Abstract
Passive sinking of particulate organic matter (POM) is the main mechanism through which the biological pump transports surface primary production to the ocean interior. However, the contribution and variability of different biological sources to vertical export is not fully understood. Here, we use DNA metabarcoding of the 18S rRNA gene and particle interceptor traps (PITs) to characterize the taxonomic composition of particles sinking out of the photic layer in the California Current Ecosystem (CCE), a productive system with high export potential. The PITs included formalin-fixed and ‘live’ traps to investigate eukaryotic communities involved in the export and remineralization of sinking particles. Sequences affiliated with Radiolaria dominated the eukaryotic assemblage in fixed traps (90%), with Dinophyta and Metazoa making minor contributions. The prominence of Radiolaria decreased drastically in live traps, possibly due to selective consumption by copepods, heterotrophic nanoflagellates, and phaeodarians that were heavily enriched in these traps. These patterns were consistent across the water masses surveyed extending from the coast to offshore, despite major differences in productivity and trophic structure of the epipelagic plankton community. Our findings identify Radiolaria as major actors in export fluxes in the CCE.
Subject terms: Water microbiology, Microbial ecology
Introduction
The main mechanisms of the biological pump include the gravitational sinking of particles, the active transport associated with zooplankton, and the mixing and diffusive transport of dissolved and particulate organic matter (DOM and POM) [1]. Among them, particle sinking is the main process contributing to carbon export and is responsible for 5–21 PgC y−1 [2–4]. Phytoplankton community structure and food-web processes determine the fraction of net primary production exported as well as the size and chemical characteristics of sinking material [5–7]. The fraction of exported particles that reaches the ocean interior is further controlled by biotic (mainly microbial and zooplankton) transformations during their downward transit, which affects remineralization rates and particulate organic carbon (POC) flux attenuation with depth [8–10].
Sinking particles are composed of zooplankton fecal pellets [11], organic aggregates of the various source (‘marine snow’) including mucilaginous structures of larger plankton [12, 13] and intact phytoplankton cells [14–16]. Drifting particle interceptor traps (PITs) are the most common approach for quantifying vertical fluxes of particles [17]. However, the material collected in PITs is often partially degraded and heavily transformed by biological activity, which hampers its taxonomic identification based on morphological attributes and limits the ability to distinguish biological sources and export mechanisms. Moreover, export fluxes from sediment traps do not usually match estimated metabolic demands in the ocean twilight zone [10, 18, 19], suggesting that other organic fluxes besides those typically considered from phytoplankton and fecal pellets contribute significantly to vertical export.
Recently developed in situ imaging methods have proven useful for identifying larger particles [6, 20], but they often fall short in resolving the composition of smaller particles and complex aggregates. On the other hand, DNA sequencing analysis coupled with accurate reference databases offer a powerful alternative for extracting detailed taxonomic information from partially degraded material and complex communities. Amacher et al. [21] pioneered this approach in sediment traps, using clone libraries to quantify the relative contributions of protist groups to downward particle fluxes in the eastern subtropical North Atlantic. More recently, a metagenomic approach based on 454 pyrosequencing technology was used to investigate microbial communities associated with sinking particles in the oligotrophic North Pacific Subtropical Gyre (NPSG) [22]. However, analogous information for productive systems with high export potential is lacking.
The California Current Ecosystem (CCE) is a coastal upwelling biome characterized by high production and strong advective fields that transport high-nutrient, high-biomass coastal waters to oligotrophic offshore areas [23]. This gradient is reflected in phytoplankton composition and productivity [24], food-web interactions [25], and export fluxes [7, 26]. The present study aims at characterizing the composition of eukaryotic communities involved in the export and remineralization of sinking particles in the CCE. We capitalize on the capability of DNA metabarcoding of the 18S rRNA gene to retrieve taxonomic information from complex environmental samples such as those collected by PITs. Our specific objectives are to assess (1) the taxonomic compositions of POM sinking out of the euphotic zone and (2) the compositional changes associated with degradation and consumption processes transforming this POM below the euphotic zone. Towards these goals, we deployed formalin-fixed (i.e., fixed) and preservative-free (i.e., live) traps at the base of the euphotic zone, on the premise that microbial activity and degradation of POM would be inhibited in formalin-fixed traps [27] but allowed in live traps [28, 29].
Materials and methods
Study area and sampling strategy
Hydrographic and biological data were collected during the CCE-P1408 Process Cruise on the R/V Melville (6 Aug–4 Sept, 2014), as part of the CCE LTER (Long Term Ecosystem Research) program. A quasi-Lagrangian strategy was adopted to sample representative water parcels from coastal to offshore conditions over 3-days sampling periods called ‘Cycles’. We used a satellite-tracked drifting array with a drogue at 15 m to follow the water parcels and sampled the water column daily at the array for a suite of physical, chemical and biological measurements [25]. Cycles 1–3 were initiated in more productive coastal waters around Point Conception, while Cycles 4 and 5 represented typical oligotrophic offshore conditions (Fig. 1). Hydrographic data and water samples were acquired at 6–8 depths from a CTD-rosette system with 10-L Niskin bottles with Teflon-coated springs.
Fig. 1.
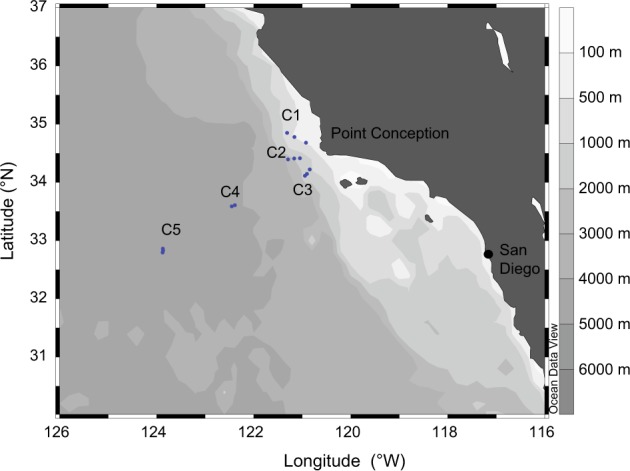
Map showing the location of the different water masses surveyed. Each water parcel was tracked for three consecutive days called Cycles (C1–C5). Blue dots represent the position of the water parcel during the predawn CTD carried out daily during the cycles
Sediment traps and export measurements
A second drifting array with VERTEX-style drifting sediment traps [17] was deployed during each cycle to collect sinking particles and assess export fluxes. The sediment trap consisted of arrays of 8–12 PITs in a cross-like layout attached to the wire at 2–3 depths below the euphotic zone (100 m, 150 m, and base of the euphotic zone if shallower than 100 m). The tubes were filled with a brine solution of 0.1 µm filtered seawater with 50 g L−1 NaCl and 80 mg L−1 of SrCl2 added to prevent mixing with in situ water and dissolution of acantharian skeletons [30], respectively. Most tubes were fixed with formaldehyde (0.4% final concentration, fixed traps), to minimize decomposition and consumption of organic matter [27], while selected tubes were not fixed (live traps) to allow these biotic processes to continue [28].
Upon recovery, the interface of brine and in situ water was visually determined in each tube and the upper layer removed gently by suction. For fixed traps, mesozooplankton swimmers were removed under a dissecting scope, before the sample was mixed, split and subsampled for particulate organic carbon (POC) and nitrogen (PON), chlorophyll a (Chl a), and phaeopigment analyses as described in [31]. Previous calibration of our PITs using 238U:234Th disequilibrium methods suggest that they are accurately collecting sinking particles [32]. For live traps, swimmers were not removed, and samples were directly split and filtered to minimize processing time and potential degradation. For DNA flux and diversity analysis, a known fraction of fixed- and live- traps was vacuum filtered in parallel through 0.8 and 8 µm Supor membrane filters (Pall Life Sciences, Port Washington, NY, USA) to obtain replicate independent samples for > 0.8 µm and > 8 µm particles. DNA fluxes from fixed traps were estimated by dividing the DNA concentration by an extraction efficiency factor of 0.153 [33].
Water column chemical and biological analysis
Water-column dissolved inorganic nutrients (DIN) and total and size-fractioned Chl a were obtained from http://oceaninformatics.ucsd.edu/datazoo/catalogs/ccelter/datasets, where sampling and analytical methods are described in detail. For DIN, seawater was filtered directly from the Niskin bottle using a Suporcap filter capsule (0.1 µm pore size) and major nutrient concentrations (NO3− + NO2−, NO2−, PO43−, NH4+, and SiOH3) analyzed by autoanalyzer using standard methods [34]. For total Chl a analysis, the samples were filtered onto 25 mm GF/F filters (Whatman, Maidstone, UK), and pigments were extracted in 90% acetone at −18 °C in the dark for 24 h and quantified on a calibrated 10 AU fluorometer (Turner Designs, Sunnyvale, CA, USA) using the acidification method. For size-fractioned Chl a, 0.10–0.25 L samples from the surface mixed layer were filtered through a series of filters with different pore sizes (20 µm Nitex mesh, 8 µm, 3 µm, and 1 µm Nucleopore and GF/F filters, Whatman, Maidstone, UK) and analyzed fluorometrically as above.
Water-column samples for DNA analysis were collected in two ways: (1) seawater was collected from the Niskin spigot into an acid-washed and Milli-Q rinsed polypropylene bottle, with a known volume (1.5–3 L) then filtered on a 0.8 µm Supor filter (i.e., called small-fraction). (2) We also emptied entire Niskin bottles into 10-L carboys by opening the bottom closure and using a large funnel to collect larger organisms [35]. These samples were then concentrated to 250 mL using a 5 µm mesh plankton net and subsequently vacuum-filtered onto 8 µm Supor filters (i.e. called large size-fraction).
Genomic DNA extraction, PCR amplification, and library preparation
DNA was extracted with the Nucleospin Plant kit (Macherey-Nagel, Düren, Germany), mini version for the water-column 0.8-µm filters and the midi version for 8.0-µm filters, and PIT samples (Supplementary Table S1). The V4 region of the 18S rRNA gene was amplified using the eukaryotic primers V4F_illum (5′-CCAGCASCYGCGGTAATTCC-3′) and V4R_illum (5′-ACTTTCGTTCTTGATYRATGA-3′) with Illumina overhang adapters (Forward 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-3′ and Reverse 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-3′) for attaching Nextera indexes [36]. PCR amplifications were done in triplicate for each sample. PCR products were visualized on 1.5% agarose gel and pooled together before purification with Agencourt AMPure XP purification system (Beckman Coulter, Brea, CA, USA). Randomly selected 20 purified amplicons were sized and validated using the Agilent High Sensitivity DNA Assay in the 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). For 6 distinct samples, the purified PCR product was split into three subsamples and also included in the library to investigate the reproducibility of the sequencing step. A second PCR step to attach index and Illumina adapters was conducted with the Nextera DNA library Preparation Kit (Illumina), followed by additional AMPure purification and library validation. The library was then quantified and prepared for 2 × 250 bp sequencing on a MiSeq platform. For Cycle 1, only fixed-trap samples were available for sequencing, while the > 0.8 µm live-trap samples from Cycle 3 were lost during processing. We sequenced a total of 54 samples, which included water-column samples from mixed-layer, deep chlorophyll maximum and below (n = 22), live-trap samples from 60, 100, and 150 m (n = 21), and fixed-trap samples from 100 m (n = 11) (Supplementary Table S1). Raw sequence have been deposited to GenBank under Bioproject number PRJNA432581.
Processing and taxonomic assignation of sequencing reads
Fastq files were checked using FastQC on the Galaxy platform [37] for sequence length (L) and Quality Score (Q). Forward reads with L >200 and Q >20 over 75% of the sequence were retained. For reverse reads, the first 85 bases were of very bad quality. These 85 bases were removed and sequences with L > 150 and Q >20 in 50% of the sequence were retained. Unpaired sequences were removed. All further processing was performed using mothur v.1.33.0 [38]. First, contigs were assembled from forward and reverse reads, keeping only contigs free of ambiguity. Singletons were removed and sequences were aligned to 18S rRNA from the Silva reference database (https://mothur.org/wiki/Silva_reference_files). Sequences with two nucleotide differences were pre-clustered, and chimeras were removed using UCHIME [39] as implemented in mothur. Only pre-clusters with > 10 sequences were retained. Pre-clustered sequences were taxonomically annotated using classify.seqs against the PR2 reference database [40] version 4.4 available from https://github.com/pr2database/pr2database/releases. Sequences were clustered using the average neighbour algorithm to determine OTUs at 97% similarity level. OTUs were taxonomically assigned using classify.otu and BLASTed against GenBank to confirm the assignation provided by mothur and to get their percentage of similarity to existing sequences. Since the community composition for triplicated amplicons was not substantially different (Supplementary Figure S1), sequences from replicated amplicons were pooled for subsequent analysis. Similarly, sequences from water-column and live-trap samples obtained at different depths were combined for downstream analysis. The OTU abundance table and OTU sequences are available as supplementary material on Figshare (10.6084/m9.figshare.5944291).
Statistics and diversity analysis of sequence data
Data analyses and statistics were done using R version 3.2.4 (R Core Team, 2016) with the vegan 2.4-3 [41], ggplot2 [42], treemap [43] packages, GraphPad 5.0 (GraphPad Software, La Jolla, CA, USA) and the Paleontological Statistics software [44]. OTU richness was estimated with rarefy function (vegan package) on a random subsample of size determined by the minimum number of sequences found among the samples compared (e.g., fixed vs. live traps).
Results
Water-column physical, chemical, and biological conditions
During the sampling cycles (C1 to C5), we explored 5 water parcels (Fig. 1) with physical and chemical properties that reflected primarily their coastal vs. offshore characteristics (Table 1, Supplementary Figure S2a). A hierarchical dendrogram based on both physico-chemical and biological properties clusters coastal C1 and C2 together, separated from offshore C4 and C5, with intermediate conditions for C3 (Supplementary Figure S2b). C1 and C2 exhibited colder and saltier surface waters and shallower nitracline depth indicative of coastal upwelling (Table 1). Surface Chl a concentration was also higher with large phytoplankton cells (> 20 μm) accounting for ~40% of Chl a for C1 and C2, while the contribution of large cells was minor (~2%) for C4 and C5 (Table 1, Supplementary Figure S2c).
Table 1.
Water column physical, chemical and biological properties averaged from 7 CTD casts conducted throughout the 3-days duration of each Cycle
| C1 | C2 | C3 | C4 | C5 | |
|---|---|---|---|---|---|
| Surface Salinity | 33.47 ± 0.0 | 33.42 ± 0.0 | 33.33 ± 0.0 | 33.02 ± 0.0 | 33.09 ± 0.0 |
| Surface Temperature (°C) | 16.5 ± 0.0 | 16.9 ± 0.0 | 18.6 ± 0.1 | 19.2 ± 0.0 | 19.8 ± 0.0 |
| Mixed layer depth (m) | 17.6 ± 4.3 | 29.3 ± 5.7 | 17.3 ± 5.9 | 24.3 ± 4.8 | 28.6 ± 4.4 |
| Nitracline depth (m) | 30.9 ± 5.8 | 40.4 ± 14.0 | 33.1 ± 6.0 | 70.6 ± 2.1 | 91.6 ± 5.7 |
| Surface Chl a (µg/L) | 0.59 ± 0.15 | 0.69 ± 0.08 | 0.21 ± 0.03 | 0.10 ± 0.02 | 0.08 ± 0.01 |
| Surface Chl a > 20 µm (%) | 42.6 ± 5.0 | 39.0 ± 5.7 | 3.4 ± 1.2 | 1.9 ± 0.3 | 1.8 ± 0.1 |
POC and PON fluxes were higher at coastal cycles and decreased offshore (Fig. 2). DNA fluxes measured in fixed traps at 100 m followed the same trend (Fig. 2a) and were significantly correlated with POC fluxes (r = 0.88, p < 0.05, n = 5). The Chl a to phaeopigment ratio of sediment trap material indicated that pigment flux during all cycles were dominated by fecal-derived material (Fig. 2b). The higher values observed at the coastal C1 and C2 cycles, however, indicated a higher contribution of ‘fresh’ algae compared to offshore C4 and C5 (Fig. 2b).
Fig. 2.
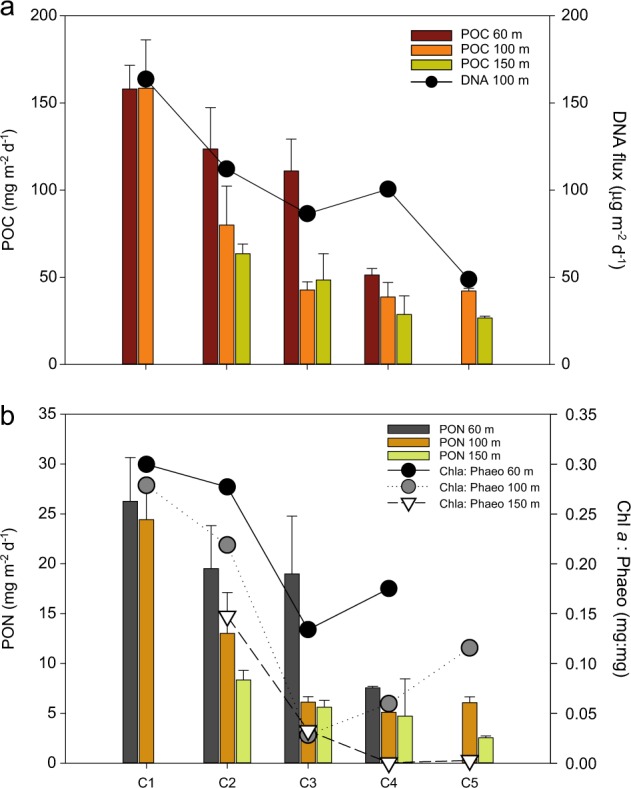
a POC and DNA fluxes in standard fixed-sediment traps. b PON fluxes and Chlorophyll to Phaeopigments ratio in the traps. Error bars represent the standard deviation of POC and PON fluxes (n = 3) in traps deployed at 60, 100, and 150 m
18S V4 OTU distribution and diversity patterns
Over 18 million paired reads were obtained from the sequencing run, half of which were removed after filtering based on quality and length (Supplementary Table S2). After processing, we obtained 2662 OTUs (97% similarity) corresponding to 2,802,466 sequences (Supplementary Table S2).
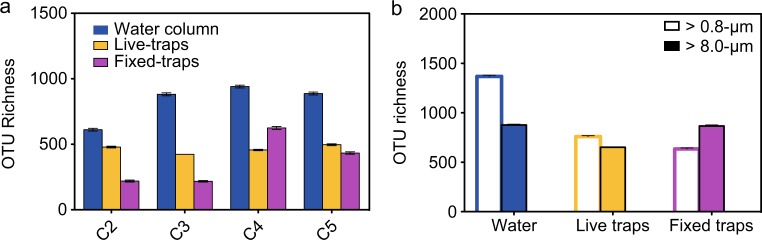
For protists, rarefaction curves for different stations were not saturated for all samples, although the analysis of pooled sequences from different sample types suggest an adequate recovery of epipelagic protist diversity for cycles C2–C5 (Supplementary Figure S3a). Cycle C1 was excluded from this analysis because only fixed-trap samples were available. Protistan OTU richness increased from coastal to oceanic locations, being higher in water-column compared to fixed- and live-trap samples for all cycles (Fig. 3a). More protistan OTUs were recovered in live compared to fixed traps for C2 and C3, while fixed traps had higher or similar OTU numbers compared to live traps for C4 and C5 (Fig. 3a). OTU richness for water-column protists was higher in the smaller compared to the larger size-fraction samples (Fig. 3b). This difference decreased in sediment traps, particularly in fixed traps, where protistan richness was similar or higher in the larger fraction (Fig. 3b). Similar diversity patterns were observed for all eukaryotic OTUs, including metazoans (Supplementary Figure S3b).
Fig. 3.
Protistan OTU richness calculated for the minimum sampling effort common to all samples compared. a Water column, live- and fixed-sediment trap samples comparison across different cycles. b Small and large size fraction comparison in different sample types. Colors refer to samples collected from the water column (blue), live (yellow), and fixed traps (purple)
Non-metric multidimensional scaling (NMDS) analysis, based on protistan OTUs abundance and Bray–Curtis distances, ordinated samples into three main clusters corresponding to the different sampling methods (Fig. 4). Compositional changes in water-column samples were significantly correlated with surface temperature, salinity, nitracline depth, and Chl a, with size fraction (NMDS1, r2 = 0.61, P = 0.001) and cycle (NMDS2, r2= 0.30, P = 0.02) being the primary ordination factors (Supplementary Figure S4). Sampling cycle emerged as the primary ordination factor for the correlations of trap samples with physico-chemical and biogeochemical variables (Supplementary Figure S4).
Fig. 4.
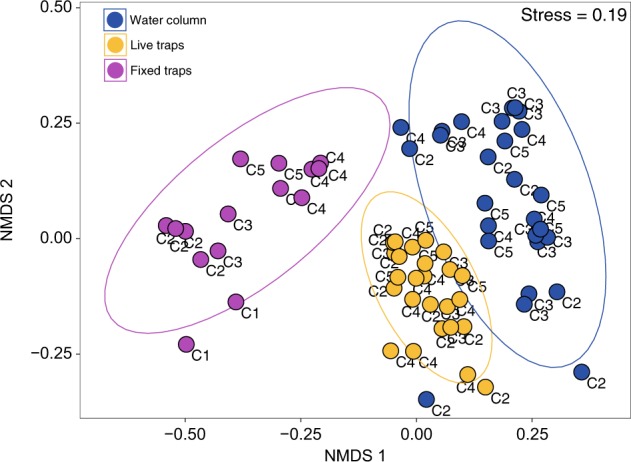
Non-parametric multidimensional scaling ordination (nMDS) plot in 2-dimension configuration (K = 2) based on Bray–curtis dissimilarity between protist OTU community composition of all samples. 95% confidence ellipses for each method type are represented
Taxonomic composition of the water-column eukaryotic community
OTU taxonomy was assigned based on the curated 18 S rRNA database PR2 [40] which uses 8 different taxonomical levels from Kingdom to Species. Protistan sequences dominated the community across all cycles and size fractions (58 ± 27% of eukaryotic sequences), with a substantial contribution of metazoan sequences (39 ± 26%, Supplementary Figure S5).
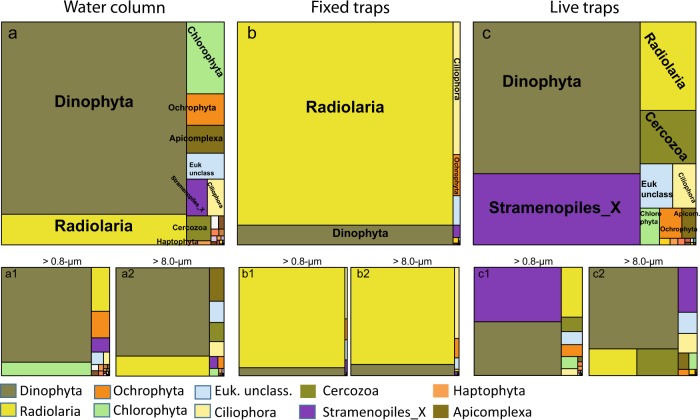
Among protists, Dinophyta was the most abundant group (72 ± 13% of protistan sequences) followed by Radiolaria (12 ± 13%) and Chlorophyta (4.7 ± 7.1%) (Fig. 5). Ochrophyta (2.1 ± 2.0%) contributed less on average but represented a substantial percentage of the protistan community at times (Figs. 5 and 6). Dinophyta included sequences affiliated with Syndiniales and Dinophyceae in similar relative abundances (~20%, Fig. 6), although their distributional pattern and partitioning between size fractions differed markedly (Fig. 7). In addition to Ptychodiscus noctiluca, uncultured Dinophyceae and Syndiniales OTUs were among the most abundant in water-column samples (Supplementary Table S3 and Figure S6).
Fig. 5.
Mean percentage of 18S rDNA reads affiliated to protistan divisions and major taxonomic groups in samples from the water column (a), fixed (b) and live traps (c). Total (>0.8 μm) and large (>8 μm) size-fractions are presented at the bottom left (1) and right (2) of each sample type, respectively
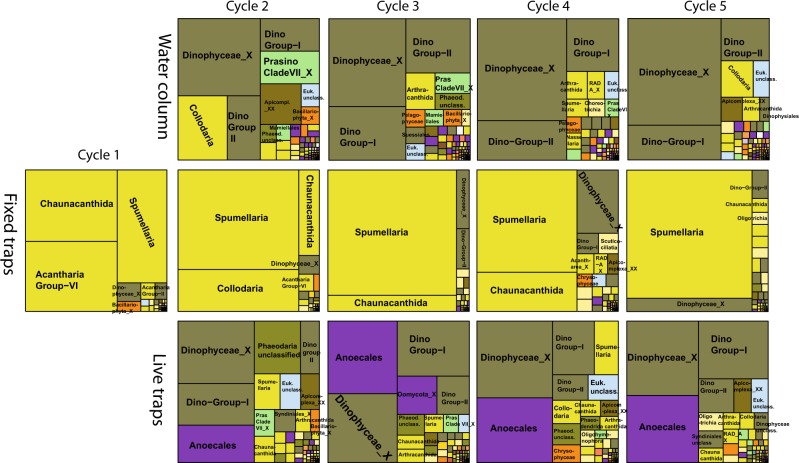
Fig. 6.
Mean percentage of 18S rDNA reads affiliated to protistan taxonomic groups (classes and orders mainly) in water-column, fixed- and live-trap samples across different sampling cycles. The area represents the mean percentage of reads affiliated to each protistan group (only groups with >1% mean contribution are labeled). Color codes, as in Fig. 5, represent the taxonomic affiliation, mainly at the division level
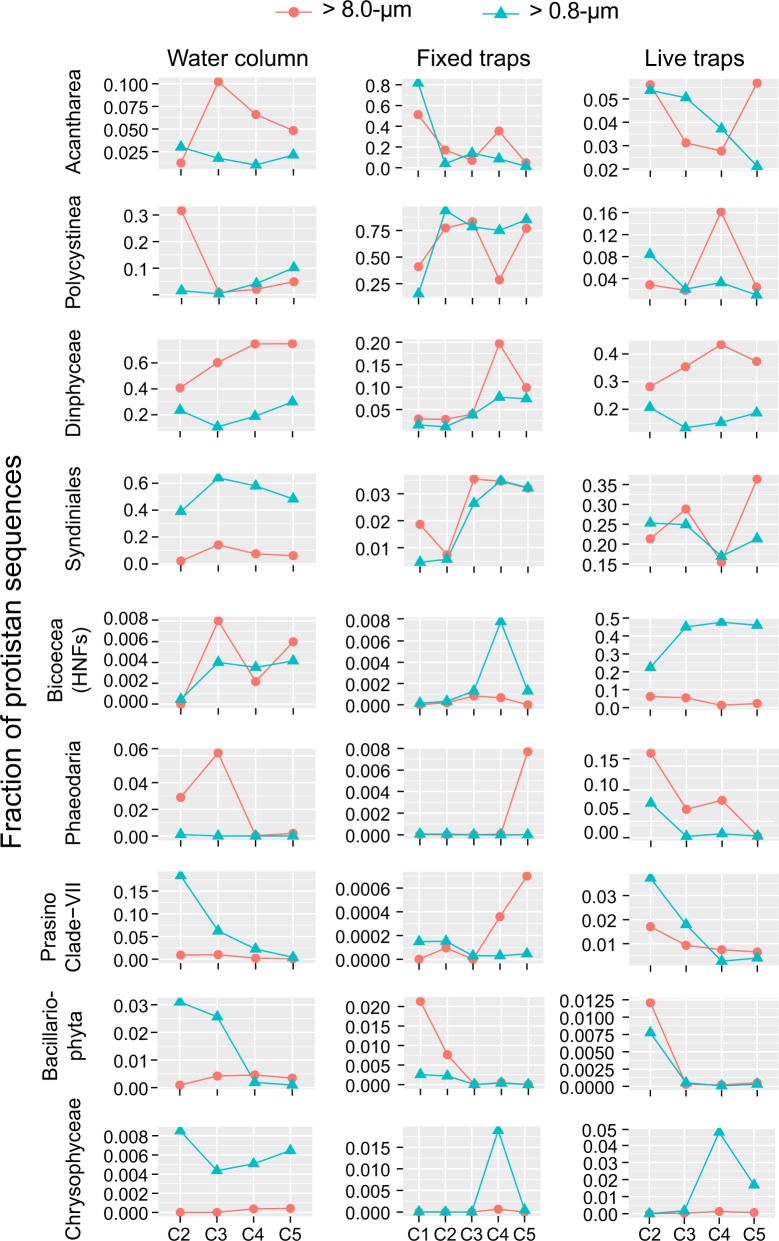
Fig. 7.
Mean relative abundance of 18S rRNA sequences of most abundant protistan groups in the total (> 0.8 μm) and large (> 8 μm) size-fraction samples from the upper water column (0–100 m), fixed and livetraps, across different cycles
Most radiolarian sequences were affiliated with Collodaria and Acantharia, although groups such as Spumellaria and RAD-A were occasionally important (Fig. 8). Chlorophyta (green algae) were mostly picoplanktonic belonging to Chloropicophyceae (Chloroparvula, previously prasinophytes clade VII-B1, [45]) and Mamiellophyceae (Bathycoccus) (Fig. 6). Both picoplanktonic groups peaked in coastal waters (e.g., Chloropicophyceae represented ca. 20% of protistan sequences in C2), and decreased offshore (Figs. 6 and 7). Ochrophyta (photosynthetic stramenopiles) sequences were also more abundant in the smaller size fraction (Fig. 5) and comprised mainly of sequences assigned to pelagophytes and diatoms (Fig. 6), with the latter reaching ~5-fold higher relative abundance toward the coast (Fig. 7). The contribution of Phaeodaria, a rhizarian group related to Radiolaria, also peaked in coastal waters, but in the larger size fraction (Fig. 7).
Fig. 8.
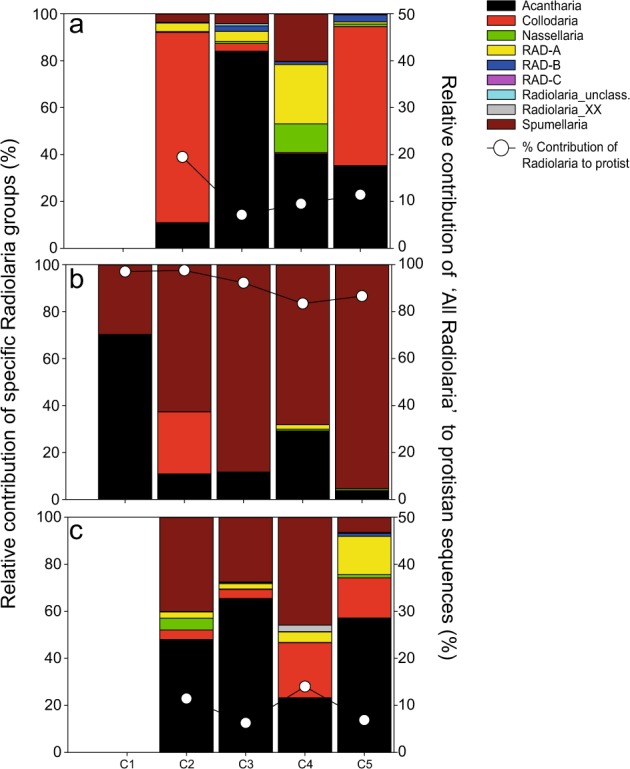
Relative contribution of Radiolaria to total protistan sequences (solid points) and percentage contribution of specific radiolarian groups in the water column (a), fixed (b) and live (c) trap samples across different cycles
Taxonomic composition of eukaryotic community in fixed traps
Fixed-trap samples were overwhelmingly dominated by protistan sequences, mainly Radiolaria (88 ± 8.6% of eukaryotic sequences) and Dinophyta (8.6 ± 6.3%) (Fig. 5 and Supplementary Figure S5). Metazoan sequences accounted for 6.1 ± 5.0% and were dominated by Crustacea, with Mollusca and Cnidaria contributing less on average (Supplementary Figure S5).
The dominance of Radiolaria was a consistent feature across cycles (Fig. 6), with contributions from a diverse suite of OTUs from the four major radiolarian groups (Acantharia, Collodaria, Nassellaria, and Spumellaria) (Fig. 8, Supplementary Table S3). Overall, Spumellaria was the most abundant group (60 ± 22% of radiolarian sequences) followed by Acantharia (22 ± 28%) and Collodaria (4.8 ± 11%) (Fig. 8). Among Dinophyta, Dinophyceae (true dinoflagellates) were relatively more abundant than Syndiniales, and their contribution increased in offshore cycles (Fig. 7) with uncultured Dinophyceae and P. noctiluca being the most abundant OTUs (Supplementary Table S3 and Figure S6). Diatoms comprised most Ochrophyta sequences in fixed traps from cycles C1 and C2 (Fig. 6), belonging mainly to the same genera (Pseudo-nitzschia and Chaetoceros) as those in the water column (Supplementary Figure S6). They virtually disappeared offshore, where sequences affiliated with the heterotrophic nanoflagellate (HNF) Paraphysomonas imperforata (Chrysophyceae) became the dominant Ochrophyta group (Fig. 6).
Taxonomic composition of eukaryotic community in live traps
Metazoan sequences dominated live-trap samples (78 ± 18% of eukaryotic sequences, Supplementary Figure S5). Among them, Crustaceans belonging to different copepod genera, notably Metridia spp., contributed most to the eukaryotic community followed by Cnidaria and Mollusca (Supplementary Table S3).
Among protists, Dinophyta (51 ± 5.7% of protistan sequences) and Stramenopiles_X (heterotrophic stramenopiles, 24 ± 7.2%) were the dominant groups, with Radiolaria (9.6 ± 3.7%) and Phaeodaria (5.4 ± 5.5%) contributing less (Figs. 5 and 6). In addition to the dinoflagellate P. noctiluca, the HNFs Caecitellus parvus and C. paraparvulus (order Anoecales) were the most abundant protistan species in live traps (Supplementary Table S3 and Figure S6), accounting for the high proportion of Stramenopiles_X. The abundance of these HNFs decreased significantly in the > 8 μm fraction, suggesting that they are free-living or feeding on particles and aggregates to which they are loosely attached (Figs 6 and 7). Diatoms and chrysophytes recovered from live traps showed the same spatial and size-fraction distribution as in fixed traps, although the contribution of chrysophytes in offshore cycles was higher in live traps (Fig. 7). Most radiolarian sequences recovered from live traps belonged to Acantharia and Spumellaria (Fig. 8). RAD-A and Collodaria were detected in all cycles but showed higher contribution in offshore cycles C4 and C5 (Fig. 8). Phaeodaria, mostly affiliated with Aulacantha spp. (Supplementary Table S3), were more abundant in coastal cycles C1 and C2, where they represented up to 20% of protistan sequences (Fig. 7).
Discussion
Overall, the protistan communities in water-column and trap samples were markedly dissimilar (Figs. 5 and 6), showing higher similarity in sinking material across cycles than between traps and the overlying euphotic zone (Fig. 4). Such pattern has been observed in previous molecular-based studies [22, 33] and suggests either that a common suite of organisms is responsible for export despite dramatic differences in ambient microbial communities or that these organisms are resistant to degradation of the genetic material. Radiolaria were identified as the most abundant group in POM sinking out of the euphotic zone, in addition to phytoplankton and metazoan zooplankton, usually considered as having a major role in vertical export in the CCE (Figs. 5 and 6). The observed dominance of radiolarian sequences in fixed traps (Figs. 5 and 6) is also consistent with clone library analysis of trap samples from the eastern subtropical North Atlantic, where radiolarian clones were most abundant [21], and water column samples from the Sargasso sea, where high proportion of radiolarian clones were obtained below the euphotic zone [46]. In contrast to these observations, the relative abundance of radiolarians determined by 18S rRNA metabarcoding in the western Antarctic Peninsula was negatively linked to community export potential [47], suggesting that the role of this group in vertical export may differ across systems.
Metabarcoding of 18S rRNA genes is largely used for assessing the composition of aquatic microbial communities [36, 48–50] and the spatio-temporal patterns of specific taxonomic groups [51–53]. Yet, a number of well-identified limitations and potential biases are acknowledged with respect to absolute quantification of plankton groups. Along with PCR biases, the presence of multiple copies of 18S rRNA genes and its variation across taxa [54] affects the quantitative interpretation of community compositional changes from read abundance data. In this regard, the prevalence and high relative contributions of Dinophyceae and Syndiniales in this study (Figs. 6 and 7) and other meta-genetic surveys [21, 36, 48, 55, 56] would be partially due to the high DNA content and number of gene copies in these groups. Similarly, the multi-nuclear nature of Radiolaria [57– 59] and the high gene copy number observed in Collodaria [60] could explain their high relative abundances in meta-genetic surveys [48, 60, 61].
Nonetheless, a positive relationship between 18S rRNA copy number and cell length has been reported across protists spanning orders of magnitude in cell size [48, 54, 60, 62], which encourages the cautious use of read abundances to infer community composition general patterns and dynamics on a quasi-biomass basis (i.e., the larger organisms have proportionately more DNA reads). Further support for an ecological, rather than bias, interpretation of compositional changes between water-column and trap samples (Figs. 4 and 5) and group-specific relative-abundance spatial patterns (Figs. 6 and 7) comes from a recent study that reported good agreement between relative picoeukaryotic cell and environmental sequence abundances [63]. The community composition obtained by parallel sequencing of replicated PCR products was virtually identical (Fig. S1), suggesting that potential errors linked to sequencing were not responsible for the differences observed among samples. Moreover, the comparative approach adopted here provides a robust framework for interpreting diversity and compositional changes between fixed and live traps from an ecological and biogeochemical perspective.
The prevalence of radiolarian sequences in sinking POM observed in this study (Fig. 6) is remarkable, considering the major differences in productivity and trophic structure of the epipelagic communities along the CCE environmental gradient (Table 1 and Supplementary Figure S2). Analysis of our preserved samples under epifluorescence microscopy (data not shown) confirmed the presence of Radiolaria, although the dissolution of hard structures observed for this, and other taxonomic groups (e.g., diatoms), precluded reliable estimates of biomass and finer taxonomic assignment. Analysis of DNA sequences, however, shows that different radiolarian groups (Fig. 8) and OTUs (Supplementary Table S3 and Figure S6) contribute broadly to this result, highlighting the functional diversity of the group. Previous microscopical and geochemical analyses of sediment trap material have stressed the importance of major radiolarian groups to export [35, 64–67]. The enrichment of Radiolaria in sediment traps may reflect both the high contribution of this group to particle export and the inadequacy of CTD and net tow sampling to capture these fragile and patchy amoeboid organisms [35, 68, 69].
Several characteristics of radiolarian cell structure and ecology are consistent with their high export potential. The silica or strontium sulfate skeletons of most Polycystines (Collodaria, Nasellaria, and Spumellaria) and Acantharia, respectively, provide substantial mineral ballast [67, 69]. In addition, their amoeboid nature and sticky pseudopodia can catalyze the formation of aggregates with high sinking velocities [70]. In contrast to recent genomic studies pointing to Acantharia and Collodaria as the key ‘export’ taxa in tropical and subtropical oceans [22, 71], we found that Spumellaria are the most important export contributors in the CCE (Fig. 8). Gowing [72] also showed Spumellaria to be the dominant radiolarian group in microscopical analyses of sediment trap material from the oligotrophic NPSG VERTEX station. In addition to Acantharia, which represented >50% of radiolarian sequences in C1 and C4, we also detected a substantial contribution by Collodaria in C2 (Figs. 6 and 8). Considering the patchy distributions and large size (μm-to-cm) of single-celled and colonial Collodaria, it is difficult to assess their contributions to export accurately, based only on discrete sediment trap analysis [35]. However, the high relative abundance of Collodaria in the CCE region inferred from molecular (Figs. 6 and 8) and in situ image analysis (Underwater Vision Profiler 5, UVP5) during this and previous cruises [73, 74] supports their important role in export flux.
Dinoflagellates were the most abundant non-radiolarian protists in both water-column and fixed-trap samples from cycles C1 and C2 in this region (Figs. 6 and 7). The warm anomaly that developed in the NE Pacific during the 2013–2014 winter [75], colloquially referred to as ‘the blob’, had already hit the California coast at the time of our cruise [76] and was responsible for the weak summer upwelling that we encountered. Off the Oregon coast, changes in plankton community composition associated with ‘the blob’ included higher dinoflagellates abundances and penetration of open-ocean copepod species onto the continental shelf region [77]. Increased abundance of autotrophic dinoflagellates has also been observed in the Point Conception region during years of delayed upwelling [24]. Consistent with this, our molecular survey revealed high sequence abundance of P. noctiluca (Supplementary Table S3 and Figure S6), a dinoflagellate species with distinctive cell covering characteristics and widespread distribution [78] that is generally less common in the CCE. Among green algae, the dominance of prasinophytes clade VII-B1 (Chloropicophyceae), a group typical of offshore waters [45], over Mamiellophyceae, characteristics of coastal waters, is another indication of the oceanic characteristics of the epipelagic zone. Collodaria is typically associated to warm oligotrophic waters [68, 74], and the observed abundance in mesotrophic coastal waters of C2 (Figs. 6 and 8) could have been favored by the warm anomaly. It therefore seems likely that the relatively low contribution of diatoms to the water-column assemblage and export fluxes was a consequence of anomalous conditions. Whether the high contribution of Radiolaria indicated by our molecular analyses was enhanced by these 2014 conditions cannot be directly addressed without comparable data from years with ‘normal’ conditions, although several lines of evidence argue against this idea.
On one hand, Radiolaria have been shown to dominate clone libraries recovered from sediment traps despite the dominance of diatoms in the upper water column in the eastern subtropical North Atlantic [21]. On the other hand, although higher fluxes of polycystine radiolarians (living + empty skeletons) have been found in coastal upwelling compared to oligotrophic offshores waters of the CCE, a clear relationship between primary production and these fluxes could not be established [66]. In the subarctic Pacific, however, a positive relationship between polycystine radiolarian fluxes and primary production was reported at station PAPA [70], suggesting differences in the export role of these group among systems. In our 2014 CCE study, microscopical counts of large phaeodarians from fixed traps and in UVP5 profiles, yielded lower abundances compared to previous and following year estimates for this group of rhizarians [79, 80]. Nonetheless, our study showed that phaeodarians represented a significant fraction of the protistan sequences in live traps (Figs. 5 and 7). Altogether, these findings argue against the idea that Radiolaria dominance was due to anomalous conditions and support the key role of rhizarians in export fluxes as a general feature of the CCE, and potentially other eastern boundary upwelling systems.
One striking result was the markedly lower contribution of Radiolaria taxa in live traps (Figs. 5 and 6), indicating rapid remineralization of organic matter associated with this group. Sinking particles and aggregates serve as natural ‘hot spots’ for both microbial and metazoan activity [28, 81]. Live traps were highly enriched with sequences from larger copepods like Eucalanus and Metridia spp. (Supplementary Table S3 and Figure S6), consistent with the potential of copepods to consume and transform sinking organic particles [9, 82, 83].
Rather than ingesting sinking particles directly, detritivorous zooplankton may benefit from the microbial growth enhanced by particle fragmentation [84]. Such “microbial gardening” would be consistent with the dramatic increase of opportunistic HNFs such as Caecitellus spp. and P. imperforata (Supplementary Table S3 and Supplementary Figure S6), which dramatically increase in enrichment cultures [85]. Heterotrophic protists are known to thrive around marine snow particles [12, 86, 87], but whether this microbial boost is directly exploited by larger metazooplankton or consumed first by small microzooplankton is unclear. The increase of ciliates in both fixed (1.8% of protistan sequences) and live traps (2.2%) relative to the water column (0.66 %) in this and previous studies [21], indicates that they likely prey on HNF, which function as a trophic link in these rich microenvironments. Further evidence of heterotrophic protistan activity associated with the degradation of sinking organic material is indicated by the enrichment of Phaeodaria in live compared to fixed traps (Figs. 5 and 7). This amoeboid group has been shown to feed on HNF and eukaryotic algae and proposed to consume marine snow particles [72, 88]. Data from the UVP5 have shown large (> 600 µm) species of phaeodarian forming a high-density layer below the euphotic zone [73], although their abundances were notably lower during this cruise compared to previous ones [79, 80]. The high relative abundance of Phaeodaria in live traps (Fig. 5), together with their trophic biology and vertical distribution in the CCE, further supports the important role that this group may have in the cycling of particulate organic matter sinking below the euphotic zone.
Conclusion
While phytoplankton and crustacean zooplankton are generally viewed as the major biological sources of vertical export in productive systems, our results, showing the abundance and diversity of rhizarian sequences in particulate material sinking below the euphotic zone, indicate an important role for this group in the export and cycling of particulate organic matter in the CCE. Recent studies using advanced molecular and imaging technologies have revealed unprecedented global abundances of Rhizaria [74, 89, 90], but the functions and impacts of this group in pelagic ecosystems have yet to be accurately quantified. The high abundance and diversity of Radiolaria reported here, and their prevalence from coastal upwelling to oceanic oligotrophic conditions, stress the need to better characterize the group’s functional diversity to improve understanding of biological controls on vertical fluxes.
Supplementary information
Acknowledgements
We would like to acknowledge Estelle Bigeard for her help during the DNA extraction and Illumina library preparation, Ralf Goericke for the Chl a and nutrient data, and the crew and research technicians of RV Melville for their assistance during the CCE-P1408 Process Cruise. CCE-LTER ship and science support were provided by NSF grants OCE-1026607 and OCE-1637632. We are grateful to the Genomer and ABIMS platforms for access to the Illumina sequencer and bioinformatic cluster, Brittany region SAD grant SYMBIOX for funding AGR research and Sorbonne Universités Emergence grant to FN, NIWA for its support to AGR during data analysis and writing of the manuscript via New Zealand Strategic Science Investment Funding to the National Coasts & Oceans Centre, and Scott Nodder (NIWA) for support and fruitful discussions. Funds for ALS was provided by FONDECYT grant PiSCO South (N1171802).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0322-7) contains supplementary material, which is available to authorized users.
References
- 1.Turner JT. Zooplankton fecal pellets, marine snow, phytodetritus and the ocean’s biological pump. Prog Oceanogr. 2015;130:205–48. [Google Scholar]
- 2.Eppley RW, Peterson BJ. Particulate organic -matter flux and planktonic new production in the deep ocean. Nature. 1979;282:677–80. [Google Scholar]
- 3.Henson SA, Sanders R, Madsen E, Morris PJ, Le Moigne F, Quartly GD. A reduced estimate of the strength of the ocean’s biological carbon pump. Geophys Res Lett. 2011;38:1–5. [Google Scholar]
- 4.Laws EA, Falkowski PG, Smith WO, Ducklow H, Mccarthyl JJ. Temperature effects on export production in the open ocean. Glob Biogeochem Cycles. 2000;14:1231–46. [Google Scholar]
- 5.Boyd PW, Newton PP. Does planktonic community structure determine downward particulate organic carbon flux in different oceanic provinces? Deep Sea Res Part I-Oceanogr Res Pap. 1999;46:63–91. [Google Scholar]
- 6.Guidi L, Stemmann L, Jackson GA. Effects of phytoplankton community on production,size and export of large aggregates: A world-ocean analysis. Limnol Oceanogr. 2009;54:1951–63. [Google Scholar]
- 7.Stukel MR, Landry MR, Benitez-Nelson CR, Goericke R. Trophic cycling and carbon export relationships in the California Current Ecosystem. Limnol Oceanogr. 2011;56:1866–78. [Google Scholar]
- 8.Giering SLC, Sanders R, Lampitt RS, Anderson TR, Tamburini C, Boutrif M, et al. Reconciliation of the carbon budget in the ocean’s twilight zone. Nature. 2014;507:480–517. doi: 10.1038/nature13123. [DOI] [PubMed] [Google Scholar]
- 9.Lampitt RS, Noji T, Vonbodungen B. What happens to zooplankton fecal pellets - implications for material flux. Mar Biol. 1990;104:15–23. [Google Scholar]
- 10.Steinberg DK, Van Mooy BAS, Buesseler KO, Boyd PW, Kobari T, Karl DM. Bacterial vs. zooplankton control of sinking particle flux in the ocean’s twilight zone. Limnol Oceanogr. 2008;53:1327–38. [Google Scholar]
- 11.Steinberg DK, Landry MR. Zooplankton and the Ocean Carbon Cycle. Ann Rev Mar Sci. 2017;9:413–44. doi: 10.1146/annurev-marine-010814-015924. [DOI] [PubMed] [Google Scholar]
- 12.Caron DA, Davis PG, Madin LP, Sieburth JM. Enrichment of microbial- populations in macroaggregates (marine snow) from surface waters of the North- Atlantic. J Mar Res. 1986;44:543–65. [Google Scholar]
- 13.Alldredge AL, Silver MW. Characteristics, dynamics and significance of marine snow. Prog Oceanogr. 1988;20:41–82. [Google Scholar]
- 14.Agusti S, González-Gordillo JI, Vaqué D, Estrada M, Cerezo MI, Salazar G, et al. Ubiquitous healthy diatoms in the deep sea confirm deep carbon injection by the biological pump. Nat Commun. 2015;6:7608. doi: 10.1038/ncomms8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin P, Lampitt RS, Perry MJ, Sanders R, Lee C, D’Asaro E. Export and mesopelagic particle flux during a North Atlantic spring diatom bloom. Deep Sea Res Part I-Oceanogr Res Pap. 2011;58:338–49. [Google Scholar]
- 16.Smetacek V, Klaas C, Strass VH, Assmy P, Montresor M, Cisewski B, et al. Deep carbon export from a Southern Ocean iron-fertilized diatom bloom. Nature. 2012;487:313–9. doi: 10.1038/nature11229. [DOI] [PubMed] [Google Scholar]
- 17.Knauer GA, Martin JH, Bruland KW. Fluxes of particulate carbon, nitrogen, and phosphorus in the upper water column of the Northeast Pacific. Deep Sea Res. 1979;26:97–108. [Google Scholar]
- 18.Burd AB, Hansell DA, Steinberg DK, Anderson TR, Aristegui J, Baltar F, et al. Assessing the apparent imbalance between geochemical and biochemical indicators of meso- and bathypelagic biological activity: What the @$#! is wrong with present calculations of carbon budgets? Deep Sea Res Part II-Top Stud Oceanogr. 2010;57:1557. [Google Scholar]
- 19.Herndl GJ, Reinthaler T. Microbial control of the dark end of the biological pump. Nat Geosci. 2013;6:718–24. doi: 10.1038/ngeo1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bochdansky AB, Jericho MH, Herndl GJ. Development and deployment of a point-source digital inline holographic microscope for the study of plankton and particles to a depth of 6000 m. Limnol Oceanogr. 2013;11:28–40. [Google Scholar]
- 21.Amacher J, Neuer S, Anderson I, Massana R. Molecular approach to determine contributions of the protist community to particle flux. Deep Sea Res Part I Oceanogr Res Pap. 2009;56:2206–15. [Google Scholar]
- 22.Fontanez KM, Eppley JM, Samo TJ, Karl DM, DeLong EF. Microbial community structure and function on sinking particles in the North Pacific Subtropical Gyre. Front Microbiol. 2015;6:1–14. doi: 10.3389/fmicb.2015.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohman M, Barbeau K, Franks P, Goericke R, Landry M, Miller A. Ecological Transitions in a Coastal Upwelling Ecosystem. Oceanography. 2013;26:210–9. [Google Scholar]
- 24.Taylor AG, Landry MR, Selph KE, Wokuluk JJ. Temporal and spatial patterns of microbial community biomass and composition in the Southern California Current Ecosystem. Deep Sea Res Part II Top Stud Oceanogr. 2015;112:117–28. [Google Scholar]
- 25.Landry MR, Ohman MD, Goericke R, Stukel MR, Tsyrklevich K. Lagrangian studies of phytoplankton growth and grazing relationships in a coastal upwelling ecosystem off Southern California. Prog Oceanogr. 2009;83:208–16. [Google Scholar]
- 26.Stukel MR, Aluwihare LI, Barbeau KA, Chekalyuk AM, Goericke R, Miller AJ, et al. Mesoscale ocean fronts enhance carbon export due to gravitational sinking and subduction. Proc Natl Acad Sci USA. 2017;114:1252–7. doi: 10.1073/pnas.1609435114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knauer GA, Karl DM, Martin JH, Hunter CN. In situ effects of selected preservatives on total carbon, nitrogen and metals collected in sediment traps. J Mar Res. 1984;42:445–62. [Google Scholar]
- 28.Karl DM, Knauer GA. Detritus-microbe interactions in the marine pelagic environment - selected results from the VERTEX experiment. Bull Mar Sci. 1984;35:550–6. [Google Scholar]
- 29.Lee C, Hedges JI, Wakeham SG, Zhu N. Effectiveness of various treatments in retarding microbial activity in sediment trap material and their effects on the collection of swimmers. Limnol Oceanogr. 1992;37:117–30. [Google Scholar]
- 30.Beers JR, Stewart GL. Preservation of acantharians in fixed plankton samples. Limnol Oceanogr. 1970;15:825–7. [Google Scholar]
- 31.Stukel MR, Ohman MD, Benitez-Nelson CR, Landry MR. Contributions of mesozooplankton to vertical carbon export in a coastal upwelling system. Marine Ecology Progress Series. 2013;491:47–65. [Google Scholar]
- 32.Stukel MR, Kahru M, Benitez‐Nelson CR, Décima M, Goericke R, Landry MR, et al. Using Lagrangian‐based process studies to test satellite algorithms of vertical carbon flux in the eastern North Pacific Ocean. J Geophys Res Ocean. 2015;120:7208–22. [Google Scholar]
- 33.Amacher J, Neuer S, Lomas M. DNA-based molecular fingerprinting of eukaryotic protists and cyanobacteria contributing to sinking particle flux at the Bermuda Atlantic time-series study. Deep Sea Res Part II Top Stud Oceanogr. 2013;93:71–472. [Google Scholar]
- 34.Gordon LI, Jennings JC, Ross AA, and Krest JM. A suggested protocol for continuous flow automated analysis of seawater nutrients in the WOCE hydrographic program and the Joint Global Ocean Fluxes Study. Grp Tech Rpt 92-1, OSU Coll Oceanogr Descr Chem Oc. 1992.
- 35.Michaels AF, Caron DA, Swanberg NR, Howse FA, Michaels CM. Planktonic sarcodines (Acantharia, Radiolaria, Foraminifera) in surface waters near Bermuda: abundance, biomass and vertical flux. J Plankton Res. 1995;17:131–63. [Google Scholar]
- 36.Piredda R., Tomasino M. P., D'Erchia A. M., Manzari C., Pesole G., Montresor M., Kooistra W. H. C. F., Sarno D., Zingone A. Diversity and temporal patterns of planktonic protist assemblages at a Mediterranean Long Term Ecological Research site. FEMS Microbiology Ecology. 2016;93(1):fiw200. doi: 10.1093/femsec/fiw200. [DOI] [PubMed] [Google Scholar]
- 37.Goecks J, Nekrutenko A, Taylor J. Lessons learned from Galaxy, a web-based platform for high-throughput genomic analyses. 2012;1–6. 10.1109/eScience.2012.6404442.
- 38.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl Env Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar R, Haas B, Clemente J, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guillou L, Bachar D, Audic S, Bass D, Berney C, Bittner L, et al. The Protist Ribosomal Reference database (PR2): a catalog of unicellular eukaryote small sub- unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2013;41:D597–604. doi: 10.1093/nar/gks1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. Vegan: Community Ecology Package. R package version 2.4-5. 2017. https://CRAN.R-project.org/package=vegan.
- 42.Wickham H. ggplot2: Elegant Graphics for Data Analysis. NewYork: Springer-Verlag; 2009. [Google Scholar]
- 43.Tennekes M. Treemap: Treemap Visualization. R package version 2.4-2. 2017.
- 44.Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. In: Harper DAT (ed). Palaeontol Electron 2001;4:1–9.
- 45.Lopes dos Santos A, Pollina T, Gourvil P, Corre E, Marie D, Garrido JL, et al. Chloropicophyceae, a new class of picophytoplanktonic prasinophytes. Sci Rep. 2017;7:14019. doi: 10.1038/s41598-017-12412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Not F, Gausling R, Azam F, Heidelberg JF, Worden AZ. Vertical distribution of picoeukaryotic diversity in the Sargasso Sea. Environ Microbiol. 2007;9:1233–52. doi: 10.1111/j.1462-2920.2007.01247.x. [DOI] [PubMed] [Google Scholar]
- 47.Lin Y, Cassar N, Marchetti A, Moreno C, Ducklow H, Li Z. Specific eukaryotic plankton are good predictors of net community production in the Western Antarctic Peninsula. Sci Rep. 2017;7:14845. doi: 10.1038/s41598-017-14109-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Vargas C., Audic S., Henry N., Decelle J., Mahe F., Logares R., Lara E., Berney C., Le Bescot N., Probert I., Carmichael M., Poulain J., Romac S., Colin S., Aury J.-M., Bittner L., Chaffron S., Dunthorn M., Engelen S., Flegontova O., Guidi L., Horak A., Jaillon O., Lima-Mendez G., Luke J., Malviya S., Morard R., Mulot M., Scalco E., Siano R., Vincent F., Zingone A., Dimier C., Picheral M., Searson S., Kandels-Lewis S., Acinas S. G., Bork P., Bowler C., Gorsky G., Grimsley N., Hingamp P., Iudicone D., Not F., Ogata H., Pesant S., Raes J., Sieracki M. E., Speich S., Stemmann L., Sunagawa S., Weissenbach J., Wincker P., Karsenti E., Boss E., Follows M., Karp-Boss L., Krzic U., Reynaud E. G., Sardet C., Sullivan M. B., Velayoudon D. Eukaryotic plankton diversity in the sunlit ocean. Science. 2015;348(6237):1261605–1261605. doi: 10.1126/science.1261605. [DOI] [PubMed] [Google Scholar]
- 49.Pearman JK, Casas L, Merle T, Michell C, Irigoien X. Bacterial and protist community changes during a phytoplankton bloom. Limnol Oceanogr. 2016;61:198–213. [Google Scholar]
- 50.Lopes dos Santos A, Gourvil P, Tragin M, Noel MH, Decelle J, Romac S, et al. Diversity and oceanic distribution of prasinophytes clade VII, the dominant group of green algae in oceanic waters. ISME J. 2017;11:512–28. doi: 10.1038/ismej.2016.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Egge ES, Johannessen TV, Andersen T, Eikrem W, Bittner L, Larsen A, et al. Seasonal diversity and dynamics of haptophytes in the Skagerrak, Norway, explored by high-throughput sequencing. Mol Ecol. 2015;24:3026–42. doi: 10.1111/mec.13160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ichinomiya M, dos Santos AL, Gourvil P, Yoshikawa S, Kamiya M, Ohki K, et al. Diversity and oceanic distribution of the Parmales (Bolidophyceae), a picoplanktonic group closely related to diatoms. ISME J. 2016;10:2419–34. doi: 10.1038/ismej.2016.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zouari AB, Hassen MB, Balague V, Sahli E, Ben Kacem MY, Akrout F, et al. Picoeukaryotic diversity in the Gulf of Gabes: variability patterns and relationships to nutrients and water masses. Aquat Microb Ecol. 2018;81:37–53. [Google Scholar]
- 54.Zhu Fei, Massana Ramon, Not Fabrice, Marie Dominique, Vaulot Daniel. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiology Ecology. 2005;52(1):79–92. doi: 10.1016/j.femsec.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 55.Guillou L, Viprey M, Chambouvet A, Welsh RM, Kirkham AR, Massana R, et al. Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata) Env Microbiol. 2008;10:3349–65. doi: 10.1111/j.1462-2920.2008.01731.x. [DOI] [PubMed] [Google Scholar]
- 56.Lie AAY, Kim DY, Schnetzer A, Caron DA. Small-scale temporal and spatial variations in protistan community composition at the San Pedro Ocean Time-series station off the coast of southern California. Aquat Microb Ecol. 2013;70:93–110. [Google Scholar]
- 57.Anderson OR. Radiolaria. New York: Springer; 1983. [Google Scholar]
- 58.Suzuki N, Ogane K, Aita Y, Kato M, Sakai S, Kurihara T, et al. Distribution patterns of the radiolarian nuclei and symbionts using DAPI-fluorescence. Bull Natl Mus Nat Sci Ser B. 2009;169–82.
- 59.Suzuki N, Aita Y. Radiolaria: achievements and unresolved issues: taxonomy and cytology. Plankt Benthos Res. 2011;6:69–91. [Google Scholar]
- 60.Biard Tristan, Bigeard Estelle, Audic Stéphane, Poulain Julie, Gutierrez-Rodriguez Andres, Pesant Stéphane, Stemmann Lars, Not Fabrice. Biogeography and diversity of Collodaria (Radiolaria) in the global ocean. The ISME Journal. 2017;11(6):1331–1344. doi: 10.1038/ismej.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Decelle J, Romac S, Sasaki E, Not F, Mahé F. Intracellular diversity of the V4 and V9 regions of the 18S rRNA in marine protists (radiolarians) assessed by high-throughput sequencing. PLoS One. 2014;9:e104297. doi: 10.1371/journal.pone.0104297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Godhe A, Asplund ME, Harnstrom K, Saravanan V, Tyagi A, Karunasagar I. Quantification of diatom and dinoflagellate biomasses in coastal marine seawater samples by real-time PCR. Appl Env Microbiol. 2008;74:7174–82. doi: 10.1128/AEM.01298-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Giner CR, Forn I, Romac S, Logares R, De Vargas C, Massana R. Environmental sequencing provides reasonable estimates of the relative abundance of specific picoeukaryotes. Appl Environ Microbiol. 2016;82:4757–66. doi: 10.1128/AEM.00560-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boltovskoy D. Vertical distribution patterns of Radiolaria Polycystina (Protista) in the World Ocean: living ranges, isothermal submersion and settling shells. J Plankton Res. 2017;39:330–49. [Google Scholar]
- 65.Decelle J, Martin P, Paborstava K, Pond DW, Tarling G, Mahé F, et al. Diversity, Ecology and Biogeochemistry of Cyst-Forming Acantharia (Radiolaria) in the Oceans. PLoS One. 2013;8:e53598. doi: 10.1371/journal.pone.0053598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gowing MM, Coale SL. Fluxes of living radiolarians and their skeletons along a northeast Pacific transect from coastal upwelling to open ocean waters. Deep Sea Res Part A-Oceanogr Res Pap. 1989;36:561–76. [Google Scholar]
- 67.Takahashi K. Radiolaria - sinking population, standing stock, and production-rate. Mar Micropaleontol. 1983;8:171–81. [Google Scholar]
- 68.Dennett MR, Caron DA, Michaels AF, Gallager SM, Davis CS. Video plankton recorder reveals high abundances of colonial Radiolaria in surface waters of the central North Pacific. J Plankton Res. 2002;24:797–805. [Google Scholar]
- 69.Suzuki N, Not F. Biology and Ecology of Radiolaria. In: Ohtsuka S, Suzaki T, Horiguchi T, Suzuki N, Not F (eds). pp. 179–222, Springer: Japan; 2015.
- 70.Takahashi K. Radiolarian flux and seasonality: climatic and El Nino response in the subarctic Pacific, 1982-4. Glob Biogeochem Cycles. 1987;1:213–31. [Google Scholar]
- 71.Guidi L, Chaffron S, Bittner L, Eveillard D, Larhlimi A, Roux S, et al. Plankton networks driving carbon export in the oligotrophic ocean. Nature. 2016;532:465–70. doi: 10.1038/nature16942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gowing MM. Trophic biology of phaeodarian radiolarians and flux of living radiolarians in the upper 2000-m of the North Pacific Central Gyre. Deep Sea Res Part A- Oceanogr Res Pap. 1986;33:655–74. [Google Scholar]
- 73.Ohman MD, Powell JR, Picheral M, Jensen DW. Mesozooplankton and particulate matter responses to a deep-water frontal system in the southern California Current System. J Plankton Res. 2012;34:815–27. [Google Scholar]
- 74.Biard T, Stemmann L, Picheral M, Mayot N, Vandromme P, Hauss H, et al. In situ imaging reveals the biomass of giant protists in the global ocean. Nature. 2016;532:504–7. doi: 10.1038/nature17652. [DOI] [PubMed] [Google Scholar]
- 75.Bond NA, Cronin MF, Freeland H, Mantua N. Causes and impacts of the 2014 warm anomaly in the NE Pacific. Geophys Res Lett. 2015;42:3414–20. [Google Scholar]
- 76.Gentemann CL, Fewings MR, García-Reyes M. Satellite sea surface temperatures along the West Coast of the United States during the 2014-6 northeast Pacific marine heat wave. Geophys Res Lett. 2017;44:312–9. [Google Scholar]
- 77.Peterson WT, Fisher JL, Strub PT, Du X, Risien C, Peterson J, et al. The pelagic ecosystem in the Northern California Current off Oregon during the 2014-6 warm anomalies within the context of the past 20 years. J Geophys Res Ocean. 2017;122:7267–90. doi: 10.1002/2017jc012952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gómez F, Qiu D, Dodge JD, Lopes RM, Lin S. Morphological and molecular characterization of Ptychodiscus noctiluca revealed the polyphyletic nature of the order Ptychodiscales (Dinophyceae) J Phycol. 2016;52:793–805. doi: 10.1111/jpy.12438. [DOI] [PubMed] [Google Scholar]
- 79.Stukel Michael R., Biard Tristan, Krause Jeffrey, Ohman Mark D. Large Phaeodaria in the twilight zone: Their role in the carbon cycle. Limnology and Oceanography. 2018;63(6):2579–2594. [Google Scholar]
- 80.Biard T, Krause JW, Stukel MR, Ohman MD. The significance of giant phaeodarians (Rhizaria) to biogenic silica export in the California Current Ecosystem. Glob Biogeochem Cycles. 2018;32:987–1004. [Google Scholar]
- 81.Taylor GT, Karl DM, Pace ML. Impact of bacteria and zooflagellates on the composition of sinking particles: an in situ experiment. Marine Ecology Progress Series. 1986;29:141–155. [Google Scholar]
- 82.Iversen MH, Poulsen LK. Coprorhexy, coprophagy, and coprochaly in the copepods Calanus helgolandicus, Pseudocalanus elongatus, and Oithona similis. Mar Ecol Prog Ser. 2007;350:79–89. [Google Scholar]
- 83.Noji TT. The influence of macrozooplankton on vertical particulate flux. Sarsia. 1991;76:1–9. [Google Scholar]
- 84.Mayor DJ, Sanders R, Giering SL, Anderson TR. Microbial gardening in theocean’s twilight zone: detritivorous metazoans benefit from fragmenting, rather than ingesting, sinking detritus: fragmentation of refractory detritus by zooplankton beneath the euphotic zone stimulates the harvestable productio. Bioessays. 2014;36:1132–7. doi: 10.1002/bies.201400100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lim EL, Dennett MR, Caron DA. The ecology of Paraphysomonas imperforate based on studies employing oligonucleotide probe identification in coastal water samples and enrichment cultures. Limnol Oceanogr. 1999;44:37–51. [Google Scholar]
- 86.Caron DA, Davis PG, Madin LP, Sieburth JM. Heterotrophic bacteria and bacterivorous protozoa in oceanic macro-aggregates. Science. 1982;218:795–7. doi: 10.1126/science.218.4574.795. [DOI] [PubMed] [Google Scholar]
- 87.Silver MW, Gowing MM, Brownlee DC, Corliss JO. Ciliated protozoa associated with oceanic sinking detritus. Nature. 1984;309:246–8. [Google Scholar]
- 88.Gowing MM, Bentham WN. Feeding ecology of phaeodarian radiolarians at the VERTEX North Pacific time-series site. J Plankton Res. 1994;16:707–19. [Google Scholar]
- 89.Not F, del Campo J, Balagué V, De Vargas C, Massana R. New insights into the diversity of marine picoeukaryotes. PLoS One. 2009;4:e7143. doi: 10.1371/journal.pone.0007143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stemmann L, Youngbluth M, Robert K, Hosia A, Picheral M, Paterson H, et al. Global zoogeography of fragile macrozooplankton in the upper 100–1000 m inferred from the underwater video profiler. ICES J Mar Sci. 2008;65:433–42. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.