Abstract
Pericyclic reactions are a distinct class of reactions that have wide synthetic utility. Before the recent discoveries described in this review, enzyme-catalyzed pericyclic reactions were not widely known to be involved in biosynthesis. This situation is changing rapidly. We define the scope of pericyclic reactions, give a historical account of their discoveries as biosynthetic reactions, and provide evidence that there are many enzymes in Nature that catalyze pericyclic reactions. These enzymes, the “pericyclases,” are the subject of this review.
Graphical Abstract

1. Introduction
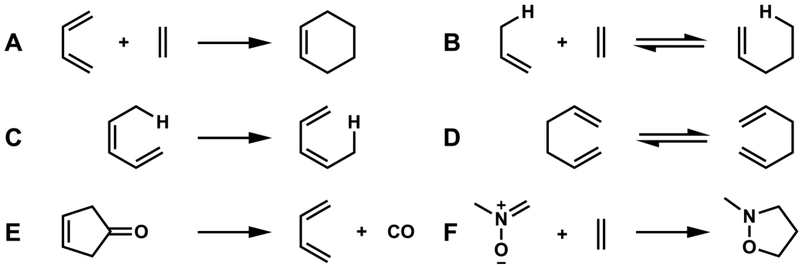
Woodward and Hoffmann introduced the concept of pericyclic reactions in their classic book, “The Conservation of Orbital Symmetry” that was first published in Angewandte Chemie in 1969.1 Following the definitions of concerted electrocyclic reactions, cycloadditions, sigmatropic shifts, cheletropic reactions and related processes (Figure 1), Woodward and Hoffmann rationalized previously puzzling experimental data based on what they named the Principle of Conservation of Orbital Symmetry. Woodward and Hoffmann also defined pericyclic reactions: “reactions in which all first-order changes in bonding relationships take place in concert on a closed curve.”2
Figure 1.
. A. Cycloaddition (Diels-Alder reaction). B. Alder-ene reaction (a Miscellaneozation). C. 1,5-Sigmatropic hydrogen shift, D. [3,3]-Sigmatropic shift (Cope rearrangement). E. Cheletropic reaction. F. A 1,3-dipolar cycloaddition, a hetero-pericyclic reaction.
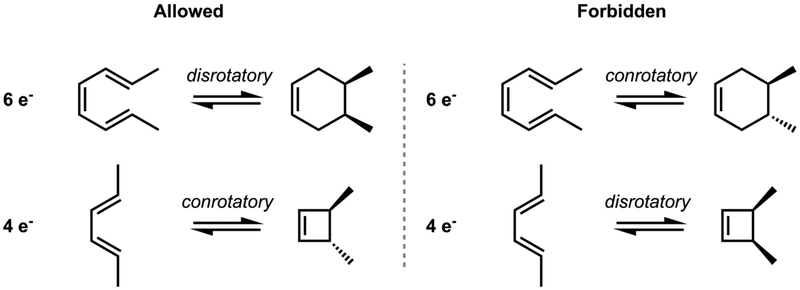
The examples in Figure 1 are all of unsubstituted systems with six electrons involved in bonding changes, while the other orbitals have only minor perturbations such as rehybridization. For each reaction, there are numerous examples of substituted cases. These are examples of hydrocarbon reactions, or of a ketone in Figure 1E, and Figure 1F is another example of a vast number of pericyclic reactions involving heteroatoms. Substitution influences activation barriers and sometimes even the mechanism, but the basic theoretical conclusion that these reactions are “allowed” to be pericyclic does not change. In each case, there are stereochemical restrictions on allowed pericyclic transition states, and the number of electrons is also important. For example, Figure 2 shows allowed and forbidden six-electron and four-electron electrocyclic reactions, and how the stereochemistry changes with the number of electrons involved.
Figure 2.
Allowed and forbidden electrocyclic reactions involving 6 and 4 electrons. In each case, only the allowed pathways are observed. Conrotatory, both termini rotate in the same direction; disrotatory, two termini rotate in the opposite direction.
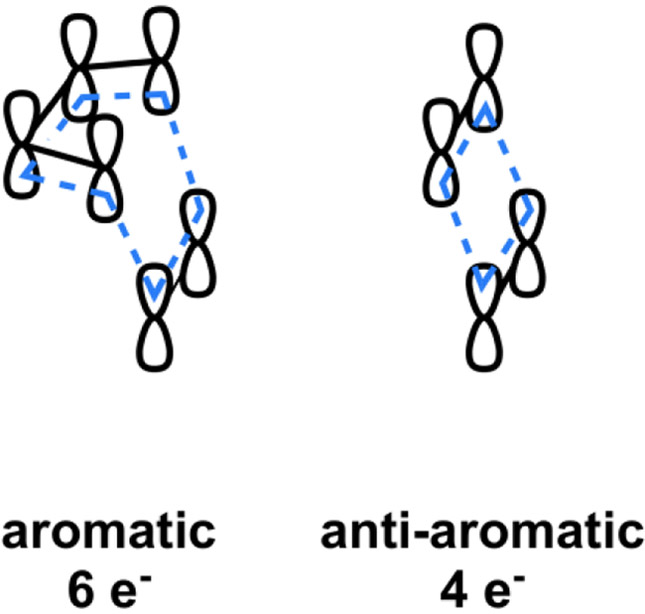
The selection rules were derived in various ways, including consideration of frontier molecular orbital interactions or the use of orbital correlation diagrams showing the behavior of orbitals along a reaction pathway. The simplest approach to understand what causes an allowed reaction to be favored, and forbidden to be disfavored, is to consider the aromaticity or anti-aromaticity of the transition state (Figure 3). The Diels-Alder transition state, like benzene, has a cyclic array of 6 electrons, while the [2+2] concerted transition state has a cyclic array of 4 electrons, like the anti-aromatic cyclobutadiene. Aromatic molecules (and transition states) are highly stabilized, more stable than linear conjugated systems (or acyclic transition states), while anti-aromatic molecules are destabilized.
Figure 3.
Schematic representation of the aromaticity of the allowed Diels-Alder transition state and anti-aromaticity of the [2+2] cycloaddition transition state.
2. Mechanistic Considerations: What is a Pericyclase?
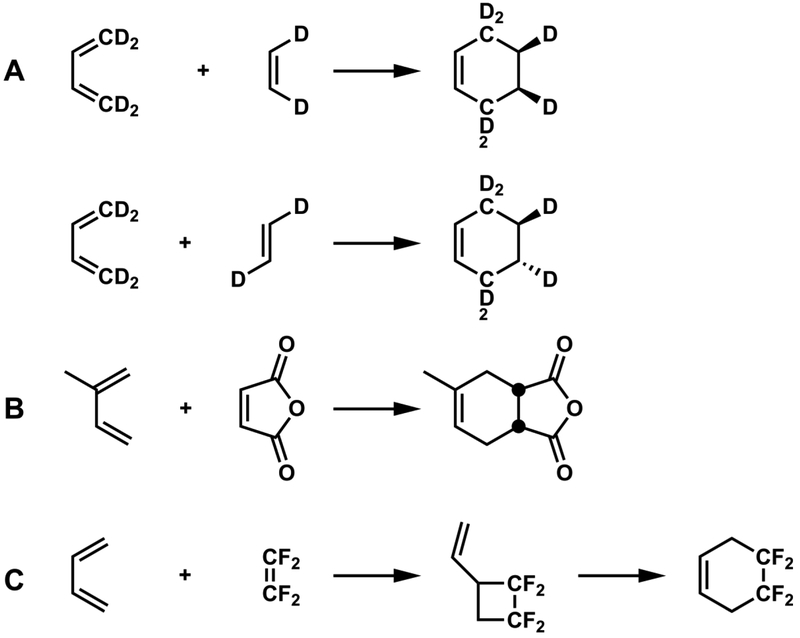
Some reactions that are allowed to be concerted pericyclic processes can occur by other mechanisms. For example, while most Diels-Alder reactions are concerted, there are some that, by virtue of substitution, occur by stepwise mechanisms (Figure 4). The definition of “concerted” has also been the subject of discussion and has evolved. A concerted reaction was originally taken to mean a reaction in which all bonding changes occur in concert in one step, while a stepwise reaction occurs in several steps involving one or more intermediates.
Figure 4.
Diels-Alder reactions: Concerted proven by (A) stereospecificity3 and (B) kinetic isotope effects.4 Stepwise proven by (C) identification of an intermediate cyclobutane.5
When reliable quantum mechanically computed transition geometries became available in the 1990s, it became more apparent that concerted reactions could involve symmetrical (synchronous) or unsymmetrical (asynchronous) bond formation. Baldwin defined “bonding concerted” and “bonding stepwise” based on whether several bonds are formed all at once or sequentially.6 Our group defined “dynamically concerted” as reactions in which both bonds are formed with a time gap of <60 femtoseconds (fs). This quantity, 60fs, is the time of passage over a potential energy maximum in Eyring’s transition state theory and is a time comparable to the lifetime of a CC vibration (~ 30 fs).7 Dynamically stepwise reactions are energetically concerted with only one potential energy maximum but with a bonding time gap greater than 60 fs. Such reactions are found on flat potential energy surfaces as found for several Diels-Alderases discussed later. Dynamically concerted and dynamically stepwise reactions can both be pericyclic when the rate-determining TS has partial bonding on a “closed curve”.
We define pericyclases as those enzymes that catalyze pericyclic reactions, reactions that have transition states with bonds partially made or broken in a cyclic array. It is this cyclic array of bonding changes in the rate-determining transition state that is special for a pericyclic reaction, rather than the detailed shape of the potential surface. A time gap between formation of the two new bonds of <60 fs is the definition of dynamically concerted, but we consider reactions as pericyclic as long as the reaction is energetically concerted, with a rate-determining TS that has a cyclic array of bonding interactions that eventually lead to products.
3. Synthetically Useful Pericyclic Reactions
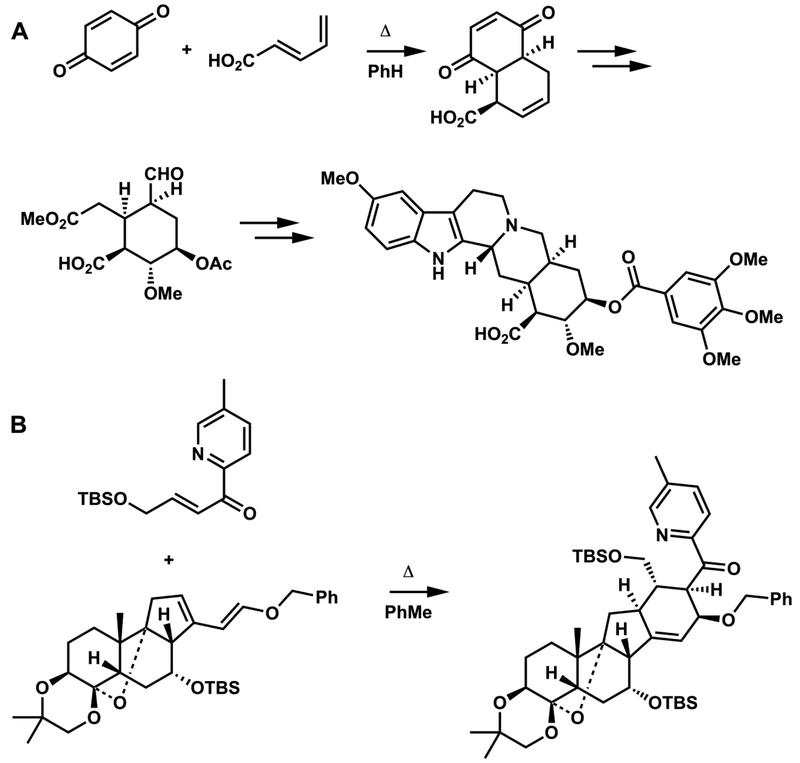
The Diels-Alder reaction has been applied to many elegant syntheses. Woodward’s reserpine synthesis involved a first step where three new stereocenters are formed stereoselectively. A double bond in the product served as the locus of two additional stereocenters. (Figure 5A).8 This synthesis has long been considered a classic of “substrate-controlled” stereoselective synthesis.9
Figure 5.
Diels-Alder steps in (A) Woodward’s reserpine synthesis (1956) and (B) Stork’s approach to the synthesis of 4-methylenegermine (2017).
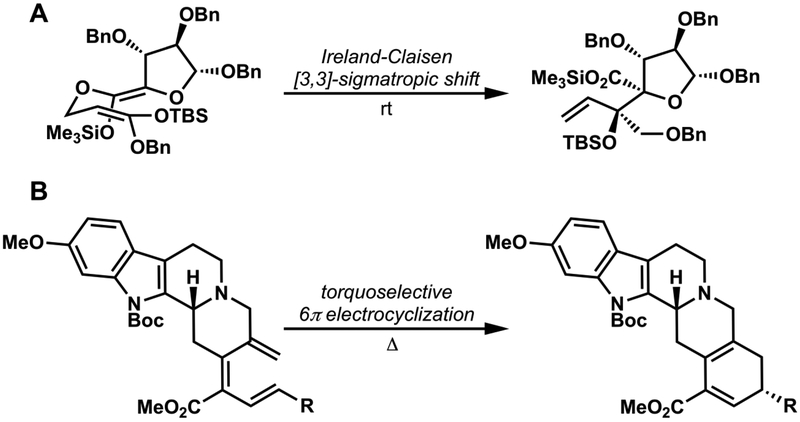
Many Diels-Alder reactions are included in Nicolaou and Sorensen’s “Classics in Organic Synthesis”.9 In Gilbert Stork’s last paper, published shortly before he died in 2017, a complicated Diels-Alder reaction was used to create a cyclohexene with complete control of stereochemistry, both exo/endo and facial selectivity (Figure 5B), leading to four new stereocenters, properly fixed with respect to the pre-existing stereocenters in the diene.10 Other types of pericyclic reaction are also commonly used in synthesis, such as the Ireland-Claisen rearrangement. Figure 6 shows this [3,3]-sigmatropic shift used in Rizzacasa’s (+)-zaragozic acid C synthesis from Zakarian’s review on the use of these reactions in synthesis11 and an electrocyclization used in Kwon’s approach to the synthesis of reserpine,12 respectively. The Ang Li group at SIOC has developed a general synthetic route to highly substituted aromatics, involving the electrocyclic reactions of hexatrienes. His group has used this electrocyclization to good advantage for the synthesis of a number of daphniphyllium alkaloids.13,14 The Maulide group in Austria has harnessed stereoselective cyclobutene ring-opening reactions for the synthesis of diene units of various macrolides.15 It is fair to single out pericyclic reactions as principle weapons in the arsenal of synthetic strategians to conquer complex stereochemical issues.
Figure 6.
Examples of (A) a [3,3]-sigmatropic shift11 and (B) an electrocyclization12 involved in synthesis.
4. The Emergence of Diels-Alderases
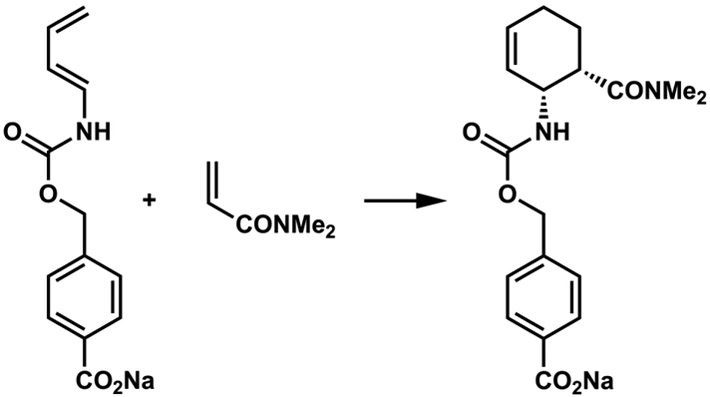
The first authenticated Diels-Alderases might be considered to be catalytic antibodies and designed enzymes, since these proteins are indeed catalysts of pericyclic reactions. The designed unnatural enzyme by Baker and Houk, which catalyzes the reaction shown in Figure 7, is often highlighted as the first designed enzyme that catalyzes a bimolecular Diels-Alder reaction.16 However, this reaction had been catalyzed earlier by a catalytic antibody produced in response to inoculation with a transition state analog, by the Lerner and Houk groups.17 Indeed, a number of groups have developed antibodies that catalyze a variety of Diels-Alder reactions.18,19
Figure 7.
The Diels-Alder reaction catalyzed by a catalytic antibody17 and later by a computationally designed enzyme.16
The groups of Baker and Houk used the computational “inside-out” approach to design the enzyme for the reaction in Figure 7 that is both stereospecific and accelerates the reaction 100-fold with respect to the reaction in water.16 Fold-It players on Baker’s on-line game improved this further by designing a loop that improves substrate binding,20 while Hilvert improved catalysis even further by directed evolution.21
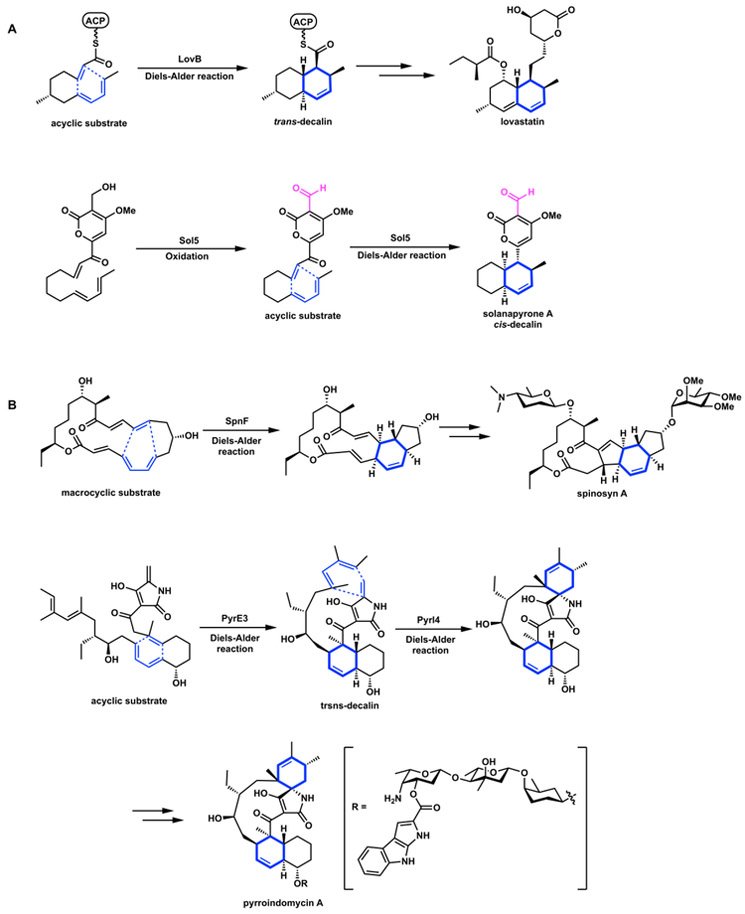
There are many natural products containing cyclohexenes or substituted cyclohexanes, and there has been much speculation that there might be Diels-Alderases in Nature.22 Three of these Diels-Alderases, macrophomate synthase, lovastatin nonaketide synthase (LovB) and solanapyrone synthase (Sol5) were reported during late 1990s to early 2000s.23–27 LovB and Sol5 catalyze the intramolecular Diels-Alder reactions that form trans- and cis-decalin scaffolds (Figure 9A), respectively. These enzymes catalyze the formation of the acyclic precursor, as well as the subsequent regioselective and stereoselective Diels-Alder reactions. Since LovB and Sol5 have additional activities other than those of Diels-Alderases, it has been difficult experimentally to prove that these enzymes catalyze the cycloaddition events. For example, although LovB has been shown to be required to afford the desired decalin stereoisomer in dihydromonacolin L,26 the exact domain of the megasynthase that catalyzes this reaction has not been identified.
Figure 9.
Examples of enzyme-catalyzed Diels-Alder reaction. (A) Multifunctional enzymes that have Diels-Alderases activities, LovB and Sol5. (B) Stand-alone Diels-Alderases, SpnF from spinosyn A biosynthesis, and PyrE3 and Pyl4 from pyrroindomycin A biosynthesis.
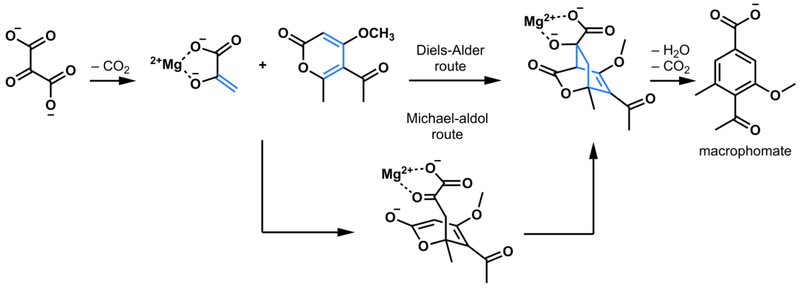
Perhaps the first Diels-Alderase was proposed by Watanabe et al. in 2000, who provided evidence for the multistep catalysis by macrophomate synthase, including formation of a Diels-Alder adduct as an intermediate followed by decarboxylation and dehydration to form macrophomate (Figure 8).27
Figure 8.
Reaction scheme comparing the concerted and stepwise Diels-Alder route to macrophomate catalyzed by macrophomate synthase27
A crystal structure of the multifunctional macrophomate synthase was obtained by Ose and Watanabe et al. in 2003 and 2004,28 which allowed Jorgensen et al. to carry out a detailed computational-mechanistic study that showed the Diels-Alder reaction occurs in two distinct steps, Michael addition to give a stable intermediate, and a subsequent aldol reaction to give a less stable intermediate that is then decarboxylated and dehydrated by the enzyme (Figure 8).29 Because of the stepwise nature of this process, Jorgensen concluded that this was not a Diels-Alderase. This finding received publicity in the chemical press,30 and is widely quoted. According to our definitions, however, macrophomate synthase is a Diels-Alderase, since it does catalyze the reaction of a diene and an alkene to form a cyclohexene; overall a Diels-Alder reaction occurs. However, in line with Jorgensen’s finding, macrophomate synthase is not a pericyclase, since the mechanism has been clearly established by Jorgensen et al. to be a stepwise Michael-aldol reaction, not a pericyclic reaction.
In 2011, Liu and co-workers reported the characterization of Diels–Alderase SpnF from insecticide spinosyn A biosynthetic gene cluster.31 This is widely touted as the first enzyme that catalyzes only the Diels-Alder cycloaddition, rather than synthesis of the precursor and (perhaps) the cycloaddition itself, as often is the case. Although SpnF was originally annotated by primary sequence comparisons to be an apparent S-adenosyl-L-methionine (SAM)-dependent methyltransferase, similar to LepI that is described later, SpnF indeed catalyzes and accelerates the rate of Diels–Alder conversion of an α,β,γ,δ-unsaturated macrolactone precursor to the tricyclic cyclohexene-containing product by approximately 500-fold (Figure 9B).31
After the discovery of SpnF, the advancement of genome sequencing and searching tools for secondary metabolite gene clusters has led to the identification of numerous stand-alone Diels-Alderases, especially from bacterial biosynthesis pathways. For example, the pyrroindomycin biosynthetic pathway involves two sequentially unrelated enzymes acting consecutively to perform tandem Diels-Alder reactions, a decalin-forming Diels-Alder reaction by PyrE3 and then a spirotetronate-forming Diels-Alder reaction by Pyrl4 enzyme (Figure 9B).32 Gene encoding Pyrl4 homologues VstJ,33 and AbyU34 are also found in the biosynthetic gene clusters of other spirotetronate-containing natural products.
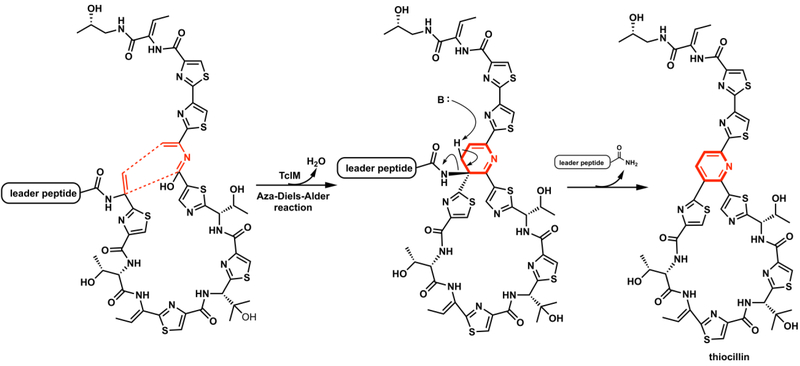
Recently, an enzymatic aza-Diels-Alder reaction was confirmed in the final step of thiazolyl peptide thiocillin biosynthesis.31 The single enzyme TclM, originally annotated as a dehydratase, was found to catalyze the formation of the trisubstituted pyridine core of thiocillin, through a Diels-Alder reaction between dihydroalanine residues (Figure 10). In the last two years, there have been several reviews of the many new Diels-Alderases identified in Nature.36–39
Figure 10.
Enzymatic intramolecular hetero Diels–Alder reaction in formation of thiocillin.
5. Recently Discovered Diels-Alderases and Ambimodal Pericyclases
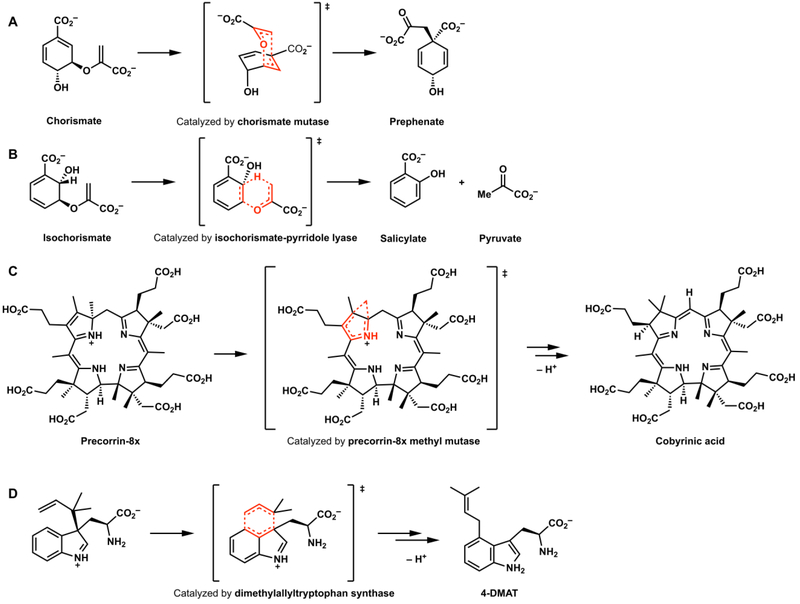
There have been a few pericyclases discovered that catalyze pericyclic processes other than the Diels-Alder reaction. Chorismate mutase is now recognized as the first documented “pericyclase”; this enzyme catalyzes the Claisen rearrangement of chorismate to prephenate in primary metabolism (Figure 10A).40 Tantillo has characterized enzymatic carbocation [1,2]-shifts in natural product biosynthesis.41 Isochorismate-pyruvate lyase,42,43 precorrin-8x methyl mutase,44 and dimethylallyltryptophan synthase45 were later reported and proposed to involve pericyclic reactions (Figure 11B–11D). In the past few years, multiple putative pericyclase-catalyzed 1,3-dipolar cycloadditions have been proposed.46
Figure 11.
A. The Claisen rearrangement ([3,3]-sigmatropic shift) catalyzed by chorismate mutase; reactions putatively catalyzed by (B) isochorismate-pyrridole lyase, (C) precorrin-8x methyl mutase, and (D) dimethylallyltryptophane synthase.
In this section, we review some of the recently discovered enzymes from our labs and others that catalyze prototypical pericyclic reactions. These involve primarily cycloaddition and [3,3]-sigmatropic rearrangement reactions. We have discovered that some of these enzymes involve “ambimodal” transition states, where a single transition state connects with multiple products via a post-transition state bifurcation.47
In addition to the Diels-Alderases described above, both pericyclic and stepwise, a number of additional Diels-Alderases have been discovered recently in our labs. These are described here.
5.1. MycB; Another Decalin Forming Diels-Alderase
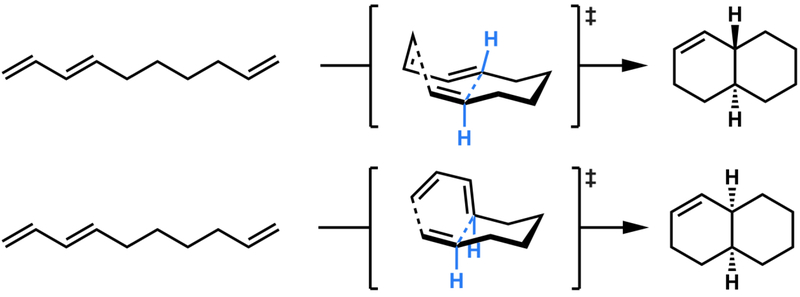
The decalin motif is frequently observed in natural products produced by bacteria and fungi.38 It has been found to involve an intramolecular Diels-Alder reaction, the simplest example of which is shown in Figure 12. Substituted versions of the decatriene backbone motif are synthesized from polyketide synthases (PKSs), especially bacterial multimodular and fungal iterative PKSs.49 In organic synthesis, decalins can be synthesized from intramolecular Diels-Alder reactions (IMDA). The parent system, studied both experimentally and theoretically, shows no significant stereoselectivity, but substitution can alter the intrinsic preference.50
Figure 12.
Intramolecular Diels-Alder reactions to form cis- and trans-octahydrodecalins.46
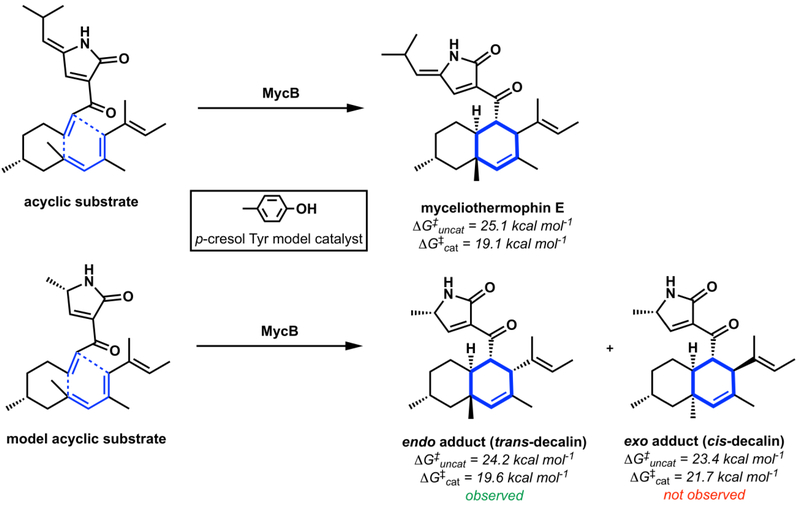
Decalins are often derived in biosynthesis from acyclic polyketide precursors. During PKS-catalyzed synthesis of acyclic precursors, programming rules of the PKSs strategically leave double bonds unreduced in the polyketide chains at precise positions to generate a diene and a dienophile interrupted by four contiguous sp3 carbons; the decatriene units can undergo Diels-Alder cyclizations.36,37,48 Indeed, the hypothesis is that Diels-Alder reactions are involved for many, if not all, of the decalins formed in biosynthesis. Bioinformatics analysis and genetic evidence have suggested that a class of lipocalin-like enzymes such as CghA and Fsa2 may be involved in the formation of the decalin ring systems of Sch210972 and equisetin, respectively.51 In vitro characterization of these enzymes using acyclic substrates was unsuccessful, partly due to the inability to capture these substrates, which are prone to undergo spontaneous uncatalyzed cycloaddition reactions to form a mixture of decalin stereoisomers.52,53 In late 2016, we serendipitously captured an overly oxidized acyclic substrate for lipocalin-like enzyme MycB from the myceliothermophins biosynthetic gene cluster.54 Myceliothermophins, including myceliothermophin A and E, are cytotoxic compounds isolated from the thermophilic fungus Myceliophthora thermophilia.55 In vitro characterization of MycB using the acyclic substrate showed that MycB is responsible for the formation of the trans-decalin scaffold from the substrate (Figure 13A). The uncatalyzed Diels-Alder reaction of the acyclic substrate is predicted to be very slow with ΔGuncat⧧ = 25.1 kcal/mol, which would lead to a rate constant of ∼10−5 s−1 at room temperature according to transition state theory. In contrast, the spontaneous Diels-Alder reaction in the biosynthesis of Sch210972 is very fast, with ΔGuncat⧧ ~ 12 kcal/mol, which accounts for our inability to isolate the acyclic substrate for CghA.55 Quantum mechanical calculations on a model substrate for the MycB reaction predicted that the uncatalyzed reactions will produce a mixture of diastereomers with the unobserved cis-adduct being the major isomer (Figure 13B). This occurs because the exo-transition state (cis-decalin) barrier is lower than the endo (trans-decalin). MycB must stabilize the thermodynamically unfavorable endo transition state in the active site to facilitate the exclusive formation of the trans-decalin structure. Calculations performed with p-cresol to mimic the effects of a catalytic tyrosine residue in the enzyme active site, predict the endo adduct will be favored. Our calculations showed that the endo transition state of MycB-catalyzed cyclization is asynchronous but concerted,7 indicating that MycB is a pericyclase that catalyzes the IMDA reaction.
Figure 13.
The MycB-catalyzed Diels-Alder reaction with calculated free energy barriers for spontaneous and model-catalyzed reactions.
5.2. SpnF; ambimodal [4+2]/[6+4]-cycloaddition and Cope rearrangement
As mentioned previously, SpnF is the first characterized monofunctional Diels-Alderase discovered in Nature.31 SpnF accelerates a nonenzymatic Diels-Alder cycloaddition reaction of the macrocyclic precursor by 500-fold to form the 5,6-bicyclic system during the biosynthesis of spinosyn A. In Smentek and Hess’ theoretical study, the SpnF-catalyzed nonenzymatic cycloaddition was described as highly asynchronous.56 Liu and coworkers discussed the possibility that the catalytic cycle involves a concerted pericyclic or alternative stepwise cyclization mechanism, but no direct evidence for mechanism was obtained in the original experimental studies.31
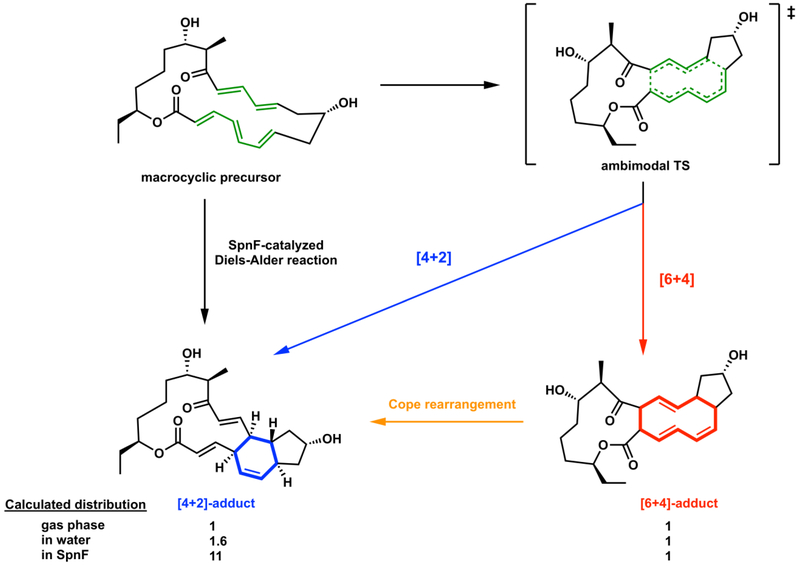
Recently, extensive quantum mechanical computations and dynamic simulations by our group showed that the transition state for the nonenzymatic reaction is an ambimodal transition state that leads directly to the observed Diels−Alder and to an unobserved [6+4]-cycloadduct (Figure 14).57,58 The unobserved [6+4]-adduct has higher free energy than the Diels-Alder adduct and is predicted to readily convert to the Diels-Alder adduct via a Cope rearrangement. We predict that both [4+2] and [6+4] adducts are formed via a bis-pericyclic59 ambimodal transition state, but the [6+4] adduct is rapidly converted to the observed [4+2] Diels-Alder adduct.57 Dynamics simulations indicate that both dynamically concerted and dynamically stepwise trajectories occur from the single ambimodal transition state.58 Since the single transition state contains a cyclic array of breaking and forming bonds, these are indeed pericyclic reactions. We have also developed an environment-perturbed transition-state sampling (EPTSS) method based on QM/MM molecular dynamics. The EPTSS method makes it possible to understand the role of solvent or SpnF enzyme structure on control of the reaction pathways.58 The EPTSS method has also been used for free-energy calculations and kinetic isotope effect calculations in solvents and enzymes. Molecular dynamics (MD) simulations show that trajectories passing through the ambimodal transition state subsequently bifurcate to the [6+4]-adduct and the Diels–Alder adduct with a ratio of 1:1 in the gas phase, 1:1.6 in water, and 1:11 in the enzyme (Figure 14).58 From gas phase to water, a trend toward [4+2]-adduct was observed, indicating that increase in solvent polarity promotes the formation of the [4+2]-adduct over the [6+4]-adduct. The enzyme SpnF alters the ambimodal transition state geometry and the post-TS bifurcation dynamics in the active site by perturbing the energy surface to favor formation of the [4+2]-adduct. We are still investigating if SpnF catalyzes the Cope rearrangement of the [6+4]-byproduct to [4+2]-adduct, but this seems likely due to the similarities of the cycloaddition and Cope transition states.
Figure 14.
SpnF-catalyzed transannular cycloaddition reactions of the macrocyclic precursor via the single ambimodal transition state to form the [4+2] and [6+4] adducts. Ratios given are from MD simulations, although the rapid Cope rearrangement converts the [6+4] adduct to the more stable Diels-Alder adduct for thermodynamic reasons.
5.3. LepI; ambimodal DA/HDA and retro-Claisen rearrangement
The inverse-electron-demand hetero-Diels Alder reaction has been proposed as a key biotransformation to give a dihydropyran, which is a frequent structural feature in natural products including cytotoxic leporin B.61–64 Our groups recently studied the leporin B biosynthetic pathway and discovered a new pericyclase that can catalyze another important pericyclic reaction, the hetero-Diels-Alder (HDA) reaction. The synthesis of the dihydropyran core in leporin without the enzyme involves the E/Z geometric mixture of the unstable ο-quinone methide intermediate generated from the dehydration of a 2-pyridone alcohol precursor.65 The uncatalyzed process gives a mixture of only minor amounts of the naturally formed HDA adduct leporin C and mostly other regio- and stereoisomeric IMDA and HDA adducts that are not observed in the catalyzed process. Therefore, an enzyme must be encoded in the leporin biosynthetic gene cluster to catalyze the HDA reaction in a stereoselective fashion and to suppress the IMDA reaction to afford the dihydropyran core in leporin.
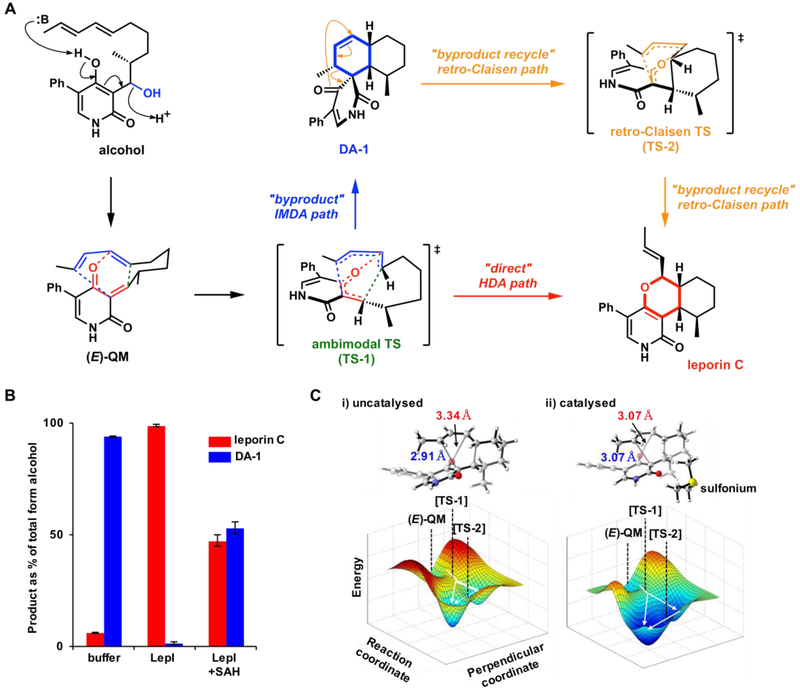
Although the biosynthetic gene cluster of leporin B was reported by Cary and coworkers, no candidate enzyme encoded in the cluster that could catalyze the HDA reaction was evident in the first pass.61 Our investigation of dihydropyran core construction led to the discovery of a multifunctional pericyclase LepI that is solely responsible for the pyran formation.66 This protein was annotated as a SAM-dependent O-methyltransferase, but we found that it catalyzes the stereoselective dehydration of the alcohol to (E)-quinone methide (QM) and then catalyzes three pericyclic transformations: IMDA and HDA via a single ambimodal transition state,47 and also a retro-Claisen rearrangement (Figure 15). The HDA reaction catalyzed by LepI to generate the pyran product leporin C is the most direct pathway to go from (E)-QM to leporin C. Alternatively, the enzyme-bound (E)-QM can also undergo a competing IMDA cyclization to give a spirobicyclic product that can be released from the enzyme. This IMDA reaction and the HDA reactions are in competition, indicated by computational results to occur from a single ambimodal transition state (Figure 15A, TS-1). Periselectivity is therefore controlled by the post-transition state bifurcation of the ambimodal transition state. In the case of the IMDA route, the spirobicyclic intermediate can then be recaptured by LepI and converted to the final product leporin C by a retro-Claisen rearrangement, the first enzymatic example of this type of pericyclic reaction. The bifurcating fate of the initially dehydrated quinone methide intermediate thus reveals three pericyclic transformations that take place in the LepI active site.
Figure 15.
A. Reactions catalyzed by LepI in leporin C biosynthesis. B. Summary of cascade of LepI-catalyzed reactions. C. Ambimodal transition state structures and asymmetrical bifurcating PES for the formation of leporin C and DA-1 from (E)-QM.
Molecular dynamics simulations were performed on the spontaneous reaction and a simple electrostatic catalysis trimethylsulfonium ion model, to probe possible modes of catalysis by SAM. The sulfonium catalysis model switches reaction periselectivity to favor leporin C formation; switching HDA:IMDA ratios of 0:100 (spontaneous) to 83:17 (catalyzed). Furthermore, the sulfonium model lowers the retro-Claisen barrier by 2.4 kcal mol−1 (100-fold rate acceleration). LepI-catalyzed pericyclic reactions are the first well-documented examples of SAM-catalyzed pericyclic reactions.
5.4. Stig cyclase
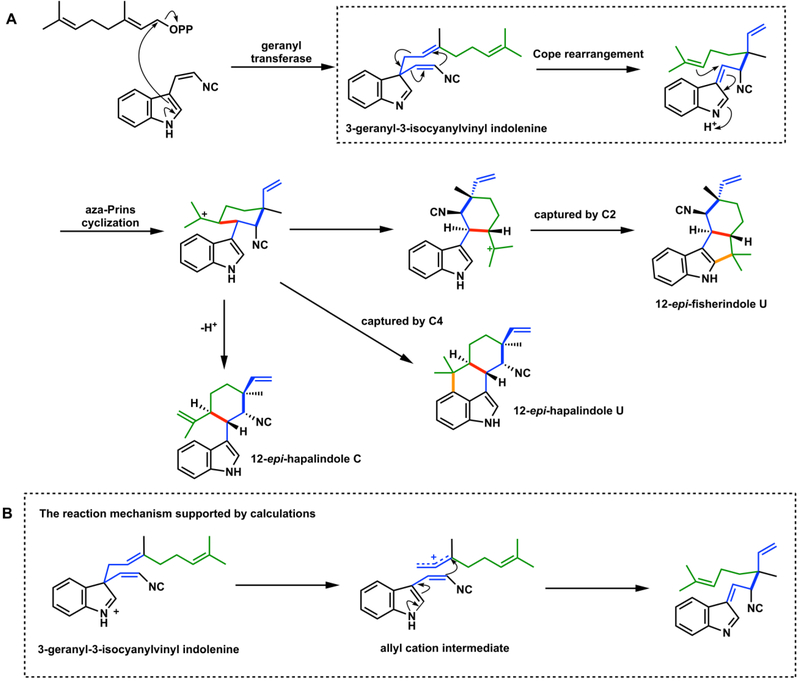
The Cope rearrangement is another very common [3,3]-sigmatropic rearrangement, frequently used in synthesis.67,68 Until recently, there has not been mechanistic evidence of an enzymatic Cope rearrangement using a purified enzyme. Both the Sherman and Liu groups reported that an enzymatic Cope rearrangement occurs at an early step in the formation of a set of prenylated cyanobacterial indole monoterpene scaffolds, including hapalindoles, fischerindoles, ambiguines, and welwitindoles.69,70,72 Interestingly, the formation of polycyclic scaffolds of the hapalindole family are catalyzed by a related set of calcium dependent dimeric cycloisomerases, namely the Stig cyclases from Stigonentales cyanobacteria. The Stig cyclases are proposed to perform the Cope rearrangement of the 3-geranyl-3-isocyanylvinyl indolenine, followed by aza-Prins cyclization and a proposed three-way partition to product families shown in Figure 16.
Figure 16.
A. The Cope rearrangement involved in hapalindole and fischerindole biosynthesis. B. The likely acid-catalyzed stepwise (nonpericyclic) mechanism.70,72
Theoretical studies by our group indicate that the pericyclic Cope rearrangement mechanism is accelerated by hydrogen-bonding groups,71 while full protonation of the indoline will cause the mechanism to change to a stepwise dissociative process involving an allyl cation intermediate (Figure 16B).69 The mechanism of this reaction has not been established experimentally, so it is not certain if this enzyme is a true pericyclase or catalyzes a dissociation and recombination of a protonated intermediate. This mechanistic detail is surprisingly difficult to experimentally tease out. Similar issues have been raised for prenylation of lysergic acid or enzyme-catalyzed formation of 4-dimethylallyl tryptophan (4-DMAT). These reactions can proceed directly by C4 prenylation or indirectly via prenylation at the more nucleophilic C3-position followed by a Cope rearrangement, deprotonation and rearomatization.87 This is the putative Cope pathway for catalytic formation of 4-DMAT by 4-DMAT synthase.87 Therefore, the discovery of a true pericyclase that catalyzes Cope rearrangement remains an attractive topic for discovery.
6. Other Pericyclases to be Discovered in Nature
While enzyme catalyzed Diels-Alder reactions have been an attractive target for chemists due to the prominent usefulness in organic synthesis fields25,26,31,51,52,74,75, other pericyclic reactions such as inverse electron demand Diels-Alder reaction (IEDDA),76,77 Alder-ene reaction,78,79 [3,3]-sigmatropic rearrangement81,82 and electrocyclic reactions9,83 are also widely utilized in synthetic chemistry. Given this and our recent discoveries of enzymatic ambimodal pericyclic reactions and retro-Claisen rearrangement,66 we expect that more pericyclic biosynthetic enzymatic transformations remain to be discovered in naturally occurring enzymes.
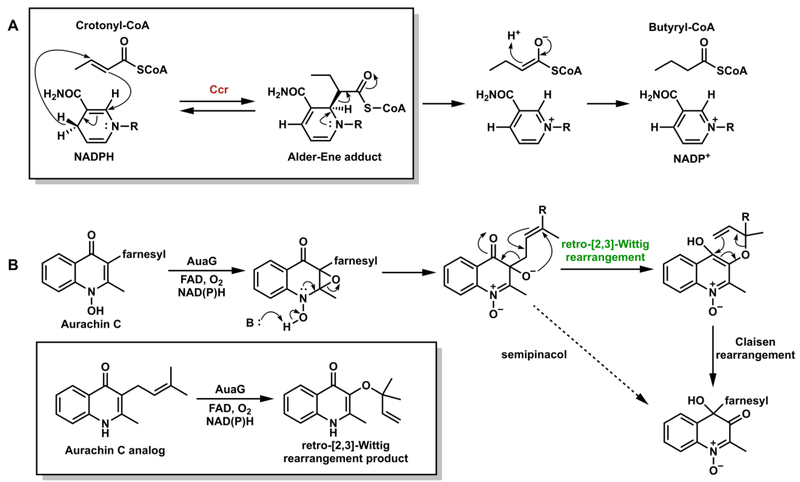
Recently, Erb and coworkers reported the identification of a transient covalent ene intermediate between NADPH and the acyl-CoA thioester during the catalysis of crotonyl-CoA carboxylase/reductase (Ccr, Figure 17A).84 They proposed that the ene adduct is formed through an intermolecular electrocyclic Alder-ene reaction mechanism; this was supported by structure of a Ccr homolog cocrystallized with NADP+ and its 2-enoyl-CoA substrate. Nay and co-workers proposed that flavin-dependent monooxygenase AuaG catalyzes the tandem retro-[2,3]-Wittig rearrangement/Claisen rearrangement in addition to the epoxidation during the aurachin B biosynthesis (Figure 17B).85 Although it still remains unclear whether the reaction mechanisms of Ccr and AuaG are indeed pericyclic mechanisms or alternative stepwise mechanism, these enzymes certainly provide new opportunities for pericyclases in Nature.
Figure 17.
Proposed pericyclic (A) Alder-ene reaction in catalysis of Ccr;84 (B) retro-[2,3]-Wittig and Claisen rearrangements catalyzed by AuaG.85
The discovery of SpnF and LepI encourages us to find other plausible intramolecular ambimodal pericyclic reactions. For example, intramolecular ambimodal pericyclic reactions would occur in substrates bearing two dienolic components as seen in LepI-catalyzed reactions, which can undergo formal Diels-Alder or inverse electron demand Diels-Alder reactions.86 Intermediates in natural product biosynthetic pathways that have this structural hallmark are likely to contain pericyclases that can catalyze ambimodal pericyclic reactions.
7. Conclusion and Outlook
The number of known enzymes that can catalyze pericyclic reactions rapidly increased in the past five years.29,30,87 There will always be questions about whether the formal pericyclic reaction is truly concerted or not, and this provides a challenging area for experimental and theoretical mechanistic analysis.31 Regardless of detailed mechanism, an enzyme that catalyzes a Diels-Alder reaction or Cope rearrangement is a “Diels-Alderase” or “Copease.” We have described here that it is correct to consider a reaction in which the rate-determining transition state has a cyclic array of bonding interactions as a pericyclic reaction. This fits the original Woodward-Hoffmann definition and takes into account the possibility of a whole spectrum of asynchronicities, including those that include “entropic intermediates” with lifetimes of hundreds of femtoseconds. Pericyclases catalyze reactions that do not have long-lived observable intermediates. The Woodward-Hoffmann rules govern the possible products of such reactions. Genomic and bioinformatic analysis will continue to be a fruitful approach to the discovery of new pericyclases in cryptic biosynthetic pathways.
Acknowledgements
We are grateful to the National Science Foundation (NSF CHE-1806581) and the National Institutes of General Medical Sciences, National Institutes of Health (GM118056) for financial support of this research.
Footnotes
Conflicts of interest
There are no conflicts to declare.
References
- 1. a).Hoffmann R and Woodward RB, Acc. Chem. Res, 1968, 1, 17–22. [Google Scholar]; b) Woodward RBand Hoffmann R, Angew. Chem. Int. Ed. Engl 1969, 8, 781; Verlag Chemie BmgH, Academic Press, 1970. [Google Scholar]
- 2.Ref. 1, page 169.
- 3.Houk KN, Lin Y-T, and Brown FK, J. Am. Chem. Soc, 1986, 108, 554. [DOI] [PubMed] [Google Scholar]
- 4.Beno BR, Houk KN, and Singleton DA, J. Am. Chem. Soc, 1996, 118, 9984–9985. [Google Scholar]
- 5.Getty SJ and Borden WT, J. Am. Chem. Soc, 113, 4334–4335 (1991). [Google Scholar]
- 6.Baldwin JE and Fleming RH, Dynamic Stereochemistry, Springer, Berlin, Heidelberg, 1970, pp. 281–310. [Google Scholar]
- 7.Black K, Liu P, Xu L, Doubleday C, and Houk KN, Proc. Natl. Acad. Sci. USA, 2012, 109, 12860–12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woodward RB, Bader FE, Bickel H, Frey AJ, Kierstead RW, J. Am. Chem. Soc. 1956, 78, 2023–2025; [Google Scholar]; Woodward RB, Bader FE, Bickel H, Frey AJ, Kierstead RW, J. Am. Chem. Soc. 1956, 78, 2657; [Google Scholar]; Woodward RB, Bader FE, Bickel H, Frey AJ, Kierstead RW, Tetrahedron 1958, 2, 1–57. [Google Scholar]
- 9.Nicolaou KC and Sorensen EJ, Classics in Total Synthesis: Targets, Strategies, Mechanisms. Weinheim, VCH; 1996; see also: [Google Scholar]; Nicolaou KC, Snyder SA, Montagnon T, Vassilikogiannakis GE, Angew. Chem. Int. Ed 2002, 41, 1668–1698. [DOI] [PubMed] [Google Scholar]
- 10.Stork G, Yamashita A, Hanson RM, Phan L, Phillips E, Dubé D, Bos PH, Clark AJ, Gough M, Greenlee ML, Jiang Y, Jones K, Kitamura M, Leonard J, Liu T, Parsons PJ, and Venkatesan AM, Org. Lett. 2017, 19, 5150–5153. [DOI] [PubMed] [Google Scholar]
- 11.Ilardi EA, Stivala CE, and Zakarian A, Chem. Soc. Rev,. 2009, 38, 3133–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barcan GA, Patel A, Houk KN, and Kwon O, Org. Lett. 2012, 14, 5388–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. a).Lu Z, Li Y, Deng J and Li A, Nature Chem. 2013, 5, 679–684. [DOI] [PubMed] [Google Scholar]; b) Chen Y, Zhang W, Ren L, Li J and Li A, Angew. Chem. Int. Ed, 2018, 57, 952–956. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Ding M, Li J, Guo Z, Lu M, Chen Y, Liu L, Shen Y-H and Li A, J. Am. Chem. Soc, 2018, 140, 4227–4231. [DOI] [PubMed] [Google Scholar]
- 15.Souris C, Misale A, Chen Y, Luparia Y, and Maulide N, Org. Lett, 2015, 17, 4486–4489; [DOI] [PubMed] [Google Scholar]; Misale A, Niyomchon S, and Maulide N, Acc. Chem. Res, 2016, 49, 2444–2458. [DOI] [PubMed] [Google Scholar]
- 16.Siegel JB, Zanghellini A, Lovick HM, Kiss G, Lambert AR, St JL. Clair JL Gallaher D Hilvert MH Gelb BL Stoddart KN Houk FE Michael, and Baker D, Science 2010, 329, 309–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gouverneur VE, Houk KN, de Pascual-Teresa B, Beno B, Janda KD, Lerner RA, Science 1993, 262, 204–208. [DOI] [PubMed] [Google Scholar]
- 18.Chen J, Deng Q, Wang R, Houk KN, and Hilvert D, ChemBioChem 2000, 1, 255–261. [DOI] [PubMed] [Google Scholar]
- 19.Kim SP, Leach AG, Houk KN, J. Org. Chem. 2002, 67, 4250. [DOI] [PubMed] [Google Scholar]
- 20.Eiben CB, Siegel JB, Bale JB, Cooper S, Khatib F, Shen BW, Players F, Stoddard BL, Popovic Z, and Baker D, Nature biotechnology, 2012, 30, 190–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Preiswerk N, Beck T, Schulz JD, Milovník P, Mayer C, Siegel JB, Baker D and Hilvert D, Proc. Natl. Acad. Sci, 2014, 111, 8013–8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. a).Oikawa H, Tokiwano T, Nat. Prod. Rep. 2004, 21, 321–352. [DOI] [PubMed] [Google Scholar]; b) Oikawa H, Edited by Liu H-W, and Mander L, 1st Edition, Elsevier, 5 Mar. 2010 [Google Scholar]
- 23.Auclair K, Sutherland A, Kennedy J, Witter DJ, Van den Heever JP, Hutchinson CR and Vederas JC, J. Am. Chem. Soc, 2000, 122, 11519–11520. [Google Scholar]
- 24.Oikawa H, Katayama K, Suzuki Y and Ichihara A, J. Chem. Soc. Chem. Commun, 1995, 0, 1321–1322. [Google Scholar]
- 25.Katayama K, Kobayashi T, Oikawa H, Honma M and Ichihara A, Biochim. Biophys. Acta BBA - Protein Struct. Mol. Enzymol, 1998, 1384, 387–395. [DOI] [PubMed] [Google Scholar]
- 26.Xu W, Chooi Y-H, Choi JW, Li S, Vederas JC, Da Silva NA and Tang Y, Angew. Chem. Int. Ed, 2013, 52, 6472–6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe K, Mie T, Ichihara A, Oikawa H, Honma M, J. Biol. Chem. 2000, 49, 38393–38401. [DOI] [PubMed] [Google Scholar]
- 28. a).Ose T, Watanabe K, Mie T, Honma M, Watanabe H, Yao M, Oikawa H, Tanaka I, Nature 2003, 422, 185–189. [DOI] [PubMed] [Google Scholar]; b) Ose T, Watanabe K, Yao M, Honma M, Oikawa H, Tanaka I, Acta Crystallogr. 2004, D60, 1187–1197. [DOI] [PubMed] [Google Scholar]
- 29.Guimarães CR, Udier-Blagović M, Jorgensen WL, J Am Chem Soc, 2005, 127, 3577–3588. [DOI] [PubMed] [Google Scholar]
- 30.Wilson E, Chem. Eng. News, 2005, 83, 38. [Google Scholar]
- 31.Kim HJ, Ruszczycky MW, Choi S, Liu Y and Liu H, Nature, 2011, 473, 109–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian Z, Sun P, Yan Y, Wu Z, Zheng Q, Zhou S, Zhang H, Yu F, Jia X, Chen D, Mándi A, Kurtán T and Liu W, Nat. Chem. Biol, 2015, 11, 259–265. [DOI] [PubMed] [Google Scholar]
- 33.Hashimoto T, Hashimoto J, Teruya K, Hirano T, Shin-ya K, Ikeda H, Liu H, Nishiyama M and Kuzuyama T, J. Am. Chem. Soc, 2015, 137, 572–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Byrne MJ, Lees NR, Han L-C, van der Kamp MW, Mulholland AJ, Stach JEM, Willis CL and Race PR, J. Am. Chem. Soc, 2016, 138, 6095–6098. [DOI] [PubMed] [Google Scholar]
- 35.Wever WJ, Bogart JW, Baccile JA, Chan AN, Schroeder FC and Bowers AA, J. Am. Chem. Soc, 2015, 137, 3494–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minami A and Oikawa H, J. Antibiot. (Tokyo), 2016, 69, 500–506. [DOI] [PubMed] [Google Scholar]
- 37.Jeon B, Wang S-A, Ruszczycky MW and Liu H, Chem. Rev, 2017, 117, 5367–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klas K, Tsukamoto S, Sherman DH and Williams RM, J. Org. Chem, 2015, 80, 11672–11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng Q, Tian Z and Liu W, Curr. Opin. Chem. Biol, 2016, 31, 95–102. [DOI] [PubMed] [Google Scholar]
- 40.Andrews PR, Smith GD and Young IG, Biochemistry (Mosc.), 1973, 12, 3492–3498. [DOI] [PubMed] [Google Scholar]
- 41. a).Hong YJ, and Tantillo DJ, J. Am. Chem. Soc, 2009, 131, 7999–8015 [DOI] [PubMed] [Google Scholar]; b) Hong YJ, and Tantillo DJ, Org. Biomol. Chem 2010, 8, 4589–4600. [DOI] [PubMed] [Google Scholar]; c) Hong YJ, and Tantillo DJ, Nature Chem, 2014, 6, 104–111. [DOI] [PubMed] [Google Scholar]
- 42.DeClue MS, Baldridge KK, Künzler DE, Kast P and Hilvert D, J. Am. Chem. Soc, 2005, 127, 15002–15003. [DOI] [PubMed] [Google Scholar]
- 43.Lamb AL, Biochemistry, 2011, 50, 7476–7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shipman LW, Li D, Roessner CA, Scott AI and Sacchettini JC, Structure, 2001, 9, 587–596. [DOI] [PubMed] [Google Scholar]
- 45.Luk LYP, Qian Q and Tanner ME, J. Am. Chem. Soc, 2011, 133, 12342–12345. [DOI] [PubMed] [Google Scholar]
- 46. a).Krenske EH, Patel A, and Houk KN, J. Am. Chem. Soc, 2013, 135, 17638–17642. [DOI] [PubMed] [Google Scholar]; b) Payne KAP, White MD, Fisher K, Khara B, Bailey SS, Parker D, Rattray NJW, Trivedi DK, Goodacre R, Beveridge R, Barran P, Rigby SEJ, Scrutton NS, Hay S, and Leys D, Nature, 2017, 522, 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ferguson KL, Eschweiler JD, Ruotolo BT, Marsh ENG, J. Am. Chem. Soc, 2017, 139, 10972–10975. [DOI] [PubMed] [Google Scholar]
- 47. a).Ess DH, Wheeler SE, Iafe RG, Xu L, Çelebi‐Ölçüm N and Houk KN, 2008, Angew. Chem. Int. Ed, 47, 7592–7601. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hare SR, Tantillo DJ, Pure Appl. Chem. 2017, 89, 679–698. [Google Scholar]
- 48.Li G, Kusari S and Spiteller M, Nat. Prod. Rep, 2014, 31, 1175–1201. [DOI] [PubMed] [Google Scholar]
- 49.Fischbach MA and Walsh CT, Chem. Rev, 2006, 106, 3468–3496. [DOI] [PubMed] [Google Scholar]
- 50.Diedrich MK, Klärner F-G, Beno BR, Houk KN, Senderowitz H, and Still WC, J. Am. Chem. Soc, 1997, 119, 10255–10259. [Google Scholar]
- 51.Healy AR and Westwood NJ, Org. Biomol. Chem, 2015, 13, 10527–10531. [DOI] [PubMed] [Google Scholar]
- 52.Sato M, Yagishita F, Mino T, Uchiyama N, Patel A, Chooi Y-H, Goda Y, Xu W, Noguchi H, Yamamoto T, Hotta K, Houk KN, Tang Y and Watanabe K, ChemBioChem, 2015, 16, 2294–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kato N, Nogawa T, Hirota H, Jang J-H, Takahashi S, Ahn JS and Osada H, Biochem. Biophys. Res. Commun, 2015, 460, 210–215. [DOI] [PubMed] [Google Scholar]
- 54.Li L, Yu P, Tang M-C, Zou Y, Gao S-S, Hung Y-S, Zhao M, Watanabe K, Houk KN and Tang Y, J. Am. Chem. Soc, 2016, 138, 15837–15840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang Y-L, Lu C-P, Chen M-Y, Chen K-Y, Wu Y-C and Wu S-H, Chem. - Eur. J, 2007, 13, 6985–6991. [DOI] [PubMed] [Google Scholar]
- 56.Hess BA, Smentek L, Org. Biomol Chem 2012. 10, 7503–7509 [DOI] [PubMed] [Google Scholar]
- 57.Patel A, Chen Z, Yang Z, Gutiérrez O, Liu H, Houk KN and Singleton DA, J. Am. Chem. Soc, 2016, 138, 3631–3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Z, Yang S, Yu P, Li Y, Doubleday C, Park J, Patel A, Jeon B, Russell WK, Liu H, Russell DH and Houk KN, Proc. Natl. Acad. Sci, 2018, 115, E848–E855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Caramella P, Quadrelli P, and Toma L, J. Am. Chem. Soc 2002, 124, 1130–1131. [DOI] [PubMed] [Google Scholar]
- 60.Desimoni G and Tacconi G, Chem. Rev, 1975, 75, 651–692. [Google Scholar]
- 61.Cary JW, Uka V, Han Z, Buyst D, Harris-Coward PY, Ehrlich KC, Wei Q, Bhatnagar D, Dowd PF, Martens SL, Calvo AM, Martins JC, Vanhaecke L, Coenye T, De Saeger S and Di Mavungu JD, Fungal Genet. Biol, 2015, 81, 88–97. [DOI] [PubMed] [Google Scholar]
- 62.Stocking EM and Williams RM, Angew. Chem. Int. Ed, 2003, 42, 3078–3115. [DOI] [PubMed] [Google Scholar]
- 63.Oikawa H and Tokiwano T, Nat. Prod. Rep, 2004, 21, 321–352. [DOI] [PubMed] [Google Scholar]
- 64.Jessen HJ and Gademann K, Nat. Prod. Rep, 2010, 27, 1168–1185. [DOI] [PubMed] [Google Scholar]
- 65.Snider BB and Lu Q, J. Org. Chem, 1996, 61, 2839–2844. [DOI] [PubMed] [Google Scholar]
- 66.Ohashi M, Liu F, Hai Y, Chen M, Tang M, Yang Z, Sato M, Watanabe K, Houk KN and Tang Y, Nature, 2017, 549, 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cope AC and Hardy EM, J. Am. Chem. Soc, 1940, 62, 441–444. [Google Scholar]
- 68.Ilardi EA, Stivala CE, and Zakarian A, Chem Soc Rev. 2009, 38, 3133–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li S, Lowell AN, Yu F, Raveh A, Newmister SA, Bair N, Schaub JM, Williams RM and Sherman DH, J. Am. Chem. Soc, 2015, 137, 15366–15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li S, Lowell AN, Newmister SA, Yu F, Williams RM and Sherman DH, Nat. Chem. Biol, 2017, 13, 467–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Newmister SA, Li S, Garcia-Borràs M, Sanders JN, Yang S, Lowell AN, Yu F, Smith JL, Williams RM, Houk KN and Sherman DH, bioRxiv, 2017, 173674. [Google Scholar]
- 72.Zhu Q and Liu X, Angew. Chem. Int. Ed, 2017, 56, 9062–9066. [DOI] [PubMed] [Google Scholar]
- 73.Lutz RP, Chem. Rev, 1984, 84, 205–247. [Google Scholar]
- 74.Nicolaou KC, Snyder SA, Montagnon T and Vassilikogiannakis G, Angew. Chem. Int. Ed, 2002, 41, 1668–1698. [DOI] [PubMed] [Google Scholar]
- 75.Nicolaou KC, Vourloumis D, Winssinger N and Baran PS, Angew. Chem. Int. Ed, 2000, 39, 44–122. [PubMed] [Google Scholar]
- 76.Jiang X and Wang R, Chem. Rev, 2013, 113, 5515–5546. [DOI] [PubMed] [Google Scholar]
- 77.Png ZM, Zeng H, Ye Q and Xu J, Chem. – Asian J, 2017, 12, 2142–2159. [DOI] [PubMed] [Google Scholar]
- 78.Hoffmann HMR, Angew. Chem. Int. Ed. Engl, 1969, 8, 556–577. [Google Scholar]
- 79.Snider BB and Ron E, J. Am. Chem. Soc, 1985, 107, 8160–8164. [Google Scholar]
- 80.Mikami K and Shimizu M, Chem. Rev, 1992, 92, 1021–1050. [Google Scholar]
- 81.Nubbemeyer U, Synthesis, 2003, 2003, 0961–1008. [Google Scholar]
- 82.Ilardi EA, Stivala CE and Zakarian A, Chem. Soc. Rev, 2009, 38, 3133–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bian LL, and Ding H, Synthesis, 2017, 28, 4383–4413. [Google Scholar]
- 84.Rosenthal RG, Ebert M-O, Kiefer P, Peter DM, Vorholt JA and Erb TJ, Nat. Chem. Biol, 2014, 10, 50–55. [DOI] [PubMed] [Google Scholar]
- 85.Katsuyama Y, Li X-W, Müller R and Nay B, ChemBioChem, 2014, 15, 2349–2352. [DOI] [PubMed] [Google Scholar]
- 86.Yang Z, Dong X, Yu Y, Yu P, Li Y, Jamieson C and Houk KN, J. Am. Chem. Soc, 2018, 140, 3061–3067. [DOI] [PubMed] [Google Scholar]
- 87.Walsh CT and Tang Y, Biochemistry, 2018, 57, 3087–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]