Abstract
Seagrasses thrive in anoxic sediments where sulphide can accumulate to phytotoxic levels. So how do seagrasses persist in this environment? Here, we propose that radial oxygen loss (ROL) from actively growing root tips protects seagrasses from sulphide intrusion not only by abiotically oxidising sulphides in the rhizosphere of young roots, but also by influencing the abundance and spatial distribution of sulphate-reducing and sulphide-oxidising bacteria. We used a novel multifaceted approach combining imaging techniques (confocal fluorescence in situ hybridisation, oxygen planar optodes, and sulphide diffusive gradients in thin films) with microbial community profiling to build a complete picture of the microenvironment of growing roots of the seagrasses Halophila ovalis and Zostera muelleri. ROL was restricted to young root tips, indicating that seagrasses will have limited ability to influence sulphide oxidation in bulk sediments. On the microscale, however, ROL corresponded with decreased abundance of potential sulphate-reducing bacteria and decreased sulphide concentrations in the rhizosphere surrounding young roots. Furthermore, roots leaking oxygen had a higher abundance of sulphide-oxidising cable bacteria; which is the first direct observation of these bacteria on seagrass roots. Thus, ROL may enhance both abiotic and bacterial sulphide oxidation and restrict bacterial sulphide production around vulnerable roots, thereby helping seagrasses to colonise sulphide-rich anoxic sediments.
Subject terms: Plant ecology, Environmental microbiology
Introduction
The health and productivity of seagrass ecosystems across the world is linked with the sulphur cycle. The reasons for this lies in the high availability of dissolved sulphate in seawater combined with the reduced nature of seagrass sediments [1]. As such, the mineralisation of organic matter and release of plant available nutrients (N and P) in the sediment is typically coupled to microbial sulphate reduction [2, 3]. In the process of liberating nutrients from organic matter, sulphate reduction also results in the production of sulphides such as H2S [4]. H2S is a known phytotoxin of plants as it disables energy production by inhibiting the activity of cytochrome c oxidase and other metal containing enzymes [4]. This paradox leads us to question how seagrasses survive in reduced sediments with high dissolved sulphide concentrations.
At present, the survival of seagrasses in sulphide-rich sediments has been attributed, in part, to the radial diffusion of oxygen from within roots to the rhizosphere (commonly termed radial oxygen loss, or ROL) [5, 6]. This ROL can protect seagrasses by abiotically oxidising sulphide to less harmful forms, thus reducing entry of toxic sulphide into roots [7]. However, the presence of sulphide-oxidising bacteria on seagrass roots may prove a more efficient strategy for the removal of sulphides from the sediment than abiotic removal, as bacterially mediated sulphide oxidation can be 1000—10,000 times faster than abiotic oxidation [8]. The idea of plant protection by microbial sulphide oxidation is not new, and the presence of sulphide-oxidising bacteria inside the gills of Lucinid clams have been associated with decreased sulphide concentration in seagrass sediments [9]. Free-living sulphide-oxidising Beggiatoa have also been shown to decrease sulphide concentrations in the soils of waterlogged rice seedlings [10]. However, similar mutually favourable interactions between free-living sulphide oxidisers and seagrass roots are yet to be established.
One group of free-living sulphide-oxidising bacteria that are suited to living in the oxic–anoxic microzones of seagrass roots are the filamentous bacteria within the Desulfobulbaceae known as ‘cable bacteria’ [11]. Cable bacteria span the oxic–anoxic interphase and transport electrons along their filaments to connect sulphide oxidation in the anoxic layer to reduction of oxygen in the oxic layer [12]. Recently, cable bacteria have been found in a diversity of habitats that have characteristically high sulphide levels, including sediments in salt marshes and adjacent to seagrass beds [13]. However, their presence has yet to be confirmed in the seagrass rhizosphere. If cable bacteria do occur in association with seagrass roots, benefits would exist for the seagrass (as an additional defence against sulphide intrusion) and for the cable bacteria (via the creation of oxic microzones, and hence generation of electron acceptors in an otherwise anoxic and high-sulphide environment).
As the primary electron acceptor, O2 leaking from seagrass roots has the potential to influence the balance between sulphide-oxidising bacteria (sulphide consumption) and sulphate-reducing bacteria (sulphide production) within the rhizosphere, but this essential link has not been explored. Hence, although ROL has some capacity to directly oxidise sulphide in the rhizosphere, we propose that the oxic microenvironment generated around seagrass roots could also enhance the oxidisation of sulphides by encouraging the colonisation of sulphide-oxidising bacteria through the provision of electron acceptors (O2, but also NO2− or NO3− via nitrification), and limit sulphide production by creating an environment that is not favourable for sulphate-reducing bacteria [14].
Here, we use a novel multifaceted approach to link microscale patterns of ROL and sulphide concentrations in rhizospheres of Halophila ovalis and Zostera muelleri, to colonisation patterns of sulphide-oxidising and sulphate-reducing bacteria. This approach enabled us to show, for the first time, the spatial extent and influence of ROL on colonisation of sulphate reducers and sulphide oxidisers on seagrass roots in situ and, ultimately, provided new evidence for how seagrass survive in sulphide-rich sediments.
Materials and methods
Seagrass and sediment collection
H. ovalis, Z. muelleri, and sediments were collected from a water depth of ~ 1 m using Perspex corers (10 cm i.d., 30 cm depth) from the Swan River estuary (approximately 4 km from the river mouth; − 32.03, 115.76), Western Australia in November 2016 (Austral Spring). H. ovalis is the dominant seagrass in the Swan estuary and its growth has previously been found to be constrained by conditions of the river sediment (high concentrations of acid volatile sulphide and organic matter) [15]. High sulphide intrusion into roots of Z. muelleri has also been correlated with reduced biomass and shoot density at sites in an estuary in the south-east of Australia [16].
The two seagrass species were growing as a mixed meadow in salinity close to that of seawater (~ 30 – 32 psu). Maximum time-integrated sediment sulphide concentrations in the meadow measured using sulphide diffusive gradient in thin films (DGTs; see section below and supplementary) were 172 µM during the day and 84 µM during the night (Fig. S1). Intact cores of seagrasses and sediment were kept in water in insulated boxes and transported back to the University of Western Australia within an hour of collection. On return to the laboratory, plants were carefully separated from the sediment by rinsing in water collected from the sample site. Ramets consisting of three shoots with intact apicals were immediately planted into rhizoboxes. Each rhizobox (20 cm (h) × 20 cm (l) × 2 cm (w)) consisted of two clear acrylic sheets (2 mm thickness) with an open square spacer of water-resistant foam between them (Fig. S2). One side of the rhizobox was glued permanently to the foam, whereas the other side was sealed using non-toxic silicone grease and held together with rubber bands to make the side detachable, yet water tight. Each rhizobox was fitted with oxygen planar optodes (see section below) and filled with sieved (< 2 mm) sediment from the sample site. The sieved and homogenised sediment was a silty sand with a bulk density of 1.50 ± 0.09 g cm−3, a porosity of 52.8 ± 2.02 % and organic matter content of 3.26 ± 0.05 %. The rhizoboxes were then placed within larger (25 L) temperature controlled (24 °C) recirculating glass aquaria filled with aerated filtered seawater (~ 30 psu). Rhizoboxes were covered with black plastic to avoid interference from ambient light and tilted at 45° to encourage root growth along the optode wall (Fig. S2). Plants were grown under an irradiance of 250 µmol photons m−2 s−1 (measured with a LI-COR underwater quantum sensor LI−192 connected to a LI−250A light meter) and a 14:10 h light:dark cycle. All measurements were performed on new roots that had grown into the sediments after transplanting.
Oxygen planar optode measurements
The two-dimensional O2 distribution in seagrass rhizospheres was mapped using planar oxygen optodes consisting of a 1% (wt) O2 quenchable platinum(II) meso-tetra(pentafluorophenyl)porphine combined with a 1% (wt) reference dye (Macrolex © fluorescence yellow 10GN) immobilised in 4% (wt) polystyrene coated on a transparent 100 µm polyester support [17]. The optode was covered with a layer of grey silicone (biocide-free) for insulation and to minimise soil auto-fluorescence [18]. Both dye and silicone layers were applied using a K-Hand coater (50 µm wet film; RK Print Coat Instruments, UK). Planar optodes were taped to the side of the rhizoboxes using waterproof tape and a thin water film, while carefully avoiding air bubbles.
The oxygen planar optodes were imaged using a colour ratiometric approach, where seven blue LED’s (λ-peak = 447.5 nm; Luxeonstar, Canada) equipped with a dichroic blue filter (CDB-5051; UQC optics, UK) excited both dyes to create duel emission; red luminescence from the O2-sensitive dye and green luminescence from the reference dye. Emission was captured using a modified digital SLR (Canon EOS 1000D) equipped with a macro lens (Sigma 50 mm F2.8) and a long-pass emission filter (OG 530, Schott; UQG optics, UK). The LEDs and SLR camera were controlled via USB using the software ‘Look@RGB’ (imaging.fish-n-chips.de), which also splits images into red, green, and blue channels. The intensity ratio (red channel–green channel)/green channel) was used to measure O2 concentration at the surface of the optode using previously obtained calibration curves. These calibration curves were obtained with and without a strip of DGT (see below) by purging with N2 and ambient air in filtered seawater (same salinity and temperature as the experiment) and monitored continuously with a dissolved O2 meter (YSI ProODO). Two-point calibrations were subsequently performed throughout the experiment using anoxic sediment (0 µmol L−1 O2; 0% air saturation) and the overlying water column (260 µmol L−1 O2; 100% air saturation) as a reference to check and correct for any sensor drift over time. Further details regarding calibration calculations and image capture and processing can be found in Morten et al. [17]. All images were processed using the free software ImageJ (version 1.48) (https://rsbweb.nih.gov/ij/).
Oxygen imaging and root sampling
The oxygen planar optodes were sufficiently transparent to discern roots growing behind them, which allowed the continual monitoring of root growth and subsequent ROL. Oxygen concentration, width, and length of the oxic area around roots were used as indicators of the extent of ROL using tools in ImageJ. To corroborate the ROL patterns observed using O2 planar optodes, roots of Z. muelleri and H. ovalis were also examined for ROL using the methylene blue agar method [19]. Detailed methodology on methylene blue staining can be found in the supplementary.
At the end of the 4 weeks, roots were sampled for both Fluorescence in situ hybridisation (FISH) analysis and 16 S rRNA sequencing (see section below). For FISH sampling, ROL from the roots of three replicate plants in separate rhizoboxes of H. ovalis and Z. muelleri was imaged and immediately quantified. Rhizoboxes were then carefully opened and roots were cut using the planar optode image as a reference to identify whole roots that were exhibiting ROL (deemed ‘leaking’) and those exhibiting no ROL (deemed ‘non-leaking’). H. ovalis has only one root per rhizome node and the apical root consistently exhibited ROL, whereas no ROL was seen from roots on older nodes. Hence, we collected the root from the second node as the ‘non-leaking’ root. For Z. muelleri, multiple roots grow from each rhizome node and hence we selected roots that exhibited ROL or no ROL from the apical shoot based on the optode image. Both leaking and non-leaking roots were cut at the root tip (~ 5 mm, which includes the root cap and elongation zone with no root hairs) and at the root hair zone (~ 20–40 mm from the root tip). Root hairs were absent in the elongation zone for both leaking and non-leaking roots. The unwashed root segments were immediately placed into a 4% formaldehyde (v/v) solution in phosphate-buffered saline (PBS) and fixed over night at 4 °C. Roots were then washed in 1 × PBS three times and stored in a 1x PBS/96 % EtOH (40:60) at − 20 °C until hybridisations (see below) [20].
For DNA sampling, Z. muelleri roots were sampled in the same way as those for FISH, that is, ROL from roots of four replicate plants were imaged and quantified, leaking and non-leaking unwashed roots were removed from the sediment, cut into sections defined above and immediately frozen at − 80 °C. Bulk sediment DNA were also collected from rhizoboxes. A schematic of the sampling design can be found in Supplementary Figure 3.
Sulphide DGT
The two-dimensional distribution of sulphide in the rhizosphere was measured using the DGT technique. This approach consists of a polyacrylamide hydrogel impregnated with silver iodide (AgI(s)) that reacts with dissolved sulphide to form a black Ag2S(s) precipitate, which is quantified using computer-imaging densitometry [21]. To minimise the blurring of the oxygen distributions measured by the planar optodes, the AgI DGT gels were prepared at ~ 150 µm thickness (i.e., much thinner than the conventional AgI DGT gels, which are ~ 800 µm thick), using a method modified from Robertson et al. [22]. Rectangles (6 × 10 cm) of polyethersulfone filter membrane (0.45 µm pore size, 100 µm thick; Sterlitech) were dip-coated in a solution containing 31.2 mL of deionised water, 18.8 mL of acrylamide/bisacrylamide solution (19:1, 40% (w/w); Sigma Aldrich), 6 mL of 2 mol L−1 silver nitrate and 140 μL of 10% (w/w) ammonium persulfate. The membranes were then sandwiched between two sheets of 250 µm thick polyvinyl chloride and allowed to polymerise at room temperature for at least 30 min. The membrane-supported gels were then immersed in a solution of 0.4 mol L−1 potassium iodide over night, to allow the precipitation of silver iodide within the gel. The membrane-supported AgI gels were then rinsed in deionised water and any loosely adhered AgI precipitate removed by wiping the gels with lint-free laboratory tissue (Kimwipes). Gels were stored at 4 °C in deionised water, but were placed in seawater (same salinity as the tanks) 24 h prior to deployment to condition the gels to the same ionic strength. AgI DGT gels were deployed at midday on four separate rhizoboxes; two containing Z. muelleri and two containing H. ovalis. At each application time, the front plates of the rhizoboxes were removed under water and replaced with a blank acrylic plate to keep the sediments and plants submerged and in place and to prevent re-oxidation of sulphides prior to DGT deployment. DGT gels were then mounted onto the optode with tape, carefully avoiding air bubbles, and overlain with a NucleoporeTM membrane (0.2 µm pore size, 10 µm thick) to prevent any direct contact between the gels and the sediments and roots. Once, the front plates of the rhizoboxes were carefully replaced and resealed, images of the oxygen-optode-DGT were then taken every 10 min to monitor oxygen distribution and ensure sediments remained anoxic during deployment (total deployment time was 1 h).
FISH and confocal laser scanning microscopy
Standard FISH was conducted on root segments (~ 0.5 cm) of either root tips or root hair zones in 500 µL micro-tubes according to [20, 23]. Root segments were incubated in 150 µL of hybridisation buffer (900 mM NaCl, 20 mM Tris-HCl, 35% formamide (v/v), and 10% SDS (v/v)) and 15 µL of up to four probes (50 ng µL−1) were added to each tube (Table 1). Doubly labelled probes were used for Cy5 and 6-Fam to increase fluorescence signals above background auto-fluorescence [24], whereas the new-generation Atto dyes provide brighter fluorescence and greater photo-stability and hence was applied as a single labelled probe [25]. All probes were synthesised by Biomers (Ulm, Germany) and were hybridised at a formamide concentration of 35% at 46 °C for 4 h. Root sections were carefully transferred to 50 mL of pre-warmed washing buffer (70 mM NaCl, 20 mM Tris-HCl, and 5 mM EDTA) and incubated for 15 min at 48 °C. Root pieces were then transferred to cold MQ H2O for 1 min before mounting in VectaShield AF1 (Vector Laboratories, Burlingame, CA) on a glass slide. Spacers of carbon tape were used between the slide and coverslip to prevent damage to the roots.
Table 1.
Fluorescently labelled oligonucleotide probes applied in this study
| Probe sequence (5′–3′) | Probe name | Target group | Fluorophore | Reference |
|---|---|---|---|---|
| GCTGCCTCCCGTAGGAGT | EUB338 (I)a | Most bacteria | Cy5-Cy5c | [63] |
| GCAGCCACCCGTAGGTGT | EUB338 (II)a | Planctomycetales | Cy5-Cy5c | [64] |
| GCTGCCACCCGTAGGTGT | EUB338 (III)a | Verrucomicrobiales | Cy5-Cy5c | [64] |
| AGTTAGCCGGTGCTTCCT | Delta495ab | Most Deltaproteobacteria and most Gemmatimonadetes | 6-Fam-6-Famc | [65] |
| AGTTAGCCGGCGCTTCCT | Delta495bb | Deltaproteobacteria | 6-Fam-6-Famc | [65] |
| AATTAGCCGGTGCTTCCT | Delta495cb | Deltaproteobacteria | 6-Fam-6-Famc | [65] |
| ACCGGTATTCCTCCCGAT | DSB706 | Desulfobulbaceae (cable bacteria) and Thermodesulforhabdus | Atto 565 | [66] |
| ACTCCTACGGGAGGCAGC | NONEUB | None (negative control probe) | Cy5-Cy5c, 6-Fam-6-Famc, Atto 565 | [67] |
With exception of the probe with the fluorescent tag Atto 565, all probes were doubly labelled at the 5′ and 3′ end
aProbes applied as an equimolar mix (EUB mix)
bProbes applied as an equimolar mix (Delta mix) with a competitor probe mix (Delta comp. mix)
cDoubly labelled at both 5′ and 3′ end
Confocal laser scanning microscopy (CLSM) was performed on a Nikon Ti-E inverted microscope with a Nikon A1Si spectral detector. Fluorescent dyes 6-Fam, Atto 565, and Cy5 were sequentially excited with 488, 561, and 640 nm laser lines, respectively. Laser power was set at 4% and photomultiplier gain and offset were individually optimised for each channel and every field of view. For each sample, the upper root surface was located and 15 random images were acquired using a × 60 Plan Apo VC Oil lens with 1.5 × and 2 × zoom with four line averaging. Manual counting of cells hybridised in each laser channel was performed using ImageJ using the Cell Counter plug-in (https://imagej.nih.gov/ij/plugins/cell-counter.html). To account for non-specific binding, manual cell counting was also performed on the negative controls (using the probe NONEUB; Table 1), and any cell numbers counted in the negative controls were subtracted from the corresponding sample counts (Fig. S4).
DNA extraction, 16 S rRNA sequencing, and bioinformatics
DNA was extracted from unwashed roots using the MoBio PowerSoil DNA extraction kit according the manufacturer’s instructions. A fast prep 24 (MP Biomedicals) set at 4.5 m s−1 for 60 sec was used to disrupt the soft seagrass roots and lyse cells with DNA eluted in nuclease-free water. Microbial communities were profiled using the primers 341 F—806 R that target the V3–V4 hypervariable region of the 16 S rRNA gene and have lower off-target amplification of plant chloroplasts [26–28]. Sequencing of pooled amplicons was performed by the Australian Genome Research Facility (AGRF) on the Illumina MiSeq platform, using Nextera XT v2 indices and 300 bp paired end sequencing chemistry. Bioinformatic analysis of sequence reads was largely completed following the pipeline developed by [29]. Paired end reads were assembled by aligning the forward and reverse reads using PEAR (version 0.9.1) [30]. Quality metrics were checked using FastQC (version 0.11.5) and filtered using tools in FASTX (version 0.7) and BBmap (version 35.84). Ambiguous and chimeric sequences were identified and removed using VSEARCH (version 1.4.0) with the RDP as reference [31]. All downstream analyses were performed in QIIME (version 1.9.1) [32]. Open-reference operational taxonomic unit picking was performed using the SortMeRNA method [33] with a minimum identify of 97%. Taxonomy was assigned using UCLUST (version 1.2.22) [34] with the SILVA rRNA as reference (release 128) and sequences were aligned using PyNAST (version 1.2.2) [35]. OTUs identified in < 0.1% of the reads were removed as well as reads identified as chloroplast or mitochondria. Archaeae were not filtered out. General sequencing statistics are provided in supplementary Table 1.
Data analysis
Bacterial diversity of Z. muelleri roots within samples (alpha diversity) was estimated using Faith’s Phylogenetic Diversity. Weighted Unifrac of relative abundances of rarefied data were used to construct dissimilarity matrices of the communities (beta diversity) and visualised using PCoA. Stratified permutational multivariate analysis of variance (PERMANOVA; R vegan function adonis) with 999 permutations was conducted to explore the percentage of variance that could be explained by the differences in beta diversity among leaking and non-leaking roots and root regions of Z. muelleri.
Differences in microbial colonisation and bacterial alpha diversity among root regions (e.g., root cap, elongation, and root hairs) for each seagrass species were tested using either linear mixed effects or generalised linear models, with individual plants included as a random effect to account for measures taken from the same root/plant. Differential abundance of OTUs between leaking and non-leaking Z. muelleri roots was performed on variance stabilised data using the DESeq2 package [36]. Significance was determined by Benjamini–Hochberg corrected p values < 0.05. Desulfobulbaceae sequences were re-aligned using SINA aligner (v1.2.11) with a minimum identity of 0.9 and a sequence cutoff of 0.7. A maximum-likelihood tree was then constructed using FastTree (General Time Reversible model and gamma rate) to show the phylogenetic relationship between cable bacteria sequences recovered in our study to those of known cable bacteria (Fig. S8).
All statistical testing was carried out in R studio (version 1.0.153) using the Phyloseq (version 1.14.0) [37], ggplot2 (version 2.2.1), vegan (version 2.4-3) and nlme (version 3.1−137) packages. All raw 16 S rRNA sequences have been uploaded to NCBI Sequence Read Archive under submission number SRP133137.
Results
Spatiotemporal patterns of ROL and sulphide in seagrass rhizospheres
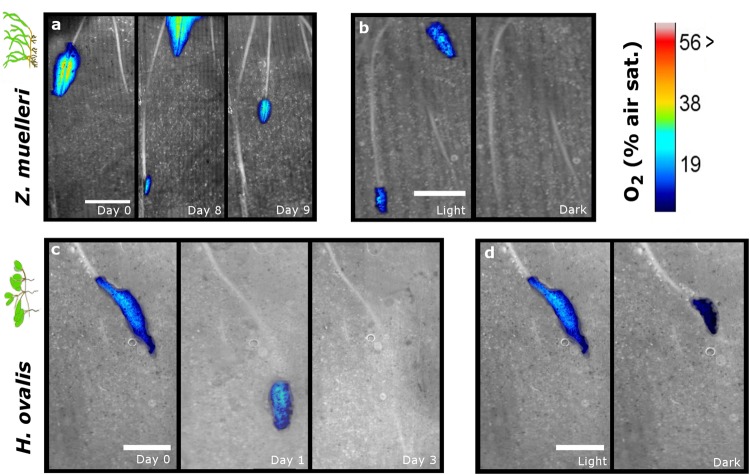
Oxygen planar optodes and methylene blue staining demonstrated that ROL was spatially restricted to immediately behind the root tips of young apical roots (Fig. 1, Fig. S5). The zone of ROL extended to an average length of 10.6 ± 4.8 mm for H. ovalis and 4.2 ± 2.7 mm for Z. muelleri and an average width of 1.5 ± 0.7 mm for H. ovalis and 1.3 ± 0.8 mm for Z. muelleri (Table S2), generally corresponding to the region of root elongation. In this region, the average oxygen concentration measured from nine replicate plants was 59 ± 19 and 59 ± 21.7 µmol L−1 (22.6 ± 7.3 and 22.5 ± 8.3 % of air equilibrium (air = 20.6 kPa oxygen)) during the day for H. ovalis and Z. muelleri, respectively, which is within concentrations previously reported for belowground tissues of other. Zostera species and H. ovalis [38, 39]. ROL was related to the age of the root (ceasing within 5–9 days of root growth for these fast growing species) and depended on available light to leaves and shoots; ROL was reduced by > 50 % in length and oxygen concentration within 30 min of darkness (Fig. 1 and Table S2).
Fig. 1.
Spatiotemporal patterns in ROL from the roots of Zostera muelleri and Halophila ovalis. a Oxygen loss from Z. muelleri roots over 9 days of growth. A second new root can be seen leaking oxygen in day 8 and 9. b Oxygen loss in roots of Z. muelleri during light (250 µmol photons m−2 s−1) and dark transitions (30 min darkness). c Oxygen loss from H. ovalis root over 3 days. d Oxygen loss in H. ovalis root during light and dark transitions. Images are overlays of transparent optode images on bw images of the roots. Nine replicate plants were measured for each species. Days are relative to when ROL was first observed (i.e., when the root grew to the optode wall). Scale bar is 5 mm
Sulphide DGTs combined with oxygen planar optodes revealed that ROL of Z. muelleri resulted in an oxidised band surrounding the root tip and a decrease in the concentration of sulphides within this band (Fig. 2a). Conversely, for those roots not exhibiting ROL, there was an increase in sulphide concentration along the length of the root (Fig. 2b).
Fig. 2.
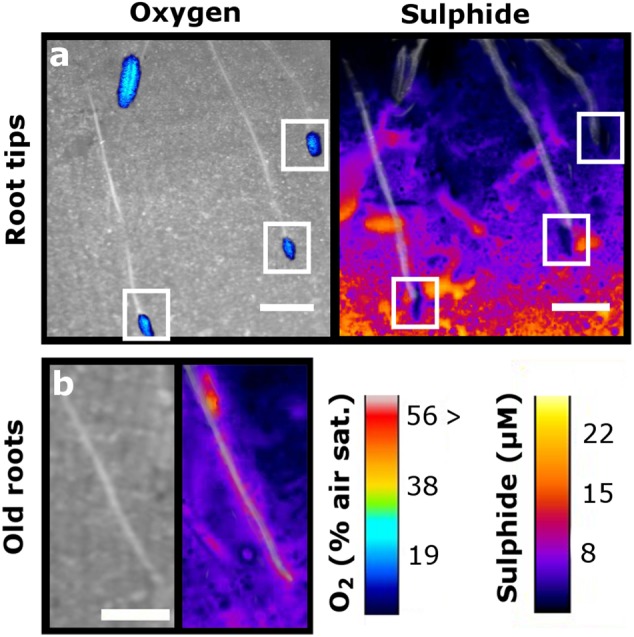
2-D distribution of oxygen and sulphide in the rhizosphere of Zostera muelleri during full light (250 µmol photons m−2 s−1). a Oxygen (left) and corresponding reduction in sulphides (right) around the leaking root tips (overlain with bw image of roots and marked with white squares where oxidation has occurred). b Lack of oxygen loss around older roots (left) and corresponding increase in sediment sulphides (right) along the length of the root (overlain with bw image of roots). Scale bar is 5 mm
Colonisation by root-attached bacteria within oxic–anoxic microsites
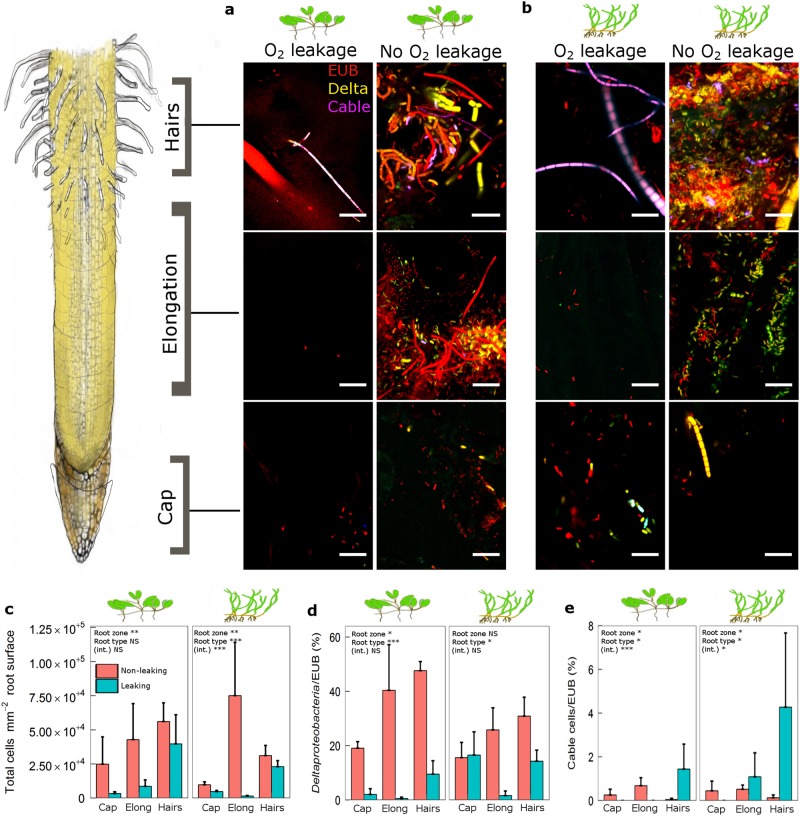
To obtain insights into colonisation patterns of key bacterial groups within the oxic and anoxic microsites of seagrasses, we sampled both roots leaking oxygen and those not leaking oxygen, hybridised these root pieces with specific fluorescent bacterial DNA probes and made observations using CLSM (Fig. S3). For H. ovalis, colonisation by bacteria (cells mm−2 root surface) was higher in the root hair zone versus the root cap, but there was no significant difference between roots exhibiting ROL (termed ‘leaking roots’) and roots exhibiting no ROL (termed ‘non-leaking’ roots) (Figs. 3a, c). Conversely, for Z. muelleri, colonisation by bacteria (cells mm−2 root surface) differed among root zones and between leaking roots and non-leaking roots (Figs. 3b, c). Non-leaking roots had higher numbers of bacterial cells than leaking roots (Fig. 3c). For leaking roots, the highest number of bacterial cells was in the root hair zone where no ROL was observed compared with the elongation zone and root cap (Fig. 3c). Conversely for non-leaking roots, total bacterial cell numbers did not differ among the three root zones (cap, elongation zone, and root hair zone) (Fig. 3c). There was a significant reduction in the proportion of δ-proteobacterial cells in leaking roots versus non-leaking roots for H. ovalis and in the elongation zone (region of ROL) for Z. muelleri (Fig. 3d). Filamentous sulphide-oxidising cable bacteria were detected on the root surface of both H. ovalis and Z. muelleri, where they were significantly higher in abundance in the root hair zone of leaking roots (Fig. 3e). Cable bacteria were absent from the root cap of leaking roots for both seagrass species and from the elongation zone for H. ovalis (Fig. 3e).
Fig. 3.
Colonisation patterns of microbial populations on oxic–anoxic microsites of H. ovalis and Z. muelleri root surfaces. a, b Representative confocal laser scanning microscopy (CLSM) micrographs showing bacterial colonisation of root zones (root hairs, elongation, and root cap) of a H. ovalis, with younger oxygen leaking roots on the left panel and older non-leaking roots on the right panel, and b Zostera muelleri, with younger oxygen leaking roots on the left panel and older non-leaking roots on the right panel. Bacteria were stained with oligonucleotide probes where red = bacteria (probe EUB I–III), yellow = δ-proteobacteria (probe Delta 495a-c), blue/purple = cable bacteria (probe DSB706). Scale bar is 10 µm. Fifteen images of each replicate root were examined per root region. c–e Mean numbers of bacterial groups (N = 3, SE ± 1) detected on the surface of root zones (root cap, elongation, root hairs) c Bacteria (probe EUB I–III; red) d δ-proteobacteria expressed as a proportion of total bacteria (probe Delta 495a-c; yellow) and e Cable bacteria expressed as a proportion of total bacteria (probe DSB706; purple). The significance of fixed factors (root zone, root type, and their interaction) are displayed in each plot, where * = 0.05, ** < 0.01, *** < 0.001, and NS = not significant
Diversity and community structure of root-attached bacteria within oxic–anoxic microsites
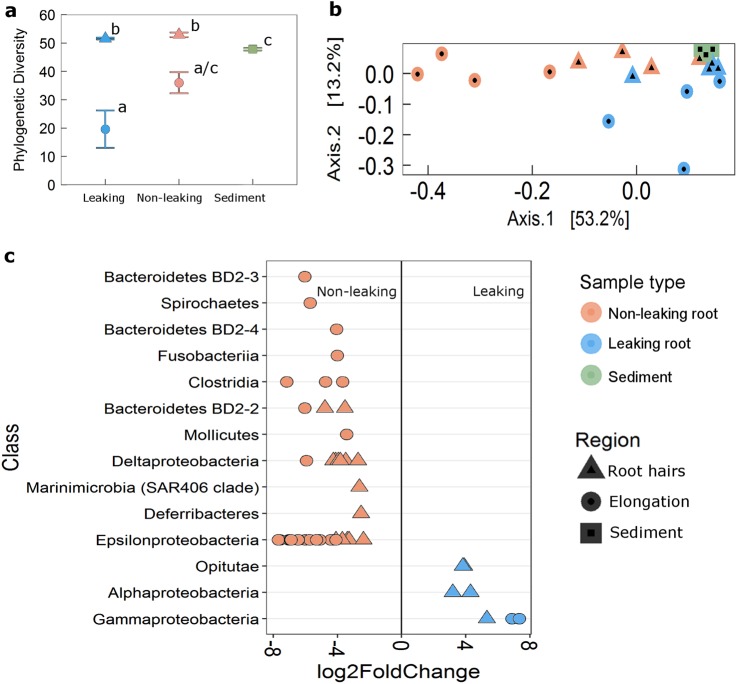
To further investigate the influence of oxic microsites on root microbial community composition, we sampled roots of Z. muelleri leaking oxygen and those not leaking oxygen, extracted DNA from the root tip (including both elongation zone and root cap), and root hair zone, as well as from bulk sediment and performed microbial community profiling using 16 S rRNA (V3–V4) sequencing (Fig. S3). A 16 S rRNA community profiling largely confirmed the influence of ROL on root-associated microbial communities (Fig. 4). Diversity (Faith’s phylogenetic diversity) differed by both root region (F2,15 = 41.6, p < 0.001) and root type (F1,15 = 8.5, p = 0.01), which was lower on the root tip (the region of ROL) relative to the root hair zone and the bulk sediment (Fig. 4a). The composition of the Z. muelleri root microbiome was strongly affected by the type of root (i.e., leaking vs non-leaking) and differed by root region as revealed by PCoA of weighted UniFrac distances (PERMANOVA; interaction term F1,19 = 3.52, p = 0.02). The largest differentiation among microbial communities was detected in the root tip (corresponding to the region of ROL) between leaking and non-leaking roots (Fig. 4b, Fig. S6, Fig. S7), confirming the patterns observed in FISH-CLSM (Fig. 3). Putative sulphide-oxidising bacteria within the ε-proteobacteria (e.g. Sulfurimonas, Nitratifractor, and Arcobacter) were also found to be more differentially abundant on non-leaking roots relative to leaking roots (Fig. 4c, Fig. S6). Numerous genera of putative sulphate-reducing bacteria within the δ-proteobacteria were more abundant on non-leaking roots than on leaking roots (Fig. S7).
Fig. 4.
16 S rRNA sequencing of root regions of oxygen leaking and non-leaking roots of Z. muelleri roots and bulk sediment. a Mean estimated alpha diversity (Faith’s Phylogenetic Diversity) ± 1 SE (N = 4; points annotated with different letters differ at p < 0.05). b Principal coordinates analysis (PCoA) of weighted UniFrac distances. c Differentially abundant OTUs (significance of p < 0.05) between root regions of oxygen leaking and non-leaking roots. Sediment was excluded from differential abundance analysis. OTUs are arranged by increasing significance of their adjusted p values and the direction of the log2 fold change. Greater than 0 log2 fold change indicates OTUs more differentially abundant in leaking roots. Roots were sampled during full light (250 µmol photons m−2 s−1) supply to the shoots, and immediately after oxygen loss was quantified
Several sequences aligned with the candidate cable bacteria species Candidatus Electrothrix communis (Fig. S8). Other sequences recovered from our 16 S rDNA data also grouped within the monophyletic Candidatus Electrothrix cluster, suggesting that these OTUs are potentially novel cable bacteria species (Fig. S8). No sequences aligned to the freshwater cable bacteria Candidatus Electronema.
Discussion
ROL in seagrasses is temporally and spatially restricted
Oxygenation of the seagrass rhizosphere appears restricted to areas surrounding the tips of young growing roots. However, the degree to which this occurred was species specific, with H. ovalis exhibiting ROL over a greater length of root than Z. muelleri. The differences in ROL between these two species and among roots of different ages could be due to the variation in the development of barriers to oxygen loss as roots age. For example, we found evidence of suberisation of the outer walls of mature root sections for Z. muelleri (Fig. S9), thus promoting longitudinal diffusion rather than radial loss and ensuring oxygen reaches metabolically active apical meristems [40]. Conversely, there was no evidence of suberisation of outer cell walls in H. ovalis roots (Fig. S9), suggesting that either suberin was not obvious in this species, or that an alternative barrier to oxygen loss exists [41]. Evidence that H. ovalis develops a barrier to ROL along the basal portion of the root has been shown from one other study [41], though root anatomy was not evaluated. Furthermore, differences in the proportion of root volume occupied by aerenchyma cells among seagrass species may also influence ROL [6], as well as differences in growth rates; where slower-growing root cells closer to the root tip are older than cells at the same distance of a faster growing root, and so the barrier can form closer to the tip of a slower-growing root. This may have been the case for the two species investigated in this study where average root growth rates of Z. muelleri and H. ovalis were estimated to be 4.2 and 10.5 mm d−1, respectively.
ROL in Z. muelleri corresponded to a decrease in sulphides surrounding the leaking root tip, suggesting ROL acts as a ‘barrier’ to sulphide intrusion when the plant is sufficiently oxygenated. Conversely, when roots are older and no longer showing ROL, sulphide concentrations in the rhizosphere increased, likely related to release of carbon compounds from sloughed root cells and/or exudation from the higher density of root hairs present on older roots [42]. Hence, owing to the highly restricted site of ROL in these seagrass species (spatially and temporally), we argue that while the ability of O2 loss from roots to oxidise sediment sulphides is an important protective mechanism for seagrasses, this process is not likely significant for the bulk sediment on a meadow-scale. Frederiksen and Glud estimated total root ROL for a bed of Z. marina to range from 12 to 22 mmol O2 m−2 d−1 under saturating light conditions [38], which is less than the ROL associated with fauna activity (e.g., around burrows and bioturbation), suggesting that ROL of seagrasses contributes only a minor part to total benthic O2 uptake [43]. As H. ovalis has only one root per node of the rhizome, it can be assumed that ROL from a bed of H. ovalis of equal shoot density would also contribute only a minor part to benthic O2 uptake. Hence, in the context of bulk sediment biogeochemistry, the presence of seagrasses are more likely to enhance sulphate reduction than sulphide oxidisation in sediments owing to exudation of carbon compounds from roots and the accumulation of organic matter in seagrass sediments [44].
Diversity, community structure, and colonisation of root-attached bacteria are influenced by root age and oxic–anoxic microsites
Younger seagrass roots exhibiting ROL had a lower abundance of total bacteria and lower proportion of potential sulphate-reducing bacteria, but a higher proportion of sulphide-oxidising cable bacteria than older roots exhibiting no ROL. The differences in microbial colonisation patterns between O2 leaking and non-leaking roots could be due directly to the presence of oxygen, particularly as the majority of bacteria in anoxic sediments are adapted to zero oxygen conditions. In addition, as a majority of δ-proteobacteria are sulphate-reducing bacteria [45], it is possible that oxygen loss from root tips creates an environment not suitable for many of these sulphate reducers. However, as there was also a reduction in δ-proteobacteria in the root hair zone between ROL and no ROL roots, these differences could also have been driven by differences in root exudation patterns between younger and older roots. The differences in microbial colonisation patterns could also be due to differences in carbon and nitrogen exudation among root zones (for example exudation at the root tips versus root hairs) [46, 47]. In addition, shedding of outer layers of root cap cells as roots grow through sediment could also reduce microbial colonisation on root caps. Regardless of what is driving this process, a reduction in the number of sulphate-reducing bacteria would result in lower sulphide production.
This is the first discovery of a close association of sulphide-oxidising cable bacteria with seagrass roots. The long filaments of cable bacteria (occasionally longer than 20 mm) allow them to bridge the anoxic-oxic interface on the seagrass root surface to make use of the high sulphide concentration in the anoxic root hair zone, as well as the electron acceptors (either O2 or NO3−) available in the oxic zone, to ultimately provide an additional defence against sulphide intrusion for seagrasses. Based on our oxygen planar optode images, we have estimated the average root growth (extension rates) of Z. muelleri and H. ovalis to be 4.2 and 10.5 mm d−1, respectively, which is within the range of that previously reported for Z. marina (8.7 mm d−1) [38]. The high motility of cable bacteria (up to 0.8 µm s−1, or 70 mm d−1) [48] would enable them to respond to the oxygen front that moves on a diel pattern of photosynthetic activity and as the root grows through the sediment. Cable bacteria have been shown to adjust their position in the sediment in response to a moving oxygen front produced by a photosynthetic algal biofilm [49]. Furthermore, root hairs and the region of oxygen loss in older roots may leak minute quantities of oxygen—too small to be detected with the optode film—that may be sufficient to support electrogenic sulphide oxidation in these root regions. As mentioned previously, the higher proportion of cable bacteria in the root hair zone of leaking roots could also be due to differences in carbon exudation between younger and older roots and among root zones, as cable bacteria have previously been shown to exhibit heterotrophic growth on carbon substrates [50].
The ability of cable bacteria to improve sediment conditions in seagrass rhizospheres is not limited to their ability to reduce sulphide concentrations and thus intrusion into roots, but also extends to their capacity to increase the availability of plant nutrients (Fe2+and PO43−) owing to dissolution of Fe and Ca minerals (e.g., FeS and CaCO3) from acidification of the anodic reaction (in the sulphidic zone) [51, 52]. The highest density of cable bacteria occurred in the root hair zone, which is also proposed to be the site of greatest nutrient uptake for these seagrasses owing to the increase in surface area from root hairs, and the prominent plasmodesmatal connections apparent in the root hairs [53]. This cable bacteria-mediated mineral weathering may also explain the expansion of meadows in sediments with low free phosphorus levels [54].
There was also an increased abundance of ε-proteobacteria (particularly Sulfurimonas) in older roots of Z. muelleri with no ROL. Sulfurimonas, Nitratifractor, and Arcobacter all possess the full sulphide oxidation gene cassette (sox genes) as well as the sulphide-oxidising sulphide:quinone oxidoreductase, which catalyzes the oxidation of hydrogen sulphide [55]. The increased abundance of these ε-proteobacteria in roots with no ROL is likely a reflection of the higher availability of electron donors (reduced sulphides) surrounding older roots compared to younger roots. In addition, many sulphur-oxidising ε-proteobacteria display high versatility in their metabolic pathways and are able to use alternative electron acceptors to oxygen (such as NO3− and NO2−) [56, 57]. It is also possible that these older roots may still be leaking minute amounts of oxygen that may be sufficient to support sulphide oxidisation for these bacteria, many of which are classified as microaerobic [56]. The abundance of these putative sulphide-oxidising ε-proteobacteria also correlated with the occurrence of sulphate-reducing bacteria, suggesting they are likely involved in syntrophic feeding (or ‘obligately mutualistic metabolism’) [45]. Sulphur-syntrophy between populations of microbes producing sulphides and those consuming sulphides is widespread and occurs in a diversity of habitats from photosynthetic microbial mats [58] to gutless marine worms [59]. Hence, ROL, as well as carbon and nitrogen, from seagrass roots is likely central for maintaining and fine-tuning the balance in the availability of resources between these two bacterial groups. Sulfurimonas, and more generally the Campylobacterales, appear to be a dominant microbial genus within the roots of seagrasses [60, 61], and were recently found to decline in abundance when three seagrass species (H. ovalis, Cymodocea serrulata, and Halodule uninervis) were grown under low and fluctuating light conditions [62]. Although the functional role of these bacteria for seagrass health remains to be investigated, the dominance of Sulfurimonas in seagrass roots combined with their apparent sensitivity to environmental perturbations suggests they may be useful as an indicator for declining seagrass sediment health.
Conclusions
Seagrass roots have a strong barrier to prevent the radial loss of oxygen to the rhizosphere, thus ensuring that oxygen reaches the distal, metabolically active, apical meristems. This results in a highly restricted zone of oxygen loss (~ 1.5 mm wide) surrounding tips of young roots. Hence, our findings highlight that although ROL from roots oxidising sulphides is an important protective mechanism for seagrasses on the microscale, this process is not significant for the bulk sediment sulphides, and that seagrasses may enhance sulphide production in sediments rather than alleviate it (Fig. 5). Nevertheless, despite the restricted nature of ROL, our multifaceted approach reveals a previously unrecognised role for this localised O2 loss: ROL creates spatially and temporally restricted oxic zones that may serve to protect sensitive seagrass tissues from sulphide intrusion by oxidising sulphides surrounding the growing tip, and by influencing the abundance of sulphate-reducing and sulphide-oxidising microorganisms that colonise these younger roots (Fig. 5).
Fig. 5.
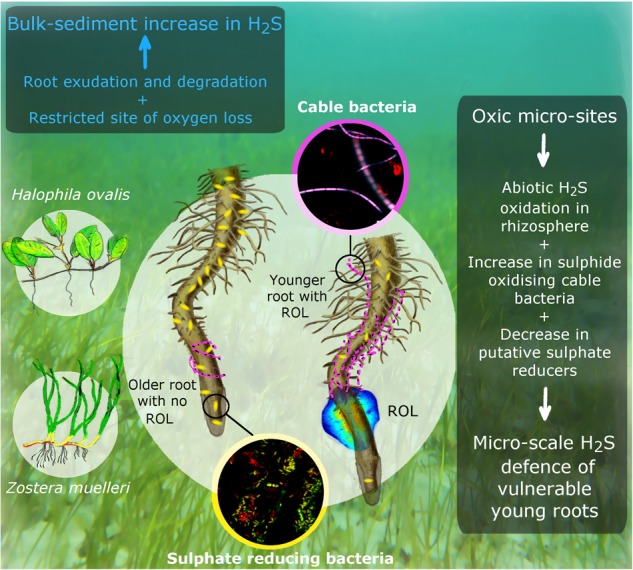
Conceptual summary of the influence of radial oxygen loss (ROL) on defence against sulphide intrusion in Halophila ovalis and Zostera muelleri. Seagrasses enhance sulphide production in bulk sediments owing to release of exudates and decomposition of underground tissue (blue text in box). On the microscale (white text in box), growing roots lose oxygen from root tips, which works to defend against sulphide intrusion into vulnerable young roots by both abiotically oxidising sulphides in the rhizosphere, as well as promoting the colonisation of sulphide-oxidising cable bacteria (shown in purple) and decreasing the colonisation of sulphate-reducing bacteria (shown in yellow)
Our study is also the first report on the direct association of sulphide-oxidising cable bacteria with seagrass roots. These specialised bacteria live in the interface of oxic–anoxic microzones and may improve sediment conditions by oxidising sulphides to enable seagrasses to colonise sulphide-rich sediments. The increased occurrence of cable bacteria in roots exhibiting ROL may prove to be a widespread phenomenon among submerged (i.e., aquatic) and semi-submerged (i.e., amphibious) and emergent wetland plants, which colonise sulphide-rich sediments, including mangroves and saltmarsh plants, and adds a new plant-microbial dimension to our understanding of plant tolerances of anoxic sediment conditions.
Electronic supplementary material
Acknowledgements
Thanks to Morten Larsen and Adam Kessler for their advice in building and optimising the oxygen-optode system and Hannes Schmidt for advice in FISH protocols, as well as PWIS and the PP for many fruitful discussions. We thank the anonymous reviewers whose detailed comments helped improve and clarify this manuscript. We acknowledge the use of the services and facilities of AGRF as well as the facilities, and the scientific and technical assistance of the Australian Microscopy & Microanalysis Research Facility at the Centre for Microscopy, Characterisation & Analysis, The University of Western Australia, a facility funded by the University, State and Commonwealth Governments. Graduate Women WA provided financial support for BCM. Megan Ryan is funded by an ARC Future Fellowship (FT140100103) and Gary Kendrick by two ARC Linkage Grants that also funded the 16 S rRNA sequencing (LP130100918, LP130100155). William Bennett was funded by an ARC Discovery Early Career Researcher Award (DE140100056).
Author contributions
BCM: study design and conception, method development, data collection data analysis, and drafting manuscript. JB: study design and conception, method development, and drafting manuscript. MR: drafting manuscript. WB: method development, data analysis, and drafting manuscript. TC: method development and drafting of manuscript. NJ: method development and drafting of manuscript. YO: method development, data analysis, and drafting manuscript. GK: study design and conception and drafting manuscript. All authors approved the final manuscript.
Compliance with ethical standards
Conflict of interest
All authors declare they have no affiliations with or involvement in any organisation or entity with any financial interest or non-financial interest in the work described.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0308-5) contains supplementary material, which is available to authorised users.
References
- 1.Terrados J, Duarte CM, Kamp-Nielsen L, Agawin NSR, Gacia E, Lacap D, et al. Are seagrass growth and survival constrained by the reducing conditions of the sediment? Aquat Bot. 1999;65:175–197. doi: 10.1016/S0304-3770(99)00039-X. [DOI] [Google Scholar]
- 2.Jørgensen BB. Mineralization of organic matter in the sea bed - the role of sulphate reduction. Nature. 1982;296:643–645. doi: 10.1038/296643a0. [DOI] [Google Scholar]
- 3.Holmer M, Andersen FO, Nielsen SL, Boschker HTS. The importance of mineralization based on sulfate reduction for nutrient regeneration in tropical seagrass sediments. Aquat Bot. 2001;71:1–17. doi: 10.1016/S0304-3770(01)00170-X. [DOI] [Google Scholar]
- 4.Lamers LPM, Govers LL, Janssen ICJM, Geurts JJM, Van der Welle MEW, Van Katwijk MM, et al. Sulfide as a soil phytotoxin-a review. Front Plant Sci. 2013;4:268. doi: 10.3389/fpls.2013.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armstrong W. Aeration in higher plants. Adv Bot Res. 1979;7:225–332. doi: 10.1016/S0065-2296(08)60089-0. [DOI] [Google Scholar]
- 6.Colmer TD. Long-distance transport of gases in plants: a perspective on internal aeration and radial oxygen loss from roots. Plant Cell Environ. 2003;26:17–36. doi: 10.1046/j.1365-3040.2003.00846.x. [DOI] [Google Scholar]
- 7.Brodersen KE, Nielsen DA, Ralph PJ, Michael K. Oxic microshield and local pH enhancement protects Zostera muelleri from sediment derived hydrogen sulphide. New Phytol. 2014;3:1–13. doi: 10.1111/nph.13124. [DOI] [PubMed] [Google Scholar]
- 8.Luther GW, Findlay AJ, MacDonald DJ, Owings SM, Hanson TE, Beinart RA, et al. Thermodynamics and kinetics of sulfide oxidation by oxygen: A look at inorganically controlled reactions and biologically mediated processes in the environment. Front Microbiol. 2011;2:1–9. doi: 10.3389/fmicb.2011.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Heide T, Govers LL, de Fouw J, Olff H, van der Geest M, van Katwijk MM, et al. A three-stage symbiosis forms the foundation of seagrass ecosystems. Science. 2012;336:1432–1434. doi: 10.1126/science.1219973. [DOI] [PubMed] [Google Scholar]
- 10.Joshi MM, Hollis JP. Interaction of Beggiatoa and rice plant: detoxification of hydrogen sulphide in the rice rhizosphere. Science. 1976;195:179–180. doi: 10.1126/science.195.4274.179. [DOI] [PubMed] [Google Scholar]
- 11.Trojan D, Schreiber L, Bjerg JT, Bøggild A, Yang T. A Taxonomic framework for cable bacteria and proposal of the candidate genera Electrothrix and Electronema. Syst Appl Microbiol. 2016;39:297–306. doi: 10.1016/j.syapm.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfeffer C, Larsen S, Song J, Dong M, Besenbacher F, Meyer RL, et al. Filamentous bacteria transport electrons over centimetre distances. Nature. 2012;491:10–13. doi: 10.1038/nature11586. [DOI] [PubMed] [Google Scholar]
- 13.Burdorf LDW, Tramper A, Seitaj D, Meire L, Hidalgo-Martinez S, Zetsche EM, et al. Long-distance electron transport occurs globally in marine sediments. Biogeosciences. 2017;14:683–701. doi: 10.5194/bg-14-683-2017. [DOI] [Google Scholar]
- 14.Kjeldsen KU, Joulian C, Ingvorsen K. Oxygen tolerance of sulfate-reducing bacteria in activated sludge. Environ Sci Technol. 2004;38:2038–2043. doi: 10.1021/es034777e. [DOI] [PubMed] [Google Scholar]
- 15.Kilminster K, Forbes V, Holmer M. Development of a ‘sediment-stress’ functional-level indicator for the seagrass Halophila ovalis. Ecol Indic. 2014;36:280–289. doi: 10.1016/j.ecolind.2013.07.026. [DOI] [Google Scholar]
- 16.Holmer M, Bennett WW, Ferguson AJP, Potts J, Hasler-Sheetal H, Welsh DT. Drivers of sulfide intrusion in Zostera muelleri in a moderately affected estuary in south-eastern Australia. Mar Freshw Res. 2017;68:2134–2144. doi: 10.1071/MF16402. [DOI] [Google Scholar]
- 17.Larsen M, Borisov SM, Grunwald B, Klimant I, Glud RN. A simple and inexpensive high resolution color ratiometric planar optode imaging approach: application to oxygen and pH sensing. Limnol Oceanogr Methods. 2011;9:348–360. doi: 10.4319/lom.2011.9.348. [DOI] [Google Scholar]
- 18.Glud RN, Ramsing NB, Gundersen JK, Klimant I, Gludl RN. Planar optrodes: a new tool for fine scale measurements of two-dimensional O2 distribution in benthic communities. Mar Ecol Prog Ser. 1996;140:217–226. doi: 10.3354/meps140217. [DOI] [Google Scholar]
- 19.Armstrong J, Armstrong W. Phragmities australis-a preliminary study of soil-oxidizing sites and internal gas transport pathways. New Phytol. 1988;108:373. doi: 10.1111/j.1469-8137.1988.tb04177.x. [DOI] [Google Scholar]
- 20.Schmidt H, Eickhorst T. Detection and quantification of native microbial populations on soil-grown rice roots by catalyzed reporter deposition-fluorescence in situ hybridization. FEMS Microbiol Ecol. 2014;87:390–402. doi: 10.1111/1574-6941.12232. [DOI] [PubMed] [Google Scholar]
- 21.Teasdale PR, Hayward S, Davison W. In situ, high-resolution measurement of dissolved sulfide using diffusive gradients in thin films with computer-imaging densitometry. Anal Chem. 1999;71:2186–2191. doi: 10.1021/ac981329u. [DOI] [PubMed] [Google Scholar]
- 22.Robertson D, Teasdale PR, Welsh DT. A novel gel-based technique for the high resolution, two-dimensional determination of iron (II) and sulfide in sediment. Limnol Oceanogr. 2008;6:502–512. doi: 10.4319/lom.2008.6.502. [DOI] [Google Scholar]
- 23.Watt M, Hugenholtz P, White R, Vinall K. Numbers and locations of native bacteria on field-grown wheat roots quantified by fluorescence in situ hybridization. (FISH) 2006;8:871–884. doi: 10.1111/j.1462-2920.2005.00973.x. [DOI] [PubMed] [Google Scholar]
- 24.Stoecker K, Dorninger C, Daims H, Wagner M. Double labeling of oligonucleotide probes for fluorescence in situ hybridization (DOPE-FISH) improves signal intensity and increases rRNA accessibility. Appl Environ Microbiol. 2010;76:922–926. doi: 10.1128/AEM.02456-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buschmann V, Weston KD, Sauer M. Spectroscopic study and evaluation of red-absorbing fluorescent dyes. Bioconjugate Chem. 2003;14:195–204. doi: 10.1021/bc025600x. [DOI] [PubMed] [Google Scholar]
- 26.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. PNAS. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muhling M, Woolven-Allen J, Murrell JC, Joint I. Improved group-specific PCR primers for denaturing gradient gel electrophoresis analysis of the genetic diversity of complex microbial communities. Isme J. 2008;2:379–392. doi: 10.1038/ismej.2007.97. [DOI] [PubMed] [Google Scholar]
- 28.Muyzer G, De Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Comeau AM, Douglas GM, Langille MGI. Microbiome Helper: a custom and streamlined workflow for microbiome research. mSystems. 2017;2:1–11. doi: 10.1128/mSystems.00127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:1–22. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kopylova E, Noé L, Touzet H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 34.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 35.Caporaso JG, Bittinger K, Bushman FD, Desantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10:1–12. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:1–11. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frederiksen MS, Glud RN. Oxygen dynamics in the rhizosphere of Zostera marina: a two-dimensional planar optode study. Limnol Oceanogr. 2006;51:1072–1083. doi: 10.4319/lo.2006.51.2.1072. [DOI] [Google Scholar]
- 39.Brodersen K, Nielsen D, Ralph PJ, Kühl M. A split flow chamber with artificial sediment to examine the below‑ground microenvironment of aquatic macrophytes. Mar Biol. 2014;161:2921–2930. doi: 10.1007/s00227-014-2542-3. [DOI] [Google Scholar]
- 40.Kotula L, Colmer TD, Nakazono M. Effects of organic acids on the formation of the barrier to radial oxygen loss in roots of hordeum marinum. Funct Plant Biol. 2014;41:187–202. doi: 10.1071/FP13178. [DOI] [PubMed] [Google Scholar]
- 41.Connell EL, Colmer TD, Walker DI. Radial oxygen loss from intact roots of Halophila ovalis as a function of distance behind the root tip and shoot illumination. Aquat Bot. 1999;63:219–228. doi: 10.1016/S0304-3770(98)00126-0. [DOI] [Google Scholar]
- 42.Martin BC, Statton J, Siebers AR, Grierson PF, Ryan MH, Kendrick GA. Colonizing tropical seagrasses increase root exudation under fluctuating and continuous low light. Limnol Oceanogr. 2018;63:381–391. doi: 10.1002/lno.10746. [DOI] [Google Scholar]
- 43.Wenzho F, Glud RN, Wenzhofer F. Small-scale spatial and temporal variability in coastal benthic O2 dynamics: Effects of fauna activity. Limnol Oceanogr. 2004;49:1471–1481. doi: 10.4319/lo.2004.49.5.1471. [DOI] [Google Scholar]
- 44.Samper-Villarreal J, Lovelock CE, Saunders MI, Roelfsema C, Mumby PJ. Organic carbon in seagrass sediments is influenced by seagrass canopy complexity, turbidity, wave height, and water depth. Limnol Oceanogr. 2016;61:938–952. doi: 10.1002/lno.10262. [DOI] [Google Scholar]
- 45.Morris BEL, Henneberger R, Huber H, Moissl-Eichinger C. Microbial syntrophy: Interaction for the common good. FEMS Microbiol Rev. 2013;37:384–406. doi: 10.1111/1574-6976.12019. [DOI] [PubMed] [Google Scholar]
- 46.Shane MW, Lambers H. Cluster roots: a curiosity in context. Plant Soil. 2005;274:101–125. doi: 10.1007/s11104-004-2725-7. [DOI] [Google Scholar]
- 47.Martin BC, George SJ, Price CA, Ryan MH, Tibbett M. The role of root exuded low molecular weight organic anions in facilitating petroleum hydrocarbon degradation: Current knowledge and future directions. Sci Total Environ. 2014;472:642–653. doi: 10.1016/j.scitotenv.2013.11.050. [DOI] [PubMed] [Google Scholar]
- 48.Bjerg JT, Damgaard L, Holm A, Schramm A, Nielsen P. Motility of electric cable bacteria. Appl Environ Microbiol. 2016;82:3816–3821. doi: 10.1128/AEM.01038-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malkin SY, Meysman FJR. Rapid redox signal transmission by ‘cable bacteria’ beneath a photosynthetic biofilm. Appl Environ Microbiol. 2015;81:948–956. doi: 10.1128/AEM.02682-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vasquez-Cardenas D, van de Vossenberg J, Polerecky L, Malkin SY, Schauer R, Hidalgo-Martinez S, et al. Microbial carbon metabolism associated with electrogenic sulphur oxidation in coastal sediments. ISME J. 2015;9:1966–1978. doi: 10.1038/ismej.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rao AMF, Malkin SY, Hidalgo-Martinez S, Meysman FJR. The impact of electrogenic sulfide oxidation on elemental cycling and solute fluxes in coastal sediment. Geochim Cosmochim Acta. 2016;172:265–286. doi: 10.1016/j.gca.2015.09.014. [DOI] [Google Scholar]
- 52.Malkin SY, Seitaj D, Burdorf LDW, Nieuwhof S, Hidalgo-Martinez S, Tramper A, et al. Electrogenic sulfur oxidation by cable bacteria in bivalve reef sediments. Front Mar Sci. 2017;4:28. doi: 10.3389/fmars.2017.00028. [DOI] [Google Scholar]
- 53.Roberts DG. Root-hair structure and development in the seagrass Halophila ovalis (R. Br.) Hook. f. Mar Freshw Res. 1993;44:85–100. doi: 10.1071/MF9930085. [DOI] [Google Scholar]
- 54.Fraser MW, Kendrick GA, Grierson PF, Fourqurean JW, Vanderklift MA, Walker DI. Nutrient status of seagrasses cannot be inferred from system-scale distribution of phosphorus in Shark Bay, Western Australia. Mar Freshw Res. 2012;63:1015–1026. doi: 10.1071/MF12026. [DOI] [Google Scholar]
- 55.Waite DW, Vanwonterghem I, Rinke C, Parks DH, Zhang Y, Takai K et al. Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to epsilonbacteraeota (phyl. nov.). Front Microbiol 2017; 8:682. [DOI] [PMC free article] [PubMed]
- 56.Campbell BJ, Engel AS, Porter ML, Takai K. The versatile epsilon-proteobacteria: key players in sulphidic habitats. Nat Rev Microbiol. 2006;4:458–468. doi: 10.1038/nrmicro1414. [DOI] [PubMed] [Google Scholar]
- 57.Han Y, Perner M. The globally widespread genus Sulfurimonas: versatile energy metabolisms and adaptations to redox clines. Front Microbiol. 2015;6:1–17. doi: 10.3389/fmicb.2015.00989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilbanks EG, Jaekel U, Salman V, Humphrey PT, Eisen JA, Facciotti MT, et al. Microscale sulfur cycling in the phototrophic pink berry consortia of the sippewissett salt marsh. Environ Microbiol. 2014;16:3398–3415. doi: 10.1111/1462-2920.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dubilier N, Mülders C, Ferdelman T, de Beer D, Pernthaler A, Klein M, et al. Endosymbiotic sulphate-reducing and sulphide-oxidizing bacteria in an oligochaete worm. Nature. 2001;411:298–302. doi: 10.1038/35077067. [DOI] [PubMed] [Google Scholar]
- 60.Fahimipour AK, Kardish MR, Eisen JA, Lang JM, Green JL, Stachowicz JJ. Global-scale structure of the eelgrass microbiome. Appl Environ Microbiol. 2017;83:1–12. doi: 10.1128/AEM.03391-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ettinger CL, Voerman SE, Lang JM, Stachowicz JJ, Eisen JA. Microbial communities in sediment from Zostera marina patches, but not the Z. marina leaf or root microbiomes, vary in relation to distance from patch edge. PeerJ. 2017;5:1–25. doi: 10.7717/peerj.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin BC, Gleeson D, Statton J, Siebers AR, Grierson P, Ryan MH, et al. Low light availability alters root exudation and reduces putative beneficial microorganisms in seagrass roots. Front Microbiol. 2018;8:1–16. doi: 10.3389/fmicb.2017.02667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Amann RI, Blinder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–25. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daims H, Brühl A, Amann R, Schleifer KH, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–44. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 65.Loy A, Lehner A, Lee N, Adamczyk J, Meier H, Ernst J, et al. Oligonucleotide microarray for 16S rRNA gene based detection of all recognized lineages of sulfate reducing prokaryotes in the environment. Appl Env Microbiol. 2002;68 SRC-:5064–5081. doi: 10.1128/AEM.68.10.5064-5081.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lucker S, Steger D, Kjeldsen KU, MacGregor BJ, Wagner M, Loy A. Improved 16S rRNA-targeted probe set for analysis of sulfate-reducing bacteria by fluorescence in situ hybridization. J Microbiol Methods. 2007;69:523–528. doi: 10.1016/j.mimet.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 67.Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.