Abstract
Water is essential for life on Earth, and an important medium for microbial energy and metabolism. Dormancy is a state of low metabolic activity upon unfavorable conditions. Many microorganisms can switch to a metabolically inactive state after water shortage, and recover once the environmental conditions become favorable again. Here, we resuscitated dormant anammox bacteria from dry terrestrial ecosystems after a resting period of >10 ka by addition of water without any other substrates. Isotopic-tracer analysis showed that water induced nitrate reduction yielding sufficient nitrite as substrate and energy for activating anammox bacteria. Subsequently, dissimilatory nitrate reduction to ammonium (DNRA) provided the substrate ammonium for anammox bacteria. The ammonium and nitrite formed were used to produce dinitrogen gas. High throughput sequencing and network analysis identified Brocadia as the dominant anammox species and a Jettenia species seemed to connect the other community members. Under global climate change, increasing precipitation and soil moisture may revive dormant anammox bacteria in arid soils and thereby impact global nitrogen and carbon cycles.
Subject terms: Climate-change ecology, Biogeochemistry
Introduction
Anammox bacteria inhabit oxygen-limited marine, freshwater, wetland, and flooded soils systems, where they contribute significantly to the global nitrogen cycle [1–3]. Environmental and climate change may alter suitable microbial ecotones into dryland with water shortage forcing anammox bacteria into dormancy. Such dormancy may enable the anammox bacteria to survive in dry or cold environments [4]. In addition, their unique properties including a separate anammoxosome and protective ladderane lipids may support to maintain water and nutrients, and thus may further help to survive under adverse conditions [5, 6]. Beside oxygen-limited aquatic environments, anammox bacteria may also inhabit dry terrestrial environments [7, 8]. So far, endospores or/and cysts have not been reported for anammox bacteria. Anammox biomass from bioreactors has been reactivated after 6 months of resting [9], however the upper time limit for dormancy was not well explored.
Therefore, the objectives of this study were (i) to explore whether anammox bacteria can be revived from dry terrestrial environments after a longtime dormancy; and (ii) to elucidate the underlying metabolic mechanism using isotopic tracing techniques, functional gene expression, and high-throughput sequencing analyses. In order to verify our hypothesis, we chose very old soils from a dry arid environment (0–63 m) with minimal human perturbation in Kalgoorlie Western Australia for the laboratory incubation experiments with water.
Materials and methods
Soil/regolith samples collection and incubation
A typical dryland soil/regolith succession (63 m depth) was selected as sampling site (6617774N, 330210E) at Mt. Pleasant near Kalgoorlie (Goldfields Esperance region, Western Australia) where an average annual rainfall of 260 mm, indicative of arid climate conditions prevails. The Western part of Australia has a long geological history of 4.4 billion years, with most sediments formed in shallow marine environments. In geologic history uplifting, plate contraction and collision lead to the development of terrestrial environments and soil formation. Rocks in the Kalgoorlie region started to develop about 2.6–2.7 billion years ago (Supplementary document 1) and formed mainly intrusives, volcanics, and volcaniclastics, with only minor sediment deposition. The first regolith, i.e., deep weathering zone, may have started to form in the Paleozoic but unconsolidated regolith material was eroded and removed upon uplift during rifting between Australia and Antarctica commencing in the Early to Middle Tertiary. Weathering processes then started again on freshly denuded landforms and reached a depth of >150 m over the Late Miocene to Pliocene, when climate became more arid. An overall uplift of Southern Australia in excess of 200 m over the Neogene prevented marine incursions and in combination with increasing aridity lead to the formation of soil substrate formed from old Proterozoic bedrocks. In the study area at Mt. Pleasant regolith development evolved from mafic to ultramafic and therefore easily weathering volcanics. Postglacial aridity preventing microbial colonization from the surface lasted for more than 10,000 years (Supplementary Table S1) as evidenced by 14C dating of organic residues. In the following these parent materials evolved into primitive soil horizons by the actions of soil biota including microorganisms and lower plants, which then developed mature surface soil horizons under the comprehensive effects of biology, climate, landform, and time. The soil/regolith succession studied had developed mature soil layers, comprising the upper humus Ah layer, the (eluvation) A horizon (about 0–3 m) to the lower (illuvation) B horizon (about 3–33 m) and parent regolith bedrock layer (about 36–63 m), with soil/regolith colors transitioning from reddish-brown to gray.
Given the high heterogeneity of soil parent material, organic matter, and genetic classification, a total of 21 triplicate soil/regolith samples were collected from different depths. The soil/regolith samples collected were stored in sterile plastic bags, then sealed and transported on ice to the laboratory. Four soil/regolith samples at the depths of 6–9, 18–21, 45–48, and 51–54 m were chosen for incubation. The former two represent reddish brown illuvium layers and the latter two grey regolith layers developed from the parent bedrock. Using accelerator mass spectrometry (AMS), five 14C radiocarbon dates derived from soil/regolith samples yielded ages between 15,675 and 10,245 years before present (Beta Analytic radiocarbon dating service sample number 418750–418753, 418756, Supplementary document 2). The spoons, tweezers, bottles, and sealing stoppers that were used in soil treatment and incubation experiments of soil samples were sterilized by autoclaving, and each sample was handled with a separate spoon and tweezers to avoid cross-contamination. During experiments the operator’s gloves were sterilized by 75% ethanol. All operations were carried out in a super clean workbench. Before experiments, 75% ethanol was used to wipe the table and an ultraviolet lamp was used to sterilize for 20 min. Subsequently, in the super clean workbench, the triplicate fresh soil/regolith samples from each depth were mixed with a spoon to form one composite sample, whereby visible roots and residues were removed with tweezers before homogenizing the soil/regolith fraction of each subsample. All composite samples were divided into two groups, a positive experiment with addition of sterile water and a negative control experiment without water addition. Each group consists of on average 7 portions, corresponding with 7 time points for incubation, i.e., from the starting point to the sixth month, one point per month. Negative control experiments were applied on 4 soil/regolith samples, respectively, employing identical procedures as in positive experiments. The resurrection of dormant anammox bacteria from ancient samples provides an opportunity to explore the evolution of the structure and function of the community.
Both fresh homogenized soil/regolith and media water were flushed with high-purity He (99.9%) for about 3 × 15 min to remove oxygen. Subsequently, the media water and glass serum bottles (300 mL) were sterilized by high pressure autoclaving at 121 °C for 20 min. Thereafter, the oxygen-free soil/regolith (about 50 g) and sterilized media water (about 50 mL) were transferred and completely mixed into sterilized glass serum bottles (300 mL). The resulting slurries were flushed with He (99.9%) three times for about 15 min, again to remove residual oxygen. The glass serum bottles were sealed with butyl rubber plug and pressed with aluminum cover. Then these bottles were placed into shaking incubator at 150 rpm at 25 °C in a dark anaerobic chamber system (Thermo, USA) filled with high-purity Ar (99.999%). Liquid paraffin was applied to the syringe seal once a week to prevent leakage. Gas samples (CO2, CH4, and N2O) were determined based on six parallel bottles at times of 1, 2, 3, 4, 5, and 6 months by replacing the gas sample by an equal volume of high-purity He (99.9%). Thereafter, destructive sampling was carried out for subsequent analysis in an anaerobic chamber system. The freshly collected samples were immediately sieved through a 2.0 mm mesh and divided into three subsamples: one subsample was used for the analysis of physico-chemical properties, the second subsample was incubated to determine various microbial N-cycle process activities, and the third subsample was frozen at −80 °C for molecular analysis. The details of each analysis were included in the special chapters below.
DNA extraction, (q) PCR and the related growth rate calculation
DNA was extracted from 0.33 g of freeze-dried soil/regolith using the FastDNA SPIN Kit for Soil (MP Biomedical, USA) with a beating time of 45 s and speed setting of 5.5. The extracted DNA was checked on 1% agarose gel and the concentration was quantified with Nanodrop ND-2000 ultraviolet–visible spectrophotometry (Thermo Scientific, USA).
The abundance of anammox bacteria was determined targeting hzsB gene using primer pairs HSBeta396F and HSBeta742R [10] by the fluorescent dye SYBR-Green approach on an ABI Prism 7500 Sequence Detection System (Applied Biosystems, USA). The (q)PCR assays were carried out in a volume of 20 µL, containing 10 µL SYBR Premix Ex TaqTM (TAKARA, Dalian, China), 4 pmol of each primer and 2 μL of 10-fold diluted DNA template. The following protocol steps were set: 3 min at 95 °C, followed by 40 cycles of 30 s at 95 °C, 30 s at 59 °C, and 30 s at 72 °C [10]. Ten-fold serial dilutions of a known copy number of the plasmid DNA were subjected to real-time qPCR in triplicate to generate an external standard curve. Melting curves were generated after each assay to check the specificity of amplification. The amplification efficiencies were 90–103% (average 92%) and only the results with correlation coefficient (R2) above 0.98 were employed. All tests were performed in triplicate. The growth rate of each anammox taxa was defined as the abundance fluctuation along the incubation time. Calculation was based on the variation of the abundance divided by the corresponding incubation days.
Illumina sequencing and network analysis
A nest-PCR was conducted to amplify hzsB gene using the primer set of HSB396F (ARGGHTGGGGHAGYTGGAAG)–HSB742R (TCGGCAGTYCCHACRTCATGVGTCTG) and barcoded primer sets HSB449F (CCDCCSTTYMGDCATGARGG)–HSB742R on a C1000 Thermal Cycler (Bio-Rad, USA) [10, 11]. The amplification mixture (50 μL) contained: 5 μL of 10× buffer, 4 μL of dNTP (2.5 mM), 1 μL of each primer (10 μM), 0.5 μL of BSA, 0.25 μL of Taq (2.5 U), 2 μL of DNA and was topped up with ddH2O to a total volume of 50 μL. The PCR operation conditions consisted of an initial 95 °C for 10 min, 35 cycles of 95 °C for 60 s, 59 °C for 60 s, and a final extension at 72 °C for 45 s [10, 11]. High-throughput sequencing of hzsB gene amplification was carried out on a Hiseq 2500 platform (Illumina, San Diego, CA, USA).
Sequencing reads were assigned to each sample according to the unique 6-bp barcode of each sample. Raw sequences from original DNA fragments were merged using FLASH [12] and filtered (i.e., with a quality score <25 and read length <200 bp filtered using the split_libraries command) using the QIIME software package [13]. Then the chimeric sequences were removed using UCHIME [14]. The FrameBot tool was used to accurately detect and correct frameshifts caused by indel sequencing errors [15]. Briefly, only the sequences containing no ambiguous bases (N), without any barcode or primer mismatches, and with the corrected frameshifts and length (about 292 bp) were included into the downstream analysis. The unique sequences from the remaining high-quality nucleic acid sequences were obtained by Mothur [16] and then translated to protein sequences. Preprocessed sequences were clustered into operational taxonomic units (OTUs) based on their sequence similarity using UCLUST (identity = 0.97) [17]. A representative sequence for each OTUs was finally aligned using the MUSCLE program [18]. A local alignment search was conducted with hzsB protein sequences using Basic Local Alignment Search Tool (BLAST).
Singletons were excluded and resampling according to the minimum sequence numbers across all samples was performed before calculation. A variety of alpha diversity indices including Chao1, Shannon, and Simpson were calculated by applying Mothur software [16]. In this study, beta diversity was determined by weighted and unweighted UniFrac [19]. The comparison between anammox bacterial community compositions was visualized using a principal coordinate analysis (PCoA) of the pairwise Bray–Curtis dissimilarity matrices of OTUs similarity across the different samples.
Bacterial and archaeal 16S rRNA genes were PCR-amplified with primer pairs, with 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) at the V3–V4 region for bacteria [20], 519F (5′-CAGCCGCCGCGGTAA-3′) and 915R (5′-GTGCTCCCCCGCCAATTCCT-3′) at V4 region for archaea [21]. The PCR was conducted on an ABI GeneAmp 9700 (ABI, USA) in replicate. The mixed PCR product was screened using 2% agarose gel electrophoresis, and purified using an AxyPrep Gel Extraction Kit (AXYGEN, USA). Sequencing of positive amplicons from sludge samples was carried out on Illumina Miseq PE250 platform. The efficient readings were selected by removing low-quality sequences (dereplication and discarding singletons) and chimeras using Trimmomatic and FLASH software. The effective sequences obtained were normalized and the trimmed sequences were grouped into OTUs using 97% identity thresholds by the Usearch software program. Rarefaction curves and Shannon–Wiener were generated with Mothur software. The alpha diversity indices were calculated applying the mother software. Taxonomy was assigned via the RDP classifier with the Silva databases. The specific sequencing and analysis above were carried out by NovoGene (Beijing, China).
The phylogenetic molecular ecological networks (pMENs) analysis was carried out using the OTU matrix of all the samples [22, 23]. In the network, the nodes in the peripherals may ecologically represent specialists, whereas, module hubs and connectors refer to the generalists, while the network hubs as super-generalists [24, 25].
The accelerator mass spectrometry method of direct 14C isotope counting for soil dating
Using the AMS method to measure the 14C concentrations or residual radioactivity of soil/regolith samples, the count rate or number of decay events per gram C was obtained. The soil/regolith dating was achieved by comparing this with modern levels of activity and using the measured half-life. Brief procedures were as follows: the sample's organic carbon was chemically separated from the sediment matrix, converted to graphite, pressed into a cathode (sample target holder) where if formed a solid graphite plug or layer and then placed into a sputter ion source of an accelerator. The CO2 produced from carbonaceous raw materials was cryogenically purified (separated from non-CO2 gases) and reduced to solid graphite for measurement in an AMS [26–28]. The measurement was performed at Beta Analytic Radiocarbon Dating Laboratory (Miami, FL, USA).
Analyzing physicochemical properties of the incubation slurry
The pH-value of the slurry was determined with a DELTA 320 pH Analyzer (Mettler Toledo, USA) in a mixture of 5 g of sieved dry soil/regolith sample and 25 mL of ddH2O (a dry soil/water ratio of 1:5). The slurry NH4+, NO2−, and NO3− were extracted from 10 g of sieved dry slurry by shaking for 1 h at room temperature with 50 mL 2 M KCl, and then measured by a Continuous Flow Analyzer (SAN plus, The Netherlands) with detection limits of 0.015 mg kg−1, 0.015 mg kg−1, and 0.03 mg kg−1, respectively. All analyses were performed on triplicate samples for quality assurance/quality control (QA/QC).
Rate measurements of anammox, denitrification, NH4+ oxidation (NH4+ → NO2−), NO2− oxidation (NO2− → NO3−), NO3− reduction (NO3− → NO2−), and NO2− reduction (NO2− → NH4+) by 15N-tracer technique
Different 15N-amended substrates were added to determine the activities of the various N cycle processes. The incubated and homogenized soil/regolith samples (about 3.3 g) together with He-purged deionized water were transferred to 12.0-mL glass vials (Exetainer, Labco, UK) for incubation at 25 °C. In this study, 0.1 mL of stock solution for each isotopic mixture (15NO3− [15N at%: 99.19], 15NH4+ [15N at%: 99.16], 15NO2− [15N at%: 98.17] or 14N, purchased from Shanghai Research Institute of Chemical Industry) of 20 mmol L−1 was added to the glass vial according to the specific treatment. This afforded a final concentration of 0.17 mmol L−1 15N (15NO3−, 15NH4+, and 15NO2−) in fresh soil/regolith. All samples were purged with high-purity He (99.99%) for 15 min, three times before incubation, to reduce O2 concentrations [8].
To determine the anammox and denitrification activities, the resulting slurries first needed to be pre-incubated under anoxic conditions to deplete residual NOx− and O2 in the soil/regolith and incubation media [8, 29, 30]. Subsequently, 100 μL of high-purity He-purged stock solution for each isotopic mixture with three treatments, i.e., (1) 15NH4+, (2) 15NH4+ + 14NO3− and (3) 14NH4+ + 15NO3−, were used to calculate the combined activities of both anammox and denitrification. The incubations were stopped at defined intervals (0, 3, 6, 12, and 24 h) by injection of 100 μL of saturated mercuric chloride solution, respectively. In cases of the slurries amended with 15NH4+ only, no significant accumulation of 15N-labeled gas (29N2 and/or 30N2) could be observed in any sample, indicating that all ambient 14NOx− had been depleted during the pre-incubations. When both 15NH4+ and 14NO3− were added, 29N2 accumulated in each soil/regolith sample without any accumulation of 30N2. This pattern was reproducible and the results showed that the anammox process was detectable in all of the examined soil/regolith samples. The slurries amended solely with 15NO3− showed anammox and denitrification rates. The potential rate and contribution to N2 production of anammox and denitrification were calculated using the produced 29N2 and 30N2, as described by Thamdrup and Dalsgaard [31].
Ammonia oxidation activities were measured as the production rate of 15NO2− from incubations with 15NH4+. First, 100 μL of N2-purged 15NH4+ isotopic stock solution was added, and incubation of the slurries was terminated at defined intervals (0, 3, 6, 12, and 24 h) by injecting 100 μL of saturated mercuric chloride solution. To assess the N-isotopic ratio of 15NO2−, 16.5 mM (final concentration) of sulfamic acid was added to reduce 15NO2− to 29N2 [8, 32, 33]. The incubation time with the sulfamic acid exceeded 12 h to ensure sufficient conversion.
Nitrite oxidation activities were detected as the production rate of 15NO3− from 15NO2−. Following injecting 100 μL of saturated mercuric chloride solution after defined intervals (0, 3, 6, 12, and 24 h), about 1.0 g sponge cadmium was added under alkaline (pH = 7.0–8.0) conditions to convert the 15NO3− produced to 15NO2−. Thereafter, 16.5 mM (final concentration) of sulfamic acid was added to the incubation for longer than 12 h to reduce 15NO2− to 29N2 completely [8, 32, 33].
Nitrate reduction to nitrite rates were detected as the production rate of 15NO2− from reduced 15NO3−. 100 μL of 15NO3− isotopic stock solution was added first, followed by injection of 100 μL of saturated mercuric chloride as an inhibitor to terminate biological activity after defined intervals (0, 3, 6, 12, and 24 h). Next, 16.5 mM (final concentration) of sulfamic acid was added and incubated for more than 12 h to reduce 15NO2− to 29N2 completely [8, 32, 33].
To assess the nitrite reduction rates, 100 μL of N2-purged 15NO2− isotopic stock solution was added. After defined intervals (0, 3, 6, 12, and 24 h), the reaction was stopped by injecting 100 μL of saturated mercuric chloride solution. Subsequently, hypobromite was added and incubation for longer than 12 h converted the 15NH4+ produced to 30N2 completely [8, 32, 33]. The final concentration of hypobromite was equal to the concentration of added 15NO2−.
To assess the DNRA rates, 100 μL of diluted 15NO2− isotopic stock solution was added. After defined intervals, the reaction was stopped and hypobromite was added and incubated for more than 12 h to convert produced 15NH4+ to 30N2 completely. The final concentration of hypobromite was equal to the concentration of added 15NO2− [8, 32, 33].
The produced 29N2 or 30N2 in the above processes were measured by IRMS (MAT253 with Gasbench II, Bremen, Germany). The minimum detectable rate in this study was calculated to 0.001 nmol N g−1 h−1. The measurements were done at the Key Laboratory of Tibetan Environment Changes and Land Surface Processes, Institute of Tibetan Plateau Research, Chinese Academy of Sciences.
The measurements of carbon dioxide, methane, and nitrous oxide gas emission
Gas chromatography (GC) with electron capture detector (ECD) and flame ionization detector (FID) at a detection limit of 1 ppb were used to measure the concentration of CO2, CH4, and N2O, respectively. All analyses were performed on triplicate samples for QA/QC.
Statistical analysis
The basic data management and arrangement was performed in Microsoft Excel. Means comparison was expressed as mean ± standard deviation (S.D.) unless specified. General statistical analysis was conducted under the statistical product and service solutions 18.0 software (SPSS Inc., USA). Pearson correlation was used to evaluate the correlation relationships between variables. In this study, all the data below detection limits were considered to be zero in the statistical analysis, and all the statistical differences were set at the α = 0.05 significance level (p ≤ 0.05) unless specified. Graphing was achieved using Origin 8.0 software.
Results
Age of sampled soils
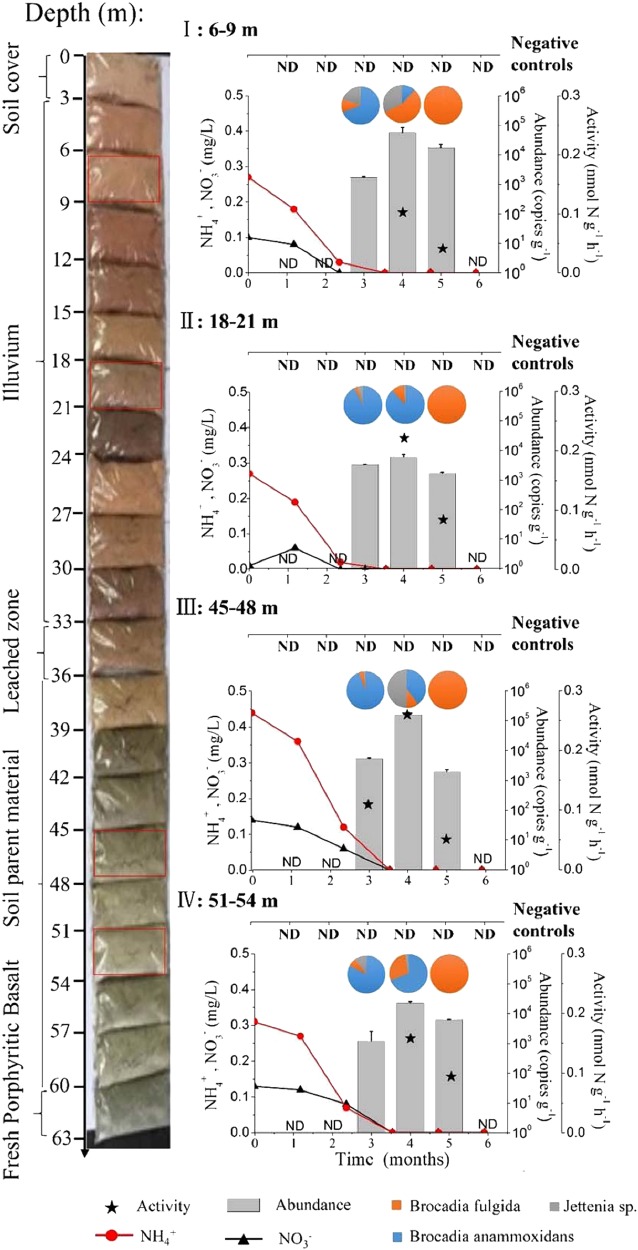
The Kalgoorlie soils studied from the surface to 63 below showed a marked profile and development (Fig. 1). The five AMS 14C radiocarbon dates determined on the soil/regolith samples ranged between 15,675 and 10,245 Cal. yr B.P. (Beta Analytic radiocarbon dating service sample number 418750–418753, 418756, supplementary documents 1). In order to experimentally test the revival of anammox bacteria after a dormancy of >10 ka, we collected dry soil/regolith samples under sterile conditions (Table S1) from different depths (6–9 m, illuvium; 18–21 m, illuvium; 45–48 m, soil parent material, and 51–54 m, soil parent material).
Fig. 1.
Anammox bacterial abundance (gray columns), activity (stellate legend), community compositions (pie chart), and substrate concentrations (black or red lines) upon water-induced incubation (0–5 months) at soil and regolith depths of 6–9 m (I), 18–21 m (II), 45–48 m (III), 51–54 m (IV), with soil/regolith succession on the left. “ND” indicates anammox abundance below detection limit (<103 copies g−1) and without positive 15N isotopic tracing for anammox rates. Data points represent triplicate experiments (mean ± SD)
Abundance and activity of anammox bacteria
As a precaution we only added sterile Milli-Q water containing no further chemicals (such as anammox substrates NH4+, NO2−, and NO3–) to the regolith samples (in a 1:1 vol/wt water:fresh soil ratio). After 2 months of incubation, real-time quantitative PCR (qPCR) showed that anammox bacterial counts were still below the limit of detection (LoD = 102 copies g−1) in all samples (Fig. 1). Nevertheless, microbial activity had already been initiated and the onset of respiratory metabolism was detected by significant CO2 production (p < 0.05; Table S2). After 3 months, the 16S rRNA and hzsB genes of anammox bacteria were detected in all samples, at an abundance level well over 103 copies g−1 dry soil. All anammox species detected showed high similarities to Candidatus Jettenia and Brocadia. The abundance of anammox bacteria at 45–48 m was the highest (1.6 ± 0.3) × 105 copies g−1 with approximately 5.2 × 103 copies g−1 d−1 of growth rate among all investigated depths at the end of the fourth month and then decreased to under the LoD. This decline should be attributed to the depletion of substrates in the incubations, which was verified by the very low level of NH4+ and NO3− in the samples.
The activity of anammox bacteria in the incubations was determined by the 15N-tracing techniques including negative controls (Fig. 1). In the third month, anammox activity (0.11 ± 0.01 nmol N g−1 h−1) could be detected in the sample from 45 to 48 m where the anammox bacteria abundance was (5.3 ± 0.4) × 103 copies g−1 dry soil (approximately 58.6 copies g−1 d−1). In contrast, anammox activity could not yet be detected in the other incubations, although the abundance of anammox was well over 103 copies g−1 dry soil. In the fourth month of incubation, anammox activity (0.26 ± 0.01 nmol N g−1 h−1) was detectable in every sample where the abundance was over 104 copies g−1 dry soil. Throughout the 5 months, no anammox bacterial abundance nor activity was detected in the negative controls.
Anammox bacterial community
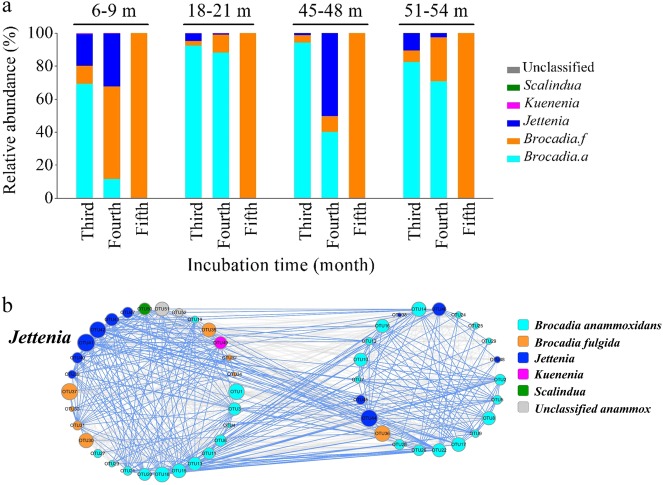
High-throughput sequencing analysis of anammox bacterial hzsB gene showed that the diversity of anammox bacterial decreased with time (Fig. 1, Fig. S1). The anammox community shifted from multiple Candidatus Jettenia, Candidatus Brocadia, and Candidatus Anammoxoglobus to a single species of Candidatus Brocadia fulgida within 4 months (Figs. 1 and 2, Fig. S1 S2). However, amongst the anammox species the growth rate of the genus Jettenia was most consistent with the overall anammox rate (r = 0.55, p = 0.012, n = 20) and abundance (r = 0.914, p < 0.001, n = 20; Table S3, Fig. S3 S4). Nitrate content had a positive relation with overall anammox bacterial growth, as well as a close relation with the growth of genus Candidatus Brocadia and Jettenia.
Fig. 2.
Overview of the anammox bacterial community and inner-relevance mechanism. a The community compositions at soil and regolith depths of 6–9 m (I), 18–21 m (II), 45–48 m (III), 51–54 m (IV) after anammox was detected. b The network of anammox bacterial community with avgCC of 0.639. Nodes are colored and shaped for phylogenetic taxa with the size proportional to node connectivity. The blue link indicates a positive interaction and the gray link negative interaction
The co-occurrence network analysis of the anammox community clustered to two significant modules with an average clustering coefficient (avgCC) of 0.639 (p < 0.01) (Fig. 2b, Fig. S5). A total of 52 nodes showed 47.9% positive and 52.1% negative interactions within total 628 interaction links. Given that nodes with higher connectivity are usually regarded as the central nodes in the network structure [34], we looked at the nodes (anammox bacterial OTUs) with high connectivity. The maximum degree of connection was found in OTU41, which was phylogenetically affiliated to Candidatus Jettenia. Amongst the Candidatus Jettenia links 67.5% (27/40) revealed positive interactions, in which 88.9% were interactions with Candidatus Brocadia anammoxidans. Thus although Candidatus Brocadia was the most dominant species, it was not the key connector.
Diversity of archaeal and bacterial community
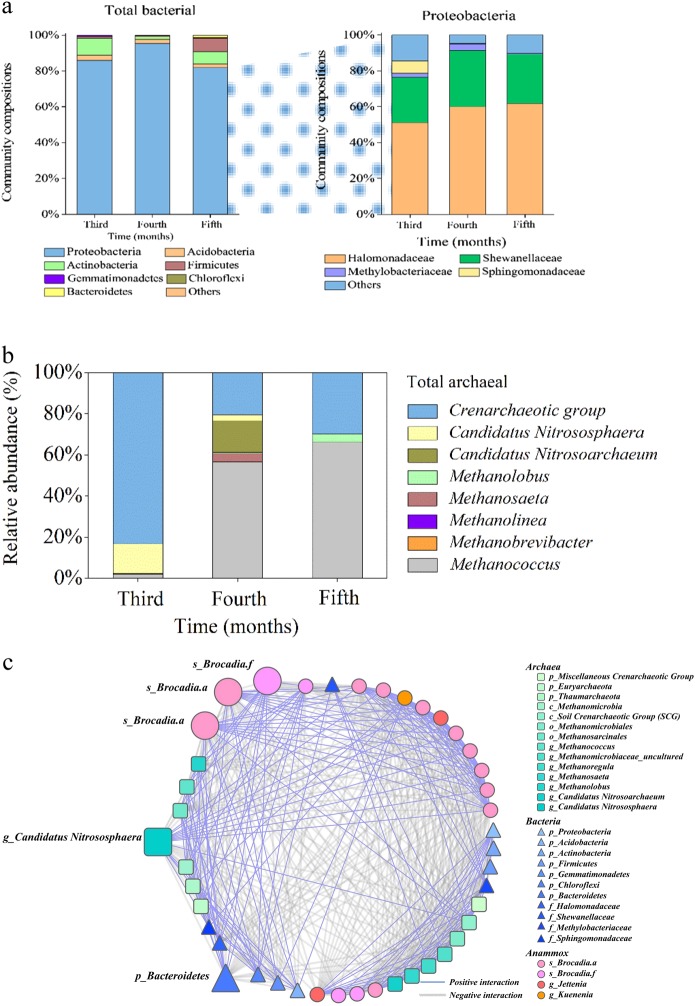
The relative abundance and microbial community structure comprising bacterial/archaeal co-occurrence with anammox bacteria was investigated by high-throughput sequencing of 16S rRNA genes derived from this extreme environment (Fig. 3). Over the entire incubation period, the bacterial community showed no significant change (Fig. 3a; Table S4), and the facultative anaerobic Proteobacteria (81.9–95.6%) dominated, of which the majority families were Halomonadaceae (50.6–57.2%, affiliated with Oceanospirillales) and Shewanellaceae (22.4–29.9% affiliated with Alteromonadales), both favoring salty marine environments [35, 36]. Further representatives of nitrite-oxidizing bacteria Nitrospira also increased from 0.05% (third month) to 0.32% (fifth month). The archaeal abundance exhibited a community shift from the Crenarchaeal group (83.2%) plus Candidatus Nitrososphaera (14.3%) in the third month to multiple Methanococcus (61.3%), Crenarchaeal group (20.5%), and Candidatus Nitrosoarchaeum (15.3%) in the fourth month and finally to Crenarchaeal groups (29.9%) combined with typical thermophilic Methanococcus (66.1%) (Fig. 3b).
Fig. 3.
Overview of the bacterial and archaeal community and relevance mechanism with anammox bacteria. Monthly variation of microbial community composition of a total bacteria and the most phylum Proteobacteria and b total archaea in incubation sample after anammox was detected. c The outer-relevance networks of anammox bacterial, archaeal, and bacterial taxa among the incubation periods of soil/regolith samples with average clustering coefficient (avgCC) of 0.704. Nodes are colored and shaped for phylogenetic taxa with the size proportional to node connectivity. The blue link indicates a positive interaction and the gray link negative interaction
For the co-occurring network of archaea, bacteria, and anammox bacteria, there was only one module (Fig. 3c, Fig. S6). This is interpreted to indicate a strong symbiosis or competition within the entire microbial community. In this module, a total of 53 nodes gave 746 interaction links in which there were 42.8% positive and 57.2% negative interactions. For the archaeal genus Candidatus Nitrososphaera, all links revealed interaction with the anammox community, and 68.8% (11/16) of the links were positive interactions. By contrast, there were only 64.7% (22/34) links of Bacteroidetes to anammox, while 58.8% (20/34) of links were negative interactions.
Interaction of anammox with other N cycling process
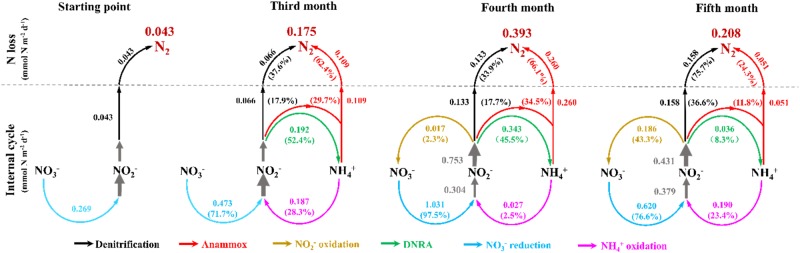
NO2− is the key intermediate in microbial N-cycling processes and the preferred electron acceptor of anammox bacteria. Hence, we determined the NO2− conversion rate of several possible N cycle processes [anammox, denitrification, NO2− oxidation and dissimilatory NO3− reduction to NH4+ (DNRA)] and the NO2− production rate [NH4+ oxidation and NO3− reduction] via the 15N isotopic-pairing technique (Fig. 4). Initially, only NO3− reduction to NO2− (0.23 ± 0.02 nmol N g−1 h−1) and denitrification (0.04 ± 0.004 nmol N g−1 h−1) were detected. After 3 months of incubation, NH4+ oxidation (0.19 ± 0.02 nmol N g−1 h−1), DNRA (0.19 ± 0.02 nmol N g−1 h−1) and anammox (0.11 ± 0.01 nmol N g−1 h−1) were also detectable. As NO2− sink, DNRA became dominant (52.4%) over anammox and denitrification, and for N-loss anammox exceeded denitrification (0.07 ± 0.008 nmol N g−1 h−1). It was noted that the potential anammox rates are probably underestimated because DNRA would lead to production of 15NH4+ from 15NO3− (or 15NO2−) and as a consequence anammox may produce 30N2 (15NH4+ + 15NO2−) and this signal may be interpreted as canonical denitrification. The prominent role of NH4+ oxidation as NO2− source in the third month shifted to NO3− reduction in the fourth month. The rate of anammox (0.26 ± 0.3 nmol N g−1 h−1), denitrification (0.13 ± 0.009 nmol N g−1 h−1) and DNRA (0.34 ± 0.03 nmol N g−1 h−1) also significantly increased and the DNRA remained the dominant pathway in NO2− consumption. After 5 months of incubation, rates of anammox (0.05 ± 0.005 nmol N g−1 h−1) and DNRA (0.036 ± 0.005 nmol N g−1 h−1) decreased, but the rate of denitrification continued to increase (0.16 ± 0.2 nmol N g−1 h−1) and became the most significant N-loss (75.7%) and NO2− sink (36.6%) pathway.
Fig. 4.
Temporal change of N-cycle microbial activity and community in soil/regolith sample under sterilized media water incubation. (I) Rates and proportions of anammox and related N cycles. The single numbers mean rates with unit of nmol N g−1 h−1. The percentage numbers (%) mean the related rates accounting in N-loss, NO2− sink or NO2− source processes
Residual oxygen of the original soil aggregates or regolith may have been responsible for NH4+ to nitrite oxidation and thus may have diminished NH4+ to very low levels when anammox was detected in the third month. However, we found the coupling of anammox to DNRA to last over the entire incubation period with DNRA most probably providing NH4+ to anammox bacteria. In the fourth month, when DNRA rate reached the highest value (0.34 ± 0.03 nmol-N g−1 h−1), the anammox rate maximized as well (0.26 ± 0.03 nmol N g−1 h−1). Comparably, when after 4 months DNRA diminished, the anammox rate declined in parallel. The decline in DNRA rate led to decreased NH4+ that was apparently insufficient to support the growth of anammox bacteria. Comparable observations have been made in substrate-limited marine oxygen minimum zone (OMZ) [37].
NO3− is the next preferred terminal electron acceptor after O2. In the oxygen-depleted incubations, NO3− not only participated in the supply of nitrite, but also constituted a key role as the main electron acceptor for respiration. During the 5 months incubation, NO3− reduction rates continuously stayed at relatively high levels (0.11–0.38 nmol-N g−1 h−1). With the relatively high background of 0.44 mg kg−1, NO3− reduction may have been the respiratory process in the soil and regolith during the first 4 months, which could supply NO2− for the growth of anammox bacteria. Following the increase of abundance and activity of anammox bacteria the first 4 months, the rate of NO2− oxidation to NO3− significantly increased from 0.02 (in the fourth month) to 0.19 nmol-N g−1 h−1 (in the fifth month) and became the major pathway of NO2− transformation even at nanomolar oxygen levels, which is in accordance with observation in the OMZs [38]. The high-throughput sequencing results further illustrated the possibility of NO2− oxidation within the Nitrospira class (0.32%) detected. Consequently, the inner NO2− sink switched from DNRA to NO2− oxidation after 4 months and the latter could supply the NO3− to maintain the energy cycle.
Discussion
Water is the main component of all living cells, and the ability to absorb and release water is a basic property [39]. However, dryland soils are an environment with very low water content. At low soil water matrix potential, the available substrate is limited, which causes dormancy and/or death of microorganisms [40]. However, it is fascinating that dormant anammox bacteria residing in dry soils of >10,000 years old were able to be revived by water addition. This demonstrates a dormancy capacity of anammox bacteria approaching that of Actinobacteria, Bacteroides and Firmicutes in dry Antarctic soils, which was reported to be 26.5 ka [41, 42] but not yet the millennia equivalents reported for endospores of clostridia or millions of years proposed for halophilic microorganisms [43, 44]. It also suggested that anammox bacteria may be able to tolerate low matric potential in the soil environment. The strong tolerance to adverse environments is a favorable condition for the wider ecological niche of anammox bacteria and contribution in terrestrial ecosystems.
During incubation experiments, we aimed to create anoxic environment for anammox bacteria by making the incubations anoxic by flushing with high-purity He gas. But as previously reported, residual oxygen will still be retained in soil aggregates [45]. The dominant Ca. “Brocadia fulgida” species in our incubations was reported to tolerate oxygen levels up to 70 μM [46]. Hence, the trace residual oxygen did not seem to affect the growth of anammox bacteria. On the other side, water addition could also stimulate other aerobic microorganisms to consume residual oxygen. As a result the observed ammonium oxidation rate driven by thaumarchaeal ammonium-oxidizers was very low (~0.003 nmol N g−1 h−1), and even several methanogens (Methanococci (67.2%) and Methanobacteria (4.0%)) appeared in the later stages of the incubations The two methanogens, Methanococci and Methanobacteria, emerged after 3 months and may have consumed undesirable organic compounds that can interfere with growth of anammox bacteria. The Methanococcus and Methanolobus belong to typical thermophilic organism which evolved from ancient marine environments [35, 36, 47], such detection may reflect the historical mark of the soil habitats. The presence of more extremophilic microorganisms probably represent the intense desiccation of the ecosystem, a process known to favor dormancy of adaptive organisms [42, 43, 48]. This study further showed that the residual oxygen may have been consumed by the growth of aerobic microorganisms after addition of water. Low soil water content might have limited microbial activity as diffusion and supply of substrate was hindered [49] Therefore, the addition of water could change the mass-transfer of anammox substrates from soils (high heterogeneity) to slurry and water column (more homogeneous), which would significantly increase the substrate availability for anammox bacteria. It is speculated that the moisture content of soil is a key environmental factor for anammox bacteria rather than oxygen.
The interaction of the anammox bacterial community with other bacterial/archaeal community members was investigated by network analysis. The analysis indicated that Jettenia was the keystone genus, rather than the abundant Brocadia. This was also in accordance with previous studies in wetlands sediments [50], and might also be due to the mixotrophic lifestyle of the genus Jettenia [51]. Furthermore, the archaeal ammonium-oxidizer Candidatus Nitrososphaera showed over 99% positive links with anammox bacteria, indicating a potential synergy in the supply of NO2−to anammox. Similar results were also reported in paddy fields [3], oceanic OMZs [52, 53], and unconfined aquifer soils [54]. In contrast, most of the negative links were shown for the Bacteroidetes that are heterotrophs frequently found on macroscopic organic matter particles, representing a significant part of free-living microbial assemblages in nutrient-rich microenvironments [55].
Nitrite, rather than nitrate, is the preferred electron donor for anammox bacteria [56]. In nature, nitrite does not accumulate and mainly derives from NH4+ oxidation in aerobic environments and from partial NO3− reduction under anoxic ones [57]. In most previous studies, NO2− mainly originated from oxidation of NH4+ in surface wetland sediments and soils as deduced from the rate of ammonia oxidation versus nitrate reduction [2, 3, 8, 58]. In this study the NH4+ oxidation rate could only be measured at the beginning of the experiments, but ammonium could no longer be detected in the later stages, indicating that then NO2− may preferentially originate from NO3− reduction under more anoxic conditions. Moreover, the anammox rate increased parallel to the NO3− reduction rate. This indicated that anammox bacteria responded positively to the NO3− reduction. In soil, ammonium maybe a limiting factor for anammox bacteria because NH4+ is bound to soil colloids with negative electrical charges and can be oxidized by ammonia oxidizing archaea and bacteria [59–61]. In contrast, nitrate can more easily diffuse and can be leached from surface soil down to deep soils. Nitrate reduction under oxygen-limited conditions may be the largest respiratory process, and will produce NO2− and NH4+ by DNRA bacteria as substrate of anammox bacteria [62, 63]. Indeed in our present study, DNRA seemed to supply NH4+ for growth of anammox bacteria as has been observed in marine ecosystems, where the coupled anammox and DNRA processes play a significant role in N loss in OMZs [33, 37, 64]. Therefore, we suggest that anammox bacteria not only inhabit water saturated soils or marine, liminic and riparian sediments but also occur, though at low metabolic rate, often approaching dormancy, in dryland soils.
This study provided experimental proof and new insight into the potential revitalization of dormant bacteria from previous and currently arid regions that account for about 40% of the global pedosphere by increasing soil moisture in the wake of global climate change. Even >10,000 years old soil, regolith, and sediment harbor dormant bacterial resting stages, which upon intensification of the hydrological cycle and higher soil moisture will have the potential to resuscitate. Although water addition is a very simple process and it will occur with rainfall in nature, it will obviously awaken a variety of microorganisms and accelerate the biogeochemical cycle process. The perspective that present climate change may potentially initiate dormant bacteria reactivation on a large (global) scale is presently not accounted for and its consequences on the nitrogen and carbon cycle are not yet explored.
Electronic supplementary material
Acknowledgements
This research was financially supported by the National Natural Science Foundation of China (Nos. 41322012, 41671471, and 91851204), Strategic Priority Research Program of the Chinese Academy of Sciences (XDB15020303), Local Innovative and Research Teams Project of Guangdong Pearl River Talents Program (2017BT01Z176), National Key R&D Program (2016YFA0602303), and a special fund from the Key Laboratory of Drinking Water Science and Technology, Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences (15Z07KLDWST and 16Z03KLDWST). The author Guibing Zhu gratefully acknowledges the Program of the Youth Innovation Promotion Association (CAS). Mike Jetten was supported by erc ag 232937 anammox, erc ag ecomom 339880, and siam 024002002. LS acknowledges support by the German Research Foundation (Schw554/25).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Guibing Zhu, Email: gbzhu@rcees.ac.cn.
Lorenz Schwark, Email: ls@gpi.uni-kiel.de.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0316-5) contains supplementary material, which is available to authorized users.
References
- 1.Lam P, Kuypers MMM. Microbial nitrogen cycling processes in oxygen minimum zones. Annu Rev Mar Sci. 2011;3:317–45. doi: 10.1146/annurev-marine-120709-142814. [DOI] [PubMed] [Google Scholar]
- 2.Zhu G, Wang S, Wang W, Wang Y, Zhou L, Jiang B, et al. Hotspots of anaerobic ammonium oxidation at land–freshwater interfaces. Nat Geosci. 2013;6:103–7. doi: 10.1038/ngeo1683. [DOI] [Google Scholar]
- 3.Zhu G, Wang S, Wang Y, Wang C, Risgaard-Petersen N, Jetten MSM, et al. Anaerobic ammonia oxidation in a fertilized paddy soils. ISME J. 2011;5:1905–12. doi: 10.1038/ismej.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu G, Xia C, Wang S, Zhou L, Liu L, Zhao S, et al. Occurrence, activity and contribution of anammox in some freshwater extreme environments. Environ Microbiol Rep. 2015;7:961–9. doi: 10.1111/1758-2229.12341. [DOI] [PubMed] [Google Scholar]
- 5.Van Niftrik L, Geerts WJC, van Donselaar EG, Humbel BM, Yakushevska A, Verkleij AJ, et al. Combined structural and chemical analysis of the anammoxosome: a membrane-bounded intracytoplasmic compartment in anammox bacteria. J Struct Biol. 2008;161:401–10. doi: 10.1016/j.jsb.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 6.Damste JSS, Strous M, Rijpstra WIC, Hopmans EC, Geenevasen JAJ, van Duin ACT, et al. Linearly concatenated cyclobutane lipids form a dense bacterial membrane. Nature. 2002;419:708–12. doi: 10.1038/nature01128. [DOI] [PubMed] [Google Scholar]
- 7.Humbert S, Tarnawski S, Fromin N, Mallet MP, Aragno M, Zopfi J, et al. Molecular detection of anammox bacteria in terrestrial ecosystems: distribution and diversity. ISME J. 2010;4:450–4. doi: 10.1038/ismej.2009.125. [DOI] [PubMed] [Google Scholar]
- 8.Zhu G, Wang S, Li Y, Zhuang L, Zhao S, Wang C, et al. Microbial pathways for nitrogen loss in an upland soil. Environ Microbiol. 2018;20:1723–38. doi: 10.1111/1462-2920.14098. [DOI] [PubMed] [Google Scholar]
- 9.Wang T, Zhang H, Yang F. Long-term storage and subsequent reactivation of Anammox sludge at 35 °C. Desalin Water Treat. 2016;57:24716–23. doi: 10.1080/19443994.2016.1145604. [DOI] [Google Scholar]
- 10.Wang Y, Zhu G, Harhangi HR, Zhu B, Jetten MSM, Yin C, et al. Co-occurrence and distribution of nitrite-dependent anaerobic ammonium and methane-oxidizing bacteria in a paddy soil. FEMS Microbiol Lett. 2012;336:79–88. doi: 10.1111/j.1574-6968.2012.02654.x. [DOI] [PubMed] [Google Scholar]
- 11.Zhao S, Zhuang L, Wang C, Li Y, Wang S, Zhu G. High-throughput analysis of anammox bacteria in wetland and dryland soils along the altitudinal gradient in Qinghai–Tibet plateau. Microbiol Open. 2017;7:e00556. doi: 10.1002/mbo3.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Magoc T, Salzberg SL. Flash: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–63. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Q, Quensen JF, Fish JA, Lee TK, Sun Y, Tiedje JM, et al. Ecological patterns of nifH genes in four terrestrial climatic zones explored with targeted metagenomics using FrameBot, a new informatics tool. mBio. 2013;4:e00592. [DOI] [PMC free article] [PubMed]
- 16.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 18.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–97. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bokulich NA, Thorngate JH, Richardson PM, Mills DA. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc Natl Acad Sci USA. 2014;111:E139–48. doi: 10.1073/pnas.1317377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vissers EW, Bodelier PLE, Muyzer G, Laanbroek HJ. A nested PCR approach for improved recovery of archaeal 16S rRNA gene fragments from freshwater samples. FEMS Microbiol Lett. 2009;298:193–8. doi: 10.1111/j.1574-6968.2009.01718.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhou J, Deng Y, Luo F, He Z, Yang Y. Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. mBio. 2011;2:e00122. [DOI] [PMC free article] [PubMed]
- 23.Zhou J, Deng Y, Luo F, He Z, Tu Q, Zhi X. Functional molecular ecological networks. mBio. 2010;1:1592–601. doi: 10.1128/mBio.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng Y, He Z, Xu M, Qin Y, Van Nostrand JD, Wu L, et al. Elevated carbon dioxide alters the structure of soil microbial communities. Appl Environ Microbiol. 2012;78:2991–5. doi: 10.1128/AEM.06924-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olesen JM, Bascompte J, Dupont YL, Jordano P. The modularity of pollination networks. Proc Natl Acad Sci USA. 2007;104:19891–6. doi: 10.1073/pnas.0706375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duplessy JC, Arnold M, Maurice P, Bard E, Duprat J, Moyes J. Direct dating of the oxygen-isotope record of the last deglaciation by 14C, accelerator mass spectrometry. Nature. 1986;320:350–2. doi: 10.1038/320350a0. [DOI] [Google Scholar]
- 27.Muller RA. Radioisotope dating with accelerators. Phys Today. 1979;32:23–30. doi: 10.1063/1.2995406. [DOI] [Google Scholar]
- 28.Hellborg R, Skog G. Accelerator mass spectrometry. Mass Spectrom Rev. 2003;70:365–72. doi: 10.1002/mas.20172. [DOI] [PubMed] [Google Scholar]
- 29.Risgaard-Petersen N. Anaerobic ammonium oxidation in an estuarine sediment. Aquat Microb Ecol. 2004;36:293–304. doi: 10.3354/ame036293. [DOI] [Google Scholar]
- 30.Engström P, Dalsgaard T, Hulth S, Aller RC. Anaerobic ammonium oxidation by nitrite (anammox): implications for N2, production in coastal marine sediments. Geochim Cosmochim Acta. 2005;69:2057–65. doi: 10.1016/j.gca.2004.09.032. [DOI] [Google Scholar]
- 31.Thamdrup B, Dalsgaard T. Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Appl Environ Microbiol. 2002;68:1312–8. doi: 10.1128/AEM.68.3.1312-1318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mcilvin MR, Altabet MA. Chemical conversion of nitrate and nitrite to nitrous oxide for nitrogen and oxygen isotopic analysis in freshwater and seawater. Anal Chem. 2005;77:5589–95. doi: 10.1021/ac050528s. [DOI] [PubMed] [Google Scholar]
- 33.Lam P, Lavik G, Jensen MM, Vossenberg JVD, Schmid M, Woebken D, et al. Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc Natl Acad Sci USA. 2009;106:4752–7. doi: 10.1073/pnas.0812444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Layeghifard M, Hwang DM, Guttman DS. Disentangling interactions in the microbiome: a network perspective. Trends Microbiol. 2017;25:217–28. doi: 10.1016/j.tim.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shnit-Orland M, Sivan A, Kushmaro A. Shewanella corallii sp nov, a marine bacterium isolated from a Red Sea coral. Int J Syst Evol Microbiol. 2010;60:2293–7. doi: 10.1099/ijs.0.015768-0. [DOI] [PubMed] [Google Scholar]
- 36.Jones WJ, Leigh JA, Mayer F, Woese CR, Wolfe RS. Methanococcus jannaschii sp-nov, an extremely thermophilic methanogen from a submarine hydrothermal vent. Arch Microbiol. 1983;136:254–61. doi: 10.1007/BF00425213. [DOI] [Google Scholar]
- 37.Jensen MM, Lam P, Revsbech NP, Nagel B, Gaye B, Jetten MSM, et al. Intensive nitrogen loss over the Omani Shelf due to anammox coupled with dissimilatory nitrite reduction to ammonium. ISME J. 2011;5:1660–70. doi: 10.1038/ismej.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuessel J, Lam P, Lavik G, Jensen MM, Holtappels M, Guenter M, et al. Nitrite oxidation in the Namibian oxygen minimum zone. ISME J. 2012;6:1200–9. doi: 10.1038/ismej.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wood JM. Osmosensing by bacteria: signals and membrane-based sensors. Microbiol Mol Biol Rev. 1999;63:230–62. doi: 10.1128/mmbr.63.1.230-262.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orchard VA, Cook FJ. Relationship between soil respiration and soil moisture. Soil Biol Biochem. 1983;15:447–53. doi: 10.1016/0038-0717(83)90010-X. [DOI] [Google Scholar]
- 41.Lennon JT, Jones SE. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol. 2011;9:119–30. doi: 10.1038/nrmicro2504. [DOI] [PubMed] [Google Scholar]
- 42.Dworkin J, Shah IM. Exit from dormancy in microbial organisms. Nat Rev Microbiol. 2010;8:890–6. doi: 10.1038/nrmicro2453. [DOI] [PubMed] [Google Scholar]
- 43.Antibus DE, Leff LG, Hall BL, Baeseman JL, Blackwood CB. Cultivable bacteria from ancient algal mats from the McMurdo Dry Valleys, Antarctica. Extremophiles. 2012;16:105–14. doi: 10.1007/s00792-011-0410-3. [DOI] [PubMed] [Google Scholar]
- 44.Stephenson K, Lewis RJ. Molecular insights into the initiation of sporulation in Gram-positive bacteria: new technologies for an old phenomenon. FEMS Microbiol Rev. 2005;29:281–301. doi: 10.1016/j.fmrre.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 45.Lefelaar PA. Water movement, oxygen supply and biological processes at the aggregate scale. Geoderma. 1993;57:143–65. doi: 10.1016/0016-7061(93)90152-B. [DOI] [Google Scholar]
- 46.Oshiki M, Satoh H, Okabe S. Ecology and physiology of anaerobic ammonium oxidizing (anammox) bacteria. Environ Microbiol. 2016;18:2784–96. doi: 10.1111/1462-2920.13134. [DOI] [PubMed] [Google Scholar]
- 47.Haba RRDL, Sánchez-Porro C, Ventosa A. Taxonomy, phylogeny, and biotechnological interest of the family Halomonadaceae. Halophiles and hypersaline environments. Berlin, Heidelberg: Springer; 2011. p. 27–64.
- 48.Van Vliet S. Bacterial dormancy: how to decide when to wake up. Curr Biol. 2015;25:753–5. doi: 10.1016/j.cub.2015.07.039. [DOI] [PubMed] [Google Scholar]
- 49.Stark JM, Firestone MK. Mechanisms for soil moisture effects on activity of nitrifying bacteria. Appl Environ Microbiol. 1995;61:218–21. doi: 10.1128/aem.61.1.218-221.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang S, Wang W, Liu L, Zhuang L, Zhao S, Su Y, et al. Microbial nitrogen cycle hotspots in the plant-bed/ditch system of a constructed wetland with N2O mitigation. Environ Sci Technol. 2018;52:6226–36. doi: 10.1021/acs.est.7b04925. [DOI] [PubMed] [Google Scholar]
- 51.Ali M, Oshiki M, Awata T, Isobe K, Kimura Z, Yoshikawa H, et al. Physiological characterization of anaerobic ammonium oxidizing bacterium ‘Candidatus Jettenia caeni’. Environ Microbiol. 2015;17:2172–89. doi: 10.1111/1462-2920.12674. [DOI] [PubMed] [Google Scholar]
- 52.Yan J, Haaijer SCM, Camp HJMOD, Niftrik LV, Stahl DA, Könneke M, et al. Mimicking the oxygen minimum zones: stimulating interaction of aerobic archaeal and anaerobic bacterial ammonia oxidizers in a laboratory-scale model system. Environ Microbiol. 2012;14:3146–58. doi: 10.1111/j.1462-2920.2012.02894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pitcher A, Villanueva L, Hopmans EC, Schouten S, Reichart GJ, Damsté JSS. Niche segregation of ammonia-oxidizing archaea and anammox bacteria in the arabian sea oxygen minimum zone. ISME J. 2011;5:1896. doi: 10.1038/ismej.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S, Radny D, Huang S, Zhuang L, Zhao S, Berg M, et al. Nitrogen loss by anaerobic ammonium oxidation in unconfined aquifer soils. Sci Rep. 2017;7:40173. doi: 10.1038/srep40173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bauer M, Kube M, Teeling H, Richter M, Lombardot T, Allers E, et al. Whole genome analysis of the marine bacteroidetes ‘gramella forsetii’ reveals adaptations to degradation of polymeric organic matter. Environ Microbiol. 2006;8:2201–13. doi: 10.1111/j.1462-2920.2006.01152.x. [DOI] [PubMed] [Google Scholar]
- 56.Strous M, Fuerst JA, Kramer EHM, Logemann S, Muyzer G, Passchoonen KTVD, et al. Missing lithotroph identified as new planctomycete. Nature. 1999;400:446–9. doi: 10.1038/22749. [DOI] [PubMed] [Google Scholar]
- 57.Kelso BHL, Smith RV, Laughlin RJ, Lennox SD. Dissimilatory nitrate reduction in anaerobic sediments leading to river nitrite accumulation. Appl Environ Microbiol. 1997;63:4679–85. doi: 10.1128/aem.63.12.4679-4685.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith RV, Burns LC, Doyle RM, Lennox SD, Kelso BHL, Foy RH, et al. Free ammonia inhibition of nitrification in river sediments leading to nitrite accumulation. J Environ Qual. 1997;26:1049–55. doi: 10.2134/jeq1997.00472425002600040016x. [DOI] [Google Scholar]
- 59.He J, Shen J, Zhang L, Zhu Y, Zheng Y, Xu M, et al. Quantitative analyses of the abundance and composition of ammonia-oxidizing bacteria and ammonia-oxidizing archaea of a Chinese upland red soil under long-term fertilization practices. Environ Microbiol. 2007;9:2364–74. doi: 10.1111/j.1462-2920.2007.01358.x. [DOI] [PubMed] [Google Scholar]
- 60.Wang S, Wang Y, Feng X, Zhai L, Zhu G. Quantitative analyses of ammonia-oxidizing archaea and bacteria in the sediments of four nitrogen-rich wetlands in China. Appl Microbiol Biotechnol. 2011;90:779–87. doi: 10.1007/s00253-011-3090-0. [DOI] [PubMed] [Google Scholar]
- 61.Zhou L, Wang S, Zou Y, Xia C, Zhu G. Species, abundance and function of ammonia-oxidizing archaea in inland waters across China. Sci Rep. 2015;5:15969. doi: 10.1038/srep15969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yin S, Chen D, Chen L, Edis R. Dissimilatory nitrate reduction to ammonium and responsible microorganisms in two Chinese and Australian paddy soils. Soil Biol Biochem. 2002;34:1131–7. doi: 10.1016/S0038-0717(02)00049-4. [DOI] [Google Scholar]
- 63.Jones ZL, Jasper JT, Sedlak DL, Sharpa JO. Sulfide-induced dissimilatory nitrate reduction to ammonium supports anaerobic ammonium oxidation (Anammox) in an open-water unit process wetland. Appl Environ Microbiol. 2017;83:e00782. [DOI] [PMC free article] [PubMed]
- 64.Kuypers MMM, Lavik G, Woebken D, Schmid M, Fuchs BM, Amann R, et al. Massive nitrogen loss from the Benguela upwelling system through anaerobic ammonium oxidation. Proc Natl Acad Sci USA. 2005;102:6478–83. doi: 10.1073/pnas.0502088102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.