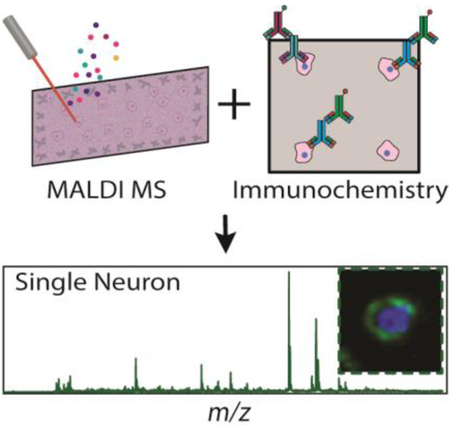
Abstract
Transcriptomics characterizes cells based on their potential molecular repertoire whereas direct mass spectrometry (MS) provides information on the actual compounds present within cells. Single cell matrix assisted laser desorption/ionization (MALDI) MS can measure the chemical contents of individual cells but spectra are difficult to correlate to conventional cell types, limiting the metabolic information obtained. We present a protocol that combines MALDI MS with immunocytochemistry to assay over a thousand individual rat brain cells. The approach entwines the wealth of knowledge obtained by immunocytochemical profiling with mass spectral information on the predominant lipids within each cell. While many lipid species showed a high degree of similarity between neurons and astrocytes, seventeen significantly differed between the two cell types, including four phosphatidylethanolamines elevated in astrocytes and nine phosphatidylcholines in neurons.
Keywords: Single Cell, Mass Spectrometry, Immunochemistry, Lipids, Neuron, Astrocyte
Graphical Abstract
The combination of single cell MALDI mass spectrometry and immunocytochemistry allows metabolic information to be directly coupled to canonical cell type. Analysis of over a thousand single cells shows that many lipid species have a high similarity between neurons and astrocytes, although seventeen lipid species differed significantly between the two cell types, including four phosphatidylethanolamines elevated in astrocytes and nine phosphatidylcholines in neurons.
Mammalian brain function critically relies on cellular heterogeneity[1] and explorations of this diversity have exploded with the advent of single cell transcriptomics.[2] The traditional metric of cell classification by morphology correlates shape to cell function.[3] Cellular chemistry also correlates to function; e.g., the biophysical properties of the cell membrane are governed by its lipid composition.[4] Recent advances in mass spectrometry (MS)[5] have ushered in a new era of single cell analysis.[6] While matrix-assisted laser desorption/ionization (MALDI) MS produces rich spectral profiles for label-free classifications,[7] the results can be difficult to correlate to canonical cell types.[8]
Immunocytochemistry (ICC) is effective for classifying cells as it can target a plethora of different molecules that distinguish cell types.[9] ICC has been combined with tissue imaging and a few examples have coupled the method to single cell MS.[10] However, fluorescence overlap of labels typically limits plexity to fewer than ten compounds,[11] and most ICC protocols require sample fixation and lipid removal, crosslinking and/or removing many compounds that are easily ionized by MALDI MS.[12] Direct hyphenation of ICC and MS to analyze the same cell was reported in 2012,[10b] but ICC was performed first, limiting the analysis to high-abundance peptides within a small number of cells. Here we describe a workflow enabling high-throughput MALDI MS analysis of thousands of cells, followed by cell classification via ICC. Performing ICC after MALDI MS expands the chemical interrogation to include lipids and small metabolites, which are essential for brain function.[13] The dataset here consists of over 1800 rodent cerebellar cells assayed using microMS, a cell finding and analysis software.[14] We demonstrate the approach by examining chemical differences between two of the most common cell types in the brain – astrocytes and neurons. Because the lipid differences between astrocytes and neurons are both poorly understood and modest, ICC classification allows us to discover statistically distinct features between these important cell types.
Mass spectra acquired from unfixed single cells contained peaks consistent with small metabolites and lipid species at high intensity but after performing the fixation procedure (as required for most ICC protocols), the resulting spectra showed weaker metabolite peak intensities (~90% lower) as well a polymer contaminant (Figure S1). Thus, we decided to perform ICC after MALDI MS (Figure 1a–c) in a manner similar to our recent paper coupling vibrational spectroscopy and MALDI MS imaging of tissue slices;[15] using MALDI MS first, we observed high-quality mass spectra of molecules, including small metabolites and lipids. For this procedure to succeed, the MALDI matrix (e.g., 2,5-dihydroxybenzoic acid), which had been thoroughly co-crystalized with the cells, had to be removed while ensuring that the cells remained anchored to the same location on the slide. Finally, the antibody must still recognize its epitope. We found that paraformaldehyde was effective in both fixing the cells and removing the matrix (Figure S2g). Importantly, a high proportion of cells (~95%) did not move during matrix removal (Figure S2, insets). While ICC requires long incubations that can lead to cell delocalization (Figure 2a), ~88% of the cells stayed in the same location, while only a few were delocalized (Figure 2a, red arrows). Relying on image registration as a metric for cell retention, we successfully matched 71±18% of cells within a 25 μm field of view.
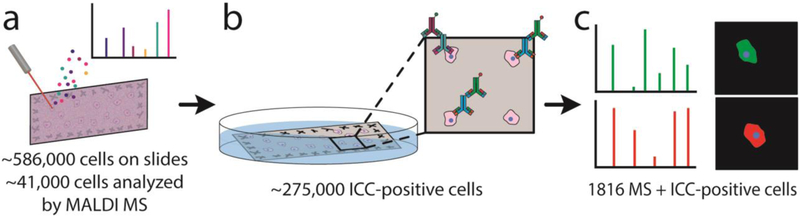
Figure 1.
Schematic of the MALDI MS-ICC approach. (a) microMS is used to localize the ~586,000 cells on the slides. Single cell MALDI MS is performed on ~41,000 cell-like structures that are at least 100 μm from other cells. (b) After MS, the MALDI matrix is removed and cells are fixed with a solution of 4% PFA in phosphate buffered solution, followed by immunostaining against GFAP and NF-L. ~275,000 cells on the slide immunostained successfully. (c) Mass spectra and ICC profiles are directly compared because cells stay in the same location. 1816 cells contained quality mass spectra and ICC profiles.
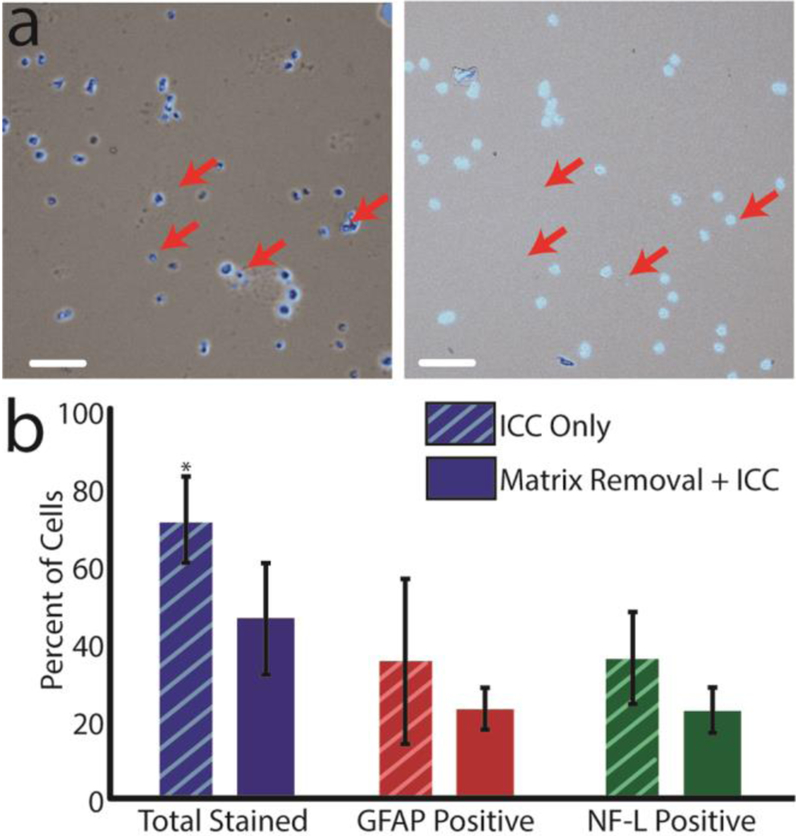
Figure 2.
(a) A subset of cells before ICC (left) and a subset of cells after ICC (right). Red arrows indicate several cells that were dislocated during the ICC procedure. Scale bar = 100 μm. (b) Graph comparing a subset of immunopositive cells in dissociated cells that were immediately fixed (ICC only, number of distinct cells = 5435, n = 6 animals) and cells that underwent MALDI matrix removal and subsequent ICC (number of distinct cells = 5646, n = 6 animals). The total number of stained cells was statistically different at the 95% confidence interval; error bars represent the standard deviation. The proportion of GFAP- to NF-L positive cells was not statistically different between the matrix-removed and ICC-only cells.
Using the optimized procedure (see Experimental Details in the Supporting Information), cells within the cerebellum were classified as glial fibrillary acidic protein (GFAP) positive, neurofilament-L (NF-L) positive, or ICC negative. While GFAP is not a universal astrocytic marker, most astrocytes within the cerebellum can be immunopositively labeled by GFAP.
Similarly, most neurons express NF-L antigens. The cells that were ICC negative may be other cell types that lack these antigens or damaged cells. Due to their ambiguous identification, ICC-negative cells were excluded from the mass spectral analysis.
Prior lipidomic studies of astrocytes and neurons have relied on cell lines or cultured primary cells to examine metabolic differences[16] as opposed to the primary isolated and mixed cell populations assayed here. The chemical nature of neurons and astrocytes results from their intertwined locations within a network of cells, information that can be lost during cell monoculture (e.g., astrocyte cell lines lose GFAP immunoreactivity). Because the dissociated cells used here were rapidly transferred from the brain, they retained many defining features; albeit, the native cell position was entirely lost. The chemical information gained from dissociated cells is similar to primary astrocyte cultures,[17] although freshly dissociated cells may reflect the native chemistry more accurately than cells after several days in culture.[18] Our protocol also allows for targeted analysis of cell types without having to purify cell-specific cultures and is adaptable to primary cultures.
To assess the effect of MALDI MS on the ICC profiles, immunostaining efficiency was compared between cells subjected to matrix removal and cells that were fixed and immunostained immediately after dissociation. In this case, the damaged cells/ bare nuclei were manually removed from the cell counts. Unsurprisingly, cells stained more efficiently when immediately fixed and stained (Figure 2b). The differences in staining could be caused by many factors, including the effects of matrix removal or laser irradiation. On control slides, 72% of cells stained, with this number decreasing to 46% following matrix removal. The discrepancy between treatments is significant (p-value = 0.002, two-tailed t-test), but if we assume the control treatment as an upper limit, more than half of the cells that could immunostain were still characterized following MALDI MS. Additionally, the relative amount of each population remained consistent between control and matrix-removed cells (p-value = 0.886 and z-value = −0.49; proportions test). The consistent proportion of GFAP- and NF-L-positive cells is important; if one population had been disproportionately affected by the MALDI MS measurement process, the derived chemical inferences may have been affected.
Using our MALDI MS-ICC approach, we can directly correlate the ICC profile, cell image, and mass spectrum for each cell (Figure 3a–d). Both NF-L-positive cells (Figure 3a, b) and GFAP-positive cells (Figure 3c, d) have the capacity to stain with high-quality mass spectra that include many lipid features that are absent from MS of fixed cells. Even after laser ablation and matrix removal, each cell remains intact (Figure 3a–d, insets) and maintains the structural features present during cell plating for easy recognition and post-analysis, except for what appears to be nuclei leakage, which sometimes occurs after MS processing (Figure 3c). In addition, the heterogeneity of each class of cells can further be visualized by t-distributed stochastic neighbor embedding (t-SNE) and hierarchical clustering analysis (HCA) (Figure 3e–f). NF-L- and GFAP-positive cells can be subsequently clustered into five and four subpopulations, respectively, based on their mass spectral features. The presence of multiple clusters demonstrates that all similarly stained cells do not possess the same mass spectral features, and thus, may belong to distinct subtypes of astrocytes/neurons. Together, our preliminary results reveal the importance of single cell measurements of the brain by MS.
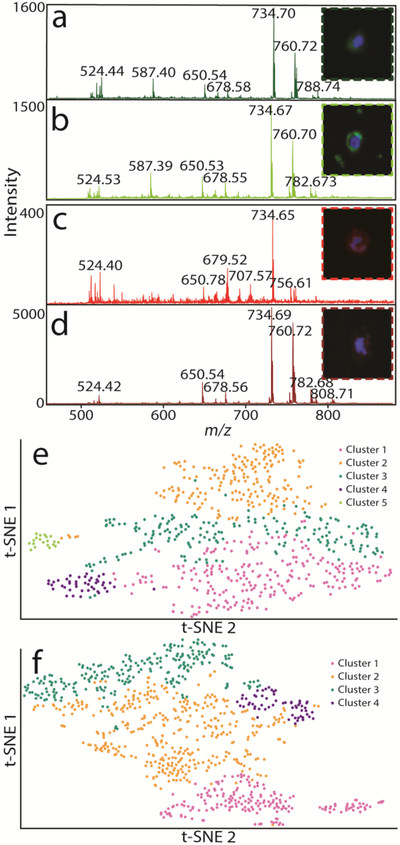
Figure 3.
High-quality single cell mass spectra and ICC images are acquired and compared for over a thousand individual cells. Single cell spectra for cells displayed in each panel are shown. (a) Single cell spectrum for an NF-L-positive cell. (b) A second NF-L positive cell with similar mass spectral features. (c) GFAP-positive single cell that is visibly different from the (d) second chosen GFAP-positive cell. Green and red fluorescence corresponds to NF-L and GFAP, respectively, while blue fluorecsence corresponds to a nuclear dye.(e) t-SNE plot of NF-L-positive cells are colored to correspond to the five clusters determined by HCA. (f) t-SNE plot of GFAP-positive cells colored to correspond to the four clusters determined by HCA.
Lipids are known to have important biological functions in the brain relating to communication, metabolism, and transport, but the specific function of many individual lipids is unknown. Reasons for this include the difficulty in measuring large numbers of lipids in complex samples and identifying the saturation points of lipids, and the limited number of databases containing lipidomic information (particularly when compared to genomics and proteomics).[19] MS is one of the only techniques capable of lipid identification in complex samples and is one of the leading methods for lipidomic analysis. We detected an average of 99 molecular features within individual cells and found that 17 lipids had statistically distinct abundances between neurons and astrocytes (Table 1). Statistical differences were determined using a rank sum test with a false discovery rate correction and high-resolution MS.
Table 1.
Lipids that are significantly different (p-value< 0.05) between GFAP- and NF-L-positive cells (n = 1547).
| Measured m/z Value | PPM Error | Assignment | Type | Relative Enrichment |
|---|---|---|---|---|
| 837.6607‡ | unknown | GFAP Positive | 61% | |
| 724.5249[20] | −0.3602 | [PE(O-34:2)+Na]+ [PE(P-34:1)+Na]+ |
GFAP Positive | 58% |
| 789.6143* | 3.3624 | [PE(38:2)+NH4]+ | GFAP Positive | 39% |
| 752.5564 | −0.0810 | [PE(O-36:2)+Na]+ [PE(P-36:1)+Na]+ |
GFAP Positive | 28% |
| 768.5880 | 0.3110 | [PC(O-34:1)+Na]+ [PC(P-34:0)+Na]+ [PE(O-37:1)+Na]+ [PE(P-37:0)+Na]+ |
GFAP Positive | 16% |
| 850.6730‡ | −1.4740 | unknown | NF-L Positive | 60% |
| 731.6060 |
−0.2064 |
[SM(d36:1)+H]+ | NF-L Positive | 30% |
| 732.5538 | 0.0246 | [PC(32:1)+H ]+ | NF-L Positive | 27% |
| 834.6003‡ | −0.5176 | [PC(40:6)+H ]+ | NF-L Positive | 26% |
| 784.5828* | 0.1580 | [PC(34:0)+Na]+ | NF-L Positive | 24% |
| 828.5504 | −1.1780 | [PC(38:6)+Na]+ | NF-L Positive | 23% |
| 756.5513[21] | −0.1004 | [PC(32:0)+Na]+ [PE(35:0) +Na]+ |
NF-L Positive | 22% |
| 808.5825* | −0.2177 | [PC(36:2)+Na]+ | NF-L Positive | 21% |
| 782.5670 | −0.0332 | [PC(34:1)+Na]+ | NF-L Positive | 18% |
| 815.6347‡ | unknown | NF-L Positive | 16% | |
| 762.5044* 762.6004* |
−0.0341 −0.4353 |
[PE(36:4)+Na]+ [PC(34:0)+H]+ |
NF-L Positive | 11% |
| 772.6218‡ | 0.4310 | [PC(O-36:2)+H]+ [PC(P-36:1)+H]+ |
NF-L Positive | 06% |
The peaks detected in the cells were mass matched to the cerebellum lipid extracts via high mass accuracy and MS/MS, unless otherwise indicated. This table does not include unlikely, isobaric lipids, such as hydroxylated lipids.
MS/MS was not possible due to insufficient isolation of the ion.
MS/MS and high mass accuracy were insufficient for identification.
Unfortunately, we cannot perform direct tandem MS of the lipids from single cells that were ICC positive because of the required fixation step. As such, we performed a lipid extract of bulk cerebral tissue for high mass accuracy and tandem MS FT-ICR MS analysis for lipid assignment. While there are several examples of multiple lipids resolved by FT-ICR MS analysis, making several assignments ambiguous (Table 1, ex. m/z value 731), most of the lipid peaks were sufficiently isolated from other spectral features, increasing confidence in the lipid assignments. All but three lipids had mass errors of less than 1 ppm and tandem MS (MS/MS) was possible for all but four of the lipids with errors less than 5 ppm (Table 1).
The 17 lipid differences detected here may not be observed in bulk tissue analysis because of their levels, instrumental dynamic range, and the blended nature of the cells within most neuronal samples. Since we performed single cell analysis and averaged groups of similarly stained cells together, we identified small, yet consistent lipid differences between astrocytes and neurons. Biological membranes tend to have similar lipids, but the differences can be important and may relate to essential functions. Determining the function of these lipids is beyond the scope of this communication; however, combining MALDI MS with ICC has narrowed targets for functional analyses down to a small number of lipids from the thousands known to be present in the brain.
The lipid differences between these cells may reflect the different functions of these cell types. Five lipids were present at higher intensity within astrocytes, four of which are phosphatidylethanolamines (PEs), such as PE(36:1). PE lipids, in particular, are important for determining membrane curvature as well as contractile ring disassembly at the cleavage furrow following cytokinesis.[22] The higher PE abundance in astrocytes may reflect their replication rate compared to neurons, and their greater curvature. PE lipids have also been implicated in vesicle fusion and release for neurotransmission, potentially explaining the presence of PEs in neurons as well as astrocytes.[22d, 23] The contributions of PE (and other lipids) that would be found at high local levels in neuronal and astrocytic processes would not be observed as our cell isolation procedures remove most processes from the isolated cell soma. Therefore, we expect that lipids enriched and found within these subcellular structures to be underrepresented in our results. Another 12 lipids had higher abundances in neurons (e.g. phosphatidylcholine (PC) (32:1) and SM(d36:1)). Many of the statistically different lipids enriched within neurons include PCs, which are common in lipid membranes and serve as the source for choline, used in neurotransmission.[24] The remaining lipids are more difficult to relate to cell physiology; however, their relative abundance may reflect the distinct shapes or functions of neurons or astrocytes. Because biological membranes are highly conserved structures, we are excited that single cell MALDI MS can detect these subtle differences and ICC allows us to connect them to cell type.
Recent single cell heterogeneity studies of brain cells have primarily been spearheaded by transcriptomics, but are inherently limited to gene expression profiles. Because of this information gap, we have developed an additional avenue for determining single cell metabolic information that can be tied directly to canonical cell types as defined by ICC. Not only can functional information be garnered from metabolites present within the cell, intrinsic chemical differences, such as lipid composition, may provide a more robust classification metric with direct links to cell phenotypes. The results obtained from MALDI MS-ICC can be used to guide targeted transcriptomic experiments to further understand how subtle chemical changes impact the global function of the brain. This information also enables targeted functional lipid experiments, which should aid our understanding of neuronal disorders as well as normal brain function. Moreover, the precise mapping of chemical details with ICC-established cell types within the brain is a significant advancement in single cell MS. Our development merges the rich chemical information found within mass spectra with the well-defined cellular classifications obtained by ICC from thousands of cells, enabling new insights into cell-to-cell differences found within the brain.
Supplementary Material
Acknowledgements
The authors acknowledge support from the National Institute of Mental Health Award No. 1U01MH109062 and the National Institute on Drug Abuse Award No. P30DA018310. E.K.N. and T.J.C. acknowledge support from the National Science Foundation Graduate Research Fellowship Program and Springborn Fellowships. The authors would also like to thank Joseph Ellis and Jennifer Mitchell for useful discussions.
Footnotes
Supporting information and the ORCID identification number(s) for the author(s) of this article can be found under: DOI: 10.1002/anie.201812892 and 10.1002/ange.201812892. Data supporting this study are openly available from the University of Illinois DataBank at https://databank.illinois.edu/ with the dataset locatable using: DOI: 10.13012/B2–3125702_V1.
References
- [1].a) Li Q, Barres BA, Nat. Rev. Immunol 2017, 18, 225; [DOI] [PubMed] [Google Scholar]; b) Morales M, Margolis EB, Nat. Rev. Neurosci 2017, 18, 73; [DOI] [PubMed] [Google Scholar]; c) Schitine C, Nogaroli L, Costa MR, Hedin-Pereira C, Front. Cell. Neurosci 2015, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Poulin J-F, Tasic B, Hjerling-Leffler J, Trimarchi JM, Awatramani R, Nat. Neurosci 2016, 19, 1131. [DOI] [PubMed] [Google Scholar]
- [3].a) Uusisaari M, Obata K, Knöpfel T, J. Neurophysiol 2007, 97, 901–911; [DOI] [PubMed] [Google Scholar]; b) Grosche J, Kettenmann H, Reichenbach A, J. Neurosci. Res 2002, 68, 138–149. [DOI] [PubMed] [Google Scholar]
- [4].a) Simons K, Vaz WLC, Annu. Rev. Biophys. Biomol. Struct 2004, 33, 269–295; [DOI] [PubMed] [Google Scholar]; b) van Meer G, Voelker DR, Feigenson GW, Nat. Rev. Mol. Cell Biol 2008, 9, 112; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Janmey PA, Kinnunen PKJ, Trends Cell Biol. 2006, 16, 538–546. [DOI] [PubMed] [Google Scholar]
- [5].Chen X, Love JC, Navin N, Pachter L, Stubbington M, Svensson V, Sweedler JV, Teichmann SA, Nat. Biotechnol 2016, 34, 1111–1118. [DOI] [PubMed] [Google Scholar]
- [6].Liu R, Zhang G, Yang Z, Chem. Commun. (Camb.) 2019, 55, 616–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].a) Cornett DS, Reyzer ML, Chaurand P, Caprioli RM, Nat. Methods 2007, 4, 828; [DOI] [PubMed] [Google Scholar]; b) Yang B, Patterson NH, Tsui T, Caprioli RM, Norris JL, J. Am. Soc. Mass Spectrom 2018, 29, 1012–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].a) Comi TJ, Do TD, Rubakhin SS, Sweedler JV, J. Am. Chem. Soc 2017, 139, 3920–3929; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Do TD, Ellis JF, Neumann EK, Comi TJ, Tillmaand EG, Lenhart AE, Rubakhin SS, Sweedler JV, Chemphyschem 2018, 19, 1180–1191; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Rubakhin SS, Romanova EV, Nemes P, Sweedler JV, Nat. Methods 2011, 8, S20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Doetsch F, Caillé I, Lim DA, García-Verdugo JM, Alvarez-Buylla A, Cell 1999, 97, 703–716. [DOI] [PubMed] [Google Scholar]
- [10].a) Li L, Romanova EV, Rubakhin SS, Alexeeva V, Weiss KR, Vilim FS, Sweedler JV, Anal. Chem 2000, 72, 3867–3874; [DOI] [PubMed] [Google Scholar]; b) Neupert S, Rubakhin SS, Sweedler JV, Chem. Biol 2012, 19, 1010–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Stack EC, Wang C, Roman KA, Hoyt CC, Methods 2014, 70, 46–58. [DOI] [PubMed] [Google Scholar]
- [12].Howat WJ, Wilson BA, Methods 2014, 70, 12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].a) Piomelli D, Sasso O, Nat. Neurosci 2014, 17, 164; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wanders RJA, Poll-The BT, Neurosci. Lett 2017, 637, 11–17; [DOI] [PubMed] [Google Scholar]; c) Bazinet RP, Layé S, Nat. Rev. Neurosci 2014, 15, 771; [DOI] [PubMed] [Google Scholar]; d) Leloup C, Allard C, Carneiro L, Fioramonti X, Collins S, Pénicaud L, Neuroscience 2016, 323, 110–120. [DOI] [PubMed] [Google Scholar]
- [14].Comi TJ, Neumann EK, Do TD, Sweedler JV, J. Am. Soc. Mass Spectrom 2017, 28, 1919–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Neumann EK, Comi TJ, Spegazzini N, Mitchell JW, Rubakhin SS, Gillette MU, Bhargava R, Sweedler JV, Anal. Chem 2018, 90, 11572–11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].a) Nieweg K, Schaller H, Pfrieger FW, J. Neurochem 2009, 109, 125–134; [DOI] [PubMed] [Google Scholar]; b) Moore SA, Yoder E, Murphy S, Dutton GR, Spector AA, J. Neurochem 1991, 56, 518–524; [DOI] [PubMed] [Google Scholar]; c) Pfrieger FW, Cell. Mol. Life Sci 2003, 60, 1158–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lange SC, Bak LK, Waagepetersen HS, Schousboe A, Norenberg MD, Neurochem. Res 2012, 37, 2569–2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Nemes P, Knolhoff AM, Rubakhin SS, Sweedler JV, ACS Chem. Neurosci 2012, 3, 782–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wenk MR, Nat. Rev. Drug Discov 2005, 4, 594. [DOI] [PubMed] [Google Scholar]
- [20].Soltwisch J, Kettling H, Vens-Cappell S, Wiegelmann M, Muthing J, Dreisewerd K, Science 2015, 348, 211–215. [DOI] [PubMed] [Google Scholar]
- [21].a) Eberlin LS, Dill AL, Golby AJ, Ligon KL, Wiseman JM, Cooks RG, Agar NY, Angew. Chem. Int. Ed. Engl 2010, 49, 5953–5956; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Jackson SN, Wang HY, Woods AS, J. Am. Soc. Mass Spectrom 2005, 16, 2052–2056. [DOI] [PubMed] [Google Scholar]
- [22].a) Vance JE, Tasseva G, Biochim. Biophys. Acta 2013, 1831, 543–554; [DOI] [PubMed] [Google Scholar]; b) Emoto K, Kobayashi T, Yamaji A, Aizawa H, Yahara I, Inoue K, Umeda M, Proc. Natl. Acad. Sci. U. S. A 1996, 93, 12867–12872; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Emoto K, Umeda M, J. Cell Biol 2000, 149, 1215–1224; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Glaser PE, Gross RW, Biochemistry 1995, 34, 12193–12203. [DOI] [PubMed] [Google Scholar]
- [23].Lohner K, Chem. Phys. Lipids 1996, 81, 167–184. [DOI] [PubMed] [Google Scholar]
- [24].Tracey TJ, Steyn FJ, Wolvetang EJ, Ngo ST, Front. Mol. Neurosci 2018, 11, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.