Abstract
Mutations in the Epidermal growth factor receptor (EGFR) kinase domain occur in 10–30% of lung adenocarcinoma and are associated with tyrosine kinase inhibitor (TKI) sensitivity. We sought to identify the immediate direct and indirect phosphorylation targets of mutant EGFRs in lung adenocarcinoma. We undertook stable isotope labeling of amino acids in cell culture (SILAC) strategy, phosphopeptide enrichment, and quantitative mass spectrometry to identify dynamic changes of phosphorylation downstream of mutant EGFRs in lung adenocarcinoma cells harboring EGFRL858R and EGFRL858R/T790M, the TKI-sensitive and -resistant mutations, respectively. Top canonical pathways that were inhibited upon erlotinib treatment in sensitive cells, but not in the resistant cells include EGFR, Insulin receptor, HGF, MAPK, mTOR, p70S6K and JAK/STAT signaling. We identified phosphosites in proteins of the autophagy network, such as ULK1 (S623) that is constitutively phosphorylated in these lung adenocarcinoma cells; phosphorylation is inhibited upon erlotinib treatment in sensitive cells, but not in resistant cells. Finally, kinase-substrate prediction analysis from our data indicated that substrates of basophilic kinase families, AGC, CAMK and STE were significantly enriched and those of proline directed kinase families, CMGC and CK were significantly depleted among substrates that exhibited increased phosphorylation upon EGF stimulation and reduced phosphorylation upon TKI inhibition. This is the first study to date to examine global phosphorylation changes upon erlotinib treatment of lung adenocarcinoma cells and results from this study provide new insights into signaling downstream of mutant EGFRs in lung adenocarcinoma.
Keywords: EGFR, NSCLC, erlotinib, phosphoproteomics, mass spectrometry, SILAC, autophagy
1. Introduction
Lung cancer is the leading cause of cancer mortality in both genders in the US. It is estimated that there were 230,000 new cancer cases and 160,000 cancer deaths in 2013 [1]. Lung adenocarcinoma is the most common histology associated with lung cancer patients. Mutations in the kinase domain of epidermal growth factor receptor (EGFR) occur in around 10% of patients with lung adenocarcinoma in the western world and 25–30% of patients in Asian countries [2–4]. Leu to Arg substitution at position 858 (EGFRL858R) and a series of deletions at the ATP binding pocket of the kinase domain of EGFR that deletes the Leu-Arg-Glu-Ala (LREA) motif (EGFRDel) account for more than 90% of mutations in EGFR. These mutations are associated with sensitivity of patients to EGFR-directed tyrosine kinase inhibitors (TKIs). Although patients harboring these TKI-sensitizing mutations respond dramatically to erlotinib, they all eventually develop secondary resistance. Around 60% of such patients develop a second site mutation in the gatekeeper residue in the ATP binding pocket of EGFR (T790M) [5–8]. Understanding the intricate signal transduction pathways that are activated by mutant EGFRs in lung adenocarcinoma is germane to developing new biomarkers of TKI sensitivity, resistance, and also developing new treatment strategies to circumvent TKI resistance.
Lung cancer-specific EGFR mutants activate the AKT and STAT signaling pathways to promote cell survival, while not affecting ERK signaling, at least in an isogenic system of immortalized mouse mammary epithelial cells and also transiently transfected COS7 cells [9]. In PC9, a lung adenocarcinoma cell line harboring EGFRDel E746-A750, gefitinib treatment caused rapid decline of pro-survival signals, pAKT and pERK1/2 levels within 4 hours and a slower but sustained increase in pro-apoptotic signal, phospho-p38 MAPK [10]. This differential attenuation of pro-survival and pro-apoptotic signals may underlie the acute sensitivity of these lung adenocarcinoma cells to mutant EGFR inhibition [11].
Recent advances in quantitative mass spectrometry and profiling of post-translational modifications, such as phosphorylation has enabled global proteome-wide analysis of normal EGFR signaling pathway [12–16]. More specifically, a global phosphoproteomic approach to identify tyrosine phosphorylated peptides has identified several oncogenic kinases such as EGFR, c-MET, PDGFRα, DDR1 and novel ALK and ROS fusions in non-small cell lung cancer (NSCLC) cell lines and tumor specimens [17]. Phosphotyrosine profiling has also been performed by mass spectrometry in lung adenocarcinoma cell lines with variable sensitivities to EGFR TKI, gefitinib. Quantitative estimation of tyrosine phosphorylation of peptides was further performed in this study in two adenocarcinoma cell lines with TKI-sensitizing EGFR mutations. This study demonstrated that there was an extensive signaling network downstream of mutant EGFRs that collapsed upon treatment with an EGFR TKI [18]. Mass spectrometry and other biochemical approaches have been used to identify dynamic phosphorylation changes in EGFR upon EGFR-TKI treatment [19–21]. We have employed quantitative mass spectrometry to identify and quantify tyrosine phosphorylation of targets of wild type or mutant EGFRs in isogenic human bronchial epithelial cells (HBECs) and identified several novel mutant EGFR targets [22]. In this study, in order to identify direct and indirect targets of mutant EGFRs in lung adenocarcinoma, we sought to identify dynamic global phosphorylation changes (primarily pSer/Thr, and also pTyr) upon immediate ligand stimulation with or without prior EGFR TKI, erlotinib inhibition of H3255 (EGFRL858R mutant), a TKI-sensitive, and H1975 (EGFRL858R/T790M mutant), a TKI-resistant lung adenocarcinoma cell lines. We posited that such analyses of dynamic phosphorylation changes in EGFR-TKI-sensitive and -resistant lung adenocarcinoma cells will identify targets of mutant and/or activated wild-type EGFR, signaling pathways responsible for TKI resistance, and possible off-target effects of erlotinib.
2. Materials and Methods
2.1 Cell Culture and preparation of lysates
Two lung adenocarcinoma cell lines, H3255 harboring EGFR L858R mutation (a kind gift from Dr. Bruce Johnson at Dana Farber Cancer Institute, Boston, MA) and H1975 with EGFR L858R/T790M mutation (purchased from American Type Culture Collection) were used in this study. These cells were cultured in RPMI SILAC media at 37°C, 5% CO2 and high humidity in accordance with the previously described protocol [12, 22, 23]. Briefly, 3-state SILAC media were prepared by supplementing different isotopic versions of 1.15 mM arginine and 0.274 mM lysine. Light media was made using light arginine and lysine, medium state contained 13C6 arginine and 4,4,5,5-D4 lysine, and heavy media contained 13C615N4 arginine and 13C615N2 lysine. All amino acids (99% pure) were purchased from either Cambridge Isotope laboratories (Andover, MA) or from Sigma-Aldrich, (St. Louis, MO). The cells were passaged at least five times and screened for labeling efficiency using mass spectrometry. Upon completion of labeling, cells were expanded into several 15 cm dishes. Cells (~5×107–108 per dish) were serum starved for 14 hours prior to treatment. Light cells were left untreated as the control, the medium labelled cells were stimulated with 100 ng/ml EGF (Millipore) for three minutes and the heavy cells were treated with erlotinib (Genentech) to a final concentration of 100 nM for one hour before stimulating the cells with 100 ng/ml EGF for three minutes. The reaction was terminated by chilling cells quickly with cold PBS; for phosphopeptide profiling, cells were lysed in urea lysis buffer containing 20 mM HEPES pH 8.0, 8 M urea, 1 mM sodium orthovanadate, 2.5 mM sodium pyrophosphate, and 1 mM β-glycerophosphate. Cells were sonicated using Branson 250 Sonifier at 20% pulse for 5 sec, 3 times on ice. Samples were centrifuged at 15,000g for 10 min and protein concentration in the supernatant was measured using modified Lowry method (BioRad). Equal amounts of protein from lysates of each state were combined. For Western blot based validation experiments unlabelled cells (control, treated with erlotinib and EGF) were lysed with modified RIPA buffer containing 50 mM Tris-HCl pH 7.4, 1% Nonidet P-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, protease inhibitor mixture tablets (Roche), 1 mM Na3VO4, and NaF 1 mM.
2.2 Tryptic digestion of proteins and purification of digested peptides
Lysates from the three SILAC states (serum starved, EGF stimulated, and TKI inhibited/EGF stimulated) were combined to constitute 20–30 mg of pooled lysate that was homogenized and centrifuged at 15,000×g followed by reduction and alkylation with 45 mM dithiothreitol (Sigma Aldrich, MO) and 100 mM iodoacetamide (Sigma Aldrich, MO), respectively [24]. Modified sequencing grade Trypsin (Promega, Madison, WI) was used to carry out digestion of the lysate at 30°C for 16 hrs. The digest was then acidified using 0.1% TFA and the tryptic peptides were cleaned using solid phase C18 extraction column (Supelco, Bellefonte, PA), dried down in a lyophilizer, and subsequently subjected to strong cation exchange (SCX) chromatography or basic reversed phase liquid chromatography (RPLC).
2.3 Strong cation exchange (SCX) chromatography for fractionation of tryptic peptides
SCX was performed using an Agilent 1100 HPLC system (Agilent Technologies) using a polysulfoethyl A SCX column (PolyLC, Columbia, MD; 200 × 2.1 mm, 5 μm, 200Å) as described earlier [25, 26]. Lyophilized tryptic peptides were resuspended in SCX Buffer A (10 mM potassium phosphate buffer containing 30% acetonitrile (ACN), pH 2.7), loaded onto SCX column and eluted over a gradient of SCX buffer B (Buffer A containing 350 mM KCl); 0–50% buffer B over a period of 30 min followed by 50–100% B for 7 min, and 100% B for 3 min. A gradient cycle time of 50 min was used. A total of 24 fractions were collected based on UV absorbance profile and the fractionated peptides were dried in a vacuum centrifuge.
2.4 Basic reversed phase liquid chromatography (RPLC) fractionation and concatenation of peptide fractions
Basic RPLC separation was performed with a XBridge C18, 250 × 4.6 mm analytical column containing 5μm particles and equipped with a 20 × 4.6 mm guard column (Waters, Milford, MA) with a flow rate of 1mL/min. The solvent consisted of 7 mM triethylammonium bicarbonate (TEABC) as mobile phase A, and 7 mM TEABC and 90% ACN as mobile phase B. Sample separation was accomplished using the following linear gradient: from 0 to 1% B in 5min, from 1 to 10% B in 5min, from 10 to 35% B in 30min, and from 35 to 100% B in 5min, and held at 100% B for an additional 3min. A total of 96 fractions were collected during the LC separation in a 96-well plate in the presence of 50μL of 1% formic acid. The collected fractions were dried in a vacuum centrifuge. Finally the samples were concatenated into 12 fractions by combining fractions 1, 13, 25, 37, 49, 61, 73, 85; and so on.
2.4 Titanium Dioxide (TiO2) Enrichment
Dried peptides dissolved in solution A containing 80% acetonitrile, 1% trifluoracetic acid and 3% 2,5-dihydroxybenzoic acid and incubated with titanium dioxide (TiO2) (Titansphere, GL Sciences) pretreated (2hrs at room temperature) with solution A [26–28]. After 12 hrs, TiO2 beads were washed thrice using solution A and twice with 80% acetonitrile containing 1% trifluoracetic acid. TiO2 bound peptides were eluted using 3% NH4OH in 40% ACN and immediately acidified using formic acid. The peptides were vacuum dried, C18 stage-tip cleaned before LC-MS/MS analysis.
2.5 LC-MS/MS analyses
Tandem mass spectrometry analysis of SILAC labelled peptides obtained from SCX chromatography followed by TiO2 enrichment were carried out on a LTQ Orbitrap XL (Thermo Scientific) mass spectrometer interfaced with a Eksigent nanoflow liquid chromatography system and an Agilent 1100 microwell plate autosampler. Magic C18 AQ 5 μm, 100Å from Michrom Bioresources was used for packing nanoflow reversed-phase columns. Peptides were loaded onto a trap column (75 μm × 2 cm) and separated on an analytical column (75μm × 15 cm) at 300 nL/min flow rate for 75–120 min duration. The data dependent LC-MS/MS analysis was carried out by acquiring FT-MS at a resolution of 60,000 at m/z 400 and MS/MS of eight most abundant ions in ion trap. Multistage activation mode was enabled with neutral loss masses of 32.66, 48.99 and 97.97. Fragmented ions were excluded dynamically for 90 seconds.
Peptides separated/fractionated by basic reversed-phase chromatography followed by TiO2 enrichment were analyzed on an LTQ-Orbitrap Elite interfaced with an Easy-nLC 1000 RPLC (Thermo Scientific, San Jose, CA). The enriched phosphopeptides were loaded onto a nano-trap column (Acclaim PepMap100 Nano Trap Column, C18, 5 μm, 100 Å, 100 μm i.d. × 2 cm) and separated on a nano-LC column (Acclaim PepMap100, C18, 3 μm, 100 Å, 75 μm i.d. × 25 cm, nanoViper). Mobile phases A and B consisted of 0.1% formic acid in water and 0.1% formic acid in 90% ACN, respectively. Peptides were eluted from the column at 300 nL/min using the following linear gradient: from 2 to 8% B in 5min, from 8 to 32% B in 100min, from 32 to 100% B in 10min, and held at 100% B for an additional 10min. The heated capillary temperature and spray voltage were 275°C and 2.2kV, respectively. Full spectra were collected from m/z 350 to 1800 in the Orbitrap analyzer at a resolution of 120,000, followed by data-dependent HCD MS/MS scans of the ten most abundant ions, using 32% collision energy and dynamic exclusion time of 30s.
2.6 Data Analysis
Peptides and proteins were identified and quantified using the Maxquant software package (version 1.3.0.5) with the Andromeda search engine [29] as well as Proteome Discoverer with Mascot and Sequest search engines (Thermo Scientific). MS/MS data were searched against the Refseq 49 human protein database and quantification was performed using default parameters for three-state SILAC in MaxQuant. Parameters for data analysis included trypsin as a protease with two allowable missed cleavages. Carbamidomethyl cysteine was specified as a fixed modification. Phosphorylation at serine, threonine and tyrosine, deamidation of asparagine and glutamine, oxidation of methionine and protein N-terminal acetylation were specified as variable modifications. The precursor mass tolerance was set to 7 ppm and fragment mass tolerance to 20 ppm. False discovery rate was calculated using a decoy database and a 1% FDR cut off was applied. Phosphorylation motifs for serine and threonine phosphorylation sites were aligned using PhosphoSite Plus [30] using the Motif-All algorithm to search the observed sites weighted by a database of known phospho serine/threonine sites.
2.7 GPS analysis
We determined putative kinase targets among all the identified phosphosites using the Group-based Prediction System 2.1.2 (GPS) [31]. Predictions of kinase-specific phosphorylation sites were done for 408 human protein kinases based on preferable peptide substrate sequences with theoretically calculated maximal false positive rate of 6% for serine-threonine kinases and of 9% for tyrosine kinases. We applied Fisher’s exact test with Benjamini-Hochberg adjustment using R statistics package to determine the enrichment or depletion of number of substrates of kinase family, subfamily, and individual kinases for the set of phosphosites hyperphosphorylated after EGF stimulation (EGF/Control, M/L > 1.5) and for the set of phosphosites sensitive to erlotinib treatment (EGF + erlotinib/EGF, H/M < 0.67) in comparison with the corresponding baselines (0.67 < M/L < 1.5 and 0.67 < H/M < 1.5). Data representation on the kinome tree was adapted from Manning et. al. [32] and the Kinome Render software [33].
2.8 Immunoblot analysis
For Western blot analysis, 50–100 μg of lysate was separated by SDS-PAGE (Invitrogen) and transferred to nitrocellulose membrane. After blocking in 5% BSA in PBST for one hour, membranes were incubated with the appropriate primary antibody followed by secondary antibody coupled with horseradish peroxidase. The primary antibodies used were against mTOR, pmTOR (S2248), RSK2, pRSK2 (S380), ERK1/2, pERK1 (T202/Y204), AKT1 and pAKT1 (S473), purchased from Cell Signaling Technology (Andover, MA). Actin was purchased from Sigma Aldrich, MO. Custom mouse monoclonal antibodies were made against EGFRL858R, that recognizes mutant EGFR, but not WT EGFR, and against EGFRL858, that recognizes wild type EGFR, but not mutant EGFR in collaboration with NanoTools (Germany). Membranes were incubated with ECL (Amersham) for 5 minutes prior to exposing to X-ray film.
3. Results
3.1 Phosphoproteomic profiling of erlotinib-sensitive and -resistant lung adenocarcinoma cells
H3255 is a lung adenocarcinoma cell line that harbors the L858R mutation in the kinase domain of EGFR. There is preferential amplification of the mutant allele of EGFR leading to overexpression of mutant EGFR protein in this cell line (Fig. S1A). These cells are exquisitely sensitive to EGFR-directed tyrosine kinase inhibitors (TKIs), such as erlotinib, with IC50 value ranging in the subnanomolar range [34]. H1975 is a lung adenocarcinoma cell line carrying two mutations in EGFR (L858R, T790M), and is resistant to first generation reversible EGFR-TKIs, such as erlotinib. Treatment of H3255 cells with erlotinib results in global decrease in tyrosine phosphorylation even in the presence of EGF treatment, while there is no significant change in global tyrosine phosphorylation in H1975 cells harbouring the resistant EGFR mutant (Fig. S1). Although inhibition of tyrosine phosphorylation is expected immediately upon erlotinib action, here we sought to identify the Serine/Theronine phosphorylation changes that occur immediately upon EGFR activation (within three minutes of EGFR stimulation) and the sites of such phosphorylation that are inhibited by erlotinib treatment. In order to examine the global signaling consequences that result from EGFR ligand stimulation and erlotinib treatment, a phosphoproteomics approach was employed to understand and identify downstream signaling events by enriching phosphopeptides for quantitative analysis.
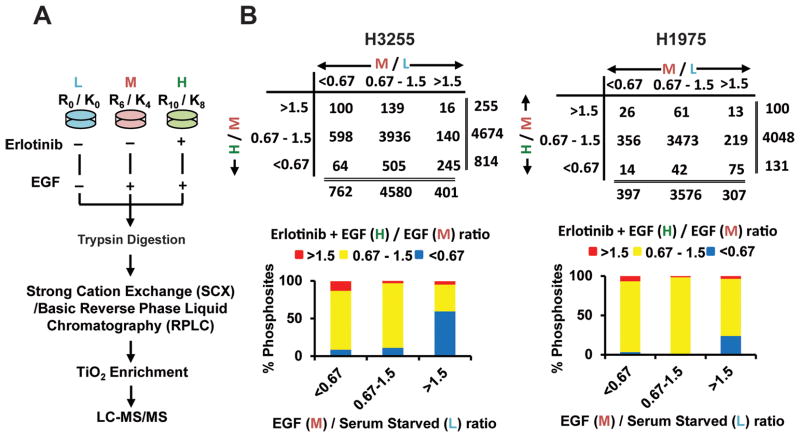
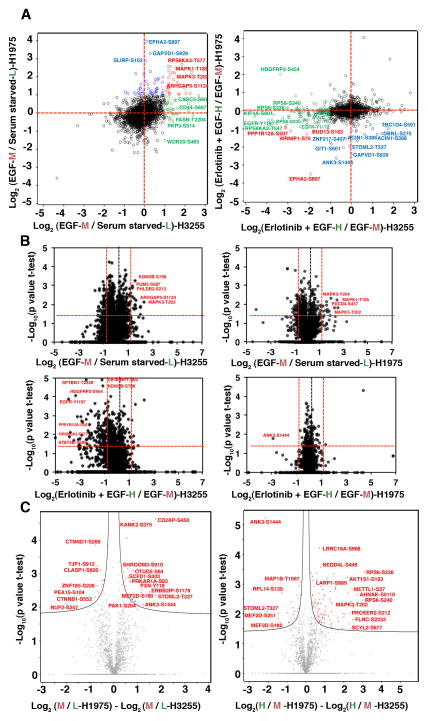
H3255 and H1975 cells were SILAC labelled with “light-L” (normal), “medium-M”, and “heavy-H” labelled amino acids to perform a “three-state” SILAC experiment. The cells were serum starved overnight. “Light-L” cells were left untreated (control), “medium-M” cells were treated with EGF for three minutes, and “heavy-H” cells were first treated with erlotinib for two hours followed by induction with EGF for three minutes. Equal amount of experimental lysates from the three states were mixed at 1:1:1 ratio and further processed for MS analysis (Fig. 1A). After digestion with trypsin, the peptides were fractionated by either strong cation exchange chromatography (SCX) or basic reversed phase liquid chromatography (RPLC) and subsequently enriched for phosphorylated peptides using titanium dioxide (TiO2). Peptides were then analyzed with high-resolution liquid chromatography-tandem mass spectrometry (nLC-MS/MS) on a hybrid linear ion trap Orbitrap instrument. Two replicates of H3255 experiment underwent SCX fractionation followed by LC-MS/MS analysis on LTQ Orbitrap XL. Three replicates of H3255 and H1975 were fractionated by basic RPLC followed with LC-MS/MS analysis on Orbitrap Elite. A total of 110 MS runs resulted in identification of 11,207 unique phosphosites (9,094 phosphoserine, 1,975 phosphotheronine and 138 phosphotyrosine sites) in 3,086 proteins. A total of 8,759 phosphosites in 2,571 proteins (7,048 phosphoserine, 1,567 phosphothreonine and 129 phosphotyrosine sites) and 6,444 phosphosites in 2,234 proteins (5,440 phosphoserine, 968 phosphothereonine and 26 phosphotyrosine sites) were identified in H3255 and H1975 cell lines, respectively. Among them, phosphorylation was quantified in 6,964 and 5,109 phosphosites in the respective cell lines of which 5,752 and 4,279 were labelled as class I phosphosites (localization probability of phosphorylation 0.75 or more) (Table S1). We used a SILAC ratio cut-off of 1.5 and 0.67 for increased and decreased phosphorylation, respectively. The distribution of the peptides based on SILAC ratios followed largely a normal distribution. The cut-off of 1.5 and 0.67 were around one standard deviation of the median of the SILAC ratios (Fig S2). Majority of phosphosites (totalling to 4,580 in H3255 and 3,473 in H1975) did not show any change upon treatment with EGF or erlotinib, which is reflective of the existence of very specific signaling cascades that are altered upon ligand stimulation or TKI inhibition. There were a total of 401 phosphosites that underwent increased phosphorylation upon EGF stimulation and 245 phosphosites were dephosphorylated upon erlotinib inhibition in H3255, the TKI sensitive cell line. However, a total of 307 phosphosites were hyper-phosphorylated on EGF stimulation in H1975, but only 75 of them were dephosphorylated with erlotinib inhibition, demonstrating the overall insensitivity of these adenocarcinoma cells to erlotinib inhibition (Fig 1B). We identified 129 and 26 phosphotyrosine sites in H3255 and H1975 cell lines, respectively, using the global phosphopeptide enrichment strategy. Among these sites, 27 sites were hyperphosphorylated upon EGF stimulation and 21 of them were dephosphorylated with erlotinib inhibition in H3255, including kinases, EGFR and MAPK3; transcription factor STAT5B; adaptor proteins DLG3 and CTNND1; adhesion proteins MLLT4 and PKP2. However, only 6 phosphotyrosine sites were found hyperphosphorylated in H1975, and they were not inhibited by erlotinib. The SILAC ratio depicting effects of EGF stimulation (M/L) and erlotinib inhibition (H/M) were plotted in a scatter plot for direct comparison of the dynamic changes in H3255 and H1975 cells (Fig 2A). Since there were five replicates of H3255 experiment and three for the H1975 experiment, we could perform statistics to identify significant changes of phosphorylation upon EGF stimulation or erlotinib inhibition for each cell line (Fig 2B). The volcano plots clearly demonstrate that while there was significant inhibition of phosphorylation in a large number of phosphosites in H3255 cells, only a handful underwent inhibition in H1975 cells which is resistant to erlotinib treatment using a more conservative cutoff of a SILAC ratio of 0.5 for erlotinib treatment (H/M) (Fig 2B, lower panel). We also compared the statistically significant differences between each of the respective SILAC ratios between the two cell lines (Fig 2C). As expected a large number of targets showed significant decrease in phosphorylation in H3255 cells upon erlotinib treatment (H/M ratio) compared to H1975 cells (Fig 2C, right panel). Specific significantly altered targets identified from these statistical analyses are shown in Table 1.
Figure 1.
SILAC-based quantitative mass spectrometry and phosphopeptides identified in groups of specific SILAC ratios. A. Flowchart showing biological treatment of SILAC-labelled cells, enrichment of phosphopeptides, and detection by tandem mass spectrometry. B. Number of class I phosphosites (localization probability >0.75) identified with SILAC ratios >1.5 (increased), 0.67–1.5 (unchanged) and <0.67 (decreased) (upper panel) and percentage of phosphosites in each of those groups (lower panel) in H3255 (left panel) and H1975 (right panel) cells. Ratio M/L is EGF/Serum starved and ratio H/M is erlotinib + EGF / EGF states.
Figure 2.
Distribution of quantified class I phosphosites and volcano plots showing significant SILAC ratios of phosphopeptides in H3255 and H1975. (A) Scatter plot showing distribution of SILAC ratios of EGF stimulation or erlotinib inhibition. Left panel is a plot against the Log 2 base ratio of EGF stimulated and serum starved cells (M/L). Right panel is a plot against the Log 2 base ratio of erlotinib inhibited before EGF stimulation and EGF stimulated cells (H/M). Each circle is a phosphorylation site. Annotated circles represent specific phosphosites with 2 fold-changes. The phosphosites that change in both H3255 and H1975, H3255 only, H1975 only are depicted in red, green, and blue circles, respectively; unaltered phosphosites between H3255 and H1975 are depicted in black. (B) Volcano plots showing significant SILAC ratios (M/L-upper panel; H/M-lower panel) of phosphosites that show hyperphosphorylation, no change or dephosphorylation with EGF and erlotinib treatment. Log2 ratios of the median of biological replicates were plotted versus −log10 of the p-values derived from a t-test. Proteins with a minimum 2-fold change combined with a p<0.05 are considered significant (red lines). Left panels are the changes in H3255; and right panels are the changes in H1975. Paired t-test was used for significance analysis from five biological replicates for H3255 and three biological replicates of H1975. (C) Volcano plots representing significant alterations of phosphorylation changes upon EGF stimulation or TKI inhibition between H3255 and H1975 cells. Differences in log2 ratios of M/L (left panel) and H/M (right panel) of H1975 and H3255 were plotted against −log10 of the p-values derived from a t-test. Hyperbolic curves separate the phosphosites significantly different between the two cell lines (red circles) from the ones unaltered between the two cell lines (grey circles).
Table 1.
Key signaling proteins and the phosphosites identified with corresponding SILAC ratios of EGF stimulation (M/L) and erlotinib inhibition (H/M) in H3255 and H1975 lung adenocarcinoma cells.
| Gene | Protein | Kinase family | Site | H3255 | H1975 | ||
|---|---|---|---|---|---|---|---|
|
| |||||||
| EGF / Control (M/L) | Erlotinib + EGF / EGF (H/M) | EGF / Control (M/L) | Erlotinib + EGF / EGF (H/M) | ||||
|
Kinases
| |||||||
| EGFR | Epidermal growth factor receptor | TK | S1064 | 1.59 | 0.48 | ||
| Y1172 | 1.36 | 0.19 | 2.08 | 0.62 | |||
| Y1197 | 2.06 | 0.07 | 2.07 | 0.61 | |||
| T725 | 1.87 | 0.62 | |||||
| S720 | 1.90 | 0.53 | |||||
| Y998 | 1.58 | 0.12 | |||||
| Y1110 | 1.15 | 0.04 | |||||
| S1057 | 1.02 | 0.19 | |||||
| S995 | 0.99 | 0.33 | |||||
| S991 | 0.72 | 0.34 | |||||
| T693 | 1.12 | 0.39 | 1.47 | 0.76 | |||
| S695 | 1.01 | 0.46 | |||||
| MAP2K2 | dual specificity mitogen-activated protein kinase kinase 2 | STE | S222 | 2.02 | 0.13 | ||
| S226 | 2.21 | 0.21 | |||||
| S295 | 1.1 | 0.44 | |||||
| T394 | 0.97 | 0.64 | 1.14 | 1.02 | |||
| MAP4K4 | mitogen-activated protein kinase kinase kinase kinase 4 | STE | S648 | 3.58 | 0.32 | ||
| S708 | 3.05 | 0.12 | |||||
| MAPK1 | mitogen-activated protein kinase 1 | CMGC | T185 | 2.37 | 0.09 | 5.61 | 0.90 |
| Y187 | 1.38 | 0.14 | 2.73 | 0.91 | |||
| MAPK3 | mitogen-activated protein kinase 3 | CMGC | T202 | 5.57 | 0.21 | 4.2 | 1.03 |
| Y204 | 1.77 | 0.13 | 3.72 | 0.89 | |||
| MKNK2 | MAP kinase-interacting serine/threonine-protein kinase 2 | CAMK | S452 | 2.23 | 0.60 | ||
| NEK9 | serine/threonine-protein kinase Nek9 | NEK | S29 | 0.98 | 0.52 | ||
| T333 | 1.2 | 0.12 | 1.62 | 0.61 | |||
| PRKCD | protein kinase C delta type | AGC | S304 | 1.67 | 0.35 | ||
| PRKD1 | serine/threonine-protein kinase D1 | CAMK | S205 | 1.5 | 0.55 | ||
| S397 | 0.93 | 0.66 | |||||
| S738 | 3.11 | 0.66 | |||||
| S742 | 1.68 | 0.41 | |||||
| S910 | 2.65 | 0.46 | |||||
| RIPK2 | receptor-interacting serine/threonine-protein kinase 2 | TKL | S178 | 0.71 | 0.40 | ||
| ROCK2 | rho-associated protein kinase 2 | AGC | S1374 | 1.53 | 0.54 | 1.03 | 0.87 |
| RPS6KA1 | ribosomal protein S6 kinase alpha-1 | AGC | S389 | 1.77 | 0.44 | ||
| RPS6KA3 | ribosomal protein S6 kinase alpha-3 | AGC | T577 | 3.78 | 0.1 | 6.26 | 0.55 |
| RPS6KB1 | ribosomal protein S6 kinase beta-1 | AGC | S427 | 1.21 | 0.22 | ||
| S441 | 1.03 | 0.56 | |||||
| TTK | dual specificity protein kinase TTK | Other | S436 | 0.73 | 0.6 | 0.97 | 0.95 |
| ULK1 | serine/threonine-protein kinase ULK1 | Other | S623 | 0.69 | 0.6 | 1.56 | 0.97 |
| S775 | 8.19 | 0.48 | |||||
| WNK1 | serine/threonine-protein kinase WNK1 | Other | S1521 | 0.7 | 0.62 | ||
| S2292 | 1.18 | 0.52 | 1.16 | 0.83 | |||
| S2530 | 2.17 | 0.61 | |||||
|
| |||||||
| Transcription factor/regulatory proteins | |||||||
|
| |||||||
| STAT5B | signal transducer and activator of transcription 5B | Y699 | 1.67 | 0.03 | |||
| MAF1 | repressor of RNA polymerase III transcription MAF1 | S75 | 2.49 | 0.26 | |||
| KHDRBS1 | KH domain-containing, RNA-binding, signal transduction-associated protein 1 | S20 | 1.58 | 0.56 | 1.12 | 0.95 | |
|
| |||||||
| Translation regulatory proteins | |||||||
|
| |||||||
| EIF4B | eukaryotic translation initiation factor 4B | T420 | 2.09 | 0.30 | |||
| S422 | 2.35 | 0.13 | |||||
| S425 | 3.09 | 0.09 | |||||
|
| |||||||
| Autophagy proteins | |||||||
|
| |||||||
| ATG16L1 | Autophagy-related protein 16-1 | S287 | 1.48 | 0.51 | 1.76 | 0.82 | |
| CAMKK2 | Calcium/calmodulin-dependent protein kinase kinase 2 | S100 | 2.15 | 0.73 | 1.11 | 1.06 | |
| RABGAP1 | Rab GTPase-activating protein 1 | T996 | 1.61 | 0.68 | |||
|
| |||||||
| Phosphatase | |||||||
|
| |||||||
| PPP2R5D | Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta | S573 | 2.0 | 0.68 | 1.66 | 1.32 | |
| PPP1R12A | Protein phosphatase 1 regulatory subunit 12A | S507 | 2.97 | 0.11 | 4.66 | 0.53 | |
| PTPN3 | Tyrosine-protein phosphatase non-receptor type 3 | S357 | 2.78 | 0.26 | |||
| PTPN11 | tyrosine-protein phosphatase non-receptor type 11 | S591 | 2.01 | 0.47 | 1.11 | 1.10 | |
| SSH2 | protein phosphatase Slingshot homolog 2 | S784 | 1.68 | 0.60 | |||
| PTPN14 | tyrosine-protein phosphatase non-receptor type 14 | S593 | 1.70 | 0.65 | 1.09 | 1.01 | |
| S594 | 1.70 | 0.65 | 1.18 | 0.93 | |||
|
| |||||||
| Proteins from PI3K/AKT/mTOR signaling pathway | |||||||
|
| |||||||
| EIF4EBP1 | Eukaryotic translation initiation factor 4E-binding protein 1 | S35 | 1.16 | 0.69 | |||
| T37 | 0.87 | 0.6 | 0.72 | 1.14 | |||
| T46 | 0.87 | 0.6 | 0.75 | 1.17 | |||
| S65 | 0.93 | 0.44 | 1.22 | 0.99 | |||
| T68 | 0.94 | 0.46 | |||||
| T70 | 0.17 | 2.6 | 0.83 | 1.15 | |||
| RPTOR | Regulatory-associated protein of mTOR isoform 1 | S863 | 0.84 | 0.58 | 0.91 | 1.02 | |
| RPS6 | 40S ribosomal protein S6 | S235 | 1.48 | 0.104 | 1.04 | 0.75 | |
| S236 | 2.66 | 0.13 | 1.17 | 0.91 | |||
| S240 | 2.48 | 0.15 | 0.7 | 1.14 | |||
| T241 | 2.75 | 0.12 | |||||
| S244 | 2.98 | 0.103 | 0.57 | 0.65 | |||
| S242 | 0.57 | 0.65 | |||||
| S247 | 0.57 | 0.65 | |||||
|
| |||||||
| Others | |||||||
|
| |||||||
| ARHGEF2 | Rho guanine nucleotide exchange factor 2 | S886 | 1.17 | 0.52 | 1.71 | 1.13 | |
| AKT1S1 | Proline-rich AKT1 substrate 1 | S183 | 0.97 | 0.24 | 1.09 | 1.02 | |
| NEDD4L | E3 ubiquitin-protein ligase NEDD4-like | S448 | 0.96 | 0.59 | 1.19 | 0.97 | |
| PEA15 | Astrocytic phosphoprotein PEA-15 | S104 | 2.53 | 0.66 | 0.86 | 1.09 | |
3.2 Specific phosphomotifs identified in the groups of regulated phosphosites
We compared the list of phosphosites having specific modes of regulation upon EGF treatment and TKI inhibition to identify potential phosphomotifs enriched among those phosphosites. All identified class I sites in three regulation groups with their neighbouring six residues on either side of the phosphorylation site were subjected to motif analysis using the Motif-All algorithm [35] in Phosphosite Plus [30]. The motifs observed were mostly arginine-enriched in the N-terminal side and proline-enriched in the C-terminal side of the phosphosites. The AGC and the CMGC protein kinase families of the human kinome represent mostly these enriched motifs (Fig S3). For the group of phosphosites that were hyperphosphorylated on EGF stimulation but hypophosphorylated with erlotinib inhibition, the motif observed was strongly representative of the RXRXXpS/T motif specific to AKT signaling [36], indicating that pretreatment of cells effectively prevents EGFR mediated activation of AKT. However, on the C-terminal side, the motif was phenylalanine-enriched in H3255 instead of proline-enriched in H1975 (Fig S3A), which is specific to the PKB/CaMKIIβ sub-family and CK2 sub-family, respectively. Erlotinib sensitive phosphosites that were not affected by EGF have a strong association with N+1 proline (Fig S3B) for both cell lines. Phosphosites that were down-regulated upon short-term exposure to EGF but upregulated with erlotinib pre-treatment showed a strong trend towards proline-dependent phosphomotif for H3255 but not H1975 (Fig S3C), such as those shared by MAPK and cyclin dependent kinases (CDK) [37]. For H1975, the motif was close to the CaMKIIα/δ group. The majority of phosphosites identified that showed no change upon either EGF stimulation or TKI inhibition had proline at N+1 and arginine at N-3 positions (Fig S3D).
3.3 Canonical pathways enriched in the groups of regulated phosphosites
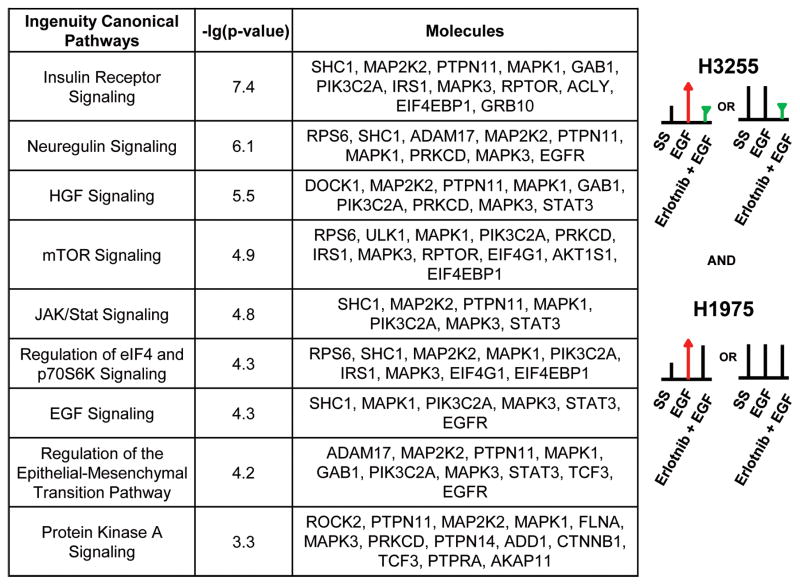
Global phosphorylation changes upon EGF stimulation or TKI inhibition are likely to identify changes in specific signaling pathways. We sought to identify canonical pathways that were altered among differentially phosphorylated proteins harbouring at least one phosphorylation site whose phosphorylation decreases upon erlotinib treatment in H3255 cells, but shows no change in H1975, an Ingenuity Pathway Analysis (IPA) was performed. The top canonical pathways that were enriched in this group included Insulin receptor, neuregulin, HGF mTOR, JAK/STAT, eIF4/p70S6K, PKA signaling, and regulation of EMT pathways (Fig 3, Fig S4). Interestingly, neuregulin signaling, including ERBB signaling was the most significant canonical pathway enriched in the group of phosphosites that were hyperphosphorylated upon EGF stimulation in both H3255 and H1975 cells and hypophosphorylated upon erlotinib inhibition in H3255 cells, but not H1975 cells (data not shown). This analysis exposes critical signaling pathways that fail to be inhibited by TKI treatment and illuminates potential mechanisms of TKI resistance. The data was also analyzed using SILAC ratios more than 2 standard deviation of the mean, which is the SILAC ratio cut-off of 2.1 and 0.42 for increased and decreased phosphorylation, respectively. The number of phosphosites regulated is much less compared to that from one standard deviation of the mean, and more phosphosites remained unchanged. However, the IPA analysis generated similar results (Fig. S5). The top canonical pathways that were enriched in this group included neuregulin, p70S6K, Insulin receptor, ERBB4, IGF-1, EGF, mTOR, eIF4/p70S6K, Rho GTPases signaling pathways.
Figure 3.
Ingenuity pathway analysis (IPA) shows top canonical pathways represented by proteins with phosphosites that are hypophosphorylated upon erlotinib inhibition in the sensitive cell line H3255, but remain unchanged in the resistant cell line H1975. The p value is a measure of the likelihood that the association between the set of phosphosites with the given pathway is due to random chance.
3.4 Changes in phosphorylation of proteins in the RAS-RAF-MAPK and PI3K-AKT-mTOR signaling pathways
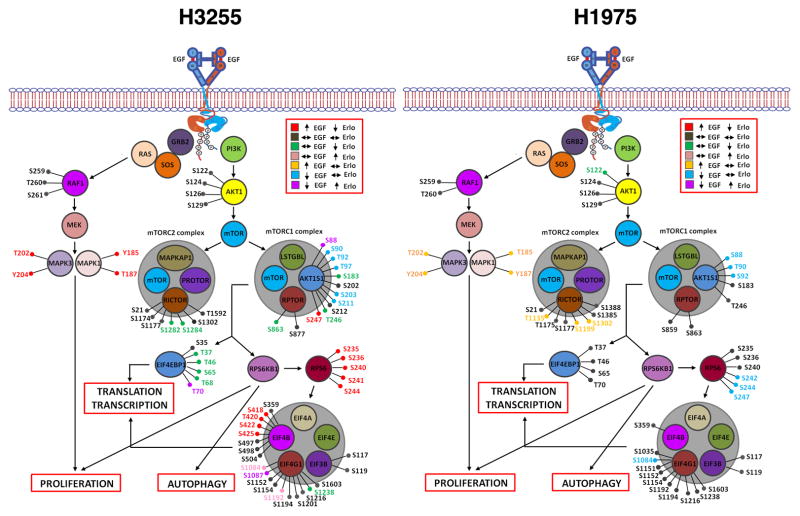
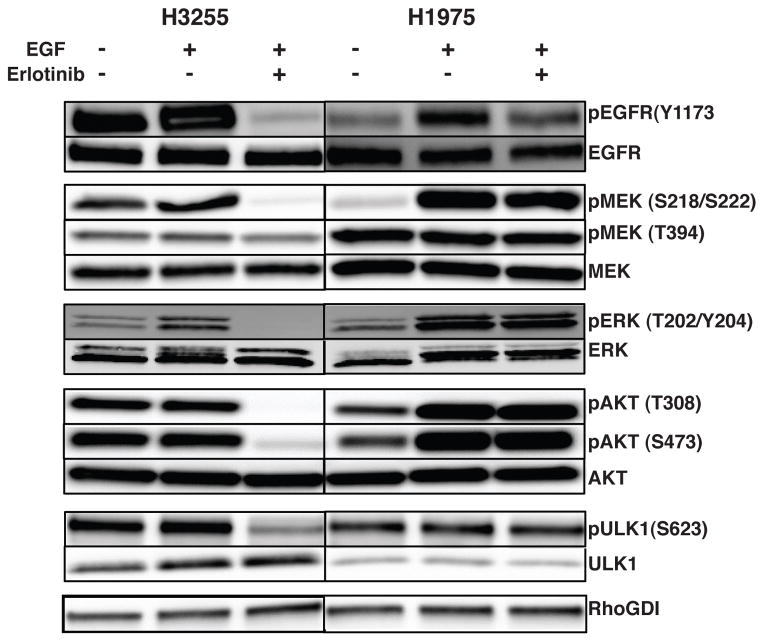
The phosphosites identified in proteins of the RAS-RAF-MAPK and the PI3K-AKT-mTOR signaling pathways and their phosphorylation changes upon EGF stimulation or TKI inhibition are shown in Fig 4. We identified several phosphorylation sites in EGFR with increased phosphorylation upon short-term exposure to EGF and decreased phosphorylation upon erlotinib inhibition in H3255 cells (Y1197, Y1172, Y998, S720, T725, S1064). Phosphorylation at two of these sites (Y1172 and Y1197) identified in H1975 increased upon EGF stimulation but had a modest decrease upon erlotinib inhibition (Table 1). An increase in phosphorylation of several autophosphorylation sites of MAPK1 (T185 and Y187) and MAPK3 (T202 and Y204) indicate that downstream MAPK signaling cascades are activated immediately upon EGF stimulation. Phosphorylation at these sites decreased in H3255 cells in the presence of erlotinib, but did not change in H1975 cells (Table 1). Several of the key sites of phosphorylation of EGFR, MEK and ERK were also validated by Western blots (Fig 5). In support of the mass spectrometry data, all of these sites had decreased phosphorylation in H3255 cells upon erlotinib treatment, while they remained unchanged in H1975 cells.
Figure 4.
Phosphosites identified in proteins of the RAS-RAF-MAPK and PI3K-AKT-mTOR signaling pathway in H3255 (left panel) and H1975 (right panel). Colors represent specific alterations in M/L (EGF/serum starved) and H/M (erlotinib+EGF/EGF) ratios as indicated.
Figure 5.
Validation of phosphorylation changes of specific phosphosites in proteins using Western blotting. Phosphorylation specific and total protein antibody blots of key proteins that are regulated upon EGF stimulation or erlotinib treatment in H3255 and H1975 cell lines. pEGFR, pAKT, pMEK, pERK and pULK1 levels were increased or not changed upon EGF stimulation and decreased upon erlotinib treatment in H3255, but not upon erlotinib treatment in H1975.
The PI3K-AKT-mTOR pathway is a commonly activated pathway in several cancers, including lung adenocarcinoma. Upstream kinases trigger the PI3K directly or via adaptors such as the IRS proteins and result in activation of AKT. Phosphorylation events at T308 and S473 have been shown to be vital for the full activation of AKT1 on EGF treatment [38]. AKT1 is phosphorylated on T308 in the activation loop by PDK1. Further phosphorylation of S473 residue located in the carboxyl hydrophobic domain is essential for the activity of AKT and likely mediated by the mTORC1 complex [39]. We could not detect AKT2 and AKT3 isoforms in our mass spectrometry analysis and could only detect certain sites on AKT1 that did not show any change on EGF or erlotinib treatment. We performed Western blot to examine the changes of phosphorylation at T308 and S473 of AKT. Phosphorylation was inhibited upon erlotinib treatment in H3255 cells while there was no change in H1975 cells (Fig 5). mTOR is a serine threonine kinase that regulates several processes such as mRNA translation, autophagy, cell cycle progression and cell survival. It is predominantly assembled into two complexes called mTORC1 (mTOR, RPTOR, AKT1S1/PRAS40, LST8/GβL and DEPTOR) and mTORC2 (mTOR, RICTOR, mSIN1, PROTOR, mLST8 and DEPTOR). mTORC1 controls cell growth in part by phosphorylating the p70S6 kinase (RPS6KB1) and the EIF4E binding protein 1(EIF4EBP1) that are key regulators of protein synthesis, while mTORC2 modulates cell survival by phosphorylating AKT1 and serum glucocorticoid regulated kinase 1 (SGK1) [40]. Several sites of phosphorylation on RICTOR were identified, of which S1282 and S1284 were dephosphorylated on treatment with erlotinib in H3255. S863 in RPTOR was found dephosphorylated on erlotinib treatment in H3255, but did not change in H1975 (Fig 4). mTORC1 activation results in phosphorylation of T37 and T46 in EIF4EBP1, which in turn results in further phosphorylation of S65 and T70. These phosphorylation events release eIF4E resulting in assembly of the eIF4F complex for cap-dependent translation [41, 42]. We identified all four of these sites in our experiments. Phosphorylation at positions T37, T46 and S65 did not increase upon EGF stimulation in both cell lines, but were inhibited upon erlotinib treatment in H3255. However, phosphorylation at position T70 was inhibited upon EGF stimulation and increased on erlotinib treatment. Further phosphorylation of ribosomal protein S6 (RPS6) and EIF4EBP1 by RPS6KB1 modulates translation of specific mRNAs [43, 44]. Interestingly, we identified five sites on RPS6 (S235 S236, S240, S241 and S244) that all exhibited increased phosphorylation upon EGF stimulation, and decreased phosphorylation upon treatment with erlotinib in H3255 (Fig 4). However, in H1975, the phosphorylation of S235 S236 and S240 remained unchanged with both EGF stimulation and erlotinib treatment. Phosphorylation of S242, S244 and S247 decreased upon short term EGF treatment and increased with erlotinib inhibition.
3.5 Serine Threonine kinases regulated by EGF stimulation or erlotinib inhibition
We identified phosphorylation sites in 145 serine/threonine kinases. 263 and 234 phosphosites were identified in 97 and 93 serine/threonine kinases from H3255 and H1975 cell lines, respectively (Table S2). Phosphosites were identified on about 30% of all kinases represented in human genome with slight enrichment of STE, CK1 and CMGC groups (more than 40%) and slight depletion of the atypical kinases (less than 20%). The phosphorylation level of most of the sites does not change upon EGF or erlotinib treatment. Phosphosites of certain kinases, such as MAPK1 (T185, Y187), MAPK3 (T202, Y204) were dephosphorylated upon TKI inhibition in H3255, but not in H1975 (Table 1 and Table S2). S218/S222 and S222/S226 of MEK1/MEK2 were hyperphosphorylated upon EGF stimulation and dephosphorylated with erlotinib in H3255. Phosphorylation at S295 and T394 of MEK2 was unaltered upon EGF stimulation, but decreased upon erlotinib treatment in H3255. However, T394 phosphorylation remained unchanged upon erlotinib treatment in H1975 cells. Specific phosphorylation sites in many other kinases, including WNK1, PRKD1, EEF2K, ROCK2 and RPS6KA1, were identified and were dephosphorylated upon erlotinib treatment in H3255 cells. Altered phosphorylation and expression of Protein Kinase C isoforms have been reported earlier in NSCLC cell lines in comparison to normal lung epithelial cells. The delta isoform PRKCD has been shown to be anti-apoptotic in function in NSCLC cells [45]. We identified a few phosphorylation sites in this protein. The phosphorylation at site S304 was stimulated with EGF and inhibited upon erlotinib treatment in H3255 cells.
3.6 Transcription/Translation related proteins regulated by EGF stimulation and erlotinib inhibition
We identified dynamic phosphorylation of specific sites of key translation regulatory proteins, such as EIF4B, EIF4EBP1, and EIF4G1 (Table S3). Phosphosites T420, S422 and S425 in EIF4B, T37, S65 and T68 in EIF4EBP1, S1238 in EIF4G1 were hyperphosphorylated on EGF stimulation, but dephosphorylated upon erlotinib treatment in H3255 cells. Three sites, including S65 and T37 in EIF4EBP1, and S1238 in EIF4G1 were also identified in H1975, but phosphorylation remained unchanged upon erlotinib treatment. The EIF4E binding family of proteins (EIF4EBPs) negatively regulates translation initiation by sequestering EIF4E, the rate limiting step in the formation of the translation initiation complex [46]. We also identified phosphorylation sites in several transcription factors, including FOXK1 (S441, S445), JUND (S100), DENND4A (S1630), RB1CC1 (S647), PHF2 (S625), and PDS5B (S1162, S1165, S1166, S1176, S1177, S1182 and S1358), which were dephosphorylated upon erlotinib treatment in H3255 cells. Among these, phosphorylation at S1177 in PDS5B and S625 in PHF2 was unaltered upon erlotinib treatment in H1975 cells (Table S1).
3.7 Components of autophagy pathway regulated by EGF stimulation and erlotinib inhibition
A large scale proteomic study has recently identified several components of the Autophagy Interaction Network (AIN) [47]. We identified 35 phosphosites in 18 proteins of the Autophagy network from both cell lines (Table S4). Phosphorylation of ATG16L1 (S287), PIK3C2A (S259), and ULK1 (S623, Fig S6) was reduced in H3255 cells, but remained unchanged in H1975 cells following erlotinib treatment. Interestingly, another phosphosite on ULK1, S775 was hyperphosphorylated upon EGF stimulation and the phosphorylation was inhibited upon erlotinib treatment in H1975 cells (Fig S6). In addition, phosphorylation of NEK9 (S29 and T333), PRKAA1 (S501, T505), PRKAA2 (S377), and RB1CC1 (S647) was inhibited upon erlotinib treatment in H3255 cells. Phosphorylation at T333 on NEK9 was inhibited upon erlotinib treatment in both cell lines, indicating that such change in phosphorylation may be an off-target effect of erlotinib. Interestingly, two phosphosites identified in ULK1 (S450 and S775) were hyperphosphorylated in H1975 upon EGF treatment, and S775 was hypophosphorylated with erlotinib inhibition. ULK1 kinase is involved in autophagy and required for autophagosome formation. It is the target of the mTOR kinase signaling pathway that regulates autophagy through the control of phosphorylation status of ATG13/KIAA0652 and ULK1 and the regulation of the ATG13-ULK1-RB1CC1 complex. ATG8/LC3, a key component of the autophagic cycle has associated proteins like SQSTM1 that function as receptors, which facilitate the engulfment of organelles or cellular proteins into the autophagosome [48] or alternatively regulate the process [47]. Four phosphosites were identified in protein SQSTM1, and they either stayed the same (S28, S272 and T269) or downregulated (S266) on EGF stimulation, but were not affected by erlotinib treatment. These dynamic phosphorylation changes in the autophagy network proteins suggest regulation of proteins involved in autophagy by mutant EGFR signaling.
3.8 Kinase substrate prediction analysis reveals CMGC group kinase substrates to be dephosphorylated upon short term EGF stimulation
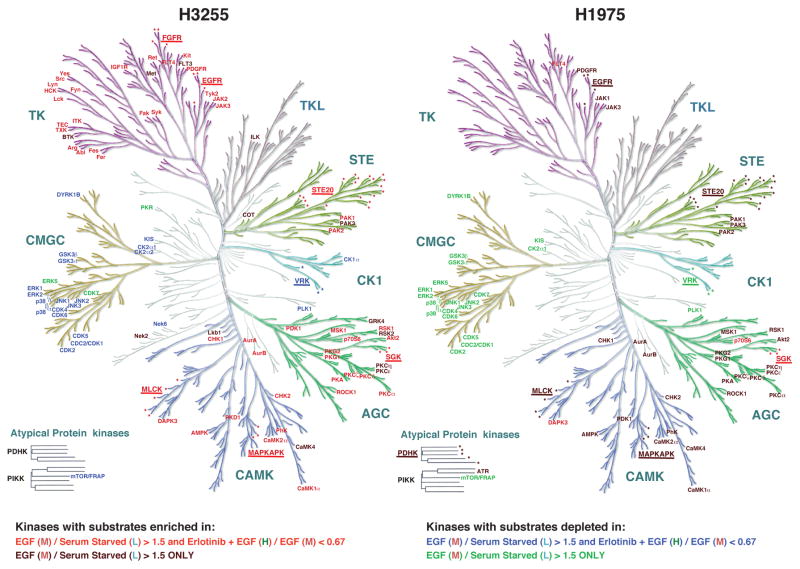
The phosphosites identified in our study are substrates of specific Ser/Thr and tyrosine kinases, many of which may be acting downstream of mutant EGFR in lung adenocarcinoma cells. We undertook a bioinformatic approach to predict specific protein kinases for all these substrates. The identified phosphosites were analyzed by GPS (Group-based Prediction System) 2.1.1 for identifying their cognate protein kinases [31]. The kinase groups predicted for each set of phosphosites sorted based on M/L and H/M SILAC ratios for H3255 and H1975 cells are shown in Fig S7. We subsequently identified statistically significant (P<0.01) “enrichment” or “depletion” of particular protein kinase groups predicted to be cognate kinases of the phosphopeptide substrates in sets of phosphosites hyperphosphorylated after EGF stimulation (M/L >1.5), or inhibited upon Erlotinib treatment (H/M<0.67) compared with the corresponding “baselines” (0.67< M/L < 1.5 and 0.67< H/M <1.5). EGF treatment in both H3255 and H1975 cells leads to increased number of phosphorylated substrates of basophilic kinases (several PKCs, PKGs, MAPKAPK, ROCK1, CAMK1a and CAMK2a), PAK and STE20 family kinases from STE group and Aurora kinases (Fig 6 and Table S5). CMGC group kinase substrates such as those of ERK, JNK and p38 as well as substrates of Casein kinases I and II were depleted among phosphosites hyperphosphorylated after EGF stimulation in both cell lines. Conversely phosphorylation levels of CMGC group kinase substrates were decreased in both cell lines, and in H3255, we identified statistically significant “enrichment” of named protein kinase substrates in sets of phosphosites dephosphorylated after EGF stimulation (M/L < 0.67) (Fig S8). Similar results were obtained by kinase-substrate prediction of groups of regulated phosphosites identified using SILAC ratio cut-offs of 2.1 and 0.42 (Table S5). It is remarkable, because there is clear decrease of phosphorylation of ERK substrates even after ERK itself became hyperphosphorylated upon EGF treatment. Interestingly, the same pattern of kinase family prediction was observed among phosphosites downregulated upon erlotinib treatment in H3255, suggesting erlotinib “on-target” effect. These kinases may be either direct or indirect targets of mutant EGFRs. In H1975, although the kinase families predicted for substrates with increased phosphorylation upon EGF stimulation was similar to those in H3255, the same pattern of kinase family distribution was not observed upon erlotinib inhibition.
Figure 6.
Group-based Prediction System (GPS) analysis reveals predicted kinases for groups of phosphosites with specific alteration upon EGF or erlotinib treatment in H3255 (left panel) and H1975 (right panel) cells. Color-coding of kinases is based on the statistically significant enrichment or depletion of kinase prediction among phosphosites in specific groups of SILAC ratios as shown. CAMK, AGC, STE and TK protein kinase groups were statistically enriched and CMGC group of kinases were depleted among phosphosites that had enhanced phosphorylation upon EGF stimulation and also among phosphosites that had reduced phosphorylation upon erlotinib inhibition in H3255 cells. In H1975 the same groups of kinases were statistically enriched or depleted, but only among phosphosites that had increased phosphorylation upon EGF stimulation. Names of kinase families are underlined and the protein kinase members are marked by asterisks for those predictions where the enrichment of a single kinase within a family did not reach significance, but the family as a whole reached statistical significance.
4 Discussion
The signal transduction pathways activated downstream of cell surface receptors, in parallel or as feedback to mutant oncogene regulate the overall tumorigenic phenotype. Here, we sought to identify the direct or indirect targets of mutant EGFR signaling in lung adenocarcinoma cells. To our knowledge, this is the first study to interrogate the global phosphorylation changes, predominantly of pSer/Thr sites, in mutant EGFR-driven lung adenocarcinoma cells after ligand stimulation or the first generation EGFR-TKI, erlotinib inhibition. We undertook a global phosphoproteomic approach to identify phosphosites downstream of lung cancer-specific mutant EGFRs. We used SILAC-based quantitative mass spectrometry to quantify the degree of phosphorylation in the identified phosphosites upon short term EGF stimulation of lung adenocarcinoma cells. Most importantly, using EGFR TKI, erlotinib-sensitive and -resistant lung adenocarcinoma cells, we demonstrated phosphorylation events that could or could not be inhibited by the mutant EGFR-directed TKI, erlotinib. This approach of quantitative phosphoproteomics is a powerful tool to examine signaling pathways activated downstream of mutant kinase oncogenes in an unbiased fashion. The use of specific kinase inhibitors in sensitive and resistant versions of lung adenocarcinoma cells allowed us to interrogate the signaling pathways that are inhibited by drug inhibition as a result of blocking the target. More importantly, the differential pathway activation between the sensitive and resistant cells may illuminate the mechanisms of resistance.
The primary outcome of our global phosphoproteomic study was the identification of specific sites of phosphorylation in key signaling proteins that appear to be direct or indirect, albeit proximal targets of mutant EGFRs (Table 1 and Table S1). For most of these differentially phosphorylated sites we do not expect changes in protein levels at three minutes of EGF stimulation or one hour of erlotinib inhibition. We estimated the protein amounts from the SILAC ratios of the non-phosphorylated peptides identified in our mass spectrometry screen and we did not detect major changes in protein amounts in our biological context (data not shown). Hence, the phosphorylation changes were a result of ligand stimulation or TKI inhibition. The use of the resistant lung adenocarcinoma cells in which the mechanism of resistance is known (i.e. a secondary mutation in the gatekeeper residue in EGFR, T790M), allowed us to identify specific phosphosites that are regulated by kinases, which are proximal targets of mutant EGFRs or by mutant EGFRs themselves (in case of the pTyr sites). These are the sites that are inhibited by erlotinib in H3255, but not in H1975 cells. It is interesting to note that the signaling pathways that were enriched in the group of proteins that have phosphosites that showed this pattern of phosphorylation upon TKI inhibition were all associated with EGFR signaling, suggesting that this global unbiased approach can be used to identify mechanisms of resistance to TKIs and chemotherapy [49, 50]. The proteins in this group overlap critical pathways, such as EGFR, Insulin receptor, HGF, mTOR, JAK/STAT, and eIF4/p70S6K signaling pathways.
We identified phosphosites in proteins that were regulated by mutant EGFR in these cells directly or mostly indirectly through other Ser/Thr kinases. The significance of this regulation is largely unknown. However, our identification of these ligand and/or TKI regulated sites in the phosphoproteome of lung adenocarcinoma raises important questions regarding the critical signaling network in play in these tumor cells. As an example, we identified twelve phosphosites in EGFR itself and phosphorylation at all of these sites was inhibited upon erlotinib treatment of TKI-sensitive H3255 cells. Only four of these are tyrosine sites and represent autophosphorylation sites. Ser/Thr kinases that phosphorylate the other regulated sites in EGFR may likely be direct targets of EGFR. Moreover, these sites may regulate normal EGFR function and signaling. Further studies are warranted to find the biological significance of the phosphosite regulation both in the context of wild-type and mutant EGFR signaling.
As predicted by the sensitivity pattern of the two lung adenocarcinoma cell lines used in this study, while 245 of all phosphosites identified in H3255 were dephosphorylated upon erlotinib inhibition, only 75 of all phosphosites were inhibited in H1975 cells that harbor the erlotinib resistant mutant EGFR under similar treatment conditions. This group of phosphosites in H1975 cells may indicate “off-target” effects of erlotinib. It is interesting to note that among these dephosphorylated sites there are key autophosphorylation sites in EGFR itself, such as Y1172 and Y1197, and also key sites in some proximal adaptors such as SHC1 (Y428), SOS1 (S-1134) and NCK1 (S85). However, the degree of inhibition of phosphorylation is still significantly more in H3255 cells compared to the resistant H1975 cells. This could be the result of inhibition of the wild-type EGFR that are expected to dimerize with EGFRL858R/T790M in the resistant lung adenocarcinoma cells. The dephosphorylation of these sites in H1975 could also be a result of “off-target” inhibition of another kinase by erlotinib. Such “off-target” kinases of erlotinib may include the SRC family of kinases [51]. Phosphorylation at specific sites of several kinases reduced upon erlotinib treatment of H1975 cells that are resistant to erlotinib. These include EPHA2 (S897), MAP4K4 (S648, S708), MAP3K1 (S-292), RPS6KA3 (T577), MKNK2 (S452), NEK9 (T333), ULK1 (S775) and others. It would be interesting to study whether phosphorylation of any of these sites regulates the function of corresponding kinase.
We identified several canonical pathways activated downstream of mutant EGFRs in lung adenocarcinoma cells upon short term EGF stimulation. These include MAPK and AKT-mTOR signaling pathways. Interestingly, in these lung adenocarcinoma cells, although MAPKs themselves were hyperphosphorylated, the MAPK targets exhibited overall decreased phosphorylation at three minutes of EGF stimulation and phosphorylation was unchanged on prior erlotinib treatment. Two separate bioinformatic analyses, the phosphomotif analysis (Fig S3) and the GPS based prediction (Fig 6, Fig S7, and Table S5) demonstrate this conclusion. Particular examples of such MAPK substrates are ZC3H13 (S77), ZMYM2 (S838), and ZYX (S344) [52]. Some of reported CDK substrates demonstrate the same dynamics such as SEP-5 (S327) [53], AAK1 (T389) [54] and ABL1 (S588) [55]. It is possible that at such short time periods after ligand stimulation, certain phosphatases are activated that dephosphorylate the targets of MAPKs so that subsequently the MAPKs themselves can re-phosphorylate those targets to a greater extent. One such candidate phosphatase identified in this study is PPP2R5D, the 56kD regulatory subunit of protein phosphatase 2A (PP2A), a phosphatase that has been shown to dephosphorylate targets of proline-directed or cAMP dependent protein kinases [56]. EGF stimulation increased and erlotinib treatment inhibited phosphorylation at S573 of this protein in the TKI sensitive H3255 cells while no alteration of phosphorylation was observed in the TKI resistant H1975 cells (Fig S9). Phosphorylation at the corresponding site by PKA has been shown to activate the phosphatase activity [57] of PP2A.
Supplementary Material
Supplementary Figure S1. (A) Western blot analysis of lysates from H3255 and H1975 shows overexpression of EGFRL858R in H3255 cells. Western blots were probed with EGFRL858R- specific (detecting specifically EGFRL858R mutant) and EGFRL858- specific (detecting corresponding wild-type EGFR, but not mutant EGFR) custom antibodies (NanoTools). tEGFR antibody (BD Bioscience) recognizes both mutant and wild-type EGFR. (B) Western blot analysis of phosphotyrosine (4G10, Millipore) from both cell extracts shows inhibition of phosphotyrosine upon erlotinib treatment of TKI-sensitive H3255 cells, but not -resistant H1975 cells. (C, D) IP-Western blot analysis of phosphotyrosine immunoprecipitates from lysates of H3255 and H1975 cells. The cells were treated with EGF (100ng/mL) for different time periods as indicated, or pretreated with erlotinib (100nM) before EGF stimulation. There is marked decrease in tyrosine phosphorylation in H3255 cells, but not in H1975 cells.
Supplementary Figure S2. Distribution of the normalized SILAC ratios with the mean +/− one standard deviation shown in dashed lines and the used cut-off of M/L (1.5) and H/M (0.67) shown in red.
Supplementary Figure S3. Phosphomotifs are preferentially enriched among differentially regulated phosphosites in H3255 and H1975. (A) Phosphomotif enriched among phosphosites with increased phosphorylation upon EGF treatment and decreased upon erlotinib inhibition. (B) Phosphorylation does not change with EGF treatment and goes down with erlotinib. (C) Phosphorylation goes down with EGF treatment and goes up with erlotinib. (D) Unchanged phosphosites with either EGF stimulation or erlotinib inhibition.
Supplementary Figure S4. Ingenuity pathway analysis (IPA) of groups of phosphosites that are inhibited upon erlotinib treatment in H3255 cells but not in H1975 cells shows that certain canonical pathways were statistically enriched. Four such pathways include Insulin receptor signaling, IGF-1 signaling, regulation of EIF4 and p70S6 Kinase, and JAK-STAT signaling pathways. Proteins with purple outline had identified peptides.
Supplementary Figure S5. Phosphosites and canonical pathways identified using SILAC ratio cut-offs more than 2 standard deviation of the mean. (A) Number of class I phosphosites (localization probability >0.75) identified with SILAC ratios >2.1 (increased), 0.42–2.1 (unchanged) and <0.42 (decreased) in H3255 (left panel) and H1975 (right panel) cells. Ratio M/L is EGF/Serum starved and ratio H/M is erlotinib + EGF / EGF states. (B) Ingenuity pathway analysis (IPA) shows top canonical pathways represented by proteins with phosphosites that were hypophosphorylated upon erlotinib inhibition in the sensitive cell line H3255, but remain unchanged in the resistant cell line H1975. The p value is a measure of the likelihood that the association between the set of phosphosites with the given pathway is due to random chance.
Supplementary Figure S6. MS and MS/MS spectra of phosphosites identified on ULK1 in H3255 (upper panel) and H1975 (middle and lower panel) cells. S623 phosphorylation was inhibited upon erlotinib treatment in H3255 cells, but showed no change in H1975 cells. S775 phosphorylation increased upon EGF stimulation of H1975 cells and the phosphorylation was inhibited upon erlotinib treatment.
Supplementary Figure S7. Distribution of GPS predictions among the major protein kinase groups for the phosphosites that are hyperphosphorylated, unchanged or dephosphorylated upon EGF treatment (A and B panels for H3255 and H1975 cell lines, respectively) or EGF and erlotinib treatment (C and D panels for H3255 and H1975 cell lines, respectively). E. GPS predictions among the major protein kinase groups for phosphosites that are hyperphosphorylated (EGF/serum starved >1.5), hypophosphorylated (EGF/Serum starved <0.67, and remain unchanged (ratio between 0.67and 1.5) at 5 minutes of EGF treatment of HeLa cells based on data published by Olsen et. al. [15].
Supplementary Figure S8. Distribution of GPS prediction count for AGC and CMGC family of kinases in H3255 (left panel) and H1975 (right panel) for the phosphosites with EGF stimulation ratio (M/L). AGC and CMGC family kinase-predictions are shown and all other predictions are groups as “others”. Number of predictions is plotted against log2(M/L) ratio. AGC family kinases are enriched among phosphosites that are hyperphosphorylated upon EGF stimulation and CMGC family kinases are enriched among phosphosites that are dephosphorylated upon EGF stimulation in both cell lines. However, since the total number of phosphosites identified is less in H1975, the total number of predictions for CMGC enrichment in H1975 does not reach statistical significance. This plot shows that the pattern of predictions for AGC and CMGC family kinases is the same for both H3255 and H1975.
Supplementary Figure S9. MS and MS/MS spectra of PPP2R5D peptide with S573 phosphorylation. Phosphorylation of S573 increased upon EGF stimulation and was inhibited upon erlotinib treatment of H3255 cells (upper panel). EGF stimulation did not alter phosphorylation at this site and erlotinib treatment increased phosphorylation in H1975 cells.
Supplementary Table 1: Phosphorylation sites identified in H3255 and H1975 cell lines.
Supplementary Table 2: Phosphorylation sites identified from protein kinases in H3255 and H1975 cell lines.
Supplementary Table 3: Selected translation and transcription factors that were dephosphorylated with erlotinib in both H3255 and H1975 cell lines.
Supplementary Table 4: Autophagy proteins identified in H3255 and H1975 cell lines.
Supplementary Table 5: Distribution of protein kinases predicted by GPS analysis to phosphorylate phosphosites that are regulated upon EGF and Erlotinib treatments. Fisher’s exact test was done with Benjamini-Hochberg adjustment for enrichment and depletion analysis of the set of phosphosites with specific SILAC ratios of EGF stimulation (M/L) and erlotinib inhibition (H/M) against phosphosites with no change of the corresponding ratios. p-values less than 0.01 are shown in bold.
Acknowledgments
This work was supported by US National Cancer Institute Center for Cancer Research (NCI-CCR) Intramural Research Program ZIA BC 011410 (U.G.) and K99 Career Transition award (K99CA140792) (U.G.). The work was also supported in part by an NIH roadmap grant for Technology Centers of Networks and Pathways (U54GM103520), NCI’s Clinical Proteomic Tumor Analysis Consortium initiative (U24CA160036) and a contract (HHSN268201000032C) from the National Heart, Lung and Blood Institute (A.P.). H.K.C.J was a recipient of a Senior Research Fellowship from Council of Scientific and Industrial Research (CSIR), India.
Footnotes
Conflict of interest: The authors declare no financial or commercial conflict of interest.
Data submission: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD001101.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Paez JG, Janne PA, Lee JC, Tracy S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Pao W, Miller V, Zakowski M, Doherty J, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pao W, Miller VA, Politi KA, Riely GJ, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 7.Oxnard GR, Arcila ME, Sima CS, Riely GJ, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in EGFR-mutant lung cancer: distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res. 2011;17:1616–1622. doi: 10.1158/1078-0432.CCR-10-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelman JA, Mukohara T, Zejnullahu K, Lifshits E, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib-sensitizing EGFR mutations in lung cancer activate anti-apoptotic pathways. Science. 2004;305:1163–1167. doi: 10.1126/science.1101637. [DOI] [PubMed] [Google Scholar]
- 10.Sharma SV, Gajowniczek P, Way IP, Lee DY, et al. A common signaling cascade may underlie “addiction” to the Src, BCR-ABL, and EGF receptor oncogenes. Cancer Cell. 2006;10:425–435. doi: 10.1016/j.ccr.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma SV, Fischbach MA, Haber DA, Settleman J. “Oncogenic shock”: explaining oncogene addiction through differential signal attenuation. Clin Cancer Res. 2006;12:4392s–4395s. doi: 10.1158/1078-0432.CCR-06-0096. [DOI] [PubMed] [Google Scholar]
- 12.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 13.Blagoev B, Kratchmarova I, Ong SE, Nielsen M, et al. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat Biotechnol. 2003;21:315–318. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, et al. Time-resolved Mass Spectrometry of Tyrosine Phosphorylation Sites in the Epidermal Growth Factor Receptor Signaling Network Reveals Dynamic Modules. Mol Cell Proteomics. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Olsen JV, Blagoev B, Gnad F, Macek B, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 16.Tong J, Taylor P, Moran MF. Proteomic analysis of the epidermal growth factor receptor (EGFR) interactome and post-translational modifications associated with receptor endocytosis in response to EGF and stress. Molecular & cellular proteomics : MCP. 2014;13:1644–1658. doi: 10.1074/mcp.M114.038596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rikova K, Guo A, Zeng Q, Possemato A, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 18.Guo A, Villen J, Kornhauser J, Lee KA, et al. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci U S A. 2008;105:692–697. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Assiddiq BF, Tan KY, Toy W, Chan SP, et al. EGFR S1166 phosphorylation induced by a combination of EGF and gefitinib has a potentially negative impact on lung cancer cell growth. Journal of proteome research. 2012;11:4110–4119. doi: 10.1021/pr3002029. [DOI] [PubMed] [Google Scholar]
- 20.Kim Y, Li Z, Apetri M, Luo B, et al. Temporal resolution of autophosphorylation for normal and oncogenic forms of EGFR and differential effects of gefitinib. Biochemistry. 2012;51:5212–5222. doi: 10.1021/bi300476v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang G, Fang B, Liu RZ, Lin H, et al. Mass spectrometry mapping of epidermal growth factor receptor phosphorylation related to oncogenic mutations and tyrosine kinase inhibitor sensitivity. Journal of proteome research. 2011;10:305–319. doi: 10.1021/pr1006203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guha U, Chaerkady R, Marimuthu A, Patterson AS, et al. Comparisons of tyrosine phosphorylated proteins in cells expressing lung cancer-specific alleles of EGFR and KRAS. Proc Natl Acad Sci U S A. 2008;105:14112–14117. doi: 10.1073/pnas.0806158105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harsha HC, Molina H, Pandey A. Quantitative proteomics using stable isotope labeling with amino acids in cell culture. Nat Protoc. 2008;3:505–516. doi: 10.1038/nprot.2008.2. [DOI] [PubMed] [Google Scholar]
- 24.Zhong J, Kim MS, Chaerkady R, Wu X, et al. TSLP signaling network revealed by SILAC-based phosphoproteomics. Mol Cell Proteomics. 2012;11:M112017764. doi: 10.1074/mcp.M112.017764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taouatas N, Altelaar AF, Drugan MM, Helbig AO, et al. Strong cation exchange-based fractionation of Lys-N-generated peptides facilitates the targeted analysis of post-translational modifications. Mol Cell Proteomics. 2009;8:190–200. doi: 10.1074/mcp.M800285-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Pinkse MW, Lemeer S, Heck AJ. A protocol on the use of titanium dioxide chromatography for phosphoproteomics. Methods Mol Biol. 2011;753:215–228. doi: 10.1007/978-1-61779-148-2_14. [DOI] [PubMed] [Google Scholar]
- 27.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJ. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Mol Cell Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 28.Thingholm TE, Jorgensen TJ, Jensen ON, Larsen MR. Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat Protoc. 2006;1:1929–1935. doi: 10.1038/nprot.2006.185. [DOI] [PubMed] [Google Scholar]
- 29.Cox J, Neuhauser N, Michalski A, Scheltema RA, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 30.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xue Y, Ren J, Gao X, Jin C, et al. GPS 2. 0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics. 2008;7:1598–1608. doi: 10.1074/mcp.M700574-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manning BD, Cantley LC. Hitting the target: emerging technologies in the search for kinase substrates. Sci STKE. 2002;2002:PE49. doi: 10.1126/stke.2002.162.pe49. [DOI] [PubMed] [Google Scholar]
- 33.Chartier M, Chenard T, Barker J, Najmanovich R. Kinome Render: a stand-alone and web-accessible tool to annotate the human protein kinome tree. PeerJ. 2013;1:e126. doi: 10.7717/peerj.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tracy S, Mukohara T, Hansen M, Meyerson M, et al. Gefitinib induces apoptosis in the EGFRL858R non-small-cell lung cancer cell line H3255. Cancer Res. 2004;64:7241–7244. doi: 10.1158/0008-5472.CAN-04-1905. [DOI] [PubMed] [Google Scholar]
- 35.He Z, Yang C, Guo G, Li N, Yu W. Motif-All: discovering all phosphorylation motifs. BMC Bioinformatics. 2011;12(Suppl 1):S22. doi: 10.1186/1471-2105-12-S1-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Obata T, Yaffe MB, Leparc GG, Piro ET, et al. Peptide and protein library screening defines optimal substrate motifs for AKT/PKB. J Biol Chem. 2000;275:36108–36115. doi: 10.1074/jbc.M005497200. [DOI] [PubMed] [Google Scholar]
- 37.Nigg EA. Targets of cyclin-dependent protein kinases. Curr Opin Cell Biol. 1993;5:187–193. doi: 10.1016/0955-0674(93)90101-u. [DOI] [PubMed] [Google Scholar]
- 38.Kumar N, Afeyan R, Sheppard S, Harms B, Lauffenburger DA. Quantitative analysis of Akt phosphorylation and activity in response to EGF and insulin treatment. Biochem Biophys Res Commun. 2007;354:14–20. doi: 10.1016/j.bbrc.2006.12.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 40.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 41.Gingras AC, Gygi SP, Raught B, Polakiewicz RD, et al. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 1999;13:1422–1437. doi: 10.1101/gad.13.11.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gingras AC, Raught B, Gygi SP, Niedzwiecka A, et al. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 2001;15:2852–2864. doi: 10.1101/gad.912401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruvinsky I, Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci. 2006;31:342–348. doi: 10.1016/j.tibs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 45.Clark AS, West KA, Blumberg PM, Dennis PA. Altered protein kinase C (PKC) isoforms in non-small cell lung cancer cells: PKCdelta promotes cellular survival and chemotherapeutic resistance. Cancer Res. 2003;63:780–786. [PubMed] [Google Scholar]
- 46.Kim YY, Von Weymarn L, Larsson O, Fan D, et al. Eukaryotic initiation factor 4E binding protein family of proteins: sentinels at a translational control checkpoint in lung tumor defense. Cancer Res. 2009;69:8455–8462. doi: 10.1158/0008-5472.CAN-09-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 49.Yoshida T, Zhang G, Smith MA, Lopez AS, et al. Tyrosine phosphoproteomics identifies both codrivers and cotargeting strategies for T790M-related EGFR-TKI resistance in non-small cell lung cancer. Clin Cancer Res. 2014;20:4059–4074. doi: 10.1158/1078-0432.CCR-13-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee BY, Hochgrafe F, Lin HM, Castillo L, et al. Phosphoproteomic profiling identifies focal adhesion kinase as a mediator of docetaxel resistance in castrate-resistant prostate cancer. Mol Cancer Ther. 2014;13:190–201. doi: 10.1158/1535-7163.MCT-13-0225-T. [DOI] [PubMed] [Google Scholar]
- 51.Weber C, Schreiber TB, Daub H. Dual phosphoproteomics and chemical proteomics analysis of erlotinib and gefitinib interference in acute myeloid leukemia cells. J Proteomics. 2012;75:1343–1356. doi: 10.1016/j.jprot.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 52.Courcelles M, Fremin C, Voisin L, Lemieux S, et al. Phosphoproteome dynamics reveal novel ERK1/2 MAP kinase substrates with broad spectrum of functions. Mol Syst Biol. 2013;9:669. doi: 10.1038/msb.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amin ND, Zheng YL, Kesavapany S, Kanungo J, et al. Cyclin-dependent kinase 5 phosphorylation of human septin SEPT5 (hCDCrel-1) modulates exocytosis. J Neurosci. 2008;28:3631–3643. doi: 10.1523/JNEUROSCI.0453-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blethrow JD, Glavy JS, Morgan DO, Shokat KM. Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc Natl Acad Sci U S A. 2008;105:1442–1447. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kipreos ET, Wang JY. Differential phosphorylation of c-Abl in cell cycle determined by cdc2 kinase and phosphatase activity. Science. 1990;248:217–220. doi: 10.1126/science.2183353. [DOI] [PubMed] [Google Scholar]
- 56.Goedert M, Jakes R, Qi Z, Wang JH, Cohen P. Protein phosphatase 2A is the major enzyme in brain that dephosphorylates tau protein phosphorylated by proline-directed protein kinases or cyclic AMP-dependent protein kinase. J Neurochem. 1995;65:2804–2807. doi: 10.1046/j.1471-4159.1995.65062804.x. [DOI] [PubMed] [Google Scholar]
- 57.Ahn JH, McAvoy T, Rakhilin SV, Nishi A, et al. Protein kinase A activates protein phosphatase 2A by phosphorylation of the B56delta subunit. Proc Natl Acad Sci U S A. 2007;104:2979–2984. doi: 10.1073/pnas.0611532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1. (A) Western blot analysis of lysates from H3255 and H1975 shows overexpression of EGFRL858R in H3255 cells. Western blots were probed with EGFRL858R- specific (detecting specifically EGFRL858R mutant) and EGFRL858- specific (detecting corresponding wild-type EGFR, but not mutant EGFR) custom antibodies (NanoTools). tEGFR antibody (BD Bioscience) recognizes both mutant and wild-type EGFR. (B) Western blot analysis of phosphotyrosine (4G10, Millipore) from both cell extracts shows inhibition of phosphotyrosine upon erlotinib treatment of TKI-sensitive H3255 cells, but not -resistant H1975 cells. (C, D) IP-Western blot analysis of phosphotyrosine immunoprecipitates from lysates of H3255 and H1975 cells. The cells were treated with EGF (100ng/mL) for different time periods as indicated, or pretreated with erlotinib (100nM) before EGF stimulation. There is marked decrease in tyrosine phosphorylation in H3255 cells, but not in H1975 cells.
Supplementary Figure S2. Distribution of the normalized SILAC ratios with the mean +/− one standard deviation shown in dashed lines and the used cut-off of M/L (1.5) and H/M (0.67) shown in red.
Supplementary Figure S3. Phosphomotifs are preferentially enriched among differentially regulated phosphosites in H3255 and H1975. (A) Phosphomotif enriched among phosphosites with increased phosphorylation upon EGF treatment and decreased upon erlotinib inhibition. (B) Phosphorylation does not change with EGF treatment and goes down with erlotinib. (C) Phosphorylation goes down with EGF treatment and goes up with erlotinib. (D) Unchanged phosphosites with either EGF stimulation or erlotinib inhibition.
Supplementary Figure S4. Ingenuity pathway analysis (IPA) of groups of phosphosites that are inhibited upon erlotinib treatment in H3255 cells but not in H1975 cells shows that certain canonical pathways were statistically enriched. Four such pathways include Insulin receptor signaling, IGF-1 signaling, regulation of EIF4 and p70S6 Kinase, and JAK-STAT signaling pathways. Proteins with purple outline had identified peptides.
Supplementary Figure S5. Phosphosites and canonical pathways identified using SILAC ratio cut-offs more than 2 standard deviation of the mean. (A) Number of class I phosphosites (localization probability >0.75) identified with SILAC ratios >2.1 (increased), 0.42–2.1 (unchanged) and <0.42 (decreased) in H3255 (left panel) and H1975 (right panel) cells. Ratio M/L is EGF/Serum starved and ratio H/M is erlotinib + EGF / EGF states. (B) Ingenuity pathway analysis (IPA) shows top canonical pathways represented by proteins with phosphosites that were hypophosphorylated upon erlotinib inhibition in the sensitive cell line H3255, but remain unchanged in the resistant cell line H1975. The p value is a measure of the likelihood that the association between the set of phosphosites with the given pathway is due to random chance.
Supplementary Figure S6. MS and MS/MS spectra of phosphosites identified on ULK1 in H3255 (upper panel) and H1975 (middle and lower panel) cells. S623 phosphorylation was inhibited upon erlotinib treatment in H3255 cells, but showed no change in H1975 cells. S775 phosphorylation increased upon EGF stimulation of H1975 cells and the phosphorylation was inhibited upon erlotinib treatment.
Supplementary Figure S7. Distribution of GPS predictions among the major protein kinase groups for the phosphosites that are hyperphosphorylated, unchanged or dephosphorylated upon EGF treatment (A and B panels for H3255 and H1975 cell lines, respectively) or EGF and erlotinib treatment (C and D panels for H3255 and H1975 cell lines, respectively). E. GPS predictions among the major protein kinase groups for phosphosites that are hyperphosphorylated (EGF/serum starved >1.5), hypophosphorylated (EGF/Serum starved <0.67, and remain unchanged (ratio between 0.67and 1.5) at 5 minutes of EGF treatment of HeLa cells based on data published by Olsen et. al. [15].
Supplementary Figure S8. Distribution of GPS prediction count for AGC and CMGC family of kinases in H3255 (left panel) and H1975 (right panel) for the phosphosites with EGF stimulation ratio (M/L). AGC and CMGC family kinase-predictions are shown and all other predictions are groups as “others”. Number of predictions is plotted against log2(M/L) ratio. AGC family kinases are enriched among phosphosites that are hyperphosphorylated upon EGF stimulation and CMGC family kinases are enriched among phosphosites that are dephosphorylated upon EGF stimulation in both cell lines. However, since the total number of phosphosites identified is less in H1975, the total number of predictions for CMGC enrichment in H1975 does not reach statistical significance. This plot shows that the pattern of predictions for AGC and CMGC family kinases is the same for both H3255 and H1975.
Supplementary Figure S9. MS and MS/MS spectra of PPP2R5D peptide with S573 phosphorylation. Phosphorylation of S573 increased upon EGF stimulation and was inhibited upon erlotinib treatment of H3255 cells (upper panel). EGF stimulation did not alter phosphorylation at this site and erlotinib treatment increased phosphorylation in H1975 cells.
Supplementary Table 1: Phosphorylation sites identified in H3255 and H1975 cell lines.
Supplementary Table 2: Phosphorylation sites identified from protein kinases in H3255 and H1975 cell lines.
Supplementary Table 3: Selected translation and transcription factors that were dephosphorylated with erlotinib in both H3255 and H1975 cell lines.
Supplementary Table 4: Autophagy proteins identified in H3255 and H1975 cell lines.
Supplementary Table 5: Distribution of protein kinases predicted by GPS analysis to phosphorylate phosphosites that are regulated upon EGF and Erlotinib treatments. Fisher’s exact test was done with Benjamini-Hochberg adjustment for enrichment and depletion analysis of the set of phosphosites with specific SILAC ratios of EGF stimulation (M/L) and erlotinib inhibition (H/M) against phosphosites with no change of the corresponding ratios. p-values less than 0.01 are shown in bold.