Abstract
Background
We examined the associations between germline variants and breast cancer mortality using a large meta-analysis of women of European ancestry.
Methods
Meta-analyses included summary estimates based on Cox models of twelve datasets using ~10.4 million variants for 96,661 women with breast cancer and 7697 events (breast cancer-specific deaths). Oestrogen receptor (ER)-specific analyses were based on 64,171 ER-positive (4116) and 16,172 ER-negative (2125) patients. We evaluated the probability of a signal to be a true positive using the Bayesian false discovery probability (BFDP).
Results
We did not find any variant associated with breast cancer-specific mortality at P < 5 × 10−8. For ER-positive disease, the most significantly associated variant was chr7:rs4717568 (BFDP = 7%, P = 1.28 × 10−7, hazard ratio [HR] = 0.88, 95% confidence interval [CI] = 0.84–0.92); the closest gene is AUTS2. For ER-negative disease, the most significant variant was chr7:rs67918676 (BFDP = 11%, P = 1.38 × 10−7, HR = 1.27, 95% CI = 1.16–1.39); located within a long intergenic non-coding RNA gene (AC004009.3), close to the HOXA gene cluster.
Conclusions
We uncovered germline variants on chromosome 7 at BFDP < 15% close to genes for which there is biological evidence related to breast cancer outcome. However, the paucity of variants associated with mortality at genome-wide significance underpins the challenge in providing genetic-based individualised prognostic information for breast cancer patients.
Subject terms: Breast cancer, Prognosis, Prognostic markers, Cancer genetics
BACKGROUND
Breast cancer is the most common cancer in the Western world and accounts for 15% of cancer-related deaths in women, with about 522,000 deaths worldwide in 2012.1 Survival after a diagnosis of breast cancer varies considerably between patients even with closely matching tumour characteristics. Models that predict the likelihood of survival after breast cancer treatment use tumour and treatment data, but currently do not take host factors into account. The identification of prognostic and predictive biomarkers inherent in the germline of the patients rather than the tumour could pinpoint mechanisms of tumour progression and help with treatment stratification to increase therapeutic benefit. Such markers include inherited genetic variation, as there is evidence for heritability of breast cancer-specific mortality in affected first-degree relatives.2–5 Germline variation may affect prognosis by affecting tumour biology, since such variants are known to be associated with risk of specific breast tumour subtypes, particularly those defined by hormone receptor status, and have different outcomes.6–8 Germline genotype could also affect the efficacy of adjuvant drug therapies9,10 or might condition the host tumour environment via vascularisation,11,12 metastatic pattern,13,14 stroma–tumour interaction15,16 and immune surveillance.17,18
The association between common germline genetic variation and breast cancer-specific mortality has been examined in many candidate gene studies,5,9,14,19–36 as well as in moderate-sized genome-wide association studies (GWAS).37–41 However, it has been difficult link GWAS results to plausible candidate genes and few have been convincingly replicated.29,42 Large studies with long follow-up and reliable data on known prognostic factors are required if novel alleles associated with prognosis in breast cancer are to be identified at a level of genome-wide significance. In the present work, we pooled genotype data from multiple breast cancer GWAS discovery and replication efforts43,44 with new genotype data obtained from a large breast cancer series genotyped using the OncoArray chip.45,46 We examined associations with risk of breast cancer-specific mortality in a total of 96,661 breast cancer patients with survival time data. We then investigated the potential functional role of the selected variants by predicting possible target genes.
Materials and methods
Breast cancer patient samples
We included data from twelve datasets (n = 96,661) in which multiple breast cancer patient cohorts were genotyped by a variety of arrays providing genome-wide coverage of common variants. An overview of the datasets with specification of the arrays used is given in Supplementary Table 1. Data from eight of these datasets have been used in previous analyses (n = 37,954).44 However, the Collaborative Oncological Gene-Environment Study (COGS) dataset from the Breast Cancer Association Consortium (BCAC) was updated to include additional follow-up and death events and additional genotype data, increasing the number of events and samples to a total of n = 29,959 patients. Two new datasets, the BCAC OncoArray and the SUCCESS A trial, comprising 58,027 samples, were added for the current analyses.
The OncoArray is a custom Illumina genotyping array designed by the Genetic Associations and Mechanisms in Oncology (GAME-ON) consortium. It includes 533,000 variants of which 260,660 form a GWAS backbone, with the remainder being custom content, details of which have been described previously.45 The SUCCESS-A Study47 is a randomised phase III study of n = 3,299 breast cancer cases. Cases from the trial were genotyped using the Illumina Human OmniExpress array. We downloaded imputed genotypes from dbGaP (data reference 6266).
COGS samples that were also genotyped on the OncoArray were removed from the COGS dataset (n = 14,426). Female patients with invasive breast cancer diagnosed at age > 18 years, and with follow-up data available were included in the analyses. BCAC data from freeze 8 was used, in which 873 COGS samples with unknown breast cancer-specific mortality status were excluded from the analyses. All stages of cancer, including metastatic, were used in the analysis. Some individual studies applied additional selection criteria such as young age or early breast cancer stage (Supplementary Table 2).
Genotype and sample quality control, ancestry analysis and imputation
The genotype and sample quality control for the datasets have been described previously.44,45,47,48 Ancestry outliers for each dataset were identified by multidimensional scaling or LAMP49 on the basis of a set of unlinked variants and HapMap2 populations. Samples of European ancestry were retained for analyses.
Ten of the datasets were imputed using the reference panel from the 1000 Genomes Project in a two-stage procedure. The 1000 Genomes project Phase 3 (October 2014) release was used as the reference panel for all the datasets apart from SUCCESS-A, which used the Phase 1 release (March 2012). Imputation for CGEMS and BPC3 was performed using the programme MACH.50 Phased genotypes were first derived using SHAPEIT51 and IMPUTE252 and then used to perform imputation on the phased data. The main analyses were based on variants that were imputed with imputation r2 > 0.3 and had minor allele frequency (MAF) > 0.01 in at least one of the datasets leading to ~10.4 million variants. To match the individual datasets in the meta-analysis we used the chromosome position. Variants were kept in the analysis as long as they were present in one of the studies. In those cases where there was ambiguity over the naming of the insertions and deletions, the MAF was used for further matching.
Statistical and bioinformatic methods
Time-to-event was calculated from the date of diagnosis. For prevalent cases with study entry after diagnosis left truncation was applied, i.e., follow-up started at the date of study entry.53 Follow-up was right censored on the date of death, on the date last known alive if death did not occur, or at 15 years after diagnosis, whichever came first. We chose the 15 years cut-off because follow-up varied between studies and after that period follow-up data became scarce. Follow-up of the cohorts is illustrated in Kaplan Meier curves (Supplementary Figure 1).
The hazard ratios (HR) for the association of genotypes with breast cancer-specific mortality were estimated using Cox proportional hazards regression54 implemented in an in-house programme written in C++. Analysis of the CGEMS and BPC3 data was conducted using ProbABEL.55 The estimates of the individual studies were combined using an inverse-variance weighted meta-analysis. Since meta-analysis results based on the Wald test have been shown to be inflated for rare variants56 we recomputed the standard errors based on the likelihood ratio test statistic (see details in Supplementary methods), using the formula:
For each dataset we included as covariates a variable number of principal components (Supplementary Table 1) from the ancestry analysis as covariates in order to control for cryptic population substructure. The Cox models were stratified by country for the OncoArray dataset and by study for the COGS dataset. Statistical tests were performed for each variant by combining the results for all the datasets using a fixed-effects meta-analysis. Inflation of the test statistics (λ) was estimated by dividing the 45th percentile of the test statistic by 0.357 (the 45th percentile for a χ2 distribution on 1 degree of freedom). Analyses were carried out for all invasive breast cancer and for oestrogen receptor (ER)-positive and ER-negative disease separately.
To assess the probability of a variant being a false positive we used a Bayesian false discovery probability (BFDP)57 test based on the P value, a prior set to 0.0001 and an upper likely HR of 1.3.
To predict potential target genes, we used Bedtools v2.26 to intersect notable variants with genomic annotation data relevant to gene regulation activity in samples derived from breast tissue. We examined features including enhancers, promoters and transcription factor binding sites identified by the Roadmap58 and ENCODE59 Projects. Expression quantitative loci (eQTL) data from GTEx60 were queried for evidence of potential cis-regulatory activity.
Results
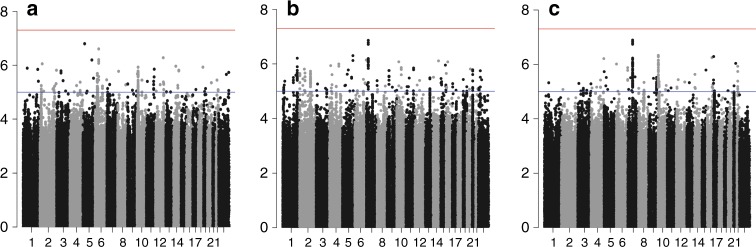
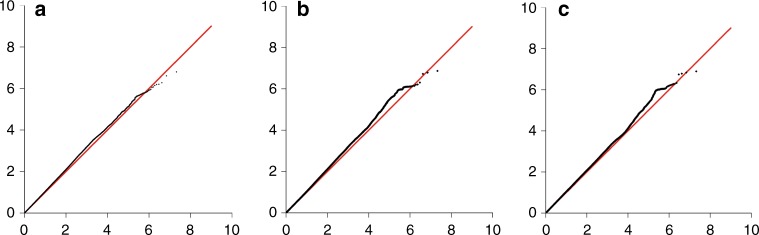
Genotype data from 96,661 breast cancer cases (64,171 ER-positive and 16,172 ER-negative) with 7697 breast cancer deaths within 15 years were included in the primary analyses. For 16,318 cases we did not have ER-status information. The average follow-up time was 6.38 years. Details of the numbers of samples and events in each dataset are given in Supplementary Table 3. Manhattan and quantile-quantile (Q–Q) plots for the associations between variants and breast cancer-specific mortality of all invasive, ER-negative and ER-positive breast cancers are shown in Fig. 1 and Fig. 2, respectively. There was some evidence of inflation of the test statistic with an inflation factor of 1.06 for all invasive and ER-positive, and 1.05 for ER-negative including all variants. These Q–Q plots showed no evidence of an association at P < 5 × 10−8; at less stringent thresholds for significance, there were an increasing number of observed associations for all three analyses (Fig. 2).
Fig. 1.
Association plot for the meta-analysis of the twelve datasets for breast cancer-specific mortality analyses (censored at 15 years) for a all breast tumours (censored at 15 years), b ER-negative tumours and c ER-positive tumours. The y-axis shows the −log10 P values of each variant analysed, and the x-axis shows their chromosome position. The red horizontal line represents P = 5 × 10−8
Fig. 2.
Q–Q plots for the meta-analysis of the twelve datasets for breast cancer-specific mortality analyses (censored at 15 years) for a all breast cancer tumours (censored at 15 years), b ER-negative tumours and c ER-positive tumours. The y-axis represents the observed −log10 P value, and the x-axis represents the expected −log10 P value. The red line represents the expected distribution under the null hypothesis of no association. Analyses were not corrected for LD-structure
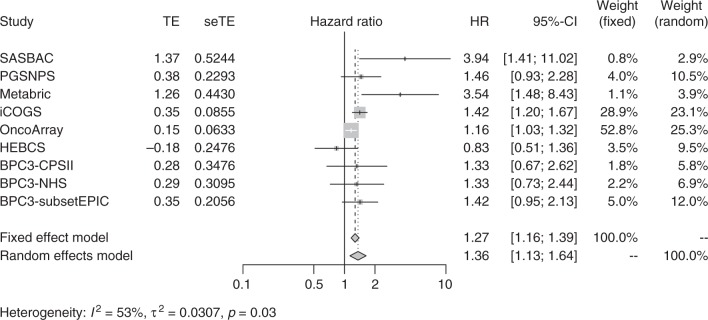
We identified three variants at BFDP < 15% associated with breast cancer-specific mortality of patients with ER-negative disease (Table 1). These variants are part of an independent set of 32 highly correlated variants61 on chromosome 7q21.1 that were associated at P < 5 × 10−6 (Supplementary Table 4). The LD matrix between these variants computed based on the 1000 European genomes,62,63 and their chromosomal positions, are shown in Supplementary Figure 1. The strongest association was for rs67918676: HR = 1.27; 95% CI = 1.16–1.39; P = 1.38 × 10−7; risk allele A frequency = 0.12 and BFDP = 11%. The imputation efficiency for this variant was high, with r2 = 0.99 for all datasets.
Table 1.
Results of the variants with BFDP < 15% in the meta-analysis of the 12 studies of breast cancer-specific mortality
| Subgroup | Variant | Chr | Position | Alt | Ref | Eaf_Ref | HR | LCL | UCL | P value | BFDP |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ER-negative | rs67918676:27445956:A:AT | 7 | 27445956 | AT | A | 0.12 | 1.27 | 1.16 | 1.39 | 1.38 × 10−7 | 0.11 |
| ER-negative | rs192185001:27448012:A:AT | 7 | 27448012 | AT | A | 0.12 | 1.27 | 1.16 | 1.39 | 1.66 × 10−7 | 0.13 |
| ER-negative | rs145963877:27473909:CAG:C | 7 | 27473909 | C | CAG | 0.11 | 1.28 | 1.17 | 1.41 | 1.91 × 10−7 | 0.15 |
| ER-positive | rs4717568:70400700:T:C | 7 | 70400700 | C | T | 0.62 | 0.88 | 0.8 | 0.92 | 1.28 × 10−7 | 0.07 |
| ER-positive | rs1917618:70396442:T:A | 7 | 70396442 | A | T | 0.62 | 0.88 | 0.84 | 0.93 | 1.46 × 10−7 | 0.08 |
| ER-positive | rs1546774:70398441:T:G | 7 | 70398441 | G | T | 0.62 | 0.88 | 0.84 | 0.93 | 1.66 × 10−7 | 0.09 |
| ER-positive | rs1546773:70398437:T:C | 7 | 70398437 | C | T | 0.62 | 0.88 | 0.84 | 0.93 | 1.81 × 10−7 | 0.10 |
| All | rs370332736:50395136:AACTT:A | 6 | 50395136 | A | AACTT | 0.09 | 1.16 | 1.10 | 1.24 | 2.48 × 10−7 | 0.13 |
The lead variant rs67918676 is located in an intron of a long intergenic non-coding RNA gene, LOC105375207 (AC004009.3), in close proximity to the HOXA gene cluster and the lncRNA HOTTIP. We tested the genes within a 500 MBp window around the 32 highly correlated variants for the association of their mRNA expression in breast tumours with recurrence-free survival using KMplotter (kmplot.com/analysis). Four of the ten closest genes with probes available showed moderate association with breast cancer survival at P < 0.005 (HOXA9, HOTTIP, EVX1 and TAX1BP1), with these associations mainly observed for ER-negative breast cancer (Supplementary Table 5A). Yet, intersecting the germline variants with several sources of genomic annotation information (e.g., chromosome conformation, enhancer–promoter correlations or gene expression) we could not find strong in silico evidence of gene regulation by the region containing the associated variants.
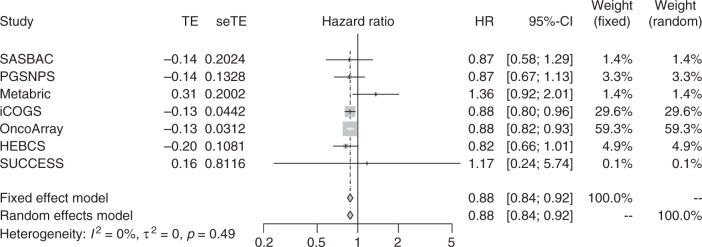
We also identified four variants at a BFDP < 15% associated with breast cancer-specific mortality of patients with ER-positive disease (Table 1). These variants were part of an independent set of 45 highly correlated variants on chromosome 7q11.22 that were associated at P < 5 × 10−6 (Supplementary Table 6). The LD matrix between these variants computed based on the 1000 European genomes,62,63 and their chromosomal positions, are shown in Supplementary Figure 3. The strongest association was for rs4717568: HR = 0.88; 95% CI:0.84–0.92; P = 1.28 × 10−7; risk allele A frequency = 0.62 and BFDP = 7%. The imputation efficiency for this variant was high, with an average r2 = 0.96 for all datasets. Two coding genes, AUTS2 and GALNT17, were located within a 500 MBp window around the 45 highly correlated variants, but the expression of neither of the two was associated with breast cancer survival in KMplotter analyses of TCGA data (Supplementary Table 5B).
The association of rs67918676 with ER-negative breast cancer was observed in eight of nine studies with no significant heterogeneity present at P < 0.01 (Fig. 3 and Supplementary Figure 4a). For ER-positive disease, the association of rs4717568 was detected in all seven studies with no heterogeneity present at P < 0.01 (Fig. 4 and Supplementary Figure 4b).
Fig. 3.
Forest plot showing the association between the ER-negative variant rs67918676 and breast cancer-specific mortality in ER-negative tumours for the datasets used in the meta-analysis. The size of the square reflects the size of the study (see also Supplementary Table 3)
Fig. 4.
Forest plot showing the association between the ER-positive variant rs4717568 and breast cancer-specific mortality in ER-positive tumours for the datasets used in the meta-analysis. The size of the square reflects the size of the study (see also Supplementary Table 3)
Apart from the 7q variants, only one isolated rare variant reached BFDP values below 15% for all tumours (Table 1). The variant, rs370332736: HR = 1.17; 95% CI: 1.10–1.24; P = 2.48 × 10−7; risk allele A frequency = 0.09 and BFDP = 13%, is located on chromosome 6 and has an average imputation efficiency of r2 = 0.96 for all datasets. In addition, there were several variants found at P < 10−6 for all three analyses (Supplementary Table 4, Supplementary Table 6 and Supplementary Table 7).
Discussion
In this large survival analysis, we report a genome-wide study for identifying genetic markers associated with breast cancer-specific mortality, involving 96,661 patients from a combined meta-analysis. We found one noteworthy region with 32 highly correlated variants on chromosome 7q21.1 for ER-negative. The lead variant rs67918676 (P = 1.38 × 10−7 and BFDP of 11% under reasonable assumptions for the prior probability of association) is located in a long intergenic non-coding RNA gene (AC004009.3). While this represents an uncharacterised transcript mainly expressed in testis and prostate, it is located about 200 kb away from a cluster of HOXA homeobox genes that has been implicated in breast cancer aetiology and prognosis.64,65 This region also contains HOTTIP, a lncRNA with prognostic value on clinical outcome in breast cancer.66 The flanking region on the opposite side contains TAX1BP1, a gene that may be involved in chemosensitivity.67 Interestingly, database mining using KMplotter revealed evidence for an association of the expression of these nearby genes with survival from ER-negative breast cancer. On the other hand, the enhancer activity at this noteworthy locus was predicted to be low based on the intersection with biofeatures characteristic of regulatory activity as no known eQTLs appear to exist in this region, suggesting that gene regulatory effects of the identified variants are limited in breast tissue or may be activated under certain untested conditions. For ER-positive tumours, we found another noteworthy region with 45 highly correlated variants at P < 5 × 10E−6 on chromosome 7q11.22. The lead variant rs4717568 (P = 1.28 × 10−7 and BFDP of 7%) is located between the AUTS2 and the GALNT17 genes. GALNT17 encodes an N-acetylgalactosaminyltransferase that may play a role in membrane trafficking.68 AUTS2 has been implicated in neurodevelopment,69 but AUTS2 overexpression in cancer has also been linked with resistance to chemotherapy and epithelial-to-mesenchymal transition.70 It has been postulated that overexpression of AUTS2 is specific for metastases,70 which may be consistent with the inconspicuous gene expression results in the TCGA database.
It is important to note the differences between the present and the previous GWAS study we had undertaken,44 the latter done in a much smaller dataset (3632 events versus 7697 events in the current study) that did not include the OncoArray study. The OncoArray study is the largest dataset used in the present meta-analysis and also the study with the highest imputation quality. The two previously reported variants (rs148760487 for all breast cancer tumours and rs2059614 for ER-negative tumours) were not associated with breast cancer-specific mortality in the current analyses (P = 1.59 × 10−3 and P = 5.41 × 10−4, respectively). The most likely explanation for this is that the original results were false-positive findings, despite the original association being nominally “genome-wide significant”. The BDFPs for the original reported associations were 54% and 16%, respectively. For the lead variants identified in the present analysis, we tested for differences in the imputation quality between the current and previous analysis. All variants had high imputation quality (~0.99) in the previous study, suggesting that the longer and more complete follow-up together with a higher number of events allowed more robust identification of breast cancer mortality associations. However, there are some weaknesses of the current meta-analysis such as heterogeneity between patient treatment over time and between countries and between datasets with different study designs that should be considered. These limitations, intrinsic to large survival meta-analyses, increase the noise and reduce the power to detect true associations.
In conclusion, we found two novel candidate regions at chromosome 7 for breast cancer survival, credible at a BFDP < 15% and associated with either ER-negative or ER-positive breast cancer-specific mortality. Concerning additional variants, we might still be underpowered to obtain a more comprehensive picture of genomic markers for breast cancer outcome. Overall, the role of germline variants in breast cancer mortality is still unclear36,37,71 and additional analyses with larger sample sizes and more complete follow-up including treatments are needed. In addition, alternative methods that integrate multiple data sources such as gene expression, protein–protein interactions or pathway analyses may be used to aggregate the effect of multiple variants with small effects.72 Such approaches could increase the power of the analyses while better explaining the underlying biological mechanisms associated with breast cancer mortality.
Supplementary information
Acknowledgements
BCAC: We thank all the individuals who took part in these studies and all the researchers, clinicians, technicians and administrative staff who have enabled this work to be carried out. We acknowledge all contributors to the COGS and OncoArray study design, chip design, genotyping and genotype analyses. ABCFS thank Maggie Angelakos, Judi Maskiell and Gillian Dite. ABCS thanks Frans Hogervorst, Sten Cornelissen and Annegien Broeks. ABCTB Investigators: Christine Clarke, Rosemary Balleine, Robert Baxter, Stephen Braye, Jane Carpenter, Jane Dahlstrom, John Forbes, Soon Lee, Debbie Marsh, Adrienne Morey, Nirmala Pathmanathan, Rodney Scott, Allan Spigelman, Nicholas Wilcken and Desmond Yip. Samples are made available to researchers on a non-exclusive basis. BBCS thanks Eileen Williams, Elaine Ryder-Mills and Kara Sargus. The BCINIS study would not have been possible without the contributions of Dr. K. Landsman, Dr. N. Gronich, Dr. A. Flugelman, Dr. W. Saliba, Dr. E. Liani, Dr. I. Cohen, Dr. S. Kalet, Dr. V. Friedman and Dr. O. Barnet of the NICCC in Haifa, and all the contributing family medicine, surgery, pathology and oncology teams in all medical institutes in Northern Israel. BIGGS thanks Niall McInerney, Gabrielle Colleran, Andrew Rowan and Angela Jones. The BREOGAN study would not have been possible without the contributions of the following: Manuela Gago-Dominguez, Jose Esteban Castelao, Angel Carracedo, Victor Muñoz Garzón, Alejandro Novo Domínguez, Maria Elena Martinez, Sara Miranda Ponte, Carmen Redondo Marey, Maite Peña Fernández, Manuel Enguix Castelo, Maria Torres, Manuel Calaza (BREOGAN), José Antúnez, Máximo Fraga and the staff of the Department of Pathology and Biobank of the University Hospital Complex of Santiago-CHUS, Instituto de Investigación Sanitaria de Santiago, IDIS, Xerencia de Xestion Integrada de Santiago—SERGAS; Joaquín González-Carreró and the staff of the Department of Pathology and Biobank of University Hospital Complex of Vigo, Instituto de Investigacion Biomedica Galicia Sur, SERGAS, Vigo, Spain. BSUCH thanks Peter Bugert, Medical Faculty Mannheim. CCGP thanks Styliani Apostolaki, Anna Margiolaki, Georgios Nintos, Maria Perraki, Georgia Saloustrou, Georgia Sevastaki and Konstantinos Pompodakis. CGPS thanks staff and participants of the Copenhagen General Population Study. For the excellent technical assistance: Dorthe Uldall Andersen, Maria Birna Arnadottir, Anne Bank and Dorthe Kjeldgård Hansen. The Danish Cancer Biobank is acknowledged for providing infrastructure for the collection of blood samples for the cases. CNIO-BCS thanks Guillermo Pita, Charo Alonso, Nuria Álvarez, Pilar Zamora, Primitiva Menendez and the Human Genotyping-CEGEN Unit (CNIO). Investigators from the CPS-II cohort thank the participants and Study Management Group for their invaluable contributions to this research. They also acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention National Programme of Cancer Registries, as well as cancer registries supported by the National Cancer Institute Surveillance Epidemiology and End Results programme. The CTS Steering Committee includes Leslie Bernstein, Susan Neuhausen, James Lacey, Sophia Wang, Huiyan Ma, and Jessica Clague DeHart at the Beckman Research Institute of City of Hope, Dennis Deapen, Rich Pinder, and Eunjung Lee at the University of Southern California, Pam Horn-Ross, Peggy Reynolds, Christina Clarke Dur and David Nelson at the Cancer Prevention Institute of California, Hoda Anton-Culver, Argyrios Ziogas, and Hannah Park at the University of California Irvine and Fred Schumacher at Case Western University. DIETCOMPLYF thanks the patients, nurses and clinical staff involved in the study. The DietCompLyf study was funded by the charity Against Breast Cancer (Registered Charity Number 1121258) and the NCRN. We thank the participants and the investigators of EPIC (European Prospective Investigation into Cancer and Nutrition). ESTHER thanks Hartwig Ziegler, Sonja Wolf, Volker Hermann, Christa Stegmaier and Katja Butterbach. FHRISK thanks NIHR for funding. GC-HBOC thanks Stefanie Engert, Heide Hellebrand, Sandra Kröber and LIFE—Leipzig Research Centre for Civilisation Diseases (Markus Loeffler, Joachim Thiery, Matthias Nüchter and Ronny Baber). The GENICA Network: Dr. Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart, and University of Tübingen, Germany [H.B. and W.Y.L.], German Cancer Consortium (DKTK) and German Cancer Research Center (DKFZ) [H.B.], Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany [Y.D.K., Christian Baisch], Institute of Pathology, University of Bonn, Germany [Hans-Peter Fischer], Molecular Genetics of Breast Cancer, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Germany [UH], Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), Bochum, Germany [Thomas Brüning, Beate Pesch, Sylvia Rabstein, Anne Lotz]; and Institute of Occupational Medicine and Maritime Medicine, University Medical Centre Hamburg-Eppendorf, Germany [Volker Harth]. HABCS thanks Michael Bremer. HEBCS thanks, Rainer Fagerholm, Kirsimari Aaltonen, Karl von Smitten, Irja Erkkilä. HUBCS thanks Shamil Gantsev. KARMA and SASBAC thank the Swedish Medical Research Counsel. KBCP thanks Eija Myöhänen, Helena Kemiläinen. kConFab/AOCS wish to thank Heather Thorne, Eveline Niedermayr, all the kConFab research nurses and staff, the heads and staff of the Family Cancer Clinics, and the Clinical Follow Up Study (which has received funding from the NHMRC, the National Breast Cancer Foundation, Cancer Australia, and the National Institute of Health (USA)) for their contributions to this resource, and the many families who contribute to kConFab. LMBC thanks Gilian Peuteman, Thomas Van Brussel, EvyVanderheyden and Kathleen Corthouts. MARIE thanks Petra Seibold, Judith Heinz, Nadia Obi, Alina Vrieling, Sabine Behrens, Ursula Eilber, Muhabbet Celik, Til Olchers and Stefan Nickels. MBCSG: Paolo Peterlongo, Bernard Peissel, Roberto Villa, Cristina Zanzottera, Irene Feroce, and the personnel of the Cogentech Cancer Genetic Test Laboratory. We thank the coordinators, the research staff and especially the MMHS participants for their continued collaboration on research studies in breast cancer. The following are NBCS Collaborators: Kristine K. Sahlberg (Ph.D.), Lars Ottestad (M.D.), Rolf Kåresen (Prof. Em.) Dr. Ellen Schlichting (M.D.), Marit Muri Holmen (M.D.), Toril Sauer (M.D.), Vilde Haakensen (M.D.), Olav Engebråten (M.D.), Bjørn Naume (M.D.), Alexander Fosså (M.D.), Cecile E. Kiserud (M.D.), Kristin V. Reinertsen (M.D.), Åslaug Helland (M.D.), Margit Riis (M.D.), Jürgen Geisler (M.D.) and OSBREAC. NHS/NHS2 would like to thank the participants and staff of the NHS and NHS2 for their valuable contributions as well as the following state cancer registries for their help: A.L., A.Z., A.R., C.A., C.O., C.T., D.E., F.L., G.A., I.D., I.L., I.N., I.A., K.Y., L.A., M.E., M.D., M.A., M.I., N.E., N.H., N.J., N.Y., N.C., N.D., O.H., O.K., O.R., P.A., R.I., S.C., T.N., T.X., V.A., W.A., W.Y. OBCS thanks Arja Jukkola-Vuorinen, Mervi Grip, Saila Kauppila, Meeri Otsukka, Leena Keskitalo and Kari Mononen for their contributions to this study. OFBCR thanks Teresa Selander and Nayana Weerasooriya. ORIGO thanks E. Krol-Warmerdam, and J. Blom for patient accrual, administering questionnaires and managing clinical information. PBCS thanks Louise Brinton, Mark Sherman, Neonila Szeszenia-Dabrowska, Beata Peplonska, Witold Zatonski, Pei Chao and Michael Stagner. The ethical approval for the POSH study is MREC/00/6/69, UKCRN ID: 1137. We thank staff in the Experimental Cancer Medicine Centre (ECMC) supported Faculty of Medicine Tissue Bank and the Faculty of Medicine DNA Banking resource. PREFACE thanks Sonja Oeser and Silke Landrith. PROCAS thanks NIHR for funding. RBCS thanks Petra Bos, Jannet Blom, Ellen Crepin, Elisabeth Huijskens, Anja Kromwijk-Nieuwlaat, Annette Heemskerk and the Erasmus MC Family Cancer Clinic. SBCS thanks Sue Higham, Helen Cramp, Dan Connley, Ian Brock, Sabapathy Balasubramanian and Malcolm W.R. Reed. We thank the SEARCH and EPIC teams. SKKDKFZS thanks all study participants, clinicians, family doctors, researchers and technicians for their contributions and commitment to this study. We thank the SUCCESS Study teams in Munich, Duessldorf, Erlangen and Ulm. We thank the SUCCESS Study teams in Munich, Duessldorf, Erlangen and Ulm. SZBCS thanks Ewa Putresza. UCIBCS thanks Irene Masunaka. UKBGS thanks Breast Cancer Now and the Institute of Cancer Research for support and funding of the Breakthrough Generations Study, and the study participants, study staff, and the doctors, nurses and other health care providers and health information sources who have contributed to the study. We acknowledge NHS funding to the Royal Marsden/ICR NIHR Biomedical Research Centre. The authors thank the WHI investigators and staff for their dedication and the study participants for making the programme possible. BCAC is funded by Cancer Research UK [C1287/A16563 and C1287/A10118], the European Union’s Horizon 2020 Research and Innovation Programme (Grant numbers 634935 and 633784 for BRIDGES and B-CAST, respectively), and by the European Community's Seventh Framework Programme under grant agreement number 223175 (Grant number HEALTH-F2-2009-223175) (COGS). The EU Horizon 2020 Research and Innovation Programme funding source had no role in study design, data collection, data analysis, data interpretation or writing of the report. Genotyping of the OncoArray was funded by the NIH Grant U19 CA148065, and Cancer UK Grant C1287/A16563 and the PERSPECTIVE project supported by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research (Grant GPH-129344) and, the Ministère de l’Économie, Science et Innovation du Québec through Genome Québec and the PSRSIIRI-701 grant, and the Quebec Breast Cancer Foundation. Funding for the iCOGS infrastructure came from: the European Community’s Seventh Framework Programme under grant agreement no. 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692 and C8197/A16565), the National Institutes of Health (CA128978) and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065 and 1U19 CA148112—the GAME-ON initiative), the Department of Defence (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, and Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund. The DRIVE Consortium was funded by U19 CA148065. ABCFS was supported by grant UM1 CA164920 from the National Cancer Institute (USA). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centres in the in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organisations imply endorsement by the USA Government or the BCFR. The ABCFS was also supported by the National Health and Medical Research Council of Australia, the New South Wales Cancer Council, the Victorian Health Promotion Foundation (Australia) and the Victorian Breast Cancer Research Consortium. J.L.H. is a National Health and Medical Research Council (NHMRC) Senior Principal Research Fellow. M.C.S. is a NHMRC Senior Research Fellow. The ABCS study was supported by the Dutch Cancer Society [Grants NKI 2007-3839; 2009-4363 and2015-7632]. The ABCTB is generously supported by the National Health and Medical Research Council of Australia, The Cancer Institute NSW and the National Breast Cancer Foundation. The work of the BBCC was partly funded by ELAN-Fond of the University Hospital of Erlangen. The BBCS is funded by Cancer Research UK and Breast Cancer Now and acknowledges NHS funding to the NIHR Biomedical Research Centre, and the National Cancer Research Network (NCRN). For the BCFR-NY, BCFR-PA, BCFR-UT this work was supported by grant UM1 CA164920 from the National Cancer Institute. For BIGGS, ES is supported by NIHR Comprehensive Biomedical Research Centre, Guy’s & St. Thomas’ NHS Foundation Trust in partnership with King’s College London, United Kingdom. IT is supported by the Oxford Biomedical Research Centre. The BREOGAN is funded by Acción Estratégica de Salud del Instituto de Salud Carlos III FIS PI12/02125/Cofinanciado FEDER; Acción Estratégica de Salud del Instituto de Salud Carlos III FIS Intrasalud (PI13/01136); Programa Grupos Emergentes, Cancer Genetics Unit, Instituto de Investigacion Biomedica Galicia Sur. Xerencia de Xestion Integrada de Vigo-SERGAS, Instituto de Salud Carlos III, Spain; Grant 10CSA012E, Consellería de Industria Programa Sectorial de Investigación Aplicada, PEME I + D e I + D Suma del Plan Gallego de Investigación, Desarrollo e Innovación Tecnológica de la Consellería de Industria de la Xunta de Galicia, Spain; Grant EC11-192. Fomento de la Investigación Clínica Independiente, Ministerio de Sanidad, Servicios Sociales e Igualdad, Spain; and Grant FEDER-Innterconecta. Ministerio de Economia y Competitividad, Xunta de Galicia, Spain. The BSUCH study was supported by the Dietmar-Hopp Foundation, the Helmholtz Society and the German Cancer Research Center (DKFZ). CCGP is supported by funding from the University of Crete. The CECILE study was supported by Fondation de France, Institut National du Cancer (INCa), Ligue Nationale contre le Cancer, Agence Nationale de Sécurité Sanitaire, de l’Alimentation, de l’Environnement et du Travail (ANSES), Agence Nationale de la Recherche (ANR). The CGPS was supported by the Chief Physician Johan Boserup and Lise Boserup Fund, the Danish Medical Research Council, and Herlev and Gentofte Hospital. The CNIO-BCS was supported by the Instituto de Salud Carlos III, the Red Temática de Investigación Cooperativa en Cáncer and grants from the Asociación Española Contra el Cáncer and the Fondo de Investigación Sanitario (PI11/00923 and PI12/00070). The American Cancer Society funds the creation, maintenance, and updating of the CPS-II cohort. The CTS was initially supported by the California Breast Cancer Act of 1993 and the California Breast Cancer Research Fund (Contract 97-10500) and is currently funded through the National Institutes of Health (R01 CA77398, UM1 CA164917 and U01 CA199277). Collection of cancer incidence data was supported by the California Department of Public Health as part of the statewide cancer reporting programme mandated by California Health and Safety Code Section 103885. The University of Westminster curates the DietCompLyf database funded by Against Breast Cancer Registered Charity No. 1121258 and the NCRN. The coordination of EPIC is financially supported by the European Commission (DG-SANCO) and the International Agency for Research on Cancer. The national cohorts are supported by: Ligue Contre le Cancer, Institut Gustave Roussy, Mutuelle Générale de l’Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); German Cancer Aid, German Cancer Research Center (DKFZ), Federal Ministry of Education and Research (BMBF) (Germany); the Hellenic Health Foundation, the Stavros Niarchos Foundation (Greece); Associazione Italiana per la Ricerca sul Cancro-AIRC-Italy and National Research Council (Italy); Dutch Ministry of Public Health, Welfare and Sports (VWS), Netherlands Cancer Registry (NKR), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF), Statistics Netherlands (The Netherlands); Health Research Fund (FIS), PI13/00061 to Granada, PI13/01162 to EPIC-Murcia, Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra, ISCIII RETIC (RD06/0020) (Spain); Cancer Research UK (14136 to EPIC-Norfolk; C570/A16491 and C8221/A19170 to EPIC-Oxford), Medical Research Council (1000143 to EPIC-Norfolk, MR/M012190/1 to EPIC-Oxford) (United Kingdom). The ESTHER study was supported by a grant from the Baden Württemberg Ministry of Science, Research and Arts. Additional cases were recruited in the context of the VERDI study, which was supported by a grant from the German Cancer Aid (Deutsche Krebshilfe). FHRISK is funded from NIHR grant PGfAR 0707-10031. The GC-HBOC is supported by the German Cancer Aid (Grant no. 110837, coordinator: Rita K. Schmutzler, Cologne). This work was also funded by the European Regional Development Fund and Free State of Saxony, Germany (LIFE—Leipzig Research Centre for Civilisation Diseases, project numbers 713-241202, 713-241202, 14505/2470 and 14575/2470). The GENICA was funded by the Federal Ministry of Education and Research (BMBF) Germany grants 01KW9975/5, 01KW9976/8, 01KW9977/0 and 01KW0114, the Robert Bosch Foundation, Stuttgart, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, the Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), Bochum, as well as the Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany. The GESBC was supported by the Deutsche Krebshilfe e. V. [70492] and the German Cancer Research Centre (DKFZ). The HABCS study was supported by the Claudia von Schilling Foundation for Breast Cancer Research, by the Lower Saxonian Cancer Society, and by the Rudolf Bartling Foundation. The HEBCS was financially supported by the Helsinki University Central Hospital Research Fund, Academy of Finland (266528), the Finnish Cancer Society, and the Sigrid Juselius Foundation. The HUBCS was supported by a grant from the German Federal Ministry of Research and Education (RUS08/017), and by the Russian Foundation for Basic Research and the Federal Agency for Scientific Organisations for support the Bioresource collections and RFBR grants 14-04-97088, 17-29-06014 and 17-44-020498. Financial support for KARBAC was provided through the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institutet, the Swedish Cancer Society, The Gustav V. Jubilee foundation and Bert von Kantzows foundation. The KARMA study was supported by Märit and Hans Rausings Initiative Against Breast Cancer. The KBCP was financially supported by the special Government Funding (EVO) of Kuopio University Hospital grants, Cancer Fund of North Savo, the Finnish Cancer Organisations, and by the strategic funding of the University of Eastern Finland. kConFab is supported by a grant from the National Breast Cancer Foundation, and previously by the National Health and Medical Research Council (NHMRC), the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia. LMBC is supported by the ‘Stichting tegen Kanker’. The MARIE study was supported by the Deutsche Krebshilfe e.V. [70-2892-BR I, 106332, 108253, 108419, 110826 and110828], the Hamburg Cancer Society, the German Cancer Research Centre (DKFZ) and the Federal Ministry of Education and Research (BMBF) Germany [01KH0402]. MBCSG is supported by grants from the Italian Association for Cancer Research (AIRC) and by funds from the Italian citizens who allocated the 5/1000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori, according to Italian laws (INT-Institutional strategic projects “5 × 1000”). The MCBCS was supported by the NIH grants CA192393, CA116167 and CA176785 an NIH Specialised Programme of Research Excellence (SPORE) in Breast Cancer [CA116201], and the Breast Cancer Research Foundation and a generous gift from the David F. and Margaret T. Grohne Family Foundation. MCCS cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 209057 and 396414, and by infrastructure provided by Cancer Council Victoria. Cases and their vital status were ascertained through the Victorian Cancer Registry (VCR) and the Australian Institute of Health and Welfare (AIHW), including the National Death Index and the Australian Cancer Database. The MEC was supported by NIH grants CA63464, CA54281, CA098758, CA132839 and CA164973. The MISS study is supported by funding from ERC-2011-294576 Advanced grant, Swedish Cancer Society, Swedish Research Council, Local hospital funds, Berta Kamprad Foundation, Gunnar Nilsson. The MMHS study was supported by NIH grants CA97396, CA128931, CA116201, CA140286 and CA177150. The work of MTLGEBCS was supported by the Quebec Breast Cancer Foundation, the Canadian Institutes of Health Research for the “CIHR Team in Familial Risks of Breast Cancer” programme—Grant # CRN-87521 and the Ministry of Economic Development, Innovation and Export Trade—grant # PSR-SIIRI-701. The NBCS has received funding from the K.G. Jebsen Centre for Breast Cancer Research; the Research Council of Norway grant 193387/V50 (to A.-L. Børresen-Dale and V.N. Kristensen) and grant 193387/H10 (to A.-L. Børresen-Dale and V.N. Kristensen), South Eastern Norway Health Authority (Grant 39346 to A.-L. Børresen-Dale) and the Norwegian Cancer Society (to A.-L. Børresen-Dale and V.N. Kristensen). The NC-BCFR and OFBCR were supported by grant UM1 CA164920 from the National Cancer Institute (USA). The NCBCS was funded by Komen Foundation, the National Cancer Institute (P50 CA058223, U54 CA156733 and U01 CA179715), and the North Carolina University Cancer Research Fund. The NHS was supported by NIH grants P01 CA87969, UM1 CA186107 and U19 CA148065. The NHS2 was supported by NIH grants UM1 CA176726 and U19 CA148065. The OBCS was supported by research grants from the Finnish Cancer Foundation, the Academy of Finland (Grant numbers 250083 and 122715, and Centre of Excellence grant number 251314), the Finnish Cancer Foundation, the Sigrid Juselius Foundation, the University of Oulu, the University of Oulu Support Foundation and the special Governmental EVO funds for Oulu University Hospital-based research activities. The ORIGO study was supported by the Dutch Cancer Society (RUL 1997-1505) and the Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL CP16). The PBCS was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA. Genotyping for PLCO was supported by the Intramural Research Programme of the National Institutes of Health, NCI, Division of Cancer Epidemiology and Genetics. The PLCO is supported by the Intramural Research Programme of the Division of Cancer Epidemiology and Genetics and supported by contracts from the Division of Cancer Prevention, National Cancer Institute, National Institutes of Health. The POSH study is funded by Cancer Research UK (Grants C1275/A11699, C1275/C22524, C1275/A19187 and C1275/A15956, and Breast Cancer Campaign grant numbers 2010PR62 and 2013PR044. PROCAS is funded from NIHR grant PGfAR 0707-10031. The RBCS was funded by the Dutch Cancer Society (DDHK 2004-3124 and DDHK 2009-4318). The SASBAC study was supported by funding from the Agency for Science, Technology and Research of Singapore (A*STAR), the US National Institute of Health (NIH) and the Susan G. Komen Breast Cancer Foundation. The SBCS was supported by Sheffield Experimental Cancer Medicine Centre and Breast Cancer Now Tissue Bank. SEARCH is funded by Cancer Research UK [C490/A10124 and C490/A16561] and supported by the UK National Institute for Health Research Biomedical Research Centre at the University of Cambridge. The University of Cambridge has received salary support for PDPP from the NHS in the East of England through the Clinical Academic Reserve. SKKDKFZS is supported by the DKFZ. The SMC is funded by the Swedish Cancer Foundation. The SZBCS was supported by Grant PBZ_KBN_122/P05/2004. The UCIBCS component of this research was supported by the NIH [CA58860, CA92044] and the Lon V Smith Foundation [LVS39420]. The UKBGS is funded by Breast Cancer Now and the Institute of Cancer Research (ICR), London. ICR acknowledges NHS funding to the NIHR Biomedical Research Centre. The USRT Study was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA. The WHI programme is funded by the National Heart, Lung, and Blood Institute, the US National Institutes of Health and the US Department of Health and Human Services (HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C and HHSN271201100004C). This work was also funded by NCI U19 CA148065-01.
Author contributions
M.K.S. and P.D.P.F. conceived the study. Q.G., M.E.G., S.K., C.J.T. and T.D. performed the data analyses. M.K.S., P.D.P.F., Q.G., M.E.G., T.D. and D.M.E. were involved in the interpretation of the data. J.D., D.F.E., P.D.P.F., S.C. and J.B. provided statistical and computational support for the data analyses. R.K., Q.W., M.K.B. and J.D. provided database support. M.E.G., Q.G., T.D., M.K.S. and P.D.P.F. wrote the first draft of the manuscript. All authors contributed data from their own studies, helped revise the manuscript and approved the final version.
Competing interests
The authors declare no competing interests.
Data availability
All estimates reported in the paper are available through the BCAC website: http://bcac.ccge.medschl.cam.ac.uk.
Ethics approval and consent to participate
The study was performed in accordance with the Declaration of Helsinki. All individual studies, from which data were used, were approved by the appropriate medical ethical committees and/or institutional review boards. All study participants provided informed consent.
Consent for publication
All authors consented to this publication.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shared first authorship: Maria Escala-Garcia, Qi Guo
Supplementary information
Supplementary information is available for this paper at 10.1038/s41416-019-0393-x.
References
- 1.IARC. http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx.
- 2.Hartman M, Lindström L, Dickman PW, Adami HO, Hall P, Czene K. Is breast cancer prognosis inherited? Breast Cancer Res. 2007;9:R39. doi: 10.1186/bcr1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindström LS, Hall P, Hartman M, Wiklund F, Grönberg H, Czene K. Familial concordance in cancer survival: a Swedish population-based study. Lancet Oncol. 2007;8:1001–6. doi: 10.1016/S1470-2045(07)70282-6. [DOI] [PubMed] [Google Scholar]
- 4.Udler M, Pharoah PD. Germline genetic variation and breast cancer survival: prognostic and therapeutic implications. Future Oncol. 2007;3:491–495. doi: 10.2217/14796694.3.5.491. [DOI] [PubMed] [Google Scholar]
- 5.Verkooijen HM, Hartman M, Usel M, Benhamou S, Neyroud-Caspar I, Czene K, et al. Breast cancer prognosis is inherited independently of patient, tumor and treatment characteristics. Int J. Cancer. 2012;130:2103–2110. doi: 10.1002/ijc.26206. [DOI] [PubMed] [Google Scholar]
- 6.Broeks A, Schmidt MK, Sherman ME, Couch FJ, Hopper JL, Dite GS, et al. Low penetrance breast cancer susceptibility loci are associated with specific breast tumor subtypes: findings from the Breast Cancer Association Consortium. Hum. Mol. Genet. 2011;20:3289–303. doi: 10.1093/hmg/ddr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang XR, Chang-Claude J, Goode EL, Couch FJ, Nevanlinna H, Milne RL, et al. Associations of breast cancer risk factors with tumor subtypes: a pooled analysis from the Breast Cancer Association Consortium studies. J. Natl. Cancer Inst. 2011;103:250–263. doi: 10.1093/jnci/djq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7:e1000279. doi: 10.1371/journal.pmed.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fagerholm R, Hofstetter B, Tommiska J, Aaltonen K, Vrtel R, Syrjäkoski K, et al. NAD(P)H:quinone oxidoreductase 1 NQO1*2 genotype (P187S) is a strong prognostic and predictive factor in breast cancer. Nat. Genet. 2008;40:844–53. doi: 10.1038/ng.155. [DOI] [PubMed] [Google Scholar]
- 10.Hoskins JM, Carey LA, McLeod HL. CYP2D6 and tamoxifen: DNA matters in breast cancer. Nat. Rev. Cancer. 2009;9:576–586. doi: 10.1038/nrc2683. [DOI] [PubMed] [Google Scholar]
- 11.Koutras A, Kotoula V, Fountzilas G. Prognostic and predictive role of vascular endothelial growth factor polymorphisms in breast cancer. Pharmacogenomics. 2015;16:79–94. doi: 10.2217/pgs.14.148. [DOI] [PubMed] [Google Scholar]
- 12.Hein A, Lambrechts D, von Minckwitz G, Häberle L, Eidtmann H, Tesch H, et al. Genetic variants in VEGF pathway genes in neoadjuvant breast cancer patients receiving bevacizumab: results from the randomized phase III GeparQuinto study. Int J. Cancer. 2015;137:2981–8. doi: 10.1002/ijc.29656. [DOI] [PubMed] [Google Scholar]
- 13.Hsieh SM, Lintell Na, Hunter KW. Germline polymorphisms are potential metastasis risk and prognosis markers in breast cancer. Breast Dis. 2007;26:157–62. doi: 10.3233/BD-2007-26114. [DOI] [PubMed] [Google Scholar]
- 14.Crawford NPS, Ziogas A, Peel DJ, Hess J, Anton-Culver H, Hunter KW. Germline polymorphisms in SIPA1 are associated with metastasis and other indicators of poor prognosis in breast cancer. Breast Cancer Res. 2006;8:R16. doi: 10.1186/bcr1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulsson J, Micke P. Prognostic relevance of cancer-associated fibroblasts in human cancer. Semin Cancer Biol. 2014;25:61–8. doi: 10.1016/j.semcancer.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Winslow S, Leandersson K, Edsjö A, Larsson C. Prognostic stromal gene signatures in breast cancer. Breast Cancer Res. 2015;17:23. doi: 10.1186/s13058-015-0530-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Van Eenoo F, et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J. Clin. Oncol. 2013;31:860–867. doi: 10.1200/JCO.2011.41.0902. [DOI] [PubMed] [Google Scholar]
- 18.Ali HR, Provenzano E, Dawson SJ, Blows FM, Liu B, Shah M, et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann. Oncol. J. Eur. Soc. Med. Oncol. 2014;25:1536–43. doi: 10.1093/annonc/mdu191. [DOI] [PubMed] [Google Scholar]
- 19.Udler M, Maia AT, Cebrian A, Brown C, Greenberg D, Shah M, et al. Common germline genetic variation in antioxidant defense genes and survival after diagnosis of breast cancer. J. Clin. Oncol. 2007;25:3015–23. doi: 10.1200/JCO.2006.10.0099. [DOI] [PubMed] [Google Scholar]
- 20.Einarsdóttir K, Darabi H, Li Y, Low YL, Li YQ, Bonnard C, et al. ESR1 and EGFgenetic variation in relation to breast cancer risk and survival. Breast Cancer Res. 2008;10:R15. doi: 10.1186/bcr1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fasching PA, Loehberg CR, Strissel PL, Lux MP, Bani MR, Schrauder M, et al. Single nucleotide polymorphisms of the aromatase gene (CYP19A1), HER2/neu status, and prognosis in breast cancer patients. Breast Cancer Res. Treat. 2008;112:89–98. doi: 10.1007/s10549-007-9822-2. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt MK, Tommiska J, Broeks A, van Leeuwen FE, Van’t Veer LJ, Pharoah PDP, et al. Combined effects of single nucleotide polymorphisms TP53 R72P and MDM2 SNP309, and p53 expression on survival of breast cancer patients. Breast Cancer Res. 2009;11:R89. doi: 10.1186/bcr2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varadi V, Brendle A, Brandt A, Johansson R, Enquist K, Henriksson R, et al. Polymorphisms in telomere-associated genes, breast cancer susceptibility and prognosis. Eur. J. Cancer. 2009;45:3008–3016. doi: 10.1016/j.ejca.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 24.Lin WY, Camp NJ, Cannon-Albright LA, Allen-Brady K, Balasubramanian S, Reed MWR, et al. A role for XRCC2 gene polymorphisms in breast cancer risk and survival. J. Med. Genet. 2011;48:477–484. doi: 10.1136/jmedgenet-2011-100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fasching PA, Pharoah PDP, Cox A, Nevanlinna H, Bojesen SE, Karn T, et al. The role of genetic breast cancer susceptibility variants as prognostic factors. Hum. Mol. Genet. 2012;21:3926–39. doi: 10.1093/hmg/dds159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrdahl M, Canzian F, Lindström S, Shui I, Black A, Hoover RN, et al. Association of breast cancer risk loci with breast cancer survival. Int J. Cancer. 2015;137:2837–2845. doi: 10.1002/ijc.29446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jamshidi M, Fagerholm R, Khan S, Aittomäki K, Czene K, Darabi H, et al. SNP–SNP interaction analysis of NF-κB signaling pathway on breast cancer survival. Oncotarget. 2015;6:37979–94. doi: 10.18632/oncotarget.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weischer M, Nordestgaard BG, Pharoah P, Bolla MK, Nevanlinna H, Van’t Veer LJ, et al. CHEK2*1100delC heterozygosity in women with breast cancer associated with early death, breast cancer-specific death, and increased risk of a second breast cancer. J. Clin. Oncol. 2012;30:4308–16. doi: 10.1200/JCO.2012.42.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pirie A, Guo Q, Kraft P, Canisius S, Eccles DM, Rahman N, et al. Common germline polymorphisms associated with breast cancer-specific survival. Breast Cancer Res. 2015;17:58. doi: 10.1186/s13058-015-0570-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ambrosone CB, Sweeney C, Coles BF, Thompson PA, McClure GY, Korourian S, et al. Polymorphisms in glutathione S-transferases (GSTM1 and GSTT1) and survival after treatment for breast cancer. Cancer Res. 2001;61:7130–5. [PubMed] [Google Scholar]
- 31.Goode EL, Dunning AM, Kuschel B, Healey CS, Day NE, Ponder BAJ, et al. Effect of germ-line genetic variation on breast cancer survival in a population-based study. Cancer Res. 2002;62:3052–7. [PubMed] [Google Scholar]
- 32.Ambrosone CB, Ahn J, Singh KK, Rezaishiraz H, Furberg H, Sweeney C, et al. Polymorphisms in genes related to oxidative stress (MPO, MnSOD, CAT) and survival after treatment for breast cancer. Cancer Res. 2005;65:1105–11. [PubMed] [Google Scholar]
- 33.Boersma BJ, Howe TM, Goodman JE, Yfantis HG, Lee DH, Chanock SJ, et al. Association of breast cancer outcome with status of p53 and MDM2 SNP309. J. Natl. Cancer Inst. 2006;98:911–9. doi: 10.1093/jnci/djj245. [DOI] [PubMed] [Google Scholar]
- 34.Thussbas C, Nahrig J, Streit S, Bange J, Kriner M, Kates R, et al. FGFR4 Arg388 allele is associated with resistance to adjuvant therapy in primary breast cancer. J. Clin. Oncol. 2006;24:3747–3755. doi: 10.1200/JCO.2005.04.8587. [DOI] [PubMed] [Google Scholar]
- 35.Decock J, Long JR, Laxton RC, Shu XO, Hodgkinson C, Hendrickx W, et al. Association of matrix metalloproteinase-8 gene variation with breast cancer prognosis. Cancer Res. 2007;67:10214–10221. doi: 10.1158/0008-5472.CAN-07-1683. [DOI] [PubMed] [Google Scholar]
- 36.Hughes S, Agbaje O, Bowen RL, Holliday DL, Shaw JA, Duffy S, et al. Matrix metalloproteinase single-nucleotide polymorphisms and haplotypes predict breast cancer progression. Clin. Cancer Res. 2007;13:6673–80. doi: 10.1158/1078-0432.CCR-07-0884. [DOI] [PubMed] [Google Scholar]
- 37.Azzato EM, Pharoah PDP, Harrington P, Easton DF, Greenberg D, Caporaso NE, et al. A genome-wide association study of prognosis in breast cancer. Cancer Epidemiol. Biomark. Prev. 2010;19:1140–1143. doi: 10.1158/1055-9965.EPI-10-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Azzato EM, Tyrer J, Fasching PA, Beckmann MW, Ekici AB, Schulz-Wendtland R, et al. Association between a germline OCA2 polymorphism at chromosome 15q13.1 and estrogen receptor-negative breast cancer survival. J. Natl. Cancer Inst. 2010;102:650–62. doi: 10.1093/jnci/djq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiyotani K, Mushiroda T, Tsunoda T, Morizono T, Hosono N, Kubo M, et al. A genome-wide association study identifies locus at 10q22 associated with clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients in Japanese. Hum. Mol. Genet. 2012;21:1665–72. doi: 10.1093/hmg/ddr597. [DOI] [PubMed] [Google Scholar]
- 40.Shu XO, Long J, Lu W, Li C, Chen WY, Delahanty R, et al. Novel genetic markers of breast cancer survival identified by a genome-wide association study. Cancer Res. 2012;72:1182–9. doi: 10.1158/0008-5472.CAN-11-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rafiq S, Tapper W, Collins A, Khan S, Politopoulos I, Gerty S, et al. Identification of inherited genetic variations influencing prognosis in early-onset breast cancer. Cancer Res. 2013;73:1883–91. doi: 10.1158/0008-5472.CAN-12-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rafiq S, Khan S, Tapper W, Collins A, Upstill-Goddard R, Gerty S, et al. A genome wide meta-analysis study for identification of common variation associated with breast cancer prognosis. PLoS One. 2014;9:e101488. doi: 10.1371/journal.pone.0101488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michailidou K, Beesley J, Lindstrom S, Canisius S, Dennis J, Lush MJ, et al. Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat. Genet. 2015;47:373–380. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo Q, Schmidt MK, Kraft P, Canisius S, Chen C, Khan S, et al. Identification of novel genetic markers of breast cancer survival. J. Natl. Cancer Inst. 2015;107:djv081–djv081. doi: 10.1093/jnci/djv081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Amos CI, Dennis J, Wang Z, Byun J, Schumacher FR, Gayther SA, et al. The OncoArray Consortium: a network for understanding the genetic architecture of common cancers. Cancer Epidemiol. Biomark. Prev. 2017;26:126–135. doi: 10.1158/1055-9965.EPI-16-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Michailidou K, Lindström S, Dennis J, Beesley J, Hui S, Kar S, et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551:92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.dbGaP (SUCCESS). https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000547. v1.p1.
- 48.van den Broek AJ, Van’t Veer LJ, Hooning MJ, Cornelissen S, Broeks A, Rutgers EJ, et al. Impact of age at primary breast cancer on contralateral breast cancer risk in BRCA1/2 mutation carriers. J. Clin. Oncol. 2016;34:409–18. doi: 10.1200/JCO.2015.62.3942. [DOI] [PubMed] [Google Scholar]
- 49.Sankararaman S, Sridhar S, Kimmel G, Halperin E. Estimating local ancestry in admixed populations. Am. J. Hum. Genet. 2008;82:290–303. doi: 10.1016/j.ajhg.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat. Methods. 2011;9:179–81. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 52.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–70. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Azzato EM, Greenberg D, Shah M, Blows F, Driver KE, Caporaso NE, et al. Prevalent cases in observational studies of cancer survival: do they bias hazard ratio estimates? Br. J. Cancer. 2009;100:1806–1811. doi: 10.1038/sj.bjc.6605062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cox DR, Hinkley D V. Theoretical Statistics. Springer US: Boston, MA, 1974 10.1007/978-1-4899-2887-0.
- 55.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 56.Ma C, Blackwell T, Boehnke M, Scott LJ, GoT2D investigators. Recommended joint and meta-analysis strategies for case–control association testing of single low-count variants. Genet. Epidemiol. 2013;37:539–50. doi: 10.1002/gepi.21742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am. J. Hum. Genet. 2007;81:208–227. doi: 10.1086/519024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–329. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aguet F, Brown AA, Castel SE, Davis JR, He Y, Jo B, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–7. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Novak P, Jensen T, Oshiro MM, Wozniak RJ, Nouzova M, Watts GS, et al. Epigenetic inactivation of the HOXA gene cluster in breast cancer. Cancer Res. 2006;66:10664–10670. doi: 10.1158/0008-5472.CAN-06-2761. [DOI] [PubMed] [Google Scholar]
- 65.Xia B, Shan M, Wang J, Zhong Z, Geng J, He X, et al. Homeobox A11 hypermethylation indicates unfavorable prognosis in breast cancer. Oncotarget. 2017;8:9794–9805. doi: 10.18632/oncotarget.14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Y, Qian J, Xiang Y, Chen Y, Qu J. The prognostic value of long noncoding RNA HOTTIP on clinical outcomes in breast cancer. Oncotarget. 2017;8:6833–6844. doi: 10.18632/oncotarget.14304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choi H, Lee SK. TAX1BP1 downregulation by EBV-miR-BART15-3p enhances chemosensitivity of gastric cancer cells to 5-FU. Arch. Virol. 2017;162:369–377. doi: 10.1007/s00705-016-3109-z. [DOI] [PubMed] [Google Scholar]
- 68.Nakayama Y, Nakamura N, Oki S, Wakabayashi M, Ishihama Y, Miyake A, et al. A putative polypeptide N-acetylgalactosaminyltransferase/Williams–Beuren syndrome chromosome region 17 (WBSCR17) regulates lamellipodium formation and macropinocytosis. J. Biol. Chem. 2012;287:32222–32235. doi: 10.1074/jbc.M112.370932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gao Z, Lee P, Stafford JM, von Schimmelmann M, Schaefer A, Reinberg D. An AUTS2–Polycomb complex activates gene expression in the CNS. Nature. 2014;516:349–354. doi: 10.1038/nature13921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Han Y, Ru GQ, Mou X, Wang H, Ma Y, He XL, et al. AUTS2 is a potential therapeutic target for pancreatic cancer patients with liver metastases. Med. Hypotheses. 2015;85:203–206. doi: 10.1016/j.mehy.2015.04.029. [DOI] [PubMed] [Google Scholar]
- 71.Kadalayil L, Khan S, Nevanlinna H, Fasching PA, Couch FJ, Hopper JL, et al. Germline variation in ADAMTSL1 is associated with prognosis following breast cancer treatment in young women. Nat. Commun. 2017;8:1632. doi: 10.1038/s41467-017-01775-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kao PYP, Leung KH, Chan LWC, Yip SP, Yap MKH. Pathway analysis of complex diseases for GWAS, extending to consider rare variants, multi-omics and interactions. Biochim. Biophys. Acta. 2017;1861:335–353. doi: 10.1016/j.bbagen.2016.11.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All estimates reported in the paper are available through the BCAC website: http://bcac.ccge.medschl.cam.ac.uk.