Abstract
RNA virus populations are diverse due to a variety of factors, including lack of proofreading of the viral RNA-dependent RNA polymerase. These diverse viral populations include defective viruses incapable of productive infection. Recent studies have determined the existence of several modes of viral transmission outside of canonical pathways, including en bloc transmission of multiple viruses into a single host cell via membrane vesicles. Additionally, it has recently been determined that viral aggregation and bacteria can facilitate the delivery of multiple viruses to a single cell. Co-infection of RNA viruses is important since it has the potential to enhance viral fitness. Furthermore, through complementation and recombination, co-infection could potentially promote “resurrection” of otherwise defective viral genomes and has the potential to expand viral diversity.
Keywords: RNA viruses, Viral Co-infection, Viral Evolution
1. Introduction
1.1. RNA viruses
RNA viruses exist as diverse populations due to the high prevalence of mutations in their genomes (Domingo and Holland, 1997; Drake, 1993). While some mutations can be advantageous, the majority of mutations within viral genomes are neutral or deleterious to the virus. RNA viruses may overcome mutation-induced defects by several genetic mechanisms. First, error-prone RNA replication can revert mutations (Domingo and Holland, 1997; Drake, 1993). Second, genetic recombination can occur when two distinct viruses co-infect the same cell and exchange genetic information (Kirkegaard and Baltimore, 1986). Recombination can combine mutations on a single viral genome, or “erase” mutations by restoring the viral consensus sequence. Third, fitness may be restored by complementation, whereby two viruses with distinct genetic defects co-infect a cell and these defects are complemented by the functional genome/protein. Fourth, fitness may be restored by reassortment. Reassortment can occur when two distinct, segmented viruses co-infect a cell and generate progeny viruses containing a mixture of segments from both viruses. Genetic mechanisms such as recombination, complementation, and reassortment all require a cell to be co-infected by two, or more, viruses. Overall, these events can promote viral diversity and may enhance pathogenesis of RNA viruses (Dolan et al., 2018; Holmblat et al., 2014; Pfeiffer and Kirkegaard, 2005; Vignuzzi et al., 2006; Xiao et al., 2017).
1.2. Poliovirus as a model to study co-infection
Since its initial discovery in 1909, poliovirus has been extensively studied, initially as a public health threat and later as a model system (Leveque and Semler, 2015; Racaniello, 2006) Poliovirus is a single-stranded non-enveloped RNA virus in the Picornaviridae family. As an enteric virus, it is spread through the fecal-oral route. Like other RNA viruses, poliovirus error frequencies during RNA replication are high: approximately one error per replication cycle (Domingo and Holland, 1997; Drake, 1993; Drake et al., 1998; Mansky and Temin, 1995). Indeed, at a high multiplicity of infection (MOI), the poliovirus recombination frequency in cultured cells was 1.3 × 10−3, meaning that approximately one of every 1,300 genomes is the product of genetic recombination (Kirkegaard and Baltimore, 1986). Recombination of poliovirus has also been observed in vivo and there is evidence of poliovirus recombination with other enteroviruses during natural infection in humans (Arita et al., 2005; Cuervo et al., 2001; Dahourou et al., 2002; Furione et al., 1993; Holmblat et al., 2014; Sergiescu et al., 1969; Simmonds and Welch, 2006). Additionally, Holmblat et al. found that defective poliovirus genomes were capable of undergoing recombination, thus restoring their fitness in vivo (Holmblat et al., 2014). In fact, individuals infected with circulating vaccine derived polioviruses (cVDPVs) are commonly infected with viruses that have undergone genetic exchange (Cherkasova et al., 2002; Cuervo et al., 2001; Dahourou et al., 2002; Furione et al., 1993; Liu et al., 2000). Apart from genetic recombination, complementation of poliovirus genomes has also been observed in infected mice (Vignuzzi et al., 2006). While genetic recombination, complementation, and reassortment of RNA viruses have been observed in vitro and in vivo, the mechanisms that promote these events have not been fully defined.
2. Aggregation-mediated viral co-infection
Early observations indicated that several viruses can form aggregates, including both enveloped and non-enveloped viruses (Floyd and Sharp, 1977, 1978, 1979; Wallis and Melnick, 1967). For poliovirus, several virions can aggregate in sewage water and viral aggregates may complicate disinfection (Floyd and Sharp, 1979; Young and Sharp, 1977). While aggregation of mammalian viruses has been observed for nearly 50 years, the implication of aggregation as a potential mode of transmission and co-infection had not been studied until recently (Aguilera et al., 2017).
A well-known assumption in virology is that a single infectious unit forms a plaque. This concept is the basis for viral quantification by plaque assay in many viral systems. Intriguingly, recent studies have shown that plaques can be generated by an infectious unit containing more than one virus, thereby creating a “chimeric plaque” (Aguilera et al., 2017; Combe et al., 2015). We found that, in addition to single viral particles, our poliovirus stocks contained aggregates ranging from 2-10 particles (Aguilera et al., 2017). More importantly, when we induced aggregation by low pH treatment, we observed an increase in the frequency of chimeric plaques (Fig 1 mechanism 1). Interestingly, the frequency of chimeric plaques was increased in viral populations with mutagenized genomes, suggesting that co-infection and subsequent recombination and/or complementation could contribute to productive infection (Aguilera et al., 2017). These results suggest that aggregation can induce viral co-infection and possibly restore viral fitness.
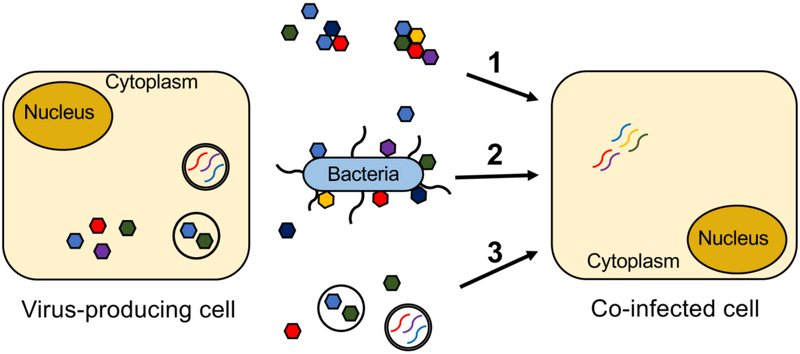
Figure 1.
Mechanisms of viral co-infection. Viral co-infection of mammalian cells can be induced by 1) Virion aggregates, 2) Bacteria bound by several viral particles, and 3) Release of viral particles or viral genomes within membrane vesicles. Genetically distinct viruses are depicted as multi-colored hexagons (virions) or curved lines (genomes).
3. Bacteria-mediated viral co-infection
3.1. Intestinal microbiota promote infection of enteric viruses
Mammalian enteric viruses encounter several barriers during infection of the host, including the relatively small number of viruses ingested as well as several physiological and immunological barriers. Therefore, initial infection of the host is likely a low MOI event due to these and other factors. In the last few years, studies from multiple groups have shown that intestinal bacteria play an important role in the infection of several unrelated RNA enteric viruses, including poliovirus, reovirus, rotavirus, mouse mammary tumor virus (MMTV) and noroviruses (Baldridge et al., 2015; Jones et al., 2014; Kane et al., 2011; Kuss et al., 2011; Uchiyama et al., 2014). Bacteria may promote viral infection through direct effects on viral particles or indirect effects on the host (Baldridge et al., 2015; Jones et al., 2014; Kane et al., 2011; Kuss et al., 2011; Li et al., 2015; Pfeiffer and Virgin, 2016; C. M. Robinson et al., 2014; Uchiyama et al., 2014). For poliovirus, the intestinal microbiota was required for efficient replication and pathogenesis in mice (Kuss et al., 2011). More specifically, bacteria increased the attachment of poliovirus to host cells and also limited virion inactivation from heat or bleach treatment in vitro (Kuss et al., 2011; C. M. Robinson et al., 2014). Our lab further determined that poliovirus binds to the surface of bacteria, indicating that direct interactions are mediating these effects (Erickson et al., 2018; Kuss et al., 2011; C. M. Robinson et al., 2014). Indirect mechanisms of bacteria-mediated enhancement of enteric viral infection include modulation of the host immune response (Baldridge et al., 2015; Kane et al., 2011). During MMTV infection, virus-bound LPS induced IL-10-mediated immune tolerance (Kane et al., 2011). For murine norovirus, bacteria may dampen IFN-λ mediated effects (Baldridge et al., 2015). Overall, bacteria facilitate infection of several unrelated RNA viruses through several mechanisms.
3.2. Intestinal bacteria promote co-infection of poliovirus
Increasing evidence supports the idea that bacteria promote infection of several mammalian viruses. While it is clear that enteric viruses interact closely with the host microbiota, whether these bacteria influence diversity of these viruses had not been studied until recently. We screened 40 bacterial strains for poliovirus binding and found that nearly all could bind the virus, and multiple virions could bind each bacterial cell (Erickson et al., 2018). Importantly, several of these bacterial strains induced the co-infection of distinct genetically marked polioviruses, even at a low MOI (Fig 1 mechanism 2). Furthermore, co-infection of viruses correlated with the ability of bacteria to adhere to host cells (Erickson et al., 2018). As a result of bacteria-mediated co-infection, genetic recombination occurred between two distinct parental strains with separate genetic defects, restoring viral fitness of progeny recombinant viruses. Additionally, we determined that bacteria can facilitate the co-infection of multiple distinct parental viruses, with up to 6 different parental viruses observed in a single plaque (Erickson et al., 2018). Overall, these findings indicate that bacteria mediate viral co-infection and may influence viral evolution.
4. Membrane vesicles containing multiple virions can promote co-infection
Several recent studies have shown that co-infection of RNA viruses can occur as a result of non-lytic, cell-to-cell transmission through membrane vesicle structures (Fig 1 mechanism 3). During infection, poliovirus can be packaged in phosphatidylserine-rich vesicles (Chen et al., 2015). This mode of packaging facilitated the transport of several viral particles, and thus co-infection, to neighboring cells in vitro (Chen et al., 2015). Importantly, Santiana et al. demonstrated that rotavirus and norovirus can be shed in stool within vesicles and that the vesicular form of the virus had enhanced disease severity in mice compared with single particles (Santiana et al., 2018). Additionally, poliovirus and coxsackievirus B3 (CVB3) can exit cells through vesicles derived from autophagosomes, a process referred to as AWOL (autophagosome-mediated exit without lysis) (Bird et al., 2014; S. M. Robinson et al., 2014). Both hepatitis A and C viruses can exit infected cells through exosomes, or vesicle-like structures, thus mediating the transfer of infectious viral particles and RNA to neighboring cells in vitro (Dreux et al., 2012; Feng et al., 2013; Longatti et al., 2015; Ramakrishnaiah et al., 2013).
5. Co-infection as a requirement for multipartite viral infection
For certain RNA viruses, such as those with segmented genomes packaged into separate particles, co-infection is a requirement for productive infection. While several mammalian viruses have segmented genomes, the segments of these viruses are packaged into a single viral particle. However, unique to certain plant and fungal viruses, some of these viruses package their genome segments into separate, individual viral particles. Productive infection of these viruses requires these individually packaged particles to infect the same cell simultaneously, a phenomena known as multipartite or multicomponent infection (Ghabrial and Suzuki, 2009; Rao, 2006). Recently, Ladner et al. determined that an animal virus has a multipartite genome similar to multipartite genomes of some plant and fungal viruses (Ladner et al., 2016). This enveloped RNA virus, Guaico Culex virus (GCXV), was isolated from Culex mosquitoes and is composed of 5 viral segments that are packaged in separate viral particles (Ladner et al., 2016). Interestingly, only 3 viral segments were required for productive infection and subsequent plaque formation (Ladner et al., 2016). While several viral particles are required for efficient infection of some RNA viruses, the mechanisms and consequences of the multicomponent infection process during transmission remain unclear.
6. Conclusions
Increasing evidence supports the idea that viral co-infection is mediated by several mechanisms and potentially at different stages of infection. Cell-to-cell transmission by virions in vesicle-like structures or aggregates could facilitate the transport of several viral particles and/or genomes from infected cells to neighboring cells. Other factors, such as the presence of bacteria, could also facilitate co-infection during inter-host transmission. These mechanisms of infection also have several implications for viral fitness of otherwise defective genomes. Overall, the studies described here highlight the existence of novel mechanisms that influence co-infection of RNA viruses and potentially promote viral evolution.
Highlights.
Emerging modes of RNA virus transmission may facilitate viral co-infection
Viral aggregates can co-infect cells and form chimeric plaques - Bacteria can facilitate viral co-infection and subsequent recombination
Multiple viruses within membrane vesicles can co-infect cells
Acknowledgements
Work in J.K.P.’s lab is funded through NIH NIAID grants R01 AI74668 and R21 AI114927, a Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Diseases Award, and a Faculty Scholar grant from the Howard Hughes Medical Institute. E.R.A. was supported in part by the National Science Foundation Graduate Research Fellowship grant 2014176649.
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguilera ER, Erickson AK, Jesudhasan PR, Robinson CM, & Pfeiffer JK (2017). Plaques Formed by Mutagenized Viral Populations Have Elevated Coinfection Frequencies. MBio, 8(2). doi: 10.1128/mBio.02020-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arita M, Zhu SL, Yoshida H, Yoneyama T, Miyamura T, & Shimizu H (2005). A Sabin 3-derived poliovirus recombinant contained a sequence homologous with indigenous human enterovirus species C in the viral polymerase coding region. J Virol, 79(20), 12650–12657. doi: 10.1128/JVI.79.20.12650-12657.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge MT, Nice TJ, McCune BT, Yokoyama CC, Kambal A, Wheadon M, … Virgin HW (2015). Commensal microbes and interferon-lambda determine persistence of enteric murine norovirus infection. Science, 347(6219), 266–269. doi: 10.1126/science.1258025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird SW, Maynard ND, Covert MW, & Kirkegaard K (2014). Nonlytic viral spread enhanced by autophagy components. Proc Natl Acad Sci U S A, 111(36), 13081–13086. doi: 10.1073/pnas.1401437111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Du W, Hagemeijer MC, Takvorian PM, Pau C, Cali A, … Altan-Bonnet N (2015). Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell, 160(4), 619–630. doi: 10.1016/j.cell.2015.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova EA, Korotkova EA, Yakovenko ML, Ivanova OE, Eremeeva TP, Chumakov KM, & Agol VI (2002). Long-term circulation of vaccine-derived poliovirus that causes paralytic disease. J Virol, 76(13), 6791–6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combe M, Garijo R, Geller R, Cuevas JM, & Sanjuan R (2015). Single-Cell Analysis of RNA Virus Infection Identifies Multiple Genetically Diverse Viral Genomes within Single Infectious Units. Cell Host Microbe, 18(4), 424–432. doi: 10.1016/j.chom.2015.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo NS, Guillot S, Romanenkova N, Combiescu M, Aubert-Combiescu A, Seghier M, … Delpeyroux F (2001). Genomic features of intertypic recombinant sabin poliovirus strains excreted by primary vaccinees. J Virol, 75(13), 5740–5751. doi: 10.1128/JVI.75.13.5740-5751.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahourou G, Guillot S, Le Gall O, & Crainic R (2002). Genetic recombination in wild-type poliovirus. J Gen Virol, 83(Pt 12), 3103–3110. doi: 10.1099/0022-1317-83-12-3103 [DOI] [PubMed] [Google Scholar]
- Dolan PT, Whitfield ZJ, & Andino R (2018). Mechanisms and Concepts in RNA Virus Population Dynamics and Evolution. Annu Rev Virol. doi: 10.1146/annurev-virology-101416-041718 [DOI] [PubMed] [Google Scholar]
- Domingo E, & Holland JJ (1997). RNA virus mutations and fitness for survival. Annu Rev Microbiol, 51, 151–178. doi: 10.1146/annurev.micro.51.1.151 [DOI] [PubMed] [Google Scholar]
- Drake JW (1993). Rates of spontaneous mutation among RNA viruses. Proc Natl Acad Sci U S A, 90(9), 4171–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW, Charlesworth B, Charlesworth D, & Crow JF (1998). Rates of spontaneous mutation. Genetics, 148(4), 1667–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreux M, Garaigorta U, Boyd B, Decembre E, Chung J, Whitten-Bauer C, … Chisari FV (2012). Short-range exosomal transfer of viral RNA from infected cells to plasmacytoid dendritic cells triggers innate immunity. Cell Host Microbe, 12(4), 558–570. doi: 10.1016/j.chom.2012.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson AK, Jesudhasan PR, Mayer MJ, Narbad A, Winter SE, & Pfeiffer JK (2018). Bacteria Facilitate Enteric Virus Co-infection of Mammalian Cells and Promote Genetic Recombination. Cell Host Microbe, 23(1), 77–88 e75. doi: 10.1016/j.chom.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, … Lemon SM (2013). A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature, 496(7445), 367–371. doi: 10.1038/nature12029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd R, & Sharp DG (1977). Aggregation of poliovirus and reovirus by dilution in water. Appl Environ Microbiol, 33(1), 159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd R, & Sharp DG (1978). Viral aggregation: effects of salts on the aggregation of poliovirus and reovirus at low pH. Appl Environ Microbiol, 35(6), 1084–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd R, & Sharp DG (1979). Viral aggregation: buffer effects in the aggregation of poliovirus and reovirus at low and high pH. Appl Environ Microbiol, 38(3), 395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furione M, Guillot S, Otelea D, Balanant J, Candrea A, & Crainic R (1993). Polioviruses with natural recombinant genomes isolated from vaccine-associated paralytic poliomyelitis. Virology, 196(1), 199–208. [DOI] [PubMed] [Google Scholar]
- Ghabrial SA, & Suzuki N (2009). Viruses of plant pathogenic fungi. Annu Rev Phytopathol, 47, 353–384. doi: 10.1146/annurev-phyto-080508-081932 [DOI] [PubMed] [Google Scholar]
- Holmblat B, Jegouic S, Muslin C, Blondel B, Joffret ML, & Delpeyroux F (2014). Nonhomologous recombination between defective poliovirus and coxsackievirus genomes suggests a new model of genetic plasticity for picornaviruses. MBio, 5(4), e01119–01114. doi: 10.1128/mBio.01119-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, … Karst SM (2014). Enteric bacteria promote human and mouse norovirus infection of B cells. Science, 346(6210), 755–759. doi: 10.1126/science.1257147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, Chervonsky AV, & Golovkina TV (2011). Successful transmission of a retrovirus depends on the commensal microbiota. Science, 334(6053), 245–249. doi: 10.1126/science.1210718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkegaard K, & Baltimore D (1986). The mechanism of RNA recombination in poliovirus. Cell, 47(3), 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, … Pfeiffer JK (2011). Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science, 334(6053), 249–252. doi: 10.1126/science.1211057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner JT, Wiley MR, Beitzel B, Auguste AJ, Dupuis AP 2nd, Lindquist ME, … Palacios G (2016). A Multicomponent Animal Virus Isolated from Mosquitoes. Cell Host Microbe, 20(3), 357–367. doi: 10.1016/j.chom.2016.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveque N, & Semler BL (2015). A 21st century perspective of poliovirus replication. PLoS Pathog, 11(6), e1004825. doi: 10.1371/journal.ppat.1004825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Breiman A, le Pendu J, & Uyttendaele M (2015). Binding to histo-blood group antigen-expressing bacteria protects human norovirus from acute heat stress. Front Microbiol, 6, 659. doi: 10.3389/fmicb.2015.00659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HM, Zheng DP, Zhang LB, Oberste MS, Pallansch MA, & Kew OM (2000). Molecular evolution of a type 1 wild-vaccine poliovirus recombinant during widespread circulation in China. J Virol, 74(23), 11153–11161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longatti A, Boyd B, & Chisari FV (2015). Virion-independent transfer of replication-competent hepatitis C virus RNA between permissive cells. J Virol, 89(5), 2956–2961. doi: 10.1128/JVI.02721-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansky LM, & Temin HM (1995). Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol, 69(8), 5087–5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer JK, & Kirkegaard K (2005). Increased fidelity reduces poliovirus fitness and virulence under selective pressure in mice. PLoS Pathog, 1(2), e11. doi: 10.1371/journal.ppat.0010011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer JK, & Virgin HW (2016). Viral immunity. Transkingdom control of viral infection and immunity in the mammalian intestine. Science, 351(6270). doi: 10.1126/science.aad5872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racaniello VR (2006). One hundred years of poliovirus pathogenesis. Virology, 344(1), 9–16. doi: 10.1016/j.virol.2005.09.015 [DOI] [PubMed] [Google Scholar]
- Ramakrishnaiah V, Thumann C, Fofana I, Habersetzer F, Pan Q, de Ruiter PE, … van der Laan LJ (2013). Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci U S A, 110(32), 13109–13113. doi: 10.1073/pnas.1221899110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao AL (2006). Genome packaging by spherical plant RNA viruses. Annu Rev Phytopathol, 44, 61–87. doi: 10.1146/annurev.phyto.44.070505.143334 [DOI] [PubMed] [Google Scholar]
- Robinson CM, Jesudhasan PR, & Pfeiffer JK (2014). Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe, 15(1), 36–46. doi: 10.1016/j.chom.2013.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SM, Tsueng G, Sin J, Mangale V, Rahawi S, McIntyre LL, … Feuer R (2014). Coxsackievirus B exits the host cell in shed microvesicles displaying autophagosomal markers. PLoS Pathog, 10(4), e1004045. doi: 10.1371/journal.ppat.1004045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiana M, Ghosh S, Ho BA, Rajasekaran V, Du WL, Mutsafi Y, … Altan-Bonnet N (2018). Vesicle-Cloaked Virus Clusters Are Optimal Units for Inter-organismal Viral Transmission. Cell Host Microbe, 24(2), 208–220 e208. doi: 10.1016/j.chom.2018.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergiescu D, Aubert-Combiescu A, & Crainic R (1969). Recombination between guanidine-resistant and dextran sulfate-resistant mutants of type 1 poliovirus. J Virol, 3(3), 326–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P, & Welch J (2006). Frequency and dynamics of recombination within different species of human enteroviruses. J Virol, 80(1), 483–493. doi: 10.1128/JVI.80.1.483-493.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama R, Chassaing B, Zhang B, & Gewirtz AT (2014). Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. J Infect Dis, 210(2), 171–182. doi: 10.1093/infdis/jiu037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, & Andino R (2006). Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature, 439(7074), 344–348. doi: 10.1038/nature04388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis C, & Melnick JL (1967). Virus aggregation as the cause of the non-neutralizable persistent fraction. J Virol, 1(3), 478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Dolan PT, Goldstein EF, Li M, Farkov M, Brodsky L, & Andino R (2017). Poliovirus intrahost evolution is required to overcome tissue-specific innate immune responses. Nat Commun, 8(1), 375. doi: 10.1038/s41467-017-00354-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DC, & Sharp DG (1977). Poliovirus aggregates and their survival in water. Appl Environ Microbiol, 33(1), 168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]