Abstract
The intracellular pathogen Mycobacterium tuberculosis (Mtb) can survive in the host and cause disease by interfering with a variety of cellular functions. The mammalian cell entry 2 (mce2) operon of Mtb has been shown to contribute to tuberculosis pathogenicity. However, little is known about the regulatory roles of Mtb Mce2 family proteins towards host cellular functions. Here we show that the Mce2 family protein Mce2E suppressed the macrophage innate immune response and promoted epithelial cell proliferation. Mce2E inhibited activation of the extracellular signal-regulated kinase (ERK) and Jun N-terminal kinase (JNK) mitogen-activated protein kinase (MAPK) signaling pathways in a non-canonical D motif (a MAPK-docking motif)-dependent manner, leading to reduced expression of TNF and IL-6 in macrophages. Furthermore, Mce2E promoted proliferation of human lung epithelium-derived lung adenoma A549 cells by inhibiting K48-linked polyubiquitination of eEF1A1 in a β strand region-dependent manner. In summary, Mce2E is a novel multifunctional Mtb virulence factor that regulates host cellular functions in a niche-dependent manner. Our data suggest a potential novel target for TB therapy.
Keywords: Mycobacterium tuberculosis, Mce2E, innate immune response, cell proliferation
Introduction
Tuberculosis (TB) remains one of the most intractable diseases in the clinic.1 Mycobacterium tuberculosis (Mtb), the causal agent of TB, uses mutagenesis to survive TB drug treatment, resulting in the emergence of drug-resistant Mtb strains.2 Additionally, as an intracellular pathogen, Mtb has developed multiple strategies to regulate host cellular functions to better survive in the hostile environment of host cells.3–7 Therefore, a better understanding of the molecular mechanisms underlying the interactions between Mtb and host cells is crucial for identifying novel targets for the development of TB treatments.
Multiple signaling pathways, such as the nuclear factor κB (NF-κB) pathway and mitogen-activated protein kinase (MAPK) cascades, including the extracellular signal-regulated kinase (ERK), Jun N-terminal kinase (JNK), and p38 MAPK pathways, are involved in a variety of essential cellular processes, such as the innate immune response as well as cellular differentiation and proliferation.8–10 Once activated by pathogens, these pathways may contribute to the expression of various pro-inflammatory genes to eliminate the pathogen.11–14 By contrast, pathogens such as Mtb can subvert these signaling pathways and cellular functions to better survive within macrophages.15, 16 In addition to macrophages, Mtb can also invade epithelial cells and has been associated with lung tumorigenesis and development.17–21 However, the specific mechanisms by which Mtb effector proteins interact with host cells, as well as the causal links between Mtb infection and lung carcinomas, remain poorly understood.
The Mtb genome contains four homologous copies of mce operons (mce1-4). Previous studies have revealed that each of the four mce operon mutant strains of Mtb show a distinct phenotype in vitro and in vivo.22–24 Furthermore, the Mce family proteins encoded by these mce operon genes exhibit differential expression profiles throughout various phases of mycobacterial infection in vivo,25 suggesting that these Mce family proteins have distinct regulatory functions during mycobacterial infection. Several Mce family proteins have been demonstrated to play pivotal roles in host immune regulation. For example, both Mce3E (also named LprM) and Mce4E (also named LprN) have been shown to suppress the host immune response by regulating the expression of cytokines in macrophages.16, 26 Previous observations have indicated that an mce2 operon mutant is attenuated in mice, suggesting that it might contribute to tuberculosis pathogenicity.24 However, the regulatory roles of individual Mce2 family proteins towards host innate immune signaling pathways and cellular functions during mycobacterial infection remain largely unknown. Here, we demonstrated that the Mce2 family protein Mce2E (also named LprL), encoded by the gene Rv0593,24, 27 inhibited the activation of the ERK and JNK MAPK signaling pathways to suppress the macrophage innate immune response. Furthermore, Mce2E blocked K48-linked polyubiquitination of eEF1A1 to promote epithelial cell proliferation. Thus, Mce2E is a novel multifunctional Mtb virulence factor that might be a potential novel target for TB therapy.
Materials and methods
Bacterial and cell culture
The bacterial strains are listed in Table S1. The M. smegmatis mc2155 strains included WT M. smegmatis, mce2E-M. smegmatis (the gene encoding mce2E constructed in the E. coli-mycobacterial shuttle vector pMV261 was introduced into M. smegmatis mc2155), and mce2E AAA-M. smegmatis (mce2E AAA-introducing M. smegmatis).15 The M. bovis BCG strains included WT BCG, BCG Δmce2E (mce2E-deleted BCG), BCG Δmce2E:mce2E (BCG Δmce2E complemented with mce2E), BCG Δmce2E:mce2E AAA (BCG Δmce2E complemented with mce2E AAA), and BCG Δmce2E:mce2E Δ67-147 (BCG Δmce2E complemented with mce2E Δ67-147). Complementation was confirmed by the PCR-sequencing method. BCG-mce3E consisted of the gene encoding mce3E constructed in the E. coli-mycobacterial shuttle vector pMV261 introduced into BCG. E. coli DH5a and E. coli BL21 (DE3) were grown in flasks using lysogeny broth medium. M. smegmatis and M. bovis BCG strains were grown in Middlebrook 7H9 broth (7H9) supplemented with 10% oleic acid–albumin–dextrose–catalase (OADC) and 0.05% Tween-80 (Sigma), or on Middlebrook 7H10 agar (BD) supplemented with 10% OADC. HEK293T (ATCC CRL-3216), HeLa (ATCC CCL-2), A549 cells (ATCC CCL-185), and RAW264.7 cells (ATCC TIB-71) were cultured in Dulbecco’s Modified Eagle’s Medium (Gibco) with 10% (v/v) heat-inactivated FBS.
Plasmids and antibodies
The pJV53 system (26904; Addgene plasmid)28 was used to create the BCG strain with deletion of the gene encoding Mce2E (BCG ∆mce2E). The integrative vector pMV306 plasmid (provided by W. R. Jacobs, Albert Einstein College of Medicine, Yeshiva University) was constructed by inserting a fusion gene, consisting of the WT or mutated mce2E gene together with its original promoter, and used to complement strain BCG ∆mce2E with WT mce2E or to create mutant strains of BCG. The Mycobacterium shuttle vector pMV261 (provided by W. Jacobs from Albert Einstein College) was used to express Mtb Mce2E in M. smegmatis. Luciferase reporter assay plasmids for pRL-TK, Gal4-Elk, Gal4-Luc, pFA-cJun, RacL61, pNF-κB-luc, RasV12, v-Raf, MEK1-ED, as well as pcDNA6A-MEK1 and pcDNA-HA-Ub were provided by Feng Shao (National Institute of Biological Sciences, Beijing, China). p3×Flag-CMV-14-ERK2 and pGEX-6p-1-ERK2 were generated in our laboratory. pcDNA-HA-Ub (K48 only) and pcDNA-HA-Ub (K63 only) were provided by Lingqiang Zhang (Beijing Institute of Radiation Medicine). The gene for Mtb mce2E was amplified from Mtb genomic DNA (P505-D3, Vazyme). For expression in mammalian cells, Mtb mce2E was cloned into pEGFP-N1 or pcDNA6A. Bacterial expression plasmids were constructed by cloning the Mtb mce2E gene into pGEX-6P-1. The gene for eEF1A1 was amplified from cDNA of A549 cells and inserted into p3xFlag-CMV14. Plasmids and oligonucleotides used in the study are listed in Table S1. The following antibodies were used in this study: anti-p-Jnk (sc-81502; Santa Cruz), anti-Jnk (9252; Cell Signaling), anti-p-p38 (sc-17852-R; Santa Cruz), anti-p38 (sc-7972; Santa Cruz), anti-p-IκBα (sc-101713; Santa Cruz), anti-IκBα (sc-371; Santa Cruz), p-ERK1/2 (9101; CST), ERK1/2 (9102; CST), p-MEK1/2 (D1A5; CST), anti-Calnexin (H-70) (sc-11397; Santa Cruz), anti-GFP (AG281; Beyotime), anti-Giantin (ab24586; Abcam), anti-Myc (sc-40; Santa Cruz), anti-GST (TA-03; ZSGB-BIO), anti-Flag (F3165; Sigma), anti-α-tubulin (T6199; Sigma), anti–Mtb Rv3134 (sc-52108; Santa Cruz), anti-eEF1A1 (BS6077; Bioworld), anti-HA (3724S; CST), anti-Ki67 (ab16667; Abcam), anti-His (TA-02; ZSGB-BIO), and anti-PARP (9542S; CST).
Cell transfection, immunoblot analysis, immunoprecipitation, and immunofluorescence confocal microscopy
HEK293T cells were transfected with polyethylenimine (DH253-1; Sigma). A549 and RAW264.7 cells were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturers’ protocol. Immunoblot analysis, immunoprecipitation and immunofluorescence confocal microscopy were performed as described previously.15
Infection of macrophage cells, colony-forming unit counting, quantitative PCR and enzyme-linked immunosorbent assay
RAW264.7 cells were infected with mycobacterial strains for colony-forming unit (CFU) counting, quantitative PCR assay and ELISA at various time points post infection as previously described.16 For quantitative PCR and ELISA, the following reagents were used: KAPA SYBR FAST qPCR Kit (KAPA Biosystems) and RayBio ELISA kits (Mouse TNF ELISA Kit: ELM-TNFa-001; Mouse IL-6 ELISA Kit: ELM-IL6-001).
Luciferase reporter assays
A dual luciferase reporter assay was performed using the Promega luciferase reporter system. To evaluate the NF-κB pathway, HEK293T cells grown in a 12-well plate were cotransfected with pNF-κB-Luc (1 µg) and pRL-TK (50 ng) in the presence or absence of Mce2E (1 µg) plasmid. Twenty-four hours later, the cells were treated with 20 ng/ml of TNF (Invitrogen) for 6 h. To measure the JNK pathway, HEK293T cells were cotransfected with Gal4-luc (0.9 mg), pFA-cJun (0.3 mg), RacL61 (0.5 mg), and pRL-TK (50 ng) with or without Mce2E (1 µg) plasmid. To measure the ERK pathway, HEK293T cells were co-transfected with Gal4-luc (0.6 µg), Gal4-Elk (0.6 µg), pRL-TK (50 ng) and RasV12 (10 ng) or v-Raf (100 ng) or MEK1-ED (100 ng) in the presence or absence of 1 µg of Mce2E plasmid.
Cellular fractionation
RAW264.7 cells infected with BCG or BCG Δmce2E for 4 h at a multiplicity of infection (MOI) of 10 were harvested at 2500 rpm for 5 min, and then, the cell pellets were washed with PBS. The pellets were resuspended in buffer containing 10 mM Tris-HCl, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 10% glycerol, 10 mM K2HPO4, 1 mM sodium vanadate, 10 mM NaF, 0.5 mM dithiothreitol (DTT) and 0.5 mM phenylmethylsulphonyl fluoride (PMSF). The cells were disrupted with 10% Nonidet P-40 and then centrifuged at 1000×g for 10 min to pellet the macrophage nuclei and BCG. The supernatant was purified from the remaining cell debris by centrifugation at 12,000×g for 30 min and collected as the cytosolic fraction of infected macrophages for immunoblot analysis.
Preparation and purification of rabbit antibodies against Mce2E
Bacterial fusion proteins were expressed in the E. coli BL21 (DE3) strain (Novagen). The Mtb mce2E gene was cloned in the prokaryotic expression vector pGEX-6P-1, resulting in an N-terminal GST-fusion to the protein. Expression of GST-tagged Mtb Mce2E was induced in the presence of 0.1–0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 16 °C up to an OD600 = 1. GST-tagged Mtb Mce2E was purified using glutathione sepharose-4B resins from GE Healthcare with affinity chromatography followed by a Superdex column with size exclusion chromatography. Purified GST-Mce2E (500 µg) was injected into rabbits, which was solubilized in Freund’s complete adjuvant (1 ml). After an interval of 15 days, three injections of 250 µg each in Freund’s incomplete adjuvant (1 ml) were administered. Ten days after the final injection, animals were bled, and titers of anti-GST-Mce2E were determined by ELISA. GST-Mce2E-specific antibody was purified from the immunized rabbit serum by affinity chromatography on protein A agarose (Santa Cruz).
Yeast two-hybrid assay
The Matchmaker Two-Hybrid System (Clontech) was used for the yeast two-hybrid assay according to the Clontech protocol. Mtb mce2E gene cloned into the bait Gal4-BD vector pGBKT7 and mouse cDNA library Mate & Plate Library (630478; Clontech Laboratories, Inc.) cloned into the GAL4-AD pACT2 vectors were transformed into Saccharomyces cerevisiae AH109 cells (Clontech) by the lithium acetate method. The transformants were then placed in medium lacking leucine and tryptophan (low-stringency conditions) and lacking adenine, histidine, leucine, and tryptophan (high-stringency conditions) to test the interactions between proteins.
Knockout cell lines
The Cas9 design target tool (http://crispr.mit.edu) was used to design the specific target sequences for sgRNA synthesis. The pairs of annealed oligos were ligated into pX458 (Addgene Plasmid #48138) after being digested with BbsI (Thermo). A549 cells were transfected with 4 µg of plasmid per well in 6-well plates using Lipofectamine 2000. Single cells were sorted using a flow cytometer (BD FACSAriaIII; BD).
Ubiquitination assay
HEK293T cells were transfected with plasmids encoding Myc-Mce2E, HA-ubiquitin (WT), HA-ubiquitin (K48 only), HA-ubiquitin (K63 only), and Flag-eEF1A1 (WT) or its mutants for 24 h, and cell lysates were immunoprecipitated with anti-HA beads. The beads were then used for immunoblot analysis with anti-HA, anti-Myc, and anti-Flag antibodies.
Mouse infection
Male and female SPF C57BL/6 mice (6–8 weeks old) were purchased from the Vital River Laboratory Animal Technology Co. Ltd. and cared for according to standard humane animal husbandry protocols. BCG strains were centrifuged at 4000×g for 5 min and resuspended in PBS plus 0.05% Tween-80 (PBST). C57BL/6 mice were infected via aerosol with 2 × 106 CFUs of BCG strain per mouse in 25 µl PBST. Lung tissues were harvested and homogenized in 1 ml 6% NaOH after infection at different time points (8, 12, 16, and 20 days) and plated onto 7H10 agar plates. Subsequently, colonies were counted to determine CFUs after 3–4 weeks of infection and incubation at 37 °C. The samples of mouse lung were fixed in 10% neutral (PBS) buffered formalin, embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E) for evaluation of pathologic changes by histology. Some samples of spleen were ground up and used for quantitative PCR.
Xenograft tumor model
Six-week-old female BALB/c nude mice were purchased from the Vital River Laboratory Animal Technology Co. Ltd. and housed under specific pathogen-free conditions. Mice were allowed to acclimate for 1 week after arrival. To study the possible effects of Mce2E on tumor growth, A549 cells were plated in 10-cm dishes and infected with BCG strains at a MOI of 10. After a 2-h incubation, the cells were washed with PBS to remove extracellular bacteria and cultured in DMEM containing gentamicin (10 mg/ml). A549 cells (1 × 107) were subcutaneously injected into the backs of BALB/c nude mice, and then, the animals were monitored daily for changes in weight, tumor size and any signs of sickness. The tumor volume was measured 2 weeks later. Tumor volumes were calculated according to the following equation: volume = L × W2 × 0.52, where L is length and W is the width. Some tumor samples were fixed in 10% neutral (PBS) buffered formalin, embedded in paraffin, sectioned, and stained with H&E or Ki-67 for evaluation of tissue pathologic changes by a pathologist.
Cell proliferation assay
A549 cells were seeded in 96-well plates at 2500 cells per well and infected with BCG strains at a MOI of 10. After a 2-h incubation, the cells were washed with PBS to remove extracellular bacteria and cultured in DMEM containing gentamicin (10 mg/ml). An aliquot of 10 µl of CCK-8 (CK04, DOJINDO) was added to each well and incubated according to the manufacturer’s instructions. The absorbance was measured at 450 nm to calculate the numbers of viable cells in each well.
Statistical analysis
Statistical analysis between groups was performed using an unpaired two-tailed Student’s t test. Data are presented as the mean ± s.e.m. P < 0.05 or P < 0.01 was considered statistically significant.
Data availability
The data that support the findings of this study are available from the authors upon request.
Results
Mtb Mce2E suppresses activation of the ERK and JNK pathways in a D motif-dependent manner
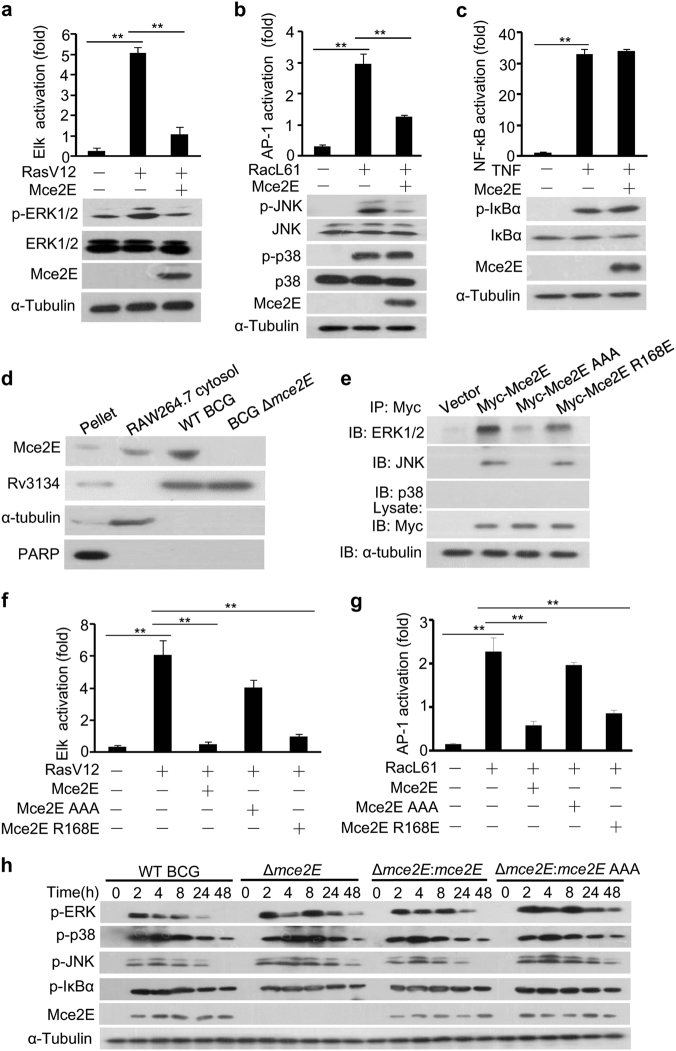
Given that the Mtb mce2 operon contributes to tuberculosis pathogenicity, little is known about the regulatory roles of individual Mtb Mce2 proteins. We previously reported that the Mce3 family protein Mce3E specifically inhibits ERK pathway activation, but it shows little effect on JNK/p38 pathway activation in mycobacteria-infected macrophage cells.16 Interestingly, we found that the Mce2 family protein Mce2E not only efficiently blocked RasV12 (a constitutively active variant of Ras)-induced ERK pathway activation in HEK293T cells but also partially suppressed RacL61 (a constitutively active variant of Rac)-induced JNK/p38 pathway activation and had no effect on TNF-stimulated NF-κB pathway activation (Fig. 1a–c, Supplementary Figure 1). Immunoblotting analysis confirmed the secretion of Mce2E into the cytosolic fraction of macrophages infected with Mycobacterium bovis Bacillus Calmette-Guerin (BCG) at 4 h post infection (Fig. 1d). Through in silico analysis (http://scansite.mit.edu/), we identified a potential non-canonical D motif (a MAPK-docking motif) in Mce2E (Supplementary Figure 2), The canonical D motif usually contains both basic and hydrophobic residues in the arrangement of (K/R)1-2-(X)2-6-ØA-X-ØB (where ØA and ØB are hydrophobic residues [Leu, Ile, or Val]). Both the basic and the hydrophobic residues of the D motif are vital in recognizing and binding MAPK isoforms, such as JNK1, ERK2 and p38α.29, 30 Thus, we replaced the conserved basic residue arginine with glutamine (Mce2E R168E) or the three hydrophobic residues (V173/L175/L177) with alanine residues (Mce2E AAA) to identify the critical residues for interactions between Mce2E and MARKs. Through co-immunoprecipitation analysis, we found that Mce2E and Mce2E R168E, but not Mce2E AAA, were able to interact with ERK and JNK, but not p38 (Fig. 1e). Consistently, Mce2E and Mce2E R168E, but not Mce2E AAA, suppressed RasV12-induced ERK pathway activation and partially suppressed RacL61-induced JNK/p38 pathway activation (Fig. 1f, g). We further explored the specific Mce2E-regulated step(s) along the Ras-Raf-MEK cascade and found that Mce2E could block ERK pathway activation in HEK293T cells stimulated with v-Raf (a constitutively active Raf) and MEK1-ED (a constitutively active MEK1) (Supplementary Figure 3), respectively. To further investigate the role of Mce2E in modulating innate immune signaling pathways during mycobacterial infection, we deleted the mce2E in BCG (BCG Δmce2E) or introduced mce2E in M. smegmatis (mce2E-M. smegmatis) to infect RAW264.7 macrophages cells. Our data showed that Mce2E, but not Mce2E AAA, substantially suppressed the phosphorylation of ERK1/2 and partially inhibited the phosphorylation of JNK, but it had little effect on the phosphorylation of p38 and IκBα, especially at 24 or 48 h after mycobacterial infection (Fig. 1h; Supplementary Figure 4). Collectively, these data suggest that Mce2E inhibited the ERK and JNK MAPK pathways in a D motif-dependent manner during mycobacterial infection.
Fig. 1.
Mtb Mce2E suppresses the ERK and JNK signaling pathways in a D motif-dependent manner. a–c Luciferase reporter assay (top) and immunoblot analysis (bottom) of Elk (a), AP-1 (b), or NF-κB (c) activation by Mtb Mce2E in HEK293T cells transfected with the empty vector or vector encoding Myc-tagged Mce2E. The ERK pathway was activated by co-expression of constitutively active RasV12. The JNK and p38 MAPK pathways were activated by co-expression of constitutively active RacL61. The NF-κB pathway was stimulated by TNF treatment. d RAW264.7 cells were infected with wild-type (WT) BCG or BCG Δmce2E at a multiplicity of infection (MOI) of 10 for 4 h. Cells were collected to obtain the cytosolic fraction, pellet containing the RAW264.7 nuclei and BCG, and bacteria for immunoblot analysis of Mce2E expression. e Immunoblot analysis of proteins immunoprecipitated with anti-Myc antibodies from lysates of HEK293T cells transfected with the empty vector or vectors encoding Myc-tagged Mce2E or its mutants. f Luciferase reporter assay of Elk activation by Mce2E or its mutants in HEK293T cells. The ERK pathway was stimulated as in c. g Luciferase reporter assay of AP-1 activation by Mce2E or its mutants in HEK293T cells. The JNK and p38 pathways were stimulated as in d. h Immunoblot analysis of phosphorylated ERK, JNK, p38, IκBα, Mce2E and total α-tubulin (loading control throughout) in RAW264.7 cells. Cells were infected with wild-type BCG (WT BCG) or BCG Δmce2E (Δmce2E) or BCG Δmce2E complemented with WT mce2E (Δmce2E:mce2E) or mce2E AAA (Δmce2E:mce2E AAA) at a MOI of 10 for 0–48 h. *P < 0.05 and **P < 0.01 (two-tailed unpaired t test). Data are representative of experiments with at least three independent biological replicates (mean and s.e.m. of n = 3 cultures)
Mtb Mce2E inhibits cytokine production of TNF and IL-6 in macrophages
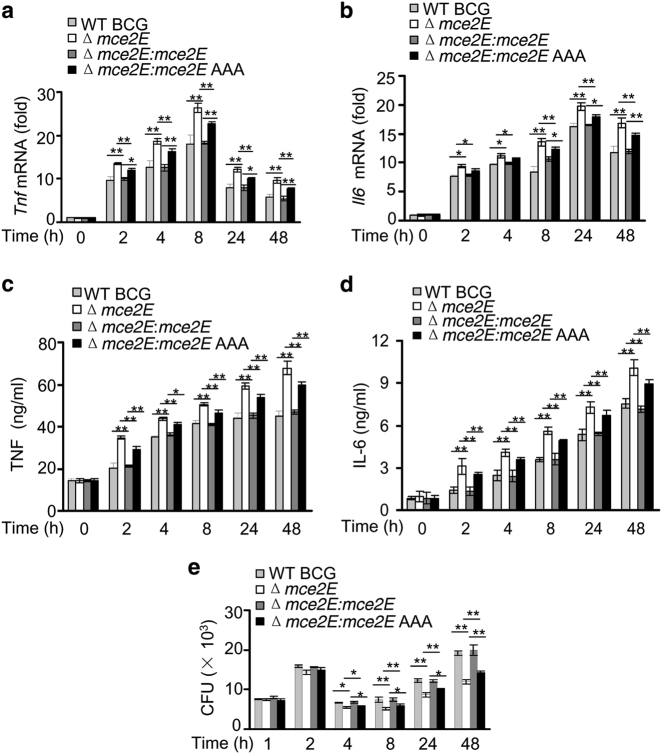
Inflammatory cytokines, such as tumor necrosis factor (TNF) and IL-6, are essential for activation of the host immune system during mycobacterial infection.31, 32 Thus, we performed a quantitative PCR (qPCR) and enzyme-linked immunosorbent assay (ELISA) to investigate whether Mce2E modulates the expression of inflammatory cytokines in macrophages. Four hours after infection, BCG Δmce2E and wild-type (WT)-M. smegmatis induced higher expression of Tnf and Il6 and fewer bacterial colony-forming units (CFUs) in RAW264.7 cells than WT BCG and mce2E-M. smegmatis. The BCG Δmce2E complemented with mce2E AAA (Δmce2E:mce2E AAA) and mce2E AAA-introducing M. smegmatis (mce2E AAA-M. smegmatis) strains exhibited similar effects as BCG Δmce2E or WT M. smegmatis (Fig. 2; Supplementary Figure 5). Previously, we reported that Mce3E inhibited ERK pathway to suppress TNF and IL-6 secretion. Thus, we sought to compare the effect of Mce2E and Mce3E on the expression of TNF and IL-6 in the same system. Because of the absence of Mce3E in the BCG strain, we introduced mce3E into BCG (mce3E-BCG) for this comparison study. Our results suggest that like Mce3E, Mce2E also inhibits TNF and IL-6 production in macrophages (Supplementary Figure 6).
Fig. 2.
Mtb Mce2E inhibits cytokine production of TNF and IL-6 in macrophages. a, b Quantitative PCR analysis of tnf mRNA (a) and il6 mRNA (b) in RAW264.7 cells. Cells were infected with WT BCG, BCG Δmce2E, BCG Δmce2E:mce2E or BCG Δmce2E:mce2E AAA at a MOI of 10 for 0–48 h. The level of each gene was normalized to that of GAPDH. c, d Enzyme-linked immunosorbent assay (ELISA) of TNF (c) and IL-6 (d) from the medium of RAW264.7 cells infected with WT BCG, BCG Δmce2E, BCG Δmce2E:mce2E or BCG Δmce2E:mce2E AAA at a MOI of 10 for 0–48 h. e Survival of BCG strains in RAW264.7 cells treated as in a and b. *P < 0.05 and **P < 0.01 (two-tailed unpaired t test). Data are representative of experiments with at least three independent biological replicates (mean and s.e.m. of n = 3 cultures)
Mtb Mce2E suppresses the host immune response to mycobacteria in mice
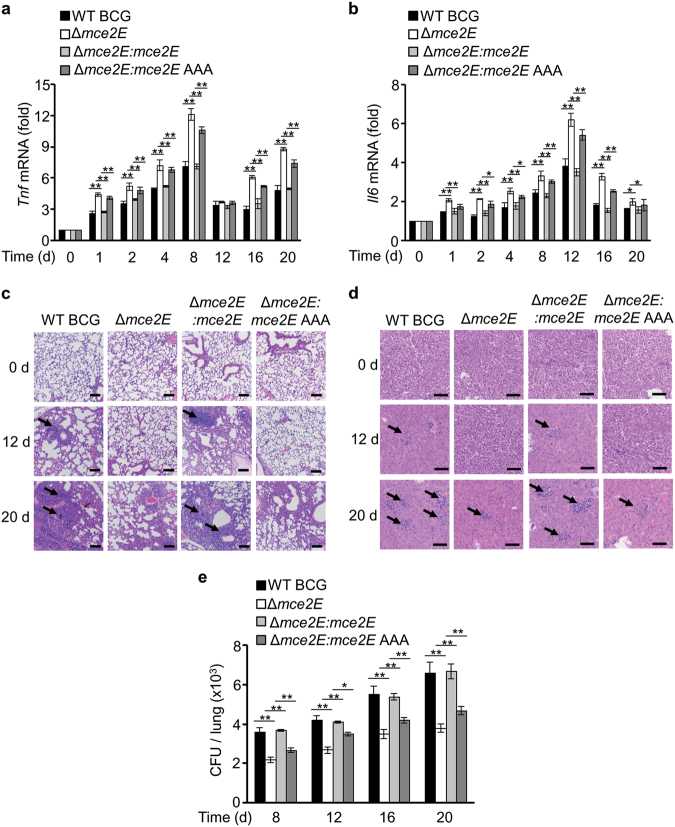
To confirm the immune suppression function of Mce2E, we challenged C57BL/6 mice intratracheally with WT BCG, BCG Δmce2E, BCG Δmce2E:Mce2E or BCG Δmce2E:Mce2E AAA. Consistently, BCG Δmce2E and BCG Δmce2E:Mce2E AAA increased the production of Tnf mRNA and Il6 mRNA in the spleen (Fig. 3a, b), inhibited cellular infiltration in the lung and liver (Fig. 3c, d), and reduced the bacterial load in the lungs of infected mice (Fig. 3e). The levels of Tnf and Il6 mRNAs induced by BCG Δmce2E and BCG Δmce2E:Mce2E AAA were much higher than those by WT BCG and BCG Δmce2E:Mce2E from 1 day after infection. The cellular infiltration foci began to appear at day 12 and became obvious at day 20 in the lung and liver of mice infected with WT BCG and BCG Δmce2E:Mce2E, but they were barely detectable in mice infected with BCG Δmce2E and BCG Δmce2E:mce2E AAA. We also compared the immuno-regulatory function of Mce2E and Mce3E in vivo. Unlike Mce2E, Mce3E exhibited minor effects on the host immune response towards mycobacterial infection in vivo (Supplementary Figure 7). Taken together, Mce2E, but not Mce3E, suppresses host immune response to mycobacteria in a D motif-dependent manner in vivo.
Fig. 3.
The D motif of Mce2E contributes to innate immune suppression during mycobacterial infection in vivo. a, b Quantitative PCR analysis of Tnf mRNA (a) and Il6 mRNA (b) in splenic cells from mycobacteria-infected mice. C57BL/6 mice were infected intratracheally with 2 × 106 of the WT BCG, BCG Δmce2E, BCG Δmce2E:mce2E, or BCG Δmce2E:mce2E AAA strain for 0–20 days. c, d Hematoxylin and eosin (H&E) staining of the lungs (c) and livers (d) of mice treated as in a and b. Arrows indicate foci of cellular infiltration. Scale bars, 200 μm. e Bacterial load in homogenates from the lungs of mice treated as in a and b. *P < 0.05 and **P < 0.01 (unpaired two-tailed Student’s t test). Data are representative of experiments with two independent biological replicates (mean and s.e.m. of n = 6 mice per group)
Mce2E suppresses the immune response to mycobacteria by trapping ERK and JNK into the ER apparatus in macrophages
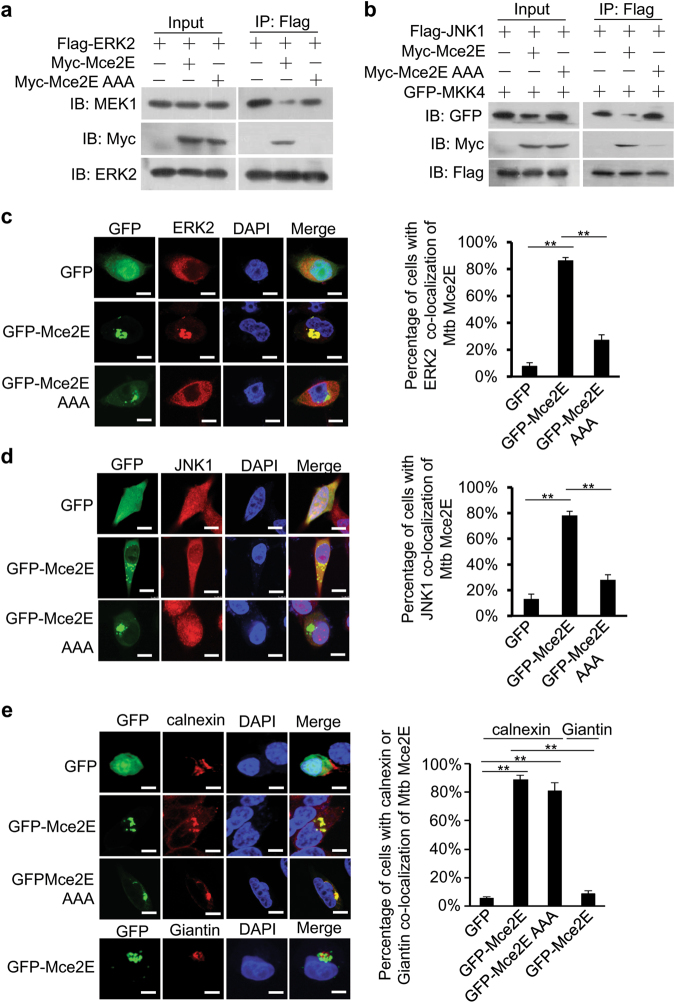
To elucidate the mechanisms by which Mce2E inhibits the ERK and JNK pathways, we performed coimmunoprecipitation and immunofluorescence assays to investigate the interactions between Mce2E and ERK/JNK proteins, as well as their subcellular localization. We found that Mce2E interacted with ERK2 and JNK1, but not p38α, in macrophage cells in a D motif-dependent manner. In addition, Mce2E interfered with the interactions between ERK and its upstream kinase MEK1, as well as the interactions between JNK and its upstream kinase MKK4. By contrast, Mce2E did not interact with p38 and had little effect on the interactions between p38 and its upstream kinase MKK3 (Fig. 4a, b; Supplementary Figure 8). Consistently, Mce2E colocalized with ERK2 and JNK1, but not p38, in macrophages in a D motif-dependent manner (Fig. 4c, d; Supplementary Figure 9). Furthermore, Mce2E colocalized with the ER marker Calnexin, but not the Golgi marker Giantin, independent of the D motif of Mce2E (Fig. 4e). Taken together, these data suggest that Mce2E might suppress the ERK and JNK MAPK pathways in at least two ways. First, Mce2E could block the phosphorylation of ERK1/2 and JNK by blocking the interactions between ERK/JNK proteins and their upstream kinases. Second, Mce2E could entrap ERK2 and JNK1 to the ER, further attenuating the activation of the ERK and JNK MAPK pathways.
Fig. 4.
Mce2E colocalizes with ERK and JNK in macrophages. a, b Immunoblot analysis of proteins immunoprecipitated with anti-Flag from lysates of HEK293T cells transfected with the empty vector or vector encoding Myc-tagged Mce2E or its mutant Mce2E AAA together with the vector encoding Flag-tagged ERK2 and MEK1 (a) or Flag-tagged JNK1 and GFP-tagged MKK4 (b). c, d Confocal microscopy analysis of Mce2E and ERK2 or JNK1 colocalization. RAW264.7 cells were transfected with the vector encoding GFP alone or GFP-tagged Mce2E or its mutant Mce2E AAA (green), and cells were then stained with an ERK2 or JNK1 antibody (red). Right, the percentage of cells with ERK2 or JNK1 colocalization with Mce2E (a total of 200 cells were counted). Nuclei were stained with DNA-binding dye (DAPI) (blue). Scale bars, 10 µm. e Confocal microscopy analysis for Mce2E subcellular localization. RAW264.7 cells were transfected as in b, and then, the ER and Golgi were stained with Calnexin and Giantin (red), respectively. Right, the percentage of cells with ER localization of Mce2E (a total of 200 cells were counted). Nuclei were stained with DNA-binding dye (DAPI) (blue). Scale bars, 10 µm. *P < 0.05 and **P < 0.01 (two-tailed unpaired ttest). Data are representative of experiments with at least three independent biological replicates (mean and s.e.m. of n = 3 cultures)
Mtb Mce2E promotes human epithelial A549 cell proliferation in a D motif-independent manner
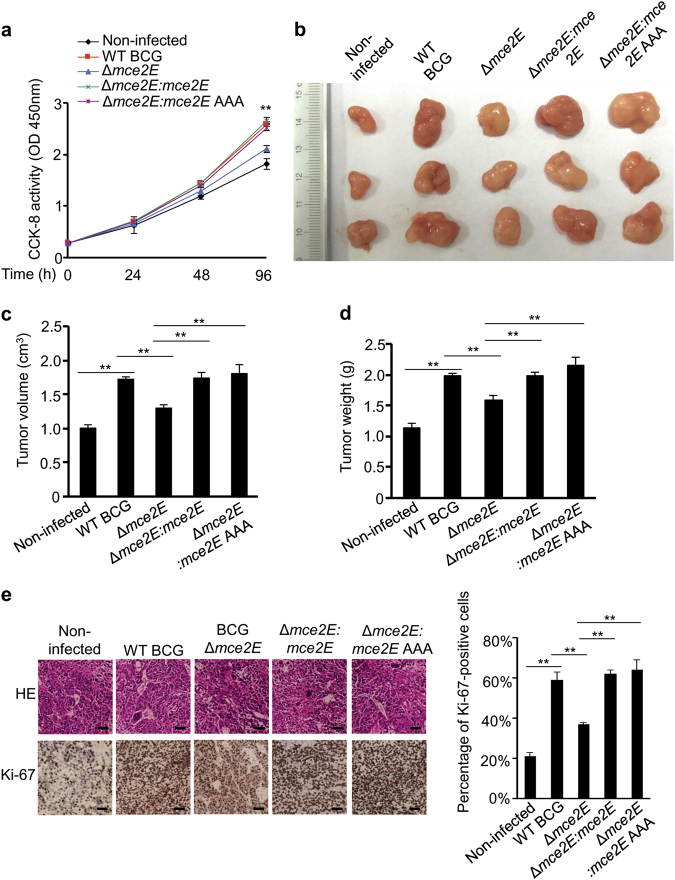
Previous studies have shown that stimulation of the ERK pathway promotes cell proliferation, and inhibition of the ERK pathway suppresses tumor cell proliferation.33–36 We have also found that the proliferation of human lung epithelium-derived lung adenoma A549 cells is attenuated by treatment with U0126 (a MEK inhibitor to block ERK pathway) (Supplementary Figure 10). Consistently, the CCK-8 assay showed that A549 cells infected with WT BCG, BCG Δmce2E:mce2E or BCG Δmce2E:mce2E AAA exhibited faster proliferation rates than cells infected with BCG Δmce2E (Fig. 5a). We also compared the effects of Mce2E and Mce3E on the proliferation of A549 cells and found that Mce3E did not show such cell proliferation-promoting functions (Supplementary Figure 11). Thus, we applied a mouse xenograft model to confirm the regulatory function of Mce2E in tumor progression in vivo, in which A549 cells were infected with WT BCG, BCG ∆mce2E, BCG Δmce2E:mce2E or BCG Δmce2E:mce2E AAA strains before inoculation into mice. At 14 days after injection, tumors were separated from nude mice for further analysis. BCG Δmce2E infection significantly decreased the size and weight of the xenograft tumors compared with the WT BCG, BCG Δmce2E:mce2E and BCG Δmce2E:mce2E AAA infection groups (Fig. 5b–d). Consistently, the percentage of Ki-67-positive cells in tumors infected with WT BCG, BCG Δmce2E:mce2E or BCG Δmce2E:mce2E AAA was much higher than in the BCG ∆mce2E infection group (Fig. 5e). Taken together, Mtb Mce2E promotes tumor cell proliferation in a D motif-independent manner, indicating that Mtb Mce2E may target other proteins or pathways to promote the proliferation of A549 epithelial cells.
Fig. 5.
Mtb Mce2E promotes A549 cell proliferation in a D motif-independent manner. a CCK-8 assay of A549 cells. Cells were infected with WT BCG, BCG Δmce2E, BCG Δmce2E:mce2E, or BCG Δmce2E:mce2E AAA at a MOI of 10 for 0–4 days, and then, the OD450nm value was measured and cell numbers were calculated. Non-infected A549 cells were used as a control. b Photograph of tumors. Tumors were derived from non-infected A549 cells or cells infected as in a in BALB/c nude mice. c Volume of tumors in nude mice. Tumor cells were injected subcutaneously into the right armpit of nude mice, the short and long diameters of the tumors were measured and the tumor volumes (cm3) were calculated at 14 days after injection. d Weight of tumors in nude mice. e Representative H&E staining histopathologic images of tumor tissues from mice (upper panels) and immunohistochemical analysis of Ki-67 (lower panels). Scale bars, 200 μm. Right, percentage of Ki-67-positive cells in tumors. Approximately 200 cells were counted. *P < 0.05 and **P < 0.01 (unpaired two-tailed Student’s t test). Data are representative of experiments with three independent biological replicates (mean and s.e.m. of n = 6 mice per group)
Mtb Mce2E inhibits K48-linked polyubiquitination of eEF1A1 to promote A549 cell proliferation
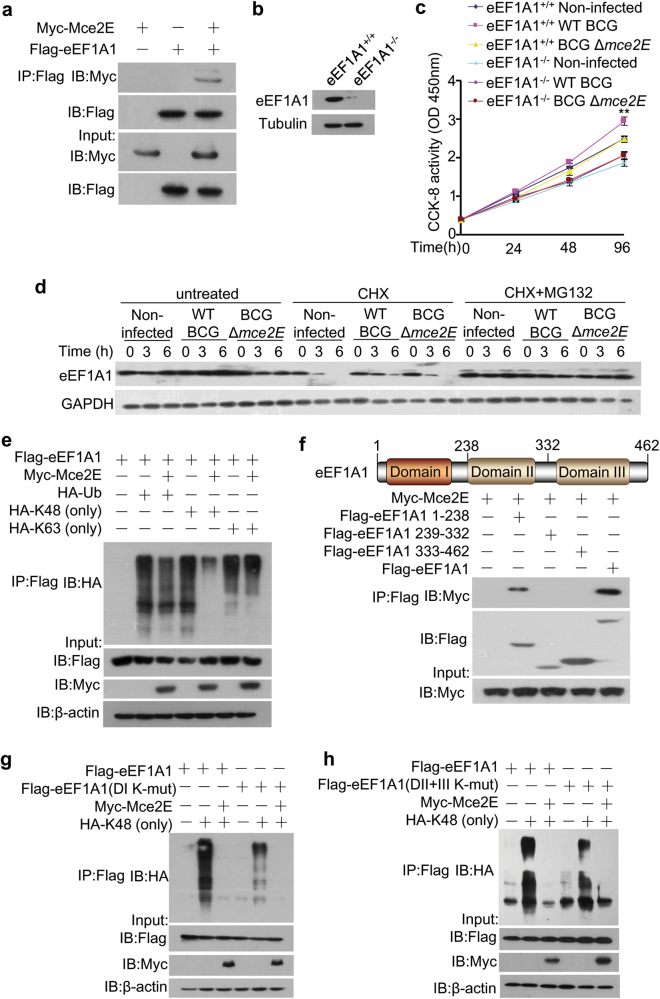
To explore the molecular mechanism of the Mce2E-promtoed cell proliferation, we performed a yeast two-hybrid assay to identify Mce2E-interacting proteins in the host and confirmed that Mce2E interacted with eukaryotic translation elongation factor 1 alpha 1 (eEF1A1) (Fig. 6a; Supplementary Figure 12), a highly conserved GTP-binding protein and a putative oncogene involved in protein synthesis and cell proliferation in mammalian cells.37, 38 We then sought to investigate whether Mce2E could regulate A549 cell proliferation by targeting eEF1A1. We first obtained eEF1A1 knockout A549 cell lines using the CRISPR-Cas9 system (Fig. 6b) for the CCK-8 assay, and our data showed that the cell proliferation-promoting effects of Mce2E were abolished in eEF1A1 KO cells (Fig. 6c). In our further efforts to elucidate the underlying mechanism by which Mce2E regulates eEF1A1 to promote A549 cell proliferation, we found that Mce2E could inhibit the proteasomal degradation of eEF1A1 protein, mainly by binding to the domain I of eEF1A1 and thus suppressing the K48-linked polyubiquitination, but not the K63-linked polyubiquitination, of lysines in domains I, II and III of eEF1A1 (Fig. 6d–h). Collectively, Mtb Mce2E inhibits K48-linked polyubiquitination and proteasomal degradation of eEF1A1 to promote the proliferation of A549 cells.
Fig. 6.
Mtb Mce2E inhibits K48-linked polyubiquitination of eEF1A1. a Immunoblot analysis of proteins immunoprecipitated with anti-Flag from lysates of HEK293T cells transfected with the vector encoding Myc-tagged Mce2E, Flag-tagged eEF1A1 or both. b Immunoblot analysis of the eEF1A1 level in WT and eEF1A1 knockout A549 cell lines. c CCK-8 assay of WT and eEF1A1 knockout A549 cell lines. Cells were infected with WT BCG or BCG Δmce2E at a MOI of 10 for 0–4 days, and then, the OD450nm value was measured. Non-infected A549 cells were used as a control. d The protein levels of eEF1A1 in A549 cells infected as in c and untreated or treated with cycloheximide (CHX, 50 µM) or both CHX (50 µM) and MG132 (20 µM) for 0–6 h. e Polyubiquitination assay of eEF1A1 in HEK293T cells transfected with Myc-Mce2E together with HA-tagged wild-type, K63 (only) or K48 (only) ubiquitin. f Immunoblot analysis of proteins immunoprecipitated with anti-Flag antibodies from lysates of HEK293T cells transfected with vectors encoding Myc-tagged Mce2E together with Flag-tagged eEF1A1 domain I (amino acids 1-238), eEF1A1 domain II (amino acids 239-332), eEF1A1 domain III (amino acids 333-462) or full-length eEF1A1. g, h K48-linked polyubiquitination assay of eEF1A1 and eEF1A1 mutants in HEK293T cells. Cells were co-transfected with the Flag-eEF1A1 or Flag-eEF1A1 mutant, DI K-mut: all lysines in domain I were replaced with arginine (g); DII+III K-mut: all lysines in domain II and III were replaced with arginine (h) together with empty vectors or the vector encoding HA-tagged K48 (only) ubiquitin, Myc-Mce2E or both HA-K48 (only) ubiquitin and Myc-Mce2E. *P < 0.05 and **P < 0.01 (two-tailed unpaired ttest). Data are representative of experiments with at least three independent biological replicates (mean and s.e.m. of n = 3 cultures)
Mtb Mce2E promotes tumor growth in a mouse xenograft model
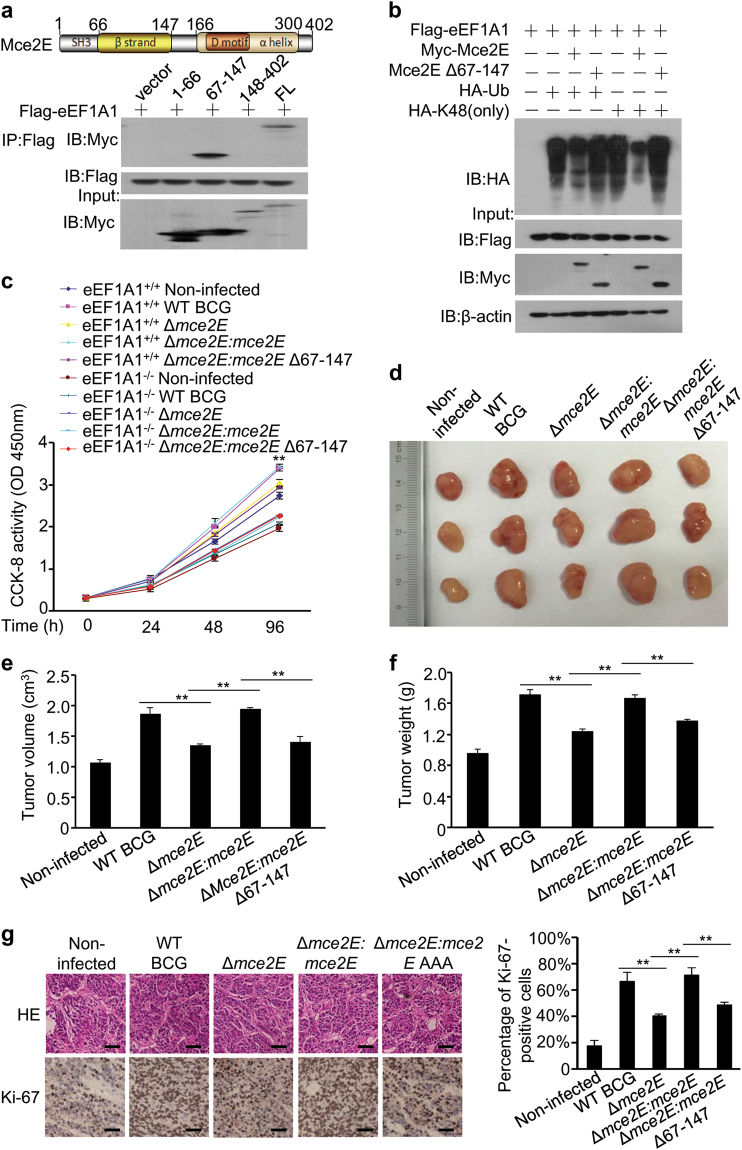
We next performed a bioinformatics analysis to identify the critical region in Mce2E that bound to eEF1A1, and we found that Mce2E was mainly composed of one β-strand, one α helix, a conserved SH3 domain and a D motif. We then confirmed that the β-strand region (amino acids 67-147) of Mce2E interacted with eEF1A1 (Fig. 7a). The truncated mutant mce2E Δ67-147, which could not efficiently interact with eEF1A1 (Supplementary Figure 13), largely lost its ability to inhibit eEF1A1 polyubiquitination and to promote A549 cell proliferation (Fig. 7b, c; Supplementary Figure 14). Consistent with the negligible effect of Mce3E on the proliferation of A549 cells, Mce3E showed little interaction with eEF1A1 and no effect on the K48-linked polyubiquitination of eEF1A1. It should be noted that although Mce3E also contains a β-strand, its amino-acid sequence is only 37.84% identical to that of Mce2E (Supplementary Figure 15). We then used the mouse xenograft model to further confirm the regulatory function of the β-strand region of Mce2E in tumor progression in vivo, and we found that infection of BCG Δmce2E or BCG Δmce2E:mce2E Δ67-147 strains significantly decreased the size and weight of the xenograft tumors (Fig. 7d–f). Consistently, the percentage of Ki-67-positive cells in tumors infected with BCG Δmce2E or BCG Δmce2E:mce2E Δ67-147 was much lower than that of the WT BCG or BCG Δmce2E:mce2E infection groups (Fig. 7g). Collectively, these results indicated that Mtb Mce2E could suppress ERK- and JNK-dependent macrophage innate immune response in a non-canonical D motif-dependent manner, while promoting epithelial cell proliferation in a β-strand region-dependent manner (Supplementary Figure 16).
Fig. 7.
Mtb Mce2E promotes A549 cell proliferation in a β-strand-dependent manner. a Immunoblot analysis of proteins immunoprecipitated with anti-myc from lysates of HEK293T cells transfected with the vector encoding myc-Mce2E and Flag-tagged eEF1A1 or its truncated forms. b Polyubiquitination assay of eEF1A1 in HEK293T cells transfected with Flag-eEF1A1 and Myc-Mce2E or Myc-Mce2E Δ67-147, together with the empty vector or vector encoding HA-tagged wild-type or K48 (only) ubiquitin. c CCK-8 assay of wild-type and eEF1A1 knockout A549 cell lines. Cells were infected with WT BCG, BCG Δmce2E, or BCG Δmce2E:mce2E, BCG Δmce2E:mce2E Δ67-147 at a MOI of 10 for 0–4 days, and then, the OD450nm value was measured. Non-infected A549 cells were used as a control. d Photograph of the tumors. Tumors were derived from non-infected A549 cells or cells infected as in b in BALB/c nude mice. e Volume (cm3) of tumors in nude mice calculated at 14 days after injection. f Weight of tumors in nude mice. g Representative H&E staining histopathologic images of tumor tissues from mice (upper panels) and immunohistochemical analysis of Ki-67 (lower panels). Scale bars, 200 μm. Right, percentage of Ki-67-positive cells in tumors. Approximately 200 cells were counted. *P < 0.05 and **P < 0.01 (unpaired two-tailed Student’s t test). Data are representative of experiments with three independent biological replicates (mean and s.e.m. of n = 6 mice per group)
Discussion
Alveolar macrophages and type II alveolar epithelium cells serve as invasive and replicative niches for Mtb.39, 40 Modulation of innate immune signaling pathways and cellular functions is critical for the successful intracellular survival of pathogens.15, 16, 41, 42 We have previously shown that Mce3E can specifically inhibit activation of the ERK signaling pathway but not the NF-κB and JNK signaling pathway in a DEF motif-dependent manner.16 Here, we reveal that another Mce family member Mce2E could suppress both the ERK and JNK signaling pathways in a non-canonical D motif-dependent manner. Thus, certain Mtb effector proteins may employ different modules to synergistically target certain important host signaling pathways, although the effects of different effector proteins could be stronger or weaker in a certain context. The seemingly contradictory phenomenon that Mtb infection induces TNF-α secretion from macrophages, while certain effector proteins, such as Mce2E and Mce3E, inhibit TNF-α secretion suggests that Mtb can produce and secrete a variety of effector proteins into host cells to either trigger or evade host immune responses in a temporal and spatial manner. In fact, considering the hostile and complicated living environment for Mtb within its host cells, effector proteins with different regulatory roles constitute a powerful arsenal, which is utilized by Mtb to adjust its regulatory roles bi-directionally and dynamically, thus ensuring its long-term intracellular survival.
Our data further reveal that Mtb Mce2E not only suppresses the host innate immune response but also promotes the proliferation of tumor cells in vitro and in vivo. Previous studies have demonstrated that Mtb-causing chronic TB infection can induce lung squamous cell carcinoma in a mouse model,19 and the human monocytic cell line (THP-1) infected with Mtb induces EMT characteristics of the lung adenocarcinoma cell line during co-culture.43 BCG has also been shown to trigger the survival of A549 cells from TNF-induced apoptosis, thereby promoting tumorigenesis in a xenograft model.20 Moreover, we have previously demonstrated that the Mtb effector protein PtpA can promote the proliferation and migration of A549 cells in vitro and in a mouse xenograft model.21 However, there is also conflicting evidence concerning a possible association between Mtb-caused pulmonary TB and the subsequent risk of lung cancer.44–46 Here we show that Mtb Mce2E can inhibit K48-linked polyubiquitination of eEF1A1 to enhance the protein stability of eEF1A1, leading to the enhancement of tumor cell proliferation. Our findings provide new evidence for a causal link between TB and lung cancer, and they suggest that eEF1A1 may be an important target for pathogen-associated chronic inflammation-promoted tumorigenesis and progression.
Our data demonstrate that Mce2E can inhibit K48-linked polyubiquitination of domains I, II, and III of eEF1A1 by binding to domain I, indicating that Mce2E may block the E3 ubiquitin ligase binding site of eEF1A1 by competitively binding to eEF1A1. The ubiquitin system regulates almost all eukaryotic cellular processes and is frequently hijacked or attacked by many pathogens for their own benefits.47 We have previously demonstrated that Mtb PtpA directly interacts with the host ubiquitin molecule through a previously undefined ubiquitin-interacting motif-like (UIML) region, leading to the suppression of host innate immunity.15 In this study, we identified another ubiquitin system-regulating Mtb effector protein Mce2E, further strengthening the notion that the ubiquitin system is frequently targeted by multiple pathogen effector proteins through different mechanisms.
It should be noted that Mce2E blocked the ERK pathway but promoted the proliferation of A549 cells during mycobacterial infection. The ERK pathway has been shown to be involved in the regulation of cellular differentiation and proliferation,34, 48 and ERK inhibitors were found to suppress cell proliferation and promote cell apoptosis.49 The seemingly contradictory phenomena regarding the down-regulation of the ERK pathway and up-regulation of cell proliferation by Mce2E, as revealed herein, can be interpreted to indicate that Mce2E may promote A549 cell proliferation largely by inhibiting eEF1A degradation, the effects of which could be strong enough to offset the potential cell proliferation-suppressive effects associated with its inhibitory role on ERK signaling pathway in A549 cells.
Taken together, our findings suggest that during infection, Mtb may secrete multi-functional effector proteins, such as Mce2E, into host cells, which function in a niche-dependent manner, thus ensuring the intracellular survival of the pathogen while causing host pathology as an accompanying outcome. Our findings reveal a novel Mtb virulence factor that regulates host innate immunity and cellular functions in a cell context-dependent manner, providing a potential novel drug target for TB treatment.
Electronic supplementary material
Acknowledgements
This work was supported by research funding from the National Key Research and Development Program of China (Grant Nos. 2017YFA0505900 and 2017YFD0500300), the National Basic Research Programs of China (Grant No. 2014CB74440), the National Natural Science Foundation of China (Grant Nos. 81571536 and 81571954), the Beijing Natural Science Foundation (Grant No. 5162021), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDPB03), and the Youth Innovation Promotion Association CAS.
Competing interests
The authors declare no competing interests.
Supplementary information
Supplementary information accompanies this paper at (10.1038/s41423-018-0016-0).
References
- 1.World Health Organization. Global Tuberculosis Report 2017. (WHO Press, Geneva, 2017).
- 2.Dheda K, et al. The epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant, extensively drug-resistant, and incurable tuberculosis. Lancet Respir. Med. 2017;5:291–360. doi: 10.1016/S2213-2600(17)30079-6. [DOI] [PubMed] [Google Scholar]
- 3.Awuh JA, Flo TH. Molecular basis of mycobacterial survival in macrophages. Cell. Mol. Life. Sci. 2016;74:1625–1648. doi: 10.1007/s00018-016-2422-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meena LS, Rajni Survival mechanisms of pathogenic Mycobacterium tuberculosis H37Rv. FEBS J. 2010;277:2416–2427. doi: 10.1111/j.1742-4658.2010.07666.x. [DOI] [PubMed] [Google Scholar]
- 5.Liu CH, Liu H, Ge B. Innate immunity in tuberculosis: host defense vs pathogen evasion. Cell. Mol. Immunol. 2017;14:963–975. doi: 10.1038/cmi.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng G, Zhang G, Chen X. Th1 cytokines, true functional signatures for protective immunity against TB? Cell. Mol. Immunol. 2018;15:206–215. doi: 10.1038/cmi.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pearl JE, Das M, Cooper AM. Luck or something more? Considering the connections between host and environment in TB. Cell. Mol. Immunol. 2018;15:226–232. doi: 10.1038/cmi.2017.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong C, Davis RJ, Flavell RA. Map kinases in the immune response. Annu. Rev. Immunol. 2002;20:55–72. doi: 10.1146/annurev.immunol.20.091301.131133. [DOI] [PubMed] [Google Scholar]
- 9.Raman M, Chen W, Cobb MH. Differential regulation and properties of MAPKs. Oncogene. 2007;26:3100–3112. doi: 10.1038/sj.onc.1210392. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Q, Lenardo MJ, Baltimore D. 30 years of NF-kappaB: a blossoming of relevance to human pathobiology. Cell. 2017;168:37–57. doi: 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arthur JS, Ley SC. Mitogen-activated protein kinases in innate immunity. Nat. Rev. Immunol. 2013;13:679–692. doi: 10.1038/nri3495. [DOI] [PubMed] [Google Scholar]
- 12.Kasper CA, et al. Cell-cell propagation of NF-kappaB transcription factor and MAP kinase activation amplifies innate immunity against bacterial infection. Immunity. 2010;33:804–816. doi: 10.1016/j.immuni.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 13.Xu G, Wang J, Gao GF, Liu CH. Insights into battles between Mycobacterium tuberculosis and macrophages. Protein Cell. 2014;5:728–736. doi: 10.1007/s13238-014-0077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen H, Chen ZW. The crucial roles of Th17-related cytokines/signal pathways in M. tuberculosis infection. Cell. Mol. Immunol. 2018;15:216–225. doi: 10.1038/cmi.2017.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, et al. Mycobacterium tuberculosis suppresses innate immunity by coopting the host ubiquitin system. Nat. Immunol. 2015;16:237–245. doi: 10.1038/ni.3096. [DOI] [PubMed] [Google Scholar]
- 16.Li J, et al. Mycobacterium tuberculosis Mce3E suppresses host innate immune responses by targeting ERK1/2 signaling. J. Immunol. 2015;194:3756–3767. doi: 10.4049/jimmunol.1402679. [DOI] [PubMed] [Google Scholar]
- 17.Vir P, Gupta D, Agarwal R, Verma I. Immunomodulation of alveolar epithelial cells by Mycobacterium tuberculosis phosphatidylinositol mannosides results in apoptosis. APMIS. 2014;122:268–282. doi: 10.1111/apm.12141. [DOI] [PubMed] [Google Scholar]
- 18.Scordo JM, Knoell DL, Torrelles JB. Alveolar epithelial cells in Mycobacterium tuberculosis infection: active players or innocent bystanders? J. Innate Immun. 2016;8:3–14. doi: 10.1159/000439275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nalbandian A, Yan BS, Pichugin A, Bronson RT, Kramnik I. Lung carcinogenesis induced by chronic tuberculosis infection: the experimental model and genetic control. Oncogene. 2009;28:1928–1938. doi: 10.1038/onc.2009.32. [DOI] [PubMed] [Google Scholar]
- 20.Holla S, Ghorpade DS, Singh V, Bansal K, Balaji KN. Mycobacterium bovis BCG promotes tumor cell survival from tumor necrosis factor-alpha-induced apoptosis. Mol. Cancer. 2014;13:210. doi: 10.1186/1476-4598-13-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, et al. The mycobacterial phosphatase PtpA regulates the expression of host genes and promotes cell proliferation. Nat. Commun. 2017;8:244. doi: 10.1038/s41467-017-00279-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimono N, et al. Hypervirulent mutant of Mycobacterium tuberculosis resulting from disruption of the mce1 operon. Proc. Natl Acad. Sci. USA. 2003;100:15918–15923. doi: 10.1073/pnas.2433882100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Senaratne RH, et al. Mycobacterium tuberculosis strains disrupted in mce3 and mce4 operons are attenuated in mice. J. Med. Microbiol. 2008;57:164–170. doi: 10.1099/jmm.0.47454-0. [DOI] [PubMed] [Google Scholar]
- 24.Marjanovic O, Miyata T, Goodridge A, Kendall LV, Riley LW. Mce2 operon mutant strain of Mycobacterium tuberculosis is attenuated in C57BL/6 mice. Tuberculosis. 2010;90:50–56. doi: 10.1016/j.tube.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talaat AM, Lyons R, Howard ST, Johnston SA. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc. Natl Acad. Sci. USA. 2004;101:4602–4607. doi: 10.1073/pnas.0306023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang F, Xie JP. Mammalian cell entry gene family of Mycobacterium tuberculosis. Mol. Cell. Biochem. 2011;352:1–10. doi: 10.1007/s11010-011-0733-5. [DOI] [PubMed] [Google Scholar]
- 27.Kumar A, Bose M, Brahmachari V. Analysis of expression profile of mammalian cell entry (mce) operons of Mycobacterium tuberculosis. Infect. Immun. 2003;71:6083–6087. doi: 10.1128/IAI.71.10.6083-6087.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Kessel JC, Hatfull GF. Recombineering in Mycobacterium tuberculosis. Nat. Methods. 2006;4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 29.Weston CR, Lambright DG, Davis RJ. Signal transduction. MAP kinase signaling specificity. Science. 2002;296:2345–2347. doi: 10.1126/science.1073344. [DOI] [PubMed] [Google Scholar]
- 30.Garai A, et al. Specificity of linear motifs that bind to a common mitogen-activated protein kinase docking groove. Sci. Signal. 2012;5:ra74. doi: 10.1126/scisignal.2003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Crevel R, Ottenhoff THM, van der Meer JWM. Innate immunity to Mycobacterium tuberculosis. Clin. Microbiol. Rev. 2002;15:294–309. doi: 10.1128/CMR.15.2.294-309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng W, et al. Mycobacterium tuberculosis PPE family protein Rv1808 manipulates cytokines profile via co-activation of MAPK and NF-kappaB signaling pathways. Cell. Physiol. Biochem. 2014;33:273–288. doi: 10.1159/000356668. [DOI] [PubMed] [Google Scholar]
- 33.DeSilva DR, et al. Inhibition of mitogen-activated protein kinase kinase blocks T cell proliferation but does not induce or prevent anergy. J. Immunol. 1998;160:4175–4181. [PubMed] [Google Scholar]
- 34.Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6:322–327. doi: 10.1016/S1470-2045(05)70168-6. [DOI] [PubMed] [Google Scholar]
- 35.Gautam US, et al. Role of TNF in the altered interaction of dormant Mycobacterium tuberculosis with host macrophages. PLoS ONE. 2014;9:e95220. doi: 10.1371/journal.pone.0095220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keane J, et al. Infection by Mycobacterium tuberculosis promotes human alveolar macrophage apoptosis. Infect. Immun. 1997;65:298–304. doi: 10.1128/iai.65.1.298-304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mateyak MK, Kinzy TG. eEF1A: thinking outside the ribosome. J. Biol. Chem. 2010;285:21209–21213. doi: 10.1074/jbc.R110.113795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thornton S, Anand N, Purcell D, Lee J. Not just for housekeeping: protein initiation and elongation factors in cell growth and tumorigenesis. J. Mol. Med. 2003;81:536–548. doi: 10.1007/s00109-003-0461-8. [DOI] [PubMed] [Google Scholar]
- 39.Adlakha N, Vir P, Verma I. Effect of mycobacterial secretory proteins on the cellular integrity and cytokine profile of type II alveolar epithelial cells. Lung India. 2012;29:313–318. doi: 10.4103/0970-2113.102796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bruns H, Stenger S. New insights into the interaction of Mycobacterium tuberculosis and human macrophages. Future Microbiol. 2014;9:327–341. doi: 10.2217/fmb.13.164. [DOI] [PubMed] [Google Scholar]
- 41.Li S, et al. Pathogen blocks host death receptor signalling by arginine GlcNAcylation of death domains. Nature. 2013;501:242–246. doi: 10.1038/nature12436. [DOI] [PubMed] [Google Scholar]
- 42.Mukherjee S, et al. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- 43.Gupta PK, Tripathi D, Kulkarni S, Rajan MG. Mycobacterium tuberculosis H37Rv infected THP-1 cells induce epithelial mesenchymal transition (EMT) in lung adenocarcinoma epithelial cell line (A549) Cell. Immunol. 2016;300:33–40. doi: 10.1016/j.cellimm.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 44.Wu CY, et al. Pulmonary tuberculosis increases the risk of lung cancer: a population-based cohort study. Cancer. 2011;117:618–624. doi: 10.1002/cncr.25616. [DOI] [PubMed] [Google Scholar]
- 45.Engels EA, et al. Tuberculosis and subsequent risk of lung cancer in Xuanwei, China. Int. J. Cancer. 2009;124:1183–1187. doi: 10.1002/ijc.24042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang HY, et al. Facts and fiction of the relationship between preexisting tuberculosis and lung cancer risk: a systematic review. Int. J. Cancer. 2009;125:2936–2944. doi: 10.1002/ijc.24636. [DOI] [PubMed] [Google Scholar]
- 47.Li J, Chai QY, Liu CH. The ubiquitin system: a critical regulator of innate immunity and pathogen-host interactions. Cell. Mol. Immunol. 2016;13:560–576. doi: 10.1038/cmi.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mebratu Y, Tesfaigzi Y. How ERK1/2 activation controls cell proliferation and cell death: Is subcellular localization the answer? Cell Cycle. 2014;8:1168–1175. doi: 10.4161/cc.8.8.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiesenauer CA, Yip-Schneider MT, Wang Y, Schmidt CM. Multiple anticancer effects of blocking MEK-ERK signaling in hepatocellular carcinoma. J. Am. Coll. Surg. 2004;198:410–421. doi: 10.1016/j.jamcollsurg.2003.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the authors upon request.