Abstract
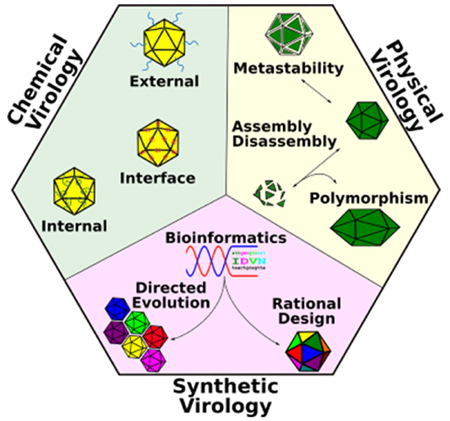
The fields of physical, chemical, and synthetic virology work in partnership to reprogram viruses as controllable nanodevices. Physical virology provides the fundamental biophysical understanding of how virus capsids assemble, disassemble, display metastability, and assume various configurations. Chemical virology considers the virus capsid as a chemically addressable structure, providing chemical pathways to modify the capsid exterior, interior, and subunit interfaces. Synthetic virology takes an engineering approach, modifying the virus capsid through rational, combinatorial, and bioinformatics-driven design strategies. Advances in these three subfields of virology aim to develop virus-based materials and tools that can be applied to solve critical problems in biomedicine and biotechnology, including applications in gene therapy and drug delivery, diagnostics, and immunotherapy. Examples discussed include mammalian viruses, such as adeno-associated virus (AAV), plant viruses, such as cowpea mosaic virus (CPMV), and bacterial viruses, such as Qβ bacteriophage. Importantly, research efforts in physical, chemical, and synthetic virology have further unravelled the design principles foundational to the form and function of viruses.
Graphical/Visual Abstract and Caption
Physical, chemical, and synthetic virology work synergistically to engineer viruses for important biomedical and biotechnological applications.
Introduction
The virus is nature’s tool for delivering foreign genetic material into a living organism. Many viruses are icosahedral or helical in structure and are composed of nucleic acids encapsidated in a protein shell. The protein shells are made up of multiple repeating subunits encoded by the viral genome. Some viruses are also enveloped, possessing an additional lipid membrane outside the protein capsid. Depending on the virus, the genome can be single- or double-stranded and composed of DNA or RNA. The protein capsid contains subunits ranging from tens to hundreds in number and can self-assemble spontaneously in some viruses. Altering the make-up of the individual subunits or the interaction between these subunits can lead to reprogramming of virus behavior. Viruses are often referred to as virus nanoparticles (VNPs) if they have been modified chemically or genetically to obtain some property that is different from that of the wild-type form, and virus-like particles (VLPs) if they have had their genetic material removed and are non-infectious (Steinmetz, 2010).
In this review, we describe some basic approaches used in the fields of physical, chemical, and synthetic virology that have allowed us to reprogram viruses into controllable nanodevices. We describe the discoveries in the field of physical virology that have established the basis for our understanding of how virus capsids assemble, disassemble, and assume different configurations. Application of this knowledge within chemical and synthetic virology has allowed us to develop viruses as biocomputing nanoplatforms with controllable targeting and switchable behavior. Specifically, chemical virology uses bioconjugation techniques to expand the functionality of the virus whereas synthetic virology applies rational design-based genetic modifications, directed evolution, and bioinformatics-driven design strategies.
PHYSICAL VIROLOGY
Physical virology can be broadly defined as the study of virus structure and dynamics. The viral capsid plays an important role in carrying and protecting the genome of the virus and, thus, assembly of the capsid is pivotal to its propagation. Crick and Watson proposed that a spherical virus may take on an icosahedral shape to enclose a large volume with small repeating subunits comprised of one or a few repeating protein sequences arranged in a highly symmetrical manner (Crick & Watson, 1956). This was shown to be true by Capsar in 1956 with his observation of the icosahedral bushy stunt virus (D. L. D. Caspar, 1956). Icosahedrons require 60 identical subunits with identical interactions with the neighboring subunits (Figure 1), however, viruses with more than 60 subunits have been observed. Caspar and Klug proposed in 1962 the theory of quasi-equivalence, which explained how capsids with more than 60 subunits can still form an icosahedral shape (D. L. Caspar & Klug, 1962). Due to this quasi-equivalence in subunit-subunit interaction, the same initial protein subunits may also display conformational polymorphism in order to fit into the icosahedral capsid that can be classified using triangulation numbers (T) (Caspar & Klug, 1962; J. E. Johnson & Speir, 1997).
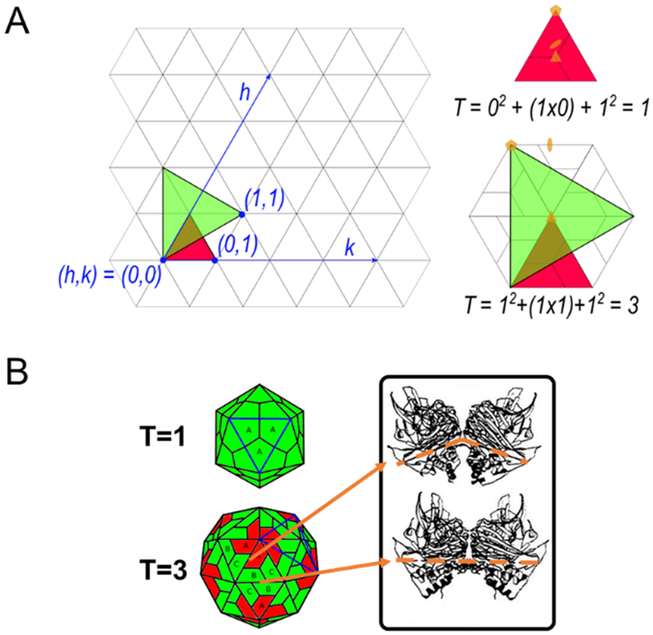
Figure 1:
Quasi-equivalence and triangulation numbers of icosahedrons. (A) The icosahedron can be displayed as a hexagonal lattice. The arrangement of the 5-fold symmetry axes on this lattice gives the icosahedral shape its triangulation number, given as T = h2 + hk + k2, where h and k are vector coordinates () defining the points of the 5-fold axes. (B) As an icosahedral shape gets larger, there are more subunits inside each equilateral triangle (blue outline), as shown by the T=1 and T=3 icosahedrons. All symmetrical contacts within the T=1 are equivalent, while those in higher T numbers are not (J. E. Johnson & Speir, 1997). This quasi-equivalence allows for conformational polymorphism of the subunits in higher T numbered icosahedral capsids. For example, in the T=3 diagram even though A, B, and C subunits are virtually identical, the quasi-equivalence of A-C and B-B contacts indicated in the figure have bent and flat contact angles, respectively. This gives the pentamers (red subunits) a bent surface and the hexamers (green subunits) a flat surface to form the icosahedral shape. Adapted with permission from “Functional implications of quasi-equivalence in a T = 3 icosahedral animal virus established by cryo-electron microscopy and X-ray crystallography,” by R. H. Cheng et al, 1994, Structure (Cheng et al., 1994).
The field of physical virology is foundational to the creation of programmable virus-based materials. Viruses and their capsids are currently being reprogrammed for use in numerous applications, ranging from gene therapy, drug delivery, diagnostics, and immunotherapy. Each of these applications may have different requirements on capsid stability, metastability, and shape (Mateu, 2011). The study of capsid assembly and disassembly pathways, metastability, conformational switching behaviors, and discovery of polymorphic structures provide insights into what may be possible. The quantitative design rules uncovered in physical virology can enable the proper engineering of synthetic viruses with completely new functionalities that are controllable and predictable.
Capsid Assembly and Disassembly
Capsid formation is a process that differs among viruses. For example, tobacco mosaic viruses (TMV) can spontaneously form infectious viral particles in the presence of the capsid proteins and RNA genome in vitro. Other viruses, such as some serotypes of adeno-associated virus (AAV), require assembly inside host cell nuclei with the help of assembly-activating protein (AAP) (Earley et al., 2017). Once the capsid is assembled, the AAV genome is inserted into the shell by non-structural viral proteins in order to form infectious units (Earley et al., 2017; Sonntag et al., 2011). These examples illustrate that there are different pathways of capsid assembly, e.g. capsid assembly around a genomic template, or genome insertion into completed capsids. Unlike AAV serotypes that require the aid of AAP as described above, several other viruses (e.g. simian virus 40 (SV40), minute virus of mice (MVM), cowpea chlorotic mottle virus (CCMV), and hepatitis B virus (HBV)) can spontaneously form empty capsids from their viral proteins (VPs) in vitro. This spontaneous in vitro assembly of viral capsids has allowed the examination of different viral assembly kinetics.
Many techniques have been used to visualize virus capsids in order to study the progression of capsid assembly and to measure the kinetics of spontaneous capsid assembly in vitro, such as size exclusion chromatography (SEC) (J. M. Johnson et al., 2005; Singh & Zlotnick, 2003; Zlotnick, Aldrich, Johnson, Ceres, & Young, 2000), X-ray and light scattering (Casini, Graham, Heine, Garcea, & Wu, 2004; C. Chen, Kao, & Dragnea, 2008; J. M. Johnson et al., 2005; J. S. Johnson et al., 2010; Kler et al., 2012), cryo-electron microscopy (cryo-EM) (Berthet-Colominas, Cuillel, Koch, Vachette, & Jacrot, 1987; Prevelige, Thomas, & King, 1993), and atomic force microscopy (AFM) (Bernaud et al., 2018; Castellanos, Pérez, Carrillo, de Pablo, & Mateu, 2012; Medrano et al., 2016). It has been a challenge to capture the entire capsid assembly process, and many studies were only able to detect two-states of the capsid assembly process: complete capsids or small oligomers referred to as assembly units (AUs) (Katen & Zlotnick, 2009a). The size of these AUs is subject to variation depending on the virus. For example, HBV, human rhinovirus, and MVM appear to have dimers, pentamers, and trimers for AU, respectively (Medrano et al., 2016; Zlotnick & Fane, 2010).
Due to the transient nature of the intermediate species of viral assembly, computational models and simulations have been valuable in elucidating the assembly pathways of different viruses. For example, a combination of in silico and in vitro experiments revealed that the ratio of complete capsid concentration to free AU concentration follows the law of mass action’s equilibrium concentration condition (Ceres & Zlotnick, 2002; Hagan, 2014; Perlmutter & Hagan, 2015). Since capsid intermediates are not readily observed experimentally, the concentration of intermediates can be considered negligible, and the relationship between completely assembled capsid and excess free floating assembly units AUs can be modelled with the two-state approximation, considering only the two states - complete capsids and free AUs (Katen & Zlotnick, 2009a).
| (1) |
Equation 1 shows the equilibrium condition between the two states with [capsid] as the concentration of fully assembled capsid, Kcapsid as the global kinetic constant for capsid association, [AU] as the concentration of free assembly units, and N as the number of AUs in a completed capsid. The fraction of total AU that becomes incorporated into complete capsids, f, can therefore be given by
| (2) |
with [AUT] being the total concentration of assembly units. Based on equation 1, the completed capsid concentration scales with the power of N. The steep concentration dependence on [AU] resembles a critical concentration – a crystallization concept that describes the concentration below which no capsids are formed, and above which a “phase-transition” would occur where all AUs would assemble into capsids. However, as a consequence of the equilibrium condition in equation 1, there does not seem to be a maximum concentration limit for the [AU], and capsid assembly would be possible at even low concentration. Therefore, the capsid assembly process is more accurately described using a pseudo-critical concentration, [AU*] – below which most [AU] are free (f ≪ 1), and above which most AUs would assemble into complete capsids (f ~ 1 – [AU*]/[AUT]) (Perlmutter & Hagan, 2015; Zlotnick & Fane, 2010). The pseudo-critical concentration also enables the calculation of Gibbs free energy of assembly, which can be used as a measurement of how stable the viral capsid is thermodynamically with the law of mass action:
| (3) |
We can then use van’t Hoff analysis to extract enthalpic and entropic contributions from the Gibbs free energy to find the driving force of capsid formation (Ceres & Zlotnick, 2002). Through this analysis, it was discovered that HBV capsid formation is driven by entropy and that the weak interactions between the viral subunits are sufficient to induce stable capsid formation (Ceres & Zlotnick, 2002; Katen & Zlotnick, 2009b).
To fully elucidate the steps that lead to complete capsid formation, Zlotnick and colleagues have formulated a system of rate equations that follow the nucleation and elongation steps of polymer or crystal formation (Zlotnick, 1994, 2005; Zlotnick, Johnson, Wingfield, Stahl, & Endres, 1999) which allows for the prediction of kinetic traps of capsid formation, as well as the hysteresis of assembly and disassembly (Singh & Zlotnick, 2003; Zlotnick, 2005; Zlotnick, Johnson, et al., 1999). This model also helps explain when and why capsid assembly can fail. If the association energy is high enough to prevent dissociation of AUs, the transient intermediates become too stable and the thermodynamics of the system becomes a barrier resulting in many half-formed capsids without sufficient numbers of free AUs to complete the formation of capsids, leading to a lower proportion of completed capsids. Alternatively, if the nucleation reaction happens too quickly, then there are too many starting sites and insufficient free AUs to complete all the partially formed capsids (Katen & Zlotnick, 2009b; A. Zlotnick & Fane, 2010). Due to this insight, in silico modification to the forward reaction rate coefficient or association energies can rescue the capsid formation from the kinetic traps, helping to identify, predict, and perhaps optimize, the reaction conditions (e.g. protein and salt concentration, pH) required for successful in vitro capsid formation. Additionally, the nucleation step is fully compatible with Caspar’s autostery mechanism, where the free subunits of a capsid undergo a conformational change before becoming “activated” for assembly (D. L. Caspar, 1980). For a more rigorous review of kinetics and thermodynamics models of virus assembly please refer to (Hagan, 2014; Katen & Zlotnick, 2009a; Perlmutter & Hagan, 2015; A. Zlotnick & Fane, 2010).
The thermodynamic-kinetic analysis uncovered great insights into the pathway of capsid assembly. However, this treatment of capsid assembly is often difficult to recapitulate experimentally as many viruses are unable to form capsids spontaneously in vitro. Therefore, it has also been important to study capsid disassembly and measure capsid energies in these viruses. However, experiments have shown that completed capsids do not disassemble to the same fraction of AU that forms capsid, fc, that the assembly process would have reached given the same concentration of total AUs. In fact, capsid disassembly needs to be induced using mechanical indentation (Castellanos et al., 2012), thermal denaturation (Horowitz et al., 2013), or chaotrophic salt solutions (Singh & Zlotnick, 2003). The process of disassembly appears to require more energy than the assembly process, and the measured free energy for disassembly is often higher than the free energy of assembly. This hysteresis between assembly and disassembly can be explained by the kinetic model of capsid assembly. While the global thermodynamics of the capsid drives disassembly, the local kinetics of the disassociated AUs favors association over disassociation, indicating that the capsid is thermodynamically unstable but kinetically stable, as was shown in HBV (Morozov, Bruinsma, & Rudnick, 2009; Singh & Zlotnick, 2003).
Recently, AFM indentation and thermal denaturation experiments have shown the accuracy of theoretical models in predicting the pathway of capsid disassembly for MVM, a small T=1 icosahedral virus (Castellanos et al., 2012; Medrano et al., 2016). For a more in-depth review of mechanical indentation and perturbation of viral capsids using AFM, refer to the review by Roos et al. (Roos, Bruinsma, & Wuite, 2010). The disassembly of virus capsids appears to be dependent on serotypes, as various studies demonstrate that the capsid disassembly profile can be used to identify specific serotypes of the virus (Bennett et al., 2017; Pacouret et al., 2017).
In summary, by using simple systems of rate equations, researchers have been able to develop a high-level view of the capsid formation and disassembly pathways which helps explain and predict capsid assembly and disassembly rates and extents. It also allows for the calculation of ΔG for capsid formation as well as subsequent thermodynamic analysis. By understanding the biophysical rules for natural capsid assembly and disassembly, currently existing viruses may be manipulated to exhibit altered assembly/disassembly behaviors, or synthetic viruses may be created from non-viral components that mimic the properties of natural viruses.
Capsid Metastability and Conformational Switching
The hypothesis that viral capsids are kinetically stable but thermodynamically unstable makes intuitive sense in the context of the virus life cycle. A virus particle must be able to stay intact in environments with very few of its constituent AUs, like inside the host organism. It must be able to protect its genome from extracellular and intracellular defences of the host but must then disassemble to deliver its genome inside the appropriate host nucleus. Capsid metastability refers to the concept that the capsid is stable but is not in its most favorable energetic state, therefore allowing for disassembly or conformational changes to happen readily under the correct conditions.
Metastability of the capsid is an important trait for many viruses. For example, metastability of the AAV capsid is critical for its infectivity. The AAV capsid is made up of three variants of viral proteins (VPs), with the common overlapping region VP3. VP2 has, in addition to the VP3 domain, a nuclear localization sequence at the N-terminus. VP1, the longest of the VPs, overlaps VP2 and has an additional phospholipase A2 domain on the N-terminus. The three variants exist in an approximate ratio of 1:1:10 (VP1, VP2, VP3)(J. S. Johnson et al., 2010). The N-termini of VP1 and VP2 subunits are not exposed on the capsid surface until a capsid conformational change occurs inside the host cell endosome. The newly externalized VP1 and VP2 domains allow for endosomal escape and nuclear localization of the virus (Bleker, Sonntag, & Kleinschmidt, 2005; Kronenberg, Böttcher, von der Lieth, Bleker, & Kleinschmidt, 2005; Sonntag, Bleker, Leuchs, Fischer, & Kleinschmidt, 2006; Thomas, Storm, Huang, & Kay, 2004). AAV’s metastability has recently been harnessed to develop a platform of viruses able to externalize exogenous peptides upon activation (Thadani, Dempsey, Zhao, Vasquez, & Suh, 2017). Interestingly, mosaicism of the capsid – differing subunits assembling to form the capsid – appears to be a requirement for obtaining robust activatable peptide display with AAV.
The metastable properties of viral capsids have been exploited to package different cargo within the capsid lumen (Loo, H. Guenther, A. Lommel, & Franzen, 2008; Ren, Wong, & Lim, 2006; Yildiz, Lee, Chen, Shukla, & Steinmetz, 2013). Interestingly, some viral capsids undergo structural swelling as the conformation of their protein subunits change under different pH or metal ion concentrations (Cao et al., 2014; Liu, Qu, Johnson, & Case, 2003; Oda et al., 2000). For example, CCMV can expand as much as 10% radially when the pH is raised from pH 5 to 7 in the absence of divalent cations (Figure 2a) (Tama & Brooks, 2002). This swelling results in the formation of larger pores in the capsid, allowing therapeutics or imaging agents to gain entry into the capsid lumen and subsequently become entrapped upon reversal of the pH (Loo et al., 2008; Tama & Brooks, 2002; Yildiz et al., 2013). Future studies investigating, harnessing, or reprogramming the viral capsid’s metastability may yield VNPs with new functional properties for biotechnological or biomedical applications.
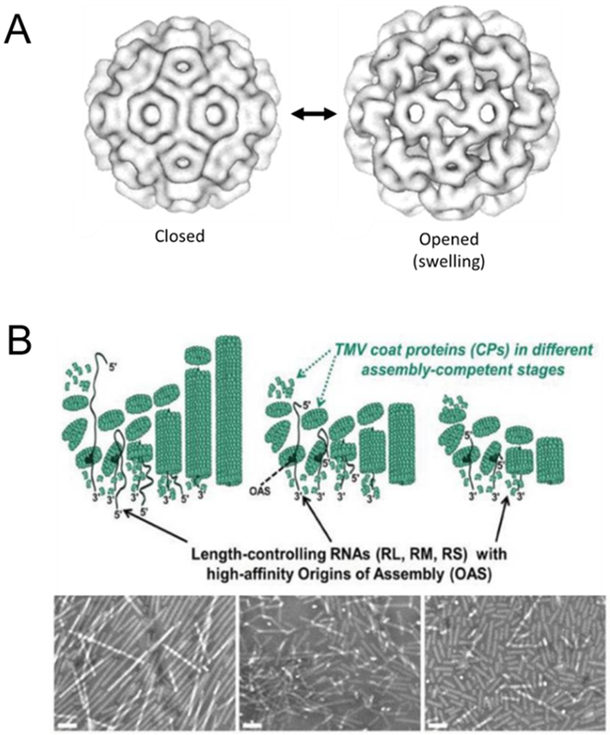
Figure 2:
Capsid metastability and polymorphism. (A) CCMV swells up when the solution pH is increased from pH 5 to pH 7 at low ionic strength without divalent cations, allowing larger pores to form on the capsid. These pores can be used to pack therapeutics or imaging agents inside the viral capsid for delivery. Figure adapted with permission from “Pseudo-atomic Models of Swollen CCMV from Cryo-electron Microscopy Data,” by H. Liu et al, 2003, Journal of Structural Biology. Copyright 2003 by Elsevier. (B) TMV particles of different lengths can be generated by assembly in vitro with RNA of different lengths with high-affinity Origins of Assembly. The TMV particles of different aspect ratios can be visualized with electron microscopy. Scale bar is 100nm. Figure adapted with permission from “The Impact of Aspect Ratio on the Biodistribution and Tumor Homing of Rigid Soft-Matter Nanorods,” by S. Shukla et al, 2015, Advanced Healthcare Materials. Copyright 2015 by John Wiley and Sons.
Capsid Polymorphisms and Geometries
Virus capsids exist as metastable structures that display polymorphic behavior. The interplay between thermodynamic and kinetic stability create an energy landscape with multiple local minima. The same capsid subunits can give rise to multiple forms, each falling into a locally stable conformation and geometry. This polymorphism has been directly observed in viruses that form spontaneously in vitro, such as HBV (Schlicksup et al., 2018; Zlotnick, Palmer, et al., 1999) and nodavirus (Bajaj & Banerjee, 2016). Using thermodynamics and kinetics (Moerman, van der Schoot, & Kegel, 2016; Nguyen, Reddy, & Brooks III, 2009), as well as molecular dynamics modelling (Hagan & Zandi, 2016; Nguyen et al., 2009) researchers are trying to further elucidate the spontaneous assembly processes of viral capsids that lead to polymorphic structures.
HBV is a well-characterized virus that displays this polymorphic property. Complete HBV capsids with different numbers of VPs can be formed with different icosahedral triangulations numbers, T=3 and T=4 (Moerman et al., 2016; Schlicksup et al., 2018; Zlotnick, Palmer, et al., 1999). While the two capsids have different amounts of VPs, their stabilities are identical through AFM indentation studies (Uetrecht et al., 2008). Recent work using coarse-grain thermodynamics to model the formation of T3/T4 HBV capsids successfully recapitulated the distribution of the HBV capsid dimorphism (Kim & Wu, 2014; Moerman et al., 2016). SV40 also exhibits capsid polymorphisms and can be manipulated to form capsids of different configuration (T=1, T=7) depending on the nucleic acid scaffold provided (Bruckman, VanMeter, & Steinmetz, 2015; Kler, Wang, Dhason, Oppenheim, & Zlotnick, 2013).
TMV is a rod-like virus that can be tuned geometrically for different applications. TMV can form infectious virion spontaneously in the presence of its RNA genome, a high-affinity origin of assembly, and free VPs (Shukla, Eber, et al., 2015; Steinmetz, 2010). By tuning the length of the RNA genome, TMV particles with different numbers of VPs can be generated, resulting in TMV rods of various lengths (Figure 2b). Interestingly, TMV particles with different aspect ratios result in different in vivo biodistribution, tumor homing, and longevity (Bruckman, Czapar, VanMeter, Randolph, & Steinmetz, 2016; Shukla, Eber, et al., 2015). TMV can also be manipulated to assume various shapes, from spherical particles to nanoboomerangs and tetrapods with each conformation exhibiting interesting properties (Bruckman et al., 2015; Eber, Eiben, Jeske, & Wege, 2015). For example, spherical TMV nanoparticles were used to create a potent MRI imaging agent delivery system (Bruckman et al., 2013, 2015). Similarly, potato virus X (PVX) can also be made into spherical nanoparticles through heat treatment (Nikitin et al., 2016). These various shapes of TMV and PVX demonstrate the ability of the viruses to assume specific configurations which can be harnessed as biological scaffolds for different applications (Koch et al., 2015).
Studying the polymorphism of viral capsids has increased our understanding of how virus capsids assemble. Further work in this area may allow us to develop an even greater repertoire of virus subunit-based nanodevices with different morphologies and configurations for packing different cargos. Through the understanding of capsid metastability, we have seen that viral capsids can respond differently in different physical (e.g. temperature) and chemical (e.g. pH) environments. To further control viral capsid’s interactions and responses to the local environment, we will now discuss chemical and genetic design strategies used in the field.
CHEMICAL VIROLOGY
Virus capsids are chemically addressable nanoscale structures. Chemical virology focuses on modifying the virus capsid using chemistry-based techniques. In viruses with a proteinaceous capsid, chemical modification of said capsid can generally be categorized into three locations: 1) exterior surface, 2) interior surface, and 3) subunit interface. In this section, various chemical methods to modify viruses at these three locations will be reviewed.
Exterior Surface
The exterior capsid surface is readily exposed to the environment and can easily undergo bioconjugation to alter virus behavior. Bioconjugation is a chemical modification that results in formation of a covalent bond between two molecules, at least one of which is a biomolecule. When activated, chemical functional groups on the proteinaceous capsid of VNPs provide sites for conjugation. These functional groups can form covalent bonds directly with a molecule of choice (direct attachment) or interact with the desired molecules through adaptors (indirect attachment). Described below are some bioconjugation techniques and their applications in creating VNPs.
Direct Conjugation to Lysines and Cysteines on Capsid
In the simplest bioconjugation scheme, non-viral components, such as proteins, peptides, and small molecules, can be chemically conjugated to the capsid surface via NHS-ester chemistry to exposed lysine (Figure 3a) or maleimide chemistry to cysteine residues (Figure 3b). The reader is referred to another review for a more thorough overview of bioconjugation techniques (Mavila, Eivgi, Berkovich, & Lemcoff, 2016). These conjugation schemes have been used for a variety of targeting purposes (Ren, Wong, & Lim, 2007; Huang, Steinmetz, Fu, Manchester, & Johnson, 2011; Lockney et al., 2011; Barwal, Kumar, Kateriya, Dinda, & Yadav, 2016). Carbodiimides such as 1-ethyl-3-[3-dimethyl-aminopropyl]carbodiimide hydrochloride (EDC), for example, has been used to conjugate folic acid to the surface of the cucumber mosaic virus to deliver the chemotherapy doxorubicin to folate receptor-overexpressing cancer cells (Zeng et al., 2013).
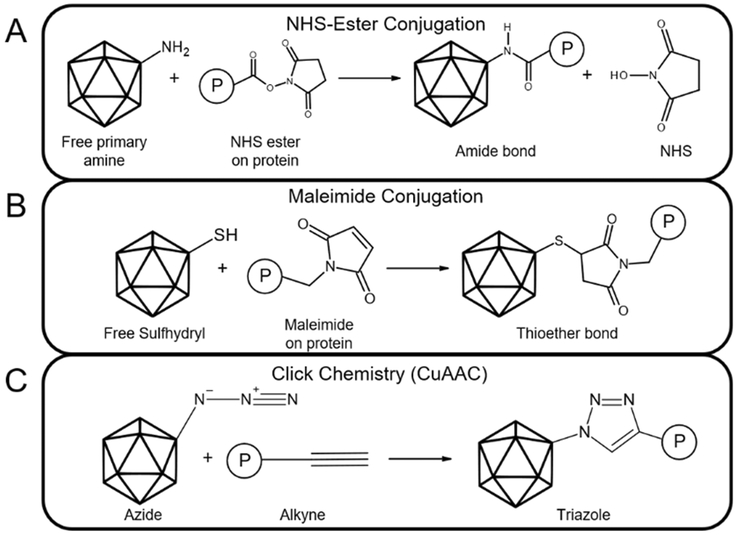
Figure 3:
Examples of capsid bioconjugation approaches. (A) Simplified scheme of NHS-ester conjugation, in which a molecule P can be attached to a free primary amine on the viral capsid through formation of an amide bond. (B) General scheme of maleimide conjugation, in which a molecule containing a maleimide group can react with a free sulfhydryl group on the protein capsid to yield a stable thioether bond. (C) Schematic of copper(I)-catalyzed alkyne-azide cycloaddition (CuAAC) click chemistry. An azide and alkyne react in the presence of a copper-based catalyst to form a triazole.
Click Chemistry
“Click” chemistries are bioconjugation reactions that are simple to perform and result in products that do not require chromatography to purify (Kolb, Finn, & Sharpless, 2001). The most commonly used click chemistry reaction is copper(I)-catalyzed azide/alkyne cycloaddition (CuAAC) (Figure 3c) (Hein & Fokin, 2010), which has been used to conjugate a variety of molecules to the virus capsid (Destito, Yeh, Rae, Finn, & Manchester, 2007; Steinmetz et al., 2011; Hovlid et al., 2012; C.-C. Chen et al., 2016; Hu et al., 2017). For example, a two-step labeling procedure was used to produce adenovirus particles bearing an N-azidoacetylglucosamine (O-GlcNAz) at Ser109 position of the fiber protein, which provides a handle for “click” chemistry attachment of folate, resulting in a ~20 fold increase in transduction of breast cancer cells (Partha Sarathi Banerjee, Ostapachuk, Hearing, & Carrico, 2010).
Click chemistry has also been used to conjugate fibrin-targeting peptides to both the icosahedral CPMV and the rod-shaped TMV carrying contrast agents to study their ability to target thrombi (Wen et al., 2015). Interestingly, the authors found that while addition of the targeting ligand to the viruses was able to increase thrombi targeting in vitro, the shape of the nanoparticle has a significantly greater effect than surface chemistry in vivo, with the elongated TMV showing the most enhanced thrombus detection regardless of the presence of a targeting ligand. Discrepancies in in vitro versus in vivo targeting is also seen with RGD-conjugated versus PEGylated PVX, where superior performance of RGD-PVX in the in vitro setting did not carry over in vivo (Shukla, DiFranco, Wen, Commandeur, & Steinmetz, 2015).
Viruses have also been altered for photodynamic therapy (PDT) using click chemistry. Photodynamic therapy (PDT) is used in conjunction with a photosensitizer, which normally leads to generation of reactive oxygen species, to kill target cells in the presence of light. The photosensitizer C60 has been conjugated to surface-exposed lysines on Qβ bacteriophage VNPs using NHS and CuAAC based techniques (Wen et al., 2012). These VNPs accumulated in human prostate cancer cells and displayed increased killing in vitro post-phototherapy compared to dark conditions. Recently, photocleavable nitroveratryl compound has been used to link doxorubicin to the external surface of bacteriophage Qβ for PDT (Z. Chen, Li, Chen, Lee, & Gassensmith, 2016). CuAAC chemistry was used to attach the photolinker and doxorubicin to surface exposed amines on Qβ. Native disulfide bonds were reduced to free thiols and re-bridged using thiol-dibromomaleimide chemistry, which allows for reformation of the disulfide bonds to maintain capsid stability while providing additional functional groups for the attachment of PEG molecules to increase solubility. This dual functionalization was required, as conjugation of the drug alone led to aggregation and precipitation. The PEGylated dox-conjugated VNP was able to kill breast cancer cells in vitro after 15 minutes of exposure to light but had minimal cytotoxicity with no light exposure.
Biotin-Based Attachment of Moieties
Biotin is a eukaryotic co-factor that has a high-affinity to the avian/amphibian protein avidin and other biotin-binding proteins such as streptavidin. The use of a biotin-avidin system provides modularity to capsid modification, as attachment of one binding partner to the virus capsid allows conjugation to nearly any molecule labeled with the other binding partner. Biotinylation is relatively simple to accomplish, as biotin is small and rarely interferes with the function of labeled molecules. Furthermore, photoactivatable biotin can attach non-specifically to nearby molecules upon exposure to ultra-violet light. Photoactivatable biotin was covalently conjugated to adenoviral vectors, which was then used to attach biotinylated targeting ligands through an avidin bridge to achieve increased transduction of target cells in vitro (J. S. Smith et al., 1999). This concept was later applied to AAV (Ponnazhagan, Mahendra, Kumar, Thompson, & Castillas, Jr., 2002). Biotin was conjugated to surface-exposed lysines on the AAV serotype 2 capsid, and fusion proteins containing streptavidin and targeting proteins human epidermal growth factor (EGF) or human fibroblast growth factor 1α (FGF1α) were prepared separately and then mixed with the vector. This modular design approach allows the targeting ligand to be swapped without requiring modification of the biotinylated vector, increasing the flexibility and ease of translating this platform for use in other applications. Similarly, streptavidin was used to couple biotinylated CCMV to biotinylated anti-SpA monoclonal antibody recognizing SpA on the surface of Staphylococcus aureus bacterial cells to create VNPs capable of penetrating biofilms to locate S. aureus (Suci, Berglund, et al., 2007). Subsequently, the authors conjugated a ruthenium-based photosensitizer to the S. aureus targeting CCMV VNPs for PDT (Suci, Varpness, Gillitzer, Douglas, & Young, 2007).
Unfortunately, chemical biotinylation of viral vectors can lead to nonspecific labeling where the conjugation does not occur evenly on a capsid that has multiple functional groups (Lesch, Kaikkonen, Pikkarainen, & Ylä-Herttuala, 2010). To gain finer control over the exact number and location of chemical reactions, genetic modification may be used to incorporate adaptors that are not found in the wild-type capsid. Thus, metabolic methods for biotinylation were developed for viral vectors using a genetically encoded biotin acceptor peptide (BAP) that is enzymatically biotinylated by holocarboxylase synthetase endogenous to human cells (Arnold, Sasser, Stachler, & Bartlett, 2006; Campos, Parrott, & Barry, 2004; Kaikkonen, Viholainen, Närvänen, Ylä-Herttuala, & Airenne, 2008; Pereboeva, Komarova, Roth, Ponnazhagan, & Curiel, 2007; Räty et al., 2004). Additionally, BAP and biotin ligase can be used in conjunction with a ketone probe to allow for conjugation of hydrazide- or hydroxylamine-functionalized targeting molecules onto AAV serotype 1 capsids, bypassing the need for an avidin/streptavidin intermediate (Kaikkonen et al., 2008; Räty et al., 2004; Stachler, Chen, Ting, & Bartlett, 2008).
Unnatural Amino Acids
Unnatural amino acid (UAA) incorporation has also been used to increase chemical diversity and introduce functionality at specific sites on the viral capsid. The amber codon suppression method is one technique for UAA incorporation, in which orthogonal transfer RNAs (tRNAs) and aminoacyl-tRNA synthetases are engineered to recognize the amber stop codon (“UAG”) and insert UAAs at that site (Mehl et al., 2003; Xie & Schultz, 2006). Successful incorporation of the UAA results in full length capsid proteins, while failure to incorporate the UAA results in a truncated and non-functional protein that is unlikely to be incorporated into the assembled virus capsid. This technique was used to introduce the unnatural amino acid p-aminophenylalanine (paF) into the capsid of bacteriophage MS2, to which aptamers targeting Jurkat leukemia T cells were subsequently attached (Tong, Hsiao, Carrico, & Francis, 2009). Porphyrin 1, a photosensitizer for PDT, was then attached to cysteine groups that had been genetically introduced to the inside of the bacteriophage using maleimide chemistry. The resulting VNP was able to selectively kill Jurkat cells in vitro within 20 minutes of exposure to light (Stephanopoulos, Tong, Hsiao, & Francis, 2010). UAA incorporation was also used to retarget AAV towards ovarian cancer cells through attachment of the RGD motif (Kelemen et al., 2016), and to attach fluorophores to AAV serotype 2 for tracking intracellular trafficking (C. Zhang, Zhou, Yao, Tian, & Zhou, 2018). UAA has been used in conjunction with β-linked N-acetylglucosamine (O-GlcNAc) to attach both the folate targeting moiety and therapeutic taxoid to adenovirus for combination therapy (Banerjee, Ostapchuk, Hearing, & Carrico, 2011; Banerjee, Zuniga, Ojima, & Carrico, 2011).
Polymers
Polymers have been conjugated to the viral capsid to improve vector properties. Polyethylene glycol (PEG) is a compound with a long history in biomaterials and polymer chemistry that was first appropriated for use in chemical virology to coat adenovirus in order to decrease viral immunogenicity (Chillón, Lee, Fasbender, & Welsh, 1998; O’Riordan et al., 1999). The presence of PEG on the virus surface drastically changes the capsid landscape, thus altering or abolishing native tropism and antibody recognition (Wonganan & Croyle, 2010). The use of PEG as crosslinkers also facilitates the attachment of other molecules by acting as a spacer between the viral capsid and the attached molecule, thereby improving conjugation efficacy and vector stability. For example, a polyethylene glycol (PEG)-based crosslinker containing an NHS ester on one end and a maleimide on the other was used to attach a cysteine-terminated epidermal growth factor receptor (EGFR) targeting peptide to surface-exposed lysines on potato virus X (PVX) (Chariou et al., 2015). This strategy resulted in increased cell uptake in EGFR-positive cancer cell lines with minimal uptake in EGFR-negative cells.
Another polymer used in conjunction with viruses is the multivalent poly-[N-(2-hydroxypropyl)methacrylamide] (pHPMA). One advantage of pHPMA over PEG is that pHPMA can be functionalized through its side-chain hydroxyl group to incorporate molecules such as targeting ligands, imaging agents, and drugs (Tucker & Sumerlin, 2014). In virus engineering, pHPMA was first used to covalently conjugate basic fibroblast growth factor (bFGF) and vascular endothelial factor (VEGF165) to the surface of adenoviral vectors, retargeting the vectors to cells expressing FGF or VEGF receptors, respectively (Fisher et al., 2001; Green et al., 2008). pHPMA has also been conjugated to AAV vectors (Carlisle et al., 2008). Interestingly, pHPMA/amine-reactive thiazolidine-2-thione copolymer is unable to react with AAV serotype 5 (AAV5) capsids due to insufficient availability of reactive amines. To fix this issue, AAV5 was modified with EDC and hexanediamine (HD) to convert free carboxyl groups on the capsid surface to amine groups to provide attachment points for the pHPMA copolymer. This modification allowed for successful coating with pHPMA, resulting in ablation of native tropism and allowing for retargeting. AAV serotype 8, on the other hand, was successfully detargeted via pHPMA copolymer coating and retargeted with various ligands without requiring EDC/HD pre-modification, likely due to a sufficient number of chemically addressable amine groups.
Atom-Transfer Radical Polymerization
Atom-transfer radical polymerization (ATRP) is a controlled polymerization technique which allows for monomers to be polymerized to small initiators that are first conjugated to the virus capsid. ATRP has been used to create a pH-responsive nanodevice based on the Qβ bacteriophage (Pokorski, Breitenkamp, Liepold, Qazi, & Finn, 2011). Azide groups were first added to capsid external amines followed by CuAAC to attach ATRP initiators, to which oligo(ethylene glycol)-methacrylate (OEGMA) and an azido-functionalized analogue (OEGMA-N3) were added. The chemotherapy drug doxorubicin was then conjugated to the azide-functionalized polymers using an alkyne-functionalized hydrazine linker. Under acidic environments (pH 5.5), the hydrazine linkers are cleaved, and the drug is released. This formulation led to a VNP that was able to achieve a similar level of toxicity in HeLa cells as free doxorubicin. But unlike free doxorubicin, this VNP was pH-sensitive and released the drug only under acidic environments, leading to targeted delivery and decreased side effects.
Interior Surface
Modifications to the interior surface of capsids is often performed to provide new functionalities within the capsid lumen (Hooker, Kovacs, & Francis, 2004), introduce non-native material such as imaging agents (Lewis et al., 2006) and therapeutic drug molecules (Flenniken et al., 2005), or turn viruses into constrained and controllable reactors for material synthesis (T. Douglas et al., 2002; Trevor Douglas & Young, 1998) (Figure 4).
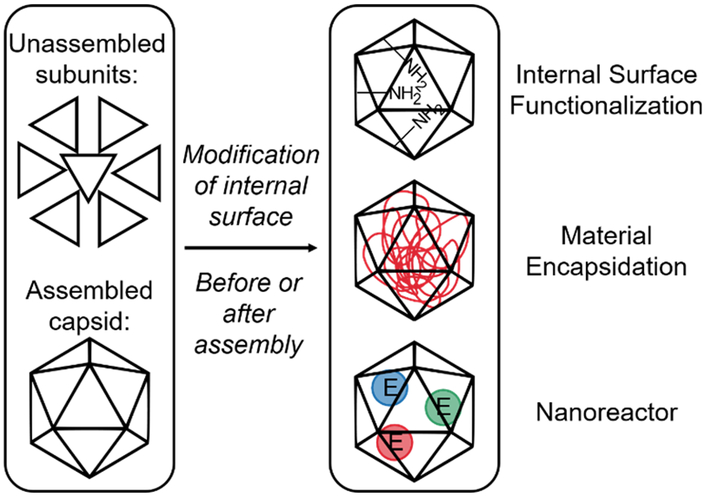
Figure 4:
Overview of internal capsid modifications. The capsid protein subunits can be modified before or after assembly for various purposes, such as internal surface functionalization, material encapsidation, and the creation of nanoreactors.
Functionalization of Capsid Interior
Modification of the capsid interior can lead to improved or new functions. An early example involved modification of the interior capsid surface of bacteriophage MS2 (Hooker et al., 2004). To functionalize the capsid interior, the native RNA genome was first removed via alkaline hydrolysis and the empty capsids were isolated via precipitation. An internally exposed tyrosine residue on the MS2 capsid was then modified using a multi-step strategy to attach an o-imino-quinone handle to serve as a highly reactive functional group. In another example, the interior of TMV, which does not have amino acid side chains commonly used in bioconjugation approaches, was functionalized by attaching amines to glutamic acid residues through a carbodiimide coupling reaction (Schlick, Ding, Kovacs, & Francis, 2005). This allowed for dual-surface modification of the virus and expands the functional capabilities of the vector. The capsid lumen of CCMV was modified using a slightly different strategy. The N-terminus of each subunit is positively charged and face towards the inside of the capsid, creating a cationic interior environment. Exploiting this localization of the N-terminus, fluorescent dye cargo was conjugated to the interior of the capsid via a bio-orthogonal vinylboronic acid handle and dipyridyl-s-tetrazine moiety (Schoonen et al., 2018). These proof-of-concept studies show that new functionalities can be introduced into the capsid lumen.
Improved Encapsidation
Viral capsids provide protection for their cargo, making them ideal delivery vectors for degradation-sensitive materials. Bacteriophage P22, for example, has an icosahedral capsid consisting of 420 coat protein monomers that are assembled with the aid of several hundred scaffolding proteins. Fusion of cargo proteins to the scaffolding proteins result in their encapsidation in the virus (Patterson, Prevelige, & Douglas, 2012; Patterson, Schwarz, Waters, Gedeon, & Douglas, 2014). ATRP with polyacrylate and polyacrylamide has also been performed in P22 (Lucon, Edwards, Qazi, Uchida, & Douglas, 2013; Lucon et al., 2012). When the initiators are conjugated to the inside of the virus, ATRP allows for the filling of capsids with synthetic polymer and without a need for working with large bulky polymers. The nature of the polymerization reaction is dependent on the initiation site; initiation at the single point mutation S39C results in polymerization that is confined to the interior of the capsid, and initiation from the K118C site results in polymerization with partial exposure on the external capsid surface and requires crosslinking to be fully constrained within the capsid. The resulting polymer network provides an abundance of functional groups that can be used as a scaffold for further conjugation reactions. Attachment of molecules such as fluorophores and contrast agents show an order of magnitude increase in incorporation compared to P22 with no internal polymerization. ATRP has also been used to fill the bacteriophage Qβ with a cationic polymer complexed with small interfering RNA, resulting in enhanced virus internalization by mammalian cells that appear to follow a different mechanism of uptake and intracellular distribution compared to wild-type capsid (Hovlid et al., 2014).
Orthogonal modification of both external and internal surfaces of the capsid can be performed to provide further functionalization, with exterior modifications altering virus behavior while interior ones alter virus cargo. Recently, P22 has been used to encapsidate cytochrome P450 enzymes (CYP) (Chauhan et al., 2018). Tamoxifen, an endocrine therapy for breast cancer, requires metabolic activation by CYP which is low in concentration at tumor sites. CYP-containing P22 that is further externally modified with a photosensitizer and an estradiol based targeting ligand is able to target ER+ breast cancer cells to allow for dual therapy through enhanced tamoxifen/CYP effects and PDT.
Molecules can also be encapsidated alongside native viral genomes. In cucumber mosaic virus, the positively-charged chemotherapeutic doxorubicin was incubated with virus containing RNA genomes, allowing doxorubicin to infuse through open pores in the capsid to associate noncovalently with the viral genome (Zeng et al., 2013). The addition of RNAse, which also appears to be able to diffuse through open pores in the capsid, caused degradation of viral RNA and greatly increased the rate of doxorubicin release from the virus. Folic acid was conjugated to surface-exposed lysines and the targeted VNP encapsidating doxorubicin showed less cardiotoxicity and enhanced antitumor effects in a mouse ovarian cancer model.
Viruses as Nanoreactors
Viruses’ ability to encapsidate molecules and provide a contained, spatially limited, and controllable interior environment allows them to serve as protein cages for material synthesis. CCMV is one early example of a viral bioreactor. In addition to having its N-terminus reside within the capsid interior, the capsid undergoes a reversible pH-dependent swelling that allows for the exchange of large molecules between the virus cavity and the bulk medium. Mineralization of polyoxometalate species in CCMV was facilitated by pH-dependent gating of capsid pores (Trevor Douglas & Young, 1998). Additionally, altering the interior surface of CCMV from cationic to anionic to promote oxidative hydrolysis can lead to size-constrained iron oxide formation within the capsid (T. Douglas et al., 2002). Since these early reports, other viruses have also been used as containers for biomineralization of inorganic metal-based nanoparticles, as reviewed elsewhere (Bain & Staniland, 2015; W. Zhang et al., 2017).
Viruses can also be converted into nanoreactors through the encapsidation of enzymes and small molecules, allowing for increased control over reaction conditions and providing a system mimicking subcellular organelles. The reversible pH-dependent swelling of the CCMV capsid can be used to encapsidate a single type of molecule, but may result in uncontrollable encapsidation behavior if multiple molecules must be incorporated (Comellas-Aragonès et al., 2007). Instead, multiple molecules can be encapsidated in a controllable manner by heterodimerization of the enzymes with capsid subunits using a coiled-coil linker and mixing this heterodimer with wild-type capsid proteins for capsid assembly (Minten, Hendriks, Nolte, & Cornelissen, 2009). Using this method to encapsidate enzymes leads to increased activity compared to free enzyme (Minten et al., 2011). Although using this heterodimerization strategy does not require the enzyme to be covalently attached to the capsid subunits, it does require the attachment of the coiled-coil protein linker, about 21 amino acids in length, to both the enzyme and the capsid subunits.
Another strategy for CCMV encapsidation of molecules involves sortase A, a gram-positive bacteria-derived enzyme, which was used to catalyze the covalent linking of an enzyme (tagged at the C-terminus with the LPXTG-motif) and CCMV subunits (tagged at the N-terminus with glycine) (Schoonen, Pille, Borrmann, Nolte, & van Hest, 2015). This resulted in attachment of small molecules in up to 58% of the subunits and of 16–18 proteins per capsid. Sortase A has also been used to attach the larger T4 lysozyme to the luminal CCMV N-terminus, with approximately 4 enzymes per capsid (Schoonen, Maassen, Nolte, & van Hest, 2017). The activity of T4 lysozyme is highly dependent on the salt concentration and pH of the environment, and the addition of nickel ions using a histidine tag originally introduced for purification purposes was required for capsid stability. This proof-of-concept study shows that large enzymes can be encapsidated in CCMV while retaining partial activity. Further studies on the assembly/disassembly cycles of such a system can yield a switchable nanoreactor that is able to shield and deliver active enzymes to targeted locations.
More recently, the CCMV capsid has been used to co-encapsidate different enzymes (Brasch et al., 2017). Two enzymes from two separate cascade systems were covalently conjugated to DNA, which served as the negatively charged template to trigger capsid formation. The DNA-enzyme molecules were co-encapsidated in a controlled manner within the same CCMV capsid. These enzymes are not covalently attached to the virus capsid. Such a system creates nanoreactors containing multiple types of molecules that can be used as a model for studying enzymatic reactions in a biological compartment. While the various methods of enzyme encapsidation all seem to report increased enzymatic activity compared to free enzyme, it is unclear how the increased activities compare between the encapsidation strategies.
Capsid Subunit Interface
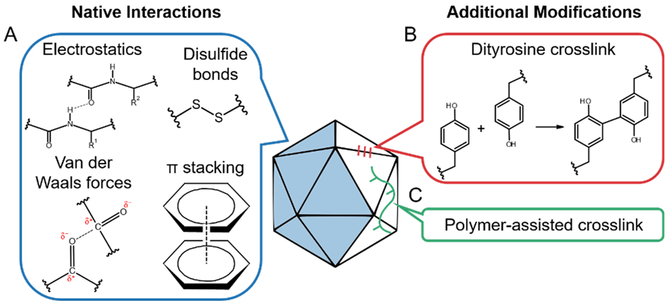
Non-covalent interactions at the subunit interface play an important role in capsid assembly and structure (Figure 5a) (Trevor Douglas & Young, 2006). Various studies have examined the impact of crosslinking key residues at the subunit interface on capsid stability. The CPMV capsid is composed of 60 copies each of two proteins that dimerize and form an icosahedral capsid. Tyrosine residues on adjacent capsid proteins across the pentameric subunit boundaries were covalently crosslinked (Figure 5b) (Meunier, Strable, & Finn, 2004). Surprisingly, no difference in stability was found between the native and crosslinked capsids. The authors postulated that increased number of crosslinking reactions and crosslinking at inherently weaker interfaces may be necessary to improve capsid stability.
Figure 5:
Examples of modifications at the capsid subunit interface. (A) Interactions that are found to play an important role in subunit-subunit interaction in the wild-type capsid include electrostatic interactions, van der Waals forces, π-stacking, and disulfide bonds. (B) Tyrosine residues can be crosslinked chemically to join nearby subunits. (C) Polymers spanning across multiple capsid subunits can also be used to crosslink the capsid.
AAV is an icosahedral virus made up of 60 subunits with a cluster of surface-exposed tyrosine residues at the two-fold symmetry axes. Interestingly, when two capsid subunits are “stitched” together at this axis using oxidative tyrosine crosslinking (Figure 5b), the resulting crosslinked AAV vector displays decreased transduction efficiency (Horowitz, Finn, & Asokan, 2012). Further investigation revealed that crosslinking at the two-fold interface hinders surface display of the VP1 subunit N-terminus, which is necessary for endosomal escape and nuclear translocation of vectors during the transduction process. These results suggest that a certain degree of flexibility is necessary for the AAV capsid to undergo the conformational changes involved in carrying out its function.
Another interaction between capsid subunits that has been explored is the role of disulfide bonds in capsid stability and function (Figure 5a). Disulfide bonds are essential for stabilizing pentamers in a simian virus 40-derived VNP (Li et al., 2014). Similarly, intersubunit disulfide bonds contribute significantly to capsid stability of bacteriophage Qβ (Fiedler et al., 2012). However, in AAV, the five cysteine residues conserved across most AAV serotypes may individually be important for virus formation and function but do not appear to form essential disulfide bonds in vitro (Pulicherla, Kota, Dokholyan, & Asokan, 2012). Thus, the importance of the disulfide bond appears dependent on the virus identity.
Polymer chains (poly(2-oxazolines)) have also been used to crosslink capsid subunits of bacteriophage Qβ via CuAAC click chemistry (Figure 5c) (Manzenrieder, Luxenhofer, Retzlaff, Jordan, & Finn, 2011). Wild-type Qβ is an icosahedron made up of 180 proteins forming noncovalently interlocked subunits each with four surface-exposed amino groups. These sites were derivatized with an adizo-NHS ester for CuAAC, by which polymer chains were attached to the exterior of the capsid and crosslinked at multiple locations along the chains. These covalent connections linked a single polymer to multiple capsid subunits, thereby crosslinking them. The polymer-linked capsids were stable after heating at 100°C, while wild-type Qβ experienced a loss of capsid integrity.
Although we are beginning to explore the subunit interface, we still do not completely understand the intricacy of interactions in the viral capsid. Further studies are necessary to address this crucial element of engineering viruses.
A major strength in capsid modification via bioconjugation lies in its simplicity and versatility. In certain situations, only slight modifications of the capsid and ligand are necessary to achieve drastic effects. In addition, multiple ligands may be incorporated onto one capsid in a controlled manner using different functional groups, allowing for the creation of multifunctional and complex VNPs. However, depending on the molecules involved, coupling conditions may yield low efficiencies and non-specific reactions. Additionally, the previously discussed methods rely to a certain degree on the location of residues native to the capsid, which may not be suitable for all applications. Other approaches to modifying the capsid, as described in the next section, may be used to further expand the use of viruses as programmable nanodevices.
SYNTHETIC VIROLOGY
Synthetic virology applies concepts and tools from engineering in order to program new functionalities into viruses. The design approaches fall broadly into three main categories: 1) rational design, 2) directed evolution, and 3) bioinformatics-driven design. Although the approaches can be distinct, there are natural synergies at the interfaces of the three resulting in combination strategies, such as using bioinformatics analysis to guide how virus libraries are generated for directed evolution paradigms. Another important perspective in synthetic virology is to conceptualize viruses as nanoscale devices that detect endogenous/exogenous inputs and produce desired output responses. The reader is referred to other reviews that focus on biomolecular computation by nanoplatforms (Evans, Thadani, & Suh, 2016) and various endogenous and exogenous stimuli that can be used to control virus function (Brun, Gomez, & Suh, 2017). We previously reviewed the synthetic virology field (Guenther et al., 2014) and this section serves as an introduction and update to that review. In particular, we will cover bioinformatics-driven design approaches in this current review. Moreover, we will focus on capsid modification of viral vectors. For in depth review of viral genome engineering aspects of synthetic virology, the reader is referred to the following reviews (Gaj, Epstein, & Schaffer, 2016; Powell et al., 2016).
Rational Capsid Design
In general, rational design strategies use prior knowledge of the virus and of functional “parts” that can be incorporated into the virus capsid in order to endow new abilities onto a viral particle. This section will focus on peptides and proteins that have been genetically integrated into virus capsids. Some of the earliest research on creating synthetic VNPs focused on targeting viruses to different cellular receptors, and the reader is referred to other reviews that cover these approaches (Waehler, Russell, & Curiel, 2007). Here, we will discuss rational design strategies used to render the virus bioactivatable in response to extracellular or intracellular stimuli.
Matrix metalloproteinases (MMP) are upregulated in sites of many diseases including various cancers (Mühlebach et al., 2010). MMP-responsive viruses have been engineered to accept the extracellular input of protease activity, resulting in the output of targeted transduction to these disease sites (Evans et al., 2016). One of the first protease-responsive viruses developed was based on the Moloney murine leukemia virus (MMLV) (Schneider et al., 2003). The virus displayed an EGFR-binding antibody fragment which was attached to the viral envelope via an MMP-cleavable sequence. These engineered MMLV vectors were sequestered to the cell surface, unable to internalize into cells, until there was sufficient MMP activity to cleave the antibody fragments off from the virus envelope, allowing the virus to internalize and transduce the host cell (Schneider et al., 2003). Therefore, the MMLV would be infectious only in an area of high MMP activity, such as at diseased sites. Subcutaneous tumor models have been shown to respond to intratumorally injected MMP-activatable MMLV vectors (Peng, Vile, Cosset, & Russell, 1999). The general concept has been expanded to create protease-activatable avian influenza virus (Szécsi et al., 2006), measles virus (Mühlebach et al., 2010), and AAV (Judd et al., 2014).
Several variants of protease-activatable AAV vectors have been developed. The general design involves insertion of a “peptide lock”, containing two MMP-cleavable sequences flanking an “inactivating sequence”, into the AAV capsid such that the peptide lock blocks the cell binding domain of the virus. The inactivating sequence, comprised of four aspartic acid residues, serves to block cellular receptor binding and is removed in the presence of high MMP levels. Initially, it was hypothesized that the negative charge of these amino acids plays an important role in electrostatically repelling the virus capsid interaction with the negatively charged cell surface heparan sulfate proteoglycans. However, further studies demonstrated amino acids of other chemical properties can also function as an inactivating motif, but the resulting vectors experience deficiencies in capsid formation (Robinson, Judd, Ho, & Suh, 2016). One problem faced by the protease-activatable AAV vectors is that transduction efficiency does not return to that of the wild-type virus once the locks are cleaved. Thus, protease-activatability is achieved at the cost of decreased gene delivery efficiency. One potential way to overcome this problem is to create mosaic vectors that have mixed capsid subunits (Ho et al., 2014). When mosaic capsid vectors with varying ratios of protease-activatable and wild-type (wt) AAV subunits were generated, it was found that increasing the ratio of wt to protease-activatable subunits results in higher transduction efficiency but at the price of higher efficiencies even in the locked state (Ho et al., 2014). Therefore, the mosaic capsid approach alone is not the optimal solution for obtaining AAV vectors that are both protease-activatable and highly efficient in transduction. Interestingly, the protease-activatable AAV vector displays a nonlinear input-output function such that majority of the inserted peptide locks need to be cleaved off the capsid to achieve half of the maximum activation level. This nonlinearity allows for the programming of logic gates, such as an AND gate that results in vectors able to deliver transgenes only when certain combinations of MMPs, such as MMP-7 and MMP-9, are present (Judd et al., 2014). Overall, protease-activatable AAV is a viable vector platform that is currently being optimized for therapeutic use and target specificity.
In addition to extracellular proteases, viruses can be designed to be responsive to other types of stimuli. For example, the intracellular AAV transduction process has been engineered to be activatable by light (Gomez, Gerhardt, Judd, Tabor, & Suh, 2016). The system uses the interaction between Phytochrome B and Phytochrome interacting factor 6 (PIF6) to create VNPs that translocate into the nucleus more efficiently under activating light conditions. Phytochrome B and PIF6 associates under red light and dissociates under far-red light. The phytochrome B used in this study was designed to display a strong nuclear localization sequence that can help PIF6-modified AAV vectors to translocate from the cytoplasm into the nucleus upon exposure to red light, thus increasing gene expression. The use of photomasks together with the engineered system allows for controlled areas of high gene expression (Gerhardt et al., 2016; Gomez et al., 2016).
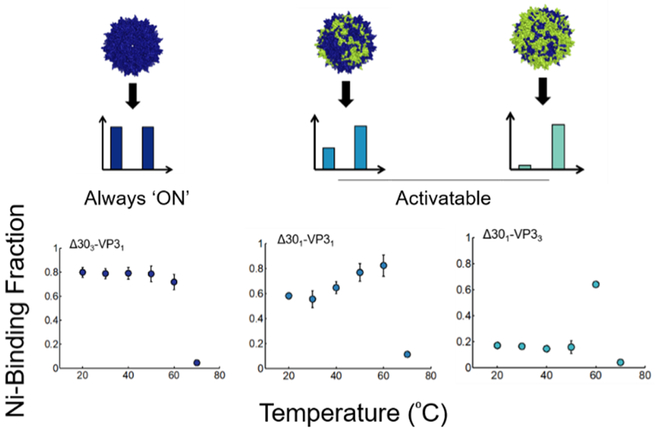
While the prior two examples demonstrate reprogramming the input of viral vectors, the output of viral vectors can also be reprogrammed in a rational manner. The AAV capsid undergoes a natural conformational change inside the host cell endosome due to low pH and other unknown factors (Venkatakrishnan et al., 2013). Specifically, the N-termini of VP1 and VP2 capsid subunits are initially hidden but then are externalized onto the capsid surface upon internalization into the cellular endosome. By creating a series of truncated VP2 capsid subunits, VNPs with variable abilities to perform activatable peptide display were generated (Figure 6) (Thadani et al., 2017). Mosaic capsids with different ratios of activatable and wt subunits display a wide range of behaviors, with some VNPs displaying peptides in a “brush-like” configuration that is always externalized and others displaying peptides only upon activation. The reprogrammed AAV vectors with variable peptide display behaviors may be used in the future to deliver peptide therapeutics and modulate vector properties such as specificity and immunogenicity.
Figure 6:
Reprogramming a conformational switch-based output of the AAV capsid. Truncated VP subunits with hexahistidine (his) tags at the N-termini were generated based on the AAV capsid. Upon incubation at different temperatures, capsids with surface-displayed his tags were captured with a nickel column. The wild-type capsid undergoes the structural conformational change upon incubation at ~60⁰C. Depending on the composition of the mutant AAV capsids, some capsids display his tags prior to activation (always “ON”) whereas other capsids display activatable peptide display (“activatable”). Reprinted with permission from “Reprogramming the Activatable Peptide Display Function of Adeno-Associated Virus Nanoparticles,” by N. Thadani et al, 2017, ACS Nano. Copyright 2017 by American Chemical Society.
Directed Evolution of Virus Capsids
Directed evolution is a method that mimics natural evolution by utilizing rounds of mutation and selection to identify mutants that meet a user-defined goal (Lutz, 2010). William Stemmer published one of the most commonly used protocols for directed evolution (Stemmer, 1994) and was the pioneer of many of its applications in bacteria (Crameri, Raillard, Bermudez, & Stemmer, 1998), retroviruses (Soong et al., 2000), and enzymes (Raillard et al., 2001). Directed evolution can be performed with various mutagenesis strategies, including random point mutation (e.g. error-prone PCR), DNA recombination (e.g. DNA shuffling), and random peptide display. For more details, the reader is referred to an in-depth review of directed evolution of viral vectors (Bartel, Weinstein, & Schaffer, 2012). For more information specifically on peptide display-based directed evolution, the reader is referred to (O’Neil & Hoess, 1995).
Several viruses have been subjected to directed evolution, including Adenovirus (Ad) (Kuhn et al., 2008), AAV (Excoffon et al., 2009), HSV (Christians, Scapozza, Crameri, Folkers, & Stemmer, 1999), MLV (Soong et al., 2000), and TEV (van den Berg, Löfdahl, Härd, & Berglund, 2006). In one recent study, the capsids of all AAV variants known to transduce the central nervous system were shuffled to create a virus that could yield efficient in vivo oligodendrocyte gene transfer (Powell et al., 2016). Cancer-specific viral therapies have also been developed using directed evolution. For example, an ovarian cancer targeting adenoviral vector, OvAd1, has been developed through directed evolution. Random mutagenesis was conducted on the Ad binding knob to create an adenovirus library which was then added to a 3D culture of ovarian cancer cells to select for a viral variant with improved infectivity (Kuhn et al., 2017).
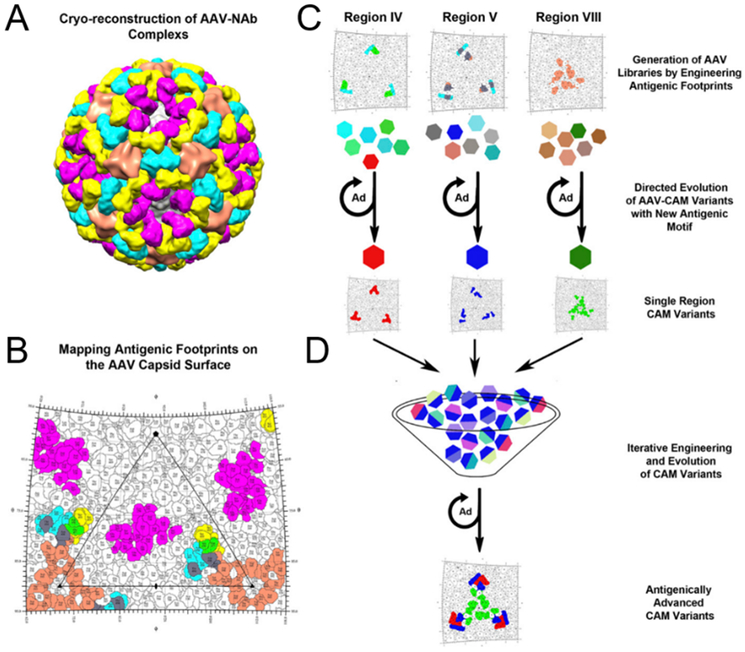
Structural information about the virus capsid can be used in combination with directed evolution to improve viral properties. Strategies involving structure-guided design depend on protein structures resolved via NMR spectroscopy, X-ray crystallography, or cryo-electron microscopy with reconstruction (reviewed in L. Zhang, Lua, Middelberg, Sun, & Connors, 2015). For example, cryo-reconstruction of the AAV1 capsid with bound neutralizing antibodies enabled the identification of antigenic epitopes on the capsid. Using this information, the amino acids within the epitopes were randomized to create a library of AAV vectors, and selection of the library led to the isolation of antigenically diversified AAV vectors called AAV-CAM variants (Figure 7) (Tse et al., 2017). In neutralizing antibody assays with immunized mouse serum, multiple AAV-CAM mutants are able to transduce cells at lower serum dilutions (i.e. higher antibody concentrations) than the parent wild-type AAV1 capsid.
Figure 7:
A combination of directed evolution and capsid structural information can enable the design of viruses with improved properties. (A) 3D cryo-reconstructed image of an AAV capsid with bound neutralizing antibodies (NAb). (B) Roadmap of where the antibodies bind to on the AAV capsid. Color-coding of each antibody are same as in panel A with overlapping residues between antibodies colored individually: green, ADK1a and 4E4; gray, 4E4 and 5H7. (C) Experimental plan for the directed evolution of three different capsid regions where antibodies bind. (D) Combination of the three viral libraries led to the generation of AAV-CAM vectors. Adapted from “Structure-guided evolution of antigenically distinct adeno-associated virus variants for immune evasion,” by L. V. Tse et al., 2017, PNAS. Creative Commons Attribution License.
Overall, directed evolution of viral capsids is a useful design strategy when not enough a priori information is available to achieve performance goals. Through generation of large libraries of viruses and application of carefully designed selection and screening procedures, new viral variants with desired phenotypes may be obtained.
Bioinformatics-Driven Capsid Design
Computational methods to aid the design of viral vectors can be categorized into sequence-based and protein structure-based tools. For example, AAVs are classified into serotypes and clades through phylogeny, genetic, and functional similarities (Gao et al., 2004). Alignment of the different AAV serotype capsid genes shows there is a high level of homology between different serotypes, and phenotypic differences in viral properties are likely caused by the small differences in their genotypes. Specifically, substantial progress over the last several decades has uncovered that phenotypic divergences in wild type AAV serotypes are correlated with hypervariable loop regions in the capsid (DiMattia et al., 2012; Zinn et al., 2015). Modification of these highly variable regions has led to the discovery of novel viral properties (Adachi, Enoki, Kawano, Veraz, & Nakai, 2014; Grimm et al., 2008). For example, an AAV capsid library was generated by altering only the variable surface loops of AAV2, and upon selection a mutant able to transduce glial cells was identified (Koerber et al., 2009).
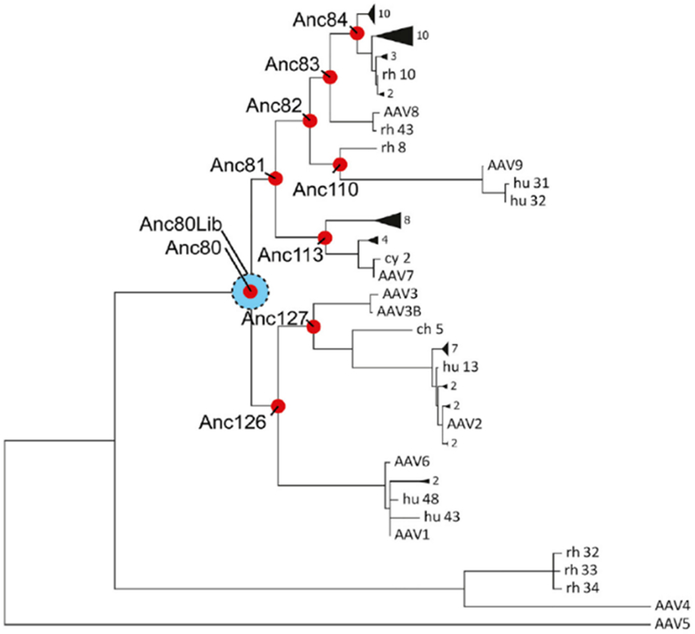
Virus capsid sequence alignments can also be combined with phylogenetic analyses to create new ancestral variants from known relatives. These algorithms were used to generate and compare a library of ancestral AAV variants which were more thermostable than all and more infectious than some of the currently available serotypes (Santiago-Ortiz et al., 2015; R. H. Smith et al., 2016). Notably, a promising AAV ancestor named Anc80 was created through an ancestral library approach (Zinn et al., 2015). Anc80 is a theoretical ancestor at the evolutionary break between AAV4 and AAV5 and the other commonly used serotypes (Figure 8). Despite being based on extant AAV serotypes, Anc80’s capsid structure is significantly divergent with a much higher melting temperature. Anc80 has been used to transduce hair cells in the cochlea with high efficiency, and as a result Anc80-based vectors look promising for treatment of the hearing impaired (Suzuki, Hashimoto, Xiao, Vandenberghe, & Liberman, 2017).
Figure 8:
Example of bioinformatics-driven capsid design. The reconstructed ancestral phylogeny of AAV was used to develop a new mutant, Anc80. Anc80 is the ancestor of currently studied AAV serotypes excluding AAV4 and AAV5. The distance between viruses in the hierarchical chart denotes relative distance of relation between the viruses. Figure reprinted with permission from “In Silico Reconstruction of the Viral Evolutionary Lineage Yields a Potent Gene Therapy Vector,” by E. Zinn et al, 2015, Cell Reports. Copyright 2015 by Elsevier.
In addition to using already available sequence data of viral variants, next generation sequencing (NGS) methods with barcoded AAV libraries can be used to provide more information regarding viral genotype-phenotype linkages. For example, almost 200 AAV capsid variants each packaging a different genetic barcode was generated, mixed together into one vector library, and the AAV barcoded library was injected in vivo (Adachi et al., 2014). This approach, named AAV Barcode-Seq, allowed for high throughput characterization of the biodistribution of each AAV variant. Further research using an approach like AAV Barcode-Seq may be able to uncover new sequence-function relationships in viral capsids on a faster timescale.
Bioinformatic tools can also be applied to protein structure instead of to protein sequence. A structure-based computational tool is SCHEMA, a method used to predict optimal cross-over points for generation of DNA shuffled libraries. Using 3D structural information, SCHEMA calculates the number of residue-to-residue contacts that are broken upon recombination. Minimizing the number of broken contacts during recombination should increase the chances for creating structurally intact viral progeny. This algorithm has been used on AAV capsids to generate new chimeric capsids (Ho, Adler, Torre, Silberg, & Suh, 2013) and mutants with new behaviors, such as the ability to transduce neural stem cells (Ojala et al., 2017). Currently, resolving viral protein structure and function is a highly developing field. As technologies improve and more fundamental information of capsid structures is revealed, structure-based design strategies will likely become more popular.
Conclusion
Much progress has been made in understanding how virus capsids assemble/disassemble, switch conformations, and assume different morphologies. Additionally, new chemical and genetic design strategies have been developed to alter viral function. Unfortunately, there are still many unknowns surrounding how viruses work. Continued efforts in studying the biophysics of virus capsids and their natural responses to environmental stimuli in a quantitative fashion will provide more insights into what design strategies may be possible in the future. The ability to specifically target certain amino-acids for functionalization would push the frontier of chemical virology and allow for further deployment of precise and useful modifications to the capsid. Synthetic virology will greatly benefit from improved models of capsid structure and function that may eventually allow for predictable virus design in silico. Overall, fully controllable and programmable viruses promise to make important contributions to many biomedical fields, including gene therapy, diagnostic imaging, and immunotherapy. As the subfields of virology discussed here – physical, chemical, and synthetic – advance our theoretical knowledge and expand our experimental toolkits, their close partnership will likely create new solutions to improve the quality of life for future generations.
Acknowledgments
This material is based upon work supported by the National Science Foundation under grant number 1611044, the National Institutes of Health under grant numbers R01CA207497 and R01HL138126, and a Hamill Foundation award to J.S.
Footnotes
No conflicts of interest to report
Contributor Information
Maria Yanqing Chen, Department of Bioengineering, Rice University.
Susan Butler, Department of Bioengineering, Rice University.
Weitong Chen, Department of Chemical and Biomolecular Engineering, Rice University.
Junghae Suh, Department of Bioengineering; Systems, Synthetic, and Physical Biology Program; Rice University.
References
- Adachi K, Enoki T, Kawano Y, Veraz M, & Nakai H (2014). Drawing a high-resolution functional map of adeno-associated virus capsid by massively parallel sequencing. Nature Communications, 5, 3075 10.1038/ncomms4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agard NJ, Baskin JM, Prescher JA, Lo A, & Bertozzi CR (2006). A Comparative Study of Bioorthogonal Reactions with Azides. ACS Chemical Biology, 1(10), 644–648. 10.1021/cb6003228 [DOI] [PubMed] [Google Scholar]
- Arnold GS, Sasser AK, Stachler MD, & Bartlett JS (2006). Metabolic biotinylation provides a unique platform for the purification and targeting of multiple AAV vector serotypes. Molecular Therapy: The Journal of the American Society of Gene Therapy, 14(1), 97–106. 10.1016/j.ymthe.2006.02.014 [DOI] [PubMed] [Google Scholar]
- Bain J, & Staniland SS (2015). Bioinspired nanoreactors for the biomineralisation of metallic-based nanoparticles for nanomedicine. Physical Chemistry Chemical Physics, 17(24), 15508–15521. 10.1039/C5CP00375J [DOI] [PubMed] [Google Scholar]
- Bajaj S, & Banerjee M (2016). In vitro assembly of polymorphic virus-like particles from the capsid protein of a nodavirus. Virology, 496, 106–115. 10.1016/j.virol.2016.05.025 [DOI] [PubMed] [Google Scholar]
- Banerjee PS, Ostapachuk P, Hearing P, & Carrico I (2010). Chemoselective attachment of small molecule effector functionality to human adenoviruses facilitates gene delivery to cancer cells. Journal of the American Chemical Society, 132(39), 13615–13617. 10.1021/ja104547x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee PS, Ostapchuk P, Hearing P, & Carrico IS (2011). Unnatural amino acid incorporation onto adenoviral (Ad) coat proteins facilitates chemoselective modification and retargeting of Ad type 5 vectors. Journal of Virology, 85(15), 7546–7554. 10.1128/JVI.00118-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee PS, Zuniga ES, Ojima I, & Carrico IS (2011). Targeted and armed oncolytic adenovirus via chemoselective modification. Bioorganic & Medicinal Chemistry Letters, 21(17), 4985–4988. 10.1016/j.bmcl.2011.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel MA, Weinstein JR, & Schaffer DV (2012). Directed evolution of novel adeno-associated viruses for therapeutic gene delivery. Gene Therapy, 19(6), 694–700. 10.1038/gt.2012.20 [DOI] [PubMed] [Google Scholar]
- Barwal I, Kumar R, Kateriya S, Dinda AK, & Yadav SC (2016). Targeted delivery system for cancer cells consist of multiple ligands conjugated genetically modified CCMV capsid on doxorubicin GNPs complex. Scientific Reports, 6 10.1038/srep37096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett A, Patel S, Mietzsch M, Jose A, Lins-Austin B, Yu JC, … Agbandje-McKenna M (2017). Thermal Stability as a Determinant of AAV Serotype Identity. Molecular Therapy - Methods & Clinical Development, 6(Supplement C), 171–182. 10.1016/j.omtm.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernaud J, Rossi A, Fis A, Gardette L, Aillot L, Büning H, … Faivre-Moskalenko C (2018). Characterization of AAV vector particle stability at the single-capsid level. Journal of Biological Physics. 10.1007/s10867-018-9488-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet-Colominas C, Cuillel M, Koch MHJ, Vachette P, & Jacrot B (1987). Kinetic study of the self-assembly of brome mosaic virus capsid. European Biophysics Journal, 15(3), 159–168. 10.1007/BF00263680 [DOI] [Google Scholar]
- Bleker S, Sonntag F, & Kleinschmidt JA (2005). Mutational Analysis of Narrow Pores at the Fivefold Symmetry Axes of Adeno-Associated Virus Type 2 Capsids Reveals a Dual Role in Genome Packaging and Activation of Phospholipase A2 Activity. Journal of Virology, 79(4), 2528–2540. 10.1128/JVI.79.4.2528-2540.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasch M, Putri RM, de Ruiter MV, Luque D, Koay MST, Castón JR, & Cornelissen JJLM (2017). Assembling Enzymatic Cascade Pathways inside Virus-Based Nanocages Using Dual-Tasking Nucleic Acid Tags. Journal of the American Chemical Society, 139(4), 1512–1519. 10.1021/jacs.6b10948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckman MA, Czapar AE, VanMeter A, Randolph LN, & Steinmetz NF (2016). Tobacco mosaic virus-based protein nanoparticles and nanorods for chemotherapy delivery targeting breast cancer. Journal of Controlled Release : Official Journal of the Controlled Release Society, 231, 103–113. 10.1016/j.jconrel.2016.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckman MA, Hern S, Jiang K, Flask CA, Yu X, & Steinmetz NF (2013). Tobacco mosaic virus rods and spheres as supramolecular high-relaxivity MRI contrast agents. Journal of Materials Chemistry B, 1(10), 1482 10.1039/c3tb00461a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckman MA, VanMeter A, & Steinmetz NF (2015). Nanomanufacturing of Tobacco Mosaic Virus-Based Spherical Biomaterials Using a Continuous Flow Method. ACS Biomaterials Science & Engineering, 1(1), 13–18. 10.1021/ab500059s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun MJ, Gomez EJ, & Suh J (2017). Stimulus-responsive viral vectors for controlled delivery of therapeutics. Journal of Controlled Release, 267, 80–89. 10.1016/j.jconrel.2017.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos SK, Parrott MB, & Barry MA (2004). Avidin-Based Targeting and Purification of a Protein IX-Modified, Metabolically Biotinylated Adenoviral Vector. Molecular Therapy : The Journal of the American Society of Gene Therapy, 9(6), 942–954. 10.1016/j.ymthe.2004.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Guenther RH, Sit TL, Opperman CH, Lommel SA, & Willoughby JA (2014). Loading and Release Mechanism of Red Clover Necrotic Mosaic Virus Derived Plant Viral Nanoparticles for Drug Delivery of Doxorubicin. Small, 10(24), 5126–5136. 10.1002/smll.201400558 [DOI] [PubMed] [Google Scholar]
- Carlisle RC, Benjamin R, Briggs SS, Sumner-Jones S, McIntosh J, Gill D, … Fisher KD (2008). Coating of adeno-associated virus with reactive polymers can ablate virus tropism, enable retargeting and provide resistance to neutralising antisera. The Journal of Gene Medicine, 10(4), 400–411. 10.1002/jgm.1161 [DOI] [PubMed] [Google Scholar]
- Casini GL, Graham D, Heine D, Garcea RL, & Wu DT (2004). In vitro papillomavirus capsid assembly analyzed by light scattering. Virology, 325(2), 320–327. 10.1016/j.virol.2004.04.034 [DOI] [PubMed] [Google Scholar]
- Caspar DL (1956). Structure of Bushy Stunt Virus. Nature, 177(4506), 475–476. 10.1038/177475a0 [DOI] [PubMed] [Google Scholar]
- Caspar DL (1980). Movement and self-control in protein assemblies. Quasi-equivalence revisited. Biophysical Journal, 32(1), 103–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar DL, & Klug A (1962). Physical principles in the construction of regular viruses. Cold Spring Harbor Symposia on Quantitative Biology, 27, 1–24. [DOI] [PubMed] [Google Scholar]
- Castellanos M, Pérez R, Carrillo PJP, de Pablo PJ, & Mateu MG (2012). Mechanical Disassembly of Single Virus Particles Reveals Kinetic Intermediates Predicted by Theory. Biophysical Journal, 102(11), 2615–2624. 10.1016/j.bpj.2012.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceres P, & Zlotnick A (2002). Weak Protein−Protein Interactions Are Sufficient To Drive Assembly of Hepatitis B Virus Capsids. Biochemistry, 41(39), 11525–11531. 10.1021/bi0261645 [DOI] [PubMed] [Google Scholar]
- Chariou PL, Lee KL, Wen AM, Gulati NM, Stewart PL, & Steinmetz NF (2015). Detection and Imaging of Aggressive Cancer Cells Using an Epidermal Growth Factor Receptor (EGFR)-Targeted Filamentous Plant Virus-Based Nanoparticle. Bioconjugate Chemistry, 26(2), 262–269. 10.1021/bc500545z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan K, Hernandez-Meza JM, Rodríguez-Hernández AG, Juarez-Moreno K, Sengar P, & Vazquez-Duhalt R (2018). Multifunctionalized biocatalytic P22 nanoreactor for combinatory treatment of ER+ breast cancer. Journal of Nanobiotechnology, 16 10.1186/s12951-018-0345-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Kao CC, & Dragnea B (2008). Self-Assembly of Brome Mosaic Virus Capsids: Insights from Shorter Time-Scale Experiments. The Journal of Physical Chemistry A, 112(39), 9405–9412. 10.1021/jp802498z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-C, Xing L, Stark M, Ou T, Holla P, Xiao K, … Cheng RH (2016). Chemically activatable viral capsid functionalized for cancer targeting. Nanomedicine, 11(4), 377–390. 10.2217/nnm.15.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Li N, Chen L, Lee J, & Gassensmith JJ (2016). Dual Functionalized Bacteriophage Qβ as a Photocaged Drug Carrier. Small, 12(33), 4563–4571. 10.1002/smll.201601053 [DOI] [PubMed] [Google Scholar]
- Cheng RH, Reddy VS, Olson NH, Fishert AJ, Baker TS, & Johnson JE (1994). Functional implications of quasi-equivalence in a T=3 icosahedral animal virus established by cryo-electron microscopy and X-ray crystallography. Structure (London, England : 1993), 2(4), 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillón M, Lee JH, Fasbender A, & Welsh MJ (1998). Adenovirus complexed with polyethylene glycol and cationic lipid is shielded from neutralizing antibodies in vitro. Gene Therapy, 5(7), 995–1002. 10.1038/sj.gt.3300665 [DOI] [PubMed] [Google Scholar]
- Christians FC, Scapozza L, Crameri A, Folkers G, & Stemmer WPC (1999). Directed evolution of thymidine kinase for AZT phosphorylation using DNA family shuffling. Nature Biotechnology, 17(3), 259–264. 10.1038/7003 [DOI] [PubMed] [Google Scholar]
- Comellas-Aragonès M, Engelkamp H, Claessen VI, Sommerdijk NAJM, Rowan AE, Christianen PCM, … Nolte RJM (2007). A virus-based single-enzyme nanoreactor. Nature Nanotechnology, 2(10), 635–639. 10.1038/nnano.2007.299 [DOI] [PubMed] [Google Scholar]
- Crameri A, Raillard S-A, Bermudez E, & Stemmer WPC (1998). DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature, 391(6664), 288–291. 10.1038/34663 [DOI] [PubMed] [Google Scholar]
- Crick FHC, & Watson JD (1956). Structure of Small Viruses. Nature, 177(4506), 473–475. 10.1038/177473a0 [DOI] [PubMed] [Google Scholar]
- Destito G, Yeh R, Rae CS, Finn MG, & Manchester M (2007). Folic acid-mediated targeting of cowpea mosaic virus particles to tumor cells. Chemistry & Biology, 14(10), 1152–1162. 10.1016/j.chembiol.2007.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMattia MA, Nam H-J, Van Vliet K, Mitchell M, Bennett A, Gurda BL, … Agbandje-McKenna M (2012). Structural insight into the unique properties of adeno-associated virus serotype 9. Journal of Virology, 86(12), 6947–6958. 10.1128/JVI.07232-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas T, Strable E, Willits D, Aitouchen A, Libera M, & Young M (2002). Protein Engineering of a Viral Cage for Constrained Nanomaterials Synthesis. Advanced Materials, 14(6), 415–418. [DOI] [Google Scholar]
- Douglas T, & Young M (1998). Host–guest encapsulation of materials by assembled virus protein cages. Nature, 393(6681), 152 10.1038/30211 [DOI] [Google Scholar]
- Douglas T, & Young M (2006). Viruses: Making Friends with Old Foes. Science, 312(5775), 873–875. 10.1126/science.1123223 [DOI] [PubMed] [Google Scholar]
- Earley LF, Powers JM, Adachi K, Baumgart JT, Meyer NL, Xie Q, … Nakai H (2017). Adeno-associated Virus (AAV) Assembly-Activating Protein Is Not an Essential Requirement for Capsid Assembly of AAV Serotypes 4, 5, and 11. Journal of Virology, 91(3). 10.1128/JVI.01980-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eber FJ, Eiben S, Jeske H, & Wege C (2015). RNA-controlled assembly of tobacco mosaic virus-derived complex structures: from nanoboomerangs to tetrapods. Nanoscale, 7(1), 344–355. 10.1039/c4nr05434b [DOI] [PubMed] [Google Scholar]
- Evans AC, Thadani NN, & Suh J (2016). Biocomputing nanoplatforms as therapeutics and diagnostics. Journal of Controlled Release, 240, 387–393. 10.1016/j.jconrel.2016.01.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffon KJDA, Koerber JT, Dickey DD, Murtha M, Keshavjee S, Kaspar BK, … Schaffer DV (2009). Directed evolution of adeno-associated virus to an infectious respiratory virus. Proceedings of the National Academy of Sciences, 106(10), 3865–3870. 10.1073/pnas.0813365106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler JD, Higginson C, Hovlid ML, Kislukhin AA, Castillejos A, Manzenrieder F, … Finn MG (2012). Engineered Mutations Change the Structure and Stability of a Virus-Like Particle. Biomacromolecules, 13(8), 2339–2348. 10.1021/bm300590x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher KD, Stallwood Y, Green NK, Ulbrich K, Mautner V, & Seymour LW (2001). Polymer-coated adenovirus permits efficient retargeting and evades neutralising antibodies. Gene Therapy, 8(5), 341 10.1038/sj.gt.3301389 [DOI] [PubMed] [Google Scholar]
- Flenniken ML, Liepold LO, Crowley BE, Willits DA, Young MJ, & Douglas T (2005). Selective attachment and release of a chemotherapeutic agent from the interior of a protein cage architecture. Chemical Communications, (4), 447 10.1039/b413435d [DOI] [PubMed] [Google Scholar]
- Gaj T, Epstein BE, & Schaffer DV (2016). Genome Engineering Using Adeno-associated Virus: Basic and Clinical Research Applications. Molecular Therapy, 24(3), 458–464. 10.1038/mt.2015.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G, Vandenberghe LH, Alvira MR, Lu Y, Calcedo R, Zhou X, & Wilson JM (2004). Clades of Adeno-associated viruses are widely disseminated in human tissues. Journal of Virology, 78(12), 6381–6388. 10.1128/JVI.78.12.6381-6388.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt KP, Olson EJ, Castillo-Hair SM, Hartsough LA, Landry BP, Ekness F, … Tabor JJ (2016). An open-hardware platform for optogenetics and photobiology. Scientific Reports, 6, 35363 10.1038/srep35363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez EJ, Gerhardt K, Judd J, Tabor JJ, & Suh J (2016). Light-Activated Nuclear Translocation of Adeno-Associated Virus Nanoparticles Using Phytochrome B for Enhanced, Tunable, and Spatially Programmable Gene Delivery. ACS Nano, 10(1), 225–237. 10.1021/acsnano.5b05558 [DOI] [PubMed] [Google Scholar]
- Green NK, Morrison J, Hale S, Briggs SS, Stevenson M, Subr V, … Fisher KD (2008). Retargeting polymer-coated adenovirus to the FGF receptor allows productive infection and mediates efficacy in a peritoneal model of human ovarian cancer. The Journal of Gene Medicine, 10(3), 280–289. 10.1002/jgm.1121 [DOI] [PubMed] [Google Scholar]
- Grimm D, Lee JS, Wang L, Desai T, Akache B, Storm TA, & Kay MA (2008). In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. Journal of Virology, 82(12), 5887–5911. 10.1128/JVI.00254-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther CM, Kuypers BE, Lam MT, Robinson TM, Zhao J, & Suh J (2014). Synthetic Virology: Engineering Viruses for Gene Delivery. Wiley Interdisciplinary Reviews. Nanomedicine and Nanobiotechnology, 6(6), 548–558. 10.1002/wnan.1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan MF (2014). Modeling Viral Capsid Assembly. In Rice SA & Dinner AR (Eds.), Advances in Chemical Physics : Volume 155 (pp. 1–68). John Wiley & Sons, Inc; 10.1002/9781118755815.ch01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan MF, & Zandi R (2016). Recent advances in coarse-grained modeling of virus assembly. Current Opinion in Virology, 18, 36–43. 10.1016/j.coviro.2016.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein JE, & Fokin VV (2010). Copper-catalyzed azide–alkyne cycloaddition (CuAAC) and beyond: new reactivity of copper(i) acetylides. Chemical Society Reviews, 39(4), 1302–1315. 10.1039/b904091a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho ML, Adler BA, Torre ML, Silberg JJ, & Suh J (2013). SCHEMA computational design of virus capsid chimeras: calibrating how genome packaging, protection, and transduction correlate with calculated structural disruption. ACS Synthetic Biology, 2(12), 724–733. 10.1021/sb400076r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho ML, Judd J, Kuypers BE, Yamagami M, Wong FF, & Suh J (2014). Efficiency of Protease-Activatable Virus Nanonodes Tuned Through Incorporation of Wild-Type Capsid Subunits. Cellular and Molecular Bioengineering; Dordrecht, 7(3), 334–343. 10.1007/s12195-014-0334-y [DOI] [Google Scholar]
- Hooker JM, Kovacs EW, & Francis MB (2004). Interior Surface Modification of Bacteriophage MS2. Journal of the American Chemical Society, 126(12), 3718–3719. 10.1021/ja031790q [DOI] [PubMed] [Google Scholar]
- Horowitz ED, Finn MG, & Asokan A (2012). Tyrosine crosslinking reveals interfacial dynamics in adeno-associated viral capsids during infection. ACS Chemical Biology, 7(6), 1059–1066. 10.1021/cb3000265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz ED, Rahman KS, Bower BD, Dismuke DJ, Falvo MR, Griffith JD, … Asokan A (2013). Biophysical and Ultrastructural Characterization of Adeno-Associated Virus Capsid Uncoating and Genome Release. Journal of Virology, 87(6), 2994–3002. 10.1128/JVI.03017-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovlid ML, Lau JL, Breitenkamp K, Higginson CJ, Laufer B, Manchester M, & Finn MG (2014). Encapsidated Atom-Transfer Radical Polymerization in Qβ Virus-like Nanoparticles. ACS Nano, 8(8), 8003–8014. 10.1021/nn502043d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovlid ML, Steinmetz NF, Laufer B, Lau JL, Kuzelka J, Wang Q, … Finn MG (2012). Guiding plant virus particles to integrin-displaying cells. Nanoscale, 4(12), 3698–3705. 10.1039/C2NR30571B [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Zhang Y, Shukla S, Gu Y, Yu X, & Steinmetz NF (2017). Dysprosium-Modified Tobacco Mosaic Virus Nanoparticles for Ultra-High-Field Magnetic Resonance and Near-Infrared Fluorescence Imaging of Prostate Cancer. ACS Nano, 11(9), 9249–9258. 10.1021/acsnano.7b04472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RK, Steinmetz NF, Fu C-Y, Manchester M, & Johnson JE (2011). Transferrin-mediated targeting of bacteriophage HK97 nanoparticles into tumor cells. Nanomedicine (London, England), 6(1), 55–68. 10.2217/nnm.10.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JE, & Speir JA (1997). Quasi-equivalent viruses: a paradigm for protein assemblies. Journal of Molecular Biology, 269(5), 665–675. 10.1006/jmbi.1997.1068 [DOI] [PubMed] [Google Scholar]
- Johnson JM, Tang J, Nyame Y, Willits D, Young MJ, & Zlotnick A (2005). Regulating Self-Assembly of Spherical Oligomers. Nano Letters, 5(4), 765–770. 10.1021/nl050274q [DOI] [PubMed] [Google Scholar]
- Johnson JS, Li C, DiPrimio N, Weinberg MS, McCown TJ, & Samulski RJ (2010). Mutagenesis of Adeno-Associated Virus Type 2 Capsid Protein VP1 Uncovers New Roles for Basic Amino Acids in Trafficking and Cell-Specific Transduction. Journal of Virology, 84(17), 8888–8902. 10.1128/JVI.00687-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd J, Ho ML, Tiwari A, Gomez EJ, Dempsey C, Van Vliet K, … Suh J (2014). Tunable Protease-Activatable Virus Nanonodes. ACS Nano, 8(5), 4740–4746. 10.1021/nn500550q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen MU, Viholainen JI, Närvänen A, Ylä-Herttuala S, & Airenne KJ (2008). Targeting and purification of metabolically biotinylated baculovirus. Human Gene Therapy, 19(6), 589–600. 10.1089/hum.2007.177 [DOI] [PubMed] [Google Scholar]
- Katen S, & Zlotnick A (2009a). Chapter 14 The Thermodynamics of Virus Capsid Assembly. In Methods in Enzymology (Vol. 6879, pp. 395–417). 10.1016/S0076-6879(08)04214-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katen S, & Zlotnick A (2009b). The Thermodynamics of Virus Capsid Assembly. Methods in Enzymology, 455, 395–417. 10.1016/S0076-6879(08)04214-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen RE, Mukherjee R, Cao X, Erickson SB, Zheng Y, & Chatterjee A (2016). A Precise Chemical Strategy To Alter the Receptor Specificity of the Adeno-Associated Virus. Angewandte Chemie International Edition, 55(36), 10645–10649. 10.1002/anie.201604067 [DOI] [PubMed] [Google Scholar]
- Kim J, & Wu J (2014). A molecular thermodynamic model for the stability of hepatitis B capsids. The Journal of Chemical Physics, 140(23), 235101 10.1063/1.4882068 [DOI] [PubMed] [Google Scholar]
- Kler S, Asor R, Li C, Ginsburg A, Harries D, Oppenheim A, … Raviv U (2012). RNA Encapsidation by SV40-Derived Nanoparticles Follows a Rapid Two-State Mechanism. Journal of the American Chemical Society, 134(21), 8823–8830. 10.1021/ja2110703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kler S, Wang JC-Y, Dhason M, Oppenheim A, & Zlotnick A (2013). Scaffold properties are a key determinant of the size and shape of self-assembled virus-derived particles. ACS Chemical Biology, 8(12). 10.1021/cb4005518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C, Wabbel K, Eber FJ, Krolla-Sidenstein P, Azucena C, Gliemann H, … Wege C (2015). Modified TMV Particles as Beneficial Scaffolds to Present Sensor Enzymes. Frontiers in Plant Science, 6, 1137 10.3389/fpls.2015.01137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerber JT, Klimczak R, Jang J-H, Dalkara D, Flannery JG, & Schaffer DV (2009). Molecular Evolution of Adeno-associated Virus for Enhanced Glial Gene Delivery. Molecular Therapy, 17(12), 2088–2095. 10.1038/mt.2009.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb HC, Finn MG, & Sharpless KB (2001). Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angewandte Chemie (International Ed. in English), 40(11), 2004–2021. [DOI] [PubMed] [Google Scholar]
- Kronenberg S, Böttcher B, von der Lieth CW, Bleker S, & Kleinschmidt JA (2005). A Conformational Change in the Adeno-Associated Virus Type 2 Capsid Leads to the Exposure of Hidden VP1 N Termini. Journal of Virology, 79(9), 5296–5303. 10.1128/JVI.79.9.5296-5303.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn I, Bauzon M, Green N, Seymour L, Fisher K, & Hermiston T (2017). OvAd1, a Novel, Potent, and Selective Chimeric Oncolytic Virus Developed for Ovarian Cancer by 3D-Directed Evolution. Molecular Therapy - Oncolytics, 4, 55–66. 10.1016/j.omto.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn I, Harden P, Bauzon M, Chartier C, Nye J, Thorne S, … Hermiston TW (2008). Directed Evolution Generates a Novel Oncolytic Virus for the Treatment of Colon Cancer. PLOS ONE, 3(6), e2409 10.1371/journal.pone.0002409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch HP, Kaikkonen MU, Pikkarainen JT, & Ylä-Herttuala S (2010). Avidin-biotin technology in targeted therapy. Expert Opinion on Drug Delivery, 7(5), 551–564. 10.1517/17425241003677749 [DOI] [PubMed] [Google Scholar]
- Lewis JD, Destito G, Zijlstra A, Gonzalez MJ, Quigley JP, Manchester M, & Stuhlmann H (2006). Viral nanoparticles as tools for intravital vascular imaging. Nature Medicine, 12(3), 354–360. 10.1038/nm1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Chen H, Ma L, Zhou K, Zhang Z-P, Meng C, … Wang Q (2014). Insights into Stabilization of a Viral Protein Cage in Templating Complex Nanoarchitectures: Roles of Disulfide Bonds. Small, 10(3), 536–543. 10.1002/smll.201300860 [DOI] [PubMed] [Google Scholar]
- Liu H, Qu C, Johnson JE, & Case DA (2003). Pseudo-atomic models of swollen CCMV from cryo-electron microscopy data. Journal of Structural Biology, 142(3), 356–363. 10.1016/S1047-8477(03)00028-5 [DOI] [PubMed] [Google Scholar]
- Lockney DM, Guenther RN, Loo L, Overton W, Antonelli R, Clark J, … Franzen S (2011). The Red clover necrotic mosaic virus Capsid as a Multifunctional Cell Targeting Plant Viral Nanoparticle. Bioconjugate Chemistry, 22(1), 67–73. 10.1021/bc100361z [DOI] [PubMed] [Google Scholar]
- Loo L, Guenther H, R., Lommel A, S., & Franzen S (2008). Infusion of dye molecules into Red clover necrotic mosaic virus. Chemical Communications, 0(1), 88–90. 10.1039/B714748A [DOI] [PubMed] [Google Scholar]
- Lucon J, Edwards E, Qazi S, Uchida M, & Douglas T (2013). Atom transfer radical polymerization on the interior of the P22 capsid and incorporation of photocatalytic monomer crosslinks. European Polymer Journal, 49(10), 2976–2985. 10.1016/j.eurpolymj.2013.06.010 [DOI] [Google Scholar]
- Lucon J, Qazi S, Uchida M, Bedwel GJ, LaFrance B, Prevelige PE, & Douglas T (2012). Using the Interior Cavity of the P22 Capsid for Site Specific Initiation of Atom Transfer Radical Polymerization with Tremendously Increased Cargo Loading. Nature Chemistry, 4(10), 781–788. 10.1038/nchem.1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz S (2010). Beyond directed evolution—semi-rational protein engineering and design. Current Opinion in Biotechnology, 21(6), 734–743. 10.1016/j.copbio.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzenrieder F, Luxenhofer R, Retzlaff M, Jordan R, & Finn MG (2011). Stabilization of Virus-Like Particles with Poly(2-oxazoline)s. Angewandte Chemie (International Ed. in English), 50(11), 2601–2605. 10.1002/anie.201006134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateu MG (2011). Virus engineering: functionalization and stabilization. Protein Engineering Design and Selection, 24(1–2), 53–63. 10.1093/protein/gzq069 [DOI] [PubMed] [Google Scholar]
- Mavila S, Eivgi O, Berkovich I, & Lemcoff NG (2016). Intramolecular Cross-Linking Methodologies for the Synthesis of Polymer Nanoparticles. Chemical Reviews, 116(3), 878–961. 10.1021/acs.chemrev.5b00290 [DOI] [PubMed] [Google Scholar]
- Medrano M, Ngel Fuertes M, Valbuena A, Carrillo PJPP, Rodríguez-Huete A, Mateu MG, … Mateu MG (2016). Imaging and Quantitation of a Succession of Transient Intermediates Reveal the Reversible Self-Assembly Pathway of a Simple Icosahedral Virus Capsid. Journal of the American Chemical Society, 138(47), 15385–15396. 10.1021/jacs.6b07663 [DOI] [PubMed] [Google Scholar]
- Mehl RA, Anderson JC, Santoro SW, Wang L, Martin AB, King DS, … Schultz PG (2003). Generation of a Bacterium with a 21 Amino Acid Genetic Code. Journal of the American Chemical Society, 125(4), 935–939. 10.1021/ja0284153 [DOI] [PubMed] [Google Scholar]
- Meunier S, Strable E, & Finn MG (2004). Crosslinking of and coupling to viral capsid proteins by tyrosine oxidation. Chemistry & Biology, 11(3), 319–326. 10.1016/j.chembiol.2004.02.019 [DOI] [PubMed] [Google Scholar]
- Minten IJ, Claessen VI, Blank K, Rowan AE, Nolte RJM, & Cornelissen JJLM (2011). Catalytic capsids: the art of confinement. Chem. Sci, 2(2), 358–362. 10.1039/C0SC00407C [DOI] [Google Scholar]
- Minten IJ, Hendriks LJA, Nolte RJM, & Cornelissen JJLM (2009). Controlled Encapsulation of Multiple Proteins in Virus Capsids. Journal of the American Chemical Society, 131(49), 17771–17773. 10.1021/ja907843s [DOI] [PubMed] [Google Scholar]
- Moerman P, van der Schoot P, & Kegel W (2016). Kinetics versus Thermodynamics in Virus Capsid Polymorphism. The Journal of Physical Chemistry B, 120(26), 6003–6009. 10.1021/acs.jpcb.6b01953 [DOI] [PubMed] [Google Scholar]
- Morozov AY, Bruinsma RF, & Rudnick J (2009). Assembly of viruses and the pseudo-law of mass action. The Journal of Chemical Physics, 131(15), 155101 10.1063/1.3212694 [DOI] [PubMed] [Google Scholar]
- Mühlebach MD, Schaser T, Zimmermann M, Armeanu S, Hanschmann K-MO, Cattaneo R, … Buchholz CJ (2010). Liver Cancer Protease Activity Profiles Support Therapeutic Options with Matrix Metalloproteinase–Activatable Oncolytic Measles Virus. Cancer Research, 70(19), 7620–7629. 10.1158/0008-5472.CAN-09-4650 [DOI] [PubMed] [Google Scholar]
- Nguyen HD, Reddy VS, & Brooks CL III (2009). Invariant Polymorphism in Virus Capsid Assembly. Journal of the American Chemical Society, 131(7), 2606–2614. 10.1021/ja807730x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin N, Ksenofontov A, Trifonova E, Arkhipenko M, Petrova E, Kondakova O, … Karpova O (2016). Thermal conversion of filamentous potato virus X into spherical particles with different properties from virions. FEBS Letters, 590(10), 1543–1551. 10.1002/1873-3468.12184 [DOI] [PubMed] [Google Scholar]
- Oda Y, Saeki K, Takahashi Y, Maeda T, Naitow H, Tsukihara T, & Fukuyama K (2000). Crystal structure of tobacco necrosis virus at 2.25 Å resolution. Journal of Molecular Biology, 300(1), 153–169. 10.1006/jmbi.2000.3831 [DOI] [PubMed] [Google Scholar]
- Ojala DS, Sun S, Santiago-Ortiz JL, Shapiro MG, Romero PA, & Schaffer DV (2017). In Vivo Selection of a Computationally Designed SCHEMA AAV Library Yields a Novel Variant for Infection of Adult Neural Stem Cells in the Subventricular Zone. Molecular Therapy, 26(1), 304–319. 10.1016/j.ymthe.2017.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil KT, & Hoess RH (1995). Phage display: protein engineering by directed evolution. Current Opinion in Structural Biology, 5(4), 443–449. 10.1016/0959-440X(95)80027-1 [DOI] [PubMed] [Google Scholar]
- O’Riordan CR, Lachapelle A, Delgado C, Parkes V, Wadsworth SC, Smith AE, & Francis GE (1999). PEGylation of Adenovirus with Retention of Infectivity and Protection from Neutralizing Antibody in Vitro and in Vivo. Human Gene Therapy, 10(8), 1349–1358. 10.1089/10430349950018021 [DOI] [PubMed] [Google Scholar]
- Oum YH, & Carrico IS (2012). Altering Adenoviral Tropism via Click Modification with ErbB Specific Ligands. Bioconjugate Chemistry, 23(7), 1370–1376. 10.1021/bc200477z [DOI] [PubMed] [Google Scholar]
- Pacouret S, Bouzelha M, Shelke R, Andres-Mateos E, Xiao R, Maurer A, … Vandenberghe LH (2017). AAV-ID: A Rapid and Robust Assay for Batch-to-Batch Consistency Evaluation of AAV Preparations. Molecular Therapy, 25(6), 1375–1386. 10.1016/j.ymthe.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson DP, Prevelige PE, & Douglas T (2012). Nanoreactors by Programmed Enzyme Encapsulation Inside the Capsid of the Bacteriophage P22. ACS Nano, 6(6), 5000–5009. 10.1021/nn300545z [DOI] [PubMed] [Google Scholar]
- Patterson DP, Schwarz B, Waters RS, Gedeon T, & Douglas T (2014). Encapsulation of an Enzyme Cascade within the Bacteriophage P22 Virus-Like Particle. ACS Chemical Biology, 9(2), 359–365. 10.1021/cb4006529 [DOI] [PubMed] [Google Scholar]
- Peng K-W, Vile RG, Cosset F-L, & Russell SJ (1999). Selective transduction of protease-rich tumors by matrix-metalloproteinase-targeted retroviral vectors. Gene Therapy, 6(9), 1552. [DOI] [PubMed] [Google Scholar]
- Pereboeva L, Komarova S, Roth J, Ponnazhagan S, & Curiel D (2007). Targeting EGFR with metabolically biotinylated fiber-mosaic adenovirus. Gene Therapy, 14(8), 627–637. 10.1038/sj.gt.3302916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmutter JD, & Hagan MF (2015). Mechanisms of Virus Assembly. Annual Review of Physical Chemistry, 66(1), 217–239. 10.1146/annurev-physchem-040214-121637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokorski JK, Breitenkamp K, Liepold LO, Qazi S, & Finn MG (2011). Functional Virus-Based Polymer–Protein Nanoparticles by Atom Transfer Radical Polymerization. Journal of the American Chemical Society, 133(24), 9242–9245. 10.1021/ja203286n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnazhagan S, Mahendra G, Kumar S, Thompson JA, & Castillas M Jr. (2002). Conjugate-Based Targeting of Recombinant Adeno-Associated Virus Type 2 Vectors by Using Avidin-Linked Ligands. Journal of Virology, 76(24), 12900–12907. 10.1128/JVI.76.24.12900-12907.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell SK, Khan N, Parker CL, Samulski RJ, Matsushima G, Gray SJ, & McCown TJ (2016). Characterization of a novel adeno-associated viral vector with preferential oligodendrocyte tropism. Gene Therapy, 23(11), 807–814. 10.1038/gt.2016.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevelige PE, Thomas D, & King J (1993). Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophysical Journal, 64(3), 824–835. 10.1016/S0006-3495(93)81443-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulicherla N, Kota P, Dokholyan NV, & Asokan A (2012). Intra- and Inter-Subunit Disulfide Bond Formation Is Nonessential in Adeno-Associated Viral Capsids. PLoS ONE, 7(2). 10.1371/journal.pone.0032163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raillard S, Krebber A, Chen Y, Ness JE, Bermudez E, Trinidad R, … Minshull J (2001). Novel enzyme activities and functional plasticity revealed by recombining highly homologous enzymes. Chemistry & Biology, 8(9), 891–898. 10.1016/S1074-5521(01)00061-8 [DOI] [PubMed] [Google Scholar]
- Räty JK, Airenne KJ, Marttila AT, Marjomäki V, Hytönen VP, Lehtolainen P, … Ylä-Herttuala S (2004). Enhanced gene delivery by avidin-displaying baculovirus. Molecular Therapy: The Journal of the American Society of Gene Therapy, 9(2), 282–291. 10.1016/j.ymthe.2003.11.004 [DOI] [PubMed] [Google Scholar]
- Ren Y, Wong SM, & Lim L-Y (2007). Folic Acid-Conjugated Protein Cages of a Plant Virus: A Novel Delivery Platform for Doxorubicin. Bioconjugate Chemistry, 18(3), 836–843. 10.1021/bc060361p [DOI] [PubMed] [Google Scholar]
- Ren Y, Wong S-M, & Lim L-Y (2006). In vitro-reassembled plant virus-like particles for loading of polyacids. Journal of General Virology, 87(9), 2749–2754. 10.1099/vir.0.81944-0 [DOI] [PubMed] [Google Scholar]
- Robinson TM, Judd J, Ho ML, & Suh J (2016). Role of Tetra Amino Acid Motif Properties on the Function of Protease-Activatable Viral Vectors. ACS Biomaterials Science & Engineering, 2(11), 2026–2033. 10.1021/acsbiomaterials.6b00439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos WH, Bruinsma R, & Wuite GJL (2010). Physical virology. Nature Physics, 6(10), nphys1797 10.1038/nphys1797 [DOI] [Google Scholar]
- Santiago-Ortiz J, Ojala DS, Westesson O, Weinstein JR, Wong SY, Steinsapir A, … Schaffer DV (2015). AAV ancestral reconstruction library enables selection of broadly infectious viral variants. Gene Therapy, 22(12), 934–946. 10.1038/gt.2015.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlick TL, Ding Z, Kovacs EW, & Francis MB (2005). Dual-Surface Modification of the Tobacco Mosaic Virus. Journal of the American Chemical Society, 127(11), 3718–3723. 10.1021/ja046239n [DOI] [PubMed] [Google Scholar]
- Schlicksup CJ, Wang JC-Y, Francis S, Venkatakrishnan B, Turner WW, VanNieuwenhze M, & Zlotnick A (2018). Hepatitis B virus core protein allosteric modulators can distort and disrupt intact capsids. ELife, 7, e31473 10.7554/eLife.31473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider RM, Medvedovska Y, Hartl I, Voelker B, Chadwick MP, Russell SJ, … Buchholz CJ (2003). Directed evolution of retroviruses activatable by tumour-associated matrix metalloproteases. Gene Therapy, 10(16), 1370 10.1038/sj.gt.3302007 [DOI] [PubMed] [Google Scholar]
- Schoonen L, Eising S, van Eldijk MB, Bresseleers J, van der Pijl M, Nolte RJM, … van Hest JCM (2018). Modular, Bioorthogonal Strategy for the Controlled Loading of Cargo into a Protein Nanocage. Bioconjugate Chemistry. 10.1021/acs.bioconjchem.7b00815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoonen L, Maassen S, Nolte RJM, & van Hest JCM (2017). Stabilization of a Virus-Like Particle and Its Application as a Nanoreactor at Physiological Conditions. Biomacromolecules, 18(11), 3492–3497. 10.1021/acs.biomac.7b00640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoonen L, Pille J, Borrmann A, Nolte RJM, & van Hest JCM (2015). Sortase A-Mediated N-Terminal Modification of Cowpea Chlorotic Mottle Virus for Highly Efficient Cargo Loading. Bioconjugate Chemistry, 26(12), 2429–2434. 10.1021/acs.bioconjchem.5b00485 [DOI] [PubMed] [Google Scholar]
- Shukla S, DiFranco NA, Wen AM, Commandeur U, & Steinmetz NF (2015). To Target or Not to Target: Active vs. Passive Tumor Homing of Filamentous Nanoparticles Based on Potato virus X. Cellular and Molecular Bioengineering, 8(3), 433–444. 10.1007/s12195-015-0388-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S, Eber FJ, Nagarajan AS, DiFranco NA, Schmidt N, Wen AM, … Steinmetz NF (2015). The Impact of Aspect Ratio on the Biodistribution and Tumor Homing of Rigid Soft-Matter Nanorods. Advanced Healthcare Materials, 4(6), 874–882. 10.1002/adhm.201400641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, & Zlotnick A (2003). Observed Hysteresis of Virus Capsid Disassembly Is Implicit in Kinetic Models of Assembly. Journal of Biological Chemistry, 278(20), 18249–18255. 10.1074/jbc.M211408200 [DOI] [PubMed] [Google Scholar]
- Smith JS, Keller JR, Lohrey NC, McCauslin CS, Ortiz M, Cowan K, & Spence SE (1999). Redirected infection of directly biotinylated recombinant adenovirus vectors through cell surface receptors and antigens. Proceedings of the National Academy of Sciences of the United States of America, 96(16), 8855–8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RH, Hallwirth CV, Westerman M, Hetherington NA, Tseng Y-S, Cecchini S, … Alexander IE (2016). Germline viral “fossils” guide in silico reconstruction of a mid-Cenozoic era marsupial adeno-associated virus. Scientific Reports, 6 10.1038/srep28965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag F, Bleker S, Leuchs B, Fischer R, & Kleinschmidt JA (2006). Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. Journal of Virology, 80(22), 11040–11054. 10.1128/JVI.01056-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag F, Köther K, Schmidt K, Weghofer M, Raupp C, Nieto K, … Kleinschmidt JA (2011). The assembly-activating protein promotes capsid assembly of different adeno-associated virus serotypes. Journal of Virology, 85(23), 12686–12697. 10.1128/JVI.05359-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soong N-W, Nomura L, Pekrun K, Reed M, Sheppard L, Dawes G, & Stemmer WPC (2000). Molecular breeding of viruses. Nature Genetics, 25(4), 436–439. 10.1038/78132 [DOI] [PubMed] [Google Scholar]
- Stachler MD, Chen I, Ting AY, & Bartlett JS (2008). Site-specific Modification of AAV Vector Particles With Biophysical Probes and Targeting Ligands Using Biotin Ligase. Molecular Therapy, 16(8), 1467–1473. 10.1038/mt.2008.129 [DOI] [PubMed] [Google Scholar]
- Steinmetz NF (2010). Viral nanoparticles as platforms for next-generation therapeutics and imaging devices. Nanomedicine: Nanotechnology, Biology and Medicine, 6(5), 634–641. 10.1016/j.nano.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz NF, Ablack AL, Hickey JL, Ablack J, Manocha B, Mymryk JS, … Lewis JD (2011). Intravital Imaging of Human Prostate Cancer Using Viral Nanoparticles Targeted to Gastrin-Releasing Peptide Receptors. Small (Weinheim an Der Bergstrasse, Germany), 7(12), 1664–1672. 10.1002/smll.201000435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer WP (1994). DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proceedings of the National Academy of Sciences of the United States of America, 91(22), 10747–10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephanopoulos N, Tong GJ, Hsiao SC, & Francis MB (2010). Dual-Surface Modified Virus Capsids for Targeted Delivery of Photodynamic Agents to Cancer Cells. ACS Nano, 4(10), 6014–6020. 10.1021/nn1014769 [DOI] [PubMed] [Google Scholar]
- Suci PA, Berglund DL, Liepold L, Brumfield S, Pitts B, Davison W, … Douglas T (2007). High-Density Targeting of a Viral Multifunctional Nanoplatform to a Pathogenic, Biofilm-Forming Bacterium. Chemistry & Biology, 14(4), 387–398. 10.1016/j.chembiol.2007.02.006 [DOI] [PubMed] [Google Scholar]
- Suci PA, Varpness Z, Gillitzer E, Douglas T, & Young M (2007). Targeting and Photodynamic Killing of a Microbial Pathogen Using Protein Cage Architectures Functionalized with a Photosensitizer. Langmuir, 23(24), 12280–12286. 10.1021/la7021424 [DOI] [PubMed] [Google Scholar]
- Suzuki J, Hashimoto K, Xiao R, Vandenberghe LH, & Liberman MC (2017). Cochlear gene therapy with ancestral AAV in adult mice: complete transduction of inner hair cells without cochlear dysfunction. Scientific Reports, 7 10.1038/srep45524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szécsi J, Drury R, Josserand V, Grange M-P, Boson B, Hartl I, … Verhoeyen E (2006). Targeted retroviral vectors displaying a cleavage site-engineered hemagglutinin (HA) through HA–protease interactions. Molecular Therapy, 14(5), 735–744. 10.1016/j.ymthe.2006.04.007 [DOI] [PubMed] [Google Scholar]
- Tama F, & Brooks CL (2002). The mechanism and pathway of pH induced swelling in cowpea chlorotic mottle virus. Journal of Molecular Biology, 318(3), 733–747. 10.1016/S0022-2836(02)00135-3 [DOI] [PubMed] [Google Scholar]
- Thadani NN, Dempsey C, Zhao J, Vasquez SM, & Suh J (2017). Reprogramming the Activatable Peptide Display Function of Adeno-Associated Virus Nanoparticles. ACS Nano, acsnano.7b07804. 10.1021/acsnano.7b07804 [DOI] [PubMed] [Google Scholar]
- Thomas CE, Storm TA, Huang Z, & Kay MA (2004). Rapid Uncoating of Vector Genomes Is the Key to Efficient Liver Transduction with Pseudotyped Adeno-Associated Virus Vectors. Journal of Virology, 78(6), 3110–3122. 10.1128/JVI.78.6.3110-3122.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong GJ, Hsiao SC, Carrico ZM, & Francis MB (2009). Viral Capsid DNA Aptamer Conjugates as Multivalent Cell Targeting Vehicles. Journal of the American Chemical Society, 131(31), 11174–11178. 10.1021/ja903857f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse LV, Klinc KA, Madigan VJ, Rivera RMC, Wells LF, Havlik LP, … Asokan A (2017). Structure-guided evolution of antigenically distinct adeno-associated virus variants for immune evasion. Proceedings of the National Academy of Sciences, 114(24), E4812–E4821. 10.1073/pnas.1704766114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker BS, & Sumerlin BS (2014). Poly(N-(2-hydroxypropyl) methacrylamide)-based nanotherapeutics. Polym. Chem, 5(5), 1566–1572. 10.1039/C3PY01279D [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetrecht C, Versluis C, Watts NR, Roos WH, Wuite GJL, Wingfield PT, … Heck AJR (2008). High-resolution mass spectrometry of viral assemblies: Molecular composition and stability of dimorphic hepatitis B virus capsids. Proceedings of the National Academy of Sciences, 105(27), 9216–9220. 10.1073/pnas.0800406105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg S, Löfdahl P-Å, Härd T, & Berglund H (2006). Improved solubility of TEV protease by directed evolution. Journal of Biotechnology, 121(3), 291–298. 10.1016/j.jbiotec.2005.08.006 [DOI] [PubMed] [Google Scholar]
- Venkatakrishnan B, Yarbrough J, Domsic J, Bennett A, Bothner B, Kozyreva OG, … Agbandje-McKenna M (2013). Structure and Dynamics of Adeno-Associated Virus Serotype 1 VP1-Unique N-Terminal Domain and Its Role in Capsid Trafficking. Journal of Virology, 87(9), 4974–4984. 10.1128/JVI.02524-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waehler R, Russell SJ, & Curiel DT (2007). Engineering targeted viral vectors for gene therapy. Nature Reviews Genetics, 8(8), 573–587. 10.1038/nrg2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen AM, Ryan MJ, Yang AC, Breitenkamp K, Pokorski JK, & Steinmetz NF (2012). Photodynamic activity of viral nanoparticles conjugated with C60. Chemical Communications, 48(72), 9044–9046. 10.1039/C2CC34695H [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen AM, Wang Y, Jiang K, Hsu GC, Gao H, Lee KL, … Steinmetz NF(2015). Shaping bio-inspired nanotechnologies to target thrombosis for dual optical-magnetic resonance imaging. Journal of Materials Chemistry. B, Materials for Biology and Medicine, 3(29), 6037–6045. 10.1039/C5TB00879D [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wonganan P, & Croyle MA (2010). PEGylated Adenoviruses: From Mice to Monkeys. Viruses, 2(2), 468–502. 10.3390/v2020468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, & Schultz PG (2006). A chemical toolkit for proteins — an expanded genetic code. Nature Reviews Molecular Cell Biology, 7(10), 775 10.1038/nrm2005 [DOI] [PubMed] [Google Scholar]
- Yildiz I, Lee KL, Chen K, Shukla S, & Steinmetz NF (2013). Infusion of imaging and therapeutic molecules into the plant virus-based carrier cowpea mosaic virus: Cargo-loading and delivery. Journal of Controlled Release, 172(2), 568–578. 10.1016/j.jconrel.2013.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Q, Wen H, Wen Q, Chen X, Wang Y, Xuan W, … Wan S (2013). Cucumber mosaic virus as drug delivery vehicle for doxorubicin. Biomaterials, 34(19), 4632–4642. 10.1016/j.biomaterials.2013.03.017 [DOI] [PubMed] [Google Scholar]
- Zhang C, Zhou X, Yao T, Tian Z, & Zhou D (2018). Precision Fluorescent Labeling of an Adeno-Associated Virus Vector to Monitor the Viral Infection Pathway. Biotechnology Journal. 10.1002/biot.201700374 [DOI] [PubMed] [Google Scholar]
- Zhang L, Lua L, L. H., Middelberg J, A. P., Sun, Y., & Connors K, N. (2015). Biomolecular engineering of virus-like particles aided by computational chemistry methods. Chemical Society Reviews, 44(23), 8608–8618. 10.1039/C5CS00526D [DOI] [PubMed] [Google Scholar]
- Zhang W, Xu C, Yin G-Q, Zhang X-E, Wang Q, & Li F (2017). Encapsulation of Inorganic Nanomaterials inside Virus-Based Nanoparticles for Bioimaging. Nanotheranostics, 1(4), 358–368. 10.7150/ntno.21384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinn E, Pacouret S, Khaychuk V, Turunen HT, Carvalho LS, Andres-Mateos E, … Vandenberghe LH (2015). In Silico Reconstruction of the Viral Evolutionary Lineage Yields a Potent Gene Therapy Vector. Cell Reports, 12(6), 1056–1068. 10.1016/j.celrep.2015.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnick A (1994). To build a virus capsid. An equilibrium model of the self assembly of polyhedral protein complexes. Journal of Molecular Biology, 241(1), 59–67. 10.1006/jmbi.1994.1473 [DOI] [PubMed] [Google Scholar]
- Zlotnick A (2005). Theoretical aspects of virus capsid assembly. Journal of Molecular Recognition: JMR, 18(6), 479–490. 10.1002/jmr.754 [DOI] [PubMed] [Google Scholar]
- Zlotnick A, Aldrich R, Johnson JM, Ceres P, & Young MJ (2000). Mechanism of Capsid Assembly for an Icosahedral Plant Virus. Virology, 277(2), 450–456. 10.1006/viro.2000.0619 [DOI] [PubMed] [Google Scholar]
- Zlotnick A, & Fane BA (2010). Chapter 10:Mechanisms of Icosahedral Virus Assembly. In Structural Virology (pp. 180–202). 10.1039/9781849732239-00180 [DOI] [Google Scholar]
- Zlotnick A, Johnson JM, Wingfield PW, Stahl SJ, & Endres D (1999). A Theoretical Model Successfully Identifies Features of Hepatitis B Virus Capsid Assembly. Biochemistry, 38(44), 14644–14652. 10.1021/bi991611a [DOI] [PubMed] [Google Scholar]
- Zlotnick A, Palmer I, Kaufman JD, Stahl SJ, Steven AC, & Wingfield PT (1999). Separation and crystallization of T = 3 and T = 4 icosahedral complexes of the hepatitis B virus core protein. Acta Crystallographica. Section D, Biological Crystallography, 55(Pt 3), 717–720. [DOI] [PubMed] [Google Scholar]