Abstract
Seminal fluid proteins (SFPs) are crucial mediators of sexual selection and sexual conflict. Recent studies have chiefly focused on environmentally induced plasticity as one source of variation in SFP expression, particularly in response to differing sperm competition levels. However, understanding the evolution of a trait in heterogenous environments requires estimates of both environmental and genetic sources of variation, as well as their interaction. Therefore, we investigated how environment (specifically mating group size, a good predictor of sperm competition intensity), genotype and genotype-by-environment interactions affect seminal fluid expression. To do so, we reared 12 inbred lines of a simultaneously hermaphroditic flatworm Macrostomum lignano in groups of either two or eight worms and measured the expression levels of 58 putative SFP transcripts. We then examined the source of variation in the expression of each transcript individually and for multivariate axes extracted from a principal component analysis. We found that mating group size did not affect expression levels according to the single transcript analyses, nor did it affect the first principal component (presumably representing overall investment in seminal fluid production). However, mating group size did affect the relative expression of different transcripts captured by the second principal component (presumably reflecting variation in seminal fluid composition). Most transcripts were genetically variable in their expression level and several exhibited genotype-by-environment interactions; relative composition also showed high genetic variation. Collectively, our results reveal the tightly integrated nature of the seminal fluid transcriptome and provide new insights into the quantitative genetic basis of seminal fluid investment and composition.
Subject terms: Evolutionary genetics, Sexual selection
Introduction
Seminal fluid proteins (SFPs), which are transferred together with sperm during mating, have numerous functions that can strongly impact on both male and female fitness. Along with their functions directly related to sperm viability such as activation and nourishment, SFPs also have many unique functions that have likely evolved as an adaptive response to post-mating sexual selection. In particular, sperm competition is a selective force that is expected to profoundly shape the seminal fluid content of the ejaculate (see reviews in Avila et al. 2011; Simmons and Fitzpatrick 2012; Perry et al. 2013). Indeed, to date, many of the best documented functions of seminal fluid are particularly related to male competitive fertilization success, such as the manipulation of egg production and egg laying rates (Chapman et al. 2003; Koene et al. 2009), inhibition of future re-mating and female attractiveness (Lung and Wolfner 2001; Chapman et al. 2003; LaFlamme et al. 2012), and displacement of rival sperm (Clark et al. 1995; Den Boer et al. 2010; Yamane et al. 2015).
Support for the notion that SFPs will often be subject to post-mating sexual selection through sperm competition (Parker 1970) comes from a range of theoretical and empirical studies at both intra- and interspecific levels. For example, studies comparing taxa with different mating strategies (e.g. monogamous versus polygamous) have demonstrated a positive relationship between the intensity of sperm competition and the strength of positive Darwinian selection on specific SFPs (e.g. Kingan et al. 2003; Dorus et al. 2004; Ramm et al. 2008; Civetta and Reimer 2014). On the other hand, at the intraspecific level, recent theoretical models predict plastic responses in SFP investment as well as their strategic allocation—depending on their different functions—according to the risk or intensity of sperm competition (Hodgson and Hosken 2006; Cameron et al. 2007; Dhole and Servedio 2014). Empirical studies have now shown some evidence that SFP production can be energetically costly (e.g. Thomsen et al. 2006; Friesen et al. 2015, though only the latter study separated the energetic costs of sperm and seminal fluid), and suggest that the production as well as allocation of SFPs can indeed be plastically adjusted in response to sperm competition. For example, in Drosophila melanogaster, when males were exposed to a competitor in their pre-mating environment, they transferred more of two key SFPs during mating (Wigby et al. 2009). In addition, males can modify the relative ratios of transcript expression levels of different SFPs in their ejaculate (Fedorka et al. 2011; see also Mohorianu et al. 2017) and transferred proportionally more of their SFP resource in response to a higher male density in a pre-mating environment, which is presumably a predictor of the expected future sperm competition level (Wigby et al. 2016). Moreover, males mating with a virgin female transfer more ovulin—a protein responsible for inducing oviposition—than males mating with a recently mated female (Sirot et al. 2011). Similarly, dynamic transcript expression of SFPs in response to sperm competition risk and intensity has recently been reported in Australian field crickets, Teleogryllus oceanicus (Simmons and Lovegrove 2017; Sloan et al. 2018). In rodents, a recent proteomics analysis that investigated several secreted SFPs in male house mice (Mus musculus) showed that total SFP production, as well as its relative composition, changes plastically in response to cues of sperm competition risk (Ramm et al. 2015). Altogether, this emerging evidence clearly suggests that males can strategically invest in SFPs according to the risk and intensity of sperm competition.
Genetic sources of variation in SFP expression could also be an extremely important factor causing the widespread variation usually observed in male sperm competitiveness and reproductive success (e.g. Simmons and Parker 1992; Lewis et al. 2012; Zhang et al. 2013). In comparison with the number of studies that have demonstrated a social environment effect on SFP production and allocation, however, to date only a few studies have investigated genetic variation in SFP expression, which can also be significant (Smith et al. 2009; Baer et al. 2012; Goenaga et al. 2015; Mangels et al. 2015). Furthermore, some studies showed that genetic polymorphism in specific SFPs is tightly linked to variation in sperm competitive ability (Fiumera et al. 2005, 2007; Zhang et al. 2013). Collectively, these studies provide good evidence for both genotypic and environmental variation in seminal fluid. This suggests that the prerequisites for genotype-by-environment interaction (GEI) effects are met, but whether there is actually genetic variation for phenotypic plasticity in SFP expression is yet to be addressed.
Genotype-by-environment interaction occurs whenever there is genotype-specific variation in reaction norms (West-Eberhard 1989; Scheiner et al. 1999). This can be further characterized according to whether one observes ‘ecological crossover’ (i.e., whether or not the reaction norms of different genotypes cross from one environment to the other) (Danielson-François et al. 2006; Kokko and Heubel 2008), implying changes in rank order of genotypes across environments. GEI is important in the context of sexual selection, where it provides a potentially widespread explanation for the maintenance of standing genetic variation in male sexually selected traits that should otherwise be rapidly depleted through strong directional selection (Kokko and Heubel 2008; Ingleby et al. 2010; Hunt and Hosken 2014). In fact, a number of studies have revealed that many other sexually selected traits do exhibit GEI (Jia et al. 2000; Danielson-François et al. 2006, 2009, Ingleby et al. 2010, 2013; Hunt and Hosken 2014), in particular male traits involved in pre-mating sexual selection (Ingleby et al. 2010; see also Hunt and Hosken 2014). Despite the fact that post-mating traits are also under strong selection—and the fact that we nevertheless often observe persistent genetic variation in these traits (e.g. Evans 2010; Immler et al. 2012; Lewis et al. 2012)—only few studies have directly addressed the existence of GEI for post-mating sexually selected traits. So far, these studies have shown GEI for testis size (Nystrand et al. 2011; Marie-Orleach et al. 2017), sperm characteristics (e.g. Ward 1998, 2000; Simmons and Kotiaho 2002; Snook et al. 2010; Evans et al. 2015) and offensive and defensive sperm competitive ability (Lewis et al. 2012).
Besides GEI, another important aspect to understand selection acting on, or predict the future evolutionary change of, SFPs is the nature of genetic correlations among them. Due to the fact that SFPs are often simultaneously produced in the same somatic tissues, and often have overlapping or complementary functions, we might expect that strong genetic correlations between SFPs could exist, and—depending on the sign of the correlation—this could either facilitate or impede evolutionary responses. For example, negative genetic correlations between SFP loci might cause antagonistic pleiotropy, and thus lead to evolutionary constraints, whilst also potentially helping to explain the maintenance of standing genetic variation. Indeed, antagonistic pleiotropy was demonstrated for some SFP genes (Fiumera et al. 2007). On the other hand, genetically variable SFP transcripts were actually found to be highly positively correlated in their expression between a set of inbred lines of D. melanogaster (Ayroles et al. 2009, 2011), suggesting that strong genetic correlations could indeed be an important factor impacting on the evolution of SFPs. This means we will often need to treat seminal fluid as a multivariate trait, and not simply investigate individual SFPs.
In our study, we sought to characterize genetic and environmental sources of variation in seminal fluid transcript expression, and their interaction, in the simultaneously hermaphroditic, free-living flatworm Macrostomum lignano. Given that it is a complex, multivariate phenotype, we firstly aimed to describe unique axes of variation in the SFP transcriptome. We then partitioned this variance into environmental and genetic components and their interaction, for both overall seminal fluid transcript investment (that is, the overall expression level of all SFP transcripts, assuming that this would correspond to overall investment in seminal fluid) and relative seminal fluid transcript expression (that is, transcript expression levels relative to each other, assuming this will likely translate into altered seminal fluid composition) that is captured by these unique axes. Moreover, we investigated each seminal fluid transcript individually to assess the contribution of each factor to individual transcript expression and calculated genetic correlations among expression level of the transcripts.
Due to the fact that we investigate these questions in a simultaneous hermaphrodite, there are several additional aspects that need to be considered. First, because simultaneous hermaphrodites can produce both sperm and eggs at the same time, there is an additional allocation decision concerning the optimal strategic investment into their male or female sex function (i.e. sex allocation) and seminal fluid investment needs to be considered as a potentially significant aspect of male allocation (Charnov 1979, 1982; Schärer 2009). Importantly in this context, theory predicts that under conditions with a small mating group size (i.e., the number of mating partners an individual has within a reproductive period, plus one—itself), the importance of competition between (related) sperm from the same individual/donor, so called ‘local sperm competition’, increases (Schärer 2009; Schärer and Pen 2013), and as a result individuals tend to invest more into their female sex function (Charnov 1979, 1982; Schärer 2009; Schärer and Pen 2013). On the other hand, when the mating group size is large, the importance of local sperm competition decreases, while the intensity of competition over fertilization between (unrelated) sperm from different individuals/donors correspondingly increases, favouring a more male-biased sex allocation. These predictions are empirically well supported by several studies, especially in terms of testis size and investment in sperm production (e.g. Schärer and Ladurner 2003; Tan et al. 2004; Schärer and Vizoso 2007; Janicke and Schärer 2009; Giannakara et al. 2016). Second, because in copulating simultaneous hermaphrodites it has been argued that individuals will often be willing to receive sperm in order to be able to donate sperm, the receiver might be especially likely to evolve post-mating mechanisms for selecting and/or removing previously received sperm (Charnov 1979; Michiels 1998; Leonard 2006; Schärer and Pen 2013). For example, adaptations for sperm removal have been shown in leeches and arrowworms (reviewed by Michiels 1998), and potentially in M. lignano (Schärer et al. 2004; Marie-Orleach et al. 2013). This could drive the evolution of SFPs with unique functions to manipulate the sperm removal behaviour of their partners that cause changes in relative investment of specific SFPs and thus affect seminal fluid composition. Finally, because each mating partner possesses both a male and female sex function in simultaneous hermaphrodites, this potentially creates unique targets for SFP action such as manipulating the sex allocation strategies of partners (Charnov 1979; Nakadera et al. 2014; Schärer 2014; Schärer and Ramm 2016).
Our study organism M. lignano is an excellent model for studying variation in seminal fluid in response to sperm competition. It is already well established that several traits which could be subject to post-mating sexual selection are plastically adjusted in response to mating group size. When the mating group size increases, male reproductive traits such as testis size (Schärer and Ladurner 2003; Janicke and Schärer 2009), testicular activity (Schärer et al. 2004), sperm production rate (Schärer and Vizoso 2007) and spermatogenesis speed (Giannakara et al. 2016) increase in response to increasing intensity of sperm competition. Notably, a recent study has also just revealed that seminal fluid transcript expression plastically changes between isolated worms and those kept in groups of octets (Ramm et al. in revision); we here sought to expand upon this insight by incorporating genetic variation and GEI, as well as by treating seminal fluid as a multivariate phenotype.
Materials and methods
Study organism
Macrostomum lignano (Rhabditophora: Macrostomida) is a reciprocally copulating flatworm in which mating partners simultaneously donate and receive ejaculates (Schärer and Ladurner 2003; Ladurner et al. 2005). Worms copulate frequently under laboratory conditions and transfer sperm together with seminal fluid via the stylet (male copulatory organ) to the partner’s female antrum (sperm storage organ) (Schärer et al. 2004; Vizoso et al. 2010). Seminal fluid is produced by prostate gland cells located around the stylet (Ladurner et al. 2005; Vizoso et al. 2010), and includes a complex mixture of proteins (Weber et al. 2018).
The worm cultures are kept in six-well tissue culture plates (Techno Plastic Products AG, Trasadingen, Switzerland) containing artificial seawater (ASW, 32‰ salinity) at 20 °C, 60% relative humidity, 14:10 light:dark cycle and fed with the algae Nitzschia curvilineata (Guillard and Ryther 1962). All animals in this experiment were taken from 12 highly inbred lines (called DV lines), namely DV1, the line with a sequenced genome (Janicke et al. 2013; Wasik et al. 2015), plus DV3, DV8, DV13, DV28, DV69 and DV71 (Sekii et al. 2013) and DV47, DV49, DV61, DV75 and DV84 (Vellnow et al. 2017). These lines belong to a larger set of inbred lines that were originally generated at the University of Innsbruck and are now maintained at the University of Basel. Briefly, each of these inbred lines was started by crossing two virgin worms extracted from outbred cultures, and kept over more than 40 generations of full- or half-sibling inbreeding (see Vellnow et al. (2017) for further details).
Mating group size experiment
We implemented a full factorial design to evaluate (i) the main effect of social environment, (ii) the main effect of genotype and (iii) the GEI effect for the overall and relative expression of SFP transcripts, treating seminal fluid as a multivariate trait by applying principal component analyses. Moreover, we evaluated the effect of environment, genotype and GEI for each SFP transcript individually.
In order to control for potential common environment and/or parental effects, we began by splitting worms from each genotype into six independent blocks (Fig S1). For each block, the genotypes were randomly distributed on two six-well tissue culture plates (Techno Plastic Products (TPP) AG, Trasadingen, Switzerland), with each block containing one complete set of the 12 genotypes. For each block × genotype combination (6 × 12 = 72 in total), the well was initiated by transferring 20 randomly chosen hatchlings of that genotype from the main stock cultures. Each well contained 5 ml 32‰ ASW and ad libitum food. Once they were adult, worms from these replicated blocks were allowed to lay eggs, and 60 offspring were collected from each well and randomly distributed into 24-well tissue culture plates (TPP AG, Trasadingen, Switzerland) to form mating groups of either pairs or octets, i.e. two or eight hatchlings per well, respectively. The mating group sizes were allocated in a way that balanced for any potential position effects on the 24-well tissue culture plates (Brauer et al. 2007; Janicke and Schärer 2009). We set up six independent replicates of each mating group per genotype × block × environment combination (6 replicates × 12 genotypes × 6 blocks × 2 environments = 864 in total). All wells contained 1 ml of 32‰ ASW and worms were fed weekly with a standard per capita amount of algae that ensured ad libitum food conditions per individual. Growing worms were transferred to freshly prepared plates every week to avoid possible group size or density changes caused by the extended presence of hatched and maturing offspring. Transferring groups to fresh plates was repeated for 7 weeks to ensure sexual maturity of all inbred lines (which differ somewhat in their maturation time). At the end of this experimental phase, seven wells (out of 864) in which we had never observed offspring, which were all from the pair treatment, were discarded from the experiment.
Before the worms were prepared for seminal fluid transcript expression level measurements, some morphological and behavioural assays were carried out. Although the data from these analyses are not reported here, for completeness, we briefly summarize these measurements. The measurements started when the worms were at age 51 days, by choosing, for each genotype, both worms from three random pairs and two randomly chosen worms from three random octets from each of the six blocks. Therefore, in total, 36 pair worms and 36 octet worms were measured per genotype, from 18 independent pair replicates and 18 independent octet replicates, respectively. Each selected individual was then paired with a standardized, colour-labelled (Marie-Orleach et al. 2013) virgin adult worm for 2 h, during which time we recorded videos to score their mating behaviours. Subsequently, every second individual was imaged to collect morphological data using standard protocols (Schärer and Ladurner 2003). Thereafter, for each genotype, six samples (one per block) out of the 18 that were recorded and imaged were transferred individually to 25 µl RNALater® solution (Ambion, Austin, TX, USA), which immediately stabilizes RNA, for SFP transcript expression measurements.
A previous study showed that worms are more female-biased in terms of their sex allocation when they were isolated compared to pairs or octets, presumably due to low allocation to sperm production when no mating can occur (Schärer and Janicke 2009). A similar pattern has also been shown for seminal fluid transcript expression in one inbred line (DV1) of M. lignano, in which expression level of putative SFP transcripts were the lowest in isolated worms (Ramm et al. in revision). In addition, sex allocation is phenotypically flexible in adult worms, and rapidly adjusted to the prevailing social conditions (Brauer et al. 2007), suggesting the possibility that worms can downregulate seminal fluid transcript expression in non-mating conditions. Therefore, after the experimental phase just described, some of the remaining individuals out of 18 replicates were removed from their social groups and kept in isolation for 6−7 weeks, to enable us to measure seminal fluid transcript expression level also under non-mating conditions. After this time, the chosen isolated samples were transferred individually to 25 µl RNALater® solution for seminal fluid transcript expression measurements. However, because the age of isolated samples was different to the pairs and octets, and moreover because they were not treated at exactly the same time or under the same conditions, we do not include the data from isolated samples in the main statistical analyses. Rather, we analysed these samples to address a secondary, supplementary question of whether there is genetic variation in baseline level of SFP transcript expression among the lines under non-mating conditions.
Transcript expression measurements
Total RNA was extracted from 184 worms where we chose one individual sample per genotype−environment combination from each block randomly and therefore we eliminated the block effect statistically by choosing one sample per block. In total, we chose samples of 71 pairs (one sample failed during RNA extraction), 72 octets, and 41 isolated (only a subset of all isolated samples) for transcript measurements. Extractions were made by using the Reliaprep™ RNA tissue miniprep system (Promega, USA) as 10 µl total RNA solution following the manufacturer’s instructions, which also includes a DNase step to remove genomic DNA contamination. Afterwards, reverse transcription was performed using 4.5 µl of total RNA with the cDNA-Synthesis Kit H Plus (VWR Peqlab, Germany).
Putative SFP transcripts were selected according to three criteria, based on an earlier study that identified prostate-limited transcripts in M. lignano (Weber et al. 2018), which was itself based on previous studies that investigated the positional (Arbore et al. 2015) and socially sensitive (Ramm et al. in revision) differential expression of transcripts. First, Arbore et al. (2015) identified 366 transcripts that show heightened expression in the tail fragment where the prostate glands (responsible for seminal fluid production) and other gland systems such as adhesive glands are located. Second, Ramm et al. (in revision) expected that if some of those transcripts represent an aspect of male allocation during mating in M. lignano, they would show a response in their expression to changing social group size (isolated vs. groups). Based on those transcripts that exhibited such differential expression between isolated and grouped worms which were measured in one inbred line, Ramm et al. (in revision), thus considered 150 out of the 366 transcripts as candidate prostate-specific components of seminal fluid. Finally, because the tail is also the location of the developing eggs and other female and male reproductive organs, Weber et al. (2018) performed a whole mount in situ hybridization screen in order to identify which of these 150 differentially expressed tail-specific candidates are expressed exclusively in the prostate gland cells, and thus most likely to putatively encode seminal fluid components. Specifically, they tested 146 out of these 150 differentially expressed tail-specific candidates, identifying 76 transcripts with prostate-limited expression, and we attempted to include all of these in our study. Moreover, Weber et al. (2018) showed that at least 64 (out of 65 which could be analysed) are predicted to be secreted proteins. Supporting the predictions and results of these earlier studies, Lengerer et al. (2018) also showed that only 11 out of 166 non-differentially expressed tail-specific candidates from Ramm et al. (in revision) that were screened in a separate effort to identify bioadhesion candidates exhibit expression in prostate gland cells. We included three of these transcripts in our study, plus a fourth additional transcript (MLRNA110815 _2216.1), which based on Arbore et al. (2015) increased expression in the testis- as well as the tail-region, but for which we had in situ hybridization evidence for prostate-limited expression (Fig. S2).
Using the software Primer 3 (Untergasser et al. 2012), we were able to design primer pairs for 58 out of these 76 transcripts as well as for all four additional prostate-limited transcripts not originally identified as seminal fluid transcript candidates as explained above; thus, in total, primer pairs were designed for 62 putative SFP transcripts (Table S1). One limiting factor to design primers for all putative SFP transcripts was that all primers should be amplified simultaneously in the instrument used for this study, which required the same specific annealing temperature (59–60 °C) for all primer pairs.
For pre-amplification of cDNA samples, we pooled primers of 62 SFP transcripts and seven housekeeping genes (see below) as internal controls. (Note that 11 primers of other tissue-limited transcripts were also included in the expression measurements but are not reported here.) Pre-amplification was performed as 14 cycles of PCR amplification using the TaqMan PreAmp Master Mix (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. The unincorporated primers were digested by adding Exonuclease I (ThermoScientific, Waltham, MA, USA) after pre-amplification. These pre-amplified samples were diluted 18-fold prior to analysis in the BioMark RealTime PCR System (Fluidigm Corporation, CA, USA). RT-qPCR was carried out using a total of four 96.96 Dynamic Array integrated fluidic circuits (IFCs) on the BioMark System (Fluidigm, CA, USA) following the manufacturer’s protocol (Spurgeon et al. 2008) and using 2× SsoFast EvaGreen Supermix with low ROX (BioRad, CA, USA). Individual samples were equally distributed in terms of experimental factors into each array to measure against all primers which were also equally distributed. Thus, each array contained one half of the samples and one half of the primers, and its complementary second array included the same half of the samples with the other half of the primers; note that all arrays included all primers of internal controls. The Ct (Cycle threshold) values of 184 experimental samples, eight negative controls without template cDNA and four samples of dilution tests against 80 primers were extracted using BioMark Real-Time PCR Analysis Software v2.0 (Fluidigm). The optimal Ct range for the Biomark system is expected to be 6−25 (Pieprzyk and High 2009). Therefore, Ct values greater than 25 and those that could not be quantified including negative controls were removed, amounting to approximately 11% of the total data. We obtained duplicate measurements of the same transcript for each sample (i.e., technical replicates). The coefficient of variation (CV) of technical replicates was calculated and samples with CV bigger than 10 (which is only 0.1% of the data) and samples with failed technical replicates (4% of the data) were assumed to be erroneous. This 4.1% erroneous technical replicates where the variation between replicates were either large or there was only one measurement might have happened because of pipetting errors or damaged chips in the array and were therefore removed from the data to decrease measurement error. These erroneous technical replicates appeared to be randomly distributed between arrays and samples.
For the remaining transcripts, a single Ct value was calculated as the mean of the two technical replicates. One practical issue with large-scale transcript expression data is that, due to technical problems (inoperative primers, defective chips on the array or excluded technical replicates), such data sets usually contain missing values, whereas subsequent analysis requires a complete data matrix. The R package ‘VIM’ was therefore used to calculate the percentage of the missing values per transcript, after removing two inoperative internal control primers, 11 other tissue-limited primers, dilution test samples and the complete set of ‘isolated’ samples (not included in the main analysis, see above). In this refined dataset including expression value of 142 samples against 62 SFP transcripts, missing values amounted to 6.2% of the potential total, but were somewhat unequally distributed across the different transcripts: more than half of these missing values were for just four transcripts, and the rest were seemingly randomly distributed across the remaining transcripts and seemed also not to be biased towards particular genotypes and/or environments. Therefore, we excluded only the four most negatively affected transcripts. To overcome the limitation of missing data, the remaining missing values were then imputed using the R package missMDA, which uses the regularized iterative principal component analysis (PCA) algorithm to estimate the missing values based on the available data (Josse and Husson 2012).
Finally, relative expression for each transcript was calculated as the efficiency-calibrated relative expression ratio (Pfaffl 2001), using the germ cell-related gene macpiwi as an internal control (Pfister et al. 2007). macpiwi was chosen from among the seven housekeeping genes—which also included macactin (Pfister et al. 2007), hsp90, actp, tubb, cox4 and gapdh (Plusquin et al. 2012). Primer pairs of gapdh and tubb that were published by Plusquin et al. (2012) did not work sufficiently with the Fluidigm instrument. To select an appropriate control from among the remaining transcripts, we first determined which of these internal control candidates was the most stably expressed across the different mating group sizes. While expression of macpiwi and hsp90 did not show significant differences between mating groups, expression of cox4 changed only slightly and the other two transcripts showed significant differential expression which makes them unreliable internal controls for this experiment (macpiwi: t(140) = 1.58, P = 0.12; hsp90: t(140) = 1.49, P = 0.14; macactin: t(140) = 2.31, P = 0.02; actp: t(140) = 2.14, P = 0.04; cox4: t(140) = 1.96, P = 0.04). Additionally, Pearson correlation coefficients were calculated between each target transcript and each potential internal control, and the average correlation was found to be highest for macpiwi (r(mean) = 0.60 ± 0.16), further supporting its relative stability. The calibrator was chosen from the isolated samples, taking the sample with the lowest Ct value from among all isolated samples and all transcripts (Pfaffl 2001). In total, we quantified the expression of 58 transcripts for 142 individual worms (70 pairs and 72 octets).
Statistical analyses
All statistical analyses were performed with the R statistical software, version 3.2.3 (R Development Core Team, 2013).
Phenotypic correlations and dimension reduction
We estimated phenotypic correlations between all pairwise combinations of the 58 transcripts with Pearson’s product−moment correlation coefficients, followed by a correlation network analysis to evaluate the collinearity of the data, generated with the R-package ‘qgraph’ (Epskamp et al. 2012). Because these analyses revealed that the different SFP transcript expression levels are highly correlated with one another, we performed PCA to reduce the dimensionality of the data and to thereby capture unique axes of multivariate variation in seminal fluid transcript expression. We carried out the PCA on the data including both environments (pairs and octets) and genotypes combined, such that extracted components are comparable across the different environments and genotypes. Additionally, we carried out a second PCA on the data that included also the isolated group as a third level of environment (see the supplementary file for details). PCAs were performed using the ‘principal()’ function in the ‘psych’ R package (Revelle 2015) and principal component (PC) scores were extracted using the correlation matrix (Jolliffe 2002).
Effect of mating group size, genotype and GEI
Considering that most of the variation in the data was described by the first four PCs (see Results), we tested these as dependent variables in order to assess the effect of mating group size (environment), genotype and GEI using general linear mixed models (LMMs), which allowed for an unbalanced design. We fitted LMMs using the R package ‘lme4’ (Bates et al. 2015) including the fixed factor mating group size as a nominal variable with two levels: pair and octet. The variance homoscedasticity was tested for mating group size by performing Levene tests (Levene 1960). The correlated random factors in the analysis were genotype and GEI. The χ² (Chi-square), AIC and P values were obtained from likelihood ratio tests by comparing model parameters. Marginal R2, which describes the proportion of variance explained by the fixed factor alone, and conditional R2, which describes the proportion of variance explained by both the fixed and random factors, were calculated for the model that includes environment as a fixed factor and GEI as a random factor (Nakagawa and Schielzeth 2013).
Generalized linear mixed models for transcript expression
In order to test the generality of our findings for seminal fluid investment and composition, generalized linear mixed models (GLMMs) were fitted using a gaussian family distribution with log-link for each of the 58 SFP transcripts individually, taking the efficiency-corrected relative expression ratio of each transcript as the dependent variable. We performed likelihood ratio tests to determine the contribution of environment, genotype and GEI to variation in each transcript, and then assessed how many of the transcripts exhibited significant environment, genotype and GEI effects. To correct for multiple hypothesis testing, we applied a Benjamini-Hochberg False Discovery Rate (FDR) correction using the R function ‘p.adjust’ in the ‘stat’ package by applying a 10% FDR cut-off (van Iterson et al. 2010).
Cross-environment genetic correlations
We also calculated cross-environment genetic correlations (rg) based on mean PC scores of genotypes, to test for the potential existence of GEI with ecological crossover, i.e. whether correlations depart significantly from +1 or −1. To test this, we calculated z-scores (), where x is the genetic correlation and σ is the standard error. Standard errors were determined from distributions of 1000 bootstrap values, in which genotype was the unit of bootstrapping. Significance tests for estimated z-scores were two-tailed for genetic correlations.
Estimation of heritability and genetic correlations among transcripts
Due to the fact that our experimental design does not allow to separate additive and non-additive genetic effects, broad sense heritability (H2) was measured as an estimate of repeatability under the assumption that all differences between genotypes are genetic. We estimated H2 of PCs both for within (across) and between (pooled) environments based on variance components of fitted linear mixed-effect models where genotype was the random effect. Heritability and its standard error estimations were quantified with the R package ‘rptR’ (Stoffel et al. 2017) by performing bootstrapping with 1000 repeating steps. We also calculated genetic correlations between each pair of transcripts by calculating Pearson’s product−moment correlation coefficients between mean efficiency-corrected relative expression ratio by pooling environments.
Discriminant analysis
While PCA reduces the dimensionality of the data by describing the largest axes of variation irrespective of treatment, there is another multivariate statistical technique, canonical linear discriminant analysis (CLDA), that instead focuses on what linear combinations of variables can best explain most of the variation in the data according to a pre-defined grouping of samples. Specifically, CLDA seeks a discriminant function that maximizes the between-group variance to the within-group variance, and its main purpose is enabling prediction of group membership of the samples based on a linear combination of the variables (McLachlan 1992). Thus, we also performed CLDA to study differences between mating group sizes by analysing whether variation in SFP transcript expression between samples differs depending on the mating group size.
Results
Seminal fluid transcripts are highly correlated in their expression
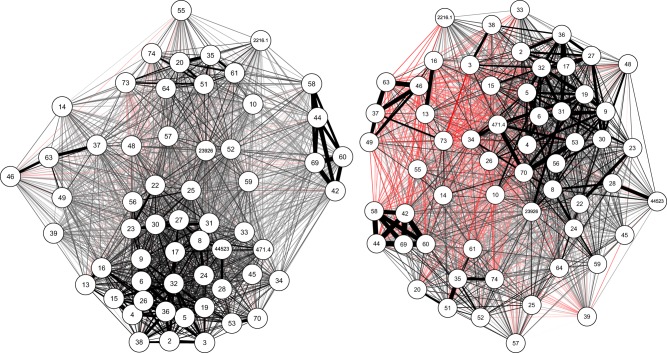
The correlation network (Fig. 1) confirmed the phenotypically highly correlated nature of seminal fluid transcript expression level: correlation coefficients for pairs of transcripts ranged between −0.31 and 1.00, with an average value of 0.38; 78% of correlations had P values smaller than 0.05 (not shown). In addition, at least four separate clusters of highly correlated transcripts can be observed on the correlation network (Fig. 1).
Fig. 1.
Phenotypic (left) and genetic (right) coexpression network of 58 seminal fluid transcripts in M. lignano based on Pearson’s product moment correlation coefficients. Nodes (numbers) represent the Mlig-pro1–Mlig-pro76 transcripts, plus four prostate-limited transcripts (2216.1, 23926, 471.4 and 44523; depicted in the figure with their transcript IDs from the MLRNA110815 transcriptome assembly). Lines represent correlation coefficients between transcripts (red: negative, black: positive), with line width scaled according to the strength of each correlation (thicker = stronger). The group of highly correlated nodes, for example, appearing to the lower part of the network means that these nodes have higher correlations between each other than with the other transcripts. Genetic correlations were estimated from Pearson’s product−moment correlation coefficients based on genotype means for each transcript (mating group sizes were pooled) (colour figure online)
The PCA identified four orthogonal (uncorrelated) PCs of seminal fluid transcript expression that individually explained more than 5% of the total variation with an eigenvalue >2. We interpreted component loadings for each transcript to these eigenvectors of higher than an absolute value of 0.44 as biologically significant at the P = 0.01 level (Table S2) (Norman and Streiner 2009). Together these PCs explained a substantial proportion (ca. 76%) of the total variation (Table 1). PC1 accounted for ca. 42% of the total variance, including positive loadings >0.44 for 39/58 transcripts; with no corresponding negative loadings, PC1 can therefore be interpreted as capturing overall investment in seminal fluid transcript expression. By contrast, PC2–PC4 each exhibited a mixture of positive and negative loadings for a smaller subset of SFP transcripts (Table 1 and Table S2); they thus describe variation in seminal fluid composition. We did not include the subsequent PCs in the main analyses since each captured variation in only one or two transcripts, and they do not therefore provide additional insight beyond the single transcript analyses (Table S2).
Table 1.
Principal component analysis of seminal fluid transcript expression in M. lignano
| Principal components | PC1 | PC2 | PC3 | PC4 |
|---|---|---|---|---|
| Variation explained (%) | 41.9 | 18.8 | 10.1 | 5.1 |
| Eigenvalue | 24.3 | 10.9 | 5.9 | 2.9 |
| No. of positive loadings | 39 | 14 | 5 | 3 |
| No. of negative loadings | 0 | 6 | 6 | 2 |
The first four principal components with eigenvalues >2, which each explained more than 5% of variation in seminal fluid transcript expression, were extracted for further analyses. Numbers of positive and negative loadings (eigenvectors), out of 58 seminal fluid transcripts in total, that have values >±0.44 were counted as biologically significant at the 1% level
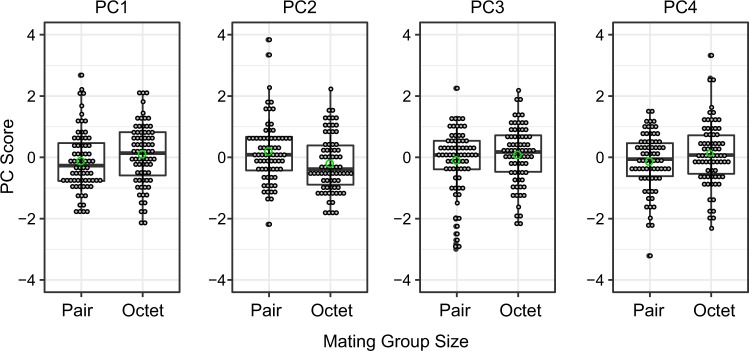
Mating group size affects seminal fluid transcript composition but not overall investment
We compared seminal fluid transcript expression levels between the two mating group sizes based on the outcome of the PCA globally, as well as for each transcript separately. We first evaluated the variances for mating group size by performing Levene’s test (Levene 1960) to confirm whether mating group sizes have equal variances. The P values of Levene tests for PC1 to 4 (0.85, 0.95, 0.92 and 0.69) indicate homoscedasticity of variances. The LMMs revealed a significant effect of mating group size only for PC2 (Table 2, Fig. 2). According to the GLMM analyses of single transcripts, at a false discovery rate of 10%, we found no transcripts exhibited significant differences in their expression level between pairs and octets (Table S3). By contrast, when we performed a separate PCA including also isolated samples, isolated individuals showed clear evidence of conserved investment in seminal fluid transcript expression (PC1), with markedly lower expression levels compared to pairs and octets (see Table S5 and Fig. S3).
Table 2.
Linear mixed-effect models for the first four principal components describing seminal fluid transcript expression variation in M. lignano
| Principal component | Factor | χ² | AIC | P values | Marginal R2/Conditional R2 |
|---|---|---|---|---|---|
| PC1 | Environment | 406.21 | 0.19 | ||
| Genotype | 2.55 | 405.66 | 0.11 | ||
| GEI | 4.32 | 405.34 | 0.11 | 0.01/0.14 | |
| PC2 | Environment | 402.12 | 0.02 | ||
| Genotype | 6.11 | 398.01 | 0.01 | ||
| GEI | 0.43 | 401.58 | 0.81 | 0.04/0.17 | |
| PC3 | Environment | 406.86 | 0.29 | ||
| Genotype | 136.87 | 271.98 | <0.01 | ||
| GEI | 1.45 | 274.53 | 0.49 | 0.01/0.74 | |
| PC4 | Environment | 405.72 | 0.14 | ||
| Genotype | 88.76 | 318.96 | <0.01 | ||
| GEI | 1.15 | 321.81 | 0.56 | 0.02/0.62 |
The mating group size was taken as a fixed effect, and genotype and genotype-by-environment interaction as correlated random effects. Test results for random effects (χ², AIC etc.) are based on comparing the models with F-tests. Marginal R2 describes the proportion of variance explained by the fixed factor alone and conditional R2 describes the proportion of variance explained by both the fixed and random factors. Bold text indicates statistically significant effects (P < 0.05)
Fig. 2.
Social environment (mating group size) effect on the first four principal components capturing variation in seminal fluid transcript expression in M. lignano. Figures show box plots of PC scores against mating group size for PC1−4. Genotypes were pooled. In all cases, the boxes represent the values between the 25th and 75th percentiles, the line whiskers represent values that are 1.5 times the interquartile range. The grey circles depict the raw data for each individual sample and the green circle is the mean (colour figure online)
We further performed CLDA to address whether and how the variation in SFP transcript expression can be differentiated according to mating group size (Fig. 3). The discriminant function indeed revealed an association between mating group size and seminal fluid transcript expression level that is effective in predicting mating group size membership. The mean discriminant scores of pairs and octets was found to be significantly different (F = 86.5, df = 1,140, P < 0.001) and an increasing pattern of seminal fluid transcript expression in octets was observed (Fig. 3).
Fig. 3.
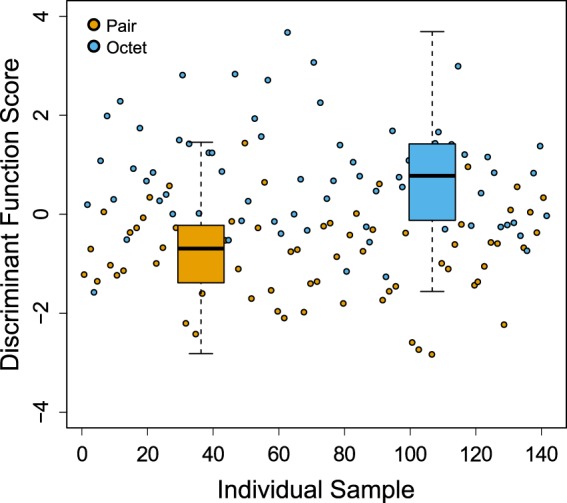
Canonical linear discriminant analysis of seminal fluid transcript expression in M. lignano grouped by mating group size. Each circle represents an individual sample from one of two mating group sizes (orange: pair; blue: octet). The box plots show the median and interquartile range of discriminant function scores, dashed whiskers extend 1.5 times beyond the interquartile range (colour figure online)
SFP transcript expression exhibited genetic variation and GEI
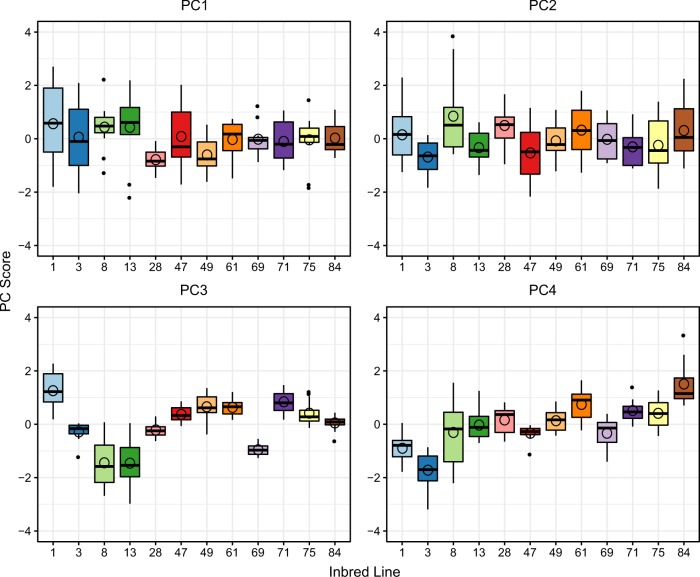
We found no genotype effect for PC1 (Table 2, Fig. 4) (although there was a genotype effect upon inclusion of isolated individuals, see Table S5, Fig S4), but highly significant genotypic variation for PC2, PC3 and PC4 (Table 2, Fig. 4). Moreover, individual seminal fluid transcript expression analyses revealed that the majority of transcripts (47/58, at FDR of 10%) exhibited a significant genotype effect (Table S3). Broad sense heritability (H2) of PCs (genotypes pooled across environments) was correspondingly significantly different from zero for PC2−PC4, but not for PC1 (Table 3). However, these heritability estimations of pooled environments did not eliminate the confounding influence of GEI; thus we also performed LMMs separately for each mating group size to estimate H2 for each PC within each environment (Table 3). Here, we found some evidence of genetic variation for PC1 in pairs but not in octets. Heritability of PC2 was not significantly different from zero in either pairs or octets; however, heritability was high and significant for both PC3 and PC4.
Fig. 4.
Genotype (inbred line) effects on the first four principal components capturing variation in seminal fluid transcript expression in M. lignano. Figures show box plots of PC scores against inbred lines for PC1−PC4. Mating group sizes (pair and octet) were pooled. The open circles represent the mean values. The boxes represent the values between the 25th and 75th percentiles, the line whiskers represent values that are 1.5 times the interquartile range
Table 3.
Bootstrapped broad sense heritability (H2) within and across pooled mating group sizes and genetic correlations (rg) between pairs of mating group sizes for the first four principal components describing SFP transcript expression variation in M. lignano
| Principal component | PC1 | PC2 | PC3 | PC4 |
|---|---|---|---|---|
| Environment | H 2 (SE) | H 2 (SE) | H 2 (SE) | H 2 (SE) |
| Pair | 0.28 (0.13) | 0.14 (0.10) | 0.75 (0.10) | 0.54 (0.14) |
| Octet | 0.00 (0.05) | 0.08 (0.09) | 0.71 (0.11) | 0.64 (0.12) |
| Pooled | 0.07 (0.06) | 0.12 (0.07) | 0.72 (0.10) | 0.59 (0.12) |
| rg (SE) | 0.16 (0.38) | 0.54 (0.16) | 0.93 (0.03) | 0.94 (0.06) |
The standard errors (in brackets) are based on distributions of 1000 bootstrapped values, in which genotype are the unit of bootstrapping. Estimations that appear in bold differ significantly from zero for heritability or from one for genetic correlations according to calculated z-scores with alpha level of 0.05.
Turning to genotype-by-environment interactions, significant GEI could not be detected for any of the four PCs according to LMMs (Table 2, see also Fig. 5) although a substantial number of individual transcripts exhibit GEI (14/58, at FDR of 10%) according to the individual GLMM analyses (Table S3). We further estimated cross-environment genetic correlations between pairs and octets for each PC: these were not significantly different from one for PC3 and PC4 (Table 3), however, significantly different from one (P < 0.01) for PC1 and PC2, indicating the possibility of GEI for these components (Table 3). Moreover, when we included the isolated group in the supplemental analyses, we found significant GEI for some components (see Table S4, S5 and Fig. S5), suggesting GEI for seminal fluid investment and composition. The varying expression pattern of all PCs between pairs and octets for each individual inbred line, including individual observations, can also be seen in Supplementary Figures S6–S9.
Fig. 5.
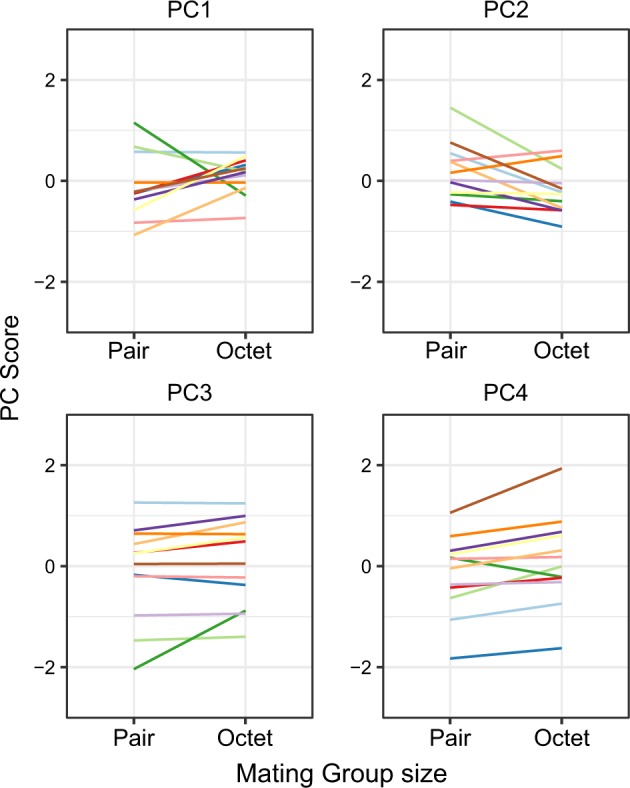
Reaction norms of 12 different genotypes (inbred lines) of M. lignano for the first four principal components capturing variation in seminal fluid transcript expression across environments (mating group sizes). For each reaction norm, PC score is on the y axis and mating group size is on the x axis. Line tips represent the mean value of that genotype in the given mating group size. Colour codes for the genotypes are the same as Fig. 4
Strong positive genetic correlations between seminal fluid transcript expression
The genetic correlations calculated based on Pearson’s product−moment correlation coefficients (Fig. 1) between the mean relative expression ratio of each transcript of each genotype (pairs and octets were pooled) ranged between −0.73 and 1.00 with a mean value of 0.21; around 29% of them had a negative value. In addition, approximately 15% of these genetic correlation coefficients were statistically significant, and only 2% of them had a negative value.
Discussion
Our study aimed to investigate the contribution of environmental and genetic factors to variation in expression level of seminal fluid transcripts in the context of sexual selection. Our results showed that pairs and octets do not differ significantly for overall seminal fluid transcript expression level in M. lignano, but we clearly demonstrated genetic variation in seminal fluid transcript expression and relative composition, together with some evidence of GEI. Here, we discuss our findings in terms of (i) the complexity of seminal fluid as a multivariate phenotype; (ii) the impact of mating group size; (iii) the genotypic and GEI effects and (iv) genetic correlations for seminal fluid transcript expression.
Complexity and dimensionality of seminal fluid
Several studies in which SFPs have been identified in large-scale transcriptomic or proteomic analyses have emphasized the complexity of seminal fluid, with tens to hundreds of different components (e.g. Pilch and Mann 2006; Dottorini et al. 2007; Findlay et al. 2008; Baer et al. 2009; Ramm et al. 2009; South et al. 2011; Goenaga et al. 2015; reviewed in Poiani 2006). Such complexity can be expected also in our model organism, and indeed 76 putative SFP transcripts have just been identified in M. lignano (Weber et al. 2018). This could imply that there are at least 76 unique dimensions to seminal fluid investment, but instead we show that many seminal fluid transcripts are, in fact, highly correlated in their expression. Hence, we established that a substantial proportion of the total variation in seminal fluid transcript expression can actually be captured by just a few unique axes. The highly correlated nature of seminal fluid transcript expression might hint that multiple SFPs are produced for a single functional goal. For example, a network of at least eight SFPs in Drosophila melanogaster was found that is required to bind a ninth SFP to sperm (Ram and Wolfner 2009; Findlay et al. 2014). For the time being, we know little about the specific functions of individual transcripts in M. lignano. However, the recently published annotated genome assembly of M. lignano gives some evidence for sequence similarities with other organisms (Wudarski et al. 2017). Additionally, we should bear in mind that a few of the different transcripts that were most highly correlated with one another could in fact represent the same underlying gene, reflecting the fragmentary and still imperfect nature of the M. lignano transcriptome assembly we employed (see Weber et al. 2018). While this might explain the high pairwise correlations for a few transcripts, it cannot account for the high correlations we found throughout the seminal fluid transcriptome, and it would not strongly affect our analyses based on extracted principal components. It is also worth noting that we as yet have no information on whether transcript expression levels correspond to protein abundance in our study organism. A recent study in another organism found significant positive correlations between secreted seminal fluid transcript expression level and protein abundance (Bonilla et al. 2015). However, it has been shown in many cell types that transcript expression level is not always highly correlated with protein amount (De Sousa Abreu et al. 2009; Vogel and Marcotte 2012) and so we should be cautious in our conclusion. Nevertheless, transcription is also a costly process and so differing expression levels should themselves reflect variation in investment.
We argued that SFPs are together a multivariate phenotype that could include multiple distinct dimensions and, therefore, that SFP evolution itself should be considered with a multidimensional approach. There are parallels here with another well-studied multivariate sexually selected phenotype, the cuticular hydrocarbons (CHCs) that play a major role in mate choice in insects. As with SFPs, CHC profile has also been defined as a highly complex phenotype that nevertheless exhibits a relatively limited number of axes of variation (Hine and Blows 2006; Dembeck et al. 2015; Finck et al. 2016). Thus, many studies that focus on the evolution of CHC expression examine overall CHC profiles rather than focusing on single CHCs (reviewed in Ingleby 2015). Those studies have highlighted the role of sexual selection on the evolution of overall CHC profile and its divergence between populations (reviewed in Steiger and Stökl 2014). By analogy, investigating the divergence of SFPs adopting a multidimensional approach is similarly necessary to understand seminal fluid evolution. For example, supporting this idea, Goenaga et al. (2015) showed that different populations have diverged along distinct multivariate dimensions of SFP abundance variation and this variation has a significant relation with male and female reproductive success, potentially causing isolation between populations.
The impact of mating group size on seminal fluid transcript composition
Sex allocation theory for simultaneous hermaphrodites predicts that the optimal investment of available resources allocated to each sex function depends on mating group size, i.e. on the relative importance of sperm competition to male reproductive success (Charnov 1979, 1982). Indeed, numerous studies have shown that, when mating group size is large, individuals increase investment in male function in M. lignano (e.g. Schärer and Ladurner 2003; Schärer et al. 2004; Schärer and Vizoso 2007; Janicke and Schärer 2009; Janicke et al. 2013; Giannakara et al. 2016). A recent study has revealed that seminal fluid transcript expression increases with increasing group size, with especially large differences in expression between isolated and pairs and somewhat smaller differences between pairs and octets for the tail-specific differentially expressed transcripts (Ramm et al. in revision). Even though this study only focused on one inbred line (DV1), the overall pattern that we found here based on PC1, is consistent. PC1, which describes the overall investment in seminal fluid transcripts, shows an increasing pattern between non-mating and two grouped treatments, and there is then only a relatively small increase in investment from pairs to octets. Although in our dataset the difference between isolated and two grouped treatments could be attributable to uncontrolled factors such as age of the worms, consistency of the results with Ramm et al. (in revision), coupled with evidence for highly flexible sex allocation (Brauer et al. 2007), suggests that worms can downregulate seminal fluid transcript expression in non-mating conditions. Besides, a previous study in M. lignano showed that virgin worms that were kept isolated are more female-biased in their sex allocation when compared with worms in pairs, with reduced testis size but similar ovary size (Schärer and Janicke 2009) supporting the idea that worms reduce investment in male reproductive function if they do not mate.
The modest increase between pairs and octets in PC1 is somewhat surprising in terms of our expectations, but the average pattern will of course have been heavily influenced by the specific genotypes we sampled, which were only a small subset of the population. Even though sperm competition theory predicts increasing investment in the ejaculate, theory further predicts that males should strategically invest in different ejaculate components depending on their specific functions (Cameron et al. 2007; Perry et al. 2013). The functions of SFPs are diverse, and therefore the costs and benefits associated with their production will also differ (Poiani 2006; Perry et al. 2013). For example, recent models predict that when males evolve to produce SFPs with functions such as those related to defence or offence ability during sperm competition, they might invest unequally in those proteins (Dhole and Servedio 2014). We can thus expect that when mating is random, all individual genotypes are equally likely to mate first or second and a trade-off might exist between investing resources in different SFPs depending on the order of the mating. The subsequent PCs in our analyses, especially PC2, which differs significantly between pairs and octets, might be relevant in this context providing evidence for changing seminal fluid transcript composition according to sperm competition. The biological interpretation of PC2 is that the individuals with positive PC2 scores tend to have higher expression for transcripts with positive loadings while having lower expression for transcripts with negative loadings, and individuals with negative PC2 scores tend to show the opposite expression pattern. Considering the nature of PC2 with positive and negative loadings of several individual transcripts, the difference of overall PC2 score between pairs and octets indicates a notable change in relative transcript expression between individuals according to mating group size. Our discriminant function analysis also identified a clear axis separating pairs from octets, providing further evidence for changing seminal fluid composition as a response to sperm competition. A similar pattern has also been shown in rodents that seminal fluid composition changes in response to social cues of sperm competition (Ramm et al. 2015), suggesting this might be a widespread phenomenon.
Significant genetic contribution to seminal fluid transcript expression variation
In our study, we found genotype effects for PC2−PC4, as well as for the majority of individual transcripts. In particular, the high genetic variation for single transcripts supports some of the few earlier studies that also found significant genetic variation in single SFPs in some insects (Smith et al. 2009; Baer et al. 2012). By contrast, PC1, and thus overall seminal fluid transcript investment, showed no genetic variation. Considering that SFP transcripts may be multifunctional by having pleiotropic effects and/or interactions with each other, leading to genetic correlations between them (Ayroles et al. 2011), strong selection on some transcripts may lead to rapid depletion of genetic variation for overall seminal fluid transcript investment. We indeed found some strong genetic correlations between expression level of transcripts. On the other hand, stabilizing selection has been shown to be much stronger on combinations of such traits than on each individual trait that contributes to the combination (e.g. Blows 2003; Kirkpatrick 2009; Walsh and Blows 2009). Strong stabilizing selection on expression could thus be one explanation for the lack of genetic variation for PC1 due to the fact that pleiotropic mutational effects have been shown to be widespread among gene expression traits (McGuigan et al. 2014). On the other hand, the inbreeding protocol followed to generate the inbred lines used in this study could have relaxed selection on male allocation (at least during the early part of the selection regime, when worms were kept in sibling pairs; see Vellnow et al. 2017) and thus partly explain the lack of genetic variance in overall seminal fluid transcript investment. Additionally, there appears to be higher genetic variance in pairs than in octets for PC1 according to heritability estimates. This could simply be attributable to the fact that worms might rarely experience low mating group sizes because of locally high density of worms or their high mating rate, and/or because relatively long sperm storage is possible (Schärer et al. 2004; Janicke and Schärer 2009). Therefore, selection on seminal fluid transcript expression level under such circumstances could be weak. On the other hand, if SFP transcription is costly, we might still expect all genotypes would minimize transcript production in pairs compared to octets. One possible alternative explanation is that a higher genetic variation for seminal fluid transcript investment in pairs might reflect an underlying trade-off: if this variation influences the ability/speed of response to changes in mating group size, the cost of maintaining ‘unnecessary’ investment when mating group size is small might simultaneously be of benefit if and when more mating opportunities arise, ensuring a sufficient supply or quicker upregulation of seminal fluid and thus a heightened sperm competitive ability. This would mean that the worms in small mating groups that maintain seminal fluid production pay an ongoing production cost but can begin/resume mating more quickly. These predictions are also supported if we look at high genetic variance in isolated worms (Fig S6); however, considering how this group was treated in our experiment, differences in genetic variance could also be attributable to some extent to other factors which we could not control, such as the age of the worms.
We found clear evidence for the existence of GEI for many individual transcripts (14 out of 58), although model comparisons did not support a GEI effect for any of the PCs. It is possible that there is actually GEI for PC1 and PC2, but we could not detect it perhaps due to insufficient statistical power. Notably, cross-environment correlations were significantly different from absolute value 1, which could be indicative of ecological crossover (crossover of the reaction norms across environments) for PC1 and PC2, and thus GEI for seminal fluid transcript investment and composition. This implies that different genotypes could be relatively better in different competitive environments, if this variation has adaptive significance. A recent study in M. lignano suggests that the situation is even more complex, because the expression of reproductive traits can be affected not just by the mating group size but also the specific genotypes of competitors in the mating group (Marie-Orleach et al. 2017). Therefore, GEI might be a possible explanation for the maintenance of variation in seminal fluid transcript expression level. Here, we also should recall that we found a very low number of significant negative genetic correlations, which may indicate that GEI is a more plausible explanation for maintaining variation in seminal fluid transcripts than trade-offs through genetic correlations between transcripts.
In D. melanogaster, genetic variation in gene expression and transcript abundance is associated with first and/or second male sperm competitive ability (Fiumera et al. 2005, 2007; Zhang et al. 2013). Similarly, it was recently shown in the seed beetle Callosobruchus maculatus that there is a significant effect of some size-fractions of SFPs on the sperm competitive ability of second males to mate (Yamane et al. 2015). Considering our findings, together with the fact that sperm displacement is known to occur in M. lignano (Marie-Orleach et al. 2014), it seems plausible that SFPs might play a similar role in sperm competitiveness in our study species. Some genotypes might be relatively better in their offensive and/or defensive abilities in different social environments depending on their strategically adjusted seminal fluid transcript investment and composition. These predictions should now be tested to see whether changing seminal fluid transcript expression does indeed impact on sperm competitiveness, and therefore male fitness, in M. lignano.
Conclusion
By experimentally manipulating mating group size in a set of inbred lines and measuring expression level of 58 putative seminal fluid transcripts of the simultaneously hermaphroditic flatworm M. lignano, we uncovered the multivariate nature of seminal fluid and then described how mating group size, thus the intensity of sperm competition, affects seminal fluid investment and composition. We found a highly significant contribution of genetic factors to variation in seminal fluid transcript expression levels, as well as some evidence that GEI and genetic correlations could play a role in shaping seminal fluid evolution. We thereby provide novel evidence of genetic and environmental sources of variation in seminal fluid transcript expression as an important class of post-mating sexually selected traits.
Data archiving
Data and analysis code are archived with Dryad; 10.5061/dryad.n3pg1hm.
Electronic supplementary material
Acknowledgements
We thank L. Schärer and D. Vizoso from the University of Basel for providing the M. lignano inbred lines used in this study. Data produced in this paper were generated in collaboration with the Genetic Diversity Centre (GDC), ETH Zurich. We are grateful to B. Lengerer, R. Pjeta, P. Ladurner (University of Innsbruck) and L. Schärer for providing the in situ hybridization data regarding transcript MLRNA110815_2216.1. We thank G. Arnqvist for his valuable statistical advice and L. Schärer and Y. Nakadera for providing valuable feedback on an early version of the manuscript. This work was funded by the Deutsche Forschungsgemeinschaft (DFG) (grant no. RA 2468/1-1 to SAR).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41437-018-0160-4) contains supplementary material, which is available to authorized users.
References
- Arbore R, Sekii K, Beisel C, Ladurner P, Berezikov E, Schärer L. Positional RNA-Seq identifies candidate genes for phenotypic engineering of sexual traits. Front Zool. 2015;12:14. doi: 10.1186/s12983-015-0106-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila FW, Sirot LK, LaFlamme B, Rubinstein CD, Wolfner MF. Insect seminal fluid proteins: identification and function. Annu Rev Entomol. 2011;56:21–40. doi: 10.1146/annurev-ento-120709-144823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles JF, Carbone MA, Stone EA, Jordan KW, Lyman RF, Magwire MM, et al. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet. 2009;41:299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroles JF, Laflamme BA, Stone EA, Wolfner MF, Mackay TFC. Functional genome annotation of Drosophila seminal fluid proteins using transcriptional genetic networks. Genet Res. 2011;93:387–395. doi: 10.1017/S0016672311000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer B, Heazlewood JL, Taylor NL, Eubel H, Millar AH. The seminal fluid proteome of the honeybee Apis mellifera. Proteomics. 2009;9:2085–2097. doi: 10.1002/pmic.200800708. [DOI] [PubMed] [Google Scholar]
- Baer B, Zareie R, Paynter E, Poland V, Millar AH. Seminal fluid proteins differ in abundance between genetic lineages of honeybees. J Proteom. 2012;75:5646–5653. doi: 10.1016/j.jprot.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67:51. [Google Scholar]
- Blows MW. Measuring nonlinear selection. Am Nat. 2003;162:815–820. doi: 10.1086/378905. [DOI] [PubMed] [Google Scholar]
- Bonilla ML, Todd C, Erlandson M, Andres J. Combining RNA-seq and proteomic profiling to identify seminal fluid proteins in the migratory grasshopper Melanoplus sanguinipes (F) BMC Genom. 2015;16:1–15. doi: 10.1186/s12864-015-2327-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer VS, Schärer L, Michiels NK. Phenotypically flexible sex allocation in a simultaneous hermaphrodite. Evolution. 2007;61:216–222. doi: 10.1111/j.1558-5646.2007.00018.x. [DOI] [PubMed] [Google Scholar]
- Cameron E, Day T, Rowe L. Sperm competition and the evolution of ejaculate composition. Am Nat. 2007;169:E158–E172. doi: 10.1086/516718. [DOI] [PubMed] [Google Scholar]
- Chapman T, Bangham J, Vinti G, Seifried B, Lung O, Wolfner MF, et al. The sex peptide of Drosophila melanogaster: Female post-mating responses analyzed by using RNA interference. Proc Natl Acad Sci USA. 2003;100:9923–9928. doi: 10.1073/pnas.1631635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnov EL. Simultaneous hermaphroditism and sexual selection. Proc Natl Acad Sci USA. 1979;76:2480–2484. doi: 10.1073/pnas.76.5.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnov EL. The theory of sex allocation. Monogr Popul Biol. 1982;18:1–355. [PubMed] [Google Scholar]
- Civetta A, Reimer A. Positive selection at a seminal fluid gene within a QTL for conspecific sperm precedence. Genetica. 2014;142:537–543. doi: 10.1007/s10709-014-9800-7. [DOI] [PubMed] [Google Scholar]
- Clark AG, Aguade M, Prout T, Harshman LG, Langley CH. Variation in sperm displacement and its association with accessory gland protein loci in Drosophila melanogaster. Genetics. 1995;139:189–201. doi: 10.1093/genetics/139.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Boer SPA, Baer B, Boomsma JJ. Seminal fluid mediates ejaculate competition in social insects. Science. 2010;327:1506–1509. doi: 10.1126/science.1184709. [DOI] [PubMed] [Google Scholar]
- Danielson-François AM, Kelly JK, Greenfield MD. Genotype × environment interaction for male attractiveness in an acoustic moth: evidence for plasticity and canalization. J Evol Biol. 2006;19:532–542. doi: 10.1111/j.1420-9101.2005.01006.x. [DOI] [PubMed] [Google Scholar]
- Danielson-François AM, Zhou Y, Greenfield MD. Indirect genetic effects and the lek paradox: inter-genotypic competition may strengthen genotype × environment interactions and conserve genetic variance. Genetica. 2009;136:27–36. doi: 10.1007/s10709-008-9297-z. [DOI] [PubMed] [Google Scholar]
- Dembeck LM, Böröczky K, Huang W, Schal C, Anholt RRH, Mackay TFC. Genetic architecture of natural variation in cuticular hydrocarbon composition in Drosophila melanogaster. eLife. 2015;4:e09861. doi: 10.7554/eLife.09861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhole S, Servedio MR. Sperm competition and the evolution of seminal fluid composition. Evolution. 2014;68:3008–3019. doi: 10.1111/evo.12477. [DOI] [PubMed] [Google Scholar]
- Dorus S, Evans PD, Wyckoff GJ, Sun SC, Lahn BT. Rate of molecular evolution of the seminal protein gene SEMG2 correlates with levels of female promiscuity. Nat Genet. 2004;36:1326–1329. doi: 10.1038/ng1471. [DOI] [PubMed] [Google Scholar]
- Dottorini T, Nicolaides L, Ranson H, Rogers DW, Crisanti A, Catteruccia F. A genome-wide analysis in Anopheles gambiae mosquitoes reveals 46 male accessory gland genes, possible modulators of female behavior. Proc Natl Acad Sci USA. 2007;104:16215–16220. doi: 10.1073/pnas.0703904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5:1512–1526. doi: 10.1039/b908315d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JP. Quantitative genetic evidence that males trade attractiveness for ejaculate quality in guppies. Proc R Soc B Biol Sci. 2010;277:3195–3201. doi: 10.1098/rspb.2010.0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JP, Rahman MM, Gasparini C. Genotype-by-environment interactions underlie the expression of pre- and post-copulatory sexually selected traits in guppies. J Evol Biol. 2015;28:959–972. doi: 10.1111/jeb.12627. [DOI] [PubMed] [Google Scholar]
- Fedorka KM, Winterhalter WE, Ware B. Perceived sperm competition intensity influences seminal fluid protein production prior to courtship and mating. Evolution. 2011;65:584–590. doi: 10.1111/j.1558-5646.2010.01141.x. [DOI] [PubMed] [Google Scholar]
- Finck J, Berdan EL, Mayer F, Ronacher B, Geiselhardt S. Divergence of cuticular hydrocarbons in two sympatric grasshopper species and the evolution of fatty acid synthases and elongases across insects. Sci Rep. 2016;6:33695. doi: 10.1038/srep33695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GD, Sitnik JL, Wang W, Aquadro CF, Clark NL, Wolfner MF. Evolutionary rate covariation identifies new members of a protein network required for Drosophila melanogaster female post-mating responses. PLoS Genet. 2014;10:e1004108. doi: 10.1371/journal.pgen.1004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GD, Yi X, MacCoss MJ, Swanson WJ. Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biol. 2008;6:1417–1426. doi: 10.1371/journal.pbio.0060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiumera AC, Dumont BL, Clark AG. Sperm competitive ability in Drosophila melanogaster associated with variation in male reproductive proteins. Genetics. 2005;169:243–257. doi: 10.1534/genetics.104.032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiumera AC, Dumont BL, Clark AG. Associations between sperm competition and natural variation in male reproductive genes on the third chromosome of Drosophila melanogaster. Genetics. 2007;176:1245–1260. doi: 10.1534/genetics.106.064915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen CR, Powers DR, Copenhaver PE, Mason RT. Size dependence in non-sperm ejaculate production is reflected in daily energy expenditure and resting metabolic rate. J Exp Biol. 2015;218:1410–1418. doi: 10.1242/jeb.120402. [DOI] [PubMed] [Google Scholar]
- Giannakara A, Schärer L, Ramm SA. Sperm competition-induced plasticity in the speed of spermatogenesis. BMC Evol Biol. 2016;16:60. doi: 10.1186/s12862-016-0629-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goenaga J, Yamane T, Rönn J, Arnqvist G. Within-species divergence in the seminal fluid proteome and its effect on male and female reproduction in a beetle. BMC Evol Biol. 2015;15:266. doi: 10.1186/s12862-015-0547-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard RRL, Ryther JH. Studies of marine planktonic diatoms: I. Cyclotella Nana Hustedt, and Detonula Confervacea (Cleve) Gran. Can J Microbiol. 1962;8:229–239. doi: 10.1139/m62-029. [DOI] [PubMed] [Google Scholar]
- Hine E, Blows MW. Determining the effective dimensionality of the genetic variance-covariance matrix. Genetics. 2006;173:1135–1144. doi: 10.1534/genetics.105.054627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson DJ, Hosken DJ. Sperm competition promotes the exploitation of rival ejaculates. J Theor Biol. 2006;243:230–234. doi: 10.1016/j.jtbi.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Hunt J, Hosken DJ (2014) Genotype-by-environment interactions and sexual selection. John Wiley & Sons, Hoboken, NJ. [DOI] [PubMed]
- Immler S, Griffth SC, Zann R, Birkhead TR. Intra-specific variance in sperm morphometry: a comparison between wild and domesticated Zebra Finches Taeniopygia guttata. Ibis. 2012;154:480–487. [Google Scholar]
- Ingleby FC. Insect cuticular hydrocarbons as dynamic traits in sexual communication. Insects. 2015;6:732–742. doi: 10.3390/insects6030732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingleby FC, Hosken DJ, Flowers K, Hawkes MF, Lane SM, Rapkin J, et al. Genotype-by-environment interactions for cuticular hydrocarbon expression in Drosophila simulans. J Evol Biol. 2013;26:94–107. doi: 10.1111/jeb.12030. [DOI] [PubMed] [Google Scholar]
- Ingleby FC, Hunt J, Hosken DJ. The role of genotype-by-environment interactions in sexual selection. J Evol Biol. 2010;23:2031–2045. doi: 10.1111/j.1420-9101.2010.02080.x. [DOI] [PubMed] [Google Scholar]
- Janicke T, Marie-Orleach L, De Mulder K, Berezikov E, Ladurner P, Vizoso DB, et al. Sex allocation adjustment to mating group size in a simultaneous hermaphrodite. Evolution. 2013;67:3233–3242. doi: 10.1111/evo.12189. [DOI] [PubMed] [Google Scholar]
- Janicke T, Schärer L. Determinants of mating and sperm-transfer success in a simultaneous hermaphrodite. J Evol Biol. 2009;22:405–415. doi: 10.1111/j.1420-9101.2008.01660.x. [DOI] [PubMed] [Google Scholar]
- Jia AF, Greenfield MD, Collins RD, Ia FENGOUJ, Reenfield MIDG. Genetic variance of sexually selected traits in waxmoths: maintenance by genotype × environment interaction. Evolution. 2000;54:953–967. doi: 10.1111/j.0014-3820.2000.tb00095.x. [DOI] [PubMed] [Google Scholar]
- Jolliffe IT. Principal component analysis. J Am Stat Assoc. 2002;98:487. [Google Scholar]
- Josse J, Husson F. Handling missing values in exploratory multivariate data analysis methods. J la Société Fr. 2012;153:79–99. [Google Scholar]
- Kingan SB, Tatar M, Rand DM. Reduced polymorphism in the chimpanzee semen coagulating protein, semenogelin I. J Mol Evol. 2003;57:159–169. doi: 10.1007/s00239-002-2463-0. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M. Patterns of quantitative genetic variation in multiple dimensions. Genetica. 2009;136:271–284. doi: 10.1007/s10709-008-9302-6. [DOI] [PubMed] [Google Scholar]
- Koene J, Hoffer J, Brouwer A. Reduced egg laying caused by a male accessory gland product opens the possibility for sexual conflict in a simultaneous hermaphrodite. Anim Biol. 2009;59:435–448. [Google Scholar]
- Kokko H, Heubel K. Condition-dependence, genotype-by-environment interactions and the lek paradox. Genetica. 2008;134:55–62. doi: 10.1007/s10709-008-9249-7. [DOI] [PubMed] [Google Scholar]
- Ladurner P, Pfister D, Seifarth C, Schärer L, Mahlknecht M, Salvenmoser W, et al. Production and characterisation of cell- and tissue-specific monoclonal antibodies for the flatworm Macrostomum sp. Histochem Cell Biol. 2005;123:89–104. doi: 10.1007/s00418-004-0722-9. [DOI] [PubMed] [Google Scholar]
- Ladurner P, Schärer L, Salvenmoser W, Rieger RM. A new model organism among the lower Bilateria and the use of digital microscopy in taxonomy of meiobenthic Platyhelminthes: Macrostomum lignano, n. sp. (Rhabditophora, Macrostomorpha) J Zool Syst Evol Res. 2005;43:114–126. [Google Scholar]
- LaFlamme BA, Ravi Ram K, Wolfner MF. The Drosophila melanogaster seminal fluid protease ‘Seminase’ regulates proteolytic and post-mating reproductive processes. PLoS Genet. 2012;8:30–32. doi: 10.1371/journal.pgen.1002435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengerer B, Wunderer J, Pjeta R, Carta G, Kao D, Aboobaker A, et al. Organ specific gene expression in the regenerating tail of Macrostomum lignano. Dev Biol. 2018;433:448–460. doi: 10.1016/j.ydbio.2017.07.021. [DOI] [PubMed] [Google Scholar]
- Leonard JL. Sexual selection: lessons from hermaphrodite mating systems. Integr Comp Biol. 2006;46:349–367. doi: 10.1093/icb/icj041. [DOI] [PubMed] [Google Scholar]
- Levene H. Levene test for equality of variances. Contrib to Probab Stat. 1960;1:278–292. [Google Scholar]
- Lewis SM, Tigreros N, Fedina T, Ming QL. Genetic and nutritional effects on male traits and reproductive performance in Tribolium flour beetles. J Evol Biol. 2012;25:438–451. doi: 10.1111/j.1420-9101.2011.02408.x. [DOI] [PubMed] [Google Scholar]
- Lung O, Wolfner MF. Identification and characterization of the major Drosophila melanogaster mating plug protein. Insect Biochem Mol Biol. 2001;31:543–551. doi: 10.1016/s0965-1748(00)00154-5. [DOI] [PubMed] [Google Scholar]
- Mangels R, Young B, Keeble S, Ardekani R, Meslin C, Ferreira Z, et al. Genetic and phenotypic influences on copulatory plug survival in mice. Heredity. 2015;115:496–502. doi: 10.1038/hdy.2015.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie-Orleach L, Janicke T, Schärer L. Effects of mating status on copulatory and postcopulatory behaviour in a simultaneous hermaphrodite. Anim Behav. 2013;85:453–461. [Google Scholar]
- Marie-Orleach L, Janicke T, Vizoso DB, Eichmann M, Schärer L. Fluorescent sperm in a transparent worm: validation of a GFP marker to study sexual selection. BMC Evol Biol. 2014;14:148. doi: 10.1186/1471-2148-14-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie-Orleach L, Vogt-Burri N, Mouginot P, Schlatter A, Vizoso DB, Bailey NW, et al. Indirect genetic effects and sexual conflicts: Partner genotype influences multiple morphological and behavioral reproductive traits in a flatworm. Evolution. 2017;71:1232–1245. doi: 10.1111/evo.13218. [DOI] [PubMed] [Google Scholar]
- McGuigan K, Collet JM, McGraw EA, Ye YH, Allen SL, Chenoweth SF, et al. The nature and extent of mutational pleiotropy in gene expression of male Drosophila serrata. Genetics. 2014;196:911–921. doi: 10.1534/genetics.114.161232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan GJ (1992) Discriminant analysis and statistical pattern recognition. John Wiley & Sons, Hoboken, NJ.
- Michiels NK (1998) Mating conflicts and sperm competition in simultaneous hermaphrodites. In: Sperm competition and sexual selection. Edited by: Birkhead TR, Møller AP pp. 219–254.
- Mohorianu II, Bretman A, Smith DT, Fowler E, Dalmay T, Chapman T. Genomic responses to socio-sexual environment in male Drosophila melanogaster exposed to conspecific rivals. RNA. 2017;23:1048–1059. doi: 10.1261/rna.059246.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakadera Y, Swart EM, Hoffer JNa, Den Boon O, Ellers J, Koene JM. Receipt of seminal fluid proteins causes reduction of male investment in a simultaneous hermaphrodite. Curr Biol. 2014;24:859–862. doi: 10.1016/j.cub.2014.02.052. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Schielzeth H. A general and simple method for obtaining R^2 from generalized linear mixed-effects models. Methods Ecol Evol. 2013;4:133–142. [Google Scholar]
- Norman GR, Streiner DL. Biostatistics: the bare essentials. JAMA. 2009;302:2261–2262. [Google Scholar]
- Nystrand M, Dowling DK, Simmons LW. Complex genotype by environment interactions and changing genetic architectures across thermal environments in the Australian field cricket, Teleogryllus oceanicus. BMC Evol Biol. 2011;11:222. doi: 10.1186/1471-2148-11-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker GA. Sperm competition and its evolutionary consequences in the insects. Biol Rev. 1970;45:525–567. [Google Scholar]
- Perry JC, Sirot L, Wigby S. The seminal symphony: how to compose an ejaculate. Trends Ecol Evol. 2013;28:414–422. doi: 10.1016/j.tree.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:45e–45e. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister D, De Mulder K, Philipp I, Kuales G, Hrouda M, Eichberger P, et al. The exceptional stem cell system of Macrostomum lignano: Screening for gene expression and studying cell proliferation by hydroxyurea treatment and irradiation. Front Zool. 2007;4:1–14. doi: 10.1186/1742-9994-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieprzyk M, High H. Fluidigm dynamic arrays provide a platform for single- cell gene expression analysis. Nat Methods. 2009;6:iii–iv. [Google Scholar]
- Pilch B, Mann M. Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biol. 2006;7:R40. doi: 10.1186/gb-2006-7-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plusquin M, DeGheselle O, Cuypers A, Geerdens E, Van Roten A, Artois T, et al. Reference genes for qPCR assays in toxic metal and salinity stress in two flatworm model organisms. Ecotoxicology. 2012;21:475–484. doi: 10.1007/s10646-011-0809-8. [DOI] [PubMed] [Google Scholar]
- Poiani A. Complexity of seminal fluid: a review. Behav Ecol Sociobiol. 2006;60:289–310. [Google Scholar]
- Ram KR, Wolfner MF. A network of interactions among seminal proteins underlies the long-term postmating response in Drosophila. Proc Natl Acad Sci USA. 2009;106:15384–15389. doi: 10.1073/pnas.0902923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramm SA, Edward DA, Claydon AJ, Hammond DE, Brownridge P, Hurst JL, et al. Sperm competition risk drives plasticity in seminal fluid composition. BMC Biol. 2015;13:87. doi: 10.1186/s12915-015-0197-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramm SA, Lengerer B, Arbore R, Pjeta R, Wunderer J, Giannakara A et al. Sex allocation plasticity on a transcriptome scale: socially-sensitive gene expression in the hermaphroditic flatworm Macrostomum lignano. In revision. [DOI] [PubMed]
- Ramm SA, McDonald L, Hurst JL, Beynon RJ, Stockley P. Comparative proteomics reveals evidence for evolutionary diversification of rodent seminal fluid and its functional significance in sperm competition. Mol Biol Evol. 2009;26:189–198. doi: 10.1093/molbev/msn237. [DOI] [PubMed] [Google Scholar]
- Ramm SA, Oliver PL, Ponting CP, Stockley P, Emes RD. Sexual selection and the adaptive evolution of mammalian ejaculate proteins. Mol Biol Evol. 2008;25:207–219. doi: 10.1093/molbev/msm242. [DOI] [PubMed] [Google Scholar]
- Revelle W (2015) Package ‘psych’—procedures for psychological, psychometric and personality research. Version 1 2 12.
- Schärer L. Tests of sex allocation theory in simultaneously hermaphroditic animals. Evolution. 2009;63:1377–1405. doi: 10.1111/j.1558-5646.2009.00669.x. [DOI] [PubMed] [Google Scholar]
- Schärer L. Evolution: don’t be so butch, dear! Curr Biol. 2014;24:R311–R313. doi: 10.1016/j.cub.2014.03.003. [DOI] [PubMed] [Google Scholar]
- Schärer L, Janicke T. Sex allocation and sexual conflict in simultaneously hermaphroditic animals. Biol Lett. 2009;5:705–708. doi: 10.1098/rsbl.2009.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schärer L, Joss G, Sandner P. Mating behaviour of the marine turbellarian Macrostomum sp.: these worms suck. Mar Biol. 2004;145:373–380. [Google Scholar]
- Schärer L, Ladurner P. Phenotypically plastic adjustment of sex allocation in a simultaneous hermaphrodite. Proc R Soc B Biol Sci. 2003;270:935–941. doi: 10.1098/rspb.2002.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schärer L, Ladurner P, Rieger RM. Bigger testes do work more: experimental evidence that testis size reflects testicular cell proliferation activity in the marine invertebrate, the free-living flatworm Macrostomum sp. Behav Ecol Sociobiol. 2004;56:420–425. [Google Scholar]
- Schärer L, Pen I. Sex allocation and investment into pre- and post-copulatory traits in simultaneous hermaphrodites: the role of polyandry and local sperm competition. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120052. doi: 10.1098/rstb.2012.0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schärer L., Ramm S.A. Encyclopedia of Evolutionary Biology. 2016. Hermaphrodites; pp. 212–224. [Google Scholar]
- Schärer L, Vizoso DB. Phenotypic plasticity in sperm production rate: there’s more to it than testis size. Evol Ecol. 2007;21:295–306. [Google Scholar]
- Scheiner SM, Schlichting CD, Pigliucci M. Phenotypic evolution: a reaction norm perspective. Am J Bot. 1999;86:145. [Google Scholar]
- Sekii K, Vizoso DB, Kuales G, De Mulder K, Ladurner P, Schärer L. Phenotypic engineering of sperm-production rate confirms evolutionary predictions of sperm competition theory. Proc R Soc B. 2013;280:20122711. doi: 10.1098/rspb.2012.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LW, Fitzpatrick JL. Sperm wars and the evolution of male fertility. Reproduction. 2012;144:519–534. doi: 10.1530/REP-12-0285. [DOI] [PubMed] [Google Scholar]
- Simmons LW, Kotiaho JS. Evolution of ejaculates: patterns of phenotypic and genotypic variation and condition dependence in sperm competition traits. Evolution. 2002;56:1622–1631. doi: 10.1111/j.0014-3820.2002.tb01474.x. [DOI] [PubMed] [Google Scholar]
- Simmons LW, Lovegrove M. Socially cued seminal fluid gene expression mediates responses in ejaculate quality to sperm competition risk. Proc R Soc B. 2017;284:20171486. doi: 10.1098/rspb.2017.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons LW, Parker GA. Individual variation in sperm competition success of yellow dung flies Scatophaga stercoraria. Evolution. 1992;46:366–375. doi: 10.1111/j.1558-5646.1992.tb02044.x. [DOI] [PubMed] [Google Scholar]
- Sirot LK, Wolfner MF, Wigby S. Protein-specific manipulation of ejaculate composition in response to female mating status in Drosophila melanogaster. Proc Natl Acad Sci USA. 2011;108:9922–9926. doi: 10.1073/pnas.1100905108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan NS, Lovegrove M, Simmons LW. Social manipulation of sperm competition intensity reduces seminal fluid gene expression. Biol Lett. 2018;14:20170659. doi: 10.1098/rsbl.2017.0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DT, Hosken DJ, French-Constant RH, Wedell N. Variation in sex peptide expression in D. melanogaster. Genet Res. 2009;91:237–242. doi: 10.1017/S0016672309000226. [DOI] [PubMed] [Google Scholar]
- Snook RR, Bacigalupe LD, Moore AJ. The quantitative genetics and coevolution of male and female reproductive traits. Evolution. 2010;64:1926–1934. doi: 10.1111/j.1558-5646.2010.00958.x. [DOI] [PubMed] [Google Scholar]
- South A, Sirot LK, Lewis SM. Identification of predicted seminal fluid proteins in Tribolium castaneum. Insect Mol Biol. 2011;20:447–456. doi: 10.1111/j.1365-2583.2011.01083.x. [DOI] [PubMed] [Google Scholar]
- Spurgeon SL, Jones RC, Ramakrishnan R. High throughput gene expression measurement with real time PCR in a microfluidic dynamic array. PLoS ONE. 2008;3:e1662. doi: 10.1371/journal.pone.0001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger S, Stökl J. The role of sexual selection in the evolution of chemical signals in insects. Insects. 2014;5:423–438. doi: 10.3390/insects5020423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffel MA, Nakagawa S, Schielzeth H. rptR: repeatability estimation and variance decomposition by generalized linear mixed-effects models. Methods Ecol Evol. 2017;8:1639–1644. [Google Scholar]
- Tan GN, Govedich FR, Burd M. Social group size, potential sperm competition and reproductive investment in a hermaphroditic leech, Helobdella papillornata (Euhirudinea: Glossiphoniidae) J Evol Biol. 2004;17:574–580. doi: 10.1111/j.1420-9101.2004.00692.x. [DOI] [PubMed] [Google Scholar]
- Thomsen R, Soltis J, Matsubara M, Matsubayashi K, Onuma M, Takenaka O. How costly are ejaculates for Japanese macaques? Primates. 2006;47:272–274. doi: 10.1007/s10329-005-0171-7. [DOI] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Iterson M, Boer JM, Menezes RX. Filtering, FDR and power. BMC Bioinform. 2010;11:450. doi: 10.1186/1471-2105-11-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellnow N, Vizoso DB, Viktorin G, Schärer L. No evidence for strong cytonuclear conflict over sex allocation in a simultaneously hermaphroditic flatworm. BMC Evol Biol. 2017;17:103. doi: 10.1186/s12862-017-0952-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizoso DB, Rieger G, Schärer L. Goings-on inside a worm: functional hypotheses derived from sexual conflict thinking. Biol J Linn Soc. 2010;99:370–383. [Google Scholar]
- Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh B, Blows MW. Abundant genetic variation + strong selection=multivariate genetic constraints: a geometric view of adaptation. Annu Rev Ecol Evol Syst. 2009;40:41–59. [Google Scholar]
- Ward PI. Intraspecific variation in sperm size characters. Heredity. 1998;80:655–659. doi: 10.1046/j.1365-2540.1998.00401.x. [DOI] [PubMed] [Google Scholar]
- Ward PI. Sperm length is heritable and sex-linked in the yellow dung fly (Scathophaga stercoraria) J Zool. 2000;251:349–353. [Google Scholar]
- Wasik K, Gurtowski J, Zhou X, Ramos OM, Delás MJ, Battistoni G, et al. Genome and transcriptome of the regeneration-competent flatworm, Macrostomum lignano. Proc Natl Acad Sci USA. 2015;112:201516718. doi: 10.1073/pnas.1516718112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Wunderer J, Lengerer B, Pjeta R, Rodrigues M, Schärer L, et al. A targeted in situ hybridization screen identifies putative seminal fluid proteins in a simultaneously hermaphroditic flatworm. BMC Evol Biol. 2018;18:81. doi: 10.1186/s12862-018-1187-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West-Eberhard MJ. Phenotypic plasticity and the origins of diversity. Annu Rev Ecol Syst. 1989;20:249–278. [Google Scholar]
- Wigby S, Perry JC, Kim YH, Sirot LK. Developmental environment mediates male seminal protein investment in Drosophila melanogaster. Funct Ecol. 2016;30:410–419. doi: 10.1111/1365-2435.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigby S, Sirot LK, Linklater JR, Buehner N, Calboli FCF, Bretman A, et al. Seminal fluid protein allocation and male reproductive success. Curr Biol. 2009;19:751–757. doi: 10.1016/j.cub.2009.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wudarski J, Simanov D, Ustyantsev K, de Mulder K, Grelling M, Grudniewska M, et al. Efficient transgenesis and annotated genome sequence of the regenerative flatworm model Macrostomum lignano. Nat Commun. 2017;8:2120. doi: 10.1038/s41467-017-02214-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane T, Goenaga J, Rönn JL, Arnqvist G. Male seminal fluid substances affect sperm competition success and female reproductive behavior in a seed beetle. PLoS One. 2015;10:1–14. doi: 10.1371/journal.pone.0123770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Clark AG, Fiumera AC. Natural genetic variation in male reproductive genes contributes to nontransitivity of sperm competitive ability in Drosophila melanogaster. Mol Ecol. 2013;22:1400–1415. doi: 10.1111/mec.12113. [DOI] [PubMed] [Google Scholar]
- Team, R. C. (2013) R: A language and environment for statistical computing.
- Epskamp S, Cramer AO, Waldorp LJ, Schmittmann VD, Borsboom D. qgraph: Network visualizations of relationships in psychometric data. J Stat Softw. 2012;48:1–18. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data and analysis code are archived with Dryad; 10.5061/dryad.n3pg1hm.