Abstract
Beetles (Coleoptera) have the highest species diversity among all orders, and they have diverse food habits. Gut microbes may have contributed to this diversification of food habits. Here, we identified the pattern of the relationship between ground-dwelling beetles and their gut microbial communities (bacteria and fungi) in the field. We collected 46 beetle species of five families from secondary deciduous forests and grasslands in Japan and extracted microbial DNA from whole guts for amplicon sequencing. The gut bacterial and fungal communities differed among all habitats and all food habits of their hosts (carnivores, herbivores, omnivores, and scavengers) except for the fungal communities between carnivores and scavengers. Specifically, the abundant bacterial group varied among food habits: Xanthomonadaceae were abundant in scavengers, whereas Enterobacteriaceae were abundant in carnivores and herbivores. Phylogenetically closely related beetles had phylogenetically similar communities of Enterobacteriaceae, suggesting that the community structure of this family is related to the evolutionary change in beetle ecology. One of the fungal groups, Yarrowia species, which has been reported to have a symbiotic relationship with silphid beetles, was also detected from various carnivorous beetles. Our results suggest that the symbiotic relationships between ground-dwelling beetles and these microbes are widespread.
Subject terms: Biodiversity, Community ecology, Metagenomics
Introduction
Beetles (Coleoptera) have the highest species diversity among all orders and comprise ~40% (350 000 species) of all insects [1]. Beetles are also ecologically diverse, with species having carnivorous, herbivorous, omnivorous, and scavenger food habits [1], and they therefore play diverse roles in ecosystems. One possibility for the diversification of food habit is that it occurred in beetles via the establishment of symbiotic relationships with gut microbes. This has been reported in gall midges (Diptera: Cecidomyiidae) [2] and bugs (Hemiptera: Pyrrhocoridae) [3, 4]. However, these relationships have not been studied well in beetles, although symbiotic relationships between several beetles and yeasts are known [e.g., ref. 5].
Community analyses have revealed that the gut bacteria of vertebrates such as fish and mammals are related to these animals’ food habits [e.g., refs. 6, 7]. For example, herbivores depend more on gut microbes for digestion than do omnivores and carnivores, and they possess unique and diverse bacterial communities. The gut bacterial communities of fish differ among habitats [8], and this may also be the case with the gut microbial communities of beetles.
Diverse ground-dwelling beetles with various food habits live in forests and grasslands [e.g., refs. 9–11]. Ground-dwelling beetles are an appropriate model group for investigating the relationship between beetles and their gut microbes, because we can obtain sympatric, ecologically diverse species. In the ground-dwelling beetle family Silphidae, whose species consume dead vertebrates and invertebrates such as earthworms [12, 13], the yeast Yarrowia lipolytica and a related species group that can break down diverse carbon sources, including hydrocarbons and lipids [14], have been isolated [15, 16]. Vogel et al. [16] also noted that the gut bacteria of a silphid species contribute to the digestive process of their hosts, and suggested that Yarrowia species also help inhibit the growth of other microbes by producing an antimicrobial substance. Although various microbes inhabit beetle guts, symbiotic relationships such as mutualism and parasitism have not been studied in most beetles in the field.
In this study, we identified the general patterns of symbiotic relationships between beetles and their gut microbes (bacteria and fungi) in the field by using ground-dwelling beetles as a model system. We examined the relationships between the food habits and habitats of ground-dwelling beetles and the diversity and structure of their gut microbial communities. We also examined the relationship between the phylogenetic relatedness among beetle species and the similarity of their gut microbial communities to infer the evolutionary relationship between host and microbe. Finally, we also specifically examined the symbiotic relationship with yeasts, because mutualism with beetles, including a few ground-dwelling silphid species, has been reported [e.g., refs. 5, 15–18].
Materials and Methods
Sampling and sequencing of beetles
We collected samples at three sites in northern Japan (Supplementary Table S1). Each site contains secondary deciduous forest and grassland adjacent to each other. At each habitat, we collected beetles over 1 day by using 50 pitfall traps made of plastic cups. The surveys were conducted once each in July and August in 2013 and 2014 and twice each in July and August in 2015 at sites 1 and 2, and once each in July and August in 2013 at site 3. Individual beetles were placed separately into a tube or vial; they were brought back to the laboratory in cooler boxes with refrigerant and kept at 4 °C until use. We also collected surface soil samples from each habitat at sites 1 and 2 for DNA extraction.
The samples collected in 2013 and 2014 were used for molecular analysis of beetles and yeasts, and those collected in 2015 were used for community analysis of gut microbes by amplicon sequencing. Species identification of beetles was performed based on the references [19–21].
Parts of the mitochondrial COI gene and 28S ribosomal RNA gene were amplified and sequenced as described previously [22] by using the primers listed in Supplementary Table S2. Sequences were deposited in DDBJ under accession numbers LC373578–LC373737.
Amplicon sequencing with MiSeq and microbial community analysis
We dissected beetles under the microscope within 2 days after sampling to minimize changes in the gut microbial community to the degree possible. Extracted guts (from foreguts to hindguts) were preserved in 99% (v/v) ethanol. DNA from soil samples (0.8 g) and whole guts with gut contents were extracted by using a FastDNA SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA, USA) following the manufacturer’s protocol. Extracted DNA was subjected to PCR amplification targeting an ~300-bp fragment of the 16S rRNA variable region 4 (EMP 515F and 806R) for bacteria [23] and an ~250-bp fragment containing rRNA ITS1/2 regions (EMP ITS1F and ITS2) for fungi [24]. Primers used for those amplifications contain barcode tags to distinguish samples (Supplementary Table S2). Amplification was performed with the GoTaq Green Master Mix (Promega KK, Tokyo, Japan) in a thermal cycler under the following conditions: 94 °C for 3 min; 25 cycles of 94 °C for 30 s, 50 °C for 60 s, and 72 °C for 90 s; and finally 72 °C for 10 min. Triplicate PCRs were performed independently for each DNA sample and mixed.
PCR products were purified using the NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s instruction and quantified using a NanoDrop 2000c (Thermo Fisher Scientific, Waltham, MA, USA). All barcoded amplicons were pooled in equal concentrations and sequenced on the MiSeq platform using the MiSeq Reagent Nano Kit v2 (500 cycles; Illumina, San Diego, CA, USA) according to the manufacturer’s recommended protocol (https://icom.illumina.com/) to produce 251-bp paired-end reads. We obtained paired-end reads (accession number: DRA006652) and removed low-quality sequence data using QIIME2 ver. 2018.04 [25] for further analysis. We removed 46 bp at the 5′-end from the ITS sequence data for fungi in order to fit our data to the reference database. We used DADA2 [26] implemented in QIIME2 to construct an operational taxonomic unit (OTU) table for OTU representative sequences.
We analyzed the microbial communities in beetle guts by using QIIME2. We used the GreenGenes 13.08 database with 99% identity [27] for bacteria and assigned taxonomies to OTU representative sequences by using RDP Classifier (conducted on 3 June 2018) [28]. We used the UNITE database released on 10 October 2017 [29]; this is the database of OTUs identified by taxonomic experts as having singleton sequences for fungi. We added the ITS1/2 sequence data of the genus Yarrowia into the database from GenBank (accession numbers: FR856611, EU343806, AM279270, NR111357, KY101920, AM279259, AM279254, NR111212, EF621566, KY105998, KF649296, KF649301, and KF425323). We assigned taxonomies to OTU representative sequences by using the q2-feature-classifier plugin with BLAST+ method. To exclude OTUs other than those of fungi, such as those of beetles and their food resources, we removed the ITS1/2 sequence data identified as “unassigned” when assigning taxonomies for community analysis.
We compared Faith’s phylogenetic diversity of bacteria and fungi among habitats and among food habits as an index of alpha diversity. We conducted a pairwise Kruskal–Wallis test implemented in QIIME2. In addition, we performed principal coordinates analysis (PCoA) of the unweighted Unifrac distance [30] for bacteria and fungi as an index of beta diversity to evaluate similarity among the gut microbial communities. We set the sampling depth to 1300 for bacteria and to 5000 for fungi in order to remove samples with low sequence reads for these analyses. We conducted permutational multivariate analyses of variance (PERMANOVAs) for the unweighted Unifrac distance matrices to compare the differences in microbial communities among food habits and among habitats of beetles by using the “adonis” command with 1 million permutations in the vegan package in R ver. 3.5.0 [31]. Beetle species and collection season (July or August) were also included as the independent variables. If these PERMANOVAs for the comparison among food habits were significant, we conducted a post hoc test using the “pairwise.perm.manova” command with 100 000 permutations, Pillai’s trace statistic, and Benjamini–Hochberg correction for P-value in the RVAideMemoire package in R ver. 3.5.0.
Yeast isolation, DNA extraction, PCR fingerprinting, and nucleotide sequencing
We isolated yeasts from beetles to assess whether the symbiotic relationship reported in a few ground-dwelling beetle species [15, 16] is a general phenomenon. A series of preliminary experiments showed that the yeast species assemblage on the exoskeleton surface of beetles was similar to that isolated from the beetle gut (Supplementary Table S3). We therefore isolated yeasts from the exoskeleton to obtain cultures of importance. After a beetle’s species had been identified, yeast was isolated from the beetle’s whole body surface (in the case of samples collected in 2013) or from the elytra (in the case of samples collected in 2014). Beetles (whole body or elytra only) were aseptically placed in 1 ml of sterile 0.03% (v/v) Tween 20 solution and vortex-mixed for 1 min. A sterile plastic spreader was used to spread the solution onto a YM agar plate supplemented with 100 ppm chloramphenicol. The lid was left open for about 3 min, and then the plate was sealed and incubated for 2–7 days at room temperature (ca. 20 °C). The texture, color, and surface of agar-grown colonies that appeared on the plates were examined to distinguish yeasts from filamentous fungal colonies based on the reference [32], where apparently fast-growing filamentous colonies such as Trichoderma-like and Aspergillus-like ones were excluded. When yeast colonies appeared, 10 of them were randomly selected and picked up on a fresh YM agar plate, and then incubated for 2–7 days. Single colony isolation was performed again on YM agar, and the resultant pure culture was used for preservation and subsequent DNA extraction. The culture was suspended in 1 ml of 10% (v/v) glycerol solution and preserved at –20 °C. For DNA extraction from yeast cultures obtained in 2013, the cell mass was suspended in 200 μl of sterile distilled water, heated in a microwave oven at 700 W for 10 s, and then kept at –20 °C until used. For yeast cultures obtained in 2014, the cell mass was suspended in 200 μl of TE buffer, heated in a metal block bath at 100 °C for 10 min, chilled on ice, and then centrifuged at 15 000 rpm for 3 min at 4 °C. One hundred microliters of the supernatant were kept at –20 °C until used.
PCR fingerprinting using a microsatellite primer (GTG)5 (5′-GTG GTG GTG GTG GTG-3′) was performed on the yeast isolates, as described previously [33]. Banding patterns after electrophoresis were grouped manually, and then a few representative isolates were selected for each group, for which PCR was performed to amplify the D1/D2 region of the large subunit ribosomal RNA gene (LSU D1/D2) and the small subunit ribosomal RNA gene (SSU) using the primers listed in Supplementary Table S2. PCR conditions for LSU D1/D2 were 40 cycles of 98 °C for 10 s, 52 °C for 30 s, and 72 °C for 45 s; those for SSU were 40 cycles of 94 °C for 20 s, 52 °C for 30 s, and 68 °C for 2 min. The subsequent procedures for nucleotide sequencing were the same as those employed for COI and 28S gene sequencing in beetles (accession numbers: LC373738–LC373914).
Phylogenetic analysis of beetles and yeasts
Haplotypes for beetles and yeasts were detected by using the “pgelimdupseq” command in Phylogears2 ver. 2.0 [34]; individuals with the same haplotypes were excluded from phylogenetic analyses. We included the sequence data of Cupedidae (Archostemata) as the beetle outgroup species, those of Zygomycota as the yeast outgroup species, and those of identified yeast species from GenBank (Supplementary Tables S4 and S5). We used MAFFT ver. 7.308 (xinsi method) for the alignment of beetle 28S and yeast SSU and LSU [35]. The alignments were inspected by eye for obvious misalignments.
Phylogenetic analyses were based on the maximum likelihood and Bayesian methods. The optimum substitution models were estimated by Kakusan4 [36]. Maximum likelihood analyses were performed using TreeFinder ver. March 2011 [37] on the basis of the models selected by AICc4 (beetles: proportional model among regions and non-partitioned model among codons, COI: TVM+G, 28S: TVM+G; yeast: proportional model among regions, SSU: GTR+G, LSU: J1+G) with 1000 bootstrap replications. Bayesian analyses were performed in BEAST ver. 1.8.4 [38] with a log-normal relaxed clock model and substitution models selected by BIC4 (beetles: COI: GTR+G, 28S: GTR+G; yeast: SSU: GTR+G, LSU: GTR+G). The COI sequences of beetles were partitioned by their codon positions without unlinking parameters among codon positions. We set constraints for each beetle family (Carabidae, Silphidae, and Tenebrionidae) to be monophyletic in Bayesian analysis. We performed a run for 100 million generations with trees sampled every 10 000 generations, and we discarded the first 2500 trees. All of the 45 estimated parameters for beetles and 34 of the 37 estimated parameters for yeasts presented effective sample-size scores > 100.
Correlated evolutionary patterns between beetles and their gut microbes
We tested the correlation between the phylogenetic distance among beetle species and the dissimilarity of gut microbial communities among beetle species for several microbial families. In this analysis, we used the microbial families for which the mean relative abundance among individuals in each beetle species was higher than 5% in more than five beetle species, because abundant microbes in several beetle species are likely to have an evolutionary relationship with the host species group. Carabidae species were used for this analysis, because many species were collected in this family. We extracted the microbial sequence data with high relative abundance. We used the unweighted Unifrac distance for bacteria and fungi as an index of community dissimilarity and set the sampling depth to 100 for bacteria and to 200 for fungi. We used the mean interspecific distance of all pairs of beetle individuals. The phylogenetic distance among beetle species was calculated by using a tree constructed by BEAST, with one individual per species and the “cophenetic.phylo” command in the ape package in R ver. 3.4.1. We conducted a Mantel test with Pearson’s correlation coefficient by using the “mantel” command with 1 million permutations in the vegan package in R ver. 3.5.0. The false discovery rate for multiple tests was controlled by using the Benjamini–Hochberg method [39]. A positive correlation of the phylogenetic distance among beetles with the dissimilarity among gut microbial communities would suggest that beetle evolution and their gut microbial communities are related.
Molecular identification of yeasts
We conducted molecular identification of yeasts by the QC auto method implemented in Claident [40] by using the eukaryote LSU and SSU gene sequence databases at the genus and species level. If the identified taxonomic levels were different among genes and among databases, we applied the result with the lowest taxonomic level. We identified the samples that were not classified at the genus or species level by Claident to the genus level based on the phylogenetic relationship with known species data (Supplementary Table S5) on the phylogenetic tree.
Results
Beetle food habit and habitat
We collected 46 beetle species representing carnivores, omnivores, herbivores, and scavengers (dead-animal feeders) from five families from 2013 to 2015 (Table 1, Supplementary Tables S6 and S7). We obtained 32 species (mainly carnivores) from forests and 30 species (mainly carnivores but with more omnivores and herbivores than in the forests) from grasslands. Most carnivorous and omnivorous species belonged to the Carabidae (Supplementary Tables S6 and S7). The herbivores consisted of Carabidae and Tenebrionidae species, whereas the scavengers belonged to the Silphidae.
Table 1.
Numbers of beetle species collected from each habitat
| Habitat | ||
|---|---|---|
| Food habit | Forest | Grassland |
| Carnivore | 22 | 14 |
| Omnivore | 2 | 8 |
| Herbivore | 5 | 7 |
| Scavenger | 2 | 1 |
| Equivocal | 1 | 0 |
| Total | 32 | 30 |
Relationship between microbial community and beetle ecology
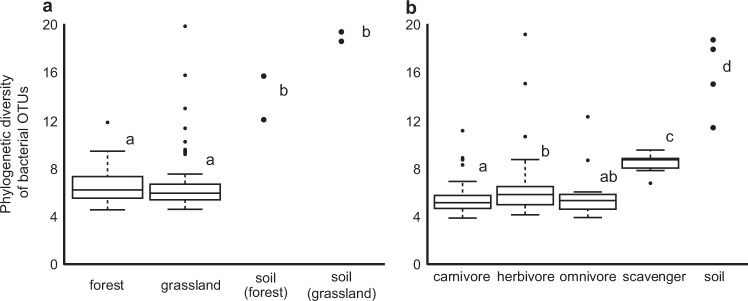
We examined the gut microbial communities of 116 beetle individuals from 32 species in four families (Supplementary Table S6). Bacterial phylogenetic diversity was significantly greater in soil than in the beetle guts, regardless of habitat (Fig. 1a, Supplementary Table S8). Bacterial phylogenetic diversity in beetle guts did not differ between habitats (Fig. 1a), but that of scavengers was significantly greater than those of the other food habits (Fig. 1b, Supplementary Table S9). The PCoA showed that bacterial community structure in beetle guts differed between habitats (R2 = 0.172, P = 0.008; Fig. 2, Supplementary Table S10). The soil bacterial community showed high similarity between habitats.
Fig. 1.
Phylogenetic diversity of bacterial OTUs collected from the guts of beetles (a) from each habitat and (b) with each food habit. Each datum is plotted for soil samples. Different letters indicate significant differences in phylogenetic diversity among habitats or food habits (P < 0.05)
Fig. 2.
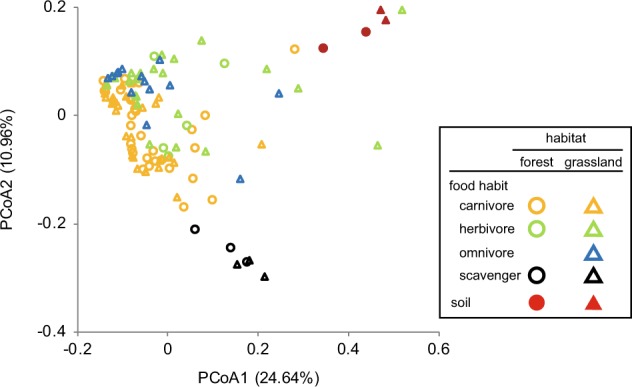
Bacterial communities in beetle guts, clustered by using PCoA based on the unweighted Unifrac distance. Circles and triangles correspond to samples collected in forest and grassland, respectively, and are colored according to beetle food habits. Eigenvalues of PCoA1 and PCoA2 are shown in parentheses
The relative abundance of sequence reads of each bacterial family in soil was distributed among more families than that in the beetle guts (mean number of families: soil: n = 4, 41.8 ± 10.8 [SD]; beetle gut: n = 111, 11.7 ± 9.9), and thus most families were classified as “others” (Fig. 3). The relative abundances of Enterobacteriaceae and Enterococcaceae were high in the guts of beetles collected from both habitats (Fig. 3a, Supplementary Figure S1). The PCoA differed significantly among all food habits (R2 = 0.172, P < 0.001; Supplementary Table S11). Food habit explained a much larger proportion of overall the variance than habitat did, although both food habit and habitat were significant factors (Supplementary Table S10). We obtained a similar pattern for PERMANOVA when we conducted the statistical analyses for the Bray–Curtis distance matrix (Supplementary Tables S12, S13). The bacterial families with high relative abundances differed among food habits. Scavengers possessed a unique bacterial community, which clustered tightly on the PCoA plot (Fig. 2) and was composed mainly of Xanthomonadaceae (Fig. 3b). Members of Enterobacteriaceae were relatively abundant in the guts of beetles with other food habits—specifically in carnivores and herbivores—although this family did not show high relative abundance in soil. Members of Enterococcaceae were also abundant in the guts of carnivores.
Fig. 3.
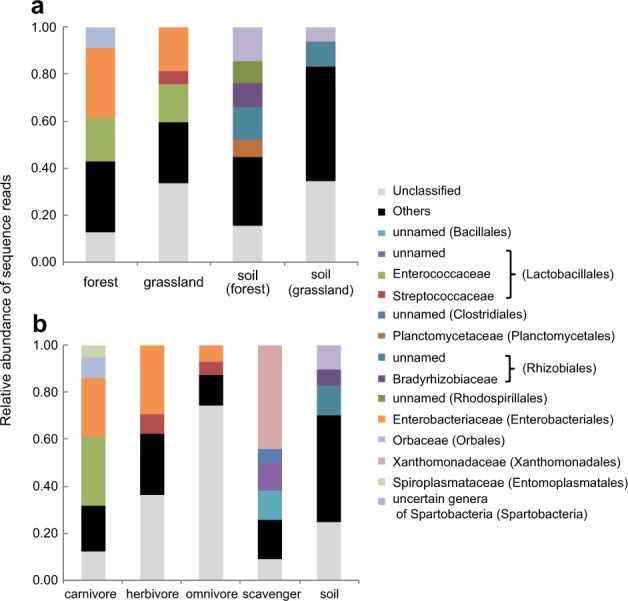
Relative abundance of each bacterial family collected from the guts of beetles (a) from each habitat and b with each food habit. Sampling depth was set to 1300 to remove samples with low sequence reads. OTUs that could not be classified into orders are summarized as “Unclassified.” Names of classified families with total abundances > 5% are shown with order names in parentheses. Other families are summarized as “Others.”
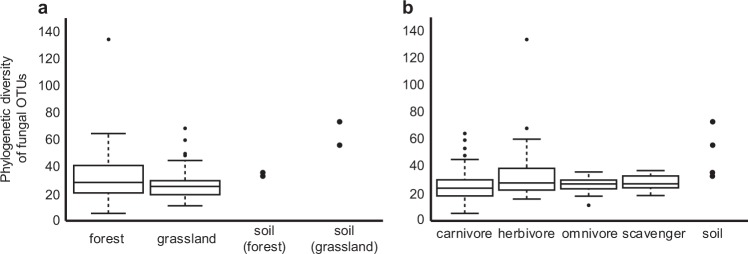
Fungal phylogenetic diversities were similar between habitats and among food habits (Fig. 4, Supplementary Table S8 and Table S9). The fungal community structure in beetle guts differed significantly between forest and grassland (R2 = 0.054, P < 0.001; Fig. 5, Supplementary Table S10). Habitat explained a slightly larger proportion of the overall variance than food habit did (Supplementary Table S10). We obtained a similar pattern for PERMANOVA when we conducted the statistical analysis for the Bray–Curtis distance matrix (Supplementary Table S12). The forest soil fungal community was similar to that in the guts of beetles collected from forest according to the PCoA plot (Fig. 5). The relative abundance of each fungal family was distributed among many families in soil and beetle guts (mean number of families: soil: n = 4, 53.5 ± 13.4 [SD]; beetle gut: n = 109, 27.9 ± 11.6), and the fungal families with high relative abundances varied among beetle species. Thus, the relative abundance of most families was less than 5%, and they were classified as “others” (Fig. 6). Members of Russulaceae were abundant in forest soil, whereas members of Mortierellaceae, an unnamed family of Hypocreales, and Pyronemataceae were abundant in grassland soil, although their relative abundances were less than 5% in beetle guts from each habitat (Fig. 6, Supplementary Figure S2). We found significant differences in fungal community structure among food habits (R2 = 0.046, P < 0.001), and significant differences were observed between food habits, except between carnivores and omnivores (Supplementary Table S11). We obtained weaker differences among food habits when we conducted the analysis for the Bray–Curtis distance matrix (Supplementary Table S13). Members of Trichocomaceae were abundant in scavengers.
Fig. 4.
Phylogenetic diversity of fungal OTUs collected from the guts of beetles (a) in each habitat and (b) with each food habit. Each datum is plotted for soil samples
Fig. 5.
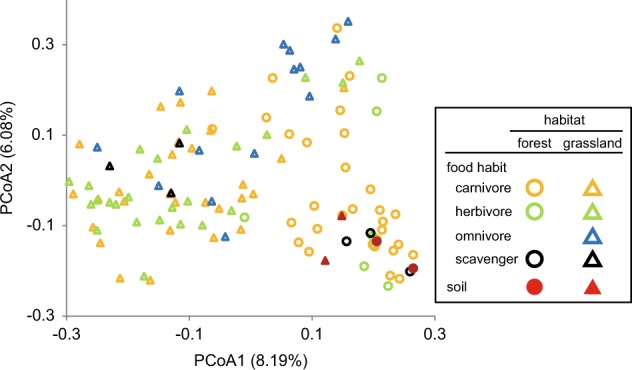
Fungal communities in beetle guts, clustered by using PCoA based on the unweighted Unifrac distance. Circles and triangles correspond to samples collected in forest and grassland, respectively, and are colored according to beetle food habits. Eigenvalues of PCoA1 and PCoA2 are shown in parentheses
Fig. 6.
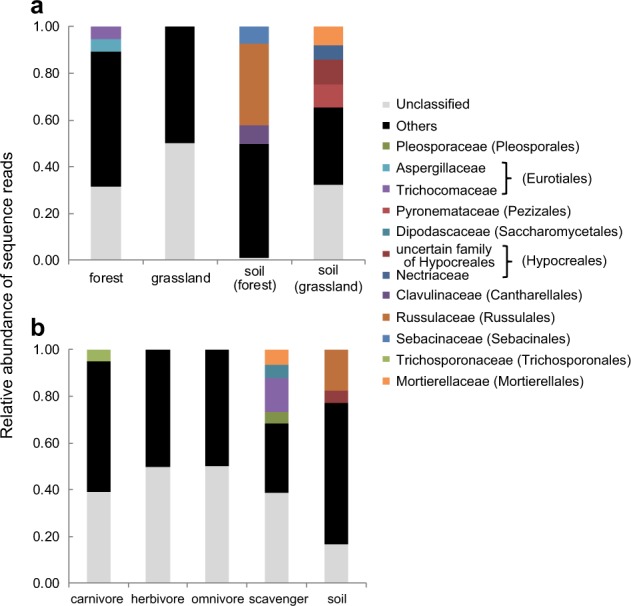
Relative abundance of each fungal family collected from the guts of beetles (a) from each habitat and b with each food habit. Sampling depth was set to 5000 to remove samples with low sequence reads. OTUs not classified into orders are summarized as “Unclassified.” Names of classified families with total abundance > 5% are shown with order names in parentheses. Other families are summarized as “Others.”
We constructed a beetle phylogenetic tree using 611 bp of the COI gene and 655 bp of the 28S gene from 165 individuals of 36 beetle species (Supplementary Figure S3). None of the fungal families showed high relative abundance (> 5%) in more than five carabid species. In one bacterial family, Enterobacteriaceae, we found a significant positive relationship between beetle phylogenetic distance and the dissimilarity of bacterial community structure in beetle guts, indicating that the Enterobacteriaceae community was more similar between more closely related beetle species (r = 0.480, P = 0.009; Fig. 7).
Fig. 7.
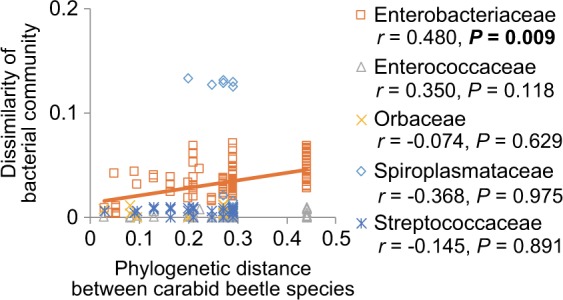
Relationships between the unweighted Unifrac distance for bacteria in beetle guts and the phylogenetic distance among beetle species (for those microbial families with high relative abundance). Statistical results of Mantel tests with Pearson’s correlation coefficients are also shown. Significant P-values after correction by using the Benjamini–Hochberg method [40] are in bold. Solid line shows the regression line for a significant relationship (P < 0.05)
Relationship between yeast and beetle ecology
We attempted to isolate yeasts from 450 beetles belonging to four families and 42 species (Supplementary Tables S7). One hundred ninety-five samples of 28 beetle species yielded yeast isolates (Supplementary Tables S7). Yeasts were obtained from scavengers and carnivores at high rates, whereas they were obtained from herbivores and omnivores at low rates. The average lengths of the nucleotide sequences for LSU D1/D2 and SSU of the isolated yeasts were 501 and 875 bp, respectively. Although both ascomycetous and basidiomycetous yeasts were detected, most isolates appeared to be members of Ascomycetes, as determined from the sequence identities inferred by Claident and the yeast phylogenetic tree. Of note, the majority of isolates were closely related to a couple of Yarrowia (Saccharomycetes, Saccharomycotina) species (Supplementary Figure S4 and Table S7). Also, isolates related to the genera Priceomyces and Starmerella/Wickerhamiella (Saccharomycetes, Saccharomycotina) were obtained from nine and five beetle species, respectively (Supplementary Figure S4 and Table S7). The relative abundance of Yarrowia species was high in the scavenger Necrophila japonica according to the fungal community analyses using the MiSeq reads (n = 6, 5.94 ± 5.41% [SE]), and Yarrowia species were detected at low rates (<0.04%) in carnivores (Synuchus melantho and Pterostichus fuligineus).
Yeast species assemblages detected from beetles were not significantly different between habitats, but they did differ among food habits (Supplementary Figure S4 and Table S7). Yeasts assumed to be Yarrowia species were frequently isolated from scavenger silphid beetles (Supplementary Table S7). Yeasts were also isolated from carnivorous beetles: Yarrowia-related isolates were readily detected and Priceomyces- and Starmerella/Wickerhamiella-related ones were less frequently found in several carnivorous beetles. In addition, basidiomycete Vanrija- and Papiliotrema-related isolates were also detected in a couple of beetle species. The frequency of yeast isolation (mainly Yarrowia and Priceomyces) was particularly high (> 50%) from carnivorous Leptocarabus arboreus, Carabus albrechti, and Pterostichus fuligineus, all of which belong to Carabidae. Yeasts were less common in herbivorous beetles. No herbivorous beetle species (excluding rare species, of which only a few individuals were collected) yielded yeast isolates with a frequency of 50% or more (Supplementary Table S7). Nonetheless, herbivorous beetles yielded minor isolates such as Starmerella/Wickerhamiella-, Kurtzmaniella-, Saitozyma-, and Moesziomyces-related yeasts, as well as Yarrowia-related ones. The former four isolates were exclusively found in herbivorous beetles. The diversity of yeast taxa was high in the herbivorous beetles Harpalus eous and Gonocephalum japanum. Very few yeast isolates were obtained from omnivorous beetles, unlike the case with carnivorous or herbivorous beetles.
Discussion
The gut microbial community obtained from related sympatric beetles differed among taxa depending on their food habit and habitat, as reported previously in vertebrates [6–8]. However, the pattern differed between bacteria and fungi. Similar bacterial families were abundant in the guts of beetles in forests and grasslands, whereas the abundant bacterial families differed among food habits. On the other hand, we did not find particularly abundant fungal families associated with either habitats or food habits (except scavengers), because diverse fungal families were detected in beetle guts and the fungal families with high relative abundances varied among beetle species. In addition, although both food habit and habitat had significant effects on bacterial and fungal community structures, the strength of these effects differed. Food habit explained a much larger proportion of the overall variance in the bacterial community than habitat did, whereas habitat explained a slightly larger proportion of overall variance in the fungal community than food habit did. These results may suggest that some bacterial groups tend to have symbiotic relationships with their host species groups in relation to beetle food habits, and they tend to become dominant in beetle guts. The gut morphology of ground-dwelling carabid beetles varies among subfamilies [41], and this variation reflects their food habits in part. The gut bacterial community may be affected by gut morphology related to food habit, and bacterial groups that find the gut environments suitable can inhabit them. Gut bacterial and fungal communities also differed between habitats. This could reflect the likelihood that the food resources of beetles already differ between habitats, even if the beetles’ food habits are the same, and the microbes that enter opportunistically with food resources and from the environment, such as from soil, are also different.
Gut bacterial communities had much lower diversity than that of soil bacterial communities, although no differences were detected in the diversities of gut fungal communities. The bacterial families with high relative abundances in beetle guts (Xanthomonadaceae, Enterobacteriaceae, and Enterococcaceae) are also abundant in a silphid species showing parental care, Nicrophorus vespilloides; these bacteria suppress the growth of competing microbes, suggesting that they play a role in preservation of the carcass for parental care [42]. These bacteria may also suppress other microbes in the guts of ground-dwelling beetles; thus microbe communities are filtered in the food resources or the guts (or both) of beetles by their activities. Our results suggest that not only silphid species with parental care but also some ground-dwelling beetles have symbiotic relationships with these bacteria. They may grow prolifically on, or in, the food resources and guts of ground-dwelling beetles and prevent beetle food resources from decaying by suppressing the proliferation of other microbes.
Phylogenetically more closely related carabid species had more phylogenetically similar communities of Enterobacteriaceae, suggesting that the community structure of this family is related to evolutionary changes in beetle ecology such as food habit, habitat, and gut morphology. Members of Enterobacteriaceae were reported to be abundant in the gut of one silphid species, N. vespilloides [42], and they normally inhabit the digestive tracts of animals, including humans; some genera are pathogenic to insects [43]. Thus, some Enterobacteriaceae groups might have cospeciated or coevolved (or both) in host–parasite relationships with beetles.
Microbes enter the beetle gut by vertical transmission from parents to their offspring or by horizontal transmission from the surrounding environment. The beetle species examined in this study, except Nicrophorus concolor [12], are not known to show parental care or any behaviors promoting vertical transmission, and thus vertical transmission is less likely to occur. However, anobiid and tenebrionid beetles transmit microbes vertically by attaching microbes to the egg surface [44, 45]. Phylogenetically closely related beetles possess phylogenetically similar communities of Enterobacteriaceae, and this family showed low relative abundance in soil (0.2%). This family may be transmitted vertically through such a process.
We also found some gut microbes that showed high relative abundances in a few particular beetle species. Silphid scavengers possessed Xanthomonadaceae with high relative abundance. Vogel et al. [16] noted that members of this family could promote carcass digestion by promoting esterase, lipase, and urease activities in the hindgut. These microbes might have mutualistic relationships within related beetle species groups. In addition, all of the OTUs assigned as Streptococcaceae, which showed high relative abundance in herbivorous beetles, were classified as Lactococcus. This genus generally inhabits the guts of animals, including humans [46]. Xanthomonadaceae and Streptococcaceae bacteria are abundant in the guts of beetles of particular food habits partly because of their ability to utilize the food resource of each type of beetle.
Yeasts were specific to each food habit of beetles regardless of habitat, indicating a symbiotic relationship between beetle and yeast that was related to beetle food habits. Yarrowia species were extremely specific to the scavengers in Silphidae: these beetle individuals possessed yeasts at high rates (59/64), and all of them belonged to Yarrowia. Vogel et al. [16] suggested that the Yarrowia species contribute not only to digestion but also to inhibition of the growth of other microbes by producing antimicrobial substances, and silphid species acquire benefits from the yeasts. Yarrowia species have also been obtained from other silphid species in Europe and the United States [15, 16], suggesting that this symbiotic relationship is universal in Silphidae and has been constructed over a long time scale. The species in Silphinae (one of two subfamilies in Silphidae), such as N. japonica in this study, do not show any parental care and related behavior, although members of the other subfamily (Nicrophorinae) show such behaviors [12, 47]. Therefore, the offspring of Silphinae may obtain these yeasts every generation from their parents, which may attach microbes to the egg surface in the same way as anobiid and tenebrionid beetles do [44, 45], or from the surrounding environment via food resources and soil. Although these yeasts contribute to the preservation of the carcasses used for parental care in silphid beetles [42], the consistent association of these yeasts with related silphid beetles that do not preserve carcasses suggests that in this beetle group they also serve only a digestive function that predates the evolution of parental care.
Some carnivorous species possessed yeasts at high rates (Leptocarabus arboreus: 8/10 individuals; Carabus albrechti: 39/46; Pterostichus fuligineus: 13/14), and Yarrowia species were isolated from most of these beetles. These results suggest some kind of symbiotic relationship between them. As in scavengers, Yarrowia species may contribute to digestion and inhibition of the growth of other microbes in these carnivorous beetles. Herbivorous beetles possessed diverse yeasts from Ascomycetes and Basidiomycetes at low rates (≤ 50%), indicating weak relationships between the beetles and these yeasts. Therefore, yeasts detected from herbivorous beetles do not appear to have symbiotic relationships with their hosts.
Trichocomaceae fungi were specifically abundant in scavengers in our fungal community analysis, although these taxa were not previously detected in a silphid species [16]. Trichocomaceae species in scavengers’ guts may be difficult to culture, because no member of this family was identified among the fungi isolated from the guts of beetles. Moreover, although Trichocomaceae species are ubiquitous in plant and soil materials [48, 49], this fungal family was rarely found in the soil in our study (< 0.63%). Entomopathogenic species are also known in this family [50], suggesting that there may be a parasitic relationship with beetles. However, it is also possible that this group enters scavengers’ guts with food resources such as dead carcasses, where they grew prolifically.
Our study revealed that the gut bacterial and fungal communities in ground-dwelling beetles were affected by the food habits and habitats of their hosts. Specifically, the abundant bacterial groups differed among food habits, and the community structure of one bacterial family, Enterobacteriaceae, was related with the beetle phylogeny. Our results indicate that some of the bacterial groups become abundant in beetle guts in relation to their food resources and gut environments (e.g. their gut morphology), which have evolutionarily differentiated among beetle lineages, suggesting symbiotic relationships between the bacteria and beetles in relation to food habit. One fungal group, Yarrowia species, was detected at high frequency from both silphid scavenger species and carnivorous beetles. This group may contribute to digestion and the inhibition of growth of other microbes in these beetles. Our results suggest that the symbiotic relationships between beetles and these microbes, including bacteria and fungi, are not limited to a few taxa such as silphid beetles, as reported previously [15, 16, 42], but are widespread in ground-dwelling beetles and perhaps other beetles as well. Beetle guts contain a diverse ecosystem of microbes [51], and there are also likely to be (as yet unknown) mutualistic or parasitic relationships. Further study is needed to elucidate the diverse symbiotic relationships between beetles and their microbes in more detail.
Electronic supplementary material
Acknowledgements
We thank the members of the Evolutionary Ecology Laboratory at Hirosaki University (S. Higuchi, S. Kosuda, H. Kusanagi, S. Ogasawara-Ohashi) for their help in collecting samples; Kôji Sasakawa for his help in identifying carabid beetles; and Akemi Yoshida for technical assistance. This study was partially supported by a Hirosaki University Grant for Exploratory Research by Young Scientists and Newly-appointed Scientists to HI and by a Japan Society for the Promotion of Science KAKENHI grant (15K18720) to RE. The doi for our data is 10.5061/dryad.mk0n0h4.
Author contributions
RK, HM, RE, and HI designed the study. RK conducted experiments. TK sequenced amplicons with MiSeq. RK, HM, and HI analyzed data. RK and HI wrote the paper. All authors discussed results and commented on the paper.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0298-3) contains supplementary material, which is available to authorized users.
References
- 1.Grimaldi D, Engel MS. Evolution of the insects. Cambridge: Cambridge University Press; 2005. [Google Scholar]
- 2.Joy JB. Symbiosis catalyses niche expansion and diversification. Proc R Soc Lond Ser B. 2013;280:20122820. doi: 10.1098/rspb.2012.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sudakaran S, Retz F, Kikuchi Y, Kost C, Kaltenpoth M. Evolutionary transition in symbiotic syndromes enabled diversification of phytophagous insects on an imbalanced diet. ISME J. 2015;9:2587–604. doi: 10.1038/ismej.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sudakaran S, Kost C, Kaltenpoth M. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol. 2017;25:375–90. doi: 10.1016/j.tim.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 5.Vega FE, Blackwell M. Insect-fungal associations: ecology and evolution. New York: Oxford University Press; 2005. [Google Scholar]
- 6.Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–51. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong S, Rawls JF. Intestinal microbiota composition in fishes is influenced by host ecology and environment. Mol Ecol. 2012;21:3100–2. doi: 10.1111/j.1365-294X.2012.05646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullam KE, Essinger SD, Lozupone CA, O’Connor MP, Rosen GL, Knight R, et al. Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Mol Ecol. 2012;21:3363–78. doi: 10.1111/j.1365-294X.2012.05552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lövei GL, Sunderland KD. Ecology and behavior of ground beetles (Coleoptera: Carabidae) Ann Rev Entomol. 1996;41:231–56. doi: 10.1146/annurev.en.41.010196.001311. [DOI] [PubMed] [Google Scholar]
- 10.Ikeda H, Homma K, Kubota K. Ground-dwelling beetle community in beech forests of Sado Island, Japan: comparison with the mainland. J Res. 2008;13:357–64. doi: 10.1007/s10310-008-0092-6. [DOI] [Google Scholar]
- 11.Ikeda H, Kubota K, Kagawa A, Sota T. Diverse diet compositions among harpaline ground beetle species revealed by mixing model analyses of stable isotope ratios. Ecol Entomol. 2010;35:307–16. doi: 10.1111/j.1365-2311.2010.01182.x. [DOI] [Google Scholar]
- 12.Scott MP. The ecology and behavior of burying beetles. Ann Rev Entomol. 1998;43:595–618. doi: 10.1146/annurev.ento.43.1.595. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda H, Kubota K, Kagaya T, Abe T. Flight capability and feeding habits of silphine beetles: are flightless species really “carrion beetles. Ecol Res. 2007;22:237–41. doi: 10.1007/s11284-006-0012-1. [DOI] [Google Scholar]
- 14.Coelho M, Amaral P, Belo I. Yarrowia lipolytica: an industrial workhorse. Cur Res, Tech Edu Top Appl Microbiol Microb Biotech. 2010;2:930–40. [Google Scholar]
- 15.Kaltenpoth M, Steiger S. Unearthing carrion beetles’ microbiome: characterization of bacterial and fungal hindgut communities across the Silphidae. Mol Ecol. 2014;23:1251–67. doi: 10.1111/mec.12469. [DOI] [PubMed] [Google Scholar]
- 16.Vogel H, Shukla SP, Engl T, Weiss B, Fischer R, Steiger S. The digestive and defensive basis of carcass utilization by the burying beetle and its microbiota. Nat Commun. 2017;8:15186. doi: 10.1038/ncomms15186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanahashi M, Kubota K, Matsushita N, Togashi K. Discovery of mycangia and the associated xylose-fermenting yeasts in stag beetles (Coleoptera: Lucanidae) Naturwissenschaften. 2010;97:311–7. doi: 10.1007/s00114-009-0643-5. [DOI] [PubMed] [Google Scholar]
- 18.Toki W, Takahashi Y, Togashi K. Fungal garden making inside bamboos by a non-social fungus-growing beetle. PLoS ONE. 2013;8:e79515. doi: 10.1371/journal.pone.0079515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueno S, Kurosawa Y, Satoh M. The Coleoptera of Japan in color. Osaka: Hoikusha; 1985. [Google Scholar]
- 20.Kurosawa Y, Hisamatsu S, Sasaji H. The Coleoptera of Japan in color. Osaka: Hoikusha; 1985. [Google Scholar]
- 21.Japanese Society of Environmental Entomology and Zoology. An illustrated guide to identify insects. Osaka: Bunkyo-Shuppan; 2013. [Google Scholar]
- 22.Kosuda S, Sasakawa K, Ikeda H. Directional mitochondrial introgression and character displacement due to reproductive interference in two closely related Pterostichus ground beetle species. J Evol Biol. 2016;29:1121–30. doi: 10.1111/jeb.12852. [DOI] [PubMed] [Google Scholar]
- 23.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–4. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGuire KL, Payne SG, Palmer MI, Gillikin CM, Keefe D, Kim SJ, et al. Digging the New York City Skyline: Soil fungal communities in green roofs and city parks. PLoS ONE. 2013;8:e58020. doi: 10.1371/journal.pone.0058020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microb. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, et al. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol. 2013;22:5271–7. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 30.Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria, 2017. http://www.R-project.org.
- 32.Kurtzman CP, Fell JW, Boekhout T, Robert V. Methods for isolation, phenotypic characterization and maintenance of yeasts. In: Kurtzman CP, Fell JW, Boekhout T (eds). The Yeasts, a Taxonomic Study. Chapter 7. Fifth edition. Elsevier: 2011; pp 87–110.
- 33.Endoh R, Suzuki M, Benno Y, Futai K. Candida kashinagacola sp. nov., C. pseudovanderkliftii sp. nov. and C. vanderkliftii sp. nov., three new yeasts from ambrosia beetle-associated sources. Antonie Van Leeuwenhoek. 2008;94:389–402. doi: 10.1007/s10482-008-9256-9. [DOI] [PubMed] [Google Scholar]
- 34.Tanabe AS Phylogears2 ver. 2.0. http://www.fifthdimension.jp/.
- 35.Katoh K, Standley DM. MAFFT Multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanabe AS. Kakusan4 and Aminosan: two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol Ecol Res. 2011;11:914–21. doi: 10.1111/j.1755-0998.2011.03021.x. [DOI] [PubMed] [Google Scholar]
- 37.Jobb G Treefinder version of March 2011. Available at http://www.treefinder.de.
- 38.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benjamini Yoav, Hochberg Yosef. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- 40.Tanabe AS, Toju H. Two new computational methods for universal DNA barcoding: A benchmark using barcode sequences of bacteria, archaea, animals, fungi, and land plants. PLoS ONE. 2013;8:e76910. doi: 10.1371/journal.pone.0076910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yahiro K. A comparative morphology of the alimentary canal in the adults of ground-beetles (Coleoptera). I. Classification into the types. ESAKIA. 1990;1:35–44. [Google Scholar]
- 42.Shukla SP, Vogel H, Heckel DG, Vilcinskas A, Kaltenpoth M. Burying beetles regulate the microbiome of carcasses and use it to transmit a core microbiota to their offspring. Mol Ecol. 2018;27:1980–91. doi: 10.1111/mec.14269. [DOI] [PubMed] [Google Scholar]
- 43.Donnenberg MS. Enterobacteriaceae. In: Mandell GL, Bennett JE, Dolin R, editors. Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. 7th edn. Philadelphia: Churchill Livingstone Elsevier; 2010. pp. 2815–34. [Google Scholar]
- 44.Breitsprecher E. Beiträge zur Kenntnis der Anobiidensymbiose.Z. Morphol Ökol Tiere. 1928;11:495–538. doi: 10.1007/BF02433460. [DOI] [Google Scholar]
- 45.Flórez LV, Scherlach K, Gaube P, Ross C, Sitte E, Hermes C, et al. Antibiotic-producing symbionts dynamically transition between plant pathogenicity and insect-defensive mutualism. Nat Commun. 2017;8:15172. doi: 10.1038/ncomms15172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Japan Society for Lactic Acid Bacteria. Science of lactic acid bacteria and bifidobacteria. Kyoto: Kyoto University Press; 2010. [Google Scholar]
- 47.Suzuki S, Nagano M, Trumbo ST. Intrasexual competition and mating behavior in Ptomascopus morio (Coleoptera: Silphidae: Nicrophorinae) J Ins Behav. 2005;18:233–42. doi: 10.1007/s10905-005-0477-8. [DOI] [Google Scholar]
- 48.McGee PA, Markovina AL, Jeong GCE, Cooper ED. Trichocomaceae in bark survive high temperatures and fire. FEMS Microbiol Ecol. 2006;56:365–71. doi: 10.1111/j.1574-6941.2006.00079.x. [DOI] [PubMed] [Google Scholar]
- 49.Houbraken J, Samson RA. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stu Mycol. 2011;70:1–51. doi: 10.3114/sim.2011.70.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang P, You Y, Song Y, Wang Y, Zhang L. First record of Aspergillus oryzae (Eurotiales: Trichocomaceae) as an entomopathogenic fungus of the locust, Locusta migratoria (Orthoptera: Acrididae) Biocon Sci Tech. 2015;25:1285–98. doi: 10.1080/09583157.2015.1049977. [DOI] [Google Scholar]
- 51.Suh SO, McHugh JV, Pollock DD, Blackwell M. The beetle gut: a hyperdiverse source of novel yeasts. Mycol Res. 2005;109:261–5. doi: 10.1017/S0953756205002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.