Abstract
Strain diversity, while now recognized as a key driver underlying partner dynamics in symbioses, is usually difficult to experimentally manipulate and image in hosts with complex microbiota. To address this problem, we have used the luminous marine bacterium Vibrio fischeri, which establishes a symbiosis within the crypts of the nascent light organ of the squid Euprymna scolopes. Competition assays in newly hatched juvenile squid have shown that symbiotic V. fischeri are either niche-sharing “S strains”, which share the light organ when co-inoculated with other S strains, or niche-dominant “D strains”, which are typically found alone in the light organ after a co-colonization. To understand this D strain advantage, we determined the minimum time that different V. fischeri strains needed to initiate colonization and used confocal microscopy to localize the symbionts along their infection track. Further, we determined whether symbiont-induced host morphogenic events also occurred earlier during a D strain colonization. We conclude that D strains colonized more quickly than S strains. Nevertheless, light-organ populations in field-caught adult squid often contain both D and S strains. We determined experimentally that this symbiont population heterogeneity might be achieved in nature by a serial encounter of different strains in the environment.
Subject terms: Symbiosis, Population dynamics
Introduction
In the field of microbial symbiosis, significant interest focuses on community-level structure and on distinguishing the contributions of different species within complex host-associated microbiomes [1–5]. Recently, an awareness of the importance of strain-level differences occurring within a colonizing bacterial species in such communities has emerged [1, 6–8]. While such strain-level differences have been hard to investigate within complex microbiomes, the squid-vibrio symbiosis is a powerful natural symbiosis that is ideally suited to explore strain-level differences within animal-bacterial relationships. Vibrio fischeri is the sole light-organ symbiont of the Hawaiian bobtail squid, Euprymna scolopes. The symbionts are provided nutrients in exchange for their bioluminescence, which is used by the nocturnal host for counterillumination [9], a common defensive behavior in the marine environment. V. fischeri cells in the ambient seawater establish themselves in this symbiotic niche within hours after the juvenile squid hatches [10]. Such single-species specificity is a feature that allows easy investigation of the dynamics of a multi-strain symbiosis.
Once the juvenile squid hatch, V. fischeri cells are harvested from the ambient seawater [11, 12] and, within two hours, begin to aggregate on the ciliated surface near the pores of the light organ. In the meantime, macrophage like cells, the host hemocytes, begin to accumulate in blood sinuses beneath the ciliated tissues of the light organ [13]. The aggregated bacteria then enter the pores of the light organ and migrate through ducts, bottlenecks, and antechambers before reaching their symbiotic niche, the epithelia-lined crypts [14]. After 12 h, the light organ of the host enters a symbiont-induced developmental program characterized by dramatic morphologic changes [10]. The symbiotic population, in turn, begins a daily cycle in which 95% of the light-organ bacteria are vented from the crypts at dawn; by nightfall, the remaining symbionts proliferate, repopulate the crypts, and produce bioluminescence, until they are vented again the next morning [15].
While the majority of investigations of the squid colonization process (for a review see ref. [16]), have been performed using a V. fischeri strain named ES114 [17], there has been an increasing interest in strain-level differences. For instance, several studies have described different V. fischeri strains isolated from squid collected around the island of Oahu, Hawaii [1, 18, 19]. While there is no biogeographic distinction between symbionts of different squid populations, various isolates are characterized by one of two different colonization behaviors [1]. When exposing juvenile squids to two strains simultaneously, a “D strain” outcompetes other co-inoculated strains for colonization, while the “S strains” eventually share the host light organ [1]. Nevertheless, both D and S strains have been isolated from same field-caught squid [19], begging the question of how such mixtures are achieved and maintained in nature.
We aimed to understand what factors the D strains’ dominant behavior relies upon, and to determine how squid become colonized by both D and S strains in the wild. While reported in some other V. fischeri strains [20], we have encountered no evidence of interference competition with the strains we tested, either during the colonization process or within the crypts of the squid light organ. Instead, our results suggest that the dominant behavior is due, at least in part, to a quicker achievement of the colonization process; however, a multi-strain symbiosis is possible if the squid encounters different strains at sequential times after hatching.
Material and methods
Bacterial strains
All the strains used in this study are V. fischeri isolated from light organs of the Hawaiian bobtail squid, E. scolopes, caught around the island of Oahu, Hawaii [18, 19]. Strains ES213, KB2B1, MB11B1, MB13B2, and MB13B3 have been described as dominant-type strains (D strains), while the others are sharing-type strains (S strains) [1]. Depending on the experiment, the strains were labeled, either (i) by harboring the plasmid pVSV102 (encoding a green fluorescent protein; GFP) or pVSV208 (encoding a red fluorescent protein; RFP) [21], or (ii) with a GFP (or RFP) and erythromycin-encoding cassette in the Tn7 site of the strains [1].
Colonization procedure
Overnight cultures were grown at 28 °C in Luria-Bertani salt (LBS) media [22] with antibiotics, when necessary, at 2.5, 5, or 100 µg ml−1 for chloramphenicol (pVSV208), erythromycin (Tn7-site insertion), and kanamycin (pVSV102), respectively. The overnight cultures were diluted a hundred time in seawater tryptone (SWT) medium broth and grown with shaking at 28 °C until mid-exponential phase (0.5 OD600 nm). Bacteria were diluted to a concentration of ~5000 cells per ml in 50 mL of filter-sterilized ocean water (FSOW) in which newly hatched juvenile squids were transferred. This concentration of bacteria allows optimal colonization efficiency and effectiveness [23]. For co-inoculation experiments, a second or third strain was added either simultaneously or at a later time, depending on the question. The final concentration of the inoculum differed somewhat from one experiment to another; however, in co-colonization inocula, the concentrations of the two strains were within 20% of each other. While this concentration of inoculum is somewhat higher than the 100–1000 CFU of V. fischeri per mL that are present in the environment surrounding squid populations [24], it is within the natural range reported in some studies [25]. Incubations were performed at room temperature, between 21 and 24 °C, as previously described [17].
To determine the exposure time needed for a strain to colonize the squid, we incubated animals with each test strain for different lengths of time. Squids were rinsed twice for 1 min, and once for 5 min, in vials containing 4 mL of FSOW. The squids were then transferred into a new vial and incubated overnight in a room with 12:12 day/night cycle. The following day, we measured the bioluminescence of each squid using a luminometer (Turner Designs, Inc.).
For experiments designed to determine the outcome of a co-colonization assay with two different strains, each strain was either GFP or RFP labeled, and the co-inoculation performed. After 24 h, individual squids were placed in vials containing 4 mL of FSOW, and the water was changed every day. Squids were frozen at −80 °C at 48 h post inoculation for experiments involving three strains, and at 72 h for those with two strains. Each squid was homogenized, different dilutions were spread onto LBS agar, and the number of colony-forming units (CFU) of each strain were calculated. GFP- and RFP- expressing CFUs were counted using a fluorescence dissecting scope. Light-organ population with > 99% of one strain were considered mono-colonized.
Confocal microscopy
Imaging was performed on a Zeiss LSM710 laser-scanning confocal microscope. To localize labeled bacteria during the colonization process, we anesthetized the squid at different times post inoculation in 2% ethanol and fixed them in 4% paraformaldehyde in mPBS (50 mM sodium phosphate buffer with 0.45 M NaCl, pH 7.4). After 48 h, the squids were rinsed four times for 30 min each, in mPBS and dissected. When appropriate, light organs tissue was counter stained overnight with 2 µM TOTO3 (Thermo Fisher Scientific, Inc.) in 1% TRITON-X100 in mPBS, and rinsed four times for 15 min in mPBS. Light organs were mounted using Vectashield mounting media (Thermo Fisher Scientific, Inc.).
To measure hemocyte trafficking into, and the length of, the light organs ciliated appendages ([13, 26]), squids colonized for 18 h were anesthetized with 2% ethanol and fixed in 4% paraformaldehyde in mPBS. After 24 h, the squids were rinsed four times for 30 min in mPBS, dissected and left overnight in 1 mL of 1% Triton-X100 containing 100 µg/mL deoxyibonuclease I, Alexa Fluor 488 conjugate (Thermo Fisher Scientific, Inc.). The following day, the light organs were rinsed four times in mPBS, and labeled overnight with 25 µg of rhodamine phalloidin per ml (Thermo Fisher Scientific, Inc.). After another four rinses in mPBS, the samples were mounted using Vectashield mounting media (Thermo Fisher Scientific, Inc.). Optical sections of the anterior appendages were analyzed using Fiji software [27]. After outlining the perimeter of the largest section of the anterior appendage, we determined the maximum longitudinal-sectioned area for each side of an organ, and the mean for each treatment was determined.
Scanning electron microscopy (SEM)
Juvenile squids were colonized, anesthetized in 2% ethanol, and fixed in 4% paraformaldehyde in mPBS at either 18 h or 41 h post inoculation, and processed for SEM as previously described [28]. Samples were examined with a Hitachi S-4800 FESEM scanning electron microscope at the University of Hawaii at Manoa Biological Electron Microscopy Facility (BEMF).
Results
D strains initiate symbiosis more quickly than S strains
We first asked whether D and S strains require the same length of incubation time to initiate a successful colonization of the host (Fig. 1). We exposed squid to individual strains at a concentration aimed at ~5000 CFU/mL, for either 30 or 60 min, then rinsed them to eliminate any bacteria in the surrounding water. The next day, we measured the squid’s bioluminescence, which is an indicator of a successful colonization. If a strain effectively colonized more than 50% of the squids within 30 min, we tried a shorter, 15 min, exposure. When 1 h was insufficient to achieve > 50% colonization, we used a 3 h inoculation, which results in 100% colonization for strain ES114 [23]. While three out of five D strains needed only 30 min of exposure to colonize > 50% of the squid, six out of seven S strains needed between 1 and 3 h (Fig. 1). This pattern followed the hierarchy of strain competitiveness for the squid colonization that was previously described [1]. For example, strain KB2B1, described as the most dominant, needed only 15 min of exposure to reach more than 50% colonized. Two of the D strains needed more than 30 min to reach this ratio, but the extent of colonized squid was at least as great as that for the S strains that required 1 h of exposure (Fig. 1). These results suggest that D strains need less time than the S strains to reach a location that was not accessible to the rinse (i.e., within the duct of the light organ). Four strains were then chosen to continue the study of differences between the D and S types: the two D strains having the strongest colonization phenotype (Fig. 1), the strongest of the S strains, and ES114, the best studied V. fischeri strain [1]. In Fig. 1, we used the name of the strains as published previously [1, 18, 19]; however, to simplify and clarify our continuing discussion, in subsequent figures we used the following pseudonyms for the four chosen strains: D1 (KB2B1), D2 (MB13B2), S1 (MB13B1) and S2 (ES114).
Fig. 1.
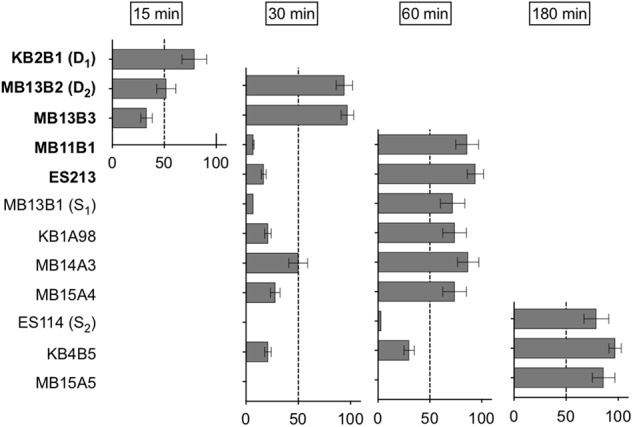
Minimum exposure time needed for V. fischeri strains to initiate a successful colonization. Squids were exposed to each bacterial strain for different periods of time, then rinsed, and colonized animals identified by their production of bioluminescence after 24 h. Strains in bold are D strains, whereas the others are S strains. The X-axis indicates the percentage of squid colonized after a given length of exposure. Strains are ranked (top to bottom) based on the minimum amount of exposure needed to colonize 50% of the animals. For each condition, between 25 and 31 animals were measured in three replicates. Error bars indicate 95% confidence intervals
D strains reach the light-organ crypts earlier than S strains
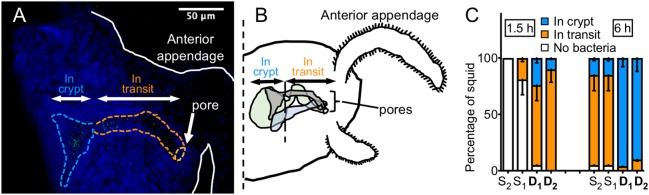
To determine whether the colonization advantage of D strains resulted from their reaching the crypts of the light organ earlier than S strains, we inoculated squid with GFP-labeled strains and, after 1.5 and 6 h, fixed the squid and localized them within the light organ by confocal microscopy. In this way we could record their position along the colonization path (Fig. 2a, b). For each squid we determined whether there were bacteria in the crypt, in transit to the crypt (pore, duct, bottleneck, or antechamber), or not yet within the tissue [16]. In most cases, no bacteria were detected in the squid 1.5 h after inoculation with an S strain. On the other hand, at that time point, both D strains were localized in the duct and, in some animals, even in the crypts. At 6 h, while most of the S strains were in transit, the D strains were already in the crypts (Fig. 2c). These results suggest that the D strains reach their niche earlier than the S strains.
Fig. 2.
Strain localization during colonization. a Confocal image of a light organ being colonized by GFP-labeled V. fischeri (green); tissue nuclei (blue) were stained with TOTO-3. Some bacteria were in transit (orange dashed area) and some were in the crypts (blue dashed area). One crypt is visualized in this optical section, but all crypts were examined for the analysis. b Schematic representation of one side of the squid light organ. V. fischeri strains enter through the pores and, after transit, reach the crypts where they grow and luminesce. c Using confocal microscopy, we identified where GFP-labeled bacterial cells were located in the light organ at different times after inoculation. For each light organ, we determined whether there was no evidence the bacteria had begun migrating into the tissues (white) or, when bacteria were present, whether they were still in transit (orange) or had reached the crypts (blue) and begun growing. Between 19 and 23 animals were analyzed in two replicates. The graph represents the mean percentages for each result, and the error bars the 95% confidence interval
Some developmental changes in host morphology occur earlier during D strain colonization
We wanted to know whether D strains triggered host morphological changes earlier than S strains; specifically, (i) hemocyte trafficking into the ciliated appendages and (ii) regression of these structures. We first looked at the number of hemocytes found in the appendages of 18-h colonized squid. While there was no distinction between the number of hemocytes per appendage between squid colonized either by D or S strains (Supplementary Figure S1), we observed that there was a significant difference in the degree of appendage regression. We measured the mean size of the anterior appendage of squid colonized by either a D or an S strain (Fig. 3a), and the results indicated that the regression of the arm occurs earlier with D strain infections. It should be noted that, because the appendage harbored the same number of hemocytes, but was reduced in volume when colonized by a D strain the tissue density of hemocytes is actually greater in the appendages of 18-h D strain colonized squid than 18-h squid colonized by an S strain. To complement this analysis, we observed by SEM the light organ of squid colonized either by a D or an S strain, at 18 and 41 h post inoculation (Fig. 3b). The morphological changes, i.e., the extent of regression of the light-organ appendages, occurred earlier with a D strain.
Fig. 3.
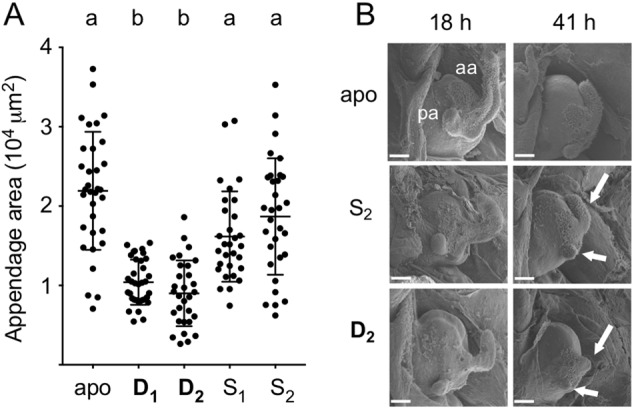
Induction of symbiont-induced regression in the host light organ. a Squids were colonized by one dominant (D1 or D2) or sharing (S1 or S2) strain for 18 h (aposymbiotic, or apo = non-colonized animals), after which the extent of regression of the organ’s appendages was determined for each condition by measuring the mean longitudinal cross-sectional area of the anterior appendage. Each dot corresponds to an individual squid. Between 29 and 35 squids were analyzed in two to three replicates. Means with standard deviations are presented. Differences between conditions were assessed by a Dunn’s test; letters above the columns indicate significantly different means (p < 0.05). b Squids were colonized by a single strain for 18 or 41 h, and observed by scanning electron microscopy to assess regression of the anterior (aa) and posterior (pa) appendages; bars = 60 mm. After colonization by strain D2, but not S2, both appendages are almost totally regressed by 41 h
D and S behavior can be affected by the timing of encounter between host and symbiont
We previously showed that, during a mixed-strain co-inoculation including a D strain, squid light organs are singly colonized [1]. However, D and S strains are often isolated from the same light organ of a field-caught adult squid [19] (e.g., D2 and S1 were isolated from the same squid [15]). We wanted to understand how, within the context of a dominant strategy, both types of strains can share a light organ. We tried to colonize the squid with both a D and an S strain using either different total inoculum levels, or different ratios between the strains; however, the outcome was uniformly similar: the D strain was the only one found in the light organ of the squid (data not shown). We then considered that, to achieve a co-colonization with the two types, the S strain might need to begin colonizing before the D strain. To test this hypothesis, we inoculated the squid with either a D or an S strain, and then we added another D or S strain at different times (Fig. 4). After 72 h, we determined whether the squids were single- or co-colonized. As expected, when a D strain was added at time 0, an S strain could not co-colonize the squid no matter when it was added. When two D strains were added at the same time, most of the squids were singly colonized by the most dominant one; however, when added serially, the D strain added initially had an increasing advantage the later the second strain was added. When two S strains were added, half of the squids were co-colonized no matter when the second strain was added. When the initial strain was an S strain, added at least 3 h before a D strain, 50% of the squids were co-colonized; to promote co-colonization, the optimal time to add the D strain was 3 h after the S strain. We concluded that, in the wild, juvenile squid may have an opportunity for a multi-strain symbiosis by encountering strains at different times.
Fig. 4.
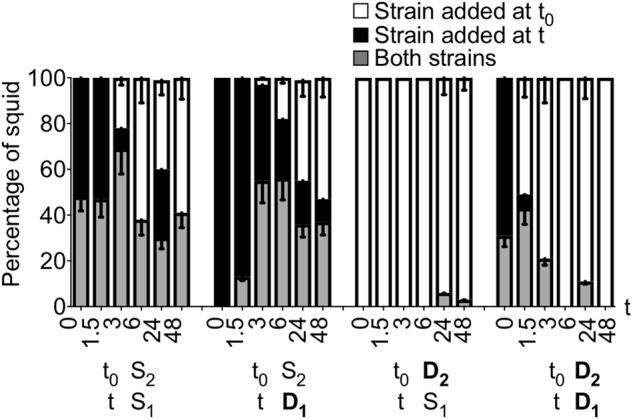
State of colonization of juvenile squid encountering two strains presented sequentially. Squids were inoculated with strain D2 or S2 at time zero (t0), and strains D1 or S1 were added into the surrounding water at different times (t, in hours; X-axis). The percentage of squids either co-colonized (gray), or colonized only by the strain added either initially (white) or later (black), were determined. Between 29 and 38 animals were analyzed in three replicates. The graph represents the mean percentages for each result, and the error bars the 95% confidence interval
Two different strains rarely share the same crypt
We also wondered whether the different types of strains in a mixed colonization were co-localized or spread between the six distinct crypts of the juvenile organ. Extensive imaging by confocal and TEM has shown that the six crypts are all independent compartments [29]; further, there is no evidence for the presence of sub-compartments within any one crypt. Specifically, all co-colonized crypts observed contained a homogenous population of the two strains: no consistent indication of partitioning between S and D strains has been reported. We inoculated squids with (i) two S, (ii) two D, or (iii) one S, with one D strain added immediately or after 3 h; we then observed the fluorescently labeled bacteria in the light organ at 48 h using confocal microscopy (Fig. 5). As expected, when squids were exposed to a D and an S strain at the same time, all the colonized crypts harbored the D strain. When squids were colonized with either two D or two S strains, we found the most dominant strain of the pair was present in more crypts than the other. Interestingly when we added a D strain 3 h after an S strain, we found more crypts to be colonized by the D strain than by the S strain, suggesting that the S strain had left “empty” crypts that could be colonized by subsequent bacteria in contrast, the D strains appeared to quickly colonize all available crypts. In any case, among over 836 colonized crypts observed under the four conditions, we rarely found a co-colonized crypt; i.e., only 10 crypts (~1%) were co-colonized. We concluded that the strains, independent of their behavior, rarely share the same location, and are instead spread among the six crypts of the squid’s light organ.
Fig. 5.
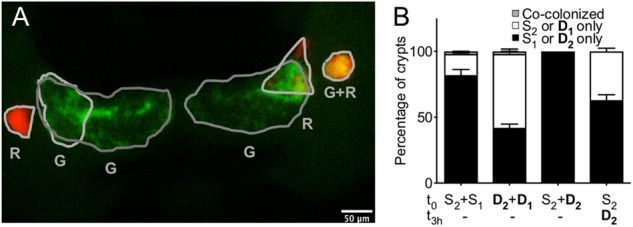
Strain localization in the crypts of the light organ. Squids were inoculated with one of four different strain combinations: (i) two S strains, (ii) two D strains, or (iii) an S strain, with a D strain added either simultaneously, or (iv) after a 3 h delay. (a) After 48 h, squids were fixed, and their light organs observed under a confocal microscope to determine the percentage of their 6 colonized crypts (gray outline) that were singly colonized by a GFP- (G) or an RFP- (R) labeled strain or were co-colonized by both of the inoculated strains (GR). (b) For each treatment, 53 or 54 animals were analyzed in four replicates, corresponding to between 187 and 217 colonized crypts observed per treatment. The bar graph represents the mean percentage of each outcome, and the error bars the 95% confidence interval. The crypts analyzed were either co-colonized (gray) by both of the inoculating strains, or were only singly colonized by one of the pair of strains (white) or the other (black)
Differential exposure of strains enables multi-strain symbiosis
Finally, we wanted to test the hypothesis that a multi-strain symbiosis can be experimentally achieved by exposing the squid to three strains at different times. Squids were inoculated with two S strains, and a D strain was added after 3 h, and the strain population was determined after 48 h (Fig. 6a). Of the squid analyzed, we found all the combinations possible for the three strains. We also obtained around 40% of the squids colonized by the three strains. Those results support the hypothesis that the diversity observed in field-caught hosts is due to the squid sequentially encountering different strains in the ambient seawater environment.
Fig. 6.
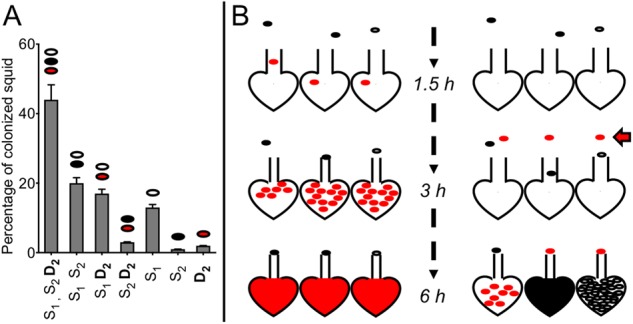
a The composition of symbiont populations in squid inoculated with three strains. Animals were first exposed to an inoculum of two S strains (S1 and S2; black and white ovals); after 3 h, strain D2 (red ovals) was added. After 72 h, we determined the percentage of colonized squid that had one of the 6 possible combinations of the three inoculated strains. One hundred squids were analyzed in three replicates. The graph represents the mean percentage, and the error bars the 95% confidence intervals. b Model for the colonization of a light organ’s crypts with a mixture of S and D strains. Two scenarios are given (left and right panels) indicating how the crypts might become colonized, depending on when the host encounters the two kinds of strains. Each scenario is illustrated using three crypts that depict different possible trajectories for the progression of colonization: (i) when all three strains (S1, S2 and D2) are added at the same time (left panel), the D strain will migrate more rapidly than the S strains, which cannot reach the deep crypts before they have been colonized, and the bottleneck has been closed by the host in response to the presence of bacteria in the crypt [30]; (ii) when there is a sequential encounter of strains (right panel), S strains have the opportunity to migrate into and colonize at least some crypts, while D strains encountered 3 h later (red arrow) can only colonize any remaining empty crypts whose bottleneck has not been closed
In our current model (Fig. 6b), when a D strain is encountered in the environment, it has the ability to reach the light-organ crypts more rapidly than an S strain, and establish a symbiosis within 3 h, thereby often pre-empting the subsequent colonization by the slower S strains. Alternatively, if an S strain arrives sufficiently ahead of any D strain, it has the opportunity to exclusively colonize the crypts. Our data indicate that, D strains do not directly interact with S strains but, instead, they rapidly signal the host that symbiosis has been achieved, triggering the constriction of the bottleneck present in the migration pathway [30], and thereby blocking slower/later strains from subsequent entry (Fig. 6b).
Discussion
The work presented here indicates that V. fischeri D strains achieve an earlier colonization of the squid light organ than S strains. Specifically, (i) exposing juvenile squid to D strains for as little as 30 min is sufficient to ensure a successful colonization; (ii) D strains traverse the colonization path to the light-organ crypts more rapidly, and (iii) in the presence of a colonizing D strain, symbiont-induced morphological changes of the host seem to be triggered earlier. In addition, this study addresses how D and S strains may be found co-existing in the light-organ populations of wild-caught animals. If newly hatched squid encounter an S strain several hours before a D strain, the host can be successfully co-colonized. However, independently of the inoculum, an individual crypt is rarely found to be co-colonized by both a D and an S strain, indicating that co-colonization is achieved through habitat segregation. Finally, a colonization by as many as three different strains can be achieved by manipulating the sequence and timing of addition of the strains, a process that may occur naturally in the field. Overall, we showed that, even within the same bacterial species, the dynamics of their behaviors can govern the degree of multi-strain colonization achieved in a population of animals.
The distinct steps of the colonization process appear to occur earlier with a D strain than with an S strain. This difference could be due to D strain-specific features, perhaps encoded within the additional 250 kb of genetic material they share [1]. Unfortunately, the dispersion of these 250 kb around the chromosomes, the proportion of hypothetical proteins and lack of obvious candidates they encode, as well as the difficulty in genetic manipulation large sections of genetic material in V. fischeri, have made a direct investigation of this genetic addition challenging. However, new genetics tools developed in this species will simplify a search of this D strain-specific genetic material [31]. In addition, because these 250 kb are present in all the D strains, but we observed a wide range in both the rate of migration (Fig. 1) and the competitiveness hierarchy [1] within this group, it seems unlikely that carriage of the 250 kb is sufficient to explain the migration phenotype.
One hypothesis for the migration dominance is that D strains have the capacity to swim faster than S strains, allowing them to reach and begin colonizing the crypts sooner. We were unable to validate this obvious possibility; specifically, D strains generally swim more slowly in soft-agar medium [19] and, when observed in liquid (SWT) suspension, no motility difference specific to the D strains was detected. Nevertheless, we cannot completely reject this hypothesis: the symbionts migrate through unique environmental conditions within the light organ, ones that are not mimicked by soft-agar medium or SWT. In addition, the bacteria may not use typical flagellar motility when colonizing the squid [32]. In any case, if the D strains actually move faster within the specific tissue environment of the light organ, that could explain their more rapid colonization (Fig. 1), and future studies should focus on this topic. Regardless of their general swimming speed, D strains may chemotax differently, either more quickly to the chitin-degradation products that attract an S strain [33] or, perhaps, to an unknown, D strain-specific chemoattractant. Kremer et al. [34] reported a “priming” of chemotaxis to chitin breakdown products by the S strain ES114, preparing the bacteria to find and gain entry into the host’s tissues [33]. This process was associated with a 3-h behavioral delay that may not be required by the D strains. Finally, D strains may be more intrinsically resistant to antimicrobial peptides or other stresses encountered on the migration route into the light-organ crypts (McFall-Ngai et al., 2010; [35, 36]); however, it is not clear how such an attribute would influence the timing of colonization.
Our present hypothesis for the typical exclusion of the S strains is neither a specific bacteria–bacteria interaction (e.g., a type VI secretion system involved in the intra-species killing of other V. fischeri strains is not specific to either D or S strains [20]), nor a host-sanctioning effect [37], because all the D and S strains used here were isolated from the light organs of adult animals [1, 18, 19]. Neither do we believe that the larger aggregations produced by some D strains [38] play a deciding role in the dominance behavior, because there is no direct correlation between the size of the aggregates and the competitiveness of the strains. Instead, we propose that the D strains arrive earlier in the crypt and signal the host that colonization has been initiated in a given crypt, which may deter the entry of other strains. This strategy is reminiscent of how, when multiple pollen tubes grow within the same style of a flower, a successful fertilization event initiates developmental processes in the plant that eliminate subsequent pollen entry [39]. However, crypt colonization is unlikely to be solely driven by priority, as has been shown for two strains of Borrelia burgdorferi co-inoculated into ticks at different times [40], because, when an S strain encounters the squid first, a D strain can still occasionally colonize secondarily (Fig. 6a). In a two species community in the zebrafish, when Vibrio cholerae is present, there is exclusion of other species by the mobility of the host gut [41]. There could be a similar mechanical exclusion of supernumerary V. fischeri strains once a D strain is in a crypt.
As we report here (Fig. 5), a light-organ crypt rarely (<1% of the time) was colonized by more than a single strain, an observation that indicates the predominance of clonal (i.e., single-bacterium) crypt colonization events. These results contrast with those of a previous study [19], in which a dual colonization of the crypts occurred 21% of the time. In this earlier study, squids were co-inoculated with two isogenic derivatives of MB13B2, carrying either a GFP- or RFP-encoding plasmid for identification. We hypothesize that the difference in the extent of co-inoculation between these two studies can be explained by the notion that two clonal strains have a higher chance of simultaneously entering the same crypt than two genetically (and phenotypically) distinct ones. This hypothesis supports the idea that colonization outcome is based on genetic differences between strains, rather than any direct strain-strain interaction. In any case, host-based mechanisms that promote a clonal population in each crypt would help avoid a scenario in which a mixed-population crypt contained a mutualistic strain and a cheater strain of V. fischeri [37, 42], a frequently occurring condition in the root nodules of certain rhizobia-plant symbioses [43]. Moreover, compartmentalization of individual symbiont strains within different crypts, also observed in the gut microbes of fungus-growing termites [44], is a strategy that minimizes cheating by ensuring high within-species relatedness within a location [45]. Thus, we propose that the squid has developed a colonization strategy that includes restricting supernumerary colonization (Fig. 6b), and that pre-emptively discourages crypt co-colonization to avoid the cost of subsequently sanctioning cheaters [37].
The competitive dominance of D strains of V. fischeri [1] raises the question: why don’t these strains sweep the V. fischeri population found in squid habitats? Indeed, all the experiments performed here use the same optimized inoculum size to ensure that each strain has an equal opportunity to encounter the squid. In the animal’s natural environment, the concentration of V. fischeri is in the hundreds to thousands of CFU per mL [25, 46]; so, one possibility is that the D strains are actually less fit in the planktonic environment through which they must pass between hosts. This hypothesis is supported by the observation that D strains persist less well when incubated in unfiltered ocean water [18], thereby increasing the probability that, upon leaving the egg, a newly hatched squid will first encounter an S strain rather than a D strain. The presence of both D and S strains in a host’s light-organ population increases symbiont diversity in the light organ, even if they are not in the same crypt; such carriage could help protect this mono-specific symbiosis from the potential of phage sweeps [47]. The daily venting behavior of the host’s light organ may also encourage the development of strain diversity because 95% of the symbiont population is lost each day of the host’s 9–12 month life span. The remaining 5% serving as the source of subsequent repopulation, producing opportunities for mutation and genetic diversification. Moreover, to our knowledge, the recruitment and colonization process occurs only once during the life of each host, and the chances of any one of the billions of vented symbionts encountering and colonizing a newly hatched juvenile is very remote; thus, it seems reasonable to imagine that there would not be an overriding selection to optimize initiation fitness over survival in the plankton. The results presented here suggest that as many as six strains may initially colonize a host organ; however, a determination of the number of strains present in an adult squid has not yet been reported, and would be complicated because of the subsequent evolution of the initial colonizers.
Studies of strain-to-strain challenge during colonization, such as those described here, are rare: most work focuses either on complex bacterial communities, or the dynamics between a mutant strain and its isogenic wild type [48, 49]. For instance, it has been shown in the oyster that an avirulent derivative can play a role in enhancing disease caused by a virulent strain [50, 51]. In contrast, some studies have described the co-existence in a host of different strains of the same species, e.g., several strains of Lactobacillus reuteri colonize the gastrointestinal tract of its hosts [8]. However, the spatial relationship of different strains remains difficult to determine. The intra-species population dynamics described here may enlighten behavior of more complex communities. For instance, is the dominant/sharing dynamic characteristic of the squid-vibrio symbiosis reflective of intra-species behaviors in other, more complex, symbioses, and how might such behaviors strengthen or weaken the robustness of a host’s complex community? To inform such studies, we look forward to learning whether D- and S strain behaviors are common to all V. fischeri strains and their symbiotic hosts or, instead, are specific to the colonization of E. scolopes.
Electronic supplementary material
Acknowledgements
We thank the Ruby and McFall-Ngai lab members, together with Émilie Koch, for their helpful discussions and support. We also thank Tina Carvalho for imaging training and use of the BEMF facility of the University of Hawaii at Manoa. Support was provided by NIH grants from NIGMS (GM099507), NIAID (AI050661) and ORIP (RR012294/OD011024).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0305-8) contains supplementary material, which is available to authorized users.
References
- 1.Bongrand C, Koch EJ, Moriano-Gutierrez S, Cordero OX, McFall-Ngai M, Polz MF, et al. A genomic comparison of 13 symbiotic Vibrio fischeri isolates from the perspective of their host source and colonization behavior. ISME J. 2016;10:2907–017. doi: 10.1038/ismej.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee KWK, Periasamy S, Mukherjee M, Xie C, Kjelleberg S, Rice SA. Biofilm development and enhanced stress resistance of a model, mixed-species community biofilm. ISME J. 2014;8:894–907. doi: 10.1038/ismej.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerwin AH, Nyholm SV. Symbiotic bacteria associated with a bobtail squid reproductive system are detectable in the environment, and stable in the host and developing eggs. Environ Microbiol. 2017;19:1463–75. doi: 10.1111/1462-2920.13665. [DOI] [PubMed] [Google Scholar]
- 4.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–30. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwong WK, Moran NA. Gut microbial communities of social bees. Nat Rev Microbiol. 2016;14:374–84. doi: 10.1038/nrmicro.2016.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McLean AHC, Parker BJ, Hrček J, Kavanagh JC, Wellham PAD, Godfray HCJ. Consequences of symbiont co-infections for insect host phenotypes. J Anim Ecol. 2018;87:478–88. doi: 10.1111/1365-2656.12705. [DOI] [PubMed] [Google Scholar]
- 7.Yawata Y, Cordero OX, Menolascina F, Hehemann JH, Polz MF, Stocker R. Competition-dispersal tradeoff ecologically differentiates recently speciated marine bacterioplankton populations. Proc Natl Acad Sci USA. 2014;111:5622–7. doi: 10.1073/pnas.1318943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duar RM, Frese SA, Lin XB, Fernando SC, Burkey TE, Tasseva G, et al. Experimental evaluation of host adaptation of Lactobacillus reuteri to different vertebrate species. Appl Environ Microbiol. 2017;83:e00132–17. doi: 10.1128/AEM.00132-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones BW, Nishiguchi MK. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda) Mar Biol. 2004;144:1151–5. doi: 10.1007/s00227-003-1285-3. [DOI] [Google Scholar]
- 10.McFall-Ngai M, Ruby E. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science. 1991;254:1491–4. doi: 10.1126/science.1962208. [DOI] [PubMed] [Google Scholar]
- 11.Nawroth JC, Guo H, Koch E, Heath-Heckman EAC, Hermanson JC, Ruby EG, et al. Motile cilia create fluid-mechanical microhabitats for the active recruitment of the host microbiome. Proc Natl Acad Sci USA. 2017;114:9510–6. doi: 10.1073/pnas.1706926114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nyholm SV, McFall-Ngai M. The winnowing: establishing the squid–vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–42. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 13.Koropatnick TA, Kimbell JR, McFall-Ngai MJ. Responses of host hemocytes during the initiation of the squid-vibrio symbiosis. Biol Bull. 2007;212:29–39. doi: 10.2307/25066578. [DOI] [PubMed] [Google Scholar]
- 14.McFall-Ngai M, Nyholm SV, Castillo MG. The role of the immune system in the initiation and persistence of the Euprymna scolopes-Vibrio fischeri symbiosis. Sem Immunol. 2010;22:48–53. doi: 10.1016/j.smim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graf J, Ruby EG. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc Natl Acad Sci USA. 1998;95:1818–22. doi: 10.1073/pnas.95.4.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McFall-Ngai MJ. The importance of microbes in animal development: lessons from the squid-vibrio symbiosis. Annu Rev Microbiol. 2014;68:177–94. doi: 10.1146/annurev-micro-091313-103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boettcher KJ, Ruby EG. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol. 1990;172:3701–6. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wollenberg MS, Ruby EG. Phylogeny and fitness of Vibrio fischeri from the light organs of Euprymna scolopes in two Oahu, Hawaii populations. ISME J. 2012;6:352–62. doi: 10.1038/ismej.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wollenberg MS, Ruby EG. Population structure of Vibrio fischeri within the light organs of Euprymna scolopes squid from Two Oahu (Hawaii) populations. Appl Environ Microbiol. 2009;75:193–202. doi: 10.1128/AEM.01792-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Speare L, Cecere AG, Guckes KR, Smith S, Wollenberg MS, Mandel MJ, et al. Bacterial symbionts use a type VI secretion system to eliminate competitors in their natural host. Proc Natl Acad Sci USA. 2018;115:8528–37. doi: 10.1073/pnas.1808302115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol. 2006;72:802–10. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graf J, Dunlap PV, Ruby EG. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol. 1994;176:6986–91. doi: 10.1128/jb.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCann J, Stabb EV, Millikan DS, Ruby EG. Population dynamics of Vibrio fischeri during Infection of Euprymna scolopes. Appl Environ Microbiol. 2003;69:5928–34. doi: 10.1128/AEM.69.10.5928-5934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee KH, Ruby EG. Symbiotic role of the viable but nonculturable state of Vibrio fischeri in Hawaiian coastal seawater. Appl Environ Microbiol. 1995;61:278–83. doi: 10.1128/aem.61.1.278-283.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones BW, Maruyama A, Ouverney CC, Nishiguchi MK. Spatial and temporal distribution of the Vibrionaceae in coastal waters of Hawaii, Australia, and France. Microb Ecol. 2007;54:314–23. doi: 10.1007/s00248-006-9204-z. [DOI] [PubMed] [Google Scholar]
- 26.Heath-Heckman EAC, McFall-Ngai MJ. The occurrence of chitin in the hemocytes of invertebrates. Zoology. 2011;114:191–8. doi: 10.1016/j.zool.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: An open-source platform for biological-image analysis. Nat Meth. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doino JA, McFall-Ngai MJ. A transient exposure to symbiosis-competent bacteria induces light organ morphogenesis in the host squid. Biol Bull. 1995;189:347–55. doi: 10.2307/1542152. [DOI] [PubMed] [Google Scholar]
- 29.Montgomery MK, McFall-Ngai MJ. Embryonic development of the light organ of the sepiolid squid Euprymna scolopes Berry. Biol Bull. 1993;184:296–308. doi: 10.2307/1542448. [DOI] [PubMed] [Google Scholar]
- 30.Sycuro LK, Ruby EG, McFall-Ngai M. Confocal microscopy of the light organ crypts in juvenile Euprymna scolopes reveals their morphological complexity and dynamic function in symbiosis. J Morphol. 2006;267:555–68. doi: 10.1002/jmor.10422. [DOI] [PubMed] [Google Scholar]
- 31.Visick KL, Hodge-Hanson KM, Tischler AH, Bennett AK, Mastrodomenico V. Tools for rapid genetic engineering of Vibrio fischeri. Appl Environ Microbiol. 2018;84:e00850–18. doi: 10.1128/AEM.00850-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kinosita Y, Kikuchi Y, Mikami N, Nakane D, Nishizaka T. Unforeseen swimming and gliding mode of an insect gut symbiont, Burkholderia sp. RPE64, with wrapping of the flagella around its cell body. ISME J. 2018;12:838–48. doi: 10.1038/s41396-017-0010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandel MJ, Schaefer AL, Brennan CA, Heath-Heckman EAC, DeLoney-Marino CR, McFall-Ngai MJ, et al. Squid-derived chitin oligosaccharides are a chemotactic signal during colonization by Vibrio fischeri. Appl Environ Microbiol. 2012;78:4620–6. doi: 10.1128/AEM.00377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kremer N, Philipp EER, Carpentier MC, Brennan CA, Kraemer L, Altura MA, et al. Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe. 2013;14:183–94. doi: 10.1016/j.chom.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schleicher TR, Nyholm SV. Characterizing the host and symbiont proteomes in the association between the bobtail squid, Euprymna scolopes, and the bacterium, Vibrio fischeri. PLoS ONE. 2011;6:e25649. doi: 10.1371/journal.pone.0025649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen F, Krasity BC, Peyer SM, Koehler S, Ruby EG, Zhang X, et al. Bactericidal permeability-increasing proteins shape host-microbe interactions. mBio. 2017;8:e00040–17. doi: 10.1128/mBio.00040-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koch EJ, Miyashiro T, McFall-Ngai MJ, Ruby EG. Features governing symbiont persistence in the squid-vibrio association. Mol Ecol. 2014;23:1624–34. doi: 10.1111/mec.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koehler S, Gaedeke R, Thompson C, Bongrand C, Visick K, Ruby E, et al. The model squid-vibrio symbiosis provides a window into the impact of strain- and species-level differences during the initial stages of symbiont engagement. Environ Microbiol. 2018 doi: 10.1111/1462-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams JH, Mazer SJ. Pollen—tiny and ephemeral but not forgotten: new ideas on their ecology and evolution. Am J Bot. 2016;103:365–74. doi: 10.3732/ajb.1600074. [DOI] [PubMed] [Google Scholar]
- 40.Devevey G, Dang T, Graves CJ, Murray S, Brisson D. First arrived takes all: Inhibitory priority effects dominate competition between co-infecting Borrelia burgdorferi strains. BMC Microbiol. 2015;15:61. doi: 10.1186/s12866-015-0381-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiles TJ, Jemielita M, Baker RP, Schlomann BH, Logan SL, Ganz J, et al. Host gut motility promotes competitive exclusion within a model intestinal microbiota. PLoS Biol. 2016;14:e1002517. doi: 10.1371/journal.pbio.1002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Visick KL, Foster J, Doino J, McFall-Ngai M, Ruby EG. Vibrio fischerilux genes play an important role in colonization and development of the host light organ. J Bacteriol. 2000;182:4578–86. doi: 10.1128/JB.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Checcucci A, Azzarello E, Bazzicalupo M, Galardini M, Lagomarsino A, Mancuso S, et al. Mixed nodule infection in Sinorhizobium meliloti–Medicago sativa symbiosis suggest the presence of cheating behavior. Front Plant Sci. 2016;7:835. doi: 10.3389/fpls.2016.00835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aanen DK, De Fine Licht HH, Debets AJM, Kerstes NAG, Hoekstra RF, Boomsma JJ. High symbiont relatedness stabilizes mutualistic cooperation in fungus growing termites. Science. 2009;326:1103–6. doi: 10.1126/science.1173462. [DOI] [PubMed] [Google Scholar]
- 45.Kuzdzal-Fick JJ, Fox SA, Strassmann JE, Queller DC. High relatedness is necessary and sufficient to maintain multicellularity in Dictyostelium. Science. 2011;334:1548–51. doi: 10.1126/science.1213272. [DOI] [PubMed] [Google Scholar]
- 46.Lee KH, Ruby EG. Detection of the light organ symbiont, Vibrio fischeri, in Hawaiian seawater by using lux gene probes. Appl Environ Microbiol. 1992;58:942–7. doi: 10.1128/aem.58.3.942-947.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Betts A, Gray C, Zelek M, MacLean RC, King KC. High parasite diversity accelerates host adaptation and diversification. Science. 2018;360:907–11. doi: 10.1126/science.aam9974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wein T, Dagan T, Fraune S, Bosch TCG, Reusch TBH, Hülter NF. Carrying capacity and colonization dynamics of Curvibacter in the hydra host habitat. Front Microbiol. 2018;9:443. doi: 10.3389/fmicb.2018.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–9. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lemire A, Goudenège D, Versigny T, Petton B, Calteau A, Labreuche Y, et al. Populations, not clones, are the unit of vibrio pathogenesis in naturally infected oysters. ISME J. 2015;9:1523–31. doi: 10.1038/ismej.2014.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Le Roux F, Wegner KM, Polz MF. Oysters and vibrios as a model for disease dynamics in wild animals. Trends Microbiol. 2016;24:568–80. doi: 10.1016/j.tim.2016.03.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.