Berghe T, Hulpiau P, Martens L, Vandenbroucke RE, Van Wonterghem E, Perry SW, Bruggeman I, Divert T, Choi SM, Vuylsteke M, Shestopalov VI, Libert C, Vandenabeele P, Passenger Mutations Confound Interpretation of All Genetically Modified Congenic Mice., Immunity. 43 (2015) 200–9. doi: 10.1016/j.immuni.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Kernohan KD, McBride A, Xi Y, Martin N, Schwartzentruber J, Dyment DA, Majewski J, Blaser S, K.M. Care4Rare Canada Consortium, Boycott KM, Chitayat D, Loss of the arginine methyltranserase PRMT7 causes syndromic intellectual disability with microcephaly and brachydactyly., Clin. Genet 91 (2017) 708–716. doi: 10.1111/cge.12884. [DOI] [PubMed] [Google Scholar]
- [89].Agolini E, Dentici MLL, Bellacchio E, Alesi V, Radio FCC, Torella A, Musacchia F, Tartaglia M, Dallapiccola B, Nigro V, Digilio MCC, Novelli A, Expanding the clinical and molecular spectrum of PRMT7 mutations: 3 additional patients and review, 93 (2018) 675–681. doi: 10.1111/cge.13137. [DOI] [PubMed] [Google Scholar]
- [90].Valenzuela I, Segura-Puimedon M, Rodríguez-Santiago B, Fernández-Alvarez P, Vendrell T, Armengol L, Tizzano E, Further delineation of the phenotype caused by loss of function mutations in PRMT7., Eur. J. Med. Genet (2018). doi: 10.1016/j.ejmg.2018.07.007. [DOI] [PubMed] [Google Scholar]
- [91].Birnbaum R, Yosha-Orpaz N, Yanoov-Sharav M, Kidron D, Gur H, Yosovich K, Lerman-Sagie T, Malinger G, Lev D, Prenatal and postnatal presentation of PRMT7 related syndrome: Expanding the phenotypic manifestations, Am. J. Med. Genet. Part A (2018). doi: 10.1002/ajmg.a.6. [DOI] [PubMed] [Google Scholar]
- [92].Verbiest V, Montaudon D, Tautu MT, Moukarzel J, Portail JP, Markovits J, Robert J, Ichas F, Pourquier P, Protein arginine (N)-methyl transferase 7 (PRMT7) as a potential target for the sensitization of tumor cells to camptothecins, FEBS Lett 582 (2008) 1483–1489. doi: 10.1016/j.febslet.2008.03.031. [DOI] [PubMed] [Google Scholar]
- [93].Buhr N, Carapito C, Schaeffer C, Kieffer E, Van Dorsselaer A, Viville S, Nuclear proteome analysis of undifferentiated mouse embryonic stem and germ cells, Electrophoresis. 29 (2008) 2381–2390. doi: 10.1002/elps.200700738. [DOI] [PubMed] [Google Scholar]
- [94].Lee S-H, Chen T-Y, Dhar SS, Gu B, Chen K, Kim YZ, Li W, Lee MG, A feedback loop comprising PRMT7 and miR-24-2 interplays with Oct4, Nanog, Klf4 and c-Myc to regulate stemness., Nucleic Acids Res 44 (2016) 10603–10618. doi: 10.1093/nar/gkw788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Wang B, Pfeiffer MJ, Drexler HCA, Fuellen G, Boiani M, Proteomic analysis of mouse oocytes identifies PRMT7 as a reprogramming factor that replaces SOX2 in the induction of pluripotent stem cells, J. Proteome Res 15 (2016) 2407–2421. doi: 10.1021/acs.jproteome.5b01083. [DOI] [PubMed] [Google Scholar]
- [96].Chen T-Y, Lee S-H, Dhar SS, Lee MG, Protein arginine methyltransferase 7-mediated microRNA-221 repression maintains Oct4, Nanog, and Sox2 levels in mouse embryonic stem cells., J. Biol. Chem 293 (2018) 3925–3936. doi: 10.1074/jbc.RA117.000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Fulton MD, Brown T, Zheng YG, Mechanisms and Inhibitors of Histone Arginine Methylation, Chem. Rec 18 (2018) 1792–1807. doi: 10.1002/tcr.201800082. [DOI] [PMC free article] [PubMed] [Google Scholar]