Abstract
The discovery of insecticidal activity in root-colonizing pseudomonads, best-known for their plant-beneficial effects, raised fundamental questions about the ecological relevance of insects as alternative hosts for these bacteria. Since soil bacteria are limited in their inherent abilities of dispersal, insects as vectors might be welcome vehicles to overcome large distances. Here, we report on the transmission of the root-colonizing, plant-beneficial and insecticidal bacterium Pseudomonas protegens CHA0 from root to root by the cabbage root fly, Delia radicum. Following ingestion by root-feeding D. radicum larvae, CHA0 persisted inside the insect until the pupal and adult stages. The emerging flies were then able to transmit CHA0 to a new plant host initiating bacterial colonization of the roots. CHA0 did not reduce root damages caused by D. radicum and had only small effects on Delia development suggesting a rather commensal than pathogenic relationship. Interestingly, when the bacterium was fed to two highly susceptible lepidopteran species, most of the insects died, but CHA0 could persist throughout different life stages in surviving individuals. In summary, this study investigated for the first time the interaction of P. protegens CHA0 and related strains with an insect present in their rhizosphere habitat. Our results suggest that plant-colonizing pseudomonads have different strategies for interaction with insects. They either cause lethal infections and use insects as food source or they live inside insect hosts without causing obvious damages and might use insects as vectors for dispersal, which implies a greater ecological versatility of these bacteria than previously thought.
Subject terms: Microbial ecology, Soil microbiology, Bacterial host response, Bacterial pathogenesis
Introduction
Every year worldwide crop production is facing major harvest losses due to plant-pathogens and pest insects. Below-ground attackers are especially difficult to tackle with chemical pesticides and adverse environmental effects of these products demand for alternative strategies such as the use of antagonistic organisms to control pest organisms, known as biological control. Root-colonizing bacteria of the Pseudomonas fluorescens species complex [1] have been extensively studied for their beneficial effects on plants, e.g., the suppression of root diseases and the promotion of plant growth [2, 3], and this research already led to several commercial products [4].
Within the P. fluorescens species complex there is a specific phylogenetic group, the Pseudomonas chlororaphis subgroup [5], comprising multi-talented bacteria of special interest for use in crop protection. These bacteria have, in addition to the plant-beneficial activity, features to colonize insects as an alternative host [6–9]. Strains of the P. chlororaphis subgroup were even found to exhibit potent oral activity against larvae of Lepidoptera [7, 9, 10] as well as against Drosophila melanogaster [11] and a P. chlororaphis toxin was found to be active against the western corn rootworm Diabrotica virgifera virgifera [12]. Several factors have been identified to contribute to insect pathogenicity: the Fit toxin, antimicrobial metabolites, secreted enzymes, lipopolysaccharide O antigen and the insecticidal protein IPD072Aa [8–19]. In-depth studies on the Fit toxin in the model strain P. protegens CHA0 revealed that the bacteria produce this insecticidal protein specifically in insects, but not on plant roots [20, 21]. Accordingly, the bacteria seem to sense their environment and regulate the production of specific compounds depending on the specific needs in the encountered habitat.
Although there is a growing body of evidence that insects represent an alternative host for P. chlororaphis subgroup bacteria, the ecology of their insect-associated lifestyle is still elusive. To date, oral insecticidal activity has been investigated only in model insects feeding on leaves [7, 9, 10, 19, 20]. However, many known strains of the P. chlororaphis subgroup are rhizosphere isolates, e.g., P. protegens CHA0 was isolated from tobacco roots, and their interaction with root-feeding insects is therefore of much greater ecological relevance. It is still unknown whether these bacteria are also pathogenic to soil insects and therefore have a potential as biocontrol organisms of root pests. Furthermore, the discovery that several plant-beneficial pseudomonads exhibit specific adaptations to a life in insects [6, 14, 15, 20] raised the hypothesis that insects might represent attractive vectors to reach new plant hosts. Rhizobacteria are limited in their inherent dispersal abilities and may largely depend on passive transport, such as water flows, to overcome large distances. Alternatively, dispersal by means of a vector is a plausible manner of attaining new habitats. Insect-mediated dispersal has been described for several plant-pathogenic bacteria [22], but data on transmission of beneficial rhizobacteria to a new host plant is scarce. Pseudomonas chlororaphis L11, an efficient root colonizer without known biocontrol activity, was found to be transmitted from plant to plant by the red-legged grasshopper, Melanoplus femurrubrum, as well as by the southern corn rootworm, Diabrotica undecimpunctata susp. howardii [23, 24], but mainly when insect vectors were feeding on L11-infested foliage. In contrast to L11, which moves from the rhizosphere into the foliage [25], P. protegens CHA0 was not found to move to above-ground plant parts [26, 27]. For the dispersal of a rhizobacterium, which is restricted to below-ground plant parts, an insect with a root-feeding larval and an above-ground flying adult stage would represent a suitable vector. A prerequisite for this kind of dispersal is persistence of the bacteria in the insect host and transstadial transmission from larva over pupa to the adult stage.
This study investigates the interaction of P. protegens CHA0 with a root-feeding pest insect, the cabbage root fly Delia radicum. Their larvae feed on brassicaceous plants, pupate in the soil and emerging adults fly to a new host plant to deposit eggs. Our first aim was to assess the oral activity of different P. chlororaphis subgroup strains against this insect. While all P. chlororaphis subgroup strains tested so far are, to a high degree, lethal to many lepidopteran insect species [7, 9], we found that the survival of D. radicum larvae was affected by certain strains of the P. chlororaphis subgroup, but not by P. protegens CHA0. The second aim was to compare the ability of model strain CHA0 to persist in little susceptible and highly susceptible insects. CHA0 was able to persist throughout different life stages in all tested insect species independently of their susceptibility level indicating that this is a rather general phenomenon. For D. radicum, we could show in addition that adult flies emerging from larvae that fed on CHA0 colonized roots transmitted the bacteria to the roots of new host plants. This provides the first direct evidence for the possibility of insect-mediated dispersal of P. protegens CHA0. Overall, our data indicate that indeed insects might be both, relevant alternative hosts and vectors for certain plant-beneficial rhizobacteria.
Materials and methods
Bacterial cultures
The bacteria used in this study are listed in Table 1. Strains with a constitutively expressed GFP tag were generated by means of the Tn7 delivery vectors pBK-miniTn7-gfp1 or pBKminiTn7-gfp2 [20]. In the results and discussion sections we always use wild type names of strains. Whether GFP-tagged variants were used is indicated in the materials and methods and in the figure legends. GFP-tagged strains did not differ in their activity from the respective wild-type strains. Bacteria were cultured in lysogeny broth (LB), supplemented with either kanamycin (25 μg/ml) or gentamicin (10 μg/ml) for GFP expressing strains, overnight at 24 °C and 180 rpm. For the cauliflower experiments with D. radicum 200 µl of LB cultures were used to inoculate King’s B (KB) agar plates [28] supplemented with gentamicin (10 μg ml−1). After one day, bacterial cultures were scraped off the plates, suspended, washed twice in sterile ddH2O, and OD600 was adjusted to the desired concentration (OD600 of 0.125 corresponds to about 108 cfu/ml). In all other experiments, LB cultures were washed twice (once for radish experiments) in sterile 0.9% NaCl solution or water before adding cell suspensions adjusted to the desired concentration to diets or roots/radish, respectively.
Table 1.
Bacterial strains used in this study
| Strain | Genotype, phenotype or relevant characteristics | Insecticidal activity | Reference or source |
|---|---|---|---|
| Pseudomonas protegens CHA0 | Wild type, isolated from tobacco roots | Yes | Jousset et al. [42]; Stutz et al. [43] |
| Pseudomonas protegens CHA0-gfp2 | CHA0::attTn7-gfp2; Gmr | Yes | Péchy-Tarr et al. [21] |
| Pseudomonas protegens CHA1176 | CHA0::attTn7-gfp2 fitD-mcherry; Gmr | Yes | Péchy-Tarr et al. [21] |
| Pseudomonas protegens PF-gfp2 | PF::attTn7-gfp2; Gmr | Yes | This study, for wild type PF see Levy et al. [44] |
| Pseudomonas sp. CMR12a-gfp1 | CMR12a::attTn7-gfp1; Kmr | Yes | This study, for wild type CMR12a see Perneel et al. [45] |
| Pseudomonas chlororaphis PCL1391-gfp2 | PCL1391::attTn7-gfp2; Gmr | Yes | This study, for wild type PCL1391 see Chin-A-Woeng et al. [46] |
| Pseudomonas thivervalensis PITR2-gfp2 | PITR2::attTn7-gfp2; Gmr | No | This study, for wild type PITR2 see Keel et al. [47] |
Gmr gentamicin resistance, Kmr kanamycin resistance
Radish experiment for testing the susceptibility of D. radicum to different Pseudomonas strains
D. radicum was reared as described by Razinger et al. [29], but larvae were fed on turnip cabbage instead of rutabaga. Greens of organically grown radishes (Migros, Switzerland) were cutoff about 0.5 cm above the bulbs. Bulbs were then washed with tap water and 70% ethanol, dried with household paper and submerged for 10 min in a bacterial suspension of OD600 of 0.47 or ddH2O as a control. Then the radishes were buried in pots (345 × 276 × 80 mm) (Bachmann Plantec AG, Switzerland) filled with sterile quartz sand. Six to eight eggs of D. radicum were deposited on top of the sand and the pots covered with aluminum foil were incubated in a climate chamber (16-h day, 20 °C, 210 μmol m−2 s−1; 8-h night cycle, 18 °C) for four weeks. Developing pupae were harvested by washing the sand over a sieve. All pupae emerging from one pot were photographed together and pupal size was measured by means of an ImageJ macro. Two weeks later, flies emerging from pupae were quantified. Flies of the CHA0-gfp2 and control treatments were checked for presence of CHA0 by incubating entire flies each in 1 ml LB supplemented with chloramphenicol (13 μg ml−1), cycloheximide (100 μg ml−1), and gentamicin (10 μg ml−1) for two days. To verify the identity of the growing bacteria, they were checked for GFP expression under a Leica DM2500 microscope (Leica Microsystems CMS GmbH, Wetzlar, Germany). The experiment was conducted twice.
Persistence/transmission of P. protegens CHA0 in experiments with cauliflower/rapeseed and D. radicum
This experiment aimed at investigating the ability of P. protegens CHA0 to persist in D. radicum throughout different life stages and the possibility of flies vectoring the bacterium from one host plant to another. Cauliflower (original host) and rapeseed (new host) were chosen for the experiments since in nature D. radicum might also move between these two brassicacean crop plants during its life-cycle. Cauliflower plants (four trays each containing twelve pots, one plant per pot) were grown with and without (control) P. protegens CHA0-gfp2 for three weeks as detailed in the Supplementary Methods. Then, five freshly hatched D. radicum larvae were added to each plant. Four weeks later, plant shoots were weighed and root systems were washed on a sieve to collect pupae as well as non-pupated larvae. Larvae were directly extracted for bacteria monitoring while pupae were kept in Petri dishes until further processing. Bacterial root colonization was assessed as described in Supplementary Methods. Pupal size was measured as described for the radish experiments. Ten to twelve pupae per treatment were extracted to assess colonization by inoculant bacteria as described below, three to four pupae were transferred to each of the transmission microcosms and the remaining pupae (44–56 per treatment) were observed to determine the rate of fly emergence. The transmission microcosms were designed to test whether flies are transmitting CHA0 to the roots of a new host plant (rapeseed) and are described in detail in the Supplementary Methods. Briefly, each transmission microcosm consisted of four rapeseed plants grown axenically on a sand-vermiculite substrate in closed plastic beakers. Little containers with three to four pupae, coming either from control or P. protegens CHA0-gfp2 treatments of the cauliflower experiment, were added to each transmission microcosm. The containers were used to prevent direct contact of the pupae with the plant and the substrate, but were open at the top to allow the flies to escape into the transmission microcosms. Nine days after flies had started to emerge and fly around inside the microcosms, roots of rapeseed plants were checked for colonization by P. protegens CHA0-gfp2 as described for cauliflower plants (Supplementary Methods). Roots of plants grown in the same beaker were pooled for analysis.
Oral toxicity of P. protegens CHA0 toward Plutella xylostella and Pieris brassicae and persistence during the insect life cycle
The experiments with P. xylostella were conducted as detailed in Flury et al. [19] and are briefly described in the Supplementary Methods. P. brassicae larvae were reared at 25 °C, 60% relative humidity and a 16-h day, 8-h night cycle and fed with Brussels sprouts variety Topline F1. During the experiments larvae were kept individually in Petri dishes lined with a moisturized filter paper and were fed with a pellet of artificial diet [30] inoculated with 10 μl of suspension of P. protegens CHA0-gfp2 or CHA0 cells at an OD600 of 20 or amended with sterile 0.9% NaCl solution (control). Larvae that did not consume the entire diet pellet were excluded from the experiment. After 24 h, larvae were transferred in groups of six into 720 ml Pint-sized BugDorms (BugDorm, Taiwan) and fed with cabbage until pupation. Twenty-four to thirty-two larvae per treatment were used for monitoring mortality. Larvae and pupae were considered dead when they did not react to poking. Further individuals (alive, crippled and dead) of each developmental stage were assessed for bacterial colonization as described below. The experiment was conducted three times.
Assessment of bacterial colonization in insects
To assess bacterial colonization, insects derived from the different experiments described above were surface-sterilized (20 s 70% ethanol, 20 s sterile ddH2O for P. xylostella and P. brassicae; 20 s 0.05% SDS, 20 s 70% ethanol, 20 s sterile ddH2O for D. radicum) and then homogenized in sterile 0.9% NaCl solution with a Polytron PT-DA 2112 blender (Kinematica, Littau, Switzerland). Efficacy of insect surface sterilization was tested as detailed in the Supplementary Methods. Dead individuals were extracted within 24 h after death occurred. The resulting suspensions were serially diluted in sterile 0.9% NaCl solution and plated onto KB agar plates supplemented with chloramphenicol (13 μg ml−1), cycloheximide (100 μg ml−1), and gentamicin (10 μg ml−1). For D. radicum, plates were additionally supplemented with ampicillin (40 μg ml−1). GFP expression of growing colonies was verified under the microscope (ex: 480/BP 40 nm, em: 527/BP 30 nm).
Microscopy
Microscopic investigations and sample preparation of P. xylostella larvae are described in Supplementary Methods.
Statistics
Data analysis was performed in RStudio version 0.98.1017 (http://www.rstudio.com) using R version 3.1.2. Data were tested for normal distribution (Shapiro–Wilk test) and homogeneity of variance and according to the results a Student’s t test or a Mann–Whitney U test (cauliflower experiments) or a Kruskal–Wallis (radish experiment) was performed. For Pieris experiments, the Log-Rank test of the Survival package of R was used to compare survival curves.
Results
Minor effects of P. chlororaphis subgroup bacteria on the root pest D. radicum
To investigate how bacteria of the P. chlororaphis subgroup, known for their insecticidal activity against various leaf-feeding insects [7, 10, 19], interact with an insect living in their natural habitat, the rhizosphere, we tested the impact of these bacteria following oral infection on the cabbage root fly D. radicum, which is an important root pest on brassicaceous crops.
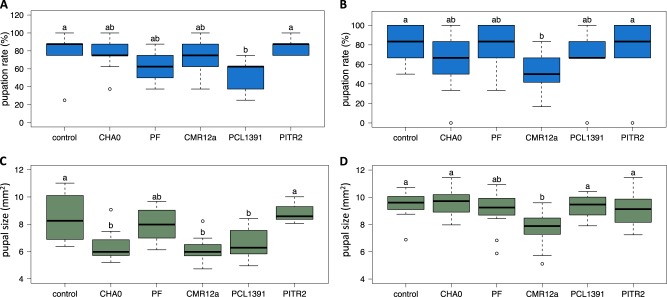
In an experiment with bacteria-treated radish bulbs, aiming at comparing different bacterial strains, P. chlororaphis PCL1391 caused a significant reduction of the pupation rate (Fig. 1a) and the pupal size (Fig. 1c) compared to the control. Moreover, in this experiment a reduction in pupal size was also observed for the strains P. protegens CHA0 and Pseudomonas sp. CMR12a (Fig. 1c). However, in a repetition of the experiment only Pseudomonas sp. CMR12a caused a significant reduction in the pupation rate (Fig. 1b) and the pupal size (Fig. 1d).
Fig. 1.
Certain strains of the Pseudomonas chlororaphis subgroup negatively affect Delia radicum pupation rate and pupal size. Pupation rate per egg (a, b) and pupal size (c, d) of D. radicum larvae fed on radishes inoculated with strains CHA0, PF, CMR12a, or PCL1391 of the P. chlororaphis subgroup, known to have insecticidal activity, or with the non-insecticidal strain PITR2 or amended with water (controls). In experiment one (a, c), nine radishes per treatment were infested each with eight D. radicum eggs, while in experiment two (b, d) twelve radishes per treatment were infested each with six eggs. Treatments with different letters significantly differed from each other (Kruskal–Wallis, p < 0.05). Control, sterile water; CHA0, Pseudomonas protegens CHA0-gfp2; PF, Pseudomonas protegens PF-gfp2; CMR12a, Pseudomonas sp. CMR12a-gfp1; PCL1391, Pseudomonas chlororaphis PCL1391-gfp2; PITR2, Pseudomonas thivervalensis PITR2-gfp2
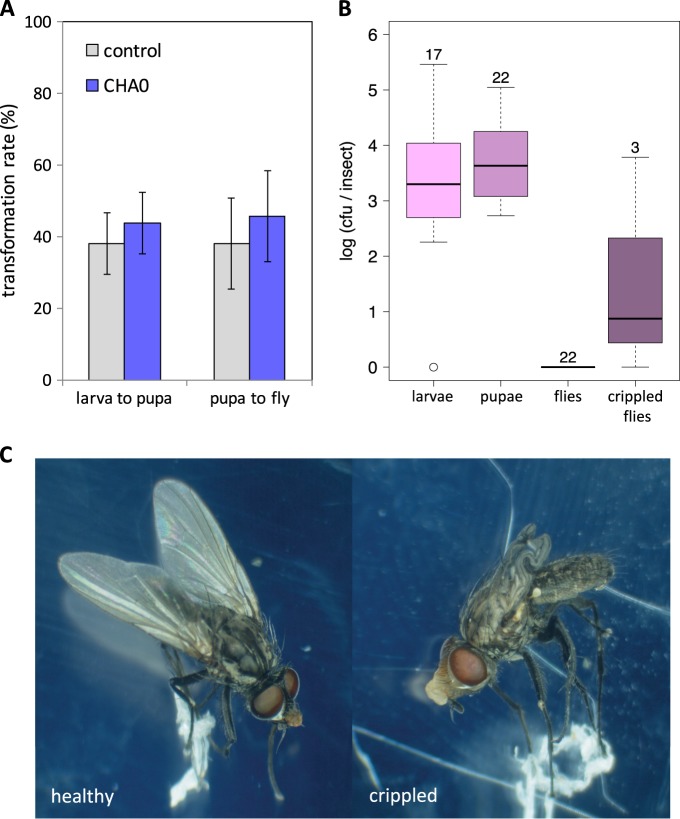
In a further experiment, larvae of D. radicum were feeding on roots of cauliflower plants inoculated with or without P. protegens CHA0 (Fig. 2). P. protegens CHA0 developed on average population sizes of 6.51 ± 0.59 (experiment 1) and 5.92 ± 0.56 (experiment 2) log10 cfu per g of root fresh-weight. In both experiments, no significant differences in pupation rate, pupal size and in the number of flies emerging from pupae could be detected between control and CHA0 treatments (Fig. 2a, Supplementary Figure S1, Supplementary Figure S2A, B). Moreover, shoot weights from plants inoculated with CHA0 did not significantly differ from those of control plants (Supplementary Figure S2C, D).
Fig. 2.
Pseudomonas protegens CHA0-gfp2 does not affect survival of Delia radicum, but it persists throughout different life stages. Five freshly hatched D. radicum larvae were added to cauliflower plants (four trays per treatment, each containing twelve pots) grown with P. protegens CHA0-gfp2 (CHA0) on the roots or without (control). a The pupation rate and the rate of flies emerging from pupae did not significantly differ between the control and the CHA0 treatment (p < 0.05; Mann–Whitney U test). Error bars depict standard deviations of the means of replicate trays. A repetition of the experiment is shown in Supplementary Figure S1. b Population sizes of P. protegens CHA0-gfp2 detected in D. radicum at different life stages after larvae fed on roots colonized by P. protegens CHA0-gfp2. No CHA0 was detected in individuals emerging from the control treatment (data not shown). Data are pooled from two experiments. Numbers above boxes indicate sample size. c Some flies in the CHA0 treatment exhibited morphological defects, e.g., crippled wings
Overall, some P. chlororaphis subgroup bacteria, particularly Pseudomonas sp. CMR12a and P. chlororaphis PCL1391, seem to affect the performance of Delia larvae leading to smaller and fewer pupae, but the effects are rather moderate and variation is high.
Transstadial transmission of P. protegens CHA0 in D. radicum and dispersal to new host plants by adult flies
In a previous study, we reported that certain strains of the P. fluorescens species complex that are not causing fatal infections are still able to persist inside larvae of the cotton leafworm Spodoptera littoralis [9]. Since no effect on survival of D. radicum larvae was observed in the present study, we were wondering whether P. protegens CHA0 was able to establish a rather commensal interaction allowing the bacterium to colonize the larvae, to persist inside the insect throughout different life stages and eventually to be transferred to new host plants by adult flies.
Persistence from larval to adult stage
Extraction of Delia larvae and pupae from the two cauliflower experiments showed that they indeed were colonized by CHA0 (Fig. 2b) at average levels of 2.7 log10 and 3.8 log10 cfu per insect. No CHA0 was detected in insects from the control treatment. In contrast to results on larvae and pupae, our method of homogenizing the flies and plating serial dilutions generally revealed no P. protegens CHA0 associated with adult flies whether these were surface-sterilized or not. However, a few flies that emerged from CHA0-treated roots showed developmental defects, most obviously malformations of the wings (Fig. 2c). Three crippled flies were extracted and remarkably two of them were colonized by CHA0 (Fig. 2b). Since healthy looking flies from the cauliflower experiments were able to transmit CHA0 to a new host plant (see results below), they must have carried the bacteria although we did not detect them. Therefore, we assessed larger amounts of Delia flies for presence of CHA0 by a second method. Entire flies emerging from control and CHA0 treatments of radish experiments were placed in selective liquid medium without prior surface disinfestation and the medium was then checked for growth of CHA0. This qualitative approach revealed that in one experiment 76% (n = 21) and in the other experiment 53% (n = 30) of the healthy flies were carrying CHA0. No CHA0 was detected on flies from the control treatment.
Dispersal by flies to a new host plant
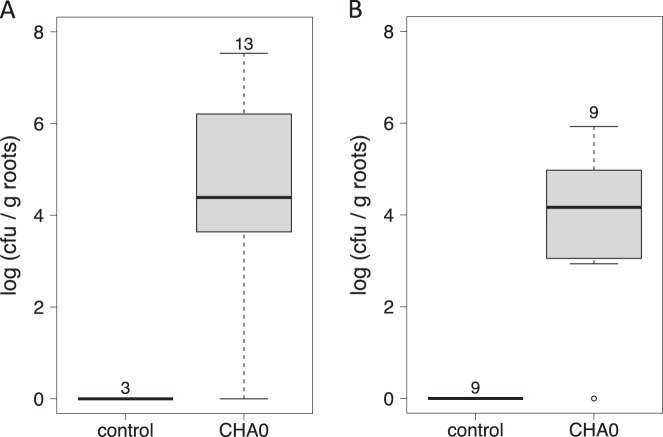
To assess, whether Delia flies which had been exposed to P. protegens CHA0 at the larval stage are able to transmit CHA0 to a new host plant, we elaborated a specific test system. Pupae that emerged from control and CHA0 treatments in the cauliflower experiments were transferred into closed plastic beakers (three to four pupae per beaker) containing rapeseed plantlets grown axenically on a sand-vermiculite substrate. To avoid transmission of bacteria by the pupae, those were kept in containments preventing direct contact with plants or substrate. Emerging flies were flying around in the transmission microcosms and in several of them they also laid eggs. Nine days after first flies started to emerge, root systems were assessed for bacterial colonization. The roots of twelve out of thirteen (experiment one) and of eight out of nine (experiment two) transmission microcosms, which had been exposed to flies emerging from the CHA0 treatment, were indeed colonized by P. protegens CHA0 (Fig. 3). In both experiments, no CHA0 could be detected on roots from transmission microcosms that had been exposed to control flies (Fig. 3). In microcosms with successful CHA0 transmission, average colonization rates in experiments one and two were 5.0 log10 and 4.3 log10 cfu per g of roots, respectively. Hence, Delia flies were able to transmit P. protegens CHA0 to a new host plant.
Fig. 3.
Pseudomonas protegens CHA0 can be dispersed to new host plants by adult Delia radicum flies. Colonization levels of CHA0-gfp2 on roots of rapeseed plants. Plants were grown in sterile soil for 18 days and were then exposed for nine days to flies, which had developed from larvae feeding on roots of control or CHA0-gfp2 treated cauliflower plants, i.e., the previous plant host. In the CHA0 treatment, in twelve out of thirteen (experiment one) (a) and in eight out of nine (experiment two) (b) systems, rapeseed roots became colonized by P. protegens CHA0-gfp2. Numbers above boxes indicate sample size
In summary, we provide first evidence that P. protegens CHA0 when ingested by larvae can be transstadially transmitted not only to the pupal stage, but even to the adult stage and that adult insects can disperse the bacterium to new host plants.
Transstadial transmission of P. protegens CHA0, a phenomenon observed in susceptible as well as in resistant insects
Since our experiments with D. radicum indicate that the relationship of CHA0 with this insect is rather of commensal than pathogenic nature we wondered to what extent the bacterium can persist in insects highly susceptible to a CHA0 infection. Thus, we monitored the persistence of CHA0 and its effects on insect development in additional insect species: the diamondback moth, Plutella xylostella, a leaf-feeder that is highly susceptible to CHA0 [9, 10]; the large white, Pieris brassicae, which is another leaf-feeding lepidopteran, for which earlier experiments suggested high susceptibility to CHA0 [6]; and finally the black vine weevil, Otiorhynchus sulcatus as a second root-feeding insect, which so far had never been investigated for its interaction with CHA0.
P. xylostella
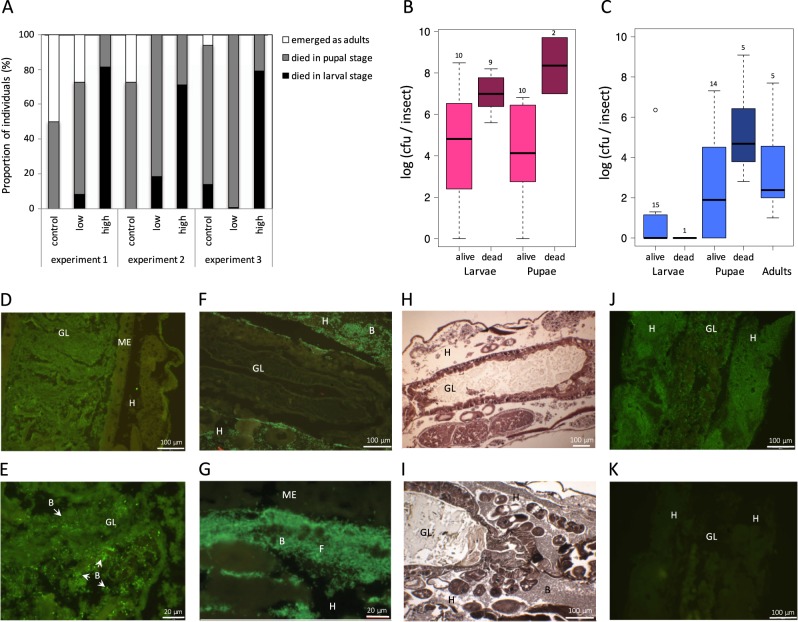
P. protegens CHA0 was found in larvae, pupae and adults of P. xylostella. Almost 80% of larvae fed on artificial diet inoculated with 10 µl of OD600 = 0.1 (high dosage) of CHA0 did not survive until pupation and the rest commonly died in the pupal stage (Fig. 4a, high). Bacterial numbers in larvae and pupae were comparable, while at both developmental stages dead individuals harbored about 100 to 1000 times more bacteria than individuals that were still alive (Fig. 4b). To investigate, whether CHA0 can persist even to the adult stage, we further extracted P. xylostella after infection with a ten times lower dosage causing almost no mortality in the larval stage anymore (Fig. 4a, low). In these infections, CHA0 was only detected in six out of fifteen larvae and at very low numbers (Fig. 4c). However, colonization levels increased in the pupal and adult stages (Fig. 4c). Generally, adult emergence was very low, also in control treatments (Fig. 4a), because the Plutella feeding assay is optimized for fast killing of larvae upon feeding on CHA0 and not for long term survival of the insects. Nevertheless, the consistent detection of CHA0 in all extracted imagines indicates transstadial transmission in P. xylostella from the larval via the pupal to the adult stage.
Fig. 4.
Infection of Plutella xylostella by Pseudomonas protegens CHA0. a–c One-week-old P. xylostella larvae (n = 32–64) were exposed to artificial diet inoculated with a low dosage (low, 10 μl of OD600 = 0.01) or a high dosage (high, 10 μl of OD600 = 0.1) of P. protegens CHA0-gfp2 or amended with sterile 0.9% NaCl solution (control). a Fraction of P. xylostella larvae dying at larval or pupal stages or emerging as adults. Three repetitions of the experiment are depicted. b Colonization of P. xylostella by P. protegens CHA0-gfp2 (high dosage). Data are pooled from experiments 2 and 3. c Colonization of P. xylostella by P. protegens CHA0-gfp2 (low dosage). Data are pooled from all three experiments. b, c Numbers above boxes indicate sample size. No P. protegens CHA0-gfp2 was detected in control insects. d–k Tracking P. protegens CHA0 in P. xylostella larvae upon oral uptake using microscopy on serial sections of fixed larvae. Larvae were infected with P. protegens CHA1176, a GFP expressing variant of P. protegens CHA0. d–g, j Sections of these larvae were stained with anti-GFP. The use of anti-GFP antibodies was necessary because fixation of larvae with Duboscq-Brazil’s alcoholic Bouin’s destroys intrinsic GFP fluorescence. h, i Sections stained with Heidenhain’s iron hematoxylin. d, e CHA0 in the gut, but not in the hemolymph. (e) is a magnification of (d). f–i CHA0 in the hemolymph and in fat body cells, but not in the gut. h Control larva fed on bacteria-free diet. j Moribund larva completely colonized by CHA0. k Consecutive section of (j) stained without adding anti-GFP antibody. B bacteria, F fat body, GL gut lumen, H hemocoel, ME midgut epithelium
Although several insect pathogenicity factors of P. protegens CHA0 have been identified [14], very little is known about the infection process inside the insect and the damage caused by CHA0. To be able to study in the same insect the localization of CHA0 and histological changes of insect tissues we established a microscopy method on thin sections of P. xylostella larvae (Supplementary Methods). Larvae fed on CHA0 containing diet were fixed and consecutive sections were either stained with anti-GFP antibodies or Heidenhain’s iron hematoxylin to visualize the bacteria or the insect tissue, respectively. Larvae from two independent experiments were analyzed and representative pictures are shown in Fig. 4d–k. They give a first insight into the colonization of P. xylostella larvae by P. protegens CHA0. In most larval samples from early infection time-points CHA0 could not be detected, but in two cases it was found in the midgut lumen (Fig. 4d, e). In contrast, at later stages of infection CHA0 was often found in the hemolymph and the fat body cells while no excessive destruction of the midgut epithelium and no bacteria in the gut could be observed (Fig. 4f, g, i). Finally, moribund larvae were always full of CHA0 all over the hemocoel and the gut and organs were not distinctively recognizable anymore (Fig. 4j).
P. brassicae
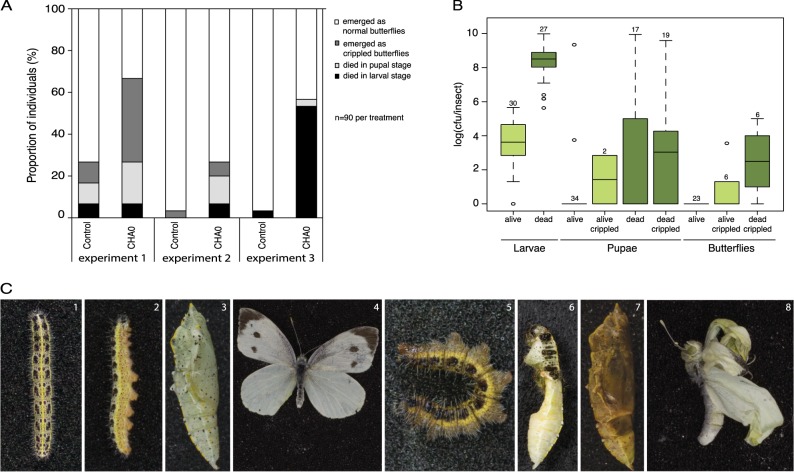
In oral infections of P. brassicae larvae with P. protegens CHA0, survival was found to be dependent on the larval stage. When 1st, 2nd, and 3rd instar larvae were fed with high dosages of CHA0, ∼ 70–95.8% of the larvae died within six days (Supplementary Figures S3A, S3B, S3C). In contrast, 4th instar larvae fed with the same number of CHA0 cells were much more resistant and showed survival rates of over 95% in most of the experiments (Supplementary Figures S3D, S4). Therefore, this instar was used to assess persistence of CHA0 throughout different developmental stages (Fig. 5). Still some CHA0-infected 4th instar larvae were unable to form intact pupae (Fig. 5c6) and some pupae with normal appearance became melanized and died (Fig. 5c7). The number of dead individuals (larvae plus pupae) was higher in the CHA0 treatment compared to the control (Fig. 5a). Moreover, 15.5% of total number of butterflies from the CHA0 treatment emerged with morphological defects, i.e., strongly deformed wings (Figs. 5a, c8), which was again higher than in the control (4.4%). Extraction of larvae, pupae, and adults revealed presence of CHA0 in nearly all living larvae at levels of around 4 log10 cfu per insect and in dead larvae even at levels as high as 9 log10 cfu per insect while in living pupae and adults with healthy appearance, CHA0 was only found exceptionally (Fig. 5b). In individuals with abnormal phenotypes, CHA0 was often detected, which indicates that in certain cases the bacterium can survive in P. brassicae until the pupal and the adult stages (Fig. 5b).
Fig. 5.
Infection of Pieris brassicae by Pseudomonas protegens CHA0. a–c Fourth instar P. brassicae larvae were fed on artificial diet inoculated with 10 µl of a P. protegens CHA0-gfp2 bacterial suspension (OD600 = 20) (CHA0) or amended with sterile 0.9% NaCl solution (control). a Fate of larvae fed with control or CHA0-treated diet. Three repetitions of the experiment are depicted. b Colonization of P. brassicae by P. protegens CHA0-gfp2. Data are pooled from three independent experiments. Numbers above boxes indicate sample size. No P. protegens CHA0-gfp2 was detected in control insects. c Phenotypical differences between insect stages developed from control (1–4) and CHA0-treated (5–8) larvae. Healthy larvae (1, 2), pupa (3) and butterfly (4); dead larva (5) and pupa (7); pupa (6), and butterfly (8) with morphological defects
O. sulcatus
Since data on the interaction of CHA0 with root-feeding insects, beside the D. radicum data presented here, is lacking, we performed first small scale experiments with a second root pest, O. sulcatus. CHA0 did not affect the survival of larvae feeding on strawberry roots (Supplementary Figure S5). Still, CHA0 was detected in two-thirds (experiment A and B) of the pupae and in two-thirds or all (experiment A and B, respectively) of the adults emerging from the CHA0-fed O. sulcatus larvae. However, the method used in these experiments only allowed a qualitative detection and no quantification of CHA0 (Supplementary Methods). The results are in line with the data obtained with D. radicum. Nevertheless, the observed persistence of CHA0 throughout different life stages of O. sulcatus warrants further investigation.
Discussion
Our results indicate that D. radicum is generally less susceptible to P. protegens and P. chlororaphis than the lepidopteran leaf-feeders tested here and in earlier studies [7, 9, 10]. Moreover, the investigated coleopteran species O. sulcatus was not affected at all after feeding on P. protegens treated roots. Thus, the pathogenicity of P. chlororaphis subgroup bacteria seems to depend on the insect species/order. When comparing susceptibility of different insect species, one has to keep in mind that root and leaf-feeders were tested in different setups. Plutella and Pieris larvae were kept in small cages and fed on artificial diet or detached leaves, an unnatural environment to the insects and thus potentially stressful. Moreover, bacterial numbers internalized by D. radicum feeding on radishes or cauliflower roots were presumably very low, since larvae burrow into the root and P. protegens CHA0 colonizes mainly the root surface [31]. However, in an earlier study by Ruffner et al. [10] PCL1391 and CHA0 were tested against three lepidopteran species in a more natural setting where bacteria were sprayed onto plant leaves. High larval or pupal mortality was obtained for all tested species when leaves were sprayed with a suspension containing 106 CFU/ml resulting in an uptake of probably only 104–105 cells, which indicates that Lepidoptera are indeed highly susceptible to these bacteria.
In our Delia experiments, we observed differences between Pseudomonas strains, which is in line with earlier studies [7, 9] where insecticidal activity also had a strain specific component. While P. protegens PF had no negative effects on D. radicum larvae, the reduction of larval survival and pupal size in the treatment with PCL1391 and CMR12a suggests that these strains can be mildly pathogenic to D. radicum. For biocontrol purposes, bacterial effects might be increased by exposing D. radicum to additional stress, e.g., by combining bacteria with organisms that could facilitate the access to the hemocoel, such as entomopathogenic fungi or nematodes.
P. protegens CHA0 did not affect survival of D. radicum larvae, but it was able to persist in the insect throughout different life stages. Thus, CHA0 seems to follow different ecological strategies depending on the insect host. It can be highly pathogenic, for instance toward P. xylostella and P. brassicae, but, as our results with D. radicum and O. sulcatus indicate, it can also live in insects as a commensal or, possibly, an opportunistic pathogen. Moreover, this study provides first evidence that in such a commensal association CHA0 could be transmitted by the insect from the roots of one plant to the roots of another plant. Hence insects might not only serve as additional hosts for P. chlororaphis subgroup bacteria, but also as vectors. In the following, we will discuss the interaction of CHA0 with non-susceptible and susceptible insect hosts.
Interaction with non/little susceptible insect hosts
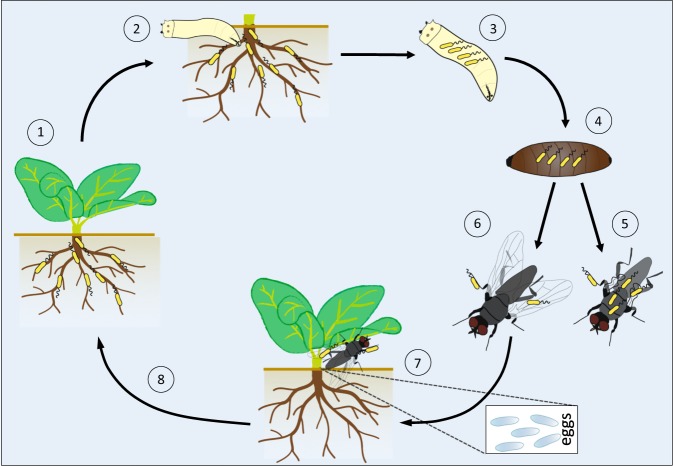
A summary of a potential association of P. protegens CHA0 with D. radicum throughout the insect life-cycle is depicted in Fig. 6. P. protegens becomes internalized by root-feeding Delia larvae, persists until the pupal stage and emerging flies can transmit the bacterium to the roots of a new host plant (Fig. 6). This could, for instance, occur when female flies lay eggs next to plant stems thereby delivering bacteria directly into a new soil habitat. In the following, the bacterium colonizes the roots of the new host plant and can again colonize larvae that hatched from deposited eggs (Fig. 6).
Fig. 6.
Pseudomonas protegens CHA0, taken up by root-feeding Delia radicum larvae, persists in the insect throughout different life stages and can be dispersed to new host plants. Roots of cauliflower plants are colonized by P. protegens CHA0 (1) and freshly hatched larvae of D. radicum are feeding on them (2). CHA0 becomes ingested by the larvae (3) and persists inside the insect also in the pupal (4) and adult stage (5, 6) (Fig. 2b). Emerging flies occasionally exhibit morphological defects (5), which affects mainly the wings (Fig. 2c). In nature, crippled flies are not able to reproduce, which will cause a decline of the insect population. However, healthy flies (6) will mate and females will search for a new host plant where they deposit their eggs in the immediate vicinity of the stem (7). Flies are able to transmit CHA0 to a new host plant resulting in bacterial colonization of the roots (8) (Fig. 3)
The results obtained with the second root feeder, O. sulcatus, suggest that the persistence of CHA0 throughout insect development in non-lethal interactions is not restricted to Delia, but might be a general trait of this bacterium. Transstadial transmission (from larva to adult) of bacteria that do not have an intracellular lifestyle, which is common for endosymbionts [32, 33], is reported for several insect species [34–37]. However, in other cases bacteria are lost during the pupal stage, before adult emergence [34, 35, 37, 38]. Our qualitative approach, where entire flies were incubated in selective medium, detected CHA0 associated with adult D. radicum in contrast to our extraction method (detection limit of the extraction method: 50 cfu/fly). Moreover, our transfer experiments showed that the bacterium was transmitted by Delia flies in about 90% of the cases. These results indicate that emerging flies regularly carry CHA0 but in very low numbers. Due to repeated molting and metamorphosis, the insect represents an unstable habitat for microbes, though bacteria might still persist in specialized crypts or paunches present in the guts of many insect species [33]. Persistence of CHA0 in insects of different orders each of which harbors specific anatomical and developmental features might rely on different strategies. Although CHA0 did not reduce Delia survival, we occasionally observed an effect on insect development, i.e., reduced pupal size and morphological defects in adults. The latter was also found for the susceptible species P. brassicae and similar observations are reported for D. melanogaster larvae infected with the related strain P. protegens Pf-5 [11, 16] and for leaffolder moths (Cnaphalocrocis medinalis) fed with rice leaves treated with a mix of P. fluorescens strains [39].
As vectors, insects would allow the bacteria to overcome large distances and to conquer new root habitats, which might considerably influence the spread of root-colonizing fluorescent pseudomonads. Insect-mediated dispersal is also known for several plant-pathogenic bacteria, but most of them do not exhibit insecticidal activity [22]; for instance Xylella fastidiosa, causing citrus variegated chlorosis and Pierce’s disease of grape, can be transmitted from one plant host to another by sharpshooter leafhoppers and spittlebugs [40]. Still, in some associations plant-pathogens are also insect-pathogenic. Pseudomonas syringae B728a can cause high mortality rates in the pea aphid and at the same time, it is excreted with honeydew and can thereby be dispersed by moving aphids [41]. Experiments carried out with sterile transmission microcosms in our study provide only a first proof of principle for the possibility of insect-mediated dispersal. In nature, a few arriving CHA0 cells might encounter considerably more difficulties to establish on the roots of a new plant host due to competition, predation or unfavorable environmental conditions. Therefore, transmission of CHA0 by insects might be a rare event under natural conditions and difficult to investigate experimentally. Nevertheless, in an experiment in which we added increasing concentrations of CHA0 cells to rapeseed seedlings grown in natural soil, we found root colonization by CHA0 to occur in 4 and 55% of the pots upon addition of as few as 50 or 500 cells, respectively (our unpublished results). This indicates that already a few transmitted CHA0 cells might be able to establish on the roots of a plant even when facing competition with a natural soil microbiome.
Interaction with susceptible insect hosts
We wondered whether CHA0 can also persist until the pupal and adult stages in insects in which the bacterium can cause lethal infections. Larvae of P. xylostella and P. brassicae are susceptible to CHA0 and become highly colonized. In P. xylostella, CHA0 generally persists in larvae and pupae but the insects seem to succumb to the infection sooner or later. However, when low dosages were fed and some individuals survived until the adult stage, we could detect CHA0 in these adults (Fig. 4c). In contrast, a considerable fraction of 4th instar P. brassicae larvae was able to eradicate CHA0 before or during pupation. However, those Pieris larvae, in which CHA0 was able to persist, seemed to succumb to the infection during pupation or exhibited major developmental defects (Figs. 5b, c5–8). Taken together, CHA0, when ingested by larvae was able to persist until the adult stage in four investigated insect species irrespective of their susceptibility pointing to the dispersal by adult insects as a possible scenario.
Finally, we wanted to take a closer look at the course of infection in a susceptible insect. During a lethal infection, P. protegens CHA0 multiplies to very high numbers as shown in Fig. 4 and reported earlier [6, 9, 10, 17]. Extraction of entire larvae does not allow any conclusion on the localization of the bacteria. The here presented microscopy method enabled the visualization of P. protegens CHA0 during the insect infection and, in parallel, the observation of histopathological changes in P. xylostella larvae. Observations indicate that P. protegens CHA0 does not colonize the gut to very high numbers and does not cause complete rupture of the midgut epithelium. Therefore, we hypothesize that P. protegens CHA0 colonizes a restricted area of the gut where it is able to enter the hemocoel. Once in the hemocoel the bacteria multiply exponentially causing a fatal septicemia. The use of the insect body as a nutrient source and a mass replication vessel is supported by the pictures of moribund larvae as well as by the very high bacteria counts in dead individuals of P. xylostella and P. brassicae. The pictures presented here give only a first insight into the colonization and infection process. How and where exactly the bacterium overcomes the gut barrier in order to invade the hemolymph remains to be discovered and requires in-depth microscopy studies.
Conclusions
This study investigated for the first time how P. chlororaphis subgroup bacteria, known for their insecticidal activity against leaf-feeding insects, interact with a root-feeding insect present in their rhizosphere habitat. While the reduction of larval survival by certain strains gives hope for potential applications as biocontrol organisms of root pests, such as D. radicum, the discovery of persistence of P. protegens CHA0 in insects throughout different developmental stages and its dispersal to a new host plant adds novel and intriguing aspects to the ecology of plant-colonizing pseudomonads. Besides being plant-beneficial rhizosphere inhabitants they can also live in insects as pathogens or commensals. Indeed, in a recent survey we could detect P. chlororaphis and P. protegens in about 10% of sampled soil arthropods (our unpublished results). Thus, these bacteria seem to be much more versatile than previously thought and we are still far from fully understanding their ecology. It remains subject to future research to discover additional habitats these bacteria might have conquered and to elucidate how they manage to switch between very different, e.g., root- and insect-associated, life styles.
Electronic supplementary material
Acknowledgements
We gratefully acknowledge Matthias Lutz, Jaka Razinger, Anne-Marie Cortesero, Martin Hommes, Syngenta Crop Protection, and the Environmental Systems Sciences group of ETH Zurich for providing insect material. Moreover, we thank Maria Péchy-Tarr and Raphël Groux for the development of the Pieris artificial diet feeding assay. We also thank Ethan Stewart for his help with the measurement of the size of D. radicum pupae by an ImageJ macro. We thank Helga Radke for technical assistance on preparation of insect samples for microscopy and Alexander Rapp from Technical University Darmstadt as well as Astrid Zimmermann for providing anti-GFP antibodies and help on the immunofluorescence microscopy. Furthermore, we acknowledge Johannes Jehle for his support to develop the collaboration between ETH Zurich and JKI Darmstadt. Finally, we thank Dylan Bär, Jana Schneider, Sandra Siegfried, Maria Haller, and Anja Taddei for technical assistance and Tobias Löser and Maria Péchy-Tarr for help with the generation of GFP-tagged strains.
Funding
This study was financed by grants obtained from the Swiss National Foundation for Scientific Research SNSF (Projects 31003A-138248, 31003A-159520, 406840-143141, and 406840-161904). The research stay of PF at JKI Darmstadt and at University of Lausanne were supported by grants obtained from Walter Hochstrasser-Stiftung, Zürich, Switzerland and by the Swiss Plant Science Web, respectively.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Contributor Information
Christoph Keel, Phone: +41-21-692-56-36, Email: christoph.keel@unil.ch.
Monika Maurhofer, Phone: +41-44-632-38-69, Email: monika.maurhofer@usys.ethz.ch.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0317-4) contains supplementary material, which is available to authorized users.
References
- 1.Garrido-Sanz D, Meier-Kolthoff JP, Goker M, Martin M, Rivilla R, Redondo-Nieto M. Genomic and genetic diversity within the Pseudomonas fluorescens complex. PLoS One. 2016;11:e0150183. doi: 10.1371/journal.pone.0150183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haas D, Défago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol. 2005;3:307–19. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- 3.Vacheron J, Desbrosses G, Bouffaud ML, Touraine B, Moenne-Loccoz Y, Muller D, et al. Plant growth-promoting rhizobacteria and root system functioning. Front Plant Sci. 2013;4:356. doi: 10.3389/fpls.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg G. Plant-microbe interactions promoting plant growth and health: perspectives for controlled use of microorganisms in agriculture. Appl Microbiol Biotechnol. 2009;84:11–8. doi: 10.1007/s00253-009-2092-7. [DOI] [PubMed] [Google Scholar]
- 5.Gomila M, Peña A, Mulet M, Lalucat J, García-Valdés E. Phylogenomics and systematics in Pseudomonas. Front Microbiol. 2015;6:214. doi: 10.3389/fmicb.2015.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kupferschmied P, Maurhofer M, Keel C. Promise for plant pest control: root-associated pseudomonads with insecticidal activities. Front Plant Sci. 2013;4:287. doi: 10.3389/fpls.2013.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rangel LI, Henkels MD, Shaffer BT, Walker FL, Davis EW, 2nd, Stockwell VO, et al. Characterization of toxin complex gene clusters and insect toxicity of bacteria representing four subgroups of Pseudomonas fluorescens. PLoS One. 2016;11:e0161120. doi: 10.1371/journal.pone.0161120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruffner B, Péchy-Tarr M, Höfte M, Bloemberg G, Grunder J, Keel C, et al. Evolutionary patchwork of an insecticidal toxin shared between plant-associated pseudomonads and the insect pathogens Photorhabdus and Xenorhabdus. BMC Genom. 2015;16:609. doi: 10.1186/s12864-015-1763-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flury P, Aellen N, Ruffner B, Pechy-Tarr M, Fataar S, Metla Z, et al. Insect pathogenicity in plant-beneficial pseudomonads: phylogenetic distribution and comparative genomics. ISME J. 2016;10:2527–42. doi: 10.1038/ismej.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruffner B, Péchy-Tarr M, Ryffel F, Hoegger P, Obrist C, Rindlisbacher A, et al. Oral insecticidal activity of plant-associated pseudomonads. Environ Microbiol. 2013;15:751–63. doi: 10.1111/j.1462-2920.2012.02884.x. [DOI] [PubMed] [Google Scholar]
- 11.Olcott MH, Henkels MD, Rosen KL, Walker FL, Sneh B, Loper JE, et al. Lethality and developmental delay in Drosophila melanogaster larvae after ingestion of selected Pseudomonas fluorescens strains. PLoS One. 2010;5:e12504. doi: 10.1371/journal.pone.0012504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schellenberger U, Oral J, Rosen BA, Wei JZ, Zhu G, Xie W, et al. A selective insecticidal protein from Pseudomonas for controlling corn rootworms. Science. 2016;354:634–7. doi: 10.1126/science.aaf6056. [DOI] [PubMed] [Google Scholar]
- 13.Jang JY, Yang SY, Kim YC, Lee CW, Park MS, Kim JC, et al. Identification of orfamide A as an insecticidal metabolite produced by Pseudomonas protegens F6. J Agric Food Chem. 2013;61:6786–91. doi: 10.1021/jf401218w. [DOI] [PubMed] [Google Scholar]
- 14.Keel C. A look into the toolbox of multi-talents: insect pathogenicity determinants of plant-beneficial pseudomonads. Environ Microbiol. 2016;18:3207–9. doi: 10.1111/1462-2920.13462. [DOI] [PubMed] [Google Scholar]
- 15.Kupferschmied P, Chai T, Flury P, Blom J, Smits TH, Maurhofer M, et al. Specific surface glycan decorations enable antimicrobial peptide resistance in plant-beneficial pseudomonads with insect-pathogenic properties. Environ Microbiol. 2016;18:4265–81. doi: 10.1111/1462-2920.13571. [DOI] [PubMed] [Google Scholar]
- 16.Loper JE, Henkels MD, Rangel LI, Olcott MH, Walker FL, Bond KL, et al. Rhizoxin, orfamide A, and chitinase production contribute to the toxicity of Pseudomonas protegens strain Pf-5 to Drosophila melanogaster. Environ Microbiol. 2016;18:3509–21. doi: 10.1111/1462-2920.13369. [DOI] [PubMed] [Google Scholar]
- 17.Péchy-Tarr M, Bruck DJ, Maurhofer M, Fischer E, Vogne C, Henkels MD, et al. Molecular analysis of a novel gene cluster encoding an insect toxin in plant-associated strains of Pseudomonas fluorescens. Environ Microbiol. 2008;10:2368–86. doi: 10.1111/j.1462-2920.2008.01662.x. [DOI] [PubMed] [Google Scholar]
- 18.Devi KK, Kothamasi D. Pseudomonas fluorescens CHA0 can kill subterranean termite Odontotermes obesus by inhibiting cytochrome c oxidase of the termite respiratory chain. FEMS Microbiol Lett. 2009;300:195–200. doi: 10.1111/j.1574-6968.2009.01782.x. [DOI] [PubMed] [Google Scholar]
- 19.Flury P, Vesga P, Pechy-Tarr M, Aellen N, Dennert F, Hofer N, et al. Antimicrobial and insecticidal: cyclic lipopeptides and hydrogen cyanide produced by plant-beneficial Pseudomonas strains CHA0, CMR12a, and PCL1391 contribute to insect killing. Front Microbiol. 2017;8:100. doi: 10.3389/fmicb.2017.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kupferschmied P, Péchy-Tarr M, Imperiali N, Maurhofer M, Keel C. Domain shuffling in a sensor protein contributed to the evolution of insect pathogenicity in plant-beneficial Pseudomonas protegens. PLoS Path. 2014;10:e1003964. doi: 10.1371/journal.ppat.1003964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Péchy-Tarr M, Borel N, Kupferschmied P, Turner V, Binggeli O, Radovanovic D, et al. Control and host-dependent activation of insect toxin expression in a root-associated biocontrol pseudomonad. Environ Microbiol. 2013;15:736–50. doi: 10.1111/1462-2920.12050. [DOI] [PubMed] [Google Scholar]
- 22.Nadarasah G, Stavrinides J. Insects as alternative hosts for phytopathogenic bacteria. FEMS Microbiol Rev. 2011;35:555–75. doi: 10.1111/j.1574-6976.2011.00264.x. [DOI] [PubMed] [Google Scholar]
- 23.Snyder WE, Tonkyn DW, Kluepfel DA. Insect-mediated dispersal of the rhizobacterium Pseudomonas chlororaphis. Phytopathology. 1998;88:1248–54. doi: 10.1094/PHYTO.1998.88.12.1248. [DOI] [PubMed] [Google Scholar]
- 24.Snyder WE, Tonkyn DW, Kluepfel DA. Transmission of a genetically engineered rhizobacterium by grasshoppers in the laboratory and field. Ecol Appl. 1999;9:245–53. doi: 10.1890/1051-0761(1999)009[0245:TOAGER]2.0.CO;2. [DOI] [Google Scholar]
- 25.Lamb TG, Tonkyn DW, Kluepfel DA. Movement of Pseudomonas aureofaciens from the rhizosphere to aerial plant tissue. Can J Microbiol. 1996;42:1112–20. doi: 10.1139/m96-143. [DOI] [Google Scholar]
- 26.Iavicoli A, Boutet E, Buchala A, Metraux JP. Induced systemic resistance in Arabidopsis thaliana in response to root inoculation with Pseudomonas fluorescens CHA0. Mol Plant-Microbe Interact. 2003;16:851–8. doi: 10.1094/MPMI.2003.16.10.851. [DOI] [PubMed] [Google Scholar]
- 27.Maurhofer M, Reimmann C, Schmidli-Sacherer P, Heeb S, Haas D, Défago G. Salicylic acid biosynthetic genes expressed in Pseudomonas fluorescens strain P3 improve the induction of systemic resistance in tobacco against tobacco necrosis virus. Phytopathology. 1998;88:678–84. doi: 10.1094/PHYTO.1998.88.7.678. [DOI] [PubMed] [Google Scholar]
- 28.King EO, Ward MK, Raney DE. 2 simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954;44:301–7. [PubMed] [Google Scholar]
- 29.Razinger J, Lutz M, Schroers HJ, Urek G, Grunder J. Evaluation of insect associated and plant growth promoting fungi in the control of cabbage root flies. J Econ Entomol. 2014;107:1348–54. doi: 10.1603/EC14004. [DOI] [PubMed] [Google Scholar]
- 30.David WAL, Gardiner BO. Rearing Pieris brassicae L. larvae on a semi-synthetic diet. Nature. 1965;207:882–3. doi: 10.1038/207882b0. [DOI] [Google Scholar]
- 31.Troxler J, Berling CH, MoenneLoccoz Y, Keel C, Defago G. Interactions between the biocontrol agent Pseudomonas fluorescens CHA0 and Thielaviopsis basicola in tobacco roots observed by immunofluorescence microscopy. Plant Pathol. 1997;46:62–71. doi: 10.1046/j.1365-3059.1997.d01-205.x. [DOI] [Google Scholar]
- 32.Kikuchi Y. Endosymbiotic bacteria in insects: their diversity and culturability. Microbes Environ. 2009;24:195–204. doi: 10.1264/jsme2.ME09140S. [DOI] [PubMed] [Google Scholar]
- 33.Engel P, Moran NA. The gut microbiota of insects - diversity in structure and function. FEMS Microbiol Rev. 2013;37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 34.Greenberg B, Klowden M. Enteric bacterial interactions in insects. Am J Clin Nutr. 1972;25:1459–66. doi: 10.1093/ajcn/25.12.1459. [DOI] [PubMed] [Google Scholar]
- 35.Moll RM, Romoser WS, Modrzakowski MC, Moncayo AC, Lerdthusnee K. Meconial peritrophic membranes and the fate of midgut bacteria during mosquito (Diptera: Culicidae) metamorphosis. J Med Entomol. 2001;38:29–32. doi: 10.1603/0022-2585-38.1.29. [DOI] [PubMed] [Google Scholar]
- 36.Chavshin AR, Oshaghi MA, Vatandoost H, Yakhchali B, Zarenejad F, Terenius O. Malpighian tubules are important determinants of Pseudomonas transstadial transmission and longtime persistence in Anopheles stephensi. Parasit Vectors. 2015;8:36. doi: 10.1186/s13071-015-0635-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radvan R. Persistence of bacteria during development in flies. Folia Microbiol. 1960;5:50–6. doi: 10.1007/BF02930896. [DOI] [Google Scholar]
- 38.Leach JG. The method of survival of bacteria in the puparia of the seedcorn maggot (Hylemyia cilicrura Rond.) J Appl Entomol. 1934;20:150–61. [Google Scholar]
- 39.Saravanakumar D, Muthumeena K, Lavanya N, Suresh S, Rajendran L, Raguchander T, et al. Pseudomonas-induced defence molecules in rice plants against leaffolder (Cnaphalocrocis medinalis) pest. Pest Manag Sci. 2007;63:714–21. doi: 10.1002/ps.1381. [DOI] [PubMed] [Google Scholar]
- 40.Chatterjee S, Almeida RPP, Lindow S. Living in two worlds: the plant and insect lifestyles of Xylella fastidiosa. Annu Rev Phytopathol. 2008;46:243–71. doi: 10.1146/annurev.phyto.45.062806.094342. [DOI] [PubMed] [Google Scholar]
- 41.Stavrinides J, McCloskey JK, Ochman H. Pea aphid as both host and vector for the phytopathogenic bacterium Pseudomonas syringae. Appl Environ Microbiol. 2009;75:2230–5. doi: 10.1128/AEM.02860-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jousset A, Schuldes J, Keel C, Maurhofer M, Daniel R, Scheu S, et al. Full-genome sequence of the plant growth-promoting bacterium Pseudomonas protegens CHA0. Genome Announc. 2014;2:e00322–00314. doi: 10.1128/genomeA.00322-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stutz EW, Défago G, Kern H. Naturally-occuring fluorescent pseudomonads involved in suppression of black root rot of tobacco. Phytopathology. 1986;76:181–5. doi: 10.1094/Phyto-76-181. [DOI] [Google Scholar]
- 44.Levy E, Gough FJ, Berlin KD, Guiana PW, Smith JT. Inhibition of Septoria tritici and other phytopathogenic fungi and bacteria by Pseudomonas fluorescens and its antibiotics. Plant Pathol. 1992;41:335–41. doi: 10.1111/j.1365-3059.1992.tb02355.x. [DOI] [Google Scholar]
- 45.Perneel M, Heyrman J, Adiobo A, De Maeyer K, Raaijmakers JM, De Vos P, et al. Characterization of CMR5c and CMR12a, novel fluorescent Pseudomonas strains from the cocoyam rhizosphere with biocontrol activity. J Appl Microbiol. 2007;103:1007–20. doi: 10.1111/j.1365-2672.2007.03345.x. [DOI] [PubMed] [Google Scholar]
- 46.Chin-A-Woeng TFC, Bloemberg GV, van der Bij AJ, van der Drift K, Schripsema J, Kroon B, et al. Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol Plant-Microbe Interact. 1998;11:1069–77. doi: 10.1094/MPMI.1998.11.11.1069. [DOI] [Google Scholar]
- 47.Keel C, Weller DM, Natsch A, Défago G, Cook RJ, Thomashow LS. Conservation of the 2,4-diacetylphloroglucinol biosynthesis locus among fluorescent Pseudomonas strains from diverse geographic locations. Appl Environ Microbiol. 1996;62:552–63. doi: 10.1128/aem.62.2.552-563.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.