Abstract
Attention-deficit/hyperactivity disorder (ADHD) is one of the most prevalent neurodevelopmental disorders in childhood and persists into adulthood in 40–65% of cases. Given the polygenic and heterogeneous architecture of the disorder and the limited overlap between genetic studies, there is a growing interest in epigenetic mechanisms, such as microRNAs, that modulate gene expression and may contribute to the phenotype. We attempted to clarify the role of microRNAs in ADHD at a molecular level through the first genome-wide integrative study of microRNA and mRNA profiles in peripheral blood mononuclear cells of medication-naive individuals with ADHD and healthy controls. We identified 79 microRNAs showing aberrant expression levels in 56 ADHD cases and 69 controls, with three of them, miR-26b-5p, miR-185-5p, and miR-191-5p, being highly predictive for diagnostic status in an independent dataset of 44 ADHD cases and 46 controls. Investigation of downstream microRNA-mediated mechanisms underlying the disorder, which was focused on differentially expressed, experimentally validated target genes of the three highly predictive microRNAs, provided evidence for aberrant myo-inositol signaling in ADHD and indicated an enrichment of genes involved in neurological disease and psychological disorders. Our comprehensive study design reveals novel microRNA–mRNA expression profiles aberrant in ADHD, provides novel insights into microRNA-mediated mechanisms contributing to the disorder, and highlights promising candidate peripheral biomarkers.
Subject terms: Predictive markers, Psychiatric disorders
Introduction
Attention-deficit/hyperactivity disorder (ADHD) is one of the most prevalent childhood onset neurodevelopmental disorders affecting around 5% of the population [1], which persists into adulthood in 40–65% of cases [2]. The disorder is characterized by severe impairments in sustaining attention, the inability to control impulses, and difficulties in modulating activity levels. With heritability estimates of around 74% [3], ADHD is considered a complex disorder caused by underlying genetic and environmental risk factors. Until today, its etiology and pathogenesis still remain largely unclarified.
Genetic association studies, pharmacological data, and/or gene–environmental interaction analyses have made a case for the involvement of genes of the dopaminergic, serotoninergic, and glutamatergic neurotransmission systems, the neurotransmitter release system, neuronal migration, and cell adhesion, among others, in the etiology of ADHD [4]. Only few genetic findings, however, were replicable, potentially due to the heterogeneity of samples, small effect sizes of individual genetic variants (in combination with limited sample sizes), and/or gene by environment interactions involved in the disorder. Given the large proportion of heritability still to be explained, there is a growing interest in the epigenetic mechanisms, such as DNA-methylation, histone modification, RNA editing, and microRNAs (miRNAs), which modulate gene expression, may provide a means to integrate effects of genetic and environmental risk factors and help explain additional phenotypic variance in psychiatric disorders like ADHD [5, 6].
miRNAs are highly conserved and non-coding small RNAs of approximately 21–25 nucleotides of length, that act as post-transcriptional regulators of gene expression. They do so through complementary base pairing with the 3′UTR of their target messenger RNAs (mRNAs), promoting the degradation or suppressing the translation of those target mRNAs [7]. The human genome encodes more than 2500 different miRNAs that play an important role in regulating diverse biological processes like cell differentiation, proliferation, and apoptosis (http://www.mirbase.org). miRNAs are known to be involved in the development of the central nervous system (CNS) and in many neurological processes including synaptic plasticity and synaptogenesis [5]. Since one single miRNA is potentially able to regulate hundreds of target genes and one single protein-coding gene is regulated by several miRNAs [8], studies focused on miRNA–mRNA interactions may elucidate novel gene-networks and biological processes underlying complex psychiatric diseases. Given the limited accessibility of the human brain for studying epigenetic modifications, miRNA profiling in peripheral blood cells is often used as a non-invasive proxy to study the transcriptional and epigenetic biosignatures, and to identify potential clinical biomarkers for psychiatric disorders [9–11]. Although, tissue specificity exists for miRNAs, a correlation of gene and miRNA expression between whole blood and CNS tissue has been observed, supporting the use of peripheral gene expression as a surrogate for expression levels in the CNS [12–14].
To date, most studies of miRNAs in ADHD have focused on the expression levels of specific miRNAs or on genetic variants in genomic regions containing miRNAs and/or their target genes [15]. Alterations in miRNA expression levels, including those of miR-let-7d, miR-18a-5p, miR-22-3p, miR-24-3p, miR-34c-3p, miR-106b-5p, miR-107, and miR-155-5p have been identified in serum or peripheral blood mononuclear cells (PBMCs) of people with ADHD, which suggests that circulating miRNA levels may serve as a potential biomarker for an ADHD diagnosis [10, 16, 17]. These data are in agreement with studies focused on the prefrontal cortex of the spontaneously hypertensive rats, an animal model for ADHD, that also point at miR-let-7d expression dysregulation involved in the disorder [18]. Association studies focused on genetic variants in miRNAs, their target genes, and genes involved in miRNAs biogenesis also support the idea that alterations affecting the expression levels, processing or target binding of miRNAs may result in functional alterations predisposing to ADHD [10, 19–21]. These studies focused largely on changes in miRNA expression but did not explore the effect of aberrant miRNA profiles on the expression of protein-coding genes.
Considering this background, we performed the first multi-step study to construct miRNA–mRNA expression profiles and evaluate the role of miRNAs in ADHD at the molecular level.
Material and methods
Study design
A multi-step approach was used to identify the role of miRNAs in ADHD by (i) assessing miRNA and mRNA expression profiles in PBMCs of ADHD cases and controls, (ii) testing the predictive performance of the differentially expressed miRNAs in an independent sample of ADHD subjects and controls, (iii) constructing paired miRNA–mRNA expression profiles and regulatory networks starting from aberrant miRNAs, (iv) identifying direct and experimentally validated relationships between miRNA–mRNA anti-correlated pairs, and (v) analyzing functional and canonical pathway over-representation and gene-networks for dysregulated miRNA target genes (Fig. 1).
Fig. 1.
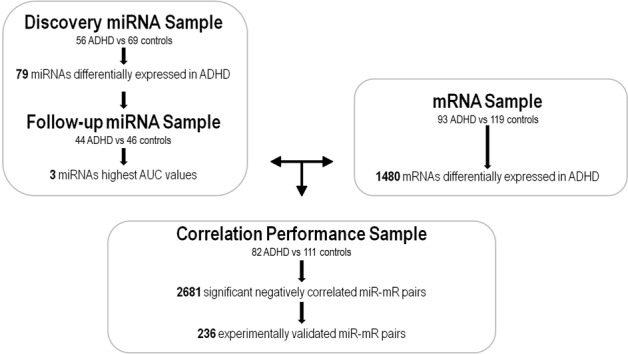
Workflow of the multi-step approach to identify the miRNA–mRNA regulatory networks in ADHD
Participants
miRNA expression profiles were assessed in a discovery sample of 56 medication-naive adult ADHD patients (average age at assessment was 37.7 years (s.d. = 11.3)) and 69 healthy controls (average age of 44.1 years (s.d. = 19.3)). The follow-up sample was composed by 44 medication-naive adult ADHD patients (average age at assessment was 30.7 years (s.d. = 10.8)) and 46 healthy controls (average age of 36.3 years (s.d. = 9.0)).
Gene expression profiles were assessed in 93 medication-naive adult ADHD subjects (average age at assessment was 19.1 years (s.d. = 11.3)) and 119 healthy controls (average age of 20.7 years (s.d. = 11.3)). There was 81% sample overlap (82 ADHD and 111 controls) between miRNA and gene expression assays.
All subjects were of European ancestry and were evaluated and recruited prospectively from a restricted geographic area in a specialized out-patient program for adult ADHD and by a single clinical group at Hospital Universitari Vall d’Hebron of Barcelona (Spain). Patients were referred to the program from primary care centers and adult community mental health services. None of them were selected retrospectively from our ADHD dataset but they were prospectively recruited according to the inclusion criteria for transcriptomic assays instead (medication-naive without other psychiatric comorbidities). They were distributed in two groups (discovery and follow-up samples) depending on the period in which they were recruited, clinically assessed, and in which the blood sample was extracted. The discovery sample consists of patients recruited from 2011 to 2015 and the follow-up sample corresponds to subjects recruited from 2015 to 2016.
The study was approved by the Clinical Research Ethics Committee (CREC) of Hospital Universitari Vall d’Hebron, all methods were performed in accordance to the relevant guidelines and regulations and written informed consent was obtained from all subjects before inclusion into the study.
Clinical assessment
The clinical assessment was conducted by structured interviews and self-reported questionnaires in two different steps: (i) assessment of ADHD diagnosis based on symptomatology using the Conner’s Adult ADHD Diagnostic Interview for DSM-IV (CAADID) by a psychiatrist and (ii) assessment of the severity of ADHD symptoms, the levels of impairment and the presence of comorbid disorders by a psychologist to increase the diagnostic accuracy and reduce the likelihood of misdiagnosis with the Conners’ ADHD Rating Scale (CAARS), the ADHD Rating Scale (ADHD-RS), the Clinical Global Impression (CGI), the Wender Utah Rating Scale (WURS), the Sheehan Disability Inventory (SDS), and the Structured Clinical Interview for DSM-IV Axis I and II Disorders (SCID-I and SCID-II). Afterwards, the psychiatrist and psychologist integrate the clinical information and self-reports for the valid assessment of symptoms and impairments. Given the clinical diagnosis of ADHD, in case of discordance between different raters of ADHD symptoms or inconsistencies between reporters in responses to items measuring similar symptoms, the clinician-identified symptoms on the CAADID prevailed.
The diagnosis of ADHD was assessed with the CAADID structured interview, which is divided into Part I and Part II that were administered separately: (i) the CAADID Part I which is a Patient History administered as a clinical interview and (ii) the CAADID Part II which is a Diagnostic Criteria Interview used to assess ADHD according to DSM-IV criteria. Diagnosis of ADHD was confirmed when patients presented six or more symptoms of inattention and/or six or more symptoms of hyperactivity–impulsivity, which persisted for at least 6 months to a degree that is considered maladaptive and inconsistent with the developmental level. When possible, a collateral interview with a family member was performed during the clinical interview, which provides valuable information that the patient may not self-report.
In addition to the clinical diagnosis, severity of ADHD symptoms and the level of impairment were assessed using self-report questionnaires that include: (i) the CAARS Scale-long version gathered from the patient (self-report; CAARS-S:L) and an observer (secondary informant; CAARS-O:L); the ADHD-RS and the CGI to measure the severity of ADHD symptoms in adulthood, (ii) the WURS to retrospectively assess ADHD symptoms in childhood, and (iii) the SDS to assess the level of functional impairment. Finally, differential diagnosis and assessment of comorbidities were assessed with SCID-I and SCID-II.
Exclusion criteria was IQ < 70; lifelong and current history of mood, psychotic, anxiety, substance abuse, and DSM-IV axis II disorders; pervasive developmental disorders; a history or the current presence of a condition or illness, including neurologic, metabolic, cardiac, liver, kidney, or respiratory disease; a chronic medication of any kind; birth weight ≤ 1.5 kg; and other neurological or systemic disorders that might explain ADHD symptoms.
The control sample consisted of unrelated healthy blood donors matched by sex with the clinical group. Individuals with ADHD symptomatology were excluded retrospectively under the following criteria: (1) not having been diagnosed with ADHD previously and (2) answering negatively to the life-time presence of the following ADHD symptoms: (a) often has trouble in keeping attention on tasks, (b) usually loses things needed for tasks, (c) often fidgets with hands or feet or squirms in seat, and (d) often gets up from seat when remaining in seat is expected.
RNA isolation and quantification
PBMCs were separated by Ficoll density gradient method immediately after blood extraction, and total mRNA, including small RNA, was isolated from PBMCs using Qiazol Lysis reagent and the RNAeasy Midi Kit (QIAgen, Hilden, Germany). RNA integrity and concentration of samples were assayed by 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA, USA).
miRNA expression analysis
Next-generation sequencing
miRNA expression was assessed using next-generation sequencing. Complementary DNA (cDNA) libraries were constructed using the Illumina’s TruSeq Small RNA protocol. After cluster generation, the library preparations were sequenced in nine different rounds on the Illumina Hiseq 2000 platform (Illumina, San Diego, CA, USA).
Bioinformatics and statistical analyses
Illumina’s adapters and reads shorter than 18 nucleotides were removed and data was filtered out for low-quality sequences using the Trimgalore algorithm (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). The remaining sequences were aligned to the reference genome (hg19) using the Bowtie2 software [22]. Preprocessing of RNA-Seq alignments for differential expression analysis was performed using the HTSeq Python package based on miRBase release 20 [23]. The relative expression level of a specific miRNA was measured from the frequency of its read counts. We considered only those miRNAs with at least 1 count-per-million (CPM) in the smallest sample sizes between cases and controls. Filtering, normalization, and summarization of raw counts were performed using the edgeR package from Bioconductor [24]. Batch effects and non-biological experimental variation (RNA integrity number (RIN), age, and gender) were adjusted for using the ComBatR algorithm [25]. Logistic regression models were applied to compare miRNA expression profiles between cases and controls in the discovery sample using the Limma R package [26]. Benjamini–Hochberg correction was applied for multiple comparisons and statistical significance was set at PBH < 0.05. miRNA enrichment analysis was performed using an online tool for annotations of human miRNAs (TAM tool) [27]. The accuracy of the differentially expressed miRNAs in discriminating subjects with ADHD from controls was evaluated in the follow-up sample with a Receiver Operating Characteristic (ROC) analysis using the pROC R package [28]. Logistic regression models were fitted in the discovery sample and the parameter estimates were then used to predict ADHD in the follow-up sample. The analysis included as covariates those clinical variables (i.e., age) showing significant differences between cases and controls (P ≤ 0.05).
Messenger-RNA expression analysis
Microarray assays
RNA was reverse transcribed using the Ambion WT Expression Kit (Life Technologies, Carlsbad, CA, USA). The cRNA was subsequently fragmented, labeled, and hybridized with the GeneChip WT Terminal Labeling and Hybridization Kit (Affymetrix, Santa Clara, CA, USA). The samples were hybridized to the GeneChip Human Gene 1.1 ST 96-Array plate (Affymetrix), covering a total of 36079 transcripts that correspond to 21014 genes. The array processing and data generation were assessed using the Gene Titan Affymetrix microarray platform.
Bioinformatics and statistical analyses
Raw data were pre-processed with the Robust Multichip Analysis (RMA) algorithm (including the background correction, normalization, and summarization of probes values) implemented in the OligoR package from Bioconductor [29]. Transcript probes corresponding to unknown genes or matching with more than one gene in the GRCh37/hg19 human genome build (release 32) were discarded, and the study was finally restricted to 19263 probes corresponding to 18475 unique genes. Batch effects and non-biological experimental variation (RIN, age, or gender) were adjusted for using the ComBatR algorithm. Similar to the assessment of dysregulated miRNAs, logistic regression models were used to compare gene expression profiles between cases and controls using the Limma R package. Benjamini–Hochberg correction was applied for multiple comparisons and statistical significance was set at PBH < 0.05.
miRNA-target genes networks
Pearson’s correlation analysis was applied to miRNAs with an area under the Receiver Operator Characteristics curve (AUC) higher than 0.70 and genes differentially expressed in a subset of 82 medication-naive adult ADHD subjects and 111 healthy controls from whom miRNA and mRNA profiles were available. Correlations were assessed using the Corrplot R package [30] and multiple testing correction was set at PBH < 0.05. Experimentally validated miRNA–mRNA interactions were identified with the Diana-Tarbase v.8.0 database [31]. Enrichment analysis of canonical pathways and downstream analyses, including diseases and functional annotations and gene networks, were performed to identify over-representation of biological and functional processes in our set of differentially expressed miRNAs with highest AUC values and their experimentally validated target genes using the Ingenuity Pathway Analysis software (Ingenuity Systems, Redwood City, CA, USA; http://www.ingenuity.com). Benjamin–Hochberg correction was used to adjust for multiple comparisons and the significance threshold was set at PBH < 0.05. Gene networks were considered of true relevance when the network score (P-score = −log10(P-value)) was over 3 (P-value = 1e−03) and the total number of focus molecules (genes from our set that passed filters and were eligible for generating networks) was above 20 out of 35.
Results
After normalization and background correction, a total of 573 miRNAs were detected in PBMC of at least 45% of individuals from the discovery sample of 59 adult subjects with ADHD and 69 controls. A total of 79 miRNAs were differentially expressed in ADHD after multiple-testing correction (PBH < 0.05), 55 were up-regulated and 24 down-regulated (Supplementary Table S1 and Supplementary Figure S1A). The up-regulated miRNAs were enriched for the cluster families miR-92a-19a-17-19b, miR-191-425, and let-7a-7b (P = 0.019, P = 0.045, and P = 0.045, respectively) and correlations between expression levels of the miRNAs within each cluster family were also detected (Supplementary Figure S2).
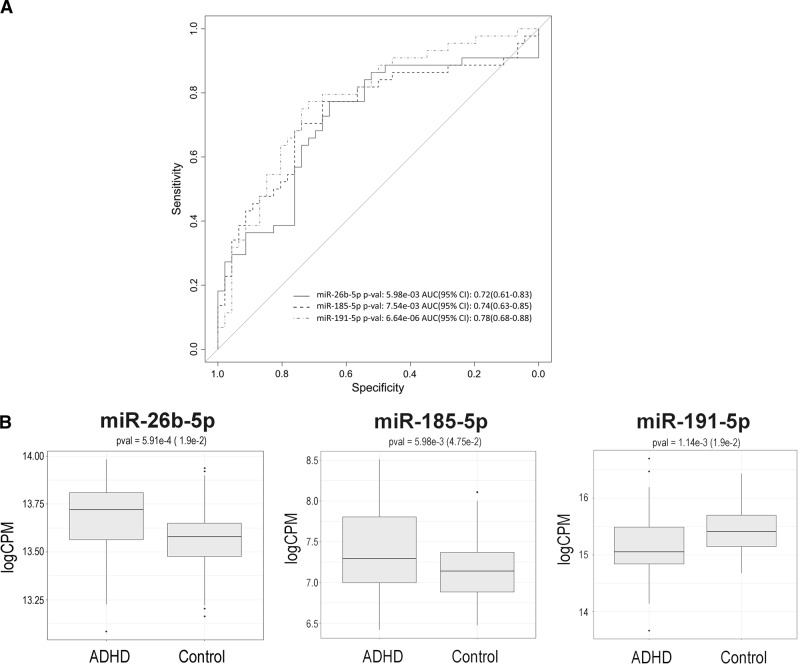
The predictive performance of the 79 differentially expressed miRNAs was assessed in the follow-up sample of 44 ADHD subjects and 46 controls. Substantial variation in the predictive capacity was observed, with area under the Receiver Operating Characteristic curve (AUC) values ranging from 0.47 to 0.78 (Supplementary Table S1). The highest AUC values were observed for miR-191-5p (AUC = 0.78; P = 6.64e−06) which was over-expressed in ADHD, and for the down-regulated miR-26b-5p (AUC = 0.72; P = 5.98e−03) and miR-185b-5p (AUC = 0.74; P = 7.54e−03) (Fig. 2). No effect of age was found in the expression levels of any of them (Supplementary Figure S3). We focused on this subset of highly predictive miRNAs (AUC > 0.7) to further identify miRNA–mRNA interaction networks dysregulated in ADHD. The transcriptome-wide expression assay assessed in PBMCs from 93 ADHD patients and 119 controls revealed 1546 differentially expressed transcripts corresponding to 1480 genes; 734 of these genes were up-regulated and 746 were down-regulated in ADHD subjects (P < 0.05). None of them, however, remained significant after Benjamin–Hochberg correction (Supplementary Figure S1B and Supplementary Table S2). Next, we explored the correlation between the expression levels of miR-26b-5p, miR-185b-5p, and miR-191-5p with those of the 1480 differentially expressed genes in a subset of 193 subjects (82 ADHD cases and 111 controls) from whom miRNA and mRNA expression data were available. The correlation analysis highlighted a total of 2681 miRNA–mRNA negative correlated pairs (PBH < 0.05), including the three miRNAs and 1400 genes. Of the latter, 236 were reported experimentally validated targets for at least one of the miRNAs (Fig. 1). Specifically, 7, 65, and 184 dysregulated genes were experimentally validated targets for miR-191-5p, miR-185b-5p, and miR-26b-5p, respectively (Supplementary Table S3).
Fig. 2.
Predictive performance for ADHD of miR-26b-5p, miR-185-5p, and miR-191-5p in the follow-up sample of 44 ADHD subjects and 46 controls: a) Receiver Operating Characteristic curves and b) box plots
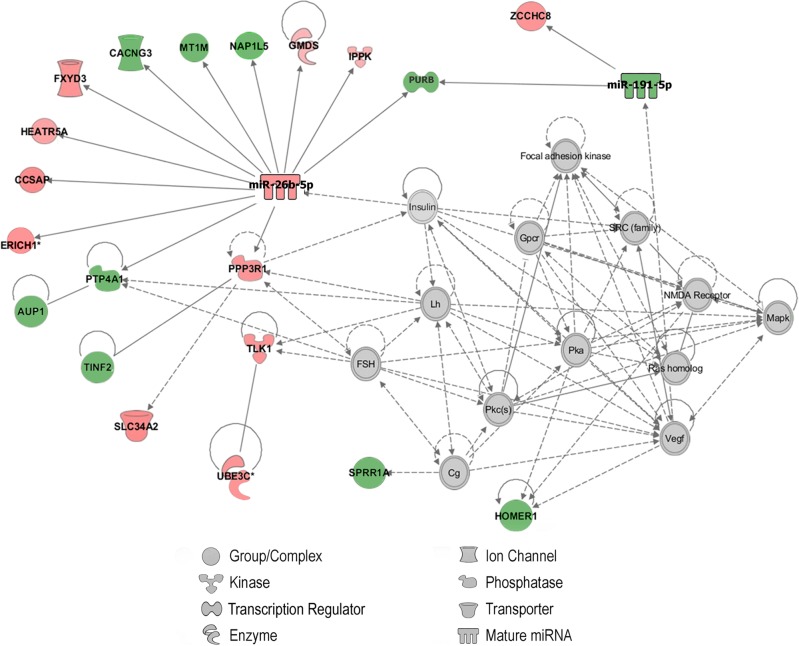
To better understand the molecular role in ADHD of the three dysregulated miRNAs with the highest predictive performance, we focused on their differentially expressed and negatively correlated experimentally validated targets. The canonical pathway enrichment analysis showed over-representation for genes related to the myo-inositol metabolism with overlap between molecules in each canonical pathway and our dataset ranging from 21.7% to 29.4%: “D-myo-inositol (1,4,5)-trisphosphate Degradation” (PBH = 2.7e−06 and 29.4% of overlap), “1D-myo-inositol Hexakisphosphate Biosynthesis II” (PBH = 3.6e–06 and 27.8% of overlap), “Superpathway of D-myo-inositol (1,4,5)-trisphosphate Metabolism” (PBH = 1.3e−05 and 21.7% of overlap), and “D-myo-inositol (1,3,4)-trisphosphate Biosynthesis” (PBH = 9.5e−05 and 22.2% of overlap). Interestingly, downstream analyses revealed an enrichment in our gene set of disease annotations related to “Neurological Disease” (P-value = 1.4e−02 to 4.9e−05, with 60 focus molecules) and “Psychological Disorders” (P-value = 1.4e−2 to 4.9e−05, with 41 focus molecules), and functional annotations such as “Schizophrenia Spectrum Disorder” (P-value = 7.6e−03, with 16 focus molecules), “Disorder of basal ganglia” (P-value = 7.6e−03, with 16 focus molecules), “Proliferation of neuroglia” (P-value = 7.8e−03, with 7 focus molecules), “Morphology of hippocampus” (P-value = 1.3e−02, with 6 focus molecules), or “Quantity of axons” (P-value = 1.3e−02, with 3 focus molecules), among others. Finally, one of the best-generated networks corresponded to “Behavior, Nervous System Development and Function, Connective Tissue Disorders” (score (−log(Fisher’s Exact test P-value)) = 38) with 22 focus molecules that include miR-26b-5p and miR-191-5p (Fig. 3).
Fig. 3.
Integrated target gene network corresponding to “Behavior, Nervous System Development and Function, Connective Tissue Disorders”, that includes miR-26b-5p, miR-191-5p, and 20 target genes. Genes from our gene set included in the network are represented with colored background, in red up-regulated genes and in green down-regulated genes in ADHD. Interacting proteins added by the Ingenuity database are represented with clear background nodes. Solid and dashed lines represent direct or indirect interaction between the molecules, respectively
Discussion
To our knowledge, this is the first study combining genome-wide miRNA expression and transcriptome profiling in a multi-step integrative analysis to identify aberrant miRNA–mRNA expression profiles in ADHD, to provide novel regulatory networks and RNA-mediated mechanisms underlying the disorder, and highlight promising candidate peripheral biomarkers for ADHD.
We found preliminary evidence of substantial dysregulation of miRNA expression in ADHD, with a subset of 79 miRNAs showing aberrant expression levels in PBMC of subjects with ADHD compared to healthy controls. Three of them, miR-26b-5p, miR-185-5p, and miR-191-5p, showed moderate predictive performance for ADHD with AUCs over 70% in an independent dataset. No evidence for their contribution to ADHD has been reported so far. However, alterations in the expression levels of the three miRNAs have earlier been found in PBMCs, lymphoblastoid cell lines, induced pluripotent stem cells, saliva samples, or postmortem brains of subjects with other comorbid psychiatric disorders sharing genetic overlap with ADHD, including schizophrenia, bipolar disorder, major depressive disorder, and autism spectrum disorder [12, 32, 33].
miRNAs exert repressive effects on gene expression via direct interaction with their target genes. The results of the present study suggest that aberrant miRNA expression in ADHD may underlie changes in the expression of genes related with D-myo-inositol (1,4,5)-trisphosphate metabolism. Notably, D-myo-inositol (1,4,5)-trisphosphate, which is found highly expressed in the brain, is a second messenger that mediates the biological response of a large number of hormones and neurotransmitters on target cells by regulating calcium release from intracellular stores [34]. D-myo-inositol (1,4,5)-trisphosphate shows altered expression levels in the brain in several psychiatric disorders, including ADHD, depression [35], or bipolar disorder [36]. In addition, differences in myo-inositol concentration were found in different brain areas of children and adolescents with ADHD when compared to controls [37–40], after 12 weeks of methylphenidate treatment in children with ADHD [41] or in untreated ADHD subjects when compared to ADHD subjects under pharmacological treatment or control individuals [42].
The three dysregulated miRNAs that best predict ADHD in our sample, miR-26b-5p, miR-185-5p, and miR-191-5p, are upstream regulators of target genes previously involved in ADHD and/or other psychiatric disorders, including ATAT1, SH3PXD2A, NTRK3, and BDNF [43–46]. While these genes did not show aberrant expression in our ADHD dataset, we found other genes with a strong background in ADHD and brain function among the subset of differentially expressed validated target genes of our three miRNAs of interest. Those included BCHE, involved in catecholamine and serotonin signaling [47], CXCR4 that regulates neuronal migration or axon wiring [48], HOMER1, a scaffolding protein localized at the post-synaptic density of glutamatergic excitatory synapses [49], NR4A2, an orphan nuclear receptor implicated in the development of dopaminergic cells [50], and YWHAE, which regulates monoamine biosynthesis and is crucial for neuronal development [51].
Although the disease-related changes in gene expression and the epigenetic mechanisms underlying them in peripheral tissues reveal potential biomarkers for ADHD, the results of the present study should be considered in the context of some limitations: First, our genome-wide miRNA and mRNA expression profiling was performed in PBMCs. Although miR-26b-5p, miR-185-5p, and miR-191-5p show similar expression patterns in blood and brain tissues [52], further evidence in the brain is required to confirm their role in the pathophysiology of ADHD. Given its capability to be non-invasive, peripheral expression profiles are proposed as a surrogate for expression profiling in the CNS. Previous work suggests that expression profiles may correlate between brain and peripheral blood, that both tissues cluster together based on their pattern of miRNA expression [13, 53] and that whole blood also shares significant gene expression similarities with multiple brain tissues [14], which strengthen the use of peripheral tissues as a proxy to study transcriptional, and epigenetic biosignatures in different psychiatric disorders. Second, we ranked differentially expressed miRNAs in ADHD subjects according to their predictive performance and focused in three of them for the downstream analyses. Given that an AUC threshold was set above 0.70, further studies with more stringent measures of their discriminatory accuracy are required. In addition, other candidate miRNAs in our genome-wide miRNA expression profiling, in particular miR-22-3p, miR-30b-5p, miR-106b-5p, miR-107, and miR-125b-5p, also showed aberrant expression levels in peripheral blood of children and adolescents with ADHD in previous studies and deserve further investigation [16]. Third, since we restricted the analysis of biological pathways and networks to experimentally validated target genes negatively correlated with the three highly predictive miRNAs for ADHD, we may have missed additional dysregulated genes contributing to the disorder. Similarly, the lack of extensive datasets of experimentally validated miRNA–mRNA interactions in multiple tissues, including PBMCs or brain, beyond those focused on oncology, may have caused some bias in the results. Future analyses are required to identify new potential targets and uncover the molecular dysregulation underlying ADHD. Fourth, although this is the first study integrating data from miRNA and gene expression profiling in ADHD performed so far, our modest sample size may have limited our power to estimate the magnitude of the observed effects of miRNAs and miRNA–gene pairs on ADHD. Fifth, recent studies reveal age-related changes in miRNAs or protein-coding gene expression and DNA methylation patterns [54, 55], which emphasize the need to explore further the influence of age on the role of miR-26b-5p, miR-185-5p, and miR-191-5p in ADHD and to identify age-related molecular pathways underlying the disorder. Lastly, expression patterns were assessed in a restricted clinical sample of medication-naive subjects with no comorbid disorders, which may facilitate the identification of novel miRNA biosignatures that might be neglected by a broader recruitment strategy. However, miRNAs showing aberrant expression in ADHD were previously related to other high comorbid psychiatric disorders that often coexist with ADHD. The pattern of psychiatric comorbidity in ADHD is highly heterogeneous and changes substantially across the lifespan, so we could not discard a possible pleiotropic effect of the identified miRNAs on other psychiatric disorders that may share common underlying etiological processes, which could explain part of the phenotypic variability as well as the symptomatic overlap between the different psychiatric conditions. Further studies are required to clarify if miR-26b-5p, miR-185-5p, and miR-191-5p have a specific role in ADHD or, alternatively, they play a pleiotropic effect on other psychiatric disorders.
In summary, we conducted a genome-wide miRNA expression analysis in ADHD subjects and explored the downstream effect of the miRNAs showing the highest predictive performance through an integrative approach that combined miRNA and gene expression profiling in PBMCs. Through this comprehensive and novel design, we showed evidence supporting new RNA-mediated mechanisms and aberrant miRNA-regulated pathways contributing to the disorder. We identified miR-26b-5p, miR-185-5p, and miR-191-5p as potential biomarkers for ADHD, we found evidence that suggests altered myo-inositol signaling in the disorder and highlighted promising candidate genes that deserve further investigation. Given that the relationship between miRNAs and mRNAs is complex and dynamic, future studies are required that provide deeper insight in the epigenetic mechanisms underlying ADHD and identify specific molecular networks that may be crucial in the etiopathogenesis of the disorder.
Funding and disclosure
Over the course of this investigation, CSM is a recipient of a Sara Borrell contract from the Instituto de Salud Carlos III, Ministerio de Economía, Industria y Competitividad, Spain (CD15/00199) and a mobility grant from the Instituto de Salud Carlos III, Ministerio de Economía, Industria y Competitividad, Spain (MV16/00039). MSA is a recipient of a contract from the Biomedical Research Networking Centre in Mental Health (CIBERSAM), Madrid, Spain. MP has been a recipient of a predoctoral fellowship from the Vall d’Hebron Research Institute (PRED-VHIR-2013) and a research grant from the Deutscher Akademischer Austauschdienst (DAAD), Germany (Research Grants - Short-Term Grants, 2017). PR is a recipient of a predoctoral fellowship from the Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR), Generalitat de Catalunya, Spain (2016FI_B 00899). MR is a recipient of a Miguel de Servet contract from the Instituto de Salud Carlos III, Ministerio de Economía, Industria y Competitividad, Spain (CP09/00119 and CPII15/00023). This investigation was supported by Instituto de Salud Carlos III (PI14/01700, PI15/01789, PI16/01505, and PI17/00289), Ministerio de Economía, Industria y Competitividad, Spain (SAF2016-80255-R and SAF2016-80255-R) and cofinanced by the European Regional Development Fund (ERDF), Agència de Gestió d’Ajuts Universitaris i de Recerca-AGAUR, Generalitat de Catalunya (2014SGR1357 and 2017SGR1461), the European College of Neuropsychopharmacology (ECNP network: “ADHD across the lifespan”), Departament de Salut, Generalitat de Catalunya, Spain, and a NARSAD Young Investigator Grant from the Brain & Behavior Research Foundation. This project also received funding from the European Union Seventh Framework Programme [FP7/2007-2013] under Grant agreement no. 602805 and from the European Union’s Horizon 2020 research and innovation programme [H2020/2014-2020] under Grant agreement nos. 728018, 667302, and 643051. This paper reflects only the authors’ views, and the European Union is not liable for any use that may be made of the information contained herein.
Dr. Sánchez-Mora, Dr. Soler Artigas, Dr. Garcia-Martínez, Mireia Pagerols, Paula Rovira, Montse Corrales, Christian Fadeuilhe, Natàlia Padilla, Dr. de la Cruz, Dr. Arias-Vásquez, and Dr. Ribasés reported no biomedical financial interests or potential conflicts of interest. Vanesa Richarte was on the speakers’ bureau and acted as a consultant for Eli-Lilly, Shire, and Rubió in the last 5 years. She also received travel awards for taking part in psychiatric meetings from Shire and Eli-Lilly. Dr. Franke has received educational speaking fees from Shire and Medice. Prof. Miguel Casas has received fees to give talks for Janssen–Cilag, Bristol–Myers Squibb, Ferrer–Brainfarma, Pfizer, Reckitt-Benckiser, Lundbeck, Otsuka, Servier, Lilly, Shire, GSK, Rovi, and Adamed. He has received financial compensation for his participation as a member of the Janssen–Cilag, Lilly, Shire, Lundbeck, Otsuka, Ferrer, and Rovi board. Dr. Ramos-Quiroga was on the speakers’ bureau and acted as a consultant for Eli-Lilly, Janssen–Cilag, Novartis, Shire, Lundbeck, Almirall, Braingaze, Sincrolab, and Rubió in the last 5 years. He also received travel awards for taking part in psychiatric meetings from Janssen–Cilag, Rubió, Shire, and Eli-Lilly. The Department of Psychiatry chaired by him received unrestricted educational and research support from the following companies in the last 5 years: Eli-Lilly, Lundbeck, Janssen–Cilag, Actelion, Shire, Ferrer, Oryzon, Roche, Psious, and Rubió.
Supplementary information
Acknowledgements
The authors are grateful to patients and controls who kindly participated in this research. The miRNA sequencing service was carried out at the Centre for Genomic Regulation (CRG), Barcelona, Spain. The microarrays service was carried out at High Technology Unit (UAT) at Vall d’Hebron Research Institute (VHIR), Barcelona (Spain), and the Statistics and Bioinformatics Unit (UEB) at the VHIR provided statistical support.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Cristina Sánchez-Mora, Phone: 0034934894162, Email: cristina.sanchez@vhir.org.
Marta Ribasés, Phone: 0034934894162, Email: marta.ribases@vhir.org.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41386-018-0297-0).
References
- 1.Polanczyk GV, Willcutt EG, Salum GA, Kieling C, Rohde LA. ADHD prevalence estimates across three decades: an updated systematic review and meta-regression analysis. Int J Epidemiol. 2014;43:434–42. doi: 10.1093/ije/dyt261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simon V, Czobor P, Balint S, Meszaros A, Bitter I. Prevalence and correlates of adult attention-deficit hyperactivity disorder: meta-analysis. Br J Psychiatry. 2009;194:204–11. doi: 10.1192/bjp.bp.107.048827. [DOI] [PubMed] [Google Scholar]
- 3.Faraone SV, Larsson H. Genetics of attention deficit hyperactivity disorder. Mol Psychiatry. 2018 Jun 11. 10.1038/s41380-018-0070-0. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 4.Bonvicini C, Faraone SV, Scassellati C. Attention-deficit hyperactivity disorder in adults: a systematic review and meta-analysis of genetic, pharmacogenetic and biochemical studies. Mol Psychiatry. 2016;21:872–84. doi: 10.1038/mp.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alural B, Genc S, Haggarty SJ. Diagnostic and therapeutic potential of microRNAs in neuropsychiatric disorders: past, present, and future. Prog Neuropsychopharmacol Biol Psychiatry. 2017;73:87–103. doi: 10.1016/j.pnpbp.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nowak JS, Michlewski G. miRNAs in development and pathogenesis of the nervous system. Biochem Soc Trans. 2013;41:815–20. doi: 10.1042/BST20130044. [DOI] [PubMed] [Google Scholar]
- 7.Cipolla GA. A non-canonical landscape of the microRNA system. Front Genet. 2014;5:337. doi: 10.3389/fgene.2014.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 9.Anitha A, Thanseem I. microRNA and autism. Adv Exp Med Biol. 2015;888:71–83. doi: 10.1007/978-3-319-22671-2_5. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Martinez I, Sanchez-Mora C, Pagerols M, Richarte V, Corrales M, Fadeuilhe C, et al. Preliminary evidence for association of genetic variants in pri-miR-34b/c and abnormal miR-34c expression with attention deficit and hyperactivity disorder. Transl Psychiatry. 2016;6:e879. doi: 10.1038/tp.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma J, Shang S, Wang J, Zhang T, Nie F, Song X, et al. Identification of miR-22-3p, miR-92a-3p, and miR-137 in peripheral blood as biomarker for schizophrenia. Psychiatry Res. 2018;265:70–6. doi: 10.1016/j.psychres.2018.03.080. [DOI] [PubMed] [Google Scholar]
- 12.Maffioletti E, Tardito D, Gennarelli M, Bocchio-Chiavetto L. Micro spies from the brain to the periphery: new clues from studies on microRNAs in neuropsychiatric disorders. Front Cell Neurosci. 2015;8:75. doi: 10.3389/fncel.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi T, Wu Y, Zeng J, Zhang F, Xue A, Jiang L, et al. Identifying gene targets for brain-related traits using transcriptomic and methylomic data from blood. Nat Commun. 2018;9:2282. doi: 10.1038/s41467-018-04558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chatterjee P, Roy D. Comparative analysis of RNA-Seq data from brain and blood samples of Parkinson’s disease. Biochem Biophys Res Commun. 2017;484:557–64. doi: 10.1016/j.bbrc.2017.01.121. [DOI] [PubMed] [Google Scholar]
- 15.Srivastav S, Walitza S, Grunblatt E. Emerging role of miRNA in attention deficit hyperactivity disorder: a systematic review. Atten Defic Hyperact Disord. 2018;10:49–63. doi: 10.1007/s12402-017-0232-y. [DOI] [PubMed] [Google Scholar]
- 16.Kandemir H, Erdal ME, Selek S, Ay OI, Karababa IF, Kandemir SB, et al. Evaluation of several micro RNA (miRNA) levels in children and adolescents with attention deficit hyperactivity disorder. Neurosci Lett. 2014;580:158–62. doi: 10.1016/j.neulet.2014.07.060. [DOI] [PubMed] [Google Scholar]
- 17.Wu LH, Peng M, Yu M, Zhao QL, Li C, Jin YT, et al. Circulating microRNA let-7d in attention-deficit/hyperactivity disorder. Neuromolecular Med. 2015;17:137–46. doi: 10.1007/s12017-015-8345-y. [DOI] [PubMed] [Google Scholar]
- 18.Wu L, Zhao Q, Zhu X, Peng M, Jia C, Wu W, et al. A novel function of microRNA let-7d in regulation of galectin-3 expression in attention deficit hyperactivity disorder rat brain. Brain Pathol. 2010;20:1042–54. doi: 10.1111/j.1750-3639.2010.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karakas U, Ay OI, Ay ME, Wang W, Sungur MA, Cevik K, et al. Regulating the regulators in attention-deficit/hyperactivity disorder: a genetic association study of microRNA biogenesis pathways. Omics. 2017;21:352–8. doi: 10.1089/omi.2017.0048. [DOI] [PubMed] [Google Scholar]
- 20.Nemeth N, Kovacs-Nagy R, Szekely A, Sasvari-Szekely M, Ronai Z. Association of impulsivity and polymorphic microRNA-641 target sites in the SNAP-25 gene. PLoS One. 2013;8:e84207. doi: 10.1371/journal.pone.0084207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez-Mora C, Ramos-Quiroga JA, Garcia-Martinez I, Fernandez-Castillo N, Bosch R, Richarte V, et al. Evaluation of single nucleotide polymorphisms in the miR-183-96-182 cluster in adulthood attention-deficit and hyperactivity disorder (ADHD) and substance use disorders (SUDs) Eur Neuropsychopharmacol. 2013;23:1463–73. doi: 10.1016/j.euroneuro.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 26.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu M, Shi B, Wang J, Cao Q, Cui Q. TAM: a method for enrichment and depletion analysis of a microRNA category in a list of microRNAs. BMC Bioinformatics. 2010;11:419. [DOI] [PMC free article] [PubMed]
- 28.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–7. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei corrplot: visualization of a correlation matrix. R package version 0.73.T. 2013. Available at: http://CRAN.R-project.org/package=corrplot.
- 31.Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, et al. DIANA-TarBasev8: a decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018;46:D239–45. doi: 10.1093/nar/gkx1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao D, Lin M, Chen J, Pedrosa E, Hrabovsky A, Fourcade HM, et al. MicroRNA profiling of neurons generated using induced pluripotent stem cells derived from patients with schizophrenia and schizoaffective disorder, and 22q11.2 Del. PLoS One. 2015;10:e0132387. doi: 10.1371/journal.pone.0132387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hicks SD, Middleton FA. A comparative review of microRNA expression patterns in autism spectrum disorder. Front Psychiatry. 2016;7:176. doi: 10.3389/fpsyt.2016.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pattni K, Banting G. Ins(1,4,5)P3 metabolism and the family of IP3-3Kinases. Cell Signal. 2004;16:643–54. doi: 10.1016/j.cellsig.2003.10.009. [DOI] [PubMed] [Google Scholar]
- 35.Chiappelli J, Rowland LM, Wijtenburg SA, Muellerklein F, Tagamets M, McMahon RP, et al. Evaluation of myo-inositol as a potential biomarker for depression in schizophrenia. Neuropsychopharmacology. 2015;40:2157–64. doi: 10.1038/npp.2015.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu W, Greenberg ML. Inositol depletion, GSK3 inhibition and bipolar disorder. Future Neurol. 2016;11:135–48. doi: 10.2217/fnl-2016-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arcos-Burgos M, Londono AC, Pineda DA, Lopera F, Palacio JD, Arbelaez A, et al. Analysis of brain metabolism by proton magnetic resonance spectroscopy (1H-MRS) in attention-deficit/hyperactivity disorder suggests a generalized differential ontogenic pattern from controls. Atten Defic Hyperact Disord. 2012;4:205–12. doi: 10.1007/s12402-012-0088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferreira PE, Palmini A, Bau CH, Grevet EH, Hoefel JR, Rohde LA, et al. Differentiating attention-deficit/hyperactivity disorder inattentive and combined types: a (1)H-magnetic resonance spectroscopy study of fronto-striato-thalamic regions. J Neural Transm. 2009;116:623–9. doi: 10.1007/s00702-009-0191-3. [DOI] [PubMed] [Google Scholar]
- 39.Soliva JC, Moreno A, Fauquet J, Bielsa A, Carmona S, Gispert JD, et al. Cerebellar neurometabolite abnormalities in pediatric attention/deficit hyperactivity disorder: a proton MR spectroscopic study. Neurosci Lett. 2010;470:60–4. doi: 10.1016/j.neulet.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 40.Tafazoli S, O’Neill J, Bejjani A, Ly R, Salamon N, McCracken JT, et al. 1H MRSI of middle frontal gyrus in pediatric ADHD. J Psychiatr Res. 2013;47:505–12. doi: 10.1016/j.jpsychires.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiguna T, Guerrero AP, Wibisono S, Sastroasmoro S. Effect of 12-week administration of 20-mg long-acting methylphenidate on Glu/Cr, NAA/Cr, Cho/Cr, and mI/Cr ratios in the prefrontal cortices of school-age children in Indonesia: a study using 1H magnetic resonance spectroscopy (MRS) Clin Neuropharmacol. 2012;35:81–5. doi: 10.1097/WNF.0b013e3182452572. [DOI] [PubMed] [Google Scholar]
- 42.Hammerness P, Biederman J, Petty C, Henin A, Moore CM. Brain biochemical effects of methylphenidate treatment using proton magnetic spectroscopy in youth with attention-deficit hyperactivity disorder: a controlled pilot study. CNS Neurosci Ther. 2012;18:34–40. doi: 10.1111/j.1755-5949.2010.00226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caputo V, Sinibaldi L, Fiorentino A, Parisi C, Catalanotto C, Pasini A, et al. Brain derived neurotrophic factor (BDNF) expression is regulated by microRNAs miR-26a and miR-26b allele-specific binding. PLoS One. 2011;6:e28656. doi: 10.1371/journal.pone.0028656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conner AC, Kissling C, Hodges E, Hunnerkopf R, Clement RM, Dudley E, et al. Neurotrophic factor-related gene polymorphisms and adult attention deficit hyperactivity disorder (ADHD) score in a high-risk male population. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1476–80. doi: 10.1002/ajmg.b.30632. [DOI] [PubMed] [Google Scholar]
- 45.Forstner AJ, Basmanav FB, Mattheisen M, Bohmer AC, Hollegaard MV, Janson E, et al. Investigation of the involvement of MIR185 and its target genes in the development of schizophrenia. J Psychiatry Neurosci. 2014;39:386–96. doi: 10.1503/jpn.130189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Smith TF, Anastopoulos AD, Garrett ME, Arias-Vasquez A, Franke B, Oades RD, et al. Angiogenic, neurotrophic, and inflammatory system SNPs moderate the association between birth weight and ADHD symptom severity. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:691–704. doi: 10.1002/ajmg.b.32275. [DOI] [PubMed] [Google Scholar]
- 47.Jacob CP, Weber H, Retz W, Kittel-Schneider S, Heupel J, Renner T, et al. Acetylcholine-metabolizing butyrylcholinesterase (BCHE) copy number and single nucleotide polymorphisms and their role in attention-deficit/hyperactivity syndrome. J Psychiatr Res. 2013;47:1902–8. doi: 10.1016/j.jpsychires.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 48.Kweon K, Shin ES, Park KJ, Lee JK, Joo Y, Kim HW. Genome-wide analysis reveals four novel loci for attention-deficit hyperactivity disorder in Korean youths. J Korean Acad Child Adolesc Psychiatry. 2008;29:62–72. doi: 10.5765/jkacap.2018.29.2.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang L, Hong Q, Zhang M, Liu X, Pan XQ, Guo M, et al. The role of Homer 1a in increasing locomotor activity and non-selective attention, and impairing learning and memory abilities. Brain Res. 2013;1515:39–47. doi: 10.1016/j.brainres.2013.03.030. [DOI] [PubMed] [Google Scholar]
- 50.Smith KM, Bauer L, Fischer M, Barkley R, Navia BA. Identification and characterization of human NR4A2 polymorphisms in attention deficit hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2005;133B:57–63. doi: 10.1002/ajmg.b.30127. [DOI] [PubMed] [Google Scholar]
- 51.Jacobsen KK, Kleppe R, Johansson S, Zayats T, Haavik J. Epistatic and gene wide effects in YWHA and aromatic amino hydroxylase genes across ADHD and other common neuropsychiatric disorders: association with YWHAE. Am J Med Genet B Neuropsychiatr Genet. 2015;168:423–32. doi: 10.1002/ajmg.b.32339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Panwar B, Omenn GS, Guan Y. miRmine: a database of human miRNA expression profiles. Bioinformatics. 2017;33:1554–60. doi: 10.1093/bioinformatics/btx019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tylee DS, Kawaguchi DM, Glatt SJ. On the outside, looking in: a review and evaluation of the comparability of blood and brain “-omes”. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:595–603. doi: 10.1002/ajmg.b.32150. [DOI] [PubMed] [Google Scholar]
- 54.Huan T, Chen G, Liu C, Bhattacharya A, Rong J, Chen BH, et al. Age-associated microRNA expression in human peripheral blood is associated with all-cause mortality and age-related traits. Aging Cell. 2018;17. [DOI] [PMC free article] [PubMed]
- 55.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–67. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.