Abstract
Meier–Gorlin syndrome (MGS) is a rare, congenital primordial microcephalic dwarfism disorder. MGS is caused by genetic variants of components of the origin recognition complex (ORC) consisting of ORC1–6 and the pre-replication complex, which together enable origin firing and hence genome replication. In addition, ORC1 has previously been shown to play a role in ciliogenesis. Here, we extend this work and investigate the function of ORC1 and two other members of the complex on cilia at an organismal level. Knockdown experiments in zebrafish confirmed the impact of ORC1 on cilia. ORC1-deficiency confers defects anticipated to arise from impaired cilia function such as formation of oedema, kidney cysts, curved bodies and left–right asymmetry defects. We found ORC1 furthermore required for cilium formation in zebrafish and demonstrate that ciliopathy phenotypes in ORC1-depleted zebrafish could not be rescued by reconstitution with ORC1 bearing a genetic variant previously identified in MGS patients. Loss-of-function of Orc4 and Orc6, respectively, conferred similar ciliopathy phenotypes and cilium shortening in zebrafish, suggesting that several, if not all, components of the ORC regulate ciliogenesis downstream to or in addition to their canonical function in replication initiation. This study presents the first in vivo evidence of an influence of the MGS genes of the ORC family on cilia, and consolidates the possibility that cilia dysfunction could contribute to the clinical manifestation of ORC-deficient MGS.
Subject terms: Neurodevelopmental disorders, Disease model
Introduction
Meier–Gorlin syndrome (MGS) is a rare disorder within the primordial microcephalic dwarfism umbrella. In addition to displaying proportional short stature and microcephaly, patients are characterised by small ears and patellar aplasia [1]. Growth retardation largely occurs during pregnancy and within the first year of life [2]. Attempts to rescue growth postnatally by growth hormone therapy, however, have shown only limited success [3]. Most MGS patients develop normal or only slightly impaired cognitive functions despite their reduced head circumference [2]. Several genes have been identified as disease loci for MGS: origin of replication complex (ORC)1 [4], ORC4 [5], ORC6 [6], CDC6 [7], CDT1 [5], MCM5 [4, 8], GMNN [9] and CDC45 [10]. These genes encode proteins which function during origin licensing or firing and hence predominantly affect replication initiation.
ORC1 is one of six ORC proteins that assemble into the ORC. ORC hexamers bind to origins of replication and license them for replication initiation through assembly of the prereplication complex (consisting of CDT1, CDC6 and MCMs 2–7) [11]. Diminished ORC1 function in patients results in reduced origin licensing and/or diminished MCM2–7 loading onto DNA [4, 12] suggesting that the clinical manifestation are a consequence of impaired replication causing reduced cell proliferation and hence growth retardation.
In a previous study, based entirely on the analysis of cultured cell lines, we proposed an additional role for ORC1 in cilia function. Interestingly, two small organelles, the centrosome and the cilium are critical for normal neuronal development and genetic variants affecting centrosomes and cilia are frequently observed in patients displaying microcephaly, one feature of ORC1-deficient MGS. Centrosomes facilitate mitotic spindle assembly and hence proper cell division of neural progenitors [13]. Centrosome anomalies result in diminished proliferation of neural progenitors, premature differentiation and apoptosis [14, 15]. In addition, the so-called mother centriole of the centrosome turns into the basal body of a cilium, once a cell exits the cell cycle. Thus, anomalies in centrosomes often lead to cilia defects [13, 16]. Cilia itself are also important regulators of neuronal progenitor cell expansion [17], underlying their causal link with microcephaly [18]. Significantly, we and others reported that impaired ORC1 function, either by knockdown (KD) or analysis of genetic variants observed in ORC1-deficient patients caused centrosome aberrations as well as structural and functional defects in primary cilia [19, 20]. However, to date this functional role has only been examined in cultured cells. Here, we aimed to gain evidence for such a role using a model system, where cilia function could be examined. We chose zebrafish for this analysis, given the well characterised features conferred by cilia dysfunction. We show that depletion of Orc1 in zebrafish results in growth deficiencies and phenotypes resembling ciliopathies. Upon Orc1 KD, zebrafish develop shorter, dysfunctional cilia. Reconstitution of Orc1 loss-of-function zebrafish embryos with wild-type (wt) ORC1 restores normal development of zebrafish, while a patient-derived amino acid change in ORC1 is unable to rescue either the ciliopathy phenotypes or the growth phenotype. To gain insight into whether this function is unique to Orc1 or requires further ORC components, we examined further Orc4 and 6, which represent additional genes causing MGS. Strikingly, depletion of Orc4 or Orc6 mimics the Orc1 phenotypes and confers similar cilia deficiencies. These findings strongly suggest that the intact ORC complex has an additional, potentially non-canonical function during ciliogenesis.
Materials and methods
Cloning
As a template for in vitro transcription of an antisense in situ probe, a 1001 bp fragment of zebrafish Orc1 (NM_199933.1) was cloned into pCRII by TOPO TA cloning (Thermo Fisher, Darmstadt, Germany). For cloning of human ORC cDNAs PCR primers were designed based on NM_004153.3 (ORC1), NM_181741.3 (ORC4) and NM_014321.3 (ORC6). The whole open reading frame of human ORC1 except for the stop codon was cloned into pCS2-GFP via BamHI and XbaI. ORC4 and ORC6 open reading frames were cloned into pCS2+ via ClaI and StuI. The MGS variants pCS2-GFP-ORC1 F89S, pCS2+-ORC4 Y174C and pCS2+-ORC6 K202R* were generated by site-directed mutagenesis of the plasmids mentioned above. The open reading frame containing plasmids were linearised with NotI and capped RNA was transcribed using the AmpliCap SP6 High Yield Message Maker Kit (Cellscript, Madison, WI, USA). All genetic variants have previously been identified in MGS patients and are deposited along with the associated clinical appearance at dbSNP: rs387906827 (ORC1 F89S) [4], rs387906847 (ORC4 Y174C) [5] and rs879255692 (ORC6 K202R*) [6].
Zebrafish husbandry and manipulation
Zebrafish were maintained in a tank rack system with automatic water recycling, water changes as well as monitoring and adjustment of water parameters (Tecniplast, Hohenpeißenberg, Germany). Fish were kept under a 14 h light and 10 h dark cycle and were fed three times a day with artemia (Aqua Schwarz, Göttingen, Germany) and pelleted dry food (Sparos Zebrafeed, Olhão, Portugal). Maintenance as well as manipulation of zebrafish were approved by the Veterinary Care Unit at Ulm University and the animal welfare commissioner of the regional board for scientific animal experiments in Tübingen, Germany. Experiments were performed according to the European Union Directive 86/609/EEC for the protection of animals used for experimental and other scientific purposes. In this study adult zebrafish (AB and EK wt strains, wt1b:GFP transgenics [21]) were naturally mated. Fertilised eggs were microinjected either at the one to two cell stage for ubiquitous manipulation of the embryo or at the 1000 cell stage for Kupffer’s vesicle (KV)-specific targeting [2]. Embryos were raised to the desired stages in an incubator set to 28.5 °C. Orc1-depleted embryos were generated by injection of 5 nl of a 0.25 mM concentrated, previously characterised antisense morpholino oligonucleotide (MO) [4]. Orc4 or Orc6 KD was achieved by injection of 5 nl of splice blocking MOs, which were diluted to 0.5 mM and have the following sequences: Orc4 exon 1 splMO: 5′- TAACATGAGGAAGGAGGACAGACCA; Orc6 exon 2 splMO: 5′-CTCTGCTTGACTGAAAACAAATGGA. Blasting against the zebrafish genome (Ensembl release 94) revealed that neither the Orc4 splMO nor the Orc6 splMO target any other sequence in the zebrafish genome. Noninjected (NI) embryos as well as those injected with a standard control MO, which does not target the zebrafish genome, were used as controls. All MOs were purchased from Gene Tools Inc (Philomath, OR, USA). Rescue injections were performed by consecutive injections of KD MO and capped RNA (500 pg per egg).
Splice-blocking verification
To verify the ability of Orc4 and Orc6 splMO, respectively in preventing regular splicing zebrafish eggs were injected at the one cell stage and allowed to develop until 24 h post fertilisation (hpf). RNA was extracted using Zymo’s Quick-RNA MiniPrep kit (Freiburg, Germany). Equal amounts of RNA were transcribed using SuperScript II (Invitrogen, Germany) and PCR was performed with the following primers: Orc4 Fwd: 5′-GAAGATGAGCAAGCGAAGGC, Orc4 Rev: 5′-GACGTTCTCCAGATGCAGCT; Orc6 Fwd: 5′-ACGCCCATTTGTGAAGCAAC, Orc6 Rev: 5′-CTCGGAGACCCAGGTTTGAC. The resulting PCR bands were sequenced to verify amplification of the right sequence and splice blocking induced by splMO injection. As housekeeping gene β-actin was used as previously described [23].
In situ hybridisation
In situ hybridisation (ISH) for the spatial detection of RNA expression was performed with fixed embryos and according to the standard protocol by Thisse et al. [24]. To generate a DIG-labelled antisense in situ probe for the detection of orc1, the plasmid containing a fragment of the Orc1 coding sequence was linearised with HindIII and transcribed using T7 RNA polymerase (NEB, Frankfurt am Main, Germany) and DIG labelling mix (Roche, Mannheim, Germany). All other probes used have been described before [25].
Imaging
Live zebrafish embryos (48 hpf) as well as those processed by whole mount ISH were imaged on a Leica M125 with an IC80HD or a MC190HD camera (Leica, Wetzlar, Germany). For the analysis of smaller heads, eyes and shorter bodies, injected embryos were compared to average NI controls. Fluorescence images of the developing kidney were acquired on a Leica M205FCA and a Leica DFC9000GT sCMOS camera. Images of KV cilia were acquired using a Leica TCS SP5II confocal microscope. Here, z-stacks were captured every 0.3 µm and processed into a 3D stack using the Leica acquisition software LAS AF. Image processing was done with the help of Adobe Photoshop CC2018 (Adobe Systems Software Ireland Limited, Dublin, Ireland). Any adjustments (brightness, contrast or colour balance) were applied equally to the whole image and equally to same treatment groups (i.e., CTRL MO vs. Orc1 MO). Final figures were assembled in Adobe Illustrator CC2018.
Cilia staining and analysis
Injected zebrafish embryos were raised until eight somite stage (ss) and fixed overnight at 4 °C in 4% PFA. Fixed embryos were dechorionated, washed with PBS containing 0.1% Tween-20 and processed for cilia staining as described [26] using an anti-acetylated tubulin antibody (Sigma, Munich, Germany, clone 6–11B-1, 1:1000). For KV detection an antibody against PKCζ (Santa Cruz Biotechnology, Heidelberg, Germany, Catalogue no. sc-216, 1:500) was used, which labels apical cell borders and allows for detection of the KV outline. Confocal stacks were imported into ImageJ and cilia length was measured by tracing the entire cilium with the help of a Wacom tablet. KV area measurements were facilitated by tracing the outline of the KV in Adobe Illustrator, filling the shape and measuring the area of the shape again in ImageJ.
Statistical analysis
Statistical analyses were performed with the help of Prism 7 (GraphPad, La Jolla, CA, USA). The respective test is indicated in the figure legends. If not otherwise noted statistical significance is indicated as follows: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns, not significant.
Results
Orc1 depletion in zebrafish leads to growth defects and a ciliopathy-like phenotype
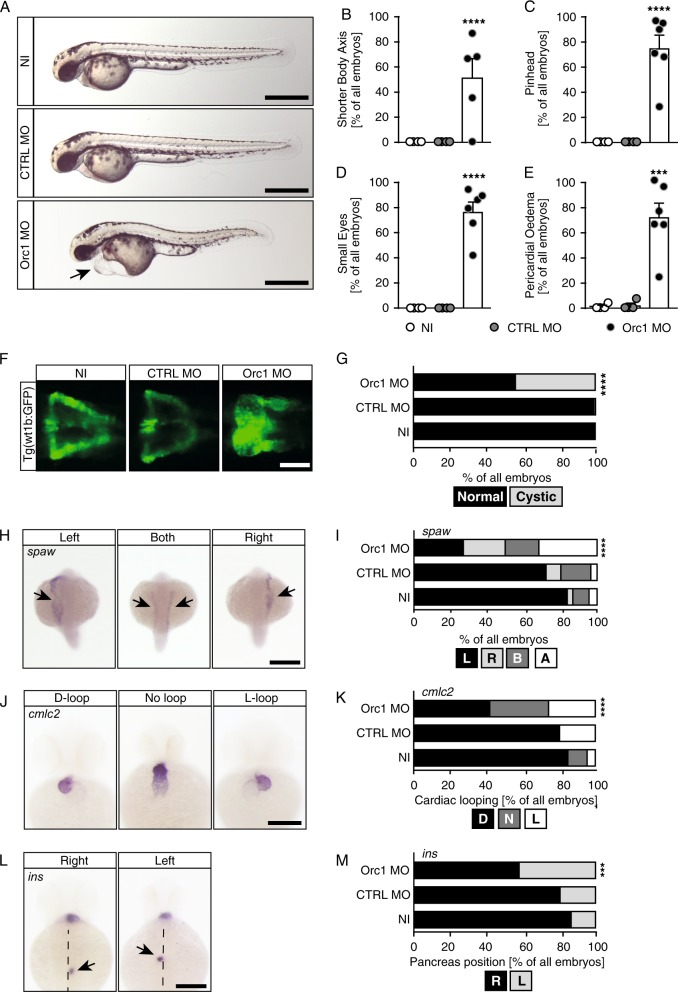
ORC1 was the first causal gene identified for MGS [4]. In order to investigate the impact of ORC1 dysfunction in vivo we made use of a previously established and validated MO-mediated KD approach [4]. Zebrafish have a single homologue of ORC1 and ubiquitous depletion of Orc1 by injection into fertilised eggs at the 1–2 cell stage results in shorter zebrafish embryos (Fig. 1a, b) [4]. In addition, anterior structures such as the head (referred to as “pinhead”) and the eyes are smaller than in control embryos upon Orc1 KD (Fig. 1c, d) consistent with the microcephaly of MGS patients. Orc1-deficient embryos also developed a distinct body curvature (Fig. 1a), which is characteristic for embryos with cilia deficiencies [27]. Furthermore, oedema such as hydrocephalus or pericardial oedema have repeatedly been reported to occur in zebrafish embryos with dysfunctional cilia [25, 28], which we also observed upon Orc1 KD (Fig. 1e). Last, but not least, we analysed the developing kidneys. A substantial fraction of Orc1 MO injected embryos displayed cystic kidneys (Fig. 1f, g), which is yet another malformation that has been strongly linked to defective cilia [29]. These data suggest that loss of Orc1 may potentially result in cilia dysfunction.
Fig. 1.
Loss of ORC1 produces ciliopathy phenotypes in zebrafish embryos. a Live images of 48 hpf zebrafish embryos. NI: uninjected controls, CTRL MO: embryos injected with a control MO, Orc1 MO: translation blocking MO against Orc1. Arrow indicates pericardial oedema. Scale bar: 500 µm. b Orc1 morphants are characterised by an overall short body as it had been shown in the original paper describing the Orc1 MO [5]. c Head size is reduced upon Orc1 depletion. d Eyes are smaller in the absence of Orc1. e Orc1 morphant embryos develop pronounced pericardial oedema. f Fluorescent images of the pronephros of control (NI, CTRL MO) and Orc1 KD embryos. To visualise the pronephros a transgenic line expressing GFP under the wt1b promoter was used (Tg(wt1b:GFP)). Scale bar: 100 µm. g Incidence of cystic pronephric tubules. Two-sided Fisher’s exact test. Number of embryos analysed: NI = 120; CTRL MO = 119; Orc1 MO = 128. h Examples of southpaw (spaw) expression in 20–22 ss embryos showing transcripts in the left or right LPM, the middle picture shows ambiguous expression. Scale bar: 300 µm. i Depletion of Orc1 randomises spaw expression. n = 5–9 experiments. Number of embryos: NI = 231; CTRL MO = 253; Orc1 MO = 114. j The heart of 48 hpf zebrafish embryos is normally looped in an S-shape with the ventricle left and above from the atrium (D-loop). When LR asymmetry is disturbed, hearts stay unlooped or show an inverse loop (L-loop). ISH for cmlc2. Scale bar: 200 µm. k Heart looping is disturbed in Orc1 morphants. n = 4–6 experiments. Number of embryos: NI = 82; CTRL MO = 67; Orc1 MO = 145. l ISH for the endocrine pancreas marker insulin (ins) reveals a normal right-sided localisation of the pancreas (right), which is reversed in asymmetry defects (left). Dashed line: midline of the embryo. Dorsal view. Scale bar: 200 µm. m Wrong-sided pancreas in Orc1 morphants. n = 4–6 experiments. Number of embryos: NI = 82; CTRL MO = 67; Orc1 MO = 145. b–e Graphs show means ± SEM with circles indicating individual experiments. One-way ANOVA with Sidak’s multiple comparisons test. n = 4–6 independent experiments. Number of embryos analysed: NI = 72; CTRL MO = 86; Orc1 MO = 151. i, k, m Data analysed using two-tailed Fisher’s exact test
Zebrafish cilia dysfunction can further be monitored by examining left–right (LR) asymmetry patterning. Early during development a ciliated vesicle (KV in zebrafish, node in mouse) temporally persists, which is crucial for the proper establishment of a left versus a right side of the body. When cilia within this vesicle form and function properly, genes of the nodal cascade are exclusively expressed on the left side from the midline. We hence analysed expression of the nodal-related gene southpaw (spaw) in 20–22 ss embryos. Regular expression of spaw arises in the left lateral plate mesoderm (LPM), while embryos with disturbed LR asymmetry development exhibit expression right from the midline or in an ambiguous fashion (Fig. 1h). Upon Orc1 KD a significant fraction of embryos failed to express spaw exclusively in the left LPM (Fig. 1i). Since aberrant leftward gene expression causes irregular placement and orientation of visceral organs, we analysed looping of the two-chambered zebrafish heart at 48 hpf. Embryos with LR defects frequently harbour inversely looped hearts or hearts that completely fail to loop, which was observed when Orc1 was depleted (Fig. 1j, k). Similarly, placement of abdominal organs as assessed by the position of the endocrine pancreas was abnormal (Fig. 1l, m). We thus conclude that loss of Orc1 confers cilia dysfunction in vivo.
Orc1 controls ciliogenesis in zebrafish
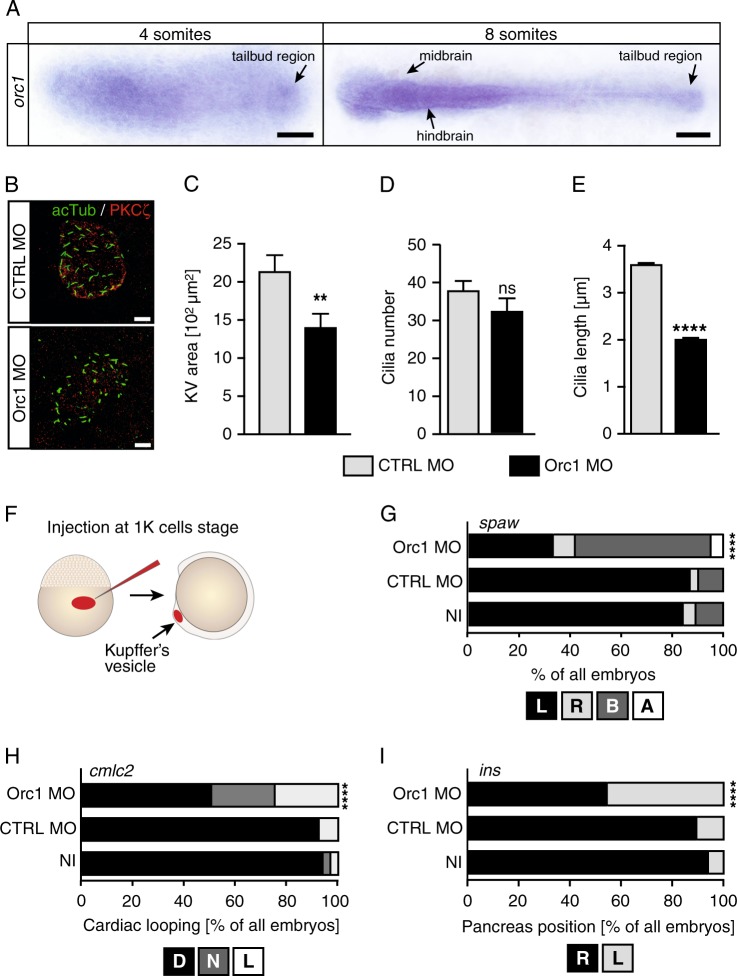
Next, we investigated the influence of Orc1 on symmetry breaking and cilia function in greater detail. Symmetry breaking occurs at a time during zebrafish development, when cells almost cease to proliferate and instead differentiate and undergo convergence–extension movements [30]. Despite there being only a few cycling cells during this stage, we detected orc1 transcripts throughout the embryo including the tailbud, where the KV forms (Fig. 2a). Closer analysis of the KV revealed that cilia do form in the absence of Orc1 (Fig. 2b). The overall size of the KV, however, was smaller than in control embryos, although Orc1 KD embryos were stage-matched (8 ss like control embryos) (Fig. 2c). As the number of cilia per KV did not change (Fig. 2d), the reduction in KV size may be attributed to an overall growth reduction in Orc1 morphants. Intriguingly, cilia were substantially shorter in Orc1 KD embryos than in control-injected embryos (Fig. 2e). To test whether Orc1 functions within the cilia-forming cells, we applied an injection strategy originally developed by Amack et al., whereby injection of MO at the 512–1000 cells stage selectively delivers the MO to the dorsal forerunner cells, the progenitor cells for the KV (Fig. 2f). Such KV-specific KD of Orc1 disrupted LR-patterning as efficiently as the ubiquitous KD (compare Figs 2g–i and 1). These data suggest that Orc1 controls ciliogenesis in vivo and functions within the ciliated cells.
Fig. 2.
Orc1 controls cilia formation and functions within ciliated cells. a Flat mounts of 4 and 8 ss embryos after orc1 ISH. Arrow indicates expression throughout the embryo, but also in the tailbud, where the KV forms. Scale bars: 150 µm. b Confocal stacks of KV cilia of 8 ss embryos injected with standard control MO or Orc1 MO. Apical cell borders to outline the KV were labelled with a PKCζ antibody (red), while cilia were stained for acetylated tubulin (green). Scale bar: 10 µm. c Smaller KVs upon Orc1 KD. p = 0.097. n = 22–30 embryos. d Cilia are formed at similar numbers in control-injected and Orc1 KD embryos. p = 0.2071. In three experiments 25 (CTRL MO) and 23 (Orc1 MO) KVs were assessed. e Shorter KV cilia after Orc1 KD. n = 901 (CTRL MO) and 669 (Orc1 MO) cilia. f Injection strategy according to Amack et al. [2] to specifically deplete Orc1 in KV cells. g KV-specific depletion randomises spaw expression in the LPM. L, left-sided expression, R, right-sided expression, B, ambiguous expression, A, no expression in the LPM. n = 4 experiments, 81–122 embryos in total. h Heart looping fails or occurs inversely upon Orc1 MO depletion in the KV. n = 4 experiments, 66–85 embryos in total. i Abdominal organ placement as shown for the endocrine pancreas is random in embryos depleted of Orc1 in the KV. n = 4–5 experiments, 66–90 embryos in total. c–e Means ± SEM. Unpaired, two-tailed t-tests with Welch’s correction. g–i Stacked bar graphs analysed using two-sided Fisher’s exact tests
ORC1 F89S, a causal genetic variant for MGS, lacks the ability to rescue the Orc1 loss-of-function phenotype in zebrafish embryos
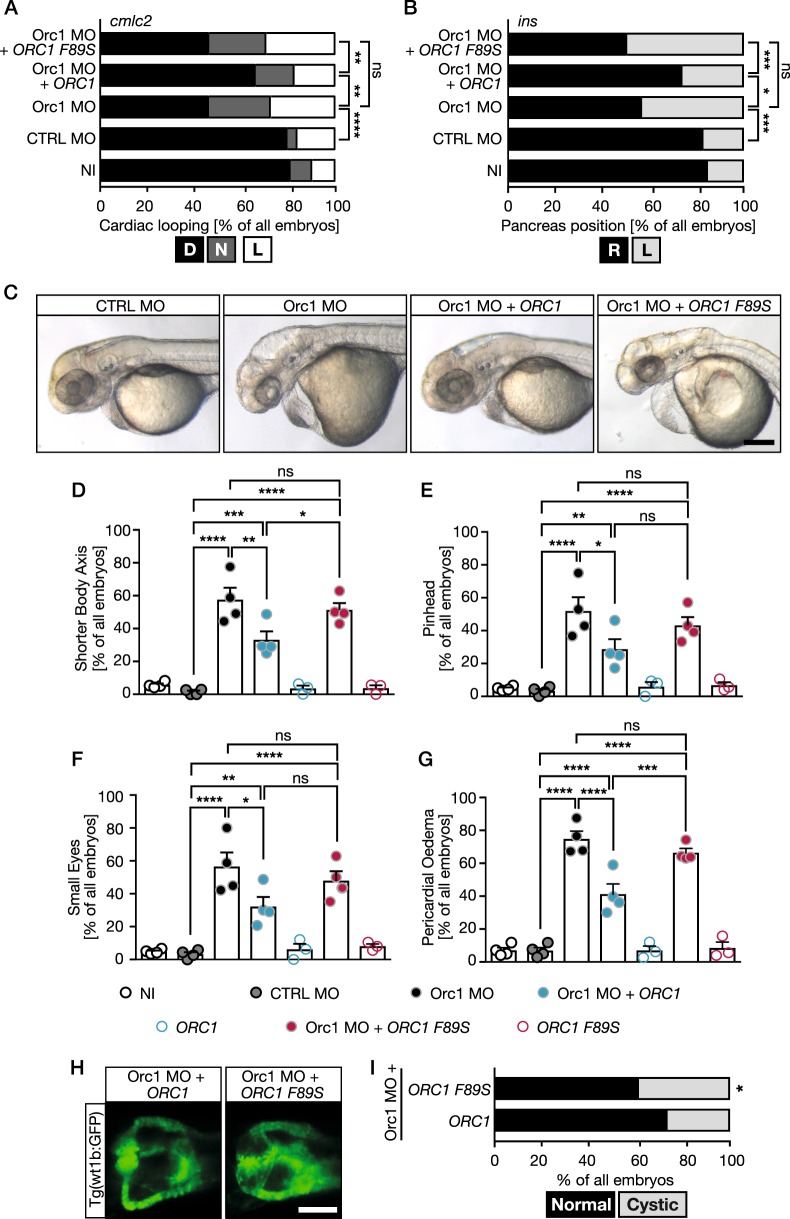
Finally, to link ORC1-dependent MGS to cilia dysfunction we performed rescue experiments in zebrafish embryos. Fertilised eggs were consecutively injected at the 1–2 cell stage with Orc1 MO and capped RNA encoding either human wt ORC1 or ORC1 F89S (c.266 T > C, rs387906827), a mutated form of ORC1 found in an MGS patient [4]. While wt ORC1 partially rescued the heart looping defects (Fig. 3a) and the randomisation of pancreas placement (Fig. 3b), no improvement could be observed by expression of ORC1 F89S. Similarly, co-injection of wt ORC1 improved the appearance of zebrafish at 48 hpf, while ORC1 F89S failed to do so (Fig. 3c). For all morphological features we assessed, namely axis shortening (Fig. 3d), head size (Fig. 3e), eye size (Fig. 3f) and pericardial oedema (Fig. 3g) we observed a significant rescue upon reconstitution with wt, but not ORC1 F89S. Last, but not least: while expression of wt ORC1 partially rescued kidney cyst formation compared to Orc1 KD alone, there was no improvement upon expression of the patient-derived F89S variant (Fig. 3h, i). We hence conclude that the MGS variant ORC1 F89S is an impacting genetic variant of ORC1 causing a phenotype similar to that conferred by cilia dysfunction.
Fig. 3.
The MGS variant F89S of ORC1 is unable to rescue ciliopathy phenotypes or microcephaly in zebrafish embryos. a Heart looping in Orc1 morphants is rescued by co-injection with RNA encoding human ORC1. Embryos were scored after cmlc2 ISH. D, correctly looped heart, N, no loop, L, inversely looped heart. b Endocrine pancreas position cannot be rescued by the MGS variant ORC1 F89S. R, pancreas right from the midline, L, pancreas left from the midline. c Live images of 48 hpf zebrafish embryos. Scale bar: 200 µm. d The shorter body axis in Orc1 MO embryos can be partially rescued by human ORC1. e Reconstitution with wt human ORC1 partially restores head growth in Orc1 morphants, while human ORC F89S does not. f Bar graph for occurrence of smaller eyes upon MO injection and rescue attempts. g Pericardial oedema can also be rescued by wt ORC1, but not by F89S. h Co-injection of ORC1 F89S RNA cannot prevent kidney cyst formation in Orc1 KD embryos. Scale bar: 100 µm. i Stacked bar graph displaying the incidence of kidney cysts. Two-sided Fisher’s exact test. n = 125 (Orc1 MO + ORC1), 123 (Orc1 MO + ORC1 F89S). a, b Stacked bar graphs (summarising 3–4 independent experiments) analysed using two-sided Fisher’s exact tests. n = 93–183 embryos. d–g Bar graphs display means ± SEM. Circles indicate means of individual experiments. n = 3–4 experiments, 80–249 embryos in total. One-way ANOVA
Depletion of other ORC components confers phenotypes reminiscent of cilium dysfunction
ORC1 is one of six ORC proteins forming the origin licensing complex needed for origin licensing. Genetic variants of ORC4 and ORC6, two additional ORCs, have also been observed in MGS patients [5, 6], suggesting that ORC dysfunction is causal for MGS. This raised the question whether any cilium dysfunction phenotype provoked by ORC1 loss-of-function could potentially arise from an overall dysfunction of the whole ORC. We hence examined whether depletion of these additional ORC proteins would phenocopy Orc1 KD. We designed splice blocking MOs (splMO) against Orc4 and Orc6, respectively. Injection of either Orc4 or Orc6 splMO interfered with regular splicing (Supplementary Fig. 1). This resulted in a frame shift causing a premature stop codon in both, orc4 and orc6 (Supplementary Fig. 1).
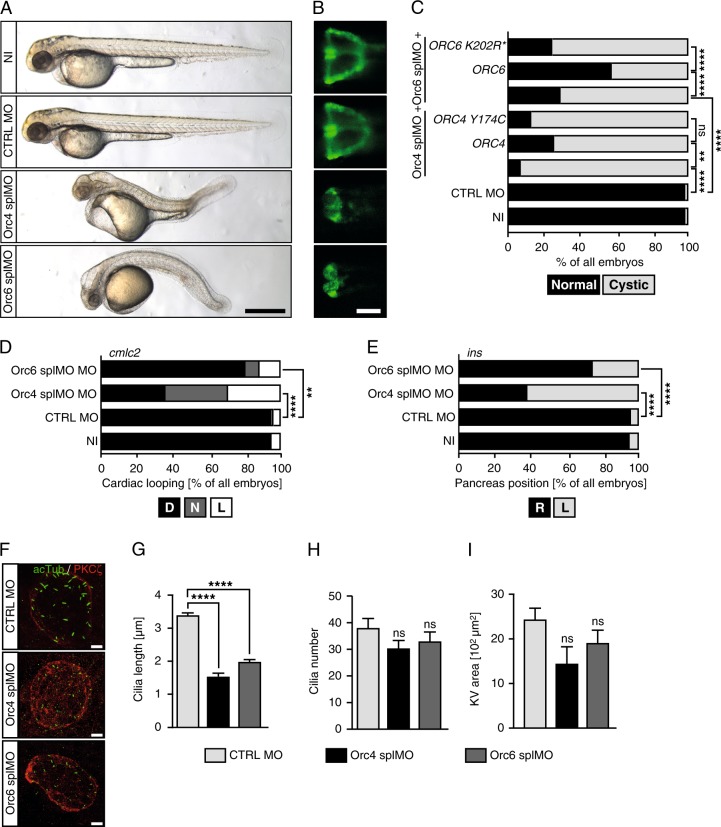
Loss of either Orc4 or Orc6 resulted in shortened embryos having smaller anterior structures, pericardiac oedema and kidney cysts as observed for Orc1 morphants (Fig. 4a–c). Importantly, rescue attempts using RNA encoding previously identified variants of ORC4 and ORC6 found in MGS patients [5, 6] were unable to rescue the kidney cysts whilst a partial, statistically significant rescue was obtained with wt RNAs (Fig. 4c). Embryos deficient in Orc4 or Orc6 also developed LR asymmetry defects such as randomised heart looping and pancreas placement (Fig. 4d, e). Taken together with the kidney cyst phenotype, these findings are consistent with an underlying cilia development defect, which was substantiated by the analysis of cilia length in the KV, which showed that injection of either splice blocking MO resulted in drastically shortened cilia (Fig. 4f, g), with no change in cilia numbers (Fig. 4h). The KV area, however, and in contrast to Orc1 morphants (Fig. 2c), was not significantly changed although a minor reduction was evident (Fig. 4i). Taken together, our findings show that three ORC components, variants of which are found in MGS patients, are required for proper ciliogenesis. This suggests that the whole ORC impacts on cilia development, providing a novel function for ORC.
Fig. 4.
Loss of additional ORC components provokes cilium dysfunction in zebrafish. a Live images of 48 hpf control and Orc4 and Orc6 depleted zebrafish embryos showing phenotypes reminding of MGS (shorter axis, smaller heads and eyes) and cilium dysfunction (pericardial oedema and body curvature). Scale bar: 500 µm. b KD of either Orc4 or Orc6 leads to cystic kidneys. Scale bar: 100 µm. c Percentage of embryos developing pronephric cysts. Co-injection of RNA encoding human ORC4 or ORC6 partially rescues cyst formation, while the MGS variants do not. n = 177 (NI), 172 (CTRL MO), 58 (Orc4 splMO), 63 (Orc4 splMO + ORC4), 63 (Orc4 splMO + ORC4 Y174C), 93 (Orc6 splMO), 108 (Orc6 splMO + ORC6) and 90 (Orc6 splMO + ORC6 K202R*) embryos. d Heart looping is randomised after Orc4 or Orc6 KD in zebrafish. n = 3 experiments with 79 NI, 93 CTRL MO, 89 Orc4 splMO and 102 Orc6 splMO embryos. e Orc4 and Orc6 deficient embryos display random placement of the endocrine pancreas. n = 3 experiments (79 NI, 93 CTRL MO, 89 Orc4 splMO and 102 Orc6 splMO embryos). f Embryos depleted of Orc4 and Orc6, respectively develop KVs with shortened cilia. Confocal stacks of KVs of 8 ss embryos stained for acetylated tubulin (green, cilia) and PKCζ (red, apical cell borders). Scale bar: 10 µm. g Cilia are shorter upon Orc4 or Orc6 KD. n = 194–292 cilia. Kruskal–Wallis test with Dunnett’s multiple comparison test. h Cilia number per KV was not affected by loss of Orc4 or Orc6. p = 0.3936 (CTRL MO vs. Orc4 splMO), p = 0.9050 (CTRL MO vs. Orc6 splMO). n = 9 (Orc4 splMO) and 10 KVs (CTRL MO, Orc6 splMO). One-way ANOVA with Dunnett’s multiple comparison test. i The KV area shows a tendency to be smaller upon KD of Orc4 and 6. p = 0.0551 (CTRL MO vs. Orc4 splMO), p = 0.03285 (CTRL MO vs. Orc6 splMO). n = 9–15 embryos in total. One-way ANOVA with Dunnett’s multiple comparison test. c–e Stacked bar graphs analysed using two-tailed Fisher’s exact tests. g–i Means ± SEM
Discussion
Previously, using human cells we found that deficiency of ORC1 as well as ORC4 and ORC6 causes cilia signalling defects, including impaired Hedgehog signalling. Cilia forming in ORC1-deficient human cells were slightly shorter than cilia forming in normal cells. Here, we focus on examining cilia function in vivo in an organism depleted of different ORC components. Similar to the situation in cultured human cells, we find that cilia forming in the KV in zebrafish are subtly shorter than those in control zebrafish. Additionally, Orc1 as well as Orc4 and Orc6 deficiency in zebrafish causes a phenotype, which includes growth retardation, LR asymmetry abnormalities and the formation of cystic kidneys, typical of cilia dysfunction. This consolidates and extends our previous findings that cilia dysfunction is a consequence of the deficiency in ORC raising the possibility that it could contribute to the clinical manifestation of ORC-dependent MGS.
A comparison of MGS and certain ciliopathy syndromes reveals overlapping phenotypic abnormalities. For example, Joubert syndrome patients with genetic variants in KIF7, a protein accumulating at the tip of cilia [31] and required for centrosomal and ciliary integrity [32], display overall growth retardation [32, 33]. Similarly, children with primary cilia dyskinesia (PCD) are smaller than healthy children and grow more slowly [34]. Furthermore, microcephaly occurs frequently in nephronophthisis, a ciliopathy with early onset renal failure due to cyst formation and fibrosis [35]. It was also reported that genetic alterations of the intraflagellar transport gene IFT172 results in growth retardation [36]. IFT172 is a ciliary protein [37] implicated in the development of the ciliopathy Bardet–Biedl syndrome [38]. Mice deficient in sperm flagellar protein 2 (SPEF2), which is associated with male infertility and PCD, are smaller, have shorter long bones and reduced osteoblast differentiation [39]. Importantly, in vitro we found reduced osteoblast differentiation upon ORC loss-of-function [20]. Vice versa, other forms of primordial dwarfism such as those linked to POC1A are associated with shorter cilia [40]. ATR, which is often mutated in Seckel syndrome, another microcephalic disorder [41], also regulates cilia formation as well as ciliary signalling [42]. Interestingly, ATR is also required during DNA replication, particularly for progression of stalled forks [43] and appears to have additional functions on ciliogenesis. Such noncanonical function in nonreplicating cells has been also observed for components of the MCM complex, which functions as the unwinding helicase during DNA replication, but appears to govern faithful ciliogenesis in quiescent cells [44].
Collectively, these data and reports argue that cilia dysfunction can also confer a clinical manifestation which overlaps that observed in MGS. Thus, a role for ORC in regulating cilia development may contribute to the clinical manifestation of ORC-deficient MGS in addition to the canonical role for ORC in replication. This novel function of the ORC could arise from an impact on the centrosome. The centrosome is indispensable for cilium formation as it comprises the basal body, from which the ciliary axoneme extends [29]. ORC1 for instance localises to the centrosome [45] and prevents reduplication events, which lead to supernumerary centrioles [19, 20, 46]. Consistently, many cilia defects have been associated with aberrant centriole number [16, 47–49].
Taken together, we provide supporting evidence that deficiency in three ORC components (ORC1, 4 and 6) confers defects in ciliogenesis with an impact on development in the model organism zebrafish. Thus, it is likely that the whole ORC complex is required for ciliogenesis rather than simply ORC1. This raises the possibility that this non-canonical function of ORC could contribute to the clinical manifestation observed in MGS.
Supplementary information
Acknowledgements
We thank Sandra Burczyk and Sophia Aicher for excellent fish care.
Authors’ contributions
LM, TCT, JG and CD performed experiments, LM, TCT, JG and MP analysed data, PAJ and MP conceived the study, designed the experiments and wrote the manuscript.
Compliance with ethical standards
Financial support
Medical Research Council (G0000502; and G0500897, both to PAJ); Deutsche Forschungsgemeinschaft [DFG PH144/4; and DFG PH144/6 to MP]; Boehringer Ingelheim Ulm University BioCenter (to MP). TCT and LDM were fellows of the International Graduate School in Molecular Medicine at Ulm University.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Lars D. Maerz, Teresa Casar Tena.
Supplementary information
The online version of this article (10.1038/s41431-019-0338-0) contains supplementary material, which is available to authorised users.
References
- 1.de Munnik SA, Hoefsloot EH, Roukema J, et al. Meier–Gorlin syndrome. Orphanet J Rare Dis. 2015;10:114. doi: 10.1186/s13023-015-0322-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Munnik SA, Otten BJ, Schoots J, et al. Meier–Gorlin syndrome: growth and secondary sexual development of a microcephalic primordial dwarfism disorder. Am J Med Genet A. 2012;158A:2733–42. doi: 10.1002/ajmg.a.35681. [DOI] [PubMed] [Google Scholar]
- 3.de Munnik SA, Bicknell LS, Aftimos S, et al. Meier–Gorlin syndrome genotype–phenotype studies: 35 individuals with pre-replication complex gene mutations and 10 without molecular diagnosis. Eur J Hum Genet. 2012;20:598–606. doi: 10.1038/ejhg.2011.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bicknell LS, Walker S, Klingseisen A, et al. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier–Gorlin syndrome. Nat Genet. 2011;43:350–5. doi: 10.1038/ng.776. [DOI] [PubMed] [Google Scholar]
- 5.Guernsey DL, Matsuoka M, Jiang H, et al. Mutations in origin recognition complex gene ORC4 cause Meier–Gorlin syndrome. Nat Genet. 2011;43:360–4. doi: 10.1038/ng.777. [DOI] [PubMed] [Google Scholar]
- 6.Shalev SA, Khayat M, Etty DS, Elpeleg O. Further insight into the phenotype associated with a mutation in the ORC6 gene, causing Meier–Gorlin syndrome 3. Am J Med Genet A. 2015;167A:607–11. doi: 10.1002/ajmg.a.36906. [DOI] [PubMed] [Google Scholar]
- 7.Yao L, Chen J, Wu X, Jia S, Meng A. Zebrafish cdc6 hypomorphic mutation causes Meier–Gorlin syndrome-like phenotype. Hum Mol Genet. 2017;26:4168–80. doi: 10.1093/hmg/ddx305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vetro A, Savasta S, Russo Raucci A, et al. MCM5: a new actor in the link between DNA replication and Meier–Gorlin syndrome. Eur J Hum Genet. 2017;25:646–50. doi: 10.1038/ejhg.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burrage LC, Charng WL, Eldomery MK, et al. De novo GMNN mutations cause autosomal-dominant primordial dwarfism associated with Meier–Gorlin syndrome. Am J Hum Genet. 2015;97:904–13. doi: 10.1016/j.ajhg.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fenwick AL, Kliszczak M, Cooper F, et al. Mutations in CDC45, encoding an essential component of the pre-initiation complex, cause Meier–Gorlin syndrome and craniosynostosis. Am J Hum Genet. 2016;99:125–38. doi: 10.1016/j.ajhg.2016.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Botchan M, Berger J. DNA replication: making two forks from one prereplication complex. Mol Cell. 2010;40:860–1. doi: 10.1016/j.molcel.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shibata E, Kiran M, Shibata Y, Singh S, Kiran S, Dutta A. Two subunits of human ORC are dispensable for DNA replication and proliferation. Elife 2016; 5:e19084. [DOI] [PMC free article] [PubMed]
- 13.Chavali P. L., Putz M., Gergely F. Small organelle, big responsibility: the role of centrosomes in development and disease. Philosophical Transactions of the Royal Society B: Biological Sciences. 2014;369(1650):20130468–20130468. doi: 10.1098/rstb.2013.0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Neill RS, Schoborg TA, Rusan NM. Same but different: pleiotropy in centrosome-related microcephaly. Mol Biol Cell. 2018;29:241–6. doi: 10.1091/mbc.E17-03-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poulton JS, Cuningham JC, Peifer M. Centrosome and spindle assembly checkpoint loss leads to neural apoptosis and reduced brain size. J Cell Biol. 2017;216:1255–65. doi: 10.1083/jcb.201607022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin CA, Ahmad I, Klingseisen A, et al. Mutations in PLK4, encoding a master regulator of centriole biogenesis, cause microcephaly, growth failure and retinopathy. Nat Genet. 2014;46:1283–92. doi: 10.1038/ng.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo J, Higginbotham H, Li J, et al. Developmental disruptions underlying brain abnormalities in ciliopathies. Nat Commun. 2015;6:7857. doi: 10.1038/ncomms8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alcantara D, O’Driscoll M. Congenital microcephaly. Am J Med Genet C Semin Med Genet. 2014;166C:124–39. doi: 10.1002/ajmg.c.31397. [DOI] [PubMed] [Google Scholar]
- 19.Hossain M, Stillman B. Meier–Gorlin syndrome mutations disrupt an Orc1 CDK inhibitory domain and cause centrosome reduplication. Genes Dev. 2012;26:1797–810. doi: 10.1101/gad.197178.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stiff T, Alagoz M, Alcantara D, et al. Deficiency in origin licensing proteins impairs cilia formation: implications for the aetiology of Meier–Gorlin syndrome. PLoS Genet. 2013;9:e1003360. doi: 10.1371/journal.pgen.1003360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perner B, Englert C, Bollig F. The Wilms tumor genes wt1a and wt1b control different steps during formation of the zebrafish pronephros. Dev Biol. 2007;309:87–96. doi: 10.1016/j.ydbio.2007.06.022. [DOI] [PubMed] [Google Scholar]
- 22.Amack JD, Yost HJ. The T box transcription factor no tail in ciliated cells controls zebrafish left-right asymmetry. Curr Biol. 2004;14:685–90. doi: 10.1016/j.cub.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Philipp M, Berger IM, Just S, Caron MG. Overlapping and opposing functions of G protein-coupled receptor kinase 2 (GRK2) and GRK5 during heart development. J Biol Chem. 2014;289:26119–30. doi: 10.1074/jbc.M114.551952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- 25.Burkhalter MD, Fralish GB, Premont RT, Caron MG, Philipp M. Grk5l controls heart development by limiting mTOR signaling during symmetry breaking. Cell Rep. 2013;4:625–32. doi: 10.1016/j.celrep.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaffe KM, Thiberge SY, Bisher ME, Burdine RD. Imaging cilia in zebrafish. Methods Cell Biol. 2010;97:415–35. doi: 10.1016/S0091-679X(10)97022-2. [DOI] [PubMed] [Google Scholar]
- 27.Sun Z, Amsterdam A, Pazour GJ, Cole DG, Miller MS, Hopkins N. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development. 2004;131:4085–93. doi: 10.1242/dev.01240. [DOI] [PubMed] [Google Scholar]
- 28.Choi SY, Chacon-Heszele MF, Huang L, et al. Cdc42 deficiency causes ciliary abnormalities and cystic kidneys. J Am Soc Nephrol. 2013;24:1435–50. doi: 10.1681/ASN.2012121236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends Genet. 2011;27:307–15. doi: 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Kendrick C, Julich D, Holley SA. Cell cycle progression is required for zebrafish somite morphogenesis but not segmentation clock function. Development. 2008;135:2065–70. doi: 10.1242/dev.022673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Endoh-Yamagami S, Evangelista M, Wilson D, et al. The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr Biol. 2009;19:1320–6. doi: 10.1016/j.cub.2009.06.046. [DOI] [PubMed] [Google Scholar]
- 32.Dafinger C, Liebau MC, Elsayed SM, et al. Mutations in KIF7 link Joubert syndrome with Sonic Hedgehog signaling and microtubule dynamics. J Clin Invest. 2011;121:2662–7. doi: 10.1172/JCI43639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tunovic S, Baranano KW, Barkovich JA, Strober JB, Jamal L, Slavotinek AM. Novel KIF7 missense substitutions in two patients presenting with multiple malformations and features of acrocallosal syndrome. Am J Med Genet A. 2015;167A:2767–76. doi: 10.1002/ajmg.a.37249. [DOI] [PubMed] [Google Scholar]
- 34.Svobodova T, Djakow J, Zemkova D, Cipra A, Pohunek P, Lebl J. Impaired growth during childhood in patients with primary ciliary dyskinesia. Int J Endocrinol. 2013;2013:731423. doi: 10.1155/2013/731423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halbritter J, Porath JD, Diaz KA, et al. Identification of 99 novel mutations in a worldwide cohort of 1056 patients with a nephronophthisis-related ciliopathy. Hum Genet. 2013;132:865–84. doi: 10.1007/s00439-013-1297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lucas-Herald AK, Kinning E, Iida A, et al. A case of functional growth hormone deficiency and early growth retardation in a child with IFT172 mutations. J Clin Endocrinol Metab. 2015;100:1221–4. doi: 10.1210/jc.2014-3852. [DOI] [PubMed] [Google Scholar]
- 37.Gorivodsky M, Mukhopadhyay M, Wilsch-Braeuninger M, et al. Intraflagellar transport protein 172 is essential for primary cilia formation and plays a vital role in patterning the mammalian brain. Dev Biol. 2009;325:24–32. doi: 10.1016/j.ydbio.2008.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bujakowska KM, Zhang Q, Siemiatkowska AM, et al. Mutations in IFT172 cause isolated retinal degeneration and Bardet–Biedl syndrome. Hum Mol Genet. 2015;24:230–42. doi: 10.1093/hmg/ddu441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lehti MS, Henriksson H, Rummukainen P, et al. Cilia-related protein SPEF2 regulates osteoblast differentiation. Sci Rep. 2018;8:859. doi: 10.1038/s41598-018-19204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaheen R, Faqeih E, Shamseldin HE, et al. POC1A truncation mutation causes a ciliopathy in humans characterized by primordial dwarfism. Am J Hum Genet. 2012;91:330–6. doi: 10.1016/j.ajhg.2012.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 42.Stiff T, Casar Tena T, O’Driscoll M, Jeggo PA, Philipp M. ATR promotes cilia signalling: links to developmental impacts. Hum Mol Genet. 2016;25:1574–87. doi: 10.1093/hmg/ddw034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bastos de Oliveira FM, Kim D, Cussiol JR, et al. Phosphoproteomics reveals distinct modes of Mec1/ATR signaling during DNA replication. Mol Cell. 2015;57:1124–32. doi: 10.1016/j.molcel.2015.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casar Tena Teresa, Maerz Lars D, Szafranski Karol, Groth Marco, Blätte Tamara J, Donow Cornelia, Matysik Sabrina, Walther Paul, Jeggo Penelope A, Burkhalter Martin D, Philipp Melanie. Resting cells rely on the DNA helicase component MCM2 to build cilia. Nucleic Acids Research. 2018;47(1):134–151. doi: 10.1093/nar/gky945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferguson RL, Pascreau G, Maller JL. The cyclin A centrosomal localization sequence recruits MCM5 and Orc1 to regulate centrosome reduplication. J Cell Sci. 2010;123:2743–9. doi: 10.1242/jcs.073098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hemerly AS, Prasanth SG, Siddiqui K, Stillman B. Orc1 controls centriole and centrosome copy number in human cells. Science. 2009;323:789–93. doi: 10.1126/science.1166745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coelho PA, Bury L, Shahbazi MN, et al. Over-expression of Plk4 induces centrosome amplification, loss of primary cilia and associated tissue hyperplasia in the mouse. Open Biol. 2015;5:150209. doi: 10.1098/rsob.150209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ogungbenro Yetunde Adesanya, Tena Teresa Casar, Gaboriau David, Lalor Pierce, Dockery Peter, Philipp Melanie, Morrison Ciaran G. Centrobin controls primary ciliogenesis in vertebrates. The Journal of Cell Biology. 2018;217(4):1205–1215. doi: 10.1083/jcb.201706095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.David A, Liu F, Tibelius A, et al. Lack of centrioles and primary cilia in STIL(−/−) mouse embryos. Cell Cycle. 2014;13:2859–68. doi: 10.4161/15384101.2014.946830. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.