Abstract
Protein acetylation and deacetylation events are finely regulated by lysine-acetyl-transferases and lysine-deacetylases and constitute an important tool for the activation or inhibition of specific cellular pathways. One of the most important lysine-acetyl-transferases is p300, which is involved in the regulation of gene expression, cell growth, DNA repair, differentiation, apoptosis, and tumorigenesis. A well-known target of p300 is constituted by the tumor suppressor protein p53, which plays a critical role in the maintenance of genomic stability and whose activity is known to be controlled by post-translational modifications, among which acetylation. p300 activity toward p53 is negatively regulated by the NAD-dependent deacetylase SIRT1, which deacetylates p53 preventing its transcriptional activation and the induction of p53-dependent apoptosis. However, the mechanisms responsible for p53 regulation by p300 and SIRT1 are still poorly understood. Here we identify the nucleosome assembly protein TSPY-Like 2 (TSPYL2, also known as TSPX, DENTT, and CDA1) as a novel regulator of SIRT1 and p300 function. We demonstrate that, upon DNA damage, TSPYL2 inhibits SIRT1, disrupting its association with target proteins, and promotes p300 acetylation and activation, finally stimulating p53 acetylation and p53-dependent cell death. Indeed, in response to DNA damage, cells silenced for TSPYL2 were found to be defective in p53 activation and apoptosis induction and these events were shown to be dependent on SIRT1 and p300 function. Collectively, our results shed new light on the regulation of p53 acetylation and activation and reveal a novel TSPYL2 function with important implications in cancerogenesis.
Subject terms: Molecular biology, Tumour-suppressor proteins, Tumour-suppressor proteins
Introduction
The tumor suppressor protein p53 plays a critical role in the maintenance of genomic stability and its loss or mutation is responsible for tumor susceptibility [1]. One of the major biological function of p53 is to regulate cell fate upon DNA lesions; indeed once activated, p53 promotes the transcription of specific genes, thus leading to the appropriate cellular outcomes, like cell cycle arrest, senescence, or apoptosis [2].
In unstressed cells, p53 levels are kept very low by the E3 ubiquitin ligase MDM2 [3]. Upon DNA lesions, the DNA damage response (DDR) kinases ATM, ATR, Chk1, and Chk2 phosphorylate p53 [4], disrupting p53-Mdm2 interaction and promoting p53 accumulation [2]. Phosphorylated p53 is then fully activated by acetylation and the most important lysine-acetyl-transferase involved is p300/CBP, which targets p53 on several residues, such as lysine-382 (K382). Acetylation of this residue competes with ubiquitination and promotes p53 stabilization and activation [3].
p53-K382 acetylation is negatively regulated by the class III NAD-dependent deacetylase SIRT1 [5], which specifically targets this residue. In unstressed cells, SIRT1 prevents apoptosis by deacetylating and inactivating p53 [5]; upon DNA damage, CCAR2 (also named DBC1 [6]), the major SIRT1 inhibitor [7, 8], represses SIRT1 function and favors p53-dependent apoptosis [9, 10].
Testis-specific protein Y-encoded-like 2 (TSPYL2, also known as DENTT, CDA1, and TSPX) is an X-linked gene [11] encoding for a nuclear protein of the TSPY-L nucleosome assembly protein-1 superfamily, characterized by a predicted nucleosome assembly protein (NAP) domain for nucleosome remodeling and gene expression regulation [12].
Recent findings suggest for TSPYL2 a tumor suppressor role and important functions in cell growth and DNA damage response regulation.
In non-small cell lung cancer cell lines, TGFβ upregulates TSPYL2 expression [13] and TSPYL2 itself enhances TGFβ signaling by interacting with the REST multisubunit transcriptional repressor complex [14]. Moreover, TSPYL2 was found mutated in endometrial carcinoma [15] and downregulated in glioma [16, 17]. In human and mouse lung primary tumors and in hepatocellular carcinoma TSPYL2 expression is significantly reduced, whereas its overexpression in lung and breast cancer cells reduces cell growth and the ability to migrate [18].
Upon camptothecin treatment, TSPYL2 mRNA and protein are strongly upregulated, whereas p53 accumulation is reduced in TSPYL2-depleted cells [19]. Moreover, in mice, TSPYL2 was implicated in G1 checkpoint maintenance upon DNA damage [20].
Here, we demonstrate, for the first time, that CCAR2 depletion induces TSPYL2 mRNA and protein accumulation and that, in human DNA-damaged cells, TSPYL2 inhibits SIRT1 deacetylase and promotes p300 acetyl-transferase activity. These events finally result in the proper acetylation/activation of p53 and induction of p53-dependent apoptosis.
Results
CCAR2 depletion promotes TSPYL2 mRNA and protein accumulation
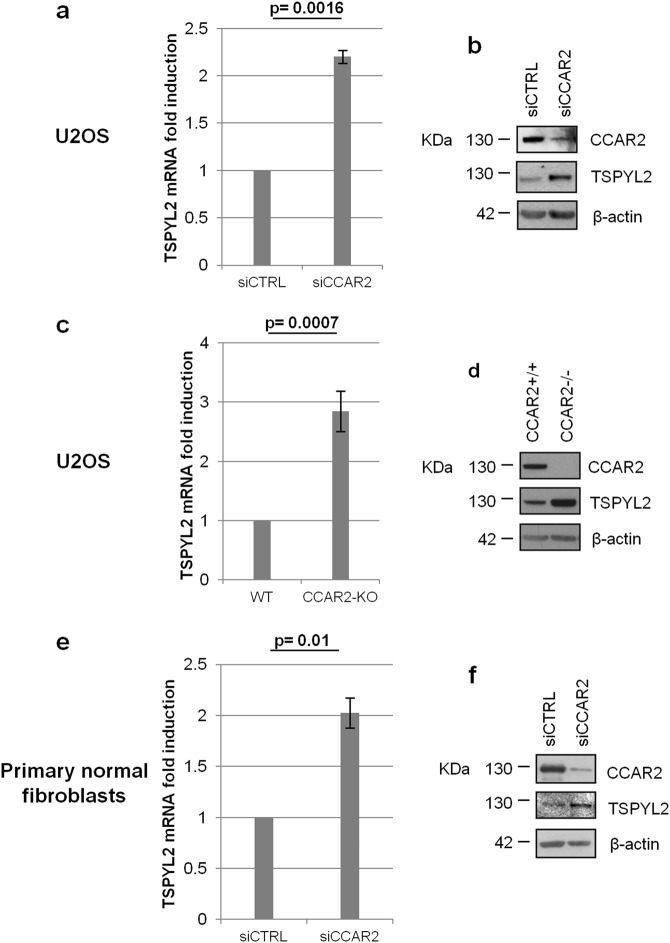
We previously performed gene expression profile analyses of human osteosarcoma U2OS cells depleted of CCAR2 for 6 days and, compared with controls, we found both upregulated and downregulated genes [21]. We focussed on TSPYL2 because its expression was significantly induced by CCAR2 depletion with a fold change of 1.78 [21] and the protein encoded by this gene was previously implicated in the DDR [19, 20]. Quantitative real-time PCR (RT-qPCR) in U2OS cells confirmed the induction of TSPYL2 expression upon 6 days of CCAR2 silencing (Fig. 1a) and western blot analyses indicated a substantial upregulation of TSPYL2 protein in CCAR2-depleted cells extracts (Fig. 1b). We also analyzed TSPYL2 mRNA and protein in U2OS-CCAR2-KO cells [22] and in primary normal fibroblasts upon 6 days of CCAR2 siRNAs transfection. RT-qPCR analyses indicated a 2.8- and a 2.0-fold induction of TSPYL2 expression, respectively, in U2OS and fibroblasts negative for CCAR2 and compared with controls (Fig. 1c, e). We then checked TSPYL2 protein levels, in the same cell lines, and found a strong increase of TSPYL2 protein in both U2OS-CCAR2-KO cells and in CCAR2-depleted fibroblasts (Fig. 1d, f), confirming that prolonged CCAR2 loss promotes TSPYL2 gene expression and protein accumulation in both cancer and normal cell lines.
Fig. 1.
CCAR2 depletion upregulates TSPYL2 mRNA and protein levels. a total RNA was extracted from U2OS cells 6 days upon control (CTRL) or CCAR2 siRNAs transfection, reverse transcribed and the levels of TSPYL2 mRNA were determined by RT-qPCR. Fold changes relative to siCTRLcells were reported in the chart. b Western blot analysis of TSPYL2 protein levels in 6 days CTRL and CCAR2-silenced U2OS cells. c Total RNA was extracted from CCAR2-WT and –KO cells, reverse transcribed and the levels of TSPYL2 mRNA were analyzed by RT-qPCR. In the chart, fold changes relative CCAR2-WT were reported. d Western blot analyses of TSPYL2 protein levels in CCAR2-WT and -KO cells. Primary fibroblasts were transfected with control or CCAR2 siRNAs and analyzed for TSPYL2 mRNA e and protein f levels, respectively, as reported in a and b
TSPYL2 regulates p53 acetylation in response to DNA lesions
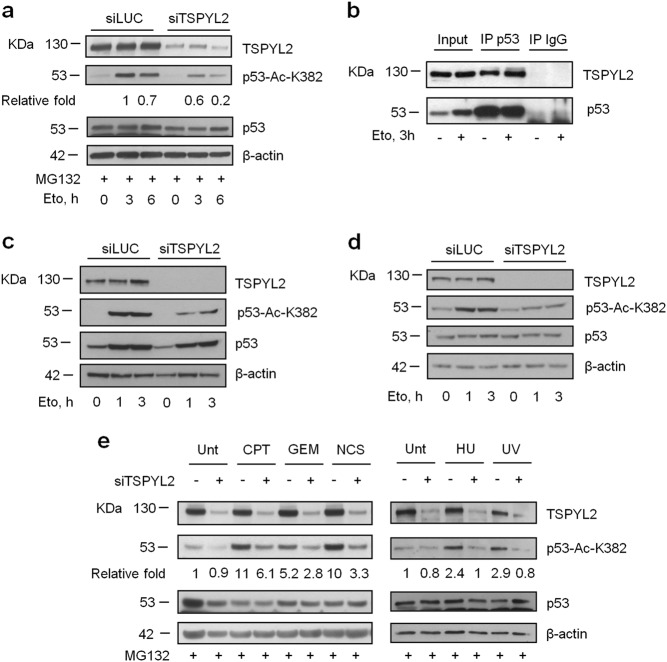
Upon DNA damage CCAR2 inhibits SIRT1 and promotes p53-K382 acetylation[7–10]; as TSPYL2 was previously linked to p53 regulation [19], we thought that it could be induced to ensure the proper SIRT1 and p53 modulation in the absence of CCAR2. To test this hypothesis, U2OS cells were transfected with a pool of two different TSPYL2 siRNAs and, 48 h later, p53 levels were stabilized by treatment with the proteasome inhibitor MG132. Acetylation of p53-K382 was then monitored at different times after exposure to etoposide, a topoisomerase II inhibitor that finally produces DNA double-strand breaks [23]. p53 acetylation was significantly reduced in TSPYL2-depleted cells at both 3 and 6 h after treatment (Fig. 2a) and these results were confirmed using single TSPYL2 siRNA sequences (Supplementary Fig. 1a). In support of these data, we found that TSPYL2 co-immunoprecipitates with p53 both before and after DNA damage (Fig. 2b). Overall, these findings suggest that, similarly to CCAR2, TSPYL2 is involved in the regulation of etoposide-induced p53 acetylation.
Fig. 2.
TSPYL2 regulates p53 acetylation in response to DNA damage. a U2OS cells were transfected with control (siLUC) or TSPYL2 siRNAs. 48 h later, cells were treated with MG132 for 20 min and then with etoposide (20 µM), for the indicated time points. p53 acetylation was assessed by western blot with a specific anti-p53-acetyl-K-382 antibody. The fold induction of acetylated p53 relative to total p53 is indicated. b western blot analysis of p53 and TSPYL2 co-immunoprecipitation in U2OS cells before and after etoposide treatment. Analyses of p53 acetylation in TSPYL2 silenced and etoposide-treated A549 c and MDA-MB-231 d cells. e Western blot analysis of p53-K382 acetylation in control and TSPYL2-silenced U2OS treated with different types of genotoxic agents for 3 h. CPT, camptothecin; GEM, gemcitabine; NCS, neocarzinostatin; HU, hydroxyurea; UV, ultraviolet radiation
To further investigate this phenomenon, we silenced TSPYL2 in the lung adenocarcinoma A549 (wild type for p53, Fig. 2c) and in the breast cancer cell line MDA-MB-231 (p53 mutated, Fig. 2d). In both cases we found that TSPYL2 depletion reduces the levels of acetylated p53 upon etoposide treatment also in the absence of proteasome inhibition, clearly demonstrating that this TSPYL2 function is not specific for U2OS cells nor for wild type p53 protein and that it is not an MG132 artifact. Then we treated control and TSPYL2-silenced U2OS cells with camptothecin (a topoisomerase-I inhibitor), gemcitabine (a DNA synthesis inhibitor [24]), neocarzinostatin (a radiomimetic compound), hydroxyurea (a DNA synthesis inhibitor), and UV radiation (which induces pyrimidine dimers formation), and we demonstrated that TSPYL2 is required for p53-K382 acetylation upon all the tested genotoxic agents (Fig. 2e). These results suggest that TSPYL2, modulating p53-K382 acetylation, participates to the general response to DNA damage.
Noteworthy, we also found that concurrent depletion of CCAR2 and TSPYL2 reduces the etoposide-induced p53 acetylation more than single silencing (Supplementary Fig. 1b), suggesting that these proteins cooperate in the regulation of p53 acetylation. Of note, in this experiment no induction of TSPYL2 protein is detectable in CCAR2-silenced cells because harvesting was performed 48 h after transfection and only prolonged CCAR2 depletion induces TSPYL2 accumulation (data not shown and Supplementary Fig. 1b and Fig. 1a, b).
TSPYL2 inhibits SIRT1 function and promotes p300 activity
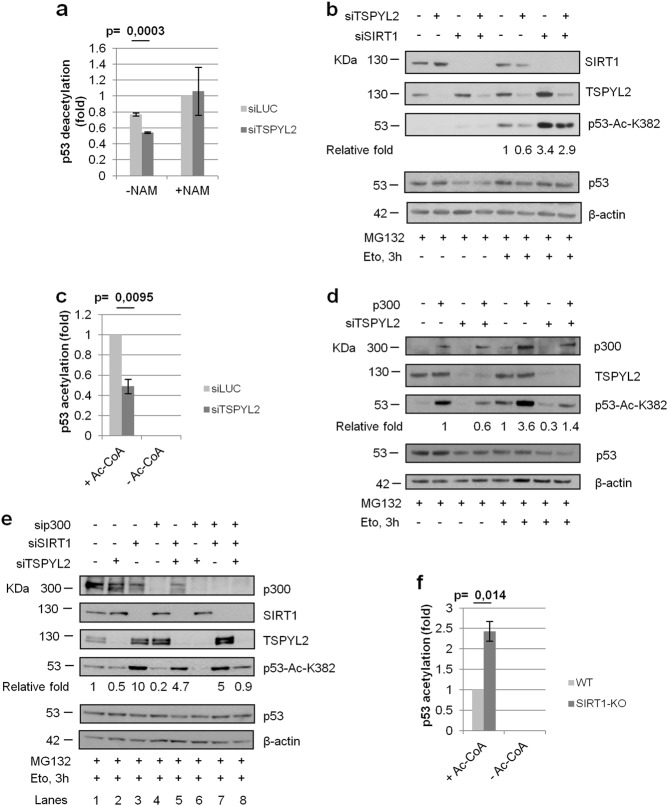
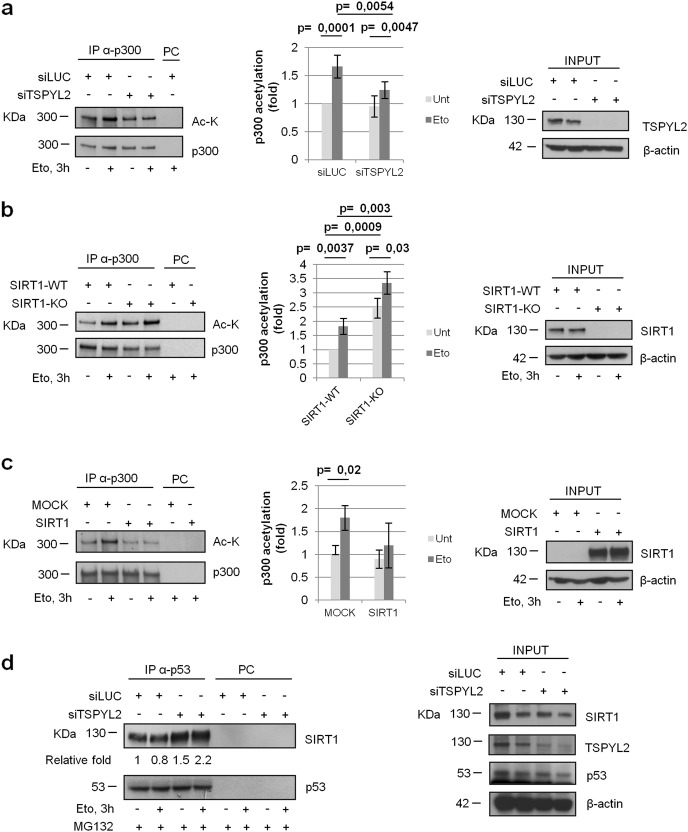
We then investigated if TSPYL2, alike CCAR2, regulates p53 acetylation by inhibiting SIRT1. To this aim, we performed in vitro deacetylation assays, analyzing the activity of SIRT1 molecules present in control and TSPYL2-depleted U2OS cells, as reported [25], on acetylated p53 protein immunoprecipitated from etoposide treated cells [7, 8]. We found that SIRT1 present in TSPYL2-silenced cells deacetylated p53 more efficiently than that present in controls and, therefore, SIRT1 is more active in the absence of TSPYL2 (Fig. 3a and Supplementary Fig. 2a).
Fig. 3.
TSPYL2 regulates p53 acetylation by inhibiting SIRT1 and promoting p300 activity. a p53 was immunoprecipitated from etoposide treated U2OS cells and incubated in vitro with cell extracts obtained from control or TSPYL2-depleted cells, also silenced for p53, in presence or absence of the SIRT1 inhibitor nicotinamide (NAM [25]). p53 acetylation/total p53 ratio was analyzed by western blot and normalized to the value of NAM-treated siLUC-transfected cells. Results of three independent experiments were reported in the chart. Error bars represent s.d. b U2OS cells were transfected with control, TSPYL2 and SIRT1 siRNAs in different combinations. After 48 h transfection, cells were treated with MG132 followed by etoposide (20 µM, 3 h) and p53 acetylation was analyzed by western blot. c p300 was immunoprecipitated from control or TSPYL2-depleted cells and its activity tested against recombinant GST-p53 in presence or absence of acetyl-coA. p53 acetylation/total p53 ratio was analyzed by western blot and normalized to the value of siLUC-transfected cells supplemented with acetyl-coA. Results of three independent experiments were reported in the chart. Error bars represent s.d. d U2OS cells were transfected with control or TSPYL2 siRNAs and MOCK or p300 expression vectors in different combinations. After 48 h, cells were treated with MG132 followed by etoposide and p53 acetylation was analyzed by western blot. e U2OS cells were transfected with control, TSPYL2, SIRT1, and p300 siRNAs in different combinations. After 48 h of transfection, cells were treated with MG132 followed by etoposide (20 µM, 3 h) and p53 acetylation was analyzed by western blot. The fold induction of acetylated p53 relative to total p53 is indicated in each western blot a–c. f p300 was immunoprecipitated from SIRT1-WT or SIRT1-KO cells and its activity tested against recombinant GST-p53 in presence or absence of acetyl-coA. p53 acetylation/total p53 ratio was analyzed by western blot and normalized to the value of SIRT1-WT cells. Results of three independent experiments were reported in the chart. Error bars represent s.d.
To confirm these results, we analyzed p53-K382 acetylation in total cell extracts of cells silenced for TSPYL2 and SIRT1, in different combinations, and exposed to etoposide for 3 h upon MG132 addition. We found that TSPYL2 depletion strongly decreased p53 acetylation, while SIRT1 loss induced it (Fig. 3b). Interestingly, we observed that, compared with single silencing, lack of both TSPYL2 and SIRT1 produces an intermediate level of p53 acetylation (Fig. 3b) and this result was also confirmed combining TSPYL2 depletion with exposure to nicotinamide (NAM), a chemical inhibitor of SIRT1 [26] (Supplementary Fig. 2b). Therefore, SIRT1 inhibition is not the only mechanism through which TSPYL2 modulates p53 acetylation.
As p300 is the acetyl-transferase responsible for the modification of p53-K382[27–29] and its activity is regulated by NAP domain containing proteins [30], we investigated whether TSPYL2 somehow affects its function. We performed in vitro acetylation assays testing the activity of p300 protein immunoprecipitated from control and TSPYL2-silenced cells on recombinant GST-p53, which is not acetylated. Notably, immunoprecipitates were washed with high salt buffers to exclude that co-immunoprecipitated proteins may affect the result. We found that p300 acetylates p53 only in presence of acetyl-coenzyme (acetyl-CoA), but its activity resulted impaired when immunoprecipitated from cells lacking TSPYL2. These results indicate that TSPYL2 loss reduces p300 activity in vitro (Fig. 3c and Supplementary Fig. 2c).
To confirm these data, we overexpressed p300 in control and TSPYL2-depleted U2OS cells and we demonstrated that ectopic p300 rescues the TSPYL2-dependent defect in p53 acetylation and that loss of TSPYL2 strongly reduces p300 function (Fig. 3d). These data collectively confirm that TSPYL2 is required for p300 activity toward p53.
To corroborate these results, we combined TSPYL2, SIRT1, and p300 silencing (Fig. 3e) and we observed that upon etoposide, both TSPYL2 and p300 depletion reduces p53 acetylation (compare lane 1 with lanes 2 and 4), whereas SIRT1 loss promotes it (lane 3). Moreover, we found that simultaneous silencing of TSPYL2 and p300 completely abrogates p53 acetylation (lane 6), possibly because of SIRT1 hyper-activation, and that contemporary depletion of SIRT1, p300, and TSPYL2 restores the basal level of p53 acetylation (compare lane 1, 6, and 8). These data confirm that, upon DNA damage, TSPYL2 regulates p53 acetylation by inhibiting SIRT1 and promoting p300 acetyl-transferase activity.
Curiously, we observed that concurrent depletion of SIRT1 and TSPYL2 or SIRT1 and p300 induces similar acetylation of p53, which is reduced if compared with SIRT1 silencing alone (Fig. 3e, compare lanes 5 and 7 with lane 3), therefore suggesting that p300 activity toward p53 may be inhibited by SIRT1.
To verify this hypothesis, we performed in vitro acetylation assays using p300 protein immunoprecipitated from control or U2OS-SIRT1-KO cells and washed with high salt buffer to prevent, when present, SIRT1 co-immunoprecipitation. We noticed that the p300 obtained from SIRT1-KO cells is more efficient in acetylating recombinant p53 than that present in WT cells (Fig. 3f and Supplementary Fig. 2d). These findings were also verified with U2OS cells depleted of SIRT1 by siRNA transfection (Supplementary Fig. 2e) and confirm that, after DNA damage, SIRT1 inhibits p300 activity.
TSPYL2 modulates SIRT1 association with target proteins and promotes p300 acetylation
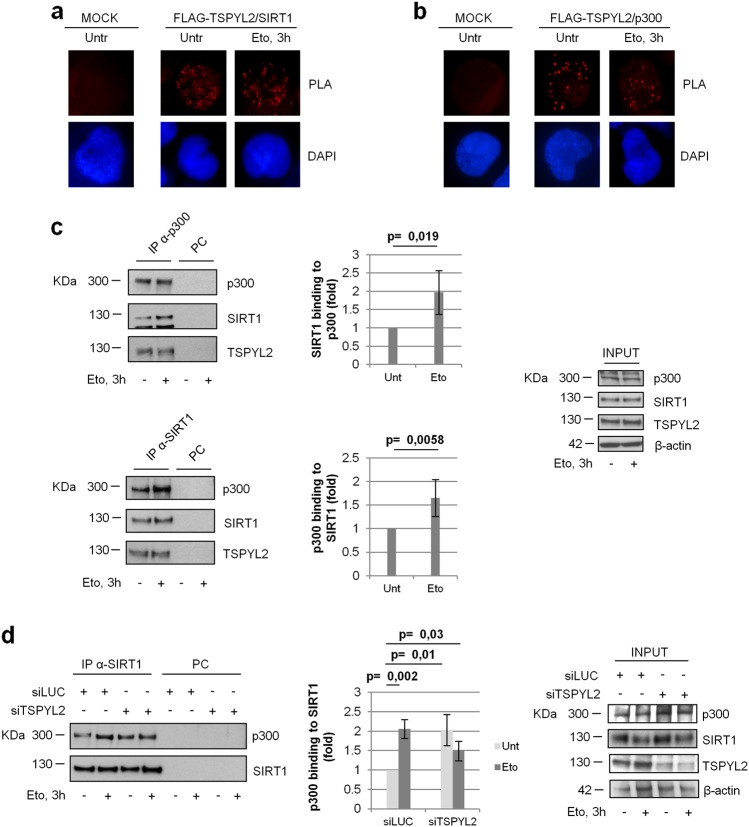
We then analyzed if TSPYL2, p300, and SIRT1 can associate in human cells. Proximity ligation assays [31] revealed that ectopic FLAG-TSPYL2 interacts with SIRT1 and p300 in the nucleus of both untreated and etoposide-treated U2OS cells (Fig. 4a, b). Coherently, co-immunoprecipitation experiments on endogenous proteins indicated that p300 and SIRT1 associate with TSPYL2 in a DNA damage-independent manner (Fig. 4c). On the contrary, SIRT1-p300 binding was induced by etoposide treatment (Fig. 4c, top and bottom panel) and the specificity of these associations was confirmed by co-immunoprecipitations performed in cells silenced for the immunoprecipitated proteins (Supplementary Fig. 3a, b). However, when we analyzed SIRT1-p300 interaction in TSPYL2-silenced cells, we found that this binding, compared with controls, is induced in both untreated and etoposide treated cells (Fig. 4d). These results clearly indicate that TSPYL2 prevents SIRT1-p300 association both before and after DNA damage.
Fig. 4.
TSPYL2 regulates p300-SIRT1 association. a, b U2OS cells were transfected with MOCK or FLAG-TSPYL2 encoding vectors and treated or not with etoposide. TSPYL2 association with SIRT1 a and p300 b was assessed by proximity ligation assay. c western blot analyses of p300 (top panel) and SIRT1 (bottom panel) immunocomplexes from U2OS cells treated or not with etoposide. IP, immunoprecipitates; PC, negative control; Input, total lysate. In the charts, densitometric quantification of SIRT1 co-immunoprecipitated with p300 (top) and p300 co-immunoprecipitated with SIRT1 (bottom). Data from etoposide treated samples were normalized to those from untreated cells. Values are mean ± standard deviation (s.d.) of three independent experiments. d SIRT1 was immunoprecipitated from control or TSPYL2-depleted cells before and after etoposide treatment. p300 presence in the immunocomplexes was evaluated by western blot. IP, immunoprecipitates; PC, negative control; Input, total lysate. The fold induction of co-immunoprecipitated p300 relative to immunoprecipitated SIRT1 is reported in the chart. Data were normalized to that from untreated siLUC cells and values are mean ± s.d. from three independent experiments
Acetylation enhances the catalytic activity of p300 [32, 33]. Therefore, we checked p300 acetylation in U2OS cells and we found that it is increased upon DNA damage (Fig. 5a, siLUC samples). Importantly, this induction cannot be observed in TSPYL2-silenced cells (Fig. 5a), where, of note, p300-SIRT1 association is augmented (Fig. 4d). These findings indicate that, upon DNA damage, TSPYL2, restricting SIRT1-p300 binding, allows the acetylation and activation of p300.
Fig. 5.
In response to DNA damage, SIRT1 inhibits p300 activity by deacetylation. a U2OS cells were transfected with siLUC or siTSPYL2 siRNAs and 48 h later p300 was immunoprecipitated before and after etoposide treatment. p300 acetylation was determined by western blot with a general anti-acetyl-lysine antibody. The mean fold induction ± s.d. of acetylated p300 relative to total p300 protein from three independent experiments is shown in the chart. IP, immunoprecipitates; PC, negative control. Input, total lysates. b p300 was immunoprecipitated from untreated and etoposide treated U2OS-SIRT1-WT or U2OS-SIRT1-KO cells. Levels of p300 acetylation were analyzed by western blot with a general anti-acetyl-lysine antibody. The mean fold induction ± SD of acetylated p300 normalized to immunoprecipitated p300 from three independent experiments is indicated in the graph. c U2OS cells were transfected with MOCK or SIRT1 encoding vectors and 48 h later p300 was immunoprecipitated before and after etoposide treatment. p300 acetylation was determined by western blot with a general anti-acetyl-lysine antibody. As above, the mean fold induction ± SD of acetylated p300 relative to immunoprecipitated p300 from three independent experiments is reported in the chart. PC, negative control. d U2OS cells were transfected with control or TSPYL2 siRNAs, treated with MG132 and then with etoposide. p53 was immunoprecipitated and its interaction with SIRT1 was analyzed by western blot. The fold induction of co-immunoprecipitated SIRT1 relative to immunoprecipitated p53 is indicated. IP, immunoprecipitates; PC, negative control; Input, total lysate
To corroborate these results, we investigated whether SIRT1 deacetylates p300. For this, we analyzed p300 acetylation in U2OS-SIRT1-WT and –KO cells both before and after etoposide treatment and we found that, compared with WT controls, the levels of p300 acetylation are significantly higher in untreated SIRT1-negative cells and further increased in response to DNA damage (Fig. 5b). Conversely, SIRT1 overexpression reduced p300 acetylation in both undamaged and etoposide treated cells (Fig. 5c). These results indicate that SIRT1 deacetylates p300 before DNA damage, as already reported [34], and that this SIRT1 activity is further promoted upon genotoxic stress.
We also investigated whether TSPYL2 itself could be regulated by acetylation, but we were not able to observe such modification on this protein (Supplementary Fig. 4).
Our finding that TSPYL2 loss increases the association between SIRT1 and p300 suggests that, alike CCAR2, TSPYL2 inhibits SIRT1 preventing its binding to substrates. To confirm this, we analyzed SIRT1–p53 interaction in control and TSPYL2-depleted cells and we showed that this association is strongly increased in both undamaged and etoposide treated TSPYL2-silenced cells (Fig. 5d).
TSPYL2 depletion reduces p53 activity and DNA damage-induced apoptosis
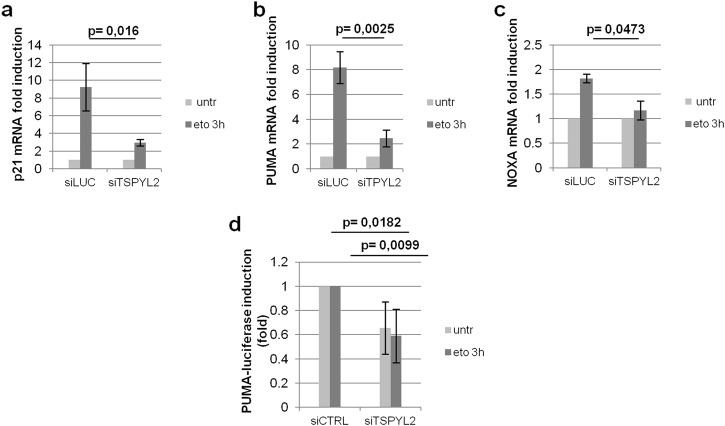
Regulation of p53-K382 acetylation by CCAR2 modulates p53 activation and apoptosis induction [7, 8]. Therefore, to investigate the physiological relevance of our findings, we evaluated the transcriptional activity of p53 in control and TSPYL2-depleted cells. RT-qPCR analyses revealed that the DNA damage-induced mRNA accumulation of the p53-target genes p21Waf1, PUMA, and NOXA was significantly reduced in TSPYL2-silenced cells, suggesting that this protein is required for the proper p53 transcriptional activity upon DNA damage (Fig. 6a–c and Supplementary Fig. 5a). To confirm these findings we performed luciferase reporter assays with control and TSPYL2-depleted cells transfected with the p53-responsive PUMA promoter-driven luciferase reporter gene. We demonstrated that PUMA promoter transactivation is strongly reduced in both untreated and etoposide treated TSPYL2-depleted cells (Fig. 6d and Supplementary Fig. 5b). Altogether, these results indicate that the DNA damage-dependent induction of p53 acetylation by TSPYL2 is required to promote p53 transcriptional activation.
Fig. 6.
TSPYL2 promotes p53 transcriptional activation upon DNA damage. a–c U2OS cells were transfected with control (siLUC) or TSPYL2 siRNAs and 48 h later, they were treated with etoposide (20 µM, 3 h). Total RNA was extracted, reverse transcribed and the levels of the p53-target genes p21, PUMA and NOXA were, respectively, evaluated by RT-qPCR. Fold changes, relative to the untreated samples, were reported in the charts. Error bars represent s.d. (d) control and TSPYL2-depleted U2OS cells were transfected with the p53-responsive PUMA-luciferase promoter. 48 h after transfection, cells were treated with etoposide (20 µM, 3 h) and PUMA promoter transactivation was determined by luciferase assays. Fold differences, relative to control (siCTRL) samples, were reported in the chart. Error bars represent s.d.
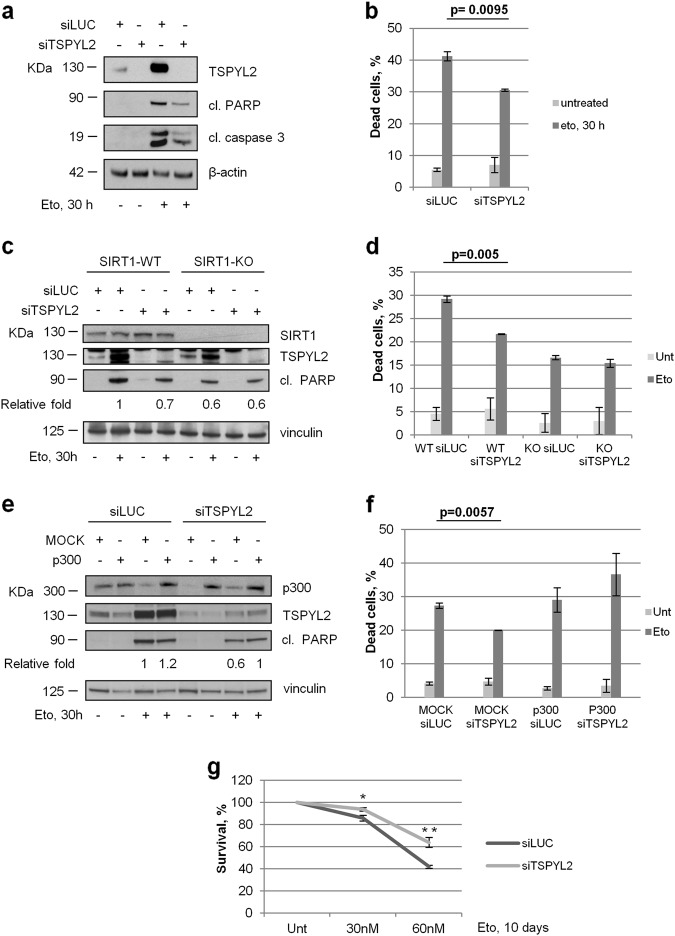
Then, we analyzed etoposide-induced apoptosis in U2OS cells silenced for TSPYL2. As reported [19], we found that prolonged exposure to genotoxic stress promotes TSPYL2 accumulation (Fig. 7a). In addition, consistently with the reduction of p53 activation and the decrease of PUMA and NOXA transcription, we demonstrated that, 30 h after DNA damage, TSPYL2 absence reduces the accumulation of pro-apoptotic markers, such as cleaved PARP and cleaved caspase-3 (Fig. 7a). Accordingly, we also demonstrated that TSPYL2 silencing causes a significant decrease of dead cells (Fig. 7b) and these results were confirmed again upon single TSPYL2 siRNAs transfection (Supplementary Fig. 6a). However, when we depleted TSPYL2 in U2OS-SIRT1-KO cells or in U2OS cells overexpressing p300, no reduction of cleaved PARP levels or of dead cells was observed upon etoposide (Fig. 7c–f). These results clearly indicate that TSPYL2 regulates apoptosis by repressing SIRT1 and promoting p300 activity.
Fig. 7.
TSPYL2 promotes DNA damage-induced apoptosis in a SIRT1- and p300- dependent manner. a U2OS cells were transfected with control or TSPYL2-specific siRNAs and the induction of pro-apoptotic markers (cleaved PARP and cleaved caspase-3) was evaluated 30 h after etoposide treatment (20 µM) by western blot. b Control and TSPYL2-depleted U2OS cells were treated or not with etoposide for 30 h and scored for viability with trypan blue staining. Values are mean ± s.d. of three independent experiments. c U2OS cells were transfected with control, TSPYL2 and SIRT1-specific siRNAs and the induction of PARP cleavage was evaluated 30 h after etoposide treatment (20 µM) by western blot. The fold induction of cleaved PARP relative to loading control (vinculin) is indicated. d U2OS cells were transfected and treated as in c and the percentage of dead cells was determined by trypan blue exclusion test. Values are mean ± s.d. from three independent experiments. e U2OS cells transfected with MOCK or p300-encoding vectors were silenced for TSPYL2 and the levels of cleaved PARP were determined 30 h after etoposide treatment (20 µM) by western blot. The fold induction of cleaved PARP relative to vinculin-loading control is reported. f U2OS cells were transfected and treated as in e and the percentage of dead cells was determined by trypan blue exclusion test. Values are mean ± s.d. from three independent experiments. g colonies formation assays were performed on U2OS cells transfected with control or TSPYL2 siRNAs and treated with increasing doses of etoposide. Colonies were counted and indicated as percentages relative to the untreated samples. * p = 0.01; **p = 0.008
Then, we found that apoptosis reduction, likewise p53 acetylation, is more evident in cells depleted of CCAR2 and TSPYL2 (Supplementary Fig. 6b), than in cells transfected with single silencing, further confirming that the two proteins collaborate in apoptosis regulation, even with different mechanism.
Importantly, the apoptotic function of TSPYL2 is not specific for etoposide, as similar results were obtained upon UV radiation or gemcitabine exposure (Supplementary Fig. 6c), and therefore TSPYL2 regulates apoptosis induction after different kinds of DNA damage.
Finally, to further investigate TSPYL2 role in the DDR, we tested the ability to form colonies of TSPYL2-depleted U2OS cells upon etoposide treatment and, as shown in Fig. 7g, we demonstrated that TSPYL2 absence significantly increases long-term clonogenic survival after DNA damage.
Discussion
Here, we demonstrate that TSPYL2, whose expression is strongly induced by CCAR2 loss, inhibits SIRT1 and promotes p300 activity, finely tuning p53 acetylation and apoptosis induction upon genotoxic stress. These results well agree with our previous findings demonstrating that, upon DNA damage, CCAR2 is not the only protein capable to inhibit SIRT1 [9] and with those from others showing that, after DNA damage, TSPYL2 regulates p53 stability [19]. Indeed, acetylation is required for p53 activation and, competing with ubiquitination, increases p53 stability [28, 35]. Therefore, our studies could provide the mechanisms at the basis of p53 regulation by TSPYL2.
We initially supposed that TSPYL2 is induced upon long-term CCAR2 depletion as an adaptation mechanism that guarantees the proper regulation of SIRT1 and p53 activities in the cells. Our findings that TSPYL2 depletion reduces p53 acetylation and apoptosis induction seem to confirm this hypothesis. However, we noticed that concurrent depletion of CCAR2 and TSPYL2 reduces the acetylation of p53 more than silencing of CCAR2 or TSPYL2 alone, therefore indicating the possibility that these proteins regulate p53 differently. Accordingly, we observed that, upon DNA damage, depletion of both SIRT1 and TSPYL2 produces an intermediate level of p53 acetylation, compared with single silencing. Subsequent analyses demonstrated that TSPYL2, beside inhibiting SIRT1, promotes p300 activity, even if we cannot exclude the possibility that it could act also on other acetyl-transferases targeting p53. Indeed, we found that concurrent TSPYL2 and p300 depletion completely abrogates p53-K382 acetylation. This phenomenon could be ascribed to the induction of SIRT1 activity consequent to TSPYL2 reduction, but also to the inhibition of another acetyl-transferase that can target p53-K382.
To further investigate the molecular mechanisms responsible for p53 regulation, we analyzed SIRT1, p300, and TSPYL2 complex formation. We observed that, although the binding of TSPYL2 with p300 and SIRT1 is not affected by DNA damage, SIRT1-p300 association is induced by genotoxic treatment in a TSPYL2-dependent manner. These results suggest that TSPYL2 interacts with SIRT1 and p300 in a separate manner, preventing SIRT1-p300 complex formation, even if we cannot exclude the possibility that these proteins may form a ternary complex. Moreover, the TSPYL2-dependent increase of p300 acetylation indicate that SIRT1 might be involved in the regulation of p300 function upon DNA damage. This hypothesis was further advised by the discovery that SIRT1 silencing induces p53 acetylation more than contemporary depletion of SIRT1 and p300 or SIRT1 and TSPYL2. Accordingly, we demonstrated that, after etoposide, SIRT1, increasing its interaction with p300, deacetylates and inhibits p300, possibly as a final attempt to prevent cell death. However, we do not know if p300 is directly or indirectly targeted by SIRT1. Importantly, this finding adds another means of regulation at p300 activity, further illustrating the cooperative function between SIRT1 and p300 that are jointly involved in different cellular processes, as they share common targets.
Our findings that, in TSPYL2-silenced cells, SIRT1-p300 association is already increased in unstressed cells (possibly because there are more SIRT1 and p300 free molecules) suggest that TSPYL2 promotes p300 acetylation by preventing its association with SIRT1. These results indicate that, alike CCAR2, TSPYL2 inhibits SIRT1 blocking its interaction with target proteins, as it was confirmed also by the analysis of p53-SIRT1 association in control and TSPYL2-depleted cells. Unfortunately, we were not able to test the effect of TSPYL2 overexpression on SIRT1 association with its targets because, as previously reported [11, 18, 19], ectopic expression of TSPYL2 arrests cell growth. However, it is also possible that TSPYL2 directly regulates p300 autoacetylation or the activity of other(s) acetyl-transferase(s) toward p300, through an unknown mechanism. Moreover, other post-translational modifications, possibly cross-talking with acetylation, may have a role in the regulation of p300 activation by TSPYL2.
p53 activity is finely regulated by post-translational modifications [28, 36] that account for its involvement in different cellular processes. As modulation of p53-K382 acetylation by CCAR2 was previously demonstrated to regulate apoptosis [7, 8], we analyzed cell death in TSPYL2-depleted cells exposed to different kinds of DNA damage. According to the reduced p53 acetylation, we found that apoptosis is downregulated in TSPYL2-silenced cells, but unaffected when TSPYL2 is depleted in SIRT1-negative or p300-overexpressing cells. These results confirm that TSPYL2 controls apoptosis induction modulating the activity of both these proteins. During these studies, we observed that TSPYL2 is strongly induced after prolonged DNA damage, as reported [19]. Therefore, it is possible, that also TSPYL2 accumulation contributes to the regulation apoptosis induction. Importantly, the role of TSPYL2 in apoptosis regulation is not specific for U2OS cells. Indeed knock-out of TSPYL2 gene in the neuroblastoma cell line Be2C significantly increases cellular survival upon 4 days of etoposide treatment (Professor SY Chan, personal communication).
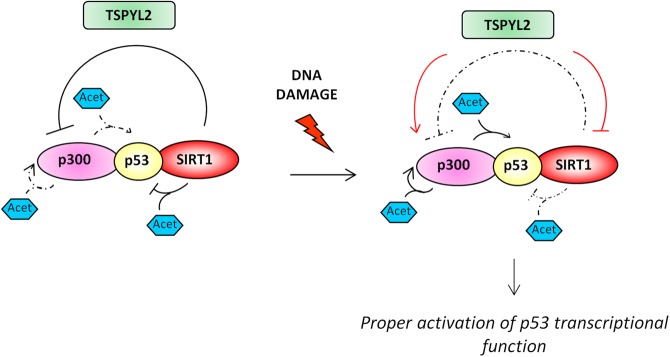
Collectively, our results suggest a model (Fig. 8) in which in unperturbed cells SIRT1 maintains p300 and p53 in a hypoacetylated state to prevent apoptosis. After genotoxic stress, TSPYL2 prevents SIRT1 association with p300 and p53 allowing their acetylation. Acetylated p300 can now target p53 for acetylation, finally leading to its transcriptional activation and induction of p53-dependent apoptosis.
Fig. 8.
Regulation of p53 activity by TSPYL2. Graphical scheme of our proposed model. In response to DNA damage, TSPYL2 contributes to the activation of p53 by inhibiting SIRT1 and promoting p300 acetylation and activation. This event facilitates p53 functional activation and apoptosis induction
Interestingly, SIRT1 inhibition is also supported by CCAR2 and other cellular molecules [37], therefore suggesting that the correct modulation of this deacetylase’s function upon DNA damage is a critical cellular process.
In summary, our findings expand previous studies indicating a TSPYL2 role in the DDR and reveal for the first time the molecular mechanisms through which TSPYL2 regulates p53 activity and apoptosis upon DNA damage. However, as both SIRT1 and p300 are involved in many cellular processes [38, 39], it is also conceivable that TSPYL2 regulates other pathways governed by p300 and SIRT1, possibly in collaboration with CCAR2.
Importantly, these studies may find an application in cancer research. Indeed the TSPYL2-dependent regulation of p53 activity could contribute to explain why reduced levels of this protein in cancer patients are associated with poor prognosis [16, 18] and suggest that, in the future, TSPYL2 may be a promising target for cancer therapy. In fact modulation of TSPYL2 expression could contribute to restore p53 acetylation and apoptosis in patients, therefore increasing their sensitivity for chemo- or radiotherapy.
Materials and methods
Cell lines
U2OS and A549 human cancer cell lines (ATCC) and human primary fibroblasts were cultured in DMEM (Lonza), supplemented with 10 % fetal bovine serum, 5000 U/ml penicillin, and 5 mg/ml streptomycin; MDA-MB-231 cells (ATCC) were cultured in RPMI (Lonza), supplemented with 10% fetal bovine serum, 5000 U/ml penicillin, and 5 mg/ml streptomycin. U2OS-SIRT1-KO cells were generated using the CRISPR/Cas9 system as reported [40], whereas U2OS-CCAR2-KO cells were previously described [22]. All cell lines were maintained at 37 °C and 5% CO2.
Cells transfections and treatments
Plasmid and siRNA transfections were performed using Lipofectamine 2000 and Lipofectamine RNAiMAX (Thermo Fisher Scientific), respectively; for p300 overexpression, TransIT-2020 (Mirus) was used. Transfections were carried out following the manufacturers’ instructions.
siRNAs against TSPYL2 and p300 were purchased from Ambion, siSIRT1 from QIAGEN, siLUC from Eurofins Genomics and siCCAR2 and siCTRL from RIBOXX.
DNA damage was induced by treating cells with etoposide (20 µM, TEVA), NCS (30 nM, Sigma), UV (20 J/m2), gemcitabine (10 µM), hydroxyurea (1 mM), and camptothecin (20 µM). MG132 (Sigma) was added 20 min before treatment, at the concentration of 25 µM. Nicotinamide was used 4 mM and was added to the cells 30 min before etoposide treatment. Viable cells after treatments were analyzed by trypan blue (Sigma) exclusion tests and counted.
Western blot and antibodies
Western blot analyses were performed using the NuPAge system (Thermo Fisher Scientific) or as described in ref. [41]. Antibodies used were: p53-DO7 (Santa Cruz Biotechnology), TSPYL2, CCAR2, (Bethyl), SIRT1, cleaved caspase-3, cleaved PARP, p53-Ac-K382, acetyl-lysine (Cell Signalling Technology), β-actin, and Vinculin (Sigma); p300 antibody from Santa Cruz Biotechnology was used for immunoprecipitation assays, whereas a p300 antibody from Bethyl was used for western blot. Densitometric analyses were performed with the Fiji software [42].
RT-qPCR
U2OS cells were transfected with control or a pool of TSPYL2 or CCAR2 siRNAs. After 48 h, or 6 days for CCAR2, total RNA was extracted from control and TSPYL2-depleted cells, purified using the miRNeasy Mini Kit (QIAGEN), according to manufacturer’s instructions and quantified using NanoDrop Microvolume spectrophotometer (Thermo). Complementary DNA (cDNA) was generated by reverse transcription of 1 μg of total RNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche) in 20 µl reactions. qPCR was performed in triplicate on 20 ng of cDNA using SYBR Green PCR Master Mix (Applied Biosystems, Thermo Fisher Scientific) and the ViiA™ 7 Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific). Reactions were performed in 10 µl of final volume. Samples were normalized using β-tubulin as reference gene. The experiment was repeated three times. Primer sequences were: TSPYL2_for AGGCACTGGAGGATATTCAG; TSPYL2_rev GAAGGGTCTTCGCATCTGGAT; PUMA_for TGAGACAAGAGGAGCAGCAG; PUMA_rev GGAGTCCCATGATGAGATTGT; NOXA_for GCAAGAACGCTCAACCGA; NOXA_rev TGCCGGAAGTTCAGTTTGTC; p21_for GGTGGACCTGGAGACTCTC; p21_rev GAAGATCAGCCGGCGTTTG; β Tubulin_for CTTCAGTGAGACGGGCGCTGGCAAGCAC; β Tubulin_rev TGATGAGCTGCTCAGGGTGGAA.
Luciferase assays
U2OS cells were silenced for TSPYL2 and then transfected with p53-responsive PUMA promoter-driven firefly luciferase reporter (PUMA-Luc) and pRL-TK (encoding Renilla luciferase) vectors. After 48 h, cells were exposed to 20 µM etoposide for 3 h or left untreated, lysed and luciferase activity was analyzed with the Dual-Luciferase Reporter Assay System (Promega), using Renilla luciferase activity for normalization.
Immunoprecipitations
Immunoprecipitations were performed as previously described [43]; In brief, cell lysates were prepared in egg lysis buffer (ELB) (150 mM NaCl, 50 mM Hepes pH 7.5, 5 mM EDTA, 0.5% NP-40). After 30 min of preclearing (PC) with protein-G (for mouse antibodies) or protein A (for rabbit antibodies) coupled sepharose resin (Sigma), proteins of interest were immunoprecipitated with specific antibodies for 3 h. After washes, immunoprecipitated proteins were detached from the resins with Laemmli buffer and analyzed by western blot.
In vitro acetylation assays
The assay was performed using bacterially purified recombinant GST-p53 and p300 acetyl-transferase immunoprecipitated from control or TSPYL2-depleted cells, treated with etoposide. p300 immunoprecipitation was carried out as described above; then the resins (IP and PC) were washed once with IP buffer (ELB) containing 0.5 M LiCl, once with IP buffer and once with acetylation buffer (20 mM Tris-HCl pH 7.5, 0.5 mM EDTA, 2 mM CaCl2 [44]. The acetylation reaction was performed in the acetylation buffer supplemented with 1 μM trichostatin A (Sigma), 5 mM nicotinamide (Sigma) and, when indicated, 10 μM acetyl-coenzyme A (Sigma). Recombinant GST-p53 was added to the resins resuspended in the indicated buffer and samples were incubated with gentle shaking at 30 °C for 30 min. The reactions were subjected to immunoblotting using the anti-p53-Ac-K382 antibody (Cell Signalling) and densitometric analyses of the bands were performed with the Fiji software [42].
In vitro deacetylation assays
U2OS cells were treated with 50 μM etoposide and p53 was immunoprecipated with anti-p53-DO7 antibody (Santa Cruz Biotechnology) and protein A sepharose beads (Sigma), as described above. Then, sepharose was washed once with IP buffer (ELB) containing 0.5 M LiCl, once with IP buffer and once with deacetylation buffer (50 mM Tris-HCl, pH 9.0, 5% glycerol, 50 mM NaCl, 4 mM MgCl2, 0.02% NP-40). Simultaneously, total cell extracts of U2OS cells were prepared as described in [25]. In brief, cells transfected with siRNAs against p53 and TSPYL2 or LUC, as control, were treated with etoposide, lysed in NETN buffer (20 mM Tris-Hcl pH8, 100 mM Nacl, 1 mM EDTA, 0.5% NP-40) and incubated 10 min at 37 °C to degrade contaminant NAD + ; then 1.5 mM DTT was added and lysates were incubated again 10 min at 37 °C. Deacetylation reactions were performed in deacetylation buffer supplemented with 1 μM trichostatin A (Sigma) and 50 mM NAD+ (Sigma). For negative controls 5 mM nicotinamide (Sigma) was also added. A total of 30 µg of U2OS lysates were incubated with the HA-p53-agarose beads resuspended in the indicated buffer and samples were maintained with gentle shaking at 30 °C for 1 h. The reactions were subjected to immunoblotting using the anti-p53-Ac-K382 antibody (Cell Signalling) and bands quantified with the Fiji software.
Proximity ligation assays
U2OS cells, transfected with MOCK or FLAG-TSPYL2 encoding vectors, were cytospinned, fixed with 4% paraformaldehyde and incubated with anti-FLAG (Sigma) and anti-SIRT1 (Sigma) or anti-p300 (Bethyl) antibodies. Proximity Ligation assays were then performed using the Duo Link In Situ reagents (Sigma) according to manufacturer’s protocol. Slides were scored by fluorescence microscopy and digital images were acquired on a Nikon Eclipse E1000 microscope equipped with a DS-U3 CCD camera.
Colony formation assays
U2OS cells were transfected with control or TSPYL2 siRNAs, plated in triplicates (500 cells/well in six-well plate) and treated with the indicated doses of etoposide. Colonies were allowed to grow for ~ 10 days, then fixed in methanol and stained with Giemsa solution (Sigma). Colonies were enumerated and expressed as percentages relative to the untreated samples.
Electronic supplementary material
Acknowledgements
This work was supported by the Italian Ministry of Health (GR-2010-2315822). M. Magni is a recipient of Umberto Veronesi Foundation fellowship.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by G. Del Sal
Electronic supplementary material
The online version of this article (10.1038/s41418-018-0168-6) contains supplementary material, which is available to authorized users.
References
- 1.Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell. 2017;170:1062–78. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roos WP, Thomas AD, Kaina B. DNA damage and the balance between survival and death in cancer biology. Nat Rev Cancer. 2016;16:20–33. doi: 10.1038/nrc.2015.2. [DOI] [PubMed] [Google Scholar]
- 3.Brooks CL, Gu W. The impact of acetylation and deacetylation on the p53 pathway. Protein Cell. 2011;2:456–62. doi: 10.1007/s13238-011-1063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zannini L, Delia D, Buscemi G. CHK2 kinase in the DNA damage response and beyond. J Mol Cell Biol. 2014;6:442–57. doi: 10.1093/jmcb/mju045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaziri H, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–59. doi: 10.1016/S0092-8674(01)00527-X. [DOI] [PubMed] [Google Scholar]
- 6.Magni M, Buscemi G, Zannini L. Cell cycle and apoptosis regulator 2 at the interface between DNA damage response and cell physiology. Mutat Res. 2018;776:1–9. doi: 10.1016/j.mrrev.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 7.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–6. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 8.Zhao W, et al. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–90. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zannini L, Buscemi G, Kim JE, Fontanella E, Delia D. DBC1 phosphorylation by ATM/ATR inhibits SIRT1 deacetylase in response to DNA damage. J Mol Cell Biol. 2012;4:294–303. doi: 10.1093/jmcb/mjs035. [DOI] [PubMed] [Google Scholar]
- 10.Magni M, et al. Chk2 and REGgamma-dependent DBC1 regulation in DNA damage induced apoptosis. Nucleic Acids Res. 2014;42:13150–60. doi: 10.1093/nar/gku1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toh BH, Tu Y, Cao Z, Cooper ME, Chai Z. Role of cell division autoantigen 1 (CDA1) in cell proliferation and fibrosis. Genes (Basel) 2010;1:335–48. doi: 10.3390/genes1030335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang GS, et al. Transcriptional modification by a CASK-interacting nucleosome assembly protein. Neuron. 2004;42:113–28. doi: 10.1016/S0896-6273(04)00139-4. [DOI] [PubMed] [Google Scholar]
- 13.Ozbun LL, Martinez A, Jakowlew SB. Differentially expressed nucleolar TGF-beta1 target (DENTT) shows tissue-specific nuclear and cytoplasmic localization and increases TGF-beta1-responsive transcription in primates. Biochim Biophys Acta. 2005;1728:163–80. doi: 10.1016/j.bbaexp.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Epping M T, Lunardi A, Nachmani D, Castillo-Martin M, Thin T H, Cordon-Cardo C, Pandolfi P P. TSPYL2 is an essential component of the REST/NRSF transcriptional complex for TGFβ signaling activation. Cell Death & Differentiation. 2015;22(8):1353–1362. doi: 10.1038/cdd.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Gallo M, et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet. 2012;44:1310–5. doi: 10.1038/ng.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim TY, Zhong S, Fields CR, Kim JH, Robertson KD. Epigenomic profiling reveals novel and frequent targets of aberrant DNA methylation-mediated silencing in malignant glioma. Cancer Res. 2006;66:7490–501. doi: 10.1158/0008-5472.CAN-05-4552. [DOI] [PubMed] [Google Scholar]
- 17.Eyler CE, et al. Glioma stem cell proliferation and tumor growth are promoted by nitric oxide synthase-2. Cell. 2011;146:53–66. doi: 10.1016/j.cell.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kandalaft LE, Zudaire E, Portal-Nunez S, Cuttitta F, Jakowlew SB. Differentially expressed nucleolar transforming growth factor-beta1 target (DENTT) exhibits an inhibitory role on tumorigenesis. Carcinogenesis. 2008;29:1282–9. doi: 10.1093/carcin/bgn087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tu Y, et al. Antiproliferative autoantigen CDA1 transcriptionally up-regulatesp21(Waf1/Cip1) by activating p53 and MEK/ERK1/2 MAPK pathways. J Biol Chem. 2007;282:11722–31. doi: 10.1074/jbc.M609623200. [DOI] [PubMed] [Google Scholar]
- 20.Tao KP, et al. TSPYL2 is important for G1 checkpoint maintenance upon DNA damage. PLoS ONE. 2011;6:e21602. doi: 10.1371/journal.pone.0021602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Restelli M, et al. A novel crosstalk between CCAR2 and AKT pathway in the regulation of cancer cell proliferation. Cell Death Dis. 2016;7:e2453. doi: 10.1038/cddis.2016.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magni M, et al. CCAR2/DBC1 is required for Chk2-dependent KAP1 phosphorylation and repair of DNA damage. Oncotarget. 2015;6:17817–31. doi: 10.18632/oncotarget.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montecucco A, Zanetta F, Biamonti G. Molecular mechanisms of etoposide. EXCLI J. 2015;14:95–108. doi: 10.17179/excli2015-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Sousa Cavalcante L, Monteiro G. Gemcitabine: metabolism and molecular mechanisms of action, sensitivity and chemoresistance in pancreatic cancer. Eur J Pharmacol. 2014;741:8–16. doi: 10.1016/j.ejphar.2014.07.041. [DOI] [PubMed] [Google Scholar]
- 25.Escande C, et al. Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. J Clin Invest. 2010;120:545–58. doi: 10.1172/JCI39319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JT, Gu W. SIRT1: regulator of p53 deacetylation. Genes Cancer. 2013;4:112–7. doi: 10.1177/1947601913484496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/S0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 28.Reed SM, Quelle DE. p53 acetylation: regulation and consequences. Cancers (Basel) 2014;7:30–69. doi: 10.3390/cancers7010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Avalos JL, et al. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol Cell. 2002;10:523–35. doi: 10.1016/S1097-2765(02)00628-7. [DOI] [PubMed] [Google Scholar]
- 30.Shikama N, et al. Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators. Mol Cell Biol. 2000;20:8933–43. doi: 10.1128/MCB.20.23.8933-8943.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koos B, et al. Analysis of protein interactions in situ by proximity ligation assays. Curr Top Microbiol Immunol. 2014;377:111–26. doi: 10.1007/82_2013_334. [DOI] [PubMed] [Google Scholar]
- 32.Thompson PR, et al. Regulation of the p300 HAT domain via a novel activation loop. Nat Struct Mol Biol. 2004;11:308–15. doi: 10.1038/nsmb740. [DOI] [PubMed] [Google Scholar]
- 33.Delvecchio M, Gaucher J, Aguilar-Gurrieri C, Ortega E, Panne D. Structure of the p300 catalytic core and implications for chromatin targeting and HAT regulation. Nat Struct Mol Biol. 2013;20:1040–6. doi: 10.1038/nsmb.2642. [DOI] [PubMed] [Google Scholar]
- 34.Bouras T, et al. SIRT1 deacetylation and repression of p300 involves lysine residues 1020/1024 within the cell cycle regulatory domain 1. J Biol Chem. 2005;280:10264–76. doi: 10.1074/jbc.M408748200. [DOI] [PubMed] [Google Scholar]
- 35.Ito A, et al. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 2002;21:6236–45. doi: 10.1093/emboj/cdf616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–22. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen R, et al. Quantitative proteomics reveals that long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation. Nucleic Acids Res. 2017;45:9947–59. doi: 10.1093/nar/gkx600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satoh A, Stein L, Imai S. The role of mammalian sirtuins in the regulation of metabolism, aging, and longevity. Handb Exp Pharmacol. 2011;206:125–62. doi: 10.1007/978-3-642-21631-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deng CX. SIRT1, is it a tumor promoter or tumor suppressor? Int J Biol Sci. 2009;5:147–52. doi: 10.7150/ijbs.5.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–6. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buscemi G, et al. Bimodal regulation ofp21(waf1) protein as function of DNA damage levels. Cell Cycle. 2014;13:2901–12. doi: 10.4161/15384101.2014.946852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zannini L, Buscemi G, Fontanella E, Lisanti S, Delia D. REGgamma/PA28gamma proteasome activator interacts with PML and Chk2 and affects PML nuclear bodies number. Cell Cycle. 2009;8:2399–407. doi: 10.4161/cc.8.15.9084. [DOI] [PubMed] [Google Scholar]
- 44.Graczyk A, Slomnicki LP, Lesniak W. S100A6 competes with the TAZ2 domain of p300 for binding to p53 and attenuates p53 acetylation. J Mol Biol. 2013;425:3488–94. doi: 10.1016/j.jmb.2013.06.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.