Abstract
Rodents engage in social communication through a rich repertoire of ultrasonic vocalizations (USVs). Recording and analysis of USVs has broad utility during diverse behavioral tests and can be performed noninvasively in almost any rodent behavioral model to provide rich insights into the emotional state and motor function of the test animal. Despite strong evidence that USVs serve an array of communicative functions, technical and financial limitations have been barriers for most laboratories to adopt vocalization analysis. Recently, deep learning has revolutionized the field of machine hearing and vision, by allowing computers to perform human-like activities including seeing, listening, and speaking. Such systems are constructed from biomimetic, “deep”, artificial neural networks. Here, we present DeepSqueak, a USV detection and analysis software suite that can perform human quality USV detection and classification automatically, rapidly, and reliably using cutting-edge regional convolutional neural network architecture (Faster-RCNN). DeepSqueak was engineered to allow non-experts easy entry into USV detection and analysis yet is flexible and adaptable with a graphical user interface and offers access to numerous input and analysis features. Compared to other modern programs and manual analysis, DeepSqueak was able to reduce false positives, increase detection recall, dramatically reduce analysis time, optimize automatic syllable classification, and perform automatic syntax analysis on arbitrarily large numbers of syllables, all while maintaining manual selection review and supervised classification. DeepSqueak allows USV recording and analysis to be added easily to existing rodent behavioral procedures, hopefully revealing a wide range of innate responses to provide another dimension of insights into behavior when combined with conventional outcome measures.
Subject terms: Motivation, Emotion
Introduction
Rodents engage in social communication through a rich repertoire of ultrasonic vocalizations (USVs, vocalizations >20 kHz). Rats and mice produce complex sequences of USVs throughout development and in a variety of social and motivational contexts [1–6]. These sequences are made up of a number of uniquely shaped syllables across a wide range of frequencies (20–115 kHz), and they appear to have a form of syntax which is contextually dependent [3, 7, 8]. Since their discovery, there has been a concerted effort to assess the significance of USVs in rats and mice and to utilize these innate responses as an indicator of the subjective experience of the animal. For example, high frequency (~50 kHz) USVs have been associated with positive affect in rats, while lower frequency (~22 kHz) USVs have been associated with negative affect [1, 9–12]. However, within rat 50 kHz USVs there is considerable variability in syllable type and sequence structure that may encode valuable information, and has yet to be deciphered [13]. Mouse USVs are less clearly tied to affective state, but the shape and sequence of syllables vary greatly across genetic strains [14–17], behavioral and social contexts [3, 18, 19], genetic manipulations [20], and development [4, 21].
Recording and analysis of USVs has broad utility and can be performed noninvasively in almost any rodent model, including those commonly used to investigate models of drug abuse, depression, fear or anxiety, neurodegenerative disease, aging, and reward processing. However, despite strong evidence that USVs serve an array of communicative functions and the extraordinary extent to which rodent behavioral models are used in neuroscience research [22], technical and financial limitations have curbed the adoption of USV analysis. While it is possible to use USVs as a relatively simple inference of positive and negative affect, it still remains unclear the extent to which syllable type and syntax may signify unique information relevant to specific biological states, affective states, or social situations. Manual classification of rat USVs often use 15 call categories [13] and semi-automated analysis of 12 different mouse strains suggested there are >100 USV categories [17]. While both of these strategies have yielded interesting behavioral insights, there is no consensus yet on exactly how to categorize USVs. Performing these analyses manually is laborious and prone to misclassification. Software such as MUPET [17] can produce automatically generated syllable repertoires of a user-defined size, but to determine that number by maximizing goodness-of-fit measures can lead to the selection of rather large repertoires that are difficult to map onto behavior. Further, the models produced by MUPET cannot be saved and applied to future datasets. There remains a need for a fully automated method of producing syllable repertoires that can be used to compare call types across experiments.
Investigator analysis of USV recordings is slow and laborious, while existing automated analysis software are vulnerable to broad spectrum noise routinely encountered in the testing environment. In order for a large portion of the neuroscience community to adopt USV recording, and to fully explore the meaning and scientific value of USVs, researchers need access to an inexpensive, accurate, and high throughput method for detecting USVs, classifying syllables, and analyzing sequences across a wide range of experiments and recording procedures. Recently, deep learning has revolutionized the field of machine hearing and vision, by allowing computers to perform human-like activities such as seeing, listening, and speaking [23–25]. Such systems are constructed from biomimetic, “deep”, artificial neural networks. Here, we present DeepSqueak, a USV detection and analysis software suite which uses regional convolutional neural networks (Faster-RCNN) [26] to increase detection rate, reduce false positives, reduce analysis time, classify calls, and perform syntax analysis automatically. While DeepSqueak is not the first software package developed for USV detection and analysis [17, 27–30], it is free, accessible, accurate, and fully featured (Table S1).
Methods
Training Faster-RCNN to detect USVs
DeepSqueak is packaged with four default detection networks: one general purpose network, one for mouse USVs, one for short rat USVs, and one for long 22 kHz rat USVs. Subsequent detection networks for novel vocalizations or different species can be generated within DeepSqueak with no programing or neural network experience. To generate the default detection networks, hundreds of rat and mouse USVs were manually isolated from our own recordings and from external labs [1, 31] using Raven Lite 2.0 (Cornell Lab of Ornithology, NY). Manually isolated calls may be imported into DeepSqueak from Raven Lite 2.0, MUPET [17], Ultravox (Noldus; Wageningen, NL), or XBAT [27]. Using the “Tools→Network Training→Create Training Data” function, individual vocalizations were transformed into sonograms. Sonogram parameters for short duration vocalization are: nfft = 0.0032 s, overlap = 0.0028 s, window = 0.0032 s. Sonogram parameters for long duration vocalizations are: nfft = 0.01 s, overlap = 0.005 s, window = 0.01 s. These sonograms were then passed to the “Tools→Network Training→Train Network” function which trains a Faster RCNN object detector neural network (Table S2). These initial networks were used to isolate thousands of USVs automatically. These new USVs were manually reviewed and used to re-train each network on a higher variety of vocalizations, resulting in networks with extremely high recall, defined as the ratio of detected true USVs to total true USVs in the file. To make detection more robust to variable recording methods, the user may also choose to augment the training set by added procedurally generated noise and variable microphone gain in order to artificially inflate its size and variability. This method was used to generate the high sensitivity rat and mouse network, and details regarding this function are outlined in the wiki.
Detecting USVs using DeepSqueak
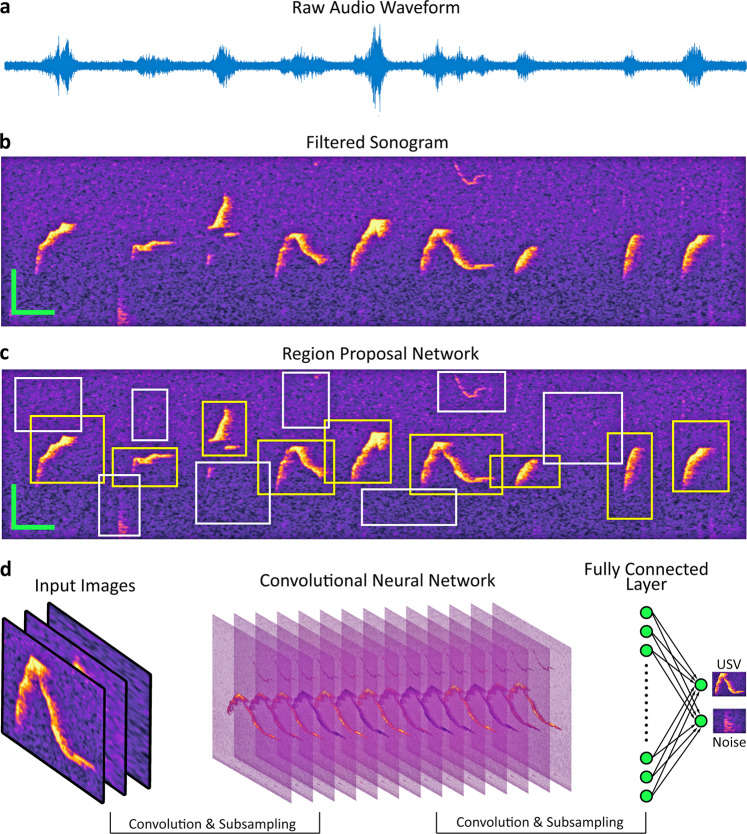
Recordings of vocalizations stored as .WAV or .FLAC files can be passed into DeepSqueak individually or in a batch (Fig. 1a). DeepSqueak will split the audio file into short segments and convert them into sonograms (Fig. 1b). These images are then passed to a Faster-RCNN object detector. Segment length, frequency range, and detection network can all be user-defined. The first stage of detection is a region proposal network, which segments the image into proposed areas of interest which may contain USVs (Fig. 1c). The sections of the image within these boxes are then passed to the classification network which determines whether the image contains a call or background noise (Fig. 1d). All USVs are saved to a detection file along with call parameters and classification confidence. These parameters can be used to automatically review USVs further.
Fig. 1.
Rapid detection of USVs using Faster RCNN. Shown is the Faster RCNN-based detection architecture. a An example waveform from a 1 s section recording containing mouse ultrasonic vocalizations. b A filtered sonogram produced from the audio segment displayed in (a). c A representation of the Faster-RCNN region proposal network determining sections of the sonogram to pass to the classification network. d The classification network receives input from the region proposal network, performs a series of convolutions and filters, and ultimately classifies the section as a call or background noise. Scale bar: Y = 20 kHz, X = 20 ms
Training a post-hoc de-noising network
The primary Faster RCNN object detector was designed to be highly sensitive to ensure that vocalizations are not missed in the audio files, at the cost of occasional false positives. Every experimental setup is subject to unique mechanical and electrical noise, a problem that plagues most automatic USV detection software. We have included a secondary detection network to identify and exclude these types of interfering noise by detecting them from several experimental conditions, and manually labeling individual detections as “Noise” or “USV” with “Tools→Call Classification→Add Custom Labels”. These manually labeled detection files were passed to “Tools→Network Training→Train Post Hoc Denoiser” which trained a neural network capable of discriminating USVs from common types of background noise (Table S3). If a user’s particular experimental setup produces noise not recognized by the included network, they may create custom de-noising networks from their own recordings.
Automatic and manual selection review
To ensure the highest possible accuracy, DeepSqueak allows for multiple methods to review detections and remove noise. The primary detection network generates a confidence score for all detections. This score, as well as spectral power and call tonality, may be used to automatically reject or accept all calls above or below user-defined values under “Tools→Automatic Review→Batch Reject by Threshold”. Furthermore, the post-hoc denoising network described above may be applied to all detection files under “Tools→Automatic Review→Post Hoc Denoising”. All rejected detections remain in the detection file but will not be analyzed while in a “rejected” state. Finally, because unsupervised clustering tends to place false positive into distinct clusters, the user may classify entire clusters of detections as noise.
While USV detection and analysis with DeepSqueak can be completely automated, it retains a complete set of manual selection review features. Each detection can be viewed as a sonogram and played back at any speed. Detections can be sorted by time or score, can be accepted or rejected, classified with user-defined labels through keyboard shortcuts, and the boxes defining calls can be redrawn.
Contour detection and call statistics
Robust contour detection is an extremely important aspect of automatic call classification, and a key feature of DeepSqueak. Each USV’s contour is constructed by first calculating the frequency at maximum amplitude for each time point in the spectrogram, and then cleaned by removing non-tonal features such as silence or broad spectrum noise. The tonality of each audio signal is calculated by dividing the geometric mean of the power spectrum by the arithmetic mean and subtracting from 1.
This is critical for extracting clean contours from noisy backgrounds, because it allows for quantification of whistle-like quality to provide clear separation between USVs and broad spectrum mechanical noise of similar amplitude. All of the statistics generated for each call are extracted from the contour. The minimum, maximum, and mean frequency, duration, slope, sinuosity, and power are all calculated based on the position of the contour. This allows us tight control over what parts of the sonogram are used to calculate these statistics and filters out any non-tonal aspects of the sonogram.
Unsupervised syllable clustering
Optimization of unguided clustering comprises two main issues. The first is how to extract from raw data, the meaningful dimensions by which discrete USV categories are encoded, and second is how to determine the quantity and distinctness of these categories. We chose to highlight how different methods of call parameterization alter clustering, and ultimately allow the user to determine which USV features were most important to include in the clustering algorithm. Call contours from any number of detection files can be loaded into “Tools→Call Classification→Unsupervised Clustering”. Currently, two unsupervised clustering algorithms are implemented in DeepSqueak, although the K-means-based approach will be used to highlight cluster optimization, as is has been shown to work exceedingly well in similar work in dolphins [32]. DeepSqueak allows the user to adjust three weighted input features: shape, frequency, and duration. The frequency of each call is reduced 10 segments, shape is defined as the first derivative of the contour at 10 segments, and duration is defined as the duration of the contour. The scale of each input is z-score normalized and divided by the number of input segments so that each dimension is initially equally weighted. These inputs are then multiplied by the user-defined input weights to alter each parameter’s effect on clustering. DeepSqueak allows the user to choose a number of clusters, or can then attempt to automatically pick the ideal number of clusters based on the elbow method. This method works by first calculating the total sum of squared error (TSS) for all USVs, and then the within-cluster sum of squared error (WSS) for clusters. When clustering a non-random dataset, this procedure produces a logarithmically decreasing curve, where the addition of new clusters initially reduces the WSS drastically, but eventually the addition of new clusters produces diminishing returns on error reduction. This point is operationalized as the elbow (inflection point) of the curve. More formally, the elbow is calculated by walking along the curve one bisection point at a time and fitting two lines, one to all the points to left of the bisection point and one to all the points to the right of the bisection point. The knee is judged to be at a bisection point which minimizes the sum of errors for the two fits (Fig. 4a). This concept of diminishing returns is very useful for USVs because it minimizes over-clustering, and allows for a reasonable number of statistical comparisons between call types.
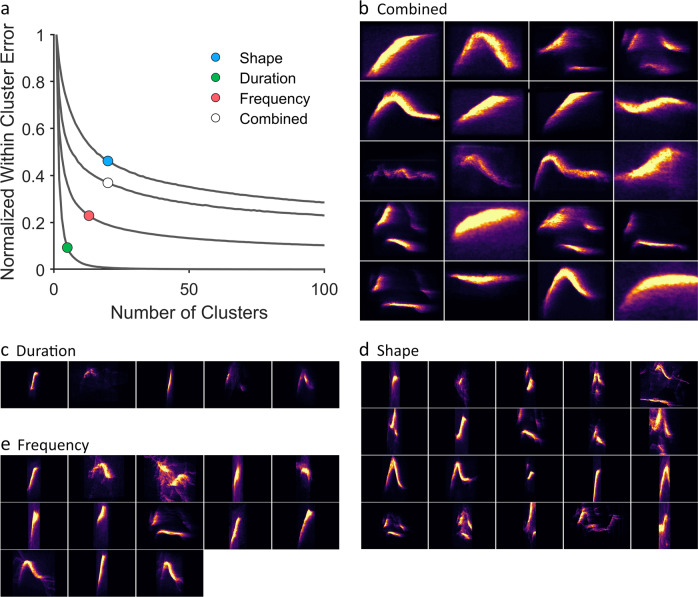
Fig. 4.
DeepSqueak attempts to optimize syllable clustering using k-means and the elbow method. a Normalized within-cluster error is logarithmically reduced by increasing the number of clusters. The elbow of each curve represents the point of diminishing returns for continuing to add clusters. This method attempts to balance reduced within-cluster error while not over-clustering. We tested 4 different parameterizations of USVs to show how input parameters effect clustering. b When clustering using combined and equally weighted shape, duration, and frequency input parameters, DeepSqueak determined the optimal number of syllables to be 20. A mean intensity projection of the top 20 USVs in each cluster is shown with varying time scales to improve clarity. This is the syllable repertoire chosen for syntax analysis in Fig. 5. c When clustering using only duration as the input parameter, DeepSqueak determined the optimal number of syllables to be 5. A mean intensity projection of the top 20 USVs in each cluster are shown on a fixed time scale. d When clustering using only shape as the input parameter, DeepSqueak determined the optimal number of syllables to be 20. A mean intensity projection of the top 20 USVs in each cluster is shown on a fixed time scale. e When clustering using only frequency as the input parameter, DeepSqueak determined the optimal number of syllables to be 13. A mean intensity projection of the top 20 USVs in each cluster are shown on a fixed time scale
The second clustering algorithm implemented is based on dynamic time-warping and an adaptive resonance theory neural network [33]. This method does not require any input for the number of clusters detected, but the thresholds can be changed for determining when a new cluster should be formed. This method is considered experimental and we have made no attempt to optimize it.
Clustering interface
Once USVs are clustered, DeepSqueak will load the clustering graphical user interface. This will generate a complete syllable repertoire, as well as allow the user to view every single call in every cluster. Calls are sorted by similarity to the cluster center and may be inspected for obvious misclassifications which may be manually removed by clicking on the individual sonograms. Clusters can be assigned names and detection files can be updated with classifications. Clusters labeled as “noise” will be automatically removed from all future analyses.
Supervised neural network-based classification
DeepSqueak allows the user to create manual classification categories and label USVs during selection review; however, it is far more efficient to use the aforementioned unsupervised clustering to categorize vocalizations. These clusters may then be manually reviewed, and misclassifications can be removed. The clusters can be named and passed to “Tools→Network Training→Train Network Classifier” which will train a classification network (Table S4). We have included a simple mouse call classification network that classifies mouse vocalizations into five categories (Split, Inverted U, Short Rise, Wave, and Step). These categories are by no means exhaustive and this network is included with DeepSqueak as an example of what is possible when researchers have large datasets of manually classified calls. We have also attempted to optimize unguided clustering but included a network that highlights DeepSqueak’s ability to perform user-guided neural network-based classification. To produce this network, DeepSqueak was used to detect ~56,000 USVs from B6D2F1 mouse recordings obtained from Mouse Tube [34]. These USVs were clustered with k-means clustering and the previously mentioned five categories of USVs were isolated, manually reviewed, labeled, and used to train the classification neural network. All USVs from the aforementioned dataset were then classified using this neural network and an analysis of male mouse syntax during exposure to male mice, female mice, and female urine was performed (Figure S1).
Syntax analysis
DeepSqueak can perform automated syntax analysis from classified USVs. This analysis may be performed on manually classified USVs, supervised classified USVs, or unsupervised classified USVs. Any number of detection files (.mat) or output statistics files (.xlsx) can be loaded into “Tools→Call Classification→Syntax Analysis” and call categories can be selected for analysis. Syntax is analyzed within bouts—bursts of USVs with short inter-call intervals. The inter bout interval may be manually specified by the user. DeepSqueak then calculates transition probabilities between call categories within call bouts, and outputs transition probability tables, heat-maps, and syntax flow paths. This type of analysis can reveal complex patterns of calling that are not evident when considering call frequency and call rate alone.
Results
Software availability and cost
DeepSqueak may be downloaded from “https://github.com/DrCoffey/DeepSqueak”. DeepSqueak is free to use and modify and alternate versions or neural networks may be shared in their own branches. While we recommend using DeepSqueak in conjunction with high-quality condenser microphones such as the UltraSoundGate CM16/CMPA (Avisoft Bioacoustics, Germany), DeepSqueak is capable of analyzing recordings from low-cost USB ultrasonic microphones such as the Ultramic250K (Dodotronic) or M500-384 (Pettersson Elektronik), without sacrificing detection accuracy. These microphones suffer from comparatively lower sensitivity and higher noise but are economical and may allow more units to be purchased by individual labs.
Analysis time
Analysis speed in DeepSqueak is fast and based on the computer’s graphics processor. On a modest NVIDIA Quadro K1100M (CUDA Cores: 384, graphics clock: 705 MHz, dedicated video memory: 2 GB GDDR5) DeepSqueak detects short USVs at least ~10× real speed and long USVs at ~20× real speed. These speeds can be increased to ~40× by using a modern high-powered GPU such as the NVIDIA 1080ti.
Systematic interrogation of detection accuracy across varying conditions
Most USV detection software performs well with high quality, high signal-to-noise recordings. However, real-life experimental conditions usually contain unpredictable noise sources from the operation of the behavioral apparatus, nearby electrical equipment, lights, ventilation, etc., and most previously available USV detection software programs lose accuracy under these conditions since the recordings are suboptimal. In order to compare detection accuracy across varying recording conditions, we measured two signal detection metrics, precision and recall. Precision refers to the ratio of detected true USVs to false positives; this measure is diminished by the over-detection of false positives. Recall refers to the ratio of detected true USVs to total true USVs in the file; this measure is reduced by failing to detect true USVs present in the file. The mean and 95% confidence intervals (CI) for each software’s recall and precision were modeled with binomial distributions.
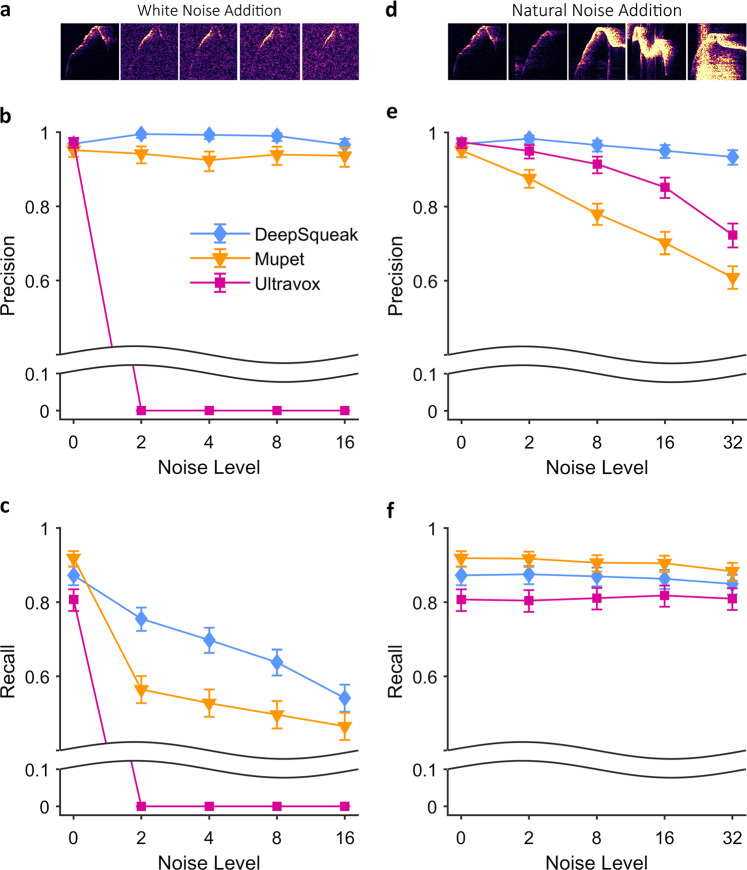
In order to test detection accuracy systematically across varying recording conditions, we manipulated two features of a recording to degrade its quality and compared detection accuracy with three automated programs: DeepSqueak, MUPET [17], and the commercially available software program Ultravox (Noldus; Wageningen, NL). We began with an ideal recording from an online resource, Mouse Tube [34] and added increasing levels of Gaussian “white” noise to the recording, replicating the effect of lower quality signal-to-noise recordings (Fig. 2a). This manipulation did not affect the precision of DeepSqueak or MUPET (Fig. 2b), while Ultravox was no longer able to detect any USVs once any noise was introduced. Lowering signal-to-noise ratio did affect recall with each software program, although DeepSqueak performed markedly better than MUPET across all Gaussian noise levels, with MUPET falling well below the 95% for DeepSqueak’s binomial distribution (Fig. 2c). Next, we progressively added real-life environmental noise recorded in a behavioral suite to the recording, such as mechanical rattling, knocking, and movement by the animals (Fig. 2d). This type of manipulation does not affect recall (Fig. 2f), but it reduced precision in all cases. Still, DeepSqueak maintained the highest precision across all noise addition levels (Fig. 2e) with both Ultravox and MUPET falling well below the 95% CI for DeepSqueak’s binomial distribution.
Fig. 2.
DeepSqueak excels under varying noise conditions. a A single USV shown with progressively increasing white noise addition. Computer generated Gaussian noise added to the entire file 4 times, with the amount of white noise doubling each time. b Adding Gaussian noise does not affect the detection precision for DeepSqueak or MUPET, but Ultravox could no longer detect any calls. c Adding Gaussian noise lowered the recall for MUPET and DeepSqueak, however DeepSqueak maintained the highest recall rate across all noise additions. d Five different calls showing representative additions of natural noise designed to mimic loud experimental noise. Natural noise was added in 4 times in progressively greater amounts and volumes. e Natural noise addition lowered the precision of MUPET and Ultravox by increasing the rate of false positive detection. Due to DeepSqueak’s noise detection neural network, it maintained the highest precision rate across all noise additions. f Natural noise addition did not affect the recall rate of any detection algorithm
Detection accuracy compared to hand scoring in representative recordings
In order to test the ability of DeepSqueak to maintain high detection accuracy across species and call type, we manually scored rat and mouse files as a “gold standard” analysis. When comparing detection accuracy between software, it is important to consider correctly detected “hits”, false negative “misses”, and false positives. We must also consider a measure of call quality. For all files, calls were ranked by tonality, which is a measure of a sound’s distinguishability from noise. The higher a vocalization’s tonality, the more distinguishable it is from background noise. For rats, we analyzed recordings from a saccharin preference test session that contain frequent ~55 kHz USVs associated with positive affect [10]. DeepSqueak maintained the highest recall rate across all tonalities (Figure S2) and also had the lowest miss and false positive rates (Figure S2d,g,j). When analyzing a very low noise mouse recording, all three software programs performed similarly well for high-frequency USVs, although DeepSqueak still had the highest recall rate (Figure S2b), with the lowest miss and false positive rates (Figure S2e,h,k). Finally, both DeepSqueak and Ultravox were equally capable of detecting long ~22 kHz vocalizations from a rat stressed by reward omission of saccharin (Figure S2c,f,i), while MUPET is not designed to detect USVs below 30 kHz.
Tonality-based contour extraction
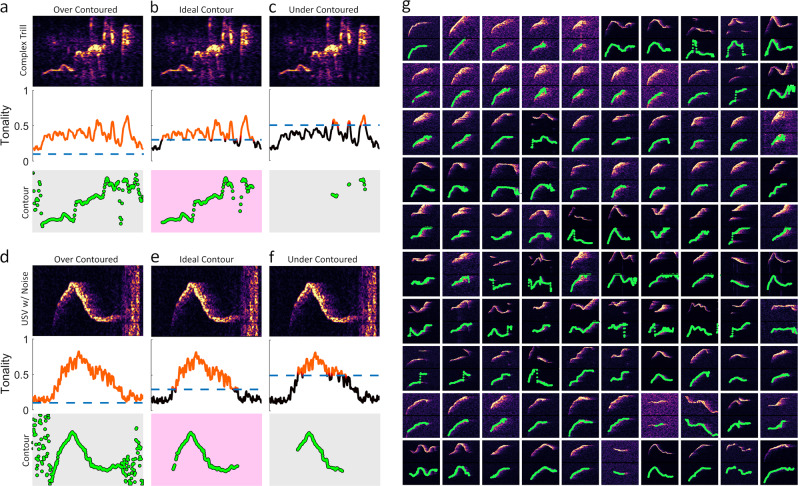
Robust contour detection in DeepSqueak is based around the mathematical concept of tonality, wherein contours are only extracted for samples in which calculated tonality is greater than a defined threshold. Accepting low tonality samples (>0.15) will extract noise (Fig. 3a, d), while accepting only high tonality samples (>0.45) will exclude some of the USV’s contour (Fig. 3c, d). We find that a simple cutoff (>0.3) is ideal to isolate calls from both silence and broad spectrum noise (Fig. 3b, e, g). While a cutoff of 0.3 works in almost all circumstances we have tested, the user retains the ability to alter the contour threshold in real time via a slider.
Fig. 3.
DeepSqueak extracts contours based on the concept of tonality. a A contour extracted from a rat “trill” using only peak amplitude. b A contour extracted from a rat “trill” using a tonality threshold of >0.3 to eliminate silence and noise. c A contour extracted from a rat “trill” using a tonality threshold of >0.45 eliminates too much of the call contour. d A contour extracted from a mouse USV using only peak amplitude. e A contour extracted from a mouse USV using a tonality threshold of >0.3 to eliminate silence and a large piece of broad spectrum noise. f A contour extracted from a mouse USV using a tonality threshold of >0.45 eliminates too much of the call contour. g The first 90 USV contours from a B6D2F1 mouse recording
Elbow optimized syllable repertoires from the B6D2F1 mouse
Syllable repertoires were generated from B6D2F1 mouse USV’s recorded by Chabout and colleagues [3] and obtained from Mouse Tube [34]. DeepSqueak was used to detect ~56,000 USVs from the aforementioned dataset. Contours extracted from these USVs were passed through DeepSqueak’s “elbow optimized” k-means based clustering algorithm with 4 different input parameters. USVs were clustered by duration, frequency, shape, and all three parameters combined and equally weighted. K-means clustering was performed repeatedly from 1 to 100 clusters, and the elbow of the within-cluster error curve was calculated (Fig. 4a). When clustering based on the combination of shape, frequency, and duration, the optimal number of clusters was 20 (Fig. 4b); when clustering based on duration, the optimal number of clusters was 5 (Fig. 4c); when clustering based on shape, the optimal number of clusters was 20 (Fig. 4d); when clustering based on frequency, the optimal number of clusters was 13 (Fig. 4e). Cluster visualizations were generated by calculating the mean intensity projection for the first 20 calls in each cluster. Clusters in Fig. 4c–e are shown with consistent durations, while clusters in Fig. 4b have variable durations, and are magnified for clarity. While there is no perfect way to parameterize USVs or optimize cluster number, we believe using elbow optimization on k-means clusters generated from the combination of shape, duration, and frequency provides an empirical and unbiased method to separate USVs into a reasonable repertoire of commonly produced syllables.
Elbow optimized syllable repertoire from Sprague Dawley rats during saccharin consumption
A syllable repertoire was generated from Sprague Dawley rats recorded during saccharin consumption. DeepSqueak was used to detect ~100,000 USVs which were passed through DeepSqueak’s “elbow optimized” k-means based clustering algorithm. When clustering based on the combination of shape, frequency, and duration, the optimal number of clusters was 18 (Figure S3). This repertoire is similar to the one presented by Wright and colleagues [13], but not identical. While most categories are represented, pure trills appear difficult to isolate via contour-based clustering. Accordingly, there is an ongoing effort to produce a classification neural network from manually labeled calls that conforms to the Wright classifications.
Syntax guided cluster optimization
Cluster number optimization in DeepSqueak is mathematically derived, and based upon reducing the within-cluster error. However, determining the optimal input parameters is not straightforward. We have chosen to explore the effect of clustering input parameters on the syntax of male mice with known behavioral outcomes. The vocalizations in this analysis were previously recorded and analyzed by Chabout and colleagues [3]. They found that mice had a higher likelihood of repeating syllables than transitioning to a new syllable. They also found that male mice exposed to anesthetized males had the highest rate of syllable repetition, while male mice exposed to female urine had the lowest rate. Therefore, our automated syllable clustering aims to maximize syllable repetition in male mice exposed to males and maximize the difference in syllable repetition between mice exposed to males and mice exposed to female urine.
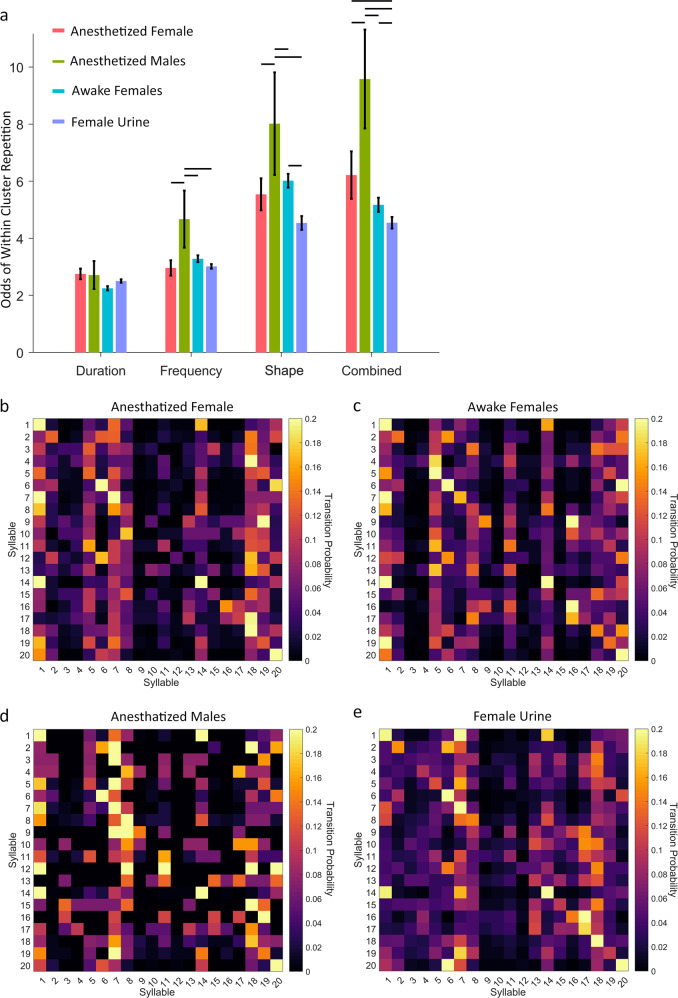
We found that clustering vocalizations based solely on duration produced a syllable repertoire with minimal odds of within-cluster repetition (Fig. 5a) and no significant difference in syllable repetition between groups. Clustering based on frequency produced an increase in within-cluster repetition, as well as an overall difference between groups (Fig. 5a; f(107) = 2.27, p = 0.08). Specifically, mice exposed to anesthetized males showed greater odds of within-cluster repetition than all other groups. Clustering based on shape also produced an increase in within-cluster repetition, as well as an even greater difference between groups (Fig. 5a; f(107) = 5.28, p = 0.002). Specifically, male mice exposed to anesthetized males showed greater odds of within-cluster repetition than all other groups, male and mice exposed to anesthetized or awake females showed greater odds of within-cluster repetition than male mice exposed to female urine. Finally, clustering with all parameters combined produced the most within-cluster repetition, as well as the greatest difference between groups (Fig. 5a; f(107) = 9.56, p < 0.001). Specifically, male mice exposed to anesthetized males showed greater odds of within-cluster repetition than all other groups, and male mice exposed to female urine showed greater odds of within-cluster repetition than all other groups. DeepSqueak also automatically generated syntax flow diagrams for each condition which can be used to visualize transitions probabilities (Figure S4). This result forms the basis for our default parameterization for clustering. We have chosen to parameterize calls with equally weighted combinations of duration, frequency, and shape. Still, DeepSqueak retains the ability for the user to change the weights of each parameter such that the individual user can cluster based around whatever feature they deem most relevant.
Fig. 5.
Behaviorally relevant syntax analysis informs input parameterization for unguided syllable clustering. a The odds of within-cluster repetition are plotted for the elbow optimized syllable repertoires generated using different input parameterizations. Male mice are known to repeat syllables more often than transition to new ones, but they tend to repeat fewer syllables when exposed to female mice or female urine. Clustering based on frequency, shape, or the combination of duration, frequency, and shape produces a significant difference in intra-cluster repetition between groups. Clustering based on combined duration, frequency, and shape input parameters produces the greatest intra-cluster repetition and the greatest difference in syntax between mice exposed to different stimuli. Horizontal lines represent significant post-hoc t-tests (p < 0.05). Transition probability tables produced from the elbow optimized syllable repertoire generated using combined duration, frequency, and shape input parameters for male mice exposed to b anesthetized females, c awake females, d anesthetized males, and e female urine. Male mice exposed to female urine produce the least syllable repetition and the greatest variability in syllable transitions
Finally, we highlighted DeepSqueak’s automated clustering and syntax analysis by replicating the results of Chabout and colleagues [3]. In that report calls were segmented into only 5 clusters, whereas our automated pipeline determined there to be 20 syllable types. Regardless, we arrived at similar conclusions to the original report. We found that males use a simpler syntax when vocalizing around other males, predominantly producing short simple USVs, and transition from complex USVs back to simple USVs (Fig. 5d). Male mice produced more complex patterns of USVs when exposed to females, with the most complex syntax occurring during exposure to female urine (Fig. 5e). This more complex structure of USV syntax has been theorized as a method of attracting females with complex courtship songs [3].
Discussion
USV recording and analysis is a valuable tool for numerous behavioral neuroscience models, including drug abuse, depression, fear or anxiety, neurodegenerative disease, aging, and reward processing. Most conventional behavioral measures such as actions, movement or place preference must be interpreted to reflect emotional state based on the experimental design, whereas USVs provide direct, ethologically relevant expressions that can be measured as an endpoint themselves or used to annotate other coincident behaviors. DeepSqueak’s ability to analyze large numbers of USVs very precisely allows for nuanced explorations of the interplay between vocalizations and behaviors.
DeepSqueak is accurate and flexible primarily because it is built around biomimetic, “deep”, artificial neural networks, allowing it to perform human quality USV detection and classification automatically and at high speeds. Neural networks are ideal for signal detection problems where the signal is variable and often embedded in noise. We have shown systematically that DeepSqueak is capable of detecting USVs with low signal or in high environmental noise recordings. These two features make DeepSqueak robust and generalizable to myriad experimental conditions. The robust nature of DeepSqueak’s detection architecture also makes recording USVs with less expensive equipment significantly more feasible. DeepSqueak is also fast compared to other automatic detection pipelines, allowing researchers to add USVs as a rapid and simple emotional assay in experiments where USVs are not the primary outcome measure. Further, DeepSqueak is flexible and customizable by the end user. If the release version of DeepSqueak is unfamiliar with a particular experimental noise and as a result detects that noise as a call, it is simple for the end user to create a new noise detection and elimination network from their own recordings. This is possible without the need for programming capabilities, or an understanding of how neural networks are trained using simple, point and click menus. The DeepSqueak interface also retains all of the features necessary for manual analyses alongside automated analyses.
One major hurdle for USV researchers, beyond detection and de-noising, has been categorizing USVs into syllable types and unraveling the meaning of their syntactic structure. Clustering USVs is an extremely difficult problem to optimize, and to date few people have tried to automate the process. One such attempt was by Van Segbroeck and colleagues, creators of MUPET [17]. In our experience, MUPET is an extremely capable software package that revolutionized automated syllable clustering. In MUPET, clustering is applied on the spectral magnitude of the segmented syllables, whereas clustering in DeepSqueak is amplitude invariant and contour-based. To compare these methods, we analyzed the same file with DeepSqueak and MUPET twice; once with a full volume file and once with the volume reduced by 50%. DeepSqueak’s clustering placed calls in the same category despite amplitude changes at a substantially higher rate than MUPET (Figure S5). This is important because most behavioral experiments include freely moving animals that produce calls of different volume, and it would be beneficial to maintain high fidelity of call categories irrespective of the animal’s position relative to the microphone. The clustering models generated by DeepSqueak may also be saved and applied to future datasets.
Using fully automated clustering with elbow optimization, we identified 20 syllables produced by the B6D2F1 mouse, whereas Van Segbroeck and colleagues identified 100–140 syllables. We favor the perspective that produces a smaller number of essential call types, but with substantial variation within classes and a lack of distinct boundaries. This lack of distinct boundaries between syllables can be visualized directly by plotting the call sonograms in t-distributed stochastic neighbor embedding (t-SNE) space (Figure S6) and is supported by the high correlation between many syllable shapes when they are separated into ~100 categories [17]. DeepSqueak provides the first fully automated and reproducible way to categorize calls into a limited number of syllables. Finally, DeepSqueak offers the first fully automated syntax analysis that can be performed on an arbitrary number of syllables. Using these features, we have shown that our automatically generated syllable repertoire for B6D2F1 mice contains behaviorally relevant syntactic information comparable to manual analysis. It is our hope that the many features of DeepSqueak will improve the accuracy, ease, reproducibility, and meaningfulness of future USV analyses.
Funding and disclosure
Supported by grants R01MH106532 and P50MH106428. The authors declare no competing interests.
Supplementary information
Acknowledgements
The authors thank Dr. David J. Barker, Dr. Aaron M. Johnson, Dr. David Euston, and Dr. Jonathan Chabout for their contribution of vocalization recordings and Dr. Michele Kelly for editing.
Author contributions
Kevin Coffey and Ruby Marx designed and coded the software, created the figures, and wrote and edited the manuscript. John Neumaier wrote and edited the manuscript.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kevin R. Coffey, Ruby E. Marx
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41386-018-0303-6).
References
- 1.Barker DJ, et al. Ultrasonic vocalizations: evidence for an affective opponent process during cocaine self-administration. Psychopharmacology. 2014;231:909–18. doi: 10.1007/s00213-013-3309-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Browning JR, Whiteman AC, Leung LY, Lu XM, Shear DA. Air-puff induced vocalizations: a novel approach to detecting negative affective state following concussion in rats. J Neurosci Methods. 2017;275:45–9. doi: 10.1016/j.jneumeth.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 3.Chabout J, Sarkar A, Dunson DB, Jarvis ED. Male mice song syntax depends on social contexts and influences female preferences. Front Behav Neurosci. 2015;9:76. doi: 10.3389/fnbeh.2015.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu RC, Miller KD, Merzenich MM, Schreiner CE. Acoustic variability and distinguishability among mouse ultrasound vocalizations. J Acoust Soc Am. 2003;114:3412–22. doi: 10.1121/1.1623787. [DOI] [PubMed] [Google Scholar]
- 5.Portfors CV. Types and functions of ultrasonic vocalizations in laboratory rats and mice. J Am Assoc Lab Anim Sci. 2007;46:28–34. [PubMed] [Google Scholar]
- 6.Seagraves KM, Arthur BJ, Egnor SE. Evidence for an audience effect in mice: male social partners alter the male vocal response to female cues. J Exp Biol. 2016;219:1437–48. doi: 10.1242/jeb.129361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chabout J, et al. A Foxp2 mutation implicated in human speech deficits alters sequencing of ultrasonic vocalizations in adult male mice. Front Behav Neurosci. 2016;10:197. doi: 10.3389/fnbeh.2016.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hernandez C, Sabin M, Riede T. Rats concatenate 22 kHz and 50 kHz calls into a single utterance. J Exp Biol. 2017;220:814–21. doi: 10.1242/jeb.151720. [DOI] [PubMed] [Google Scholar]
- 9.Borta A, Wöhr M, Schwarting R. Rat ultrasonic vocalization in aversively motivated situations and the role of individual differences in anxiety-related behavior. Behav Brain Res. 2006;166:271–80. doi: 10.1016/j.bbr.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Burgdorf J, Panksepp J, Moskal JR. Frequency-modulated 50 kHz ultrasonic vocalizations: a tool for uncovering the molecular substrates of positive affect. Neurosci Biobehav Rev. 2011;35:1831–6. doi: 10.1016/j.neubiorev.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Jelen P, Soltysik S, Zagrodzka J. 22-kHz ultrasonic vocalization in rats as an index of anxiety but not fear: behavioral and pharmacological modulation of affective state. Behav Brain Res. 2003;141:63–72. doi: 10.1016/S0166-4328(02)00321-2. [DOI] [PubMed] [Google Scholar]
- 12.Knutson B, Burgdorf J, Panksepp J. Ultrasonic vocalizations as indices of affective states in rats. Psychol Bull. 2002;128:961. doi: 10.1037/0033-2909.128.6.961. [DOI] [PubMed] [Google Scholar]
- 13.Wright JM, Gourdon JC, Clarke PB. Identification of multiple call categories within the rich repertoire of adult rat 50-kHz ultrasonic vocalizations: effects of amphetamine and social context. Psychopharmacology. 2010;211:1–13. doi: 10.1007/s00213-010-1859-y. [DOI] [PubMed] [Google Scholar]
- 14.Panksepp JB, et al. Affiliative behavior, ultrasonic communication and social reward are influenced by genetic variation in adolescent mice. PLoS ONE. 2007;2:e351. doi: 10.1371/journal.pone.0000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scattoni ML, Ricceri L, Crawley JN. Unusual repertoire of vocalizations in adult BTBR T+tf/J mice during three types of social encounters. Genes Brain Behav. 2011;10:44–56. doi: 10.1111/j.1601-183X.2010.00623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugimoto H, et al. A role for strain differences in waveforms of ultrasonic vocalizations during male–female interaction. PLoS ONE. 2011;6:e22093. doi: 10.1371/journal.pone.0022093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Segbroeck M, Knoll AT, Levitt P, Narayanan S. MUPET-mouse ultrasonic profile ExTraction: a signal processing tool for rapid and unsupervised analysis of ultrasonic vocalizations. Neuron. 2017;94:465–85. doi: 10.1016/j.neuron.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanson JL, Hurley LM. Female presence and estrous state influence mouse ultrasonic courtship vocalizations. PLoS ONE. 2012;7:e40782. doi: 10.1371/journal.pone.0040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang M, Loureiro D, Kalikhman D, Crawley JN. Male mice emit distinct ultrasonic vocalizations when the female leaves the social interaction arena. Front Behav Neurosci. 2013;7:159. doi: 10.3389/fnbeh.2013.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimsley JM, Monaghan JJ, Wenstrup JJ. Development of social vocalizations in mice. PLoS ONE. 2011;6:e17460. doi: 10.1371/journal.pone.0017460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ellenbroek B, Youn J. Rodent models in neuroscience research: is it a rat race? Dis Models Mech. 2016;9:1079–87. doi: 10.1242/dmm.026120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farabet C, Couprie C, Najman L, Lecun Y. Learning hierarchical features for scene labeling. IEEE Trans Pattern Anal Mach Intell. 2013;35:1915–29. doi: 10.1109/TPAMI.2012.231. [DOI] [PubMed] [Google Scholar]
- 24.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–44. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 25.Sainath TN, et al. Deep convolutional neural networks for large-scale speech tasks. Neural Netw. 2015;64:39–48. doi: 10.1016/j.neunet.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 26.Ren S, He K, Girshick R, Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans Pattern Anal Mach Intell. 2017;39:1137–49. doi: 10.1109/TPAMI.2016.2577031. [DOI] [PubMed] [Google Scholar]
- 27.Barker DJ, Herrera C, West MO. Automated detection of 50-kHz ultrasonic vocalizations using template matching in XBAT. J Neurosci Methods. 2014;236:68–75. doi: 10.1016/j.jneumeth.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burkett ZD, Day NF, Penagarikano O, Geschwind DH, White SA. VoICE: a semi-automated pipeline for standardizing vocal analysis across models. Sci Rep. 2015;5:10237. doi: 10.1038/srep10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reno JM, Marker B, Cormack LK, Schallert T, Duvauchelle CL. Automating ultrasonic vocalization analyses: the WAAVES program. J Neurosci Methods. 2013;219:155–61. doi: 10.1016/j.jneumeth.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zala SM, Reitschmidt D, Noll A, Balazs P, Penn DJ. Automatic mouse ultrasound detector (A-MUD): a new tool for processing rodent vocalizations. PLoS One. 2017;12:e0181200. doi: 10.1371/journal.pone.0181200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson AM, Grant LM, Schallert T, Ciucci MR. Changes in Rat 50-kHz ultrasonic vocalizations during dopamine denervation and aging: relevance to neurodegeneration. Curr Neuropharmacol. 2015;13:211–9. doi: 10.2174/1570159X1302150525122416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kershenbaum A, Sayigh LS, Janik VM. The encoding of individual identity in dolphin signature whistles: how much information is needed? PLoS One. 2013;8:e77671. doi: 10.1371/journal.pone.0077671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deecke VB, Janik VM. Automated categorization of bioacoustic signals: avoiding perceptual pitfalls. J Acoust Soc Am. 2006;119:645–53. doi: 10.1121/1.2139067. [DOI] [PubMed] [Google Scholar]
- 34.Torquet EEN. Mouse Tube. 2015. 〈https://mousetube.pasteur.fr〉.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.