Abstract
Immune tolerance is a highly regulated state and involves diverse mechanisms. Central to the induction of tolerance is the targeted modulation of T-cell activities (both effector and regulatory), in which transcription factors play a significant role. The nuclear factor kappa-B (NF-κB) family is a family of transcription factors that not only are critically involved in diverse T-cell responses but also are regulated by many mechanisms to maintain tolerance and T-cell homeostasis. NF-κB, as a transcription factor, has been extensively studied in recent decades, and the molecular mechanisms that regulate NF-κB activities have been well documented. However, recent studies have revealed exciting new roles for NF-κB; in addition to its transcriptional activity, NF-κB can also activate diverse epigenetic mechanisms that mediate extensive chromatin remodeling of target genes to regulate T-cell activities. In this review article, we highlight recent discoveries and emerging opportunities in targeting NF-κB family members as well as their associated chromatin modifiers in the induction of immune tolerance and in the clinical treatment of immune diseases.
Key words: Immune tolerance, NF-κB, chromatin modifiers
Subject terms: Immunology, Allotransplantation
Introduction
Tolerance is a hallmark of the immune system, a state that is often primarily defined based on the fundamental features of immune cells in self versus nonself recognition. Tolerance, per se, exhibits different forms and features, depending on the nature of the antigens and the time of induction as well as on the mechanisms involved.1 Tolerance to self tissues and organs is developmentally acquired, as developing T cells that are potentially aggressive toward self (i.e., autoreactive T cells) are deleted in the thymus, while self-tolerant T cells are allowed to mature and populate the periphery where they respond vigorously to nonself foreign antigens. In addition, in the thymus, a small population of T cells are selected to express the transcription factor Foxp3 (Foxp3 + Tregs), which endows these cells with suppressive properties to further enforce the tolerant state in the periphery.2–4 Tolerance can also be established to foreign antigens in a mature immune repertoire; this form of tolerance is called acquired tolerance. The mechanisms of acquired tolerance are multifaceted and involve apoptotic death, anergy, ignorance and T effector cell exhaustion, as well as the active suppression of effector cells by various types of regulatory cells.5,6
Immune tolerance can be lost (for example, in autoimmune diseases) or induced (as in transplant tolerance), and both of these events have enormous clinical implications. At the center of these events are T cells; their activation, survival, and effector functions, as well as their commitment to regulatory T cells, have a profound impact on the final outcomes of immunity versus immune tolerance.2 There are multiple signals that act contemporaneously and/or at different times in guiding T-cell activities, and those signals are triggered by T-cell receptor (TCR) activation and the engagement of T-cell costimulatory molecules and cytokine receptors as well as homing receptors and chemokine receptors. Importantly, some of the signaling pathways that activate T effector cells also promote the suppressive function of regulatory T cells, whereas others stimulate T effector cells but disarm regulatory T cells.7 This interconnected nature of functionally diverse T-cell subsets not only clearly ensures robust immunity and immune homeostasis but also provides tremendous challenges in targeting individual T-cell subsets in tolerance induction and the clinical treatment of immune-mediated diseases.
One of the striking features of T-cell activation is that signals from TCRs, costimulatory molecules, and cytokine receptors often converge to activate a myriad of transcription factors, which further coordinate the differentiation of effector T cells into a diverse repertoire of functionally different T helper subsets (e.g., Th1, Th2, Th9, Th17, and Tfh). These lineage-defining transcription factors (e.g., T-bet for Th1 cells and GATA3 for Th2 cells) are themselves the subject of complex regulation.8 It is important to note that in activated T cells, the accessibility of target genes to lineage-specific transcription factors is regulated by complex epigenetic mechanisms, wherein chromatin remodeling must occur first to make specific target genes accessible to the transcriptional machinery. Clearly, the epigenetic apparatus that assembles in activated T cells to drive chromatin remodeling is an area of considerable interest,9 as studies in this area may reveal novel targets to therapeutically modulate immune responses. However, in contrast to other areas of T-cell biology, the epigenetic mechanisms that are induced after T-cell activation and their roles in regulating the fate of various T-cell subsets are areas in which much remains to be studied. Additionally, very little is known about the potential interactions between T-cell activation signals, chromatin modifiers, and transcriptional regulators in fine-tuning effector versus regulatory programs during immune responses.
In this review article, we focus on the nuclear factor kappa-B (NF-κB) family of transcription factors because members of this family are prominently induced during early T-cell activation. We highlight recent advances in our understanding of the role of NF-κB family members in controlling T-cell tolerance. We discuss the unique roles of the noncanonical NF-κB pathway, the induction of this pathway by Tumor necrosis factor receptor superfamily (TNFR SF) costimulatory molecules, and the role of this pathway in mediating chromatin modifications during T-cell activation. We specifically highlight the observation that, in addition to their transcriptional activities, members of the NF-κB family exert profound impacts on T-cell responses by activating chromatin modifiers and thus control multiple aspects of immunity and immune tolerance. We conclude by discussing emerging opportunities for targeting NF-κB-activated chromatin modifiers in the clinical treatment of immune-mediated diseases.
The NF-κB family of transcription factors
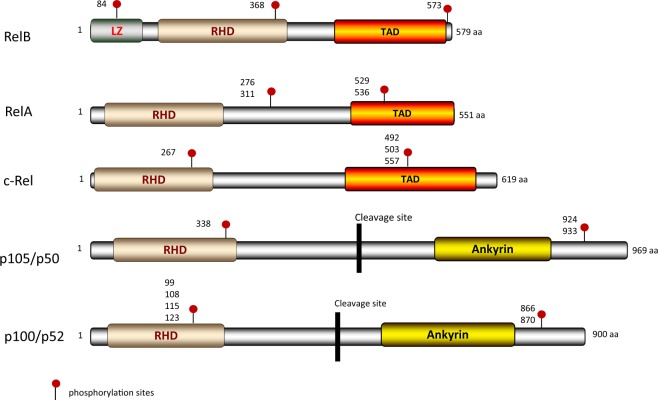
Induction of the NF-κB family of transcription factors is one of the earliest features of T-cell activation; members of this family play a significant role in the function and development of activated T cells. Traditionally, the NF-κB family consists of at least five members, namely, RelA (also called p65), RelB, c-Rel, p105, and p100.10,11 The p105 and p100 proteins require proteolytic cleavage to produce the active products p50 (NF-κB1) and p52 (NF-κB2), respectively.12 Structurally, all NF-κB family members share a Rel homology domain (RHD), which mediates homo- or heterodimerization with other NF-κB family members. The RHD domain is also responsible for the nuclear localization of NF-κB complexes as well as for their binding to the κB sites of NF-κB-responsive genes.12,13 RelA, RelB, and c-Rel also contain a transcription activation domain (TAD), which is required for activation of target gene expression. Interestingly, p105 and p100 and their cleaved products p50 and p52 do not have the TAD domain, rendering them incapable of activating gene transcription unless they form heterodimeric complexes with RelA, RelB, or c-Rel.13,14 Unlike the other family members, RelB also has a leucine zipper (LZ) domain, although the exact function of this LZ domain remains to be defined (Fig. 1). It should be noted that all NF-κB family members are subject to extensive posttranslational modifications, including phosphorylation, ubiquitination, and proteolytic cleavage, which in turn profoundly affect their biological functions.15,16
Fig. 1.
Structure of NF-κB family members. The Rel homology domain (RHD) is shared by all family members. RelA, RelB, and c-Rel contain a transactivation domain (TAD), and RelB has a leucine zipper domain (LZ). The p105 and p100 proteins have five to seven ankyrin repeats (AnkR), which can be cleaved to generate the p50 and p52 products after the activation signal is received. Regulatory phosphorylation sites are shown
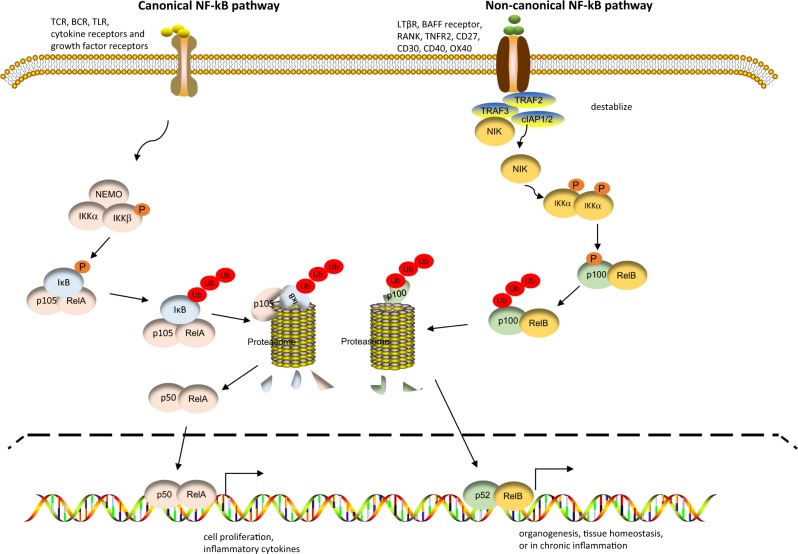
NF-κB family members have been extensively studied over the last decades for their roles as transcription factors. It has been well documented that the transcriptional activities of NF-κB family members are tightly regulated by multiple mechanisms. In resting T cells, NF-κB family members are transcriptionally inactive; they are either retained in the cytosol (e.g., RelA, p105, and p50) or segregated to prevent the formation of transcriptionally active heterodimers (e.g., RelB, p100, and p52).17 In activated T cells, there are two broadly defined pathways by which NF-κB family members are activated—the canonical and noncanonical pathways (Fig. 2), which are driven by the degradation of the NF-κB inhibitor IκB or the induction of NF-κB-inducing kinase (NIK), respectively.18 The canonical NF-κB pathway is activated by numerous cell surface receptors, including TCRs, B-cell receptors, and Toll-like receptors, as well as cytokine receptors and growth factor receptors. This pathway drives robust cell proliferation, survival, and inflammatory cytokine (i.e. interleukin (IL)-1, IL-2, IL-6, Tumor necrosis factor alpha (TNF-α) etc.) production: events that are typically seen in the acute phase of immune activation. Activation of the canonical NF-κB pathway tends to occur early and transiently during immune activation.18 At the molecular level, the engagement of cell surface receptors triggers the activation of IκB kinase (IKK), which consists of IKKα, IKKβ, and IKKγ (also called NEMO or NF-κB essential modulator). Activated IKK then phosphorylates the NF-κB inhibitor IκB, resulting in the prompt ubiquitination and proteasome-mediated degradation of IκB. IkB degradation releases RelA and p105, which allows the proteolytic cleavage of p105 to produce p50. RelA can then form heterodimers with p50, and these RelA/p50 heterodimers then translocate to the nucleus, where they bind and activate their target genes.19,20
Fig. 2.
Pathways of nuclear factor kappa-B (NF-κB) activation. Activation of the canonical NF-κB pathway depends on the IKK complex, which consists of IKKα, IKKβ, and NEMO. Activated IKK phosphorylates the NF-κB inhibitor IκB, resulting in IκB degradation, which releases RelA and p105. Upon proteolytic cleavage of p105 to p50, RelA forms heterodimers with p50, and these RelA/p50 heterodimers translocate to the nucleus, where they bind and activate target genes. Activation of the noncanonical NF-κB pathway requires the induction of NIK in the cytosol. NIK associates with and activates IKKα, which phosphorylates p100, resulting in the proteolytic cleavage of p100 to generate p52. Then, p52 forms a heterodimer with RelB, and this RelB/p52 complex translocates to the nucleus to activate its target genes
Activation of the noncanonical NF-κB pathway (RelB/p52) involves mechanisms and exhibits features very different from those of canonical NF-κB pathway activation.21 The noncanonical NF-κB pathway is activated by a selective cohort of cell surface receptors consisting primarily of receptors in the TNFR superfamily, including the lymphotoxin-β receptor (LTβR), the B-cell activating factor (BAFF) receptor, the receptor activator of NF-κB (RANK), TNFR2, CD27, CD30, CD40, and OX40 (also known as CD134) as costimulatory receptors.22 Compared to activation of the canonical NF-κB pathway, which tends to be rapid but transient, activation of the noncanonical NF-κB pathway is much slower but can be sustained for a prolonged period of time. Thus, the noncanonical NF-κB pathway is prominently involved in long-term cell programs, such as organogenesis and tissue homeostasis, or in chronic inflammation.21 Mechanistically, activation of the noncanonical NF-κB pathway is critically dependent on NIK. Under normal circumstances, NIK is constantly degraded to prevent activation of the noncanonical NF-κB pathway by this kinase. The degradation of NIK is mediated primarily by a complex consisting of TRAF2, TRAF3, cIAP1, and cIAP2 molecules, in which cIAP1/2 act as an E3 ubiquitin ligase.23 Specifically, TRAF2/3 recruits NIK to the complex, whereupon cIAP1/2 ubiquitinates NIK via K48-linked ubiquitination. This event targets NIK for proteolysis, and as a result, NIK is undetectable in resting cells. However, in activated T cells, especially after the engagement of TNFR superfamily receptors, the TRAF2/TRAF3/cIAP1/cIAP2 complex is destabilized, which prevents proteolytic degradation of NIK. This inability results in the accumulation of NIK in the cytosol of activated T cells. After accumulating to a certain level in the cytosol, NIK associates with and subsequently activates IKKα, which then phosphorylates p100, resulting in the proteolytic cleavage of p100 to generate p52. The p52 protein then forms heterodimers with RelB, and the RelB/p52 complex translocates to the nucleus to activate its target genes.24
Despite differences in their activation, biological functions, and impact, the canonical and noncanonical NF-κB pathways participate in extensive crosstalk at multiple levels. Thus, these two pathways are also interdependent and likely crossregulate each other.17,25 For example, IKKα intersects with both the canonical and noncanonical pathways. As mentioned above, IKKα is activated by NIK and plays an important role in the activation of the noncanonical NF-κB pathway through proteolytic cleavage of p100 to generate p52. However, IKKα alone cannot activate the noncanonical pathway, suggesting that NIK induction and accumulation in the cytosol is a prerequisite and that IKKα and NIK must cooperate to phosphorylate p100 in order to generate p52. Conversely, IKKα is also involved in the canonical pathway as an important component of the IKK complex, which phosphorylates IκB, resulting in IκB degradation and subsequent activation of the canonical NF-κB pathway. Furthermore, under certain conditions, IKKα can also directly phosphorylate canonical NF-κB family members and thereby promote their transcriptional activity.26–28
Another example of crosstalk concerns the dimerization of NF-κB family members. Typically, p50 forms dimers with RelA, and p52 forms dimers with RelB during canonical activation and noncanonical activation, respectively. However, under certain conditions, NF-κB components can form nontraditional dimers with other family members. It has been shown that in T cells, p52 can dimerize with c-Rel to drive prominent tissue inflammation.29 Studies using gene deletion approaches have demonstrated an unexpected interdependence involving p50 and p52. For example, RelB/p52 signaling is important for the development of secondary lymphoid organs. However, p52 knockout mice and RelB knockout mice show very different phenotypes in terms of lymph node formation and germinal center organization.30 Interestingly, deletion of p52, which usually dimerizes with RelB, results in the formation of p50/RelB heterodimers, which are not regulated by the canonical IκB complex, and this p50/RelB complex can compensate for the loss of RelB/p52 heterodimers. Similarly, p50 knockout mice show a mild deficiency in inflammatory responses that is not as severe as the deficiency seen in RelA knockout mice. The loss of p50 results in the formation of RelA/p52 heterodimers that can also respond to inflammatory stimuli, leading to the activation of target genes that are often activated by the canonical NF-κB pathway.30,31 Unexpectedly, p50 knockout mice show defects in lymph node development, and those defects are attributed to the enhanced processing of p100 to produce p52 as a binding partner for RelA. This enhanced processing drastically diminishes p100 levels in p50 knockout cells, which, in turn, indirectly inhibits noncanonical pathway activation by siphoning p52 away from RelB and the p52/RelB signaling pathway.32 Clearly, the activation and biological functions of NF-κB pathways are complex. The interdependent nature of such pathways presents both challenges and opportunities in targeting individual signaling pathways for therapeutic purposes.
Regulation of the transcriptional activities of NF-κB family members
There are multiple mechanisms that control the tempo as well as the dynamics of NF-κB pathways. In addition to those mechanisms that trigger NF-κB activation, as discussed above, several regulatory mechanisms exist to terminate NF-κB activation in a timely manner. This termination is critical to prevent pathological conditions that otherwise develop when NF-κB activation is dysregulated.
In essence, the NF-κB inhibitor IκB is the primary regulator of canonical NF-κB activation. Of particular interest is the observation that IκB is also a target gene of the canonical NF-κB pathway, as NF-κB activation leads to robust expression of IκB. Newly synthesized IκB enters the nucleus as a monomer and readily associates with p50/RelA dimers, thus preventing these dimers from engaging their target genes. This event results in the nuclear expulsion of p50/RelA dimers into the cytosol. Thus, IκB provides a potent negative feedback loop to suppress NF-κB activation.15
Another example of termination of canonical NF-κB activation involves the ubiquitin-modifying enzyme A20.33,34 A20 has dual ubiquitin-modifying activities; the carboxyl-terminal of A20 contains a zinc finger (ZnF) domain, which has ubiquitin ligase activity, while the amino-terminal OTU domain exhibits protein deubiquitinating activity. In most settings, canonical NF-κB activation results in prompt A20 expression, and a key function of A20 is to mediate the degradation of signaling molecules critical to canonical NF-κB activation (e.g., RIP1, TRAF6, and MALT1), either by removing the K63-linked polyubiquitin chains (responsible for promoting cell functions such as kinase activation and signal transduction) or by adding K48-linked polyubiquitin chains (responsible for targeting substrates for proteasome-mediated degradation).35–37 Thus, A20 expression prevents the propagation of signals that activate the IKK complex and thereby terminates canonical NF-κB activation. The importance of A20 as a negative regulator of NF-κB is clearly demonstrated in gene knockout mice, as A20-deficient mice develop severe multiorgan inflammation and cachexia and usually die within a few weeks after birth.38 A deficiency of A20 specifically in intestinal epithelial cells, myeloid cells, dendritic cells, or B cells results in organ-specific autoimmune symptoms similar to those observed in inflammatory bowel diseases, rheumatoid arthritis, and lupus-like diseases.39 Recent studies demonstrated that the CYLD and OTULIN deubiquitinases can negatively regulate NF-κB activation and that their substrates frequently overlap with those of A20.40,41 However, mice with genetic knockout of CYLD and OTULIN exhibited a phenotype substantially different from that of A20-deficient mice, suggesting that each ubiquitin-modifying enzyme may have unique functions.41 Further studies are warranted to better understand how such deubiquitinases regulate NF-κB activation and contribute to autoimmune diseases caused by dysregulated NF-κB pathways.
As mentioned above, the signaling cascade mediating the activation of the noncanonical NF-κB pathway differs significantly from that mediating canonical NF-κB activation. The induction of NIK and processing of p100 to p52 are key events in noncanonical NF-κB activation, and accordingly, NIK and p100 are the focal points of regulation. It has been shown that NLRP12 (also called Monarch1), which is a member of the nucleotide-binding domain and leucine-rich repeat-containing (NLR) family, can promote rapid degradation of NIK and consequently inhibit noncanonical NF-κB pathway activation.42 Moreover, NLRP12 directly binds TRAF3 and stabilizes its enzymatic activities, which results in NIK degradation and the suppression of noncanonical NF-κB activation.42,43 Another molecule, named OTUD7B (also known as Cezanne), was recently identified as a negative regulator of the noncanonical NF-κB pathway. OTUD7B is a deubiquitinase that exhibits sequence homology with the OTU domain of A20.44 Hu et al.44 found that upon the engagement of CD40, BAFFR, or LTβR, OTUD7B deficiency significantly enhanced noncanonical NF-κB activation, leading to increased nuclear accumulation of p52 and RelB and a reciprocal decrease in cytosolic p100 levels. Mechanistically, the engagement of CD40, BAFFR, or LTβR results in the recruitment of OTUD7B along with the TRAF2/TRAF3/cIAP1/cIAP2 complex. Via its ubiquitin-association domain, OTUD7B then associates with TRAF3. OTUD7B stabilizes TRAF3 by deubiquitinating TRAF3, and as a result, TRAF3 is hyperactivated and continuously degrades NIK to prevent noncanonical NF-κB activation.44 Clearly, OTUD7B is an attractive therapeutic target for modulating the noncanonical NF-κB pathway.
The NF-κB family is also regulated in cells by preventing the spontaneous activation of NF-κB family members. As discussed previously, RelB and p100 exist in a stable but transcriptionally inactive cytoplasmic complex called the kappaBsome. Upon the activation of immune cells, a transient complex, which contains RelB, NIK, IKKα, and p100, is formed. RelB competes with NIK and IKKα for p100 binding in this complex, with RelB promoting p100 stabilization and NIK/IKKα promoting p100 phosphorylation and processing to generate p52. If RelB outcompetes NIK and IKKα for p100 binding, p100 phosphorylation by IKKα is precluded, and p100 and RelB are maintained in the inert kappaBsome. Thus, the intrinsic function of RelB is to preserve p100, preventing spurious activation of the noncanonical pathway; only when the ratio of NIK/IKKα to RelB is sufficient to outcompete RelB can p52 and the active p52/RelB heterodimer be formed.45 Cells also prevent spontaneous NF-κB activation through the appropriate temporal separation of activating signals and receptors. For example, deletion of the ESCRT (endosomal sorting complexes required for transport) complex leads to the accumulation of LTβR and TNFR on endosomes, resulting in the spontaneous activation of both the canonical and noncanonical NF-κB pathways.46 ESCRT, therefore, plays a critical role in maintaining appropriate cellular organization to prevent aberrant signaling. Thus, cells have developed a variety of mechanisms to prevent inappropriate NF-κB activation and maintain homeostasis.
Evidence suggests that microRNAs, which are small, noncoding, single-stranded RNAs, can modulate NF-κB activity either positively or negatively. It has been shown that microRNA146 and microRNA155 negatively regulate NF-κB activation by inhibiting expression of TRAF6 and IKKβ. Conversely, microRNA-181b promotes NF-κB activation by inhibiting CYLD expression. Additionally, long noncoding RNAs (lncRNAs) are emerging as another important regulator of NF-κB signaling; lncRNAs have been shown to inhibit IκB phosphorylation and IκB downregulation.47,48 Thus, microRNAs and lncRNAs may be promising targets for modulating NF-κB activation. However, much remains to be studied before translating microRNA and lncRNA-based therapeutics from the bench to the bedside.
NF-κB family members as chromatin modifiers
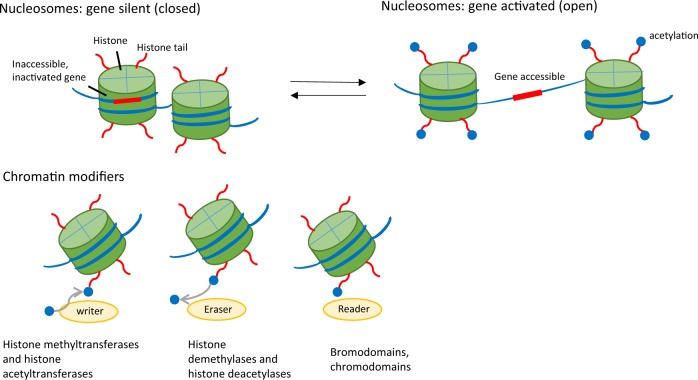
In all eukaryotic cells, cellular DNA is wound around histone proteins in the nucleus to form dense, compact chromatin structures, which can be further compacted into chromosomes. The highly compacted chromatin is called heterochromatin and is transcriptionally silent, as the gene loci in the chromatin are inaccessible to transcription factors and cofactors. Transcriptionally active chromatin is called euchromatin and is characterized by loosely linked nucleosomes where genes in the unwound DNA strands are fully accessible to the cellular transcriptional apparatus. Generally, the process that converts heterochromatin to euchromatin, or vice versa, is called chromatin remodeling and is usually driven by cellular activation (or deactivation) involving complex epigenetic mechanisms.49,50 Chromatin remodeling entails chemical modifications to both histone protein tails and DNA through acetylation (of histones) and methylation (of histones and DNA) at specific sites. There are a myriad of epigenetic enzymes, termed “chromatin modifiers,” identified so far that mediate chromatin remodeling during various stages of immune activation. Chromatin modifiers are classified into three groups, often called chromatin writers, erasers, and readers (Fig. 3). Chromatin writers are enzymes that add acetyl or methyl groups to chromatin (e.g., acetyltransferases and methyltransferases), while chromatin erasers (e.g., histone deacetylases and demethylases) remove these groups. Chromatin readers recognize and bind modified chromatin to trigger additional activities during gene expression.51 Generally, histone acetylation, especially H3K27 acetylation (H3K27ac), is associated with transcriptional activation of target genes, while histone methylation often renders target genes transcriptionally inactive.52 It is important to note that chromatin modifications precede target gene transcription, which is dependent on the correct epigenetic profile based on the coordinated action of complex chromatin modifiers. The interface between chromatin modifiers and transcription factors is an area of considerable interest because this interaction may yield new insights into mechanisms of gene regulation. Interestingly, the NF-κB family of transcription factors is well positioned at this interface and is critically involved in chromatin remodeling.53,54
Fig. 3.
Epigenetic control of gene expression. The left panel depicts tightly compacted chromatin, which is transcriptionally silent, as the gene loci are inaccessible to transcription factors and cofactors. The right panel shows loosely linked nucleosomes, which are transcriptionally active, as the unwound DNA strands are fully accessible to the cellular transcriptional apparatus
One noteworthy feature of NF-κB family members is their ability to form multimeric complexes. In addition to dimerization with other members within the NF-κB family, these proteins also complex with other proteins, including chromatin modifiers. Notably, NF-κB family members lack intrinsic enzymatic activity for modifying chromatin, but they can recruit and position chromatin modifiers onto target genes in a sequence-specific manner to mediate chromatin remodeling. Thus, depending on the specific binding partners, NF-κB family members can mediate either gene transcription (as NF-κB heterodimers) or chromatin modifications (in complex with chromatin modifiers). We recently reported that the engagement of the costimulatory receptor GITR activates the canonical NF-κB pathway (RelA/p50) in activated CD4+T cells, and under conditions of iTreg-induction (in the presence of Transforming growth factor Beta (TGF-β) and IL-2), GITR stimulation turns off iTreg-induction and redirects iTreg cells to differentiate into inflammatory Th9 cells.55 Mechanistically, we showed that p50 binds and recruits the histone deacetylases HDAC1 and Sirt1 to the promoter and CNS regions of the Foxp3 locus, which contain the p50 binding sites. Consequently, HDAC1 and Sirt1 catalyze extensive histone deacetylation to close the Foxp3 locus. This suppression of Foxp3 renders iTregs permissive to differentiation into Th9 cells,55 suggesting that p50-activated epigenetic mechanisms may convert a tolerogenic environment to an inflammatory environment. In fact, the transcription factor BATF3 can repress Foxp3 expression by recruiting the histone deacetylase Sirt1.56 This finding is consistent with other reports that p50 is capable of interacting with HDAC proteins in different cell types.57,58 It should be noted that the p50-mediated chromatin remodeling process is independent of the transcriptional activity of p50.
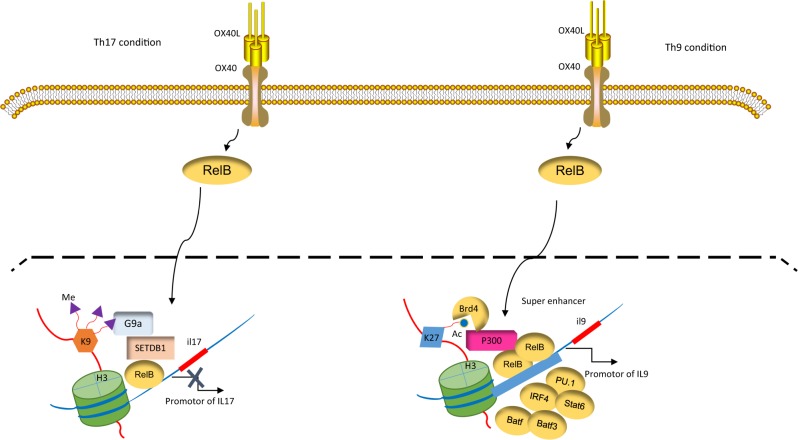
As shown in Fig. 4, RelB can also trigger extensive chromatin remodeling in activated T cells. We showed that even under Th17-inducing conditions (in the presence of TGF-β and IL-6), the engagement of the OX40 receptor strongly inhibits IL-17 expression. This inhibition is not due to the absence of Th17-specific transcription factors, such as RORγt. Rather, RORγt is expressed at high levels in OX40-stimulated T cells but fails to bind the Il17 locus.54 We found that OX40 signaling upregulates the expression of RelB and that RelB binds and recruits the histone methyltransferases G9a and SETDB1 to the κB sites at the Il17 locus. G9a and SETDB1 then catalyze the di- and trimethylation of H3K9 (i.e., H3K9me2 and H3K9me3, respectively), which are repressive chromatin marks that result in the closure of the Il17 locus and the suppression of Th17 induction.54 Interestingly, RelB also suppresses Th17 induction in p50 and p52 double-deficient T cells. Additionally, a point mutation that prevents RelB from dimerizing with p50 or p52 fails to alter the role of RelB in the suppression of Th17 cells. Furthermore, deletion of the TAD domain in RelB fails to alter RelB-mediated suppression of Th17 cells.54 Thus, the role of RelB in chromatin remodeling is strikingly different from its transcriptional activity. Our data suggest that depending on the binding partners of RelB, gene transcription and chromatin modification can be segregated. In a different model, we showed that RelB is capable of recruiting the histone acetyltransferase p300/CBP to the Il9 locus to catalyze H3K27 acetylation (an active chromatin mark), consequently mediating robust Th9 induction.59 However, the factors determining the selectivity of RelB in engaging functionally different chromatin modifiers, separate from its classic role as a transcription factor, remain unknown and warrant further investigation.
Fig. 4.
RelB activates chromatin modifiers to regulate cell fate decisions. OX40 stimulation upregulates RelB, which recruits the histone methyltransferases G9a and SETDB1 to the Il17 locus. G9a and SETDB1 trimethylate H3K9, depositing “repressive” chromatin marks and consequently repressing interleukin (IL)-17 expression. Under Th9-inducing conditions, RelB can also recruit the histone acetyltransferase p300/CBP to the Il9 locus to catalyze H3K27 acetylation. This event allows binding of the superenhancer (SE) factor BRD4 to organize the assembly of the SE complex, which in turn drives robust IL-9 expression and Th9 cell induction
Studies in other models further confirm the role of NF-κB family members in engaging chromatin modifiers to modulate cellular activities. Puto et al. reported that RelB can interact with Daxx, an apoptosis-modulating protein, which in turn recruits DNA methyltransferase 1 (Dnmt1) to target gene promoters, resulting in DNA hypermethylation and epigenetic silencing of target genes.60 The repression of target genes is RelB-dependent, as Daxx lacks domains for sequence-dependent DNA binding. The observation that the Dnmt inhibitor 5-azacitidine completely restored gene expression strongly suggests that Dnmt proteins are responsible for the repressive action of Daxx.61 Other studies showed that in certain cancer cells, RelA can be phosphorylated at serine residue 276 after TNF stimulation, leading to the recruitment of Dnmt1 to tumor suppressor genes (e.g., breast cancer metastasis suppressor 1, or BRMS1) by RelA. Assembly of the RelA/Dnmt1 complex at the BRMS1 promoter region results in gene hypermethylation and transcriptional repression, which are associated with a dramatic increase in tumor metastasis.62
Chromatin modifier-targeted interventions as potential therapeutics
NF-κB transcription factors have long been attractive therapeutic targets in the clinic, as dysregulated NF-κB pathways are implicated in numerous pathological conditions, including autoimmune diseases, inflammatory diseases, metabolic diseases, and cancer. A variety of approaches have been devised to inhibit NF-κB signaling based primarily on the premise that NF-κB family members function as transcription factors.63 The commonly studied NF-κB inhibitors target different components of the NF-κB signaling cascade, from IKK inhibition and IkB stabilization to cytoplasmic retention of NF-κB complexes and transcriptional inhibition. Despite promising results in some models, very few specific NF-κB inhibitors exist in the clinic because of the severe side effects that follow indiscriminate NF-κB pathway blockade. The recent discovery that NF-κB family members can trigger the epigenetic cascade provides exciting new opportunities for gene expression-targeted interventions in the clinic. Epigenetic events usually occur downstream of NF-κB activation and are confined to a select cohort of target genes and effector phenotypes. Therefore, targeting these epigenetic responses is likely to be more selective and specific than broadly blocking NF-κB pathways.
Accumulating evidence demonstrates the importance of the cellular epigenetic machinery in fine-tuning NF-κB-mediated immune responses. In a mouse model of allergic lung inflammation, we showed that the challenge of ovalbumin-sensitized mice induces extensive airway inflammation, especially when the OX40 receptor is concurrently stimulated.59 Further in vitro studies showed that OX40 signaling is exceptionally potent in driving the differentiation of Th9 cells, which are responsible for epithelial hyperplasia, mucin production, and mast cell accumulation in the airways. Interestingly, the induction of Th9 cells by OX40 receptor stimulation is mediated primarily by the formation of superenhancers at the Il9 locus.59 Superenhancers are very different from typical enhancers; they are clusters of enhancers that span a large chromatin segment and are densely populated with transcription factors and cofactors. Importantly, the induction of superenhancers is often associated with exceptionally high levels of gene transcription.64 In our model, the formation of the Il9 superenhancer depends on the histone acetyltransferases p300/CBP, and the recruitment of p300/CBP to the Il9 locus is mediated by RelB. The p300/CBP proteins catalyze exceptionally high levels of histone acetylation (H3K27ac) at the Il9 locus, and acetylated H3K27 provides binding sites for the chromatin reader BRD4, which is required for organizing the assembly of superenhancers.65 Thus, OX40 signaling leads to Th9 differentiation by utilizing RelB to drive histone modification and BRD4 recruitment, thereby creating a superenhancer at the Il9 locus. Of note, BRD4 belongs to the bromodomain and extra terminal domain (BET) protein family.66 JQ1 is a BET inhibitor that binds in the BRD4 pocket and usually engages acetylated H3K27 with high affinity, thus blocking BRD4 from binding to acetylated H3K27.59 In our model, JQ1 abrogates the induction of superenhancers and effectively abolishes Th9 cell induction in vitro. Importantly, JQ1 shows exceptional potency in suppressing allergic lung inflammation in vivo,59 thus demonstrating the important potential of targeting epigenetic mechanisms rather than NF-κB-induced transcriptional activities in the treatment of allergic inflammation.
Numerous other studies suggest that the inhibition of BET proteins is an attractive therapeutic strategy in a variety of disease settings. Brown et al.67 showed that upon TNF-α stimulation, NF-κB recruits BRD4 to the enhancer and promoter regions of inflammatory cytokines, establishing the role of superenhancers in endothelial cells that drive a persistent inflammatory response. These researchers also found that JQ1 can significantly attenuate endothelial cell activation during acute inflammation in vivo and ex vivo, an effect that provides opportunities for suppressing tissue inflammation through modulating chromatin structure. Moreover, the superenhancer landscape can be cell type-specific, mediating the expression of the genes that maintain cell identity in different tissues and cell lineages, which further suggests that targeting superenhancers induced by NF-κB will have more specific effects than blocking NF-κB signals, per se. Additionally, recent studies suggest that most oncogenes are under the control of superenhancers, and targeting the epigenetic mechanisms that are responsible for superenhancer formation should be of value in the treatment of certain cancers, especially those exhibiting strong NF-κB activation.67,68 Additional studies exploring the epigenetic-targeting therapeutics have demonstrated that the acetylation state of RelA dictates its neuroprotective effect in acute brain ischemia and amyotrophic lateral sclerosis. Accordingly, therapies that promote RelA acetylation, such as HDAC inhibitors, have been shown to delay brain injury in animal models.69,70 Furthermore, upon respiratory syncytial virus infection, RelA is phosphorylated at serine residue 276, this event allows the recruitment of p300/CBP, which mediates the acetylation of RelA at lysine residue 310 by p300. Acetylated RelA then interacts with BRD4 to mediate potent antiviral immunity.71 These findings clearly provide new insights into the regulation of NF-κB pathways and may lead to the development of novel clinical therapeutic approaches.
Concluding remarks
The NF-κB family is arguably one of the most thoroughly studied families of transcription factors; members of this family are ubiquitous, present in almost all cell types, and involved in diverse biological responses. NF-κB family members also play a complex role in the immune system, ranging from lymphoid organogenesis to mediating innate and adaptive immune responses. Multiple mechanisms that are involved in either activating NF-kB or terminating NF-kB activation have been identified, and dysregulation of NF-kB pathways can lead to profound autoimmune diseases, chronic inflammation, or resistance to tolerance induction.
Targeting NF-κB family members for therapeutic purposes in the clinic, especially for tolerance induction, has been fraught with challenges. Although most such approaches are designed based on the premise that NF-κB family members function as transcription factors, the side effects associated with broad and indiscriminate NF-κB blockade undoubtedly limit their clinical utility. The recent findings that members of the NF-κB family can also activate chromatin remodeling pathways, in which these proteins partner with chromatin modifiers to modulate gene expression and cellular activities, have generated tremendous enthusiasm in the field. These findings will certainly open new therapeutic opportunities in the clinic. It is important to note that mechanisms of chromatin remodeling triggered by NF-kB activation, especially those activated by RelB, can be segregated from the transcriptional activity of the involved protein. Thus, targeting chromatin remodeling pathways is likely to be more selective and specific than broad NF-kB blockade. Furthermore, the identification of superenhancers in the control of cell identity and cell fate decisions is another promising new development, and chemical inhibitors of chromatin readers associated with the assembly of superenhancers will have broad clinical implications.
However, many challenges remain. We are just beginning to comprehend the cell type- and stimulus-specific regulation of NF-κB. Further studies are needed to provide key insights into the diverse roles of NF-κB activities. Clearly, chromatin modifiers provide a promising therapeutic approach, but the mechanistic details of how NF-κB family members engage different epigenetic modifiers, along with their association with and deassociation from cell type-specific transcription factors, remain to be defined. We anticipate that future studies in this area will lead to additional encouraging developments in the field.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Van Parijs L, Abbas AK. Homeostasis and self-tolerance in the immune system: turning lymphocytes off. Science. 1998;280:243–248. doi: 10.1126/science.280.5361.243. [DOI] [PubMed] [Google Scholar]
- 2.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Sakaguchi S. Naturally arising CD4 + regulatory t cells for immunologic self-tolerance and negative control of immune responses. Annu. Rev. Immunol. 2004;22:531–562. doi: 10.1146/annurev.immunol.21.120601.141122. [DOI] [PubMed] [Google Scholar]
- 4.Zhang P, Lu Q. Genetic and epigenetic influences on the loss of tolerance in autoimmunity. Cell. Mol. Immunol. 2018;15:575–585. doi: 10.1038/cmi.2017.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz RH. Natural regulatory T cells and self-tolerance. Nat. Immunol. 2005;6:327–330. doi: 10.1038/ni1184. [DOI] [PubMed] [Google Scholar]
- 6.Wood KJ, Sakaguchi S. Regulatory T cells in transplantation tolerance. Nat. Rev. Immunol. 2003;3:199–210. doi: 10.1038/nri1027. [DOI] [PubMed] [Google Scholar]
- 7.Malissen B, Bongrand P. Early T cell activation: integrating biochemical, structural, and biophysical cues. Annu. Rev. Immunol. 2015;33:539–561. doi: 10.1146/annurev-immunol-032414-112158. [DOI] [PubMed] [Google Scholar]
- 8.Li P, Spolski R, Liao W, Leonard WJ. Complex interactions of transcription factors in mediating cytokine biology in T cells. Immunol. Rev. 2014;261:141–156. doi: 10.1111/imr.12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falvo JV, Jasenosky LD, Kruidenier L, Goldfeld AE. Epigenetic control of cytokine gene expression: regulation of the TNF/LT locus and T helper cell differentiation. Adv. Immunol. 2013;118:37–128. doi: 10.1016/B978-0-12-407708-9.00002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baldwin AS., Jr. TheNF-kappa B and I kappa B proteins: new discoveries and insights. Annu. Rev. Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 11.Siebenlist U, Franzoso G, Brown K. Structure, regulation and function of NF-kappa B. Annu. Rev. Cell. Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh G, Wang VY, Huang DB, Fusco A. NF-kappaB regulation: lessons from structures. Immunol. Rev. 2012;246:36–58. doi: 10.1111/j.1600-065X.2012.01097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smale ST. Dimer-specific regulatory mechanisms within the NF-kappaB family of transcription factors. Immunol. Rev. 2012;246:193–204. doi: 10.1111/j.1600-065X.2011.01091.x. [DOI] [PubMed] [Google Scholar]
- 15.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 16.Perkins ND. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene. 2006;25:6717–6730. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- 17.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 18.Pires, B. R. B., Silva, R., Ferreira, G. M. & Abdelhay, E. NF-kappaB: Two sides of the same coin. Genes (Basel)9,1–23 (2018). [DOI] [PMC free article] [PubMed]
- 19.Kanarek N, Ben-Neriah Y. Regulation of NF-κB by ubiquitination and degradation of the IκBs. Immunol. Rev. 2012;246:77–94. doi: 10.1111/j.1600-065X.2012.01098.x. [DOI] [PubMed] [Google Scholar]
- 20.Chen J, Chen ZJ. Regulation of NF-kappaB by ubiquitination. Curr. Opin. Immunol. 2013;25:4–12. doi: 10.1016/j.coi.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun SC. The noncanonical NF-kappaB pathway. Immunol. Rev. 2012;246:125–140. doi: 10.1111/j.1600-065X.2011.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun SC. The non-canonical NF-kappaB pathway in immunity and inflammation. Nat. Rev. Immunol. 2017;17:545–558. doi: 10.1038/nri.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Razani B, Reichardt AD, Cheng G. Non-canonical NF-kappaB signaling activation and regulation: principles and perspectives. Immunol. Rev. 2011;244:44–54. doi: 10.1111/j.1600-065X.2011.01059.x. [DOI] [PubMed] [Google Scholar]
- 24.Sun SC. Non-canonical NF-kappaB signaling pathway. Cell Res. 2011;21:71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-kappaB signaling pathways. Nat. Immunol. 2011;12:695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 26.Israel A. The IKK complex, a central regulator of NF-kappaB activation. Cold Spring Harb. Perspect. Biol. 2010;2:a000158. doi: 10.1101/cshperspect.a000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu F, Xia Y, Parker AS, Verma IM. IKKbiology. Immunol. Rev. 2012;246:239–253. doi: 10.1111/j.1600-065X.2012.01107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao G, Fong A, Sun SC. Induction of p100 processing by NF-kappaB-inducing kinase involves docking IkappaB kinase alpha (IKKalpha) to p100 and IKKalpha-mediated phosphorylation. J. Biol. Chem. 2004;279:30099–30105. doi: 10.1074/jbc.M401428200. [DOI] [PubMed] [Google Scholar]
- 29.Yu J, et al. T cell-intrinsic function of the noncanonical NF-kappaB pathway in the regulation of GM-CSF expression and experimental autoimmune encephalomyelitis pathogenesis. J. Immunol. 2014;193:422–430. doi: 10.4049/jimmunol.1303237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo JC, et al. Coordination between NF-kappaB family members p50 and p52 is essential for mediating LTbetaR signals in the development and organization of secondary lymphoid tissues. Blood. 2006;107:1048–1055. doi: 10.1182/blood-2005-06-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffmann A, Leung TH, Baltimore D. Genetic analysis of NF-kappaB/Rel transcription factors defines functional specificities. EMBO J. 2003;22:5530–5539. doi: 10.1093/emboj/cdg534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basak S, Shih VF, Hoffmann A. Generation and activation of multiple dimeric transcription factors within the NF-kappaB signaling system. Mol. Cell. Biol. 2008;28:3139–3150. doi: 10.1128/MCB.01469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 34.Shembade N, Harhaj EW. Regulation of NF-kappaB signaling by the A20 deubiquitinase. Cell. Mol. Immunol. 2012;9:123–130. doi: 10.1038/cmi.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu TT, et al. Dimerization and ubiquitin mediated recruitment of A20, a complex deubiquitinating enzyme. Immunity. 2013;38:896–905. doi: 10.1016/j.immuni.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beyaert R, Heyninck K, Van Huffel S. A20 and A20-binding proteins as cellular inhibitors of nuclear factor-kappa B-dependent gene expression and apoptosis. Biochem. Pharmacol. 2000;60:1143–1151. doi: 10.1016/S0006-2952(00)00404-4. [DOI] [PubMed] [Google Scholar]
- 37.Duwel M, et al. A20 negatively regulates T cell receptor signaling to NF-kappaB by cleaving Malt1 ubiquitin chains. J. Immunol. 2009;182:7718–7728. doi: 10.4049/jimmunol.0803313. [DOI] [PubMed] [Google Scholar]
- 38.Lee EG, et al. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Catrysse L, Vereecke L, Beyaert R, van Loo G. A20 in inflammation and autoimmunity. Trends Immunol. 2014;35:22–31. doi: 10.1016/j.it.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 40.Iwai K, Tokunaga F. Linear polyubiquitination: a new regulator of NF-kappaB activation. EMBO Rep. 2009;10:706–713. doi: 10.1038/embor.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lork M, Verhelst K, Beyaert R. CYLD, A20 and OTULIN deubiquitinases in NF-kappaB signaling and cell death: so similar, yet so different. Cell Death Differ. 2017;24:1172–1183. doi: 10.1038/cdd.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lich JD, et al. Monarch-1 suppresses non-canonical NF-kappaB activation and p52-dependent chemokine expression in monocytes. J. Immunol. 2007;178:1256–1260. doi: 10.4049/jimmunol.178.3.1256. [DOI] [PubMed] [Google Scholar]
- 43.Allen IC, et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-kappaB signaling. Immunity. 2012;36:742–754. doi: 10.1016/j.immuni.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu H, et al. OTUD7B controls non-canonical NF-kappaB activation through deubiquitination of TRAF3. Nature. 2013;494:371–374. doi: 10.1038/nature11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fusco AJ, et al. The NF-kappaB subunit RelB controls p100 processing by competing with the kinases NIK and IKK1 for binding to p100. Sci. Signal. 2016;9:ra96. doi: 10.1126/scisignal.aad9413. [DOI] [PubMed] [Google Scholar]
- 46.Maminska A, et al. ESCRT proteins restrict constitutive NF-kappaB signaling by trafficking cytokine receptors. Sci. Signal. 2016;9:ra8. doi: 10.1126/scisignal.aad0848. [DOI] [PubMed] [Google Scholar]
- 47.Ma X, Becker Buscaglia LE, Barker JR, Li Y. MicroRNAs in NF-kappaB signaling. J. Mol. Cell Biol. 2011;3:159–166. doi: 10.1093/jmcb/mjr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mao X, Su Z, Mookhtiar AK. Long non-coding RNA: a versatile regulator of the nuclear factor-kappaB signalling circuit. Immunology. 2017;150:379–388. doi: 10.1111/imm.12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dawson MA. The cancer epigenome: Concepts, challenges, and therapeutic opportunities. Science. 2017;355:1147–1152. doi: 10.1126/science.aam7304. [DOI] [PubMed] [Google Scholar]
- 50.Lim PS, Li J, Holloway AF, Rao S. Epigenetic regulation of inducible gene expression in the immune system. Immunology. 2013;139:285–293. doi: 10.1111/imm.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torres IO, Fujimori DG. Functional coupling between writers, erasers and readers of histone and DNA methylation. Curr. Opin. Struct. Biol. 2015;35:68–75. doi: 10.1016/j.sbi.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol. 2014;15:703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 53.Yang J, Tian B, Brasier AR. Targeting Chromatin Remodeling in Inflammation and Fibrosis. Adv. Protein Chem. Struct. Biol. 2017;107:1–36. doi: 10.1016/bs.apcsb.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 54.Xiao X, et al. The costimulatory receptor OX40 inhibits interleukin-17 expression through activation of repressive chromatin remodeling pathways. Immunity. 2016;44:1271–1283. doi: 10.1016/j.immuni.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao X, et al. GITR subverts Foxp3( + ) Tregs to boost Th9 immunity through regulation of histone acetylation. Nat. Commun. 2015;6:8266. doi: 10.1038/ncomms9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang X, et al. OX40 costimulation inhibits Foxp3 expression and treg induction via BATF3-dependent and independent mechanisms. Cell Rep. 2018;24:607–618. doi: 10.1016/j.celrep.2018.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhong H, May MJ, Jimi E, Ghosh S. The phosphorylation status of nuclear NF-kappa B determines its association with CBP/p300 or HDAC-1. Mol. Cell. 2002;9:625–636. doi: 10.1016/S1097-2765(02)00477-X. [DOI] [PubMed] [Google Scholar]
- 58.Yeung F, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiao X, et al. Guidance of super-enhancers in regulation of IL-9 induction and airway inflammation. J. Exp. Med. 2018;215:559–574. doi: 10.1084/jem.20170928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salomoni P, Khelifi AF. Daxx: death or survival protein? Trends Cell Biol. 2006;16:97–104. doi: 10.1016/j.tcb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 61.Puto LA, Reed JC. Daxx represses RelB target promoters via DNA methyltransferase recruitment and DNA hypermethylation. Genes Dev. 2008;22:998–1010. doi: 10.1101/gad.1632208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Y, et al. Phosphorylation of RelA/p65 promotes DNMT-1 recruitment to chromatin and represses transcription of the tumor metastasis suppressor gene BRMS1. Oncogene. 2012;31:1143–1154. doi: 10.1038/onc.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.DiDonato JA, Mercurio F, Karin M. NF-kappaB and the link between inflammation and cancer. Immunol. Rev. 2012;246:379–400. doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- 64.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Loven J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Donati B, Lorenzini E, Ciarrocchi A. BRD4 and Cancer: going beyond transcriptional regulation. Mol. Cancer. 2018;17:164. doi: 10.1186/s12943-018-0915-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brown JD, et al. NF-kappaB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol. Cell. 2014;56:219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Betancur PA, et al. A CD47-associated super-enhancer links pro-inflammatory signalling to CD47 upregulation in breast cancer. Nat. Commun. 2017;8:14802. doi: 10.1038/ncomms14802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lanzillotta A, et al. Targeted acetylation of NF-kappaB/RelA and histones by epigenetic drugs reduces post-ischemic brain injury in mice with an extended therapeutic window. Neurobiol. Dis. 2013;49:177–189. doi: 10.1016/j.nbd.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 70.Schiaffino L, et al. Acetylation state of RelA modulated by epigenetic drugs prolongs survival and induces a neuroprotective effect on ALS murine model. Sci. Rep. 2018;8:12875. doi: 10.1038/s41598-018-30659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brasier AR, et al. RelA Ser276 phosphorylation-coupled Lys310 acetylation controls transcriptional elongation of inflammatory cytokines in respiratory syncytial virus infection. J. Virol. 2011;85:11752–11769. doi: 10.1128/JVI.05360-11. [DOI] [PMC free article] [PubMed] [Google Scholar]