Abstract
BCL2A1 is an anti-apoptotic member of the BCL-2 family that contributes to chemoresistance in a subset of tumors. BCL2A1 has a short half-life due to its constitutive processing by the ubiquitin–proteasome system. This constitutes a major tumor-suppressor mechanism regulating BCL2A1 function. However, the enzymes involved in the regulation of BCL2A1 protein stability are currently unknown. Here, we provide the first insight into the regulation of BCL2A1 ubiquitination. We present evidence that TRIM28 is an E3 ubiquitin-ligase for BCL2A1. Indeed, endogenous TRIM28 and BCL2A1 bind to each other at the mitochondria and TRIM28 knock-down decreases BCL2A1 ubiquitination. We also show that TRIM17 stabilizes BCL2A1 by blocking TRIM28 from binding and ubiquitinating BCL2A1, and that GSK3 is involved in the phosphorylation-mediated inhibition of BCL2A1 degradation. BCL2A1 and its close relative MCL1 are thus regulated by common factors but with opposite outcome. Finally, overexpression of TRIM28 or knock-out of TRIM17 reduced BCLA1 protein levels and restored sensitivity of melanoma cells to BRAF-targeted therapy. Therefore, our data describe a molecular rheostat in which two proteins of the TRIM family antagonistically regulate BCL2A1 stability and modulate cell death.
Subject terms: Ubiquitylation, Ubiquitylation, Ubiquitylation, Oncogenes
Introduction
The BCL-2 protein family plays a pivotal role in the regulation of the intrinsic pathway of apoptosis by controlling the release of cytochrome c from mitochondria and thereby the activation of caspases [1]. Members of the BCL-2 family fall into pro-apototic or anti-apoptotic subgroups based on the presence of BCL-2 homology (BH) domains: in humans the six anti-apoptotic proteins BCL-2, BCL-xL, BCL-B, BCL-w, MCL-1, and BCL2A1 (also named Bfl-1/A1) contain up to four BH domains, while pro-apoptotic members belong to either the multi-BH containing BAX and BAK or the BH3-only proteins (e.g. BID, BIM, PUMA).
BCL2A1 is one of the least studied members of the BCL-2 family. In line with its anti-apoptotic activity, BCL2A1 is highly up-regulated in several hematopoietic malignancies including therapy-resistant B-cell chronic lymphocytic leukemia (B-CLL), acute myeloid leukemia (AML) with poor prognosis, and large B-cell lymphomas [2–7]. Recently, a marked overexpression of BCL2A1, or the amplification of its transcription factor MITF were shown to correlate with resistance of melanoma cells to BRAF-directed therapy [8]. Down-regulation of BCL2A1 in most of these malignancies restores sensitivity to chemotherapeutics, providing a clear therapeutic rationale for targeting BCL2A1 in cancer [8–10]. Structure-based medicinal chemistry has generated small molecule inhibitors tailored to specifically bind the BH3-binding cleft of anti-apoptotic BCL-2 proteins, blocking their survival activity and restoring the sensitivity of cancer cells towards apoptosis. To date, this strategy has not provided inhibitors of BCL2A1, possibly because of the atypical shape of BCL2A1 hydrophobic groove, which sequesters a restricted spectrum of BH3-only proteins, namely NOXA, PUMA, and BIM [11–13]. Moreover, in some cases, resistance to these BH3 mimetics developed by cancer cells involves de novo synthesis of BCL2A1 protein [14–16].
BCL2A1 is characterized by a short half-life which limits its intrinsic pro-survival activity [17, 18], a characteristic shared with its two closest relatives MCL-1 and BCL-B [19]. Importantly, reducing ubiquitin-proteasome-mediated degradation of BCL2A1 favors tumor formation in vivo [20]. BCL2A1 differs from other pro-survival BCL-2 proteins in that it contains a C-terminal helix phylogenetically unrelated to that found in other BCL-2 homologs [21] which does not fulfill the criteria of a typical transmembrane domain [22–24]. Interestingly this C-terminal segment shows features of a degron as it regulates BCL2A1 stability and includes lysine residues critical for BCL2A1 ubiquitination and degradation by the proteasome [17, 18, 20]. In addition, phosphorylation of crucial residues within the degron impairs both ubiquitination and degradation of BCL2A1 [18, 20]. Whereas the identification of several protein kinases, E3 ubiquitin-ligases and a deubiquitinase (DUB) of MCL-1 fueled intense efforts to design compounds aimed at inducing proteasomal degradation of MCL-1 [25, 26], the key determinants that regulate the ubiquitination and degradation of BCL2A1 remain unknown.
Here, we present evidence that TRIM28 is an E3 ubiquitin-ligase for BCL2A1. We also describe the existence of a “molecular rheostat” in which TRIM17 inhibits the ubiquitination and proteasomal degradation of BCL2A1 induced by TRIM28. Importantly, overexpression of TRIM28 or downregulation of TRIM17 reduced the protein level of BCLA1 and restored sensitivity to BRAF-targeted therapy in melanoma cells that exhibit a survival dependency on BCL2A1. Last, our data suggest that GSK3 is involved in the inhibition of BCL2A1 degradation.
Results
TRIM28 as the first E3 ubiquitin-ligase of BCL2A1 to be identified
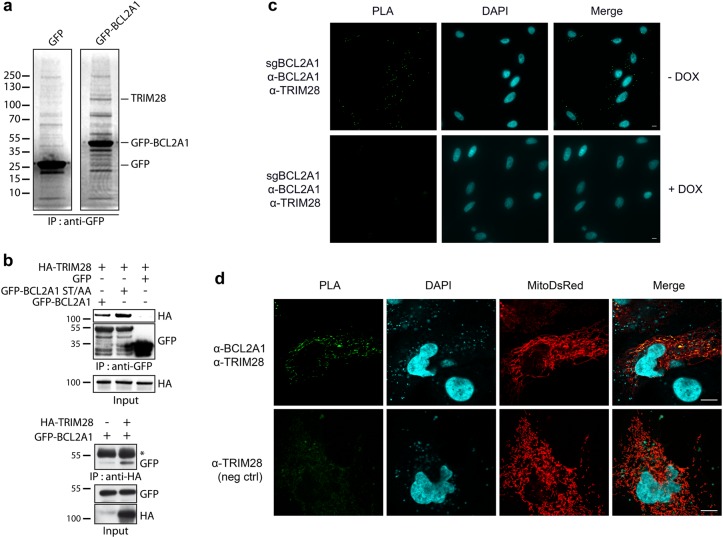
In order to identify the E3 ubiquitin-ligases that induce poly-ubiquitination of BCL2A1, we searched for binding partners of BCL2A1. Endogenous proteins from HEK293T cells co-immunoprecipitating specifically with GFP-tagged BCL2A1 were separated by SDS-PAGE and analyzed by mass spectrometry. A ∼110 kDa band corresponded to TRIM28 in GFP-BCL2A1-expressing cells but not in control GFP-expressing cells (Fig. 1a and Fig. S1a). Co-immunoprecipitation experiments confirmed that TRIM28 does interact with BCL2A1. Indeed, GFP-Trap beads precipitated HA-TRIM28 together with GFP-BCL2A1, with a marked preferential interaction of TRIM28 with the phosphorylation defective mutant BCL2A1(S152A,T156A), compared with wild type BCL2A1 (Fig. 1b, upper panel). This mutant has been previously shown to be highly ubiquitinated and labile [18, 20]. Reciprocal co-immunoprecipitations using HA-Trap beads further corroborated a physical association of these two proteins (Fig. 1b, lower panel). Then, we conducted in situ proximity ligation assay (PLA) in the SK-MEL-28 melanoma cell line that expresses high levels of endogenous BCL2A1. Close proximity was detected between endogenous TRIM28 and endogenous BCL2A1 proteins, as assessed by a PLA signal, which increased following ectopic expression of Flag-BCL2A1 (Fig. S1b) and was abolished when BCL2A1 gene expression was impaired by an inducible CRISPR/Cas9 system (Fig. 1c). As BCL2A1 is a mitochondrial protein and TRIM28 is mainly nuclear, we examined the subcellular localization of the interaction between the two endogenous proteins in HuH7 hepatocarcinoma cells. Our PLA data strongly suggest that they interact mainly at the level of mitochondria (Fig. 1d). Taken together, these results indicate that a mitochondrial pool of TRIM28 forms complexes with BCL2A1.
Fig. 1.
TRIM28 binds BCL2A1 at mitochondria. a Total protein extracts from HEK293T cells expressing either GFP-BCL2A1 or control GFP were subjected to immunoprecipitation using anti-GFP antibody. Immunoprecipitates were separated by SDS-PAGE and visualized using colloidal Coomassie-staining. b GFP-tagged BCL2A1, or its phosphorylation-defective and unstable mutant BCL2A1(S152A,T156A), were co-expressed with HA-tagged TRIM28 in HEK293T cells as indicated. GFP-BCL2A1 (upper panel) or HA-TRIM28 (lower panel) was immunoprecipitated with GFP-trap beads or HA-beads, respectively. HA-TRIM28 and GFP-BCL2A1 were detected in immunoprecipitates and input samples by western-blot using appropriate antibodies. (*) shows IgG heavy chains. c In situ proximity ligation assay (PLA) was performed in SK-MEL-28 melanoma cells expressing a DOX-inducible sgRNA targeting BCL2A1 (see Fig. 6), using anti-TRIM28 and anti-BCL2A1 antibodies. Each green bright spot indicates the very close proximity of the two endogenous proteins. A negative control was obtained by inducing the BCL2A1 sgRNA to efficiently induce InDels in the BCL2A1 locus and prevent BCL2A1 expression (see Fig. S4b). Scale bars, 10 μm. d PLA was performed in HuH7 hepatocarcinoma cells using anti-BCL2A1 and anti-TRIM28 antibodies. Mitochondria were imaged following transfection of the mitoDsRed plasmid encoding fluorescent DsRed2 fused to the mitochondrial targeting sequence from subunit VIII of human cytochrome c oxidase. A negative control was obtained by omitting the anti-BCL2A1 antibody. Scale bars, 10 μm
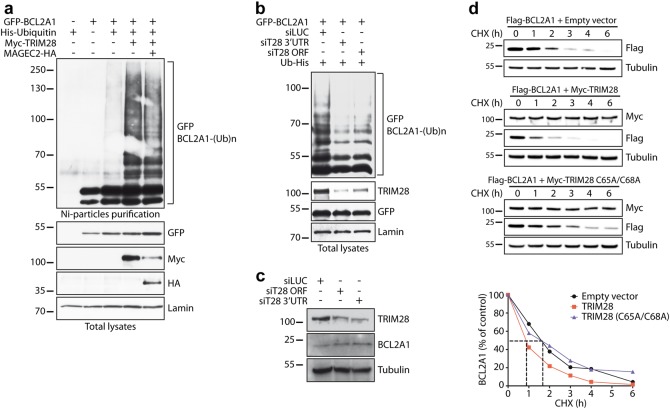
We next examined whether TRIM28 functions as a bona fide BCL2A1 E3 ubiquitin-ligase. As previous studies showed that TRIM28 E3 ubiquitin-ligase activity can be modulated by MAGE proteins [27, 28], we measured the ubiquitination level of BCL2A1 in HEK 293T cells, which do not express endogenous MAGE proteins [27]. Interestingly, BCL2A1 ubiquitination was strongly stimulated in the presence of TRIM28 regardless of MAGE-C2 co-expression (Fig. 2a). More importantly, depletion of endogenous TRIM28 by two independent siRNAs both strongly decreased the polyubiquitination of ectopically expressed BCL2A1 in HEK cells (Fig. 2b) and increased the protein level of endogenous BCL2A1 in SK-MEL-28 cells (Fig. 2c). In addition, we measured the half-life of Flag-BCL2A1, with or without co-transfected TRIM28. Notably, wild type TRIM28, but not the inactive TRIM28(C65A/C68A) RING mutant, induced a two-fold decrease in Flag-BCL2A1 half-life (Fig. 2d) indicating that TRIM28 stimulates BCL2A1 protein degradation. Moreover, this effect depends on the presence of a valid RING domain responsible for the E3 ubiquitin-ligase activity of TRIM28. Altogether, these results strongly suggest that TRIM28 is an E3 ubiquitin-ligase for BCL2A1 involved in the regulation of its stability.
Fig. 2.
TRIM28 regulates the ubiquitination and degradation of BCL2A1. a HEK293T cells were transfected with GFP-BCL2A1, Myc-TRIM28, and MAGEC2-HA constructs as indicated, together with His-tagged ubiquitin (Ub-His) for 18 h. Then cells were incubated with MG132 for 6 h. Total ubiquitinated proteins were purified using nickel beads and analyzed by western blot using anti-GFP antibody to detect poly-ubiquitinated forms of BCL2A1. Initial total lysates were analyzed for the expression of the different proteins by immunoblot. b HEK293T cells were first transfected with two different siRNAs to inhibit TRIM28 expression. 24 h later, cells were transfected with GFP-BCL2A1 and Ub-His for one additional day. Then, cells were treated and cell lysates were analyzed. c SK-MEL-28 cells were transfected with two different siRNAs for two consecutive days to inhibit TRIM28 expression. Cells were collected 48 h after the first siRNA transfection. The efficiency of TRIM28 silencing and its effect on the protein level of endogenous BCL2A1 were assessed by immunoblot. d HEK293T cells were co-transfected with Flag-tagged BCL2A1 and Myc-tagged TRIM28 or an inactive RING mutant C65/68A of TRIM28 for 24 h. Transfected cells were treated with the protein synthesis inhibitor cycloheximide (CHX, 10 μg/ml) for increasing times as indicated. Total protein extracts were analyzed by immunoblot. The protein level of Flag-BCLA1 was followed with time using anti-Flag antibody in order to measure its half-life. Anti-Myc antibody was used to verify equal expression of TRIM28 and anti-tubulin antibody to assess equal loading. Data shown are representative of three independent experiments. BCL2A1 protein level was quantified by densitometry and was expressed as a percentage of the value measured at time zero for each of the three conditions
TRIM17 enhances BCL2A1 stability by inhibiting TRIM28-mediated ubiquitination of BCL2A1
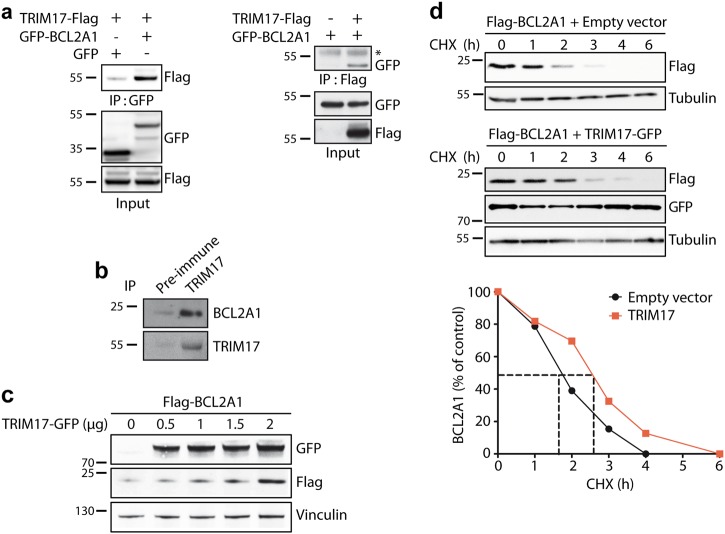
We have previously shown that TRIM17 is an E3 ubiquitin-ligase for MCL-1, the closest phylogenetic homolog of BCL2A1 [21, 29]. To test whether TRIM17 could also modulate BCL2A1 stability, we first examined whether TRIM17 binds to BCL2A1. Co-immunoprecipitation experiments showed a significant interaction between both ectopically expressed (Fig. 3a), and endogenous (Fig. 3b) TRIM17 and BCL2A1 proteins. Then, we co-expressed Flag-BCL2A1 along with increasing amounts of GFP-TRIM17 plasmid. Surprisingly, Flag-BCL2A1 significantly accumulated as TRIM17 expression increased (Fig. 3c). This accumulation was due to a stabilization of BCL2A1, as TRIM17 expression led to an increase in Flag-BCL2A1 half-life (Fig. 3d).
Fig. 3.
TRIM17 induces the stabilization of BCL2A1 protein. a HEK293Tcells were transfected with GFP-tagged BCL2A1 and TRIM17-Flag as indicated. Cell lysates were subjected to immunoprecipitation with GFP-Trap beads (left) or Flag-beads (right), and the presence of TRIM17 or BCL2A1 was detected by western blot using anti-Flag or anti-GFP antibodies respectively (*) shows IgG heavy chains. b SK-MEL-28 protein extract was subjected to immunoprecipitation using an anti-TRIM17 antibody or the corresponding pre-immune serum as an negative control, as indicated. BCL2A1 and TRIM17 proteins were detected in the immunoprecipitate by western blot using specific antibodies. c HEK293T cells were transfected with FLAG-BCL2A1 and increasing amounts of TRIM17-GFP vectors for 48 h. Total protein extracts were subjected to immunoblot analyses using the indicated antibodies. d HEK293T cells were transfected with Flag-tagged BCL2A1 in the presence or the absence of GFP-tagged TRIM17 for 24 h. Transfected cells were treated with cycloheximide (CHX, 10 μg/ml) for the indicated time periods. Total protein extracts were analyzed by immunoblot. The protein level of Flag-BCLA1 was followed using anti-Flag antibody. Anti-GFP antibody was used to verify equal expression of TRIM17 and anti-tubulin antibody to assess equal loading. Data shown are representative of three independent experiments. BCL2A1 protein level was quantified by densitometry and was expressed as a percentage of the value measured at time zero for each of the two conditions
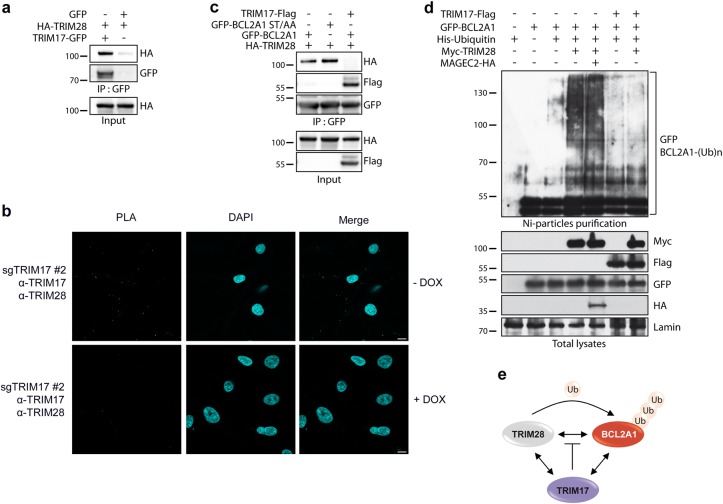
To gain insights into the mechanism of BCL2A1 stabilization by TRIM17, we first tested whether TRIM17 could bind TRIM28. Indeed, TRIM17 exhibited a strong interaction with TRIM28 as determined by co-immunoprecipitation of ectopically expressed proteins (Fig. 4a), and the detection of a specific PLA signal between the two endogenous proteins in SK-MEL-28 cells (Fig. 4b). Then, we co-expressed the three partners, BCL2A1, TRIM28, and TRIM17, and checked for the presence of TRIM proteins in BCL2A1 immunoprecipitates. Interestingly, TRIM17 completely abrogated the interaction between BCL2A1 and TRIM28, whereas the TRIM17/BCL2A1 interaction was preserved in the presence of TRIM28 (Fig. 4c). Moreover, ectopic expression of TRIM17 strongly reduced poly-ubiquitination of BCL2A1 induced by TRIM28 (Fig. 4d). Taken together, these results suggest that TRIM17 stabilizes BCL2A1 by inhibiting TRIM28-mediated ubiquitination of BCL2A1, most probably by preventing the physical interaction between BCL2A1 and its E3 ubiquitin-ligase (Fig. 4e).
Fig. 4.
TRIM17 impairs TRIM28/BCL2A1 interaction and prevents TRIM28-mediated ubiquitination of BCL2A1. a HEK293T cells were transfected with the indicated plasmids and cell lysates were subjected to immunoprecipitation with GFP-trap beads. Immunoprecipitates were analyzed by western blot using anti-HA and anti-GFP antibodies. b PLA was performed in SK-MEL-28 cells expressing a DOX-inducible sgRNA against TRIM17 (see Fig. 6), using anti-TRIM17 and anti-TRIM28 antibodies. Each green bright spot indicates the very close proximity of the two endogenous proteins. A negative control was obtained by depleting TRIM17 using DOX treatment (see Fig. S4b), scale bars, 10 μm. c HEK293T cells were transfected with GFP-BCL2A1, its labile form BCL2A1(S152A, T156A), Flag-TRIM17 and TRIM28-HA plasmids as indicated. Cell lysates were subjected to immunoprecipitation with anti-GFP antibody to pull down BCL2A1, and the presence of TRIM17 and TRIM28 in the immunoprecipitates was subsequently detected using anti-HA or anti-Flag antibodies. d HEK293T cells were transfected with the indicated plasmids in the presence or the absence of His-ubiquitin. Cells were treated with MG132 for 6 h. Total ubiquitinated proteins were purified using nickel beads and analyzed by western blot using anti-GFP antibody to detect poly-ubiquitinated forms of BCL2A1. Initial total lysates were analyzed for the expression of the different proteins by immunoblot. e Working model: the three proteins interact with each other (double arrows represent physical interactions); TRIM28 mediates BCL2A1 poly-ubiquitination; TRIM17 inhibits TRIM28-mediated ubiquitination of BCL2A1 by preventing the interaction between the E3 ubiquitin-ligase and its substrate
TRIM17/TRIM28 as a molecular rheostat modulating the survival function of BCL2A1 in melanoma cells
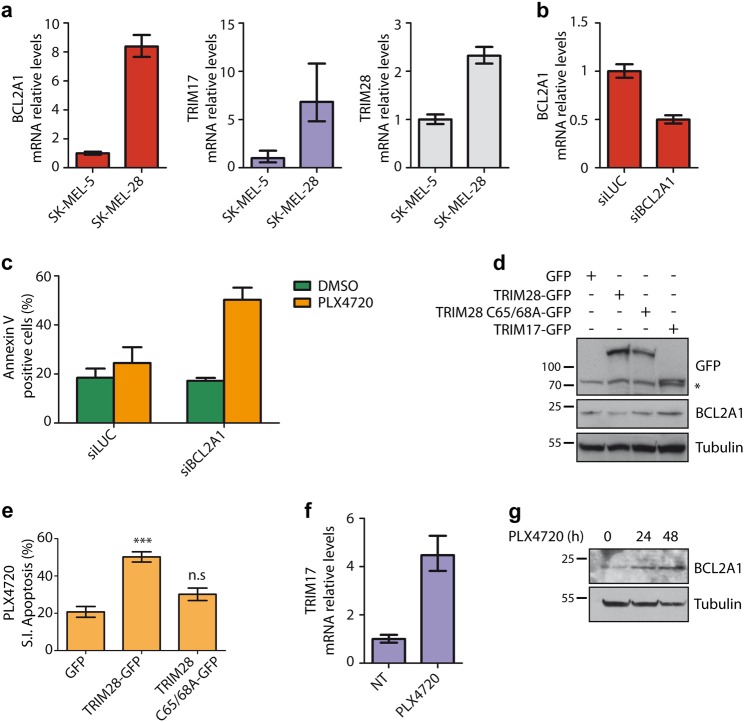
We next focused on TRIM17/TRIM28 antagonistic functions on BCL2A1 stability as a possible determinant of chemoresistance in human cancers. Previous studies demonstrated that BCL2A1 is frequently amplified in melanoma tumors and that its overexpression promotes tumorigenesis and resistance to several apoptosis-inducing drugs including some of the clinically approved BRAF inhibitors [8, 30]. Our real-time RT-PCR data show that TRIM17 and TRIM28 are expressed in SK-MEL-5 and SK-MEL-28, two cell lines harboring a BRAF(V600E) mutation and high BCL2A1 expression [8] (Fig. 5a), with a more robust expression in SK-MEL-28 cells (Fig. 5a). Strikingly, a reduction of BCL2A1 mRNA levels by half using a specific siRNA (Fig. 5b) induced a strong sensitization of SK-MEL-28 cells to the BRAF inhibitor PLX4720, with a two-fold increase in apoptotic cells (Fig. 5c). Altogether, these results establish a clear link between BRAF inhibitor resistance and BCL2A1 expression in SK-MEL-28 cells and define these cells as an appropriate model to study the effect of the TRIM17/TRIM28 balance on BCL2A1 expression and pro-survival activity.
Fig. 5.
TRIM17 and TRIM28 in BCL2A1-dependent chemoresistant melanoma cells. a Total RNA was extracted from SK-MEL-5 and SK-MEL-28 melanoma cells and mRNA levels of indicated genes were estimated by quantitative RT-PCR. b SK-MEL-28 cells were transfected with a control siRNA (siLUC) or with a specific siRNA against BCL2A1 for 24 h. Total RNAs were collected and the mRNA levels of BCL2A1 were estimated by quantitative RT-PCR. The data are the means ± SD of triplicate samples from a representative experiment. c SK-MEL-28 cells were transfected with a control siRNA (siLUC) or with a specific siRNA against BCL2A1 for 24 h. Then, cells were treated with 20 μM PLX4720 for 24 h and apoptosis was estimated by flow cytometry using AnnexinV staining. d SK-MEL-28 cells were transfected with GFP or GFP-tagged TRIM proteins, as indicated, for 24 h. Total protein lysates were analyzed by western blot for the expression of endogenous BCL2A1 and overexpressed proteins. Note that a part of SK-MEL-28 cells was not transfected and thus BCL2A1 variations were certainly underestimated. e SK-MEL-28 cells were transfected with GFP-tagged TRIM28 or TRIM28(C65A/C68A) for 24 h and subsequently treated with 20 μM PLX4720 for 24 h. Apoptosis was estimated in the GFP-positive cell population by flow cytometry using AnnexinV (APC) staining. Data are presented as % of specific induced apoptosis (SIA, see Methods) and are the means ± SEM of four independent experiments. ***p = 0.0001 significantly different from GFP-transfected cells, ns p = 0.1025 non-significantly different from GFP-transfected cells (one-way ANOVA followed by Dunnett’s multiple comparison test). f SK-MEL-28 cells were treated or not (NT) with 20 μM PLX4720 for 24 h. Total RNA was extracted and mRNA level of TRIM17 was assessed by quantitative RT-PCR. g SK-MEL-28 cells were treated or not (NT) with 20 μM PLX4720 for 24 and 48 h. BCL2A1 protein level was assessed by immunoblot
To address this latter issue, SK-MEL-28 cells were transfected with GFP-tagged versions of TRIM17 and TRIM28. Consistent with our data showing that TRIM28 accelerates BCL2A1 degradation (Fig. 2), overexpression of TRIM28, but not of its inactive RING mutant (C65A/C68A), decreased the protein level of endogenous BCL2A1 (Fig. 5d). In contrast, TRIM17 overexpression resulted in BCL2A1 accumulation (Fig. 5d), in agreement with TRIM17-mediated stabilization of BCL2A1 (Fig. 3c, d). Moreover, TRIM28 overexpression induced an increased sensitivity of SK-MEL-28 cells towards PLX4720-mediated apoptosis, whereas its inactive RING mutant had no effect (Fig. 5e, Fig. S2). Interestingly, mRNA levels of TRIM17 showed a four-fold increase in SK-MEL-28 cells treated with PLX4720 (Fig. 5f), correlating with an accumulation of BCL2A1 protein (Fig. 5g). Therefore, it is tempting to hypothesize that TRIM17 induction may participate in chemoresistance to PLX4720 by inhibiting TRIM28-mediated elimination of BCL2A1.
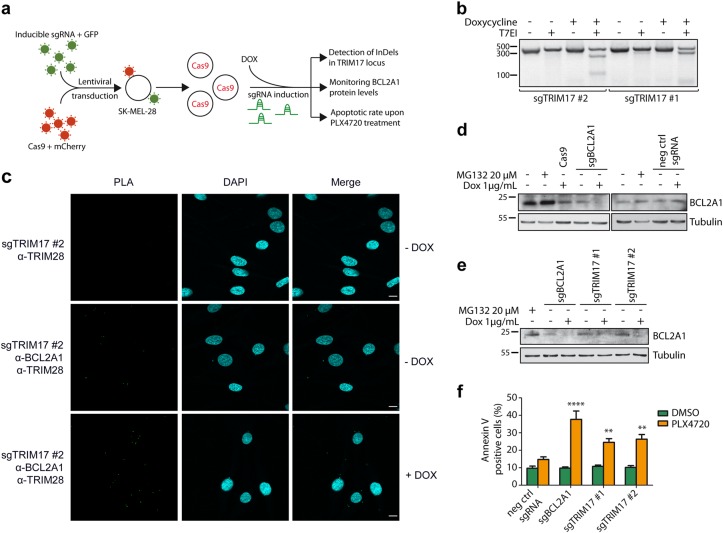
To test this hypothesis, we used an inducible CRISPR/Cas9 system [31] to induce efficient and temporally controlled depletion of TRIM17 or BCL2A1 in SK-MEL-28 cells (Fig. 6a). This system consists in two lentiviral vectors, one allowing a constitutive expression of Cas9-T2A-mCherry and the other allowing both doxycycline-inducible expression of specific single guide RNA (sgRNA) and constitutive expression of the tetracycline repressor and eGFP protein [31]. Two different sgRNAs targeting TRIM17 (sgTRIM17#1 and #2), one sgRNA against BCL2A1 (sgBCL2A1) [32] and one negative control sgRNA (targeting mouse Bim) were used. Doubly transduced SK-MEL-28 cells were selected by fluorescence-activated cell sorting (FACS) based on mCherry and eGFP expression (Fig. S3). As expected, following a 72 h doxycycline treatment, InDels (insertions and deletions of bases in the genomic DNA) were detected in exon 1 of the TRIM17 locus with both sgTRIM17#1 and sgTRIM17#2 by using the T7E1 assay (Fig. 6b) and next-generation sequencing (Fig. S4a) [31]. Doxycycline treatment resulted in an average of 72% mutation rates with sgTRIM17#1, 39% with sgTRIM17#2 and up to 95% with sgBCL2A1, whereas no InDels were detected at TRIM17 or BCL2A1 loci using the negative control sgRNA (Fig. S4). Importantly, depletion of endogenous TRIM17 using this system increased the number of PLA dots, measuring the close proximity between endogenous TRIM28 and BCL2A1, in SK-MEL-28 cells (Fig. 6c), further supporting the notion that TRIM17 restricts the TRIM28/BCL2A1 interaction. As expected, expression of sgBCL2A1 induced a strong depletion of BCL2A1 protein (Fig. 6d). Importantly, expression of the two independent sgRNA targeting TRIM17 also reduced BCL2A1 protein level (Fig. 6e) without affecting its mRNA level (Fig. S5a), thereby supporting the idea that endogenous TRIM17 regulates BCL2A1 at the post-translational level.
Fig. 6.
TRIM17 invalidation induces BCL2A1 degradation and sensitizes melanoma cells to BRAF-targeted therapy. a Method outline depicting generation of TRIM17-depleted SK-MEL-28 cells by inducible CRISPR/Cas9 method and workflow of subsequent steps of analysis. b SK-MEL-28 cells expressing Cas9 and inducible sgRNAs against TRIM17 were treated for 72 h with 1 μg/ml doxycycline. Genomic DNA of cells was amplified by PCR, around the Cas9 cleavage sites targeted by the two sgTRIM17, and PCR products were analyzed by a T7 Endonuclease I assay. The presence of InDels was visualized by the digestion of the PCR products at the Cas9 cleavage sites by T7EI. c PLA was performed in SK-MEL-28 cells expressing DOX-inducible sgRNA against TRIM17, using anti-TRIM28 and anti-BCL2A1 antibodies. Note that the number of green bright spots indicating the very close proximity between endogenous TRIM28 and BCL2A1 proteins increases when expression of endogenous TRIM17 is inhibited by doxycycline treatment. Negative control was obtained by omitting the anti-BCL2A1 antibody. Scale bars, 10 μm. d The protein level of endogenous BCL2A1 was assessed by immunoblot using anti-BCL2A1 antibody in total protein extracts from non-transduced SK-MEL-28 cells left untreated or treated with 20 μM MG132 for 6 h, or from transduced cells expressing Cas9 alone or Cas9 together with inducible sgRNAs against BCL2A1 or mouse bim (negative control) and treated or not with 1 μg/ml doxycycline for 72 h, as indicated. e Two sgRNAs against TRIM17 were induced in SK-MEL-28 cells by doxycycline treatment for 72 h and protein level of endogenous BCL2A1 was assessed by immunoblot using anti-BCL2A1 antibody. f SK-MEL-28 cells expressing Cas9 and the indicated sgRNAs were treated with 1 μg/ml doxycycline for 72 h and subsequently treated with 20 μM PLX4720 (or DMSO control) for 48 h. PLX4720-specific induced apoptosis (SIA) was assessed by annexin V staining and flow cytometry. Data are the means ± SEM of three independent experiments. ****p < 0.0001; **p = 0.0093 (sgTRIM17#1), and p = 0.0032 (sgTRIM17#2) significantly different from negative control (two-way ANOVA followed by Sidak’s multiple comparison test)
To examine whether TRIM17 plays a role in chemoresistance, we assessed the sensitivity of SK-MEL-28 cells to PLX4720 following induction of the different sgRNAs. As previously shown with RNA interference (Fig. 5b, c), PLX4720 rapidly induced apoptosis in sgBCL2A1-expressing cells (Fig. 6f). Importantly, induction of the two sgRNAs targeting TRIM17 significantly sensitized melanoma cells to PLX4720 treatment (Fig. 6f). In contrast, treatment with doxycycline alone did not trigger any cell death in sgTRIM17 and sgBCL2A1-transduced SK-MEL-28 cells for up to 72 h (Fig. S5b). Therefore, our data indicate that alteration of TRIM17 expression both reduces BCL2A1 protein levels and sensitizes melanoma cells to BRAF-directed therapy.
Role of the protein kinase GSK3 in the regulation of BCL2A1 stability
In contrast to our previous identification of TRIM17 as an E3 ubiquitin-ligase for MCL-1 [29], our present data suggest that TRIM17 counteracts BCL2A1 ubiquitination, although both proteins are the closest homologs among anti-apoptotic members of the BCL-2 family. Likewise, GSK3-induced phosphorylation promotes MCL-1 ubiquitination and degradation [33], whereas phosphorylation of BCL2A1 C-terminal alpha-9 helix prevents its protein decay [18, 20]. Consistently, the phosphorylation defective mutant BCL2A1(S152A,T156A), that is highly ubiquitinated and very unstable [18, 20] interacts more strongly with TRIM28 compared with wild type BCL2A1 (Fig. 1b). Given the opposite effect of TRIM17 and phosphorylation on BCL2A1 and MCL-1, and as GSK3 is a protein kinase for MCL-1, we examined whether GSK3 could also regulate BCL2A1 stability.
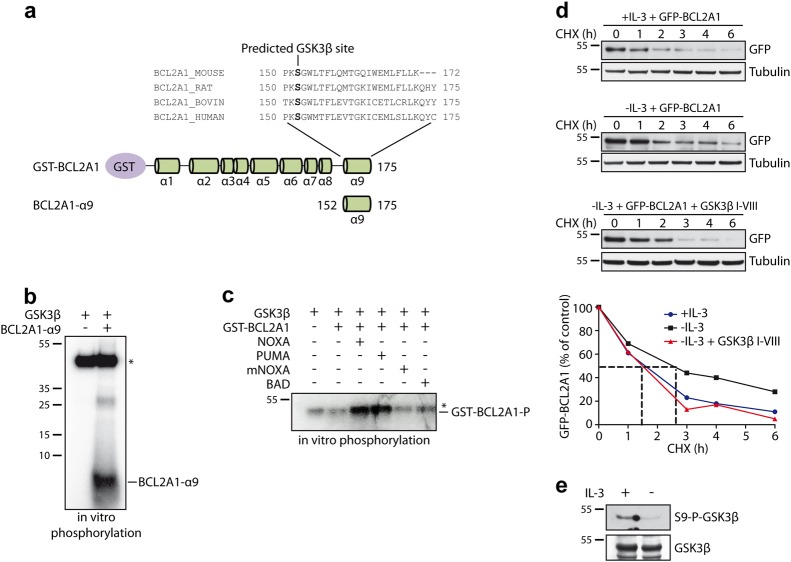
First, we found that the highly conserved serine 152 within the α9 degron sequence of BCL2A1 was predicted to be a GSK3 consensus site (Fig. 7a). Consistently, a synthetic peptide derived from the α9 helix of BCL2A1 (BCL2A1-α9) was phosphorylated in vitro with recombinant GSK3 (Fig. 7b). In contrast, no increase in phosphorylation was observed in vitro when the full-length GST-tagged BCL2A1(1–175) protein and GSK3 were incubated together (Fig. S6). However, according to a three-dimensional structure model, full-length BCL2A1(1–175) may adopt two distinct conformational states: one with the C-terminal helix α9 located within the hydrophobic BH3-binding cleft and one with the helix α9 freely accessible [34]. Based on this model, we reasoned that a BH3 peptide able to interact with the hydrophobic groove of BCL2A1 would dislodge the helix α9 from the groove and would thus expose its phosphorylation-prone residues. Therefore, we performed in vitro phosphorylation following pre-incubation of full-length GST-BCL2A1 with different BH3 peptides. Under these conditions, a strong GSK3-mediated phosphorylation of BCL2A1 was detected with BH3 peptides from NOXA and PUMA, which are known to interact with BCL2A1, whereas neither a murine NOXA-derived nor a BAD-derived BH3 peptide, both known to have poor affinity for BCL2A1 [11, 13], were able to facilitate GSK3-mediated phosphorylation of BCL2A1 (Fig. 7c).
Fig. 7.
GSK3 phosphorylates BCL2A1 in vitro and is involved in BCL2A1 stabilization in cells. a NetPhosK 1.0 software was used to predict phosphorylation sites of alpha9 helix of BCL2A1 and identified Ser152 residue as a putative phosphorylation site for GSK3. b A synthetic peptide derived from α9 helix of BCL2A1 was incubated with recombinant GSK3 in the presence of [γ-32P]-ATP. The reaction mix was separated by SDS-PAGE and radiolabeled phosphate incorporation was detected by autoradiography. *Indicates auto-phosphorylated recombinant GSK3. c Purified GST-BCL2A1 full-length recombinant protein was incubated with indicated synthetic BH3 peptides derived from indicated BH3 proteins for 2 h at 4 °C. In vitro phosphorylation has subsequently carried out by introducing recombinant GSK3 and [γ-32P]-ATP in the reaction. The protein mix was resolved by SDS-PAGE and gel was then analyzed by autoradiography. *Indicates auto-phosphorylated recombinant GSK3 which migrates approximately at the same level as phosphorylated BCL2A1. d FL5.12 cells stably expressing GFP-BCL2A1 were maintained in the presence of IL-3, or deprived of IL-3, in the presence or the absence of GSK3β I-VIII inhibitor for 4 h. Then, cycloheximide (CHX) was added to the respective culture media for the indicated time periods. Total protein extracts were analyzed by immunoblot. The protein level of GFP-BCLA1 was followed by using anti-GFP antibody. Anti-tubulin antibody was used to assess equal loading. Data shown are representative of three independent experiments. BCL2A1 protein level was quantified by densitometry and was expressed as the percentage of the value measured at time zero for each of the three conditions. e FL5.12 cells were cultured in the presence or the absence of IL-3 and endogenous GSK3 activation was assessed by estimating GSK3 dephosphorylation at Ser9 using anti Phospho Ser9 GSK3 antibody
To further examine whether endogenous GSK3 could influence BCL2A1 stability in cells, we used an interleukin-3 (IL-3)-dependent FL5.12 cell line stably expressing GFP-BCL2A1. IL-3 withdrawal is known to induce a strong GSK3 activation in FL5.12 cells [33]. Consistently, we found a substantial stabilization of BCL2A1 following IL-3 withdrawal (Fig. 7d), when GSK3 was activated (Fig. 7e). In contrast, BCL2A1 protein half-life was not significantly different from the control condition when the cells were deprived of IL-3 in the presence of a specific GSK3 inhibitor (Fig. 7d). Altogether, these data suggest that GSK3 is involved in the phosphorylation-mediated stabilization of BCL2A1.
Discussion
TRIM proteins that represent the largest class of RING-containing E3 ubiquitin-ligases [35], have various functions in cellular processes including apoptosis, autophagy, innate immunity, and carcinogenesis [36]. In the present study, we describe a molecular rheostat in which two TRIM proteins antagonistically regulate the ubiquitin-mediated degradation of BCL2A1, thereby modulating cell death. Indeed, we present several lines of evidence indicating that TRIM28 is an E3 ubiquitin-ligase for BCL2A1 that favors its degradation. Notably, overexpression of TRIM28 increased the ubiquitination level of BCL2A1 and decreased its half-life. More importantly, silencing of TRIM28 decreased the ubiquitination level of BCL2A1, indicating that endogenous TRIM28 participates in BCL2A1 ubiquitination. Interestingly, B-cell-specific TRIM28 KO mice display impaired B-cell maturation similar to that observed in transgenic mice overexpressing BCL2A1 in B-cells [37, 38]. As BCL2A1 is predominantly expressed within the hematopoietic lineage, where it plays a crucial role in B-cell maturation [23], these similar phenotypes suggest that impairing TRIM28 or overexpressing BCL2A1 has the same effect in B cells and further support the idea that TRIM28 regulates the protein level of BCL2A1.
As a predominantly nuclear protein, TRIM28 is known to regulate biological functions through transcriptional co-repression activity in association with heterochromatin-associated protein 1 (HP1) [39]. However, TRIM28 has also been described as a SUMO-ligase or ubiquitin-ligase, which can modify both nuclear and cytoplasmic proteins [27, 28, 40–42]. Consistently, our present findings indicate that the interaction between TRIM28 and BCL2A1 occurs mainly at the level of the mitochondria, suggesting that TRIM28 mediates BCL2A1 ubiquitination outside of the euchromatin context.
By inducing the ubiquitination and degradation of the prosurvival factor BCL2A1, TRIM28 may have an anti-tumoral activity in cancer cells whose survival depends on a high expression of BCL2A1. Consistently, liver-specific depletion of TRIM28 provokes hepatocarcinoma formation in mice [43]. In addition, TRIM28 has been shown to suppress the activity of the oncogenic transcription factors HIF-1α and STAT3 [44, 45], and to de-repress the transcription of major pro-apoptotic genes of the BCL-2 family including BAX, PUMA, and NOXA [46]. Conversely, TRIM28 level has been linked to poor prognosis in gastric cancer and thyroid carcinoma [47–49], and TRIM28 has been reported to ubiquitinate and eliminate p53 and AMPK, two factors that participate in tumor suppression [27, 28]. However, in the latter cases, the E3-ubiquitin ligase activity of TRIM28 requires the presence of MAGE cofactors [50], which is not the case for TRIM28-mediated ubiquitination of BCL2A1.
Our results further show that TRIM17 inhibits TRIM28-mediated ubiquitination of BCL2A1 and induces its stabilization. We propose a mechanism in which TRIM17 leads to the disruption of the TRIM28/BCL2A1 complex, thereby preventing the interaction between the E3 ubiquitin-ligase and its substrate. One possibility is that TRIM17 forms an inactive hetero-oligomer with TRIM28 that cannot bind BCL2A1. Indeed, we found that TRIM17 strongly interacts with TRIM28. Recent structural studies suggest that homo-oligomerization of TRIM proteins is crucial for their catalytic activity [51]. Therefore, it is tempting to speculate that formation of TRIM17/TRIM28 hetero-oligomers, at the expense of TRIM28 homo-oligomerization, prevents the E3 ubiquitin-ligase activity of TRIM28 and its interaction with BCL2A1. Alternatively, TRIM17 may compete with TRIM28 to bind BCL2A1. Indeed, we also found that TRIM17 is able to co-immunoprecipitate with BCL2A1. These two possibilities are not mutually exclusive and are in agreement with previous reports showing that TRIM proteins can both bind an E3-ubiquitin ligase and its substrate to prevent ubiquitination [52].
We have previously found that TRIM17 gene expression is upregulated early during neuronal apoptosis [53] and following different\cellular stresses (unpublished results). Notably, TRIM17 is induced following treatment of melanoma cells with the anti-cancer drug PLX4720. Therefore, TRIM17 induction may participate in chemoresistance, but also in tumorigenesis in cells undergoing chronic stress, by increasing BCL2A1 levels. Importantly, we have previously identified TRIM17 as an E3 ubiquitin-ligase for the anti-apoptotic protein MCL-1 in neurons [29]. Our present data thus suggest that TRIM17 may yield opposite effects in tissues concomitantly expressing the two pro-survival factors relatives MCL-1 and BCL2A1, by inducing the simultaneous down-regulation of MCL-1 and stabilization of BCL2A1. Our study further broadens this dichotomy by showing that GSK3-mediated phosphorylation of BCL2A1 prevents its degradation, whereas it is well documented that MCL-1 phosphorylation by GSK3 favors its protein decay [26, 33]. The current paradigm presents GSK3 as a pro-apoptotic protein as its expression induces cell death in neuronal and hematopoietic cells and GSK3 inhibitors protect neurons from apoptosis [29, 54–56]. Nevertheless, GSK3 expression has also been shown to have a protective function in liver cells [57]. This paradox could be resolved by the opposite effects that GSK3 can exert on anti-apoptotic proteins of the BCL-2 family, i.e. accumulation for BCL2A1 and degradation for MCL-1.
Although phylogenetically close, BCL2A1 differs from MCL-1 by its α9 C-terminal helix which is identical to a 28 amino acids stretch from the phylogenetically unrelated tumor suppressor protein HCCS-1 [22]. The duplication of this HCCS-1 sequence conferred a degron-like feature to BCL2A1 [17, 20]. Importantly, since HCCS-1 is believed to be pro-apoptotic, it is conceivable that this duplication event incidentally brought GSK3-mediated stabilization to BCL2A1 while it originally stabilized a pro-apoptotic protein. Yet, beyond these differences, BCL2A1 and MCL-1 do not share a similar pattern of tissue expression. Therefore, TRIM17 may have a pro-apoptotic effect in neurons where MCL-1 expression is crucial for survival and have an opposite anti-apoptotic effect in cell types expressing BCL2A1. Interestingly, extensive observations of transcriptional patterns underscore that MCL-1 mRNA levels are inversely correlated with BCL2A1 expression in melanoma. Notably, SK-MEL-28 cells which show high levels of BCL2A1, express almost no MCL-1 [8]. This may explain why depletion of TRIM17, using an inducible CRISPR/Cas9 system in these cells, restored their sensitivity to PLX4720-induced apoptosis, in a similar way as BCL2A1 silencing or TRIM28 overexpression. Our data thus provide a rationale for targeting TRIM17 to promote BCL2A1 degradation in order to restore sensitivity of BCL2A1-dependent cancer cells towards chemotherapeutics.
In contrast to MCL-1, which is necessary for the survival of a myriad of cell lineages [58], BCL2A1 is not indispensable for cell survival in normal physiology. Indeed, BCL2A1 knock-out mice show only minor defects in the hematopoietic compartment [59, 60]. Recent data also suggest that BCL2A1 plays a redundant role with BCL-2 and MCL-1 to maintain survival of immune cells [61]. Therefore, therapeutic targeting of BCL2A1 is expected to induce less deleterious side effects than MCL-1 targeting. Our present study provides the first insight into the regulation of BCL2A1 ubiquitination by identifying TRIM28 as one of its E3 ubiquitin-ligases, TRIM17 as a regulator of TRIM28 and GSK3 as a protein kinase regulating BCL2A1 stability. As such, our results may pave the road for the development of novel therapeutic strategies aiming at specifically down-regulating BCL2A1 in cancers that express abnormally high levels of this pro-survival protein.
Materials and methods
Cell culture
SK-MEL-28, SK-MEL-5, HEK293T, HuH7, cell lines were grown in high-glucose DMEM supplemented with 10% fetal calf serum (FCS) (PAA), 100 U/ml penicillin, 100 μg/ml streptomycin (Gibco), and 2 mM l-Glutamine.
Mouse WEHI-3B cells and the IL3-dependent mouse FL5.12 pro-B cells were maintained in RPMI-1640 medium (Sigma) supplemented with 10% fetal calf serum (FCS), 20 mM HEPES, 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml). The culture medium for FL5.12 cells was supplemented with 10% supernatant from confluent WEHI-3B cell cultures as a source of IL-3. FL5.12 cells stably expressing GFP-BCL2A1 were generated as previously described [18] and were maintained in the presence of G418 (1 μg/ml) (Gibco).
Constructs, transfection, and siRNAs
BCL2A1 cDNA sequence was synthesized by Genecust and subcloned in pFLAG-CMV2 (Sigma).
The cDNAs of human TRIM28, C-terminally fused to Myc tag in pCDNA3 MycS1 RfC or N-terminal fused to GFP in pEGFP plasmid-N1, were obtained from the ORFeome library (Montpellier Genomic Collection-MGC facility). Myc-tagged and GFP-tagged C65A/C68A RING mutants of TRIM28 were generated using Quickchange Site-Directed Mutagenesis Kit (Agilent) and were a gift of Véronique Baldin (CRBM, Montpellier). pKH3-TRIM28 plasmid expressing HA-tagged TRIM28 was obtained from Addgene.
Cells were transfected with GenJet™ in vitro transfection reagent (Ver. II) (SignaGen laboratories, Ijamsville, MD), with Lipofectamine 2000 (Invitrogen) or Lipofectamine 3000 (for SK-MEL-28 cells) according to the manufacturer’s instructions.
siRNAs transfection was performed using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer’s protocol. The sequences of the siRNAs used were as follows: siLuciferase (5′-UAUCCUCACAGCUAGUGCAGCACUGdTdT-3′), siTRIM28 3′UTR (5′-ACAGGACAGAGAACAGAGCdTdT-3′), siTRIM28 ORF (5′-UCGAAGUAUUCCGCGUACGdTdT-3′), siBCL2A1 (5′-GUUUGAAGACGGCAUCAUdTdT-3′).
Mass spectrometry
GFP-BCL2A1, or GFP as a control, was transiently expressed in HEK293T cells and immunoprecipitated using GFP-Trap system (Chromotek) in lysis buffer (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 0.5% NP-40 and protease inhibitor cocktail). Beads were washed four times in 50 mM Tris–HCl [pH 7.5], supplemented with 0.5 M NaCl and protease inhibitor cocktail. Eluted proteins were separated by SDS-PAGE and stained with coomassie blue. The band excised from the gel was subjected to reduction, carbamidomethylation, and tryptic digestion. Peptide sequences were determined by mass spectrometry using a LTQ Velos instrument (Dual Pressure Linear Ion Trap) equipped with a nanospray source (Thermo Fisher Scientific) and coupled to a U3000 nanoLC system (Thermo Fisher Scientific). A MS survey scan was acquired over the m/z range 400–1600 in enhanced resolution mode. The data dependent MS/MS scans were acquired in normal resolution mode over the m/z range 65–2000 for the 20 most intense MS ions with a charge of two or more and with a collision energy set to 35 eV. The spectra were recorded using dynamic exclusion of previously analyzed ions for 0.5 min with 50 millimass units (mmu) of mass tolerance. The peptide separation was obtained on a C18 PepMap micro-precolumn (5 μm; 100 Å; 300 μm × 5 mm; Dionex) and a C18 PepMap nanocolumn (3 μm; 100 Å; 75 μm × 150 mm; Dionex) using a linear 60 min gradient from 0% to 50% B, where solvent A was 0.1% HCOOH in H2O/CH3CN (95/5) and solvent B was 0.1% HCOOH in H2O/CH3CN (20/80) at 300 nl/min flow rate. Proteins identification was performed with the MASCOT algorithm (v2.2 Matrix Science) through the Proteome Discoverer software (v1.1 Thermo Fisher Scientific) against the Swiss-Prot Human database [UniProtKB/Swiss-Prot Release 2012_12].
BCL2A1 half-life measurement and western blot analysis
To measure the half-life of BCL2A1, cells were transfected with the indicated plasmids for 24 h and 10 μg/ml cycloheximide was added to the medium for increasing times before cell lysis and protein extraction, in order to block protein synthesis and to follow the degradation of BCL2A1 with time by immunoblot.
Total protein extracts were prepared from cell lines by lysis in RIPA buffer (25 mM Tris–HCl pH 7.6, 150 mM NaCl, 1% NP-40, 0.1% SDS, 1% sodium deoxycholate) supplemented with protease inhibitor cocktail (Roche). Protein extracts were quantified using BCA assay (Pierce), separated by 4–12% SDS-PAGE and transferred onto PVDF membranes using iBlot2 system (Invitrogen). Blocking, probing with antibodies, and chemoluminescent visualization of immunoreactive proteins were performed as previously described [53]. Antibodies used for detection of proteins were the following: GFP (Torrey Pines #TP401), Flag (Sigma, clone M2), HA (Roche, clone 3F10), BCL2A1 (Millipore, #ABC498), TRIM28 (Abcam, #ab10482), tubulin (Sigma, clone DM1A #T6199), GSK3 (cell signaling, clone D5C5Z), phosphor-Ser9 GSK3 (cell signaling, clone 5B3), vinculin (Santa-cruz #SC-55465). ImageJ software was used for optical density quantitation of western blots.
Co-immunoprecipitation
Following transfection with the indicated plasmids for 24 h, HEK293T cells were homogenized in lysis buffer A (50 mM Tris–HCl [pH 7.5], 150 mM NaCl; 0.5 mM EDTA and protease inhibitor cocktail) containing 1% NP-40 for immunoprecipitation with anti-FlagM2 beads (Sigma) and 0.5% NP-40 for immunoprecipitation with GFP-Trap-A (Chromotek), HA-beads (Biotool), or anti-TRIM17 antibody (polyclonal antibody raised against two human TRIM17 peptides, Eurogentec). For immunoprecipitation, cell lysates four times diluted in buffer B (50 mM Tris–HCl [pH 7.5], 150 mM NaCl and protease inhibitor cocktail) were incubated for 4 h at 4 °C with anti-GFP or anti-Flag beads as indicated. The beads were then recovered by centrifugation and washed four times with lysis buffer B supplemented with 0.5 M NaCl. Precipitates were then eluted by the addition of 3× Laemmli sample buffer and incubation at 95 °C for 5 min. Precipitated proteins were separated by 4–12% SDS–PAGE and analyzed by western blot as described above.
Measurement of BCL2A1 ubiquitination levels
HEK293T cells were transfected with the indicated plasmids together with His-tagged ubiquitin. 24 h after transfection, cells were treated with 20 μM MG-132 for 6 h prior cell harvesting. 10% of the cells were lysed in buffer A containing 1% NP-40 and used as total lysates. The rest of the cells were homogenized and ubiquitinated proteins were purified using nickel beads as previously described [29]. Ubiquitinated proteins and total lysate were resolved by SDS-PAGE and blotted using antibodies as indicated.
In situ PLA
HuH7 or SK-MEL-28 cells were seeded onto gelatin-coated glass coverslips. When transfection was performed, cells were transfected with pcDNA-Flag-BCL2A1 for 24 h or with the corresponding empty plasmid. Then, cells were fixed with 4% paraformaldehyde for 20 min, washed with PBS and permeabilized with 0.2% Triton X-100 in PBS for 10 min, at room temperature. The interaction between endogenous BCL2A1 or Flag-BCL2A1 and endogenous TRIM28 was detected using the Duolink® In Situ kit (Sigma), according to the manufacturer’s instructions as previously described [62], using primary antibodies against BCL2A1 (Millipore, 1:200) or TRIM28 (Abcam, clone 20C1; 1:1000) or TRIM17 (Abnova, clone 2E11, 1:1000). Images were analyzed by confocal fluorescence microscopy (Leica SP5) and ImageJ software.
sgRNA design and cloning
Constitutive Cas9 and inducible guide RNA vectors have been described previously [31]. The MIT CRISPR design software was used for the design of sgRNA (htpp://crispr.mit.edu). To clone individual sgRNAs, 24-bp oligonucleotide containing the sgRNA were synthesized (IDT). They included a 4-bp overhang for the forward (TCCC) and complementary reverse (AAAC) oligos to enable cloning into the Bsmb-I site of the lentiviral construct FgH1tUTG as previously described [31]. sgRNA sequences are as follows: TRIM17#1: 5′-CACCTTGGTCAGCAGCCGGT; TRIM17#2: 5′-GGAACTCGCCAGAAAACTGC. To target BCL2A1 we used a sgRNA guide previously described [32]. Negative control used was targeting murine sequence of Bim as previously described [31].
Viral production, transduction of cell lines, and doxycycline treatment
Lentiviral particles were produced by transient transfection of 293T and virus-containing supernatants were collected 48–72 h after transfection and passed through a 0.45 μm filter as previously described [63]. SK-MEL-28 cells were transduced by lentiviral particles as previously described [31]. In order to induce expression of the sgRNA in cell lines, doxycycline hyclate (Sigma #D9891) was added for 72 h to tissue culture medium at a final concentration of 1 μg/ml.
Detection of InDels by T7 endonuclease I assay
Genomic DNA was extracted 72 h after doxycycline treatment using QuickExtract DNA extraction solution (Epicentre). DNA fragment which comprises targeted sequences by TRIM17 sgRNAs was amplified by PCR (Gotaq, Promega) using the following primers: 5′-GAGGCTGTACAGGACGGTTG and 5′-GAAAAGCTTGAGGGGCTCGT. Amplicon was purified, denatured, and re-annealed to allow heteroduplex formation between wild-type DNA and CRISPR/Cas9-mutated DNA. Product was digested with T7 endonuclease I (NEB) and fragments separated by electrophoresis on a 2% high-resolution agarose gel.
Cell death assay and flow cytrometric analysis
Cells were seeded at 50 × 103 cells per well in a six-well plate, allowed to settle for 24 h, before treatment with 1 µg/ml doxycycline hyclate for 72 h to induce sgRNA expression. Cells were then harvested and seeded at 2 × 103 cells per well (each condition in triplicate), in a 96-well plate and allowed to settle for 24 h. Cells were then treated with either 20 µM PLX4720 or DMSO for 48 h. Cells were harvested and resuspended in Annexin V-binding buffer (0.1 M Hepes (pH 7.4), 1.4 M NaCl, 25 mM CaCl2) and Annexin V (conjugated to Alexa Fluor 647) and analyzed by flow cytometry (LSRII, Becton Dickinson).
Calculation of specific induced apoptosis (SIA)
In order to discriminate the PLX4720-specific induced apoptosis vs. spontaneous cell death due to transfection toxicity in SK-MEL-28 cells, we calculated the percentage of specific induced apoptosis (% SIA) using the following formula: % SIA = [(PLX4720-induced apoptosis − media only spontaneous apoptosis)/(100 – media only spontaneous apoptosis)] × 100.
Statistical analysis
Statistical analyses of data sets were performed using GraphPad Prism version 7.00 for MAC OS X, GraphPad Software, La Jolla, CA, USA. Unless indicated, data are presented as the mean ± SEM.
Targeted PCR and sequencing of sites of Cas9-induced InDels
Genomic DNA was prepared by resuspending cells into Direct PCR lysis buffer (Viagen) with proteinase K (Sigma-Aldrich P4850), and incubated with gentle shaking for 4–6 h followed by heat inactivation at 85 °C for 45 min. Unique primers were designed to amplify regions flanking the sgRNA-binding site (∼120 bp in total), and included sequence overhangs at the 5′ end of the forward and reverse primers as follows (FWD OH: 5′-GTGACCTATGAACTCAGGAGTC-3′; REV OH: 5′-CTGAGACTTGCACATCGCAGC-3′). The first step PCR cycling conditions were as follows; 95 °C 2 min (95 °C 30 s, 60 °C 30 s, 72 °C 30 s) × 25 cycles, 72 °C 5 min. PCR amplicons were individually purified using 1.03 Ampure Beads (Beckman Coulter). Amplicon size distribution was ascertained using the Agilent Tapestation D1000 protocol. Secondary amplification using overhang sequences and Illumina MISeq sequencing was done as previously described [31].
RNA preparation and real-time quantitative RT-PCR
Total RNA was extracted using the RNAqueous® kit (Ambion) and treated with DNase I from the DNA-free™ kit (Ambion) according to manufacturer’s instructions. RNA was used to perform a two-step reverse-transcription polymerase chain reaction (RT-PCR) as previously described [53].
The sequences of the primers used were as follows: human BCL2A1: forward 5′-ATGGATAAGGCAAAACGGAGG-3′; reverse 5′-TATGGAGTGTCCTTTCTGGTAA-3′; human TRIM17: forward 5′-GACATGGAGTACCTTCGGGA-3′; reverse 5′-GCAGTCTCCTCTTCTTCCGT-3′; human TRIM28: forward 5′-AGCTGTGAGGATAATGCCCC-3′; reverse 5′-GTTCACCATCCCGAGACTTG-3′; GAPDH forward 5′-CCATCTTCCAGGAGCGAGAT-3′; reverse 5′-GGTTCACACCCATGACGAAC-3′. Data were analyzed and relative amounts of specifically amplified cDNA were calculated with MxPro software (Agilent). Human GAPDH amplicon was used as a reference.
In vitro phosphorylation assays
Recombinant full-length human GST-tagged BCL2A1 protein was a gift of Nathalie Bonnefoy (IRCM, Montpellier). The BH3 peptides sequences used for this study were the same as previously described [13]. Peptides with free N-terminal and C-terminal were synthesized by Genecust (Luxembourg), purified by reverse-phase HPLC. They were >90% pure and were dissolved as 1 mM stock solutions in water. BCL2A1-α9 peptide purification and sequence was previously described [64]. The accession numbers on which the peptides were based are as follows: hBimL (AAC39594), hPuma (AAK39542), hBad (NP_004313), hNoxa (NP_066950), and mNoxa (NP_067426).
For in vitro phosphorylation assays, synthetic BCL2A1-α9 peptide (4 pmol), GST-BCL2A1 (400 ng), or GST-BCL2A1 pre-incubated with BH3 peptide (molar ratio 1:10) for 1 h at 4 °C, were incubated for 30 min at 30 °C in a total volume of 20 μl kinase reaction buffer (Biolabs) containing 5 μCi [γ-32P]-ATP in the presence of 100 ng of recombinant GSK3β kinase (Biolabs). The reaction was stopped by the addition of Laemmli buffer. Phosphorylation of peptide or protein was resolved by 4–12% SDS PAGE (NuPAGE, Invitrogen) and visualized by autoradiography.
Electronic supplementary material
Acknowledgements
This work was supported by grants from La Ligue contre le Cancer, regional committees of Drôme, Hérault and Lozère (to J.K.) and Gard (to S.D.), the Centre National de la Recherche Scientifique (CNRS), Leukaemia Foundation Australia, and Cancer Council Victoria Venture Grant (to M.J.H.), World Cancer Research 15-0177 (to P.R.P.). L.L. was supported by the University of Montpellier, La Ligue Nationale contre le Cancer and by the Programme de mobilité scientifique from the embassy of France in Australia. J.K. was supported by the University of Lyon and was recipient of a délégation CNRS program. The authors thank the Protein Science Facility of the SFR Biosciences Lyon for their valuable expertize and technical assistance in mass spectrometry analysis and Aurélie Cornut-Thibault for preparation of samples. The authors also thank the staff of Montpellier Genomic Collection platform for providing TRIM28 cDNA clones and the imaging facility MRI, member of the National Infrastructure France-BioImaging supported by the French National Research Agency (ANR-10-INBS-04, “Investments for the future”) for Microscopy and Cytometry Analysis. The authors are grateful to Drs. Nathalie Bonnefoy, Véronique Baldin, Olivier Coux, Damien Grégoire, Florence Cammas, and Anne-Marie Marini for fruitful discussions and reagents, Gilles Salles (HCL, Lyon-Sud) for support during the early stages of this work, and John A. Hickman for critical reading of the manuscript.
Author contributions
J.K. and S.D. conceived the study, designed the experiments, and wrote the manuscript. L.L. performed the experiments, analyzed data, and prepared figures. P.D., M.S.B, F.G., B.M., M.-A.D., S.M., I.L., and C.C. performed experiments. A.J.K. and M.P. generated and analyzed CRISPR sequencing data. R.R. produced reagents. P.R.P. contributed to experimental design and provided reagents. A.A. contributed to experimental design of the MS/MS analysis and assisted in phylogenetic discussions. M.J.H. generated reagents, designed CRISPR/Cas9 experiments, and provided invaluable access to facilities.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by A. Ashkenazi
These authors jointly supervised this work: Solange Desagher, Jérôme Kucharczak.
Contributor Information
Solange Desagher, Email: solange.desagher@igmm.cnrs.fr.
Jérôme Kucharczak, Email: jerome.kucharczak@univ-lyon1.fr.
Electronic supplementary material
The online version of this article (10.1038/s41418-018-0169-5) contains supplementary material, which is available to authorized users.
References
- 1.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis RE, Brown KD, Siebenlist U, Staudt LM. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2001;194:1861–74. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee HH, Dadgostar H, Cheng Q, Shu J, Cheng G. NF-kappaB-mediated up-regulation of Bcl-x and Bfl-1/A1 is required for CD40 survival signaling in B lymphocytes. Proc Natl Acad Sci USA. 1999;96:9136–41. doi: 10.1073/pnas.96.16.9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morales AA, Olsson A, Celsing F, Osterborg A, Jondal M, Osorio LM. High expression of bfl-1 contributes to the apoptosis resistant phenotype in B-cell chronic lymphocytic leukemia. Int J Cancer. 2005;113:730–7. doi: 10.1002/ijc.20614. [DOI] [PubMed] [Google Scholar]
- 5.Olsson A, Norberg M, Okvist A, Derkow K, Choudhury A, Tobin G, et al. Upregulation of bfl-1 is a potential mechanism of chemoresistance in B-cell chronic lymphocytic leukaemia. Br J Cancer. 2007;97:769–77. doi: 10.1038/sj.bjc.6603951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang CY, Guttridge DC, Mayo MW, Baldwin AS., Jr. NF-kappaB induces expression of the Bcl-2 homologue A1/Bfl-1 to preferentially suppress chemotherapy-induced apoptosis. Mol Cell Biol. 1999;19:5923–9. doi: 10.1128/MCB.19.9.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia L, Wurmbach E, Waxman S, Jing Y. Upregulation of Bfl-1/A1 in leukemia cells undergoing differentiation by all-trans retinoic acid treatment attenuates chemotherapeutic agent-induced apoptosis. Leukemia. 2006;20:1009–16. doi: 10.1038/sj.leu.2404198. [DOI] [PubMed] [Google Scholar]
- 8.Haq R, Yokoyama S, Hawryluk EB, Jonsson GB, Frederick DT, McHenry K, et al. BCL2A1 is a lineage-specific antiapoptotic melanoma oncogene that confers resistance to BRAF inhibition. Proc Natl Acad Sci USA. 2013;110:4321–6. doi: 10.1073/pnas.1205575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brien G, Trescol-Bierriont MC, Bonnefoy-Berard N. Downregulation of Bfl-1 protein expression sensitizes malignant B cells to apoptosis. Oncogene. 2007;26:5828–32. doi: 10.1038/sj.onc.1210363. [DOI] [PubMed] [Google Scholar]
- 10.Placzek WJ, Wei J, Kitada S, Zhai D, Reed JC, Pellecchia M. A survey of the anti-apoptotic Bcl-2 subfamily expression in cancer types provides a platform to predict the efficacy of Bcl-2 antagonists in cancer therapy. Cell Death Dis. 2010;1:e40. doi: 10.1038/cddis.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barile E, Marconi GD, De SK, Baggio C, Gambini L, Salem AF, et al. hBfl-1/hNOXA interaction studies provide new insights on the role of Bfl-1 in cancer cell resistance and for the design of novel anticancer agents. ACS Chem Biol. 2017;12:444–55. doi: 10.1021/acschembio.6b00962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Certo M, Del Gaizo Moore V, Nishino M, Wei G, Korsmeyer S, Armstrong SA, et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell. 2006;9:351–65. doi: 10.1016/j.ccr.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 13.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 14.Vogler M, Butterworth M, Majid A, Walewska RJ, Sun XM, Dyer MJ, et al. Concurrent up-regulation of BCL-XL and BCL2A1 induces approximately 1000-fold resistance to ABT-737 in chronic lymphocytic leukemia. Blood. 2009;113:4403–13. doi: 10.1182/blood-2008-08-173310. [DOI] [PubMed] [Google Scholar]
- 15.Yecies D, Carlson NE, Deng J, Letai A. Acquired resistance to ABT-737 in lymphoma cells that up-regulate MCL-1 and BFL-1. Blood. 2010;115:3304–13. doi: 10.1182/blood-2009-07-233304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esteve-Arenys A, Valero JG, Chamorro-Jorganes A, Gonzalez D, Rodriguez V, Dlouhy I, et al. The BET bromodomain inhibitor CPI203 overcomes resistance to ABT-199 (venetoclax) by downregulation of BFL-1/A1 in in vitro and in vivo models of MYC+/BCL2+ double hit lymphoma. Oncogene. 2018;37:1830–44. doi: 10.1038/s41388-017-0111-1. [DOI] [PubMed] [Google Scholar]
- 17.Herold MJ, Zeitz J, Pelzer C, Kraus C, Peters A, Wohlleben G, et al. The stability and anti-apoptotic function of A1 are controlled by its C terminus. J Biol Chem. 2006;281:13663–71. doi: 10.1074/jbc.M600266200. [DOI] [PubMed] [Google Scholar]
- 18.Kucharczak JF, Simmons MJ, Duckett CS, Gelinas C. Constitutive proteasome-mediated turnover of Bfl-1/A1 and its processing in response to TNF receptor activation in FL5.12 pro-B cells convert it into a prodeath factor. Cell Death Differ. 2005;12:1225–39. doi: 10.1038/sj.cdd.4401684. [DOI] [PubMed] [Google Scholar]
- 19.Rooswinkel RW, van de Kooij B, de Vries E, Paauwe M, Braster R, Verheij M, et al. Antiapoptotic potency of Bcl-2 proteins primarily relies on their stability, not binding selectivity. Blood. 2014;123:2806–15. doi: 10.1182/blood-2013-08-519470. [DOI] [PubMed] [Google Scholar]
- 20.Fan G, Simmons MJ, Ge S, Dutta-Simmons J, Kucharczak J, Ron Y, et al. Defective ubiquitin-mediated degradation of antiapoptotic Bfl-1 predisposes to lymphoma. Blood. 2010;115:3559–69. doi: 10.1182/blood-2009-08-236760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aouacheria A, Rech de Laval V, Combet C, Hardwick JM. Evolution of Bcl-2 homology motifs: homology versus homoplasy. Trends Cell Biol. 2013;23:103–11. doi: 10.1016/j.tcb.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ko JK, Choi KH, Pan Z, Lin P, Weisleder N, Kim CW, et al. The tail-anchoring domain of Bfl1 and HCCS1 targets mitochondrial membrane permeability to induce apoptosis. J Cell Sci. 2007;120:2912–23. doi: 10.1242/jcs.006197. [DOI] [PubMed] [Google Scholar]
- 23.Ottina E, Tischner D, Herold MJ, Villunger A. A1/Bfl-1 in leukocyte development and cell death. Exp Cell Res. 2012;318:1291–303. doi: 10.1016/j.yexcr.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogler M. BCL2A1: the underdog in the BCL2 family. Cell Death Differ. 2012;19:67–74. doi: 10.1038/cdd.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–7. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 26.Mojsa B, Lassot I, Desagher S. Mcl-1 ubiquitination: unique regulation of an essential survival protein. Cells. 2014;3:418–37. doi: 10.3390/cells3020418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doyle JM, Gao J, Wang J, Yang M, Potts PR. MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol Cell. 2010;39:963–74. doi: 10.1016/j.molcel.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pineda CT, Ramanathan S, Fon Tacer K, Weon JL, Potts MB, Ou YH, et al. Degradation of AMPK by a cancer-specific ubiquitin ligase. Cell. 2015;160:715–28. doi: 10.1016/j.cell.2015.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magiera MM, Mora S, Mojsa B, Robbins I, Lassot I, Desagher S. Trim17-mediated ubiquitination and degradation of Mcl-1 initiate apoptosis in neurons. Cell Death Differ. 2013;20:281–92. doi: 10.1038/cdd.2012.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hind CK, Carter MJ, Harris CL, Chan HT, James S, Cragg MS. Role of the pro-survival molecule Bfl-1 in melanoma. Int J Biochem Cell Biol. 2015;59:94–102. doi: 10.1016/j.biocel.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 31.Aubrey BJ, Kelly GL, Kueh AJ, Brennan MS, O’Connor L, Milla L, et al. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015;10:1422–32. doi: 10.1016/j.celrep.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Gong J.-N., Khong T., Segal D., Yao Y., Riffkin C. D., Garnier J.-M., Khaw S. L., Lessene G., Spencer A., Herold M. J., Roberts A. W., Huang D. C. S. Hierarchy for targeting prosurvival BCL2 family proteins in multiple myeloma: pivotal role of MCL1. Blood. 2016;128(14):1834–1844. doi: 10.1182/blood-2016-03-704908. [DOI] [PubMed] [Google Scholar]
- 33.Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell. 2006;21:749–60. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 34.Brien G, Debaud AL, Robert X, Oliver L, Trescol-Biemont MC, Cauquil N, et al. C-terminal residues regulate localization and function of the antiapoptotic protein Bfl-1. J Biol Chem. 2009;284:30257–63. doi: 10.1074/jbc.M109.040824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays. 2005;27:1147–57. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 36.Hatakeyama S. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci. 2017;42:297–311. doi: 10.1016/j.tibs.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 37.Santoni de Sio FR, Massacand J, Barde I, Offner S, Corsinotti A, Kapopoulou A, et al. KAP1 regulates gene networks controlling mouse B-lymphoid cell differentiation and function. Blood. 2012;119:4675–85. doi: 10.1182/blood-2011-12-401117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chuang PI, Morefield S, Liu CY, Chen S, Harlan JM, Willerford DM. Perturbation of B-cell development in mice overexpressing the Bcl-2 homolog A1. Blood. 2002;99:3350–9. doi: 10.1182/blood.V99.9.3350. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, et al. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–95. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, Schultz DC, et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell. 2007;28:823–37. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y, Fiskus W, Yong B, Atadja P, Takahashi Y, Pandita TK, et al. Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc Natl Acad Sci USA. 2013;110:6841–6. doi: 10.1073/pnas.1217692110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiao TZ, Bhatia N, Urrutia R, Lomberk GA, Simpson A, Longley BJ. MAGE I transcription factors regulate KAP1 and KRAB domain zinc finger transcription factor mediated gene repression. PLoS One. 2011;6:e23747. doi: 10.1371/journal.pone.0023747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herquel B, Ouararhni K, Khetchoumian K, Ignat M, Teletin M, Mark M, et al. Transcription cofactors TRIM24, TRIM28, and TRIM33 associate to form regulatory complexes that suppress murine hepatocellular carcinoma. Proc Natl Acad Sci USA. 2011;108:8212–7. doi: 10.1073/pnas.1101544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z, Wang D, Na X, Schoen SR, Messing EM, Wu G. The VHL protein recruits a novel KRAB-A domain protein to repress HIF-1alpha transcriptional activity. EMBO J. 2003;22:1857–67. doi: 10.1093/emboj/cdg173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsuruma R, Ohbayashi N, Kamitani S, Ikeda O, Sato N, Muromoto R, et al. Physical and functional interactions between STAT3 and KAP1. Oncogene. 2008;27:3054–9. doi: 10.1038/sj.onc.1210952. [DOI] [PubMed] [Google Scholar]
- 46.Li X, Lee YK, Jeng JC, Yen Y, Schultz DC, Shih HM, et al. Role for KAP1 serine 824 phosphorylation and sumoylation/desumoylation switch in regulating KAP1-mediated transcriptional repression. J Biol Chem. 2007;282:36177–89. doi: 10.1074/jbc.M706912200. [DOI] [PubMed] [Google Scholar]
- 47.Yokoe T, Toiyama Y, Okugawa Y, Tanaka K, Ohi M, Inoue Y, et al. KAP1 is associated with peritoneal carcinomatosis in gastric cancer. Ann Surg Oncol. 2010;17:821–8. doi: 10.1245/s10434-009-0795-8. [DOI] [PubMed] [Google Scholar]
- 48.Wang YY, Li L, Zhao ZS, Wang HJ. Clinical utility of measuring expression levels of KAP1, TIMP1 and STC2 in peripheral blood of patients with gastric cancer. World J Surg Oncol. 2013;11:81. doi: 10.1186/1477-7819-11-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martins MB, Marcello MA, Morari EC, Cunha LL, Soares FA, Vassallo J, et al. Clinical utility of KAP-1 expression in thyroid lesions. Endocr Pathol. 2013;24:77–82. doi: 10.1007/s12022-013-9245-z. [DOI] [PubMed] [Google Scholar]
- 50.Pineda CT, Potts PR. Oncogenic MAGEA-TRIM28 ubiquitin ligase downregulates autophagy by ubiquitinating and degrading AMPK in cancer. Autophagy. 2015;11:844–6. doi: 10.1080/15548627.2015.1034420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koliopoulos MG, Esposito D, Christodoulou E, Taylor IA, Rittinger K. Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity. EMBO J. 2016;35:1204–18. doi: 10.15252/embj.201593741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang NJ, Zhang L, Tang W, Chen C, Yang CS, Kornbluth S. The Trim39 ubiquitin ligase inhibits APC/CCdh1-mediated degradation of the Bax activator MOAP-1. J Cell Biol. 2012;197:361–7. doi: 10.1083/jcb.201111141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lassot I, Robbins I, Kristiansen M, Rahmeh R, Jaudon F, Magiera MM, et al. Trim17, a novel E3 ubiquitin-ligase, initiates neuronal apoptosis. Cell Death Differ. 2010;17:1928–41. doi: 10.1038/cdd.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crowder RJ, Freeman RS. Glycogen synthase kinase-3 beta activity is critical for neuronal death caused by inhibiting phosphatidylinositol 3-kinase or Akt but not for death caused by nerve growth factor withdrawal. J Biol Chem. 2000;275:34266–71. doi: 10.1074/jbc.M006160200. [DOI] [PubMed] [Google Scholar]
- 55.Hetman M, Cavanaugh JE, Kimelman D, Xia Z. Role of glycogen synthase kinase-3beta in neuronal apoptosis induced by trophic withdrawal. J Neurosci. 2000;20:2567–74. doi: 10.1523/JNEUROSCI.20-07-02567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pap M, Cooper GM. Role of glycogen synthase kinase-3 in the phosphatidylinositol 3-Kinase/Akt cell survival pathway. J Biol Chem. 1998;273:19929–32. doi: 10.1074/jbc.273.32.19929. [DOI] [PubMed] [Google Scholar]
- 57.Hoeflich KP, Luo J, Rubie EA, Tsao MS, Jin O, Woodgett JR. Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature. 2000;406:86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- 58.Perciavalle RM, Opferman JT. Delving deeper: MCL-1’s contributions to normal and cancer biology. Trends Cell Biol. 2013;23:22–29. doi: 10.1016/j.tcb.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schenk RL, Tuzlak S, Carrington EM, Zhan Y, Heinzel S, Teh CE, et al. Characterisation of mice lacking all functional isoforms of the pro-survival BCL-2 family member A1 reveals minor defects in the haematopoietic compartment. Cell Death Differ. 2017;24:534–45. doi: 10.1038/cdd.2016.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tuzlak S, Schenk RL, Vasanthakumar A, Preston SP, Haschka MD, Zotos D, et al. The BCL-2 pro-survival protein A1 is dispensable for T cell homeostasis on viral infection. Cell Death Differ. 2017;24:523–33. doi: 10.1038/cdd.2016.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carrington EM, Zhan Y, Brady JL, Zhang JG, Sutherland RM, Anstee NS, et al. Anti-apoptotic proteins BCL-2, MCL-1 and A1 summate collectively to maintain survival of immune cell populations both in vitro and in vivo. Cell Death Differ. 2017;24:878–88. doi: 10.1038/cdd.2017.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mojsa B, Mora S, Bossowski JP, Lassot I, Desagher S. Control of neuronal apoptosis by reciprocal regulation of NFATc3 and Trim17. Cell Death Differ. 2015;22:274–86. doi: 10.1038/cdd.2014.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kueh AJ, Herold MJ. Using CRISPR/Cas9 technology for manipulating cell death regulators. Methods Mol Biol. 2016;1419:253–64. doi: 10.1007/978-1-4939-3581-9_18. [DOI] [PubMed] [Google Scholar]
- 64.Valero JG, Cornut-Thibaut A, Juge R, Debaud AL, Gimenez D, Gillet G, et al. micro-Calpain conversion of antiapoptotic Bfl-1 (BCL2A1) into a prodeath factor reveals two distinct alpha-helices inducing mitochondria-mediated apoptosis. PLoS One. 2012;7:e38620. doi: 10.1371/journal.pone.0038620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.