Abstract
Along with traditional effects of aging and carcinogen exposure—inherited DNA variation has substantial contribution to cancer risk. Extraordinary progress made in analysis of common variation with GWAS methodology does not provide sufficient resolution to understand rare variation. To fulfill missing classification for rare germline variation we assembled dataset of whole exome sequences from>2000 patients (selected cases tested negative for candidate genes and unselected cases) with different types of cancers (breast cancer, colon cancer, and cutaneous and ocular melanomas) matched to more than 7000 non-cancer controls and analyzed germline variation in known cancer predisposing genes to identify common properties of disease-associated DNA variation and aid the future searches for new cancer susceptibility genes. Cancer predisposing genes were divided into non-overlapping classes according to the mode of inheritance of the related cancer syndrome or known tumor suppressor activity. Out of all classes only genes linked to dominant syndromes presented significant rare germline variants enrichment in cases. Separate analysis of protein-truncating and missense variation in this list of genes confirmed significant prevalence of protein-truncating variants in cases only in loss-of-function tolerant genes (pLI < 0.1), while ultra-rare missense variants were significantly overrepresented in cases only in constrained genes (pLI > 0.9). In addition to findings in genetically enriched cases, we observed significant burden of rare variation in unselected cases, suggesting substantial role of inherited variation even in relatively late cancer manifestation. Taken together, our findings provide reference for distribution and types of DNA variation underlying inherited predisposition to some common cancer types.
Subject terms: Cancer genetics, Cancer genetics, DNA sequencing, Genetic association study
Introduction
Discovery of over 100 germline predisposition genes in cancer have not only revolutionized identification of individuals and families at higher risk, but also provided novel mechanistic insights into the role of pathways in cancer development and helped in mitigating the risk using appropriate clinical management [1]. Common to cancer genetics approach involves studying kindred with multiple samples and searching for DNA variation segregated between affected and non-affected members of the family. However, segregation of the variants could be uniquely observed in a given kindred and do not provide compelling information about their capacity to explain cancer cases outside of kindred of interest. Multiple cohort-based studies of inherited variation in cancer with GWAS methods reached great success in identifying low to moderate risk common variants [2]. Understanding rare coding variation on population scale requires massive genome/exome sequencing data both for cases and controls. Only recently sufficient statistical power was gained to discover new cancer susceptibility genes using case–control analysis of rare germline variation [3]. However, systematic description of rare inherited variation architecture in cancer cases in comparison to control subjects has not been reported yet.
Results
In order to identify shared properties of rare germline variation in cancer 866 patients diagnosed with early onset and/or familial history either of breast cancer (MIM: [114480]), colon cancer (MIM: [114500]), cutaneous melanoma (MIM: [155600]), ocular melanoma (MIM: [155720]), Li-Fraumeni syndrome (MIM: [151623]) (Supplementary materials, Supplementary Table 1) were recruited at MGH (Boston, USA), MEEI (Boston, USA), MSKCC (New York, USA), Andreas Sygros Hospital (Athens, Greece). Written informed consent was obtained from all individuals. Patients in this cohort were subjected to initial genetic screening (Supplementary Methods) and further identified as “selected cases”. We additionally included 1754 cancer samples with matching phenotypes from The Cancer Genome Atlas with no ascertainment for family history and age of onset. This cohort was identified as “unselected cases”. Control set of 24,612 samples was assembled from dbGAP whole exome studies of non-cancer phenotypes (Supplementary Table 1). Whole exome DNA sequences from all samples were assembled in a single dataset. To ensure close ancestral matching, we performed principal component analysis (PCA; Supplementary Figure 1A) and the largest cluster of samples representing European ancestry was further subjected to relatedness analysis. We removed all duplicated samples and first-degree relatives from further analysis. Final dataset included 846 selected cases, 1496 unselected cases and 7924 matched controls. Examination of common synonymous variants (MAF > 5%) revealed a null-distribution of the Fisher’s exact test (two-sided) statistic between cases and controls with genomic inflation factor λ = 1.012 (Supplementary Figure 1B).
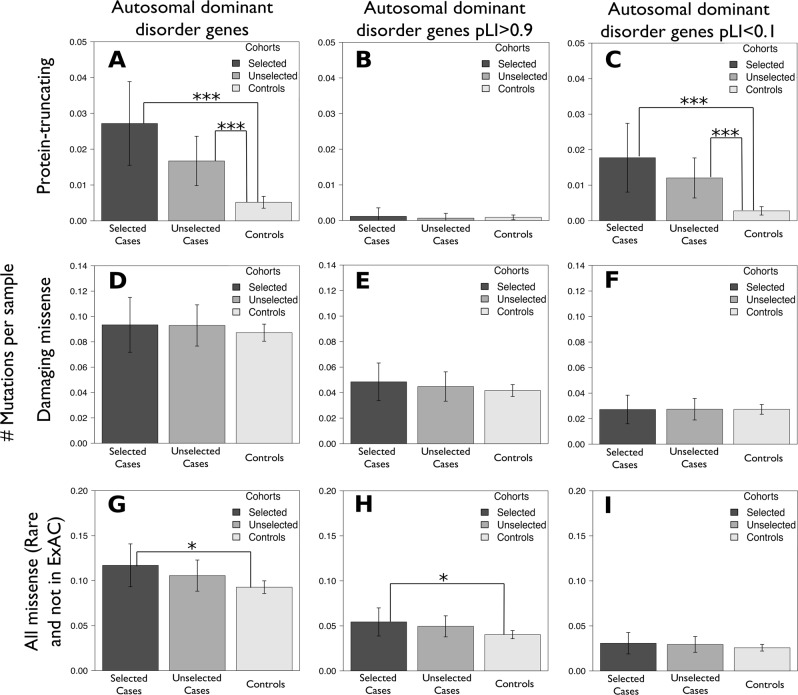
Results from Zhang et al. [4] provided good reference to known cancer susceptibility genes (germline and somatic) clustering based on dominant/recessive nature of linked cancer syndromes or known tumor-suppressor activity (Supplementary Table 2). We examined cumulative burden of rare (minor allele count of less or equal to 10) variants in cases compared to controls within each set of genes. Genes linked to dominant cancer disorders exhibited significant burden in both selected and unselected cases compared to controls. Separate analysis of damaging missense and protein-truncating variants (PTVs) established the main role of the latter in observed association signal (Supplementary Table 3A-D). Interestingly, significant abundance of risk alleles was observed both in selected (Two-sided Fisher’s test p = 3.16 × 10-8; OR = 3.62; OR CI = 2.32–5.54) and unselected cases (Two-sided Fisher’s test p = 5.95 × 10-6; OR = 2.53; OR CI = 1.69–3.74), however, burden of risk alleles in selected group was greater than in unselected group (Fig. 1a). Genes linked to breast cancer disorders carry substantial number of PTVs in controls, while genes linked to more severe phenotypes, like Li-Fraumeni syndrome (TP53), uveal and cutaneous melanomas show no or very low count of PTV carriers in control cohort (Supplementary Figure 2). Since selected cases were subjected to screening we tested whether risk genes from non-matching cancer phenotypes contribute to overall PTV-burden. BRCA1, BRCA2, and MSH2 have multiple carriers from more than 1 cancer phenotype (Supplementary Figure 3A). Once this analysis is performed jointly on selected and unselected cases PALB2 and BAP1 also have PTV carriers from non-matching cancer phenotypes (Supplementary Figure 3B), suggesting that in some cases a risk variant might be identified if additionally to known risk genes for a given cancer phenotype other knonwn risk genes are screened. Downstream examination of allele frequency spectrum for variants driving this association signal affirmed significance of singleton burden (Two-sided Fisher’s test p = 1.49 × 10-8, p = 2.36 × 10-5; OR = 5.25, OR = 3.23; OR CI = 2.99–9.01, OR CI = 1.88–5.46; selected and unselected cases, respectively) while variants with minor allele count 2–10 did not show significant enrichment (Two-sided Fisher’s test p = 0.1, p = 0.07 selected and unselected cases, respectively). Considering overrepresentation of PTVs in cases it was feasible to test genes linked to dominant cancer disorders for loss-of-function intolerance. We used probability of loss-of-function intolerance (pLI) from ExAC database [5] to separate genes into loss-of-function tolerant (pLI < 0.1) and intolerant (pLI > 0.9) groups (Supplementary Figure 4). Given that our case cohort does not have pediatric patients, for adult onset cancers we observed significant burden of singleton PTVs only in tolerant genes (Two-sided Fisher’s test p = 1.5 × 10-8, OR = 3.66, OR CI = 2.03–6.36; p = 3.0 × 10-4, OR = 2.74, OR CI = 1.57–4.65, selected and unselected cases respectively, Fig. 1b, c). While constrained genes are depleted in protein-truncating variants in cancer, we sought to test whether missense variations are uniformly distributed between constrained and tolerant genes linked to dominant cancer syndromes. We did not observe any enrichment in damaging missense variants among cases using minor allele count of 1 (MAF ~1 × 10-4) and 1–10 (MAF ~1 × 10-4 to 1 × 10-3) as a frequency cutoff (Fig. 1d–f). To examine missense variation of even lower frequency, we used non-TCGA subset of ExAC [5] database to keep for analysis only variants with MAF < 2.3 × 10-5 (not present in ExAC and singletons in ExAC non-Finnish Europeans). Ultra-rare missense variant analysis revealed significant burden in selected cases driven by loss-of-function intolerant gene contribution (Two-sided Fisher’s test p = 0.045, p = 0.025; OR = 1.26, OR = 1.44; OR CI = 1.00–1.58, OR CI = 1.03–1.97; all and constrained autosomal dominant disorder genes, respectively; Fig. 1g–i). Previous analysis of cutaneous melanoma cohort used in this study identified EBF3 (MIM: [607407]) as a new germline predisposition gene demonstrating tumor suppressor functional activity [3]. Interestingly, this gene has pLI = 1 and carried ultra-rare missense variation in conserved protein domains, consistently with our observations above.
Fig. 1.
DNA variation landscape overview. Mean DNA variant count per sample for protein truncating variants (MAC = 1; MAF~1 × 10-4) (a–c); Damaging missense variants (MAC = 1; MAF ~1 × 10-4) (d–f); Missense variants that are not or rare (MAC = 1; MAF = < 2.3 × 10-5) in non-TCGA ExAC (g–i); estimated across all genes linked to autosomal dominant disorders (a, d, g); autosomal dominant disorders linked genes with pLI > 0.9 (b, e, h); autosomal dominant disorders linked genes with pLI < 0.1 (c, f, i); *p < 0.05; ***p < 0.001
It is worth noting, however, that selected cases dataset was assembled by initial genetic screening of probands that satisfy NCCN genetic testing criteria [6]. If tested positive, they were not subsequently included in this study. Thus, genetically enriched cases have had more genetic screening and some diagnosed cases were removed before being entered in this study sample—likely attenuating the strength of association to the group with known autosomal dominant cancer predisposition genes.
Discussion
Overall, observed germline variation in both selected and unselected cases in established cancer susceptibility genes is linked to dominant cancer disorders, majorly represented by PTVs and has ultra-low frequency in population. While we observed ultra-rare missense variants enrichment in cancer, proportion of cases explained by this type of variation is likely very small. Understanding power limitations of our study and potential effects of imbalance between cancer cohort sizes, yet our results provide a reference point for allele frequencies and variation type for future search of new genes contributing to inherited cancer susceptibility through rare DNA variation. Unascertained for potential genetic enrichment “unselected” case cohort shows substantial prevalence of rare protein-truncating variation compared to controls, suggesting that cohort-based studies of rare variation in cancer might benefit from including sporadic cases. Cancer phenotypes in our case cohort potentially are of limited representation of rare cancer disorders that often have mendelian nature [7] or highly-heterogenous cancer phenotypes. We expect that overall majority of cancer cases would be explained by sporadic somatic variants and inherited polygenic risk (mostly driven by common DNA variation). On the opposite side, family-based studies of common cancer types are bound to ultra-rare DNA variation. Our results provide an informative reference for rare variation in cancer cohorts in comparison with control subjects aiding candidate variants search and prioritization.
Methods
Patient cohorts
Details could be found in Supplementary Methods. All case and control genotypes are publicly available through dbGAP database (unselected cases: phs000178.v1.p1, selected cases: phs000823.v1, phs000822.v1.p1, phs000824.v1.p1).
Exome sequencing and variant calling
Whole exome libraries were prepared using a modified version of Agilent’s Exome Capture kit and protocol, automated on the Agilent Bravo and Hamilton Starlet, followed by sequencing on the Illumina HiSeq 2000 at the Broad Institute. We used an aggregated set of samples consented for joint variant calling resulting in 37,607 samples (Supplementary Table 1). BWA-MEM algorithm (version 0.7.12-r1039) [8] and the best-practices GATK/Picard Pipeline was used for raw data alignment, followed by single batch joint variant calling using GATK v3.1-144 Haplotype Caller [9–11]. The resulting dataset had 7,094,027 distinct variants. Selected variants in CDKN2A, BRCA1, and BAP1 were confirmed with Sanger sequencing.
Principal component analysis (PCA) on common (MAF > 5%) autosomal independent SNPs was performed with Eigenstrat [12]. Relatedness analysis among Europeans was conducted with PLINK [13] as suggested in the PLINK best practices. VEP [14] was used for functional annotation of the DNA variants. Common and rare variants analyses were conducted using PLINK/SEQ (https://atgu.mgh.harvard.edu/plinkseq/.PLINK/SEQ [Internet]).
Supplementary information
Acknowledgments
This research was supported by an NHGRI grant U54 HG003067 and MSKCC Core grant NIH P30CA008748.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kenneth Offit, Phone: +1-212-639-2000, Email: offitk@mskcc.org.
Mark J. Daly, Phone: +1-617-726-5936, Email: mjdaly@atgu.mgh.harvard.edu
Supplementary information
The online version of this article (10.1038/s41431-019-0346-0) contains supplementary material, which is available to authorized users.
References
- 1.Rahman N. Realizing the promise of cancer predisposition genes. Nature. 2014;505:302–8. doi: 10.1038/nature12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sud A, Kinnersley B, Houlston RS. Genome-wide association studies of cancer: current insights and future perspectives. Nat Rev Cancer. 2017;17:692–704. doi: 10.1038/nrc.2017.82. [DOI] [PubMed] [Google Scholar]
- 3.Artomov M, Stratigos AJ, Kim I, et al. Rare variant, gene-based association study of hereditary melanoma using whole-exome sequencing. J Natl Cancer Inst. 2017;109:djx083. [DOI] [PMC free article] [PubMed]
- 4.Zhang J, Walsh MF, Wu G, et al. Germline mutations in predisposition genes in pediatric cancer. N Engl J Med. 2015;373:2336–46. doi: 10.1056/NEJMoa1508054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.National Comprehensive Cancer Network. NCCN Guidelines. 2017.
- 7.Pomerantz MM, Freedman ML. The genetics of cancer risk. Cancer J. 2011;17:416–22. doi: 10.1097/PPO.0b013e31823e5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinform. 2013;43:11.10.1–33. [DOI] [PMC free article] [PubMed]
- 10.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 13.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLaren W, Gil L, Hunt SE, et al. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.