Abstract
Fluctuating asymmetry is a classic concept linked to organismal development. It has traditionally been used as a measure of developmental instability, which is the inability of an organism to buffer environmental fluctuations during development. Developmental stability has a genetic component that influences the final phenotype of the organism and can lead to congenital disorders. According to alternative hypotheses, this genetic component might be either the result of additive genetic effects or a by-product of developmental gene networks. Here we present a genome-wide association study of the genetic architecture of fluctuating asymmetry of the skull shape in mice. Geometric morphometric methods were applied to quantify fluctuating asymmetry: we estimated fluctuating asymmetry as Mahalanobis distances to the mean asymmetry, correcting first for genetic directional asymmetry. We applied the marginal epistasis test to study epistasis among genomic regions. Results showed no evidence of additive effects but several interacting regions significantly associated with fluctuating asymmetry. Among the candidate genes overlapping these interacting regions we found an over-representation of genes involved in craniofacial development. A gene network is likely to be associated with skull developmental stability, and genes originally described as buffering genes (e.g., Hspa2) might occupy central positions within these networks, where regulatory elements may also play an important role. Our results constitute an important step in the exploration of the molecular roots of developmental stability and the first empirical evidence about its genetic architecture.
Subject terms: Quantitative trait, Development
Introduction
Environmental stressors or genetic mutations can produce perturbations of standard developmental pathways and therefore increase developmental noise. The ability of organisms to buffer their development against environmental and genetic perturbations is known as developmental stability (DS). This attribute allows the organism to develop an appropriate target phenotype, which is essential for its survival and reproduction. DS is a component of phenotypic robustness, which has been largely reviewed within the evolutionary literature and has strong clinical implications (De Visser et al. 2003; Gonzalez et al. 2016; Queitsch and Carlson 2012; Siegal and Leu 2014). In particular, the development of craniofacial traits and their stability is a topic of broad interest because of the genetic and environmental factors that may lead to congenital disorders. However, the developmental architecture of the craniofacial shape is still far from clear due to the complexity of the craniofacial genotype–phenotype map (Hallgrímsson et al. 2014).
Fluctuating asymmetry (FA) comprises random deviations from the population mean asymmetry (Palmer and Strobeck 1986) and is accepted, with some measures of caution, as a good approximation to DS (Klingenberg 2015). Over the past 60 years various authors have proposed that FA has an important genetic component (Atchley and Rutledge 1980; Van Valen 1962; Waddington 1957), which was empirically shown later in individuals affected by major genetic diseases (Bock and Bowman 2006; Miller et al. 2014; Richtsmeier et al. 2005) and through experimental set-ups (Breuker et al. 2006). DS is influenced by a genetic component that may regulate the impact of environmental conditions over the final phenotype (Mather 1953; Palmer and Strobeck 1986; Reeve 1960; Thoday 1958; Van Dongen 2006; Waddington 1957). However, the literature on DS heritability has yielded low estimates (Leamy and Klingenberg 2005; Leamy et al. 2015; Van Dongen 2006), and currently a good consensus exists about the non-additive nature of FA (Leamy and Klingenberg 2005; Leamy et al. 2015; Van Dongen 2006). In the area of craniofacial genetics, many studies about shape FA are also in agreement with this view (Fernández Iriarte et al. 2003; Leamy et al. 2000; Leamy et al. 2005; Leamy 1993; Leamy and Klingenberg 2005; Leamy et al. 2015; Leamy et al. 2001; Leamy et al. 2002). Nonetheless, the ecological (De Coster et al. 2013), genetic and developmental roots of FA remain mostly unknown, except for some molecular explanations about the general origin of asymmetric variation (Coutelis et al. 2014; Vanderberg and Levin 2013).
Despite the widespread development of genome-wide association studies (GWAS) in humans (Visscher et al. 2017) and model organisms (Flint and Eskin 2012), only few studies have applied this approach to investigate FA genetics (Guadalupe et al. 2014; Tadayon et al. 2016). To our knowledge, no previous attempt has been made to conduct a GWAS on shape FA in the context of geometric morphometrics. In model organisms (e.g., mice), GWAS can benefit from carefully designed populations with desirable genetic properties such as an unstructured pattern of relatedness (Nicod et al. 2016; Speed and Balding 2015). In addition, biologically relevant univariate measures of shape FA (Klingenberg and Monteiro 2005) simplify the use of the geometric morphometrics toolkit, which is complex and computationally expensive in the context of multivariate GWAS.
Here we collected published data on skull shape in an outbred population of mice (Pallares et al. 2015a) to explore the genetic architecture of shape FA. We tried to disentangle the association among genome and shape FA with an additive and two non-additive genetic models. Non-additivity was studied first within a locus and second as the total amount of epistatic variance associated with each locus. Finally, we have considered the loci best associated with the total amount of epistatic variance to study their pairwise interactions (Rice 1998).
Materials and methods
Samples
The sample consists of 692 Carworth Farms White mice maintained by Charles River Laboratories. These mice are a population of outbred mice derived from a small number of founders (Nicod et al. 2016). Our sample exhibits low and standardised relatedness among individuals (Parker et al. 2014) and previous studies have shown no bias in the results due to population structure (Pallares et al. 2015a; Parker et al. 2016). All data are available from Dryad (Pallares et al. 2015b), linked to the original publication by Pallares et al. (2015a).
Geometric morphometrics
The coordinates of a set of 44 tridimensional landmarks from the skull were obtained from the dataset of Pallares et al. (2015a). We performed on these landmarks a fully generalised Procrustes superimposition to standardise size, position, orientation and reflection (Dryden and Mardia 1998). The superimposition estimates the asymmetric component of each skull shape, i.e., the difference between the original and mirrored landmark configurations to the mean shape (Klingenberg et al. 2002). The asymmetric component constitutes a natural measure of the difference between the two symmetrical sides of a given biological structure. Both Procrustes superimposition and the extraction of the asymmetric component were run in MorphoJ v1.06 (Klingenberg 2011).
Because FA is randomly distributed around the mean asymmetry, the Procrustes distances from each individual asymmetric component to the population average are an acceptable univariate measure of FA (Goodall 1991; Klingenberg and McIntyre 1998). However, integration (covariation among landmarks) is a widespread feature in morphometric data (Leamy 1993) that promotes anisotropy and can bias the estimates based on Procrustes distances among individuals. Therefore, we estimated Mahalanobis distances to remove the effect of anisotropic variation (Klingenberg and Monteiro 2005). This type of distance might be especially appropriate for skulls, where integration is pervasive (Klingenberg 2013). It may be noted that a reliable estimation of Mahalanobis distances is very demanding in terms of sample size, but in our case the number of individuals is more than eleven times bigger than the effective dimensionality of the data.
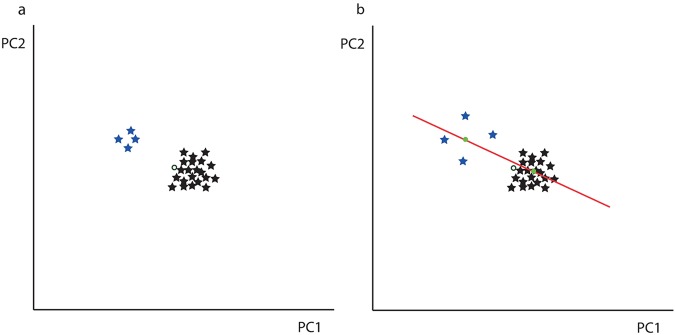
Although their usage is controversial, Procrustes distances frequently served as a measure of developmental instability (Klingenberg 2015). The fact that mice are motile organisms and therefore the differing effects of environment on each side of the skull may be negligible justify this choice. Nonetheless, for most of the genomic markers one genotype has a larger sample size, and therefore the population mean asymmetry is often closer to that genotypic population. This unbalanced sampling might bias statistical associations between genotypes and distances (Fig. 1). This effect is analogous to the presence of genetic variation for directional asymmetry (DA), which can similarly explain systematic biases on distances from each individual to the population average (Levin 2005). To control for these sources of bias, we first estimated the effect of each genotype on the mean asymmetry (i.e., DA). Thereafter, the residual distances constituted our FA measures. This procedure removes the effects of unbalanced sampling and the potential association of some markers with DA.
Fig. 1.
DA correction of the FA measure. a Unbalanced genetic sampling between two different genotypes (blue and black). The mean population (unfilled circle) is next to the black (larger) population. The effect of genotype on the variance from the mean population to the black population is therefore systematically smaller. b The red line represents a linear model of the genotype on the asymmetric component. Residuals from the means for each population (green dots) can be considered FA measures. This approach, applied in the study, removes the signal created by unbalanced sampling
Genomic data
The authors who originally collected the data followed a genotyping-by-sequencing approach and the PstI enzyme was chosen to obtain a high coverage of the genome. They multiplexed and sequenced 12 samples per lane on an Illumina HiSeq 2500 with single-end 100-bp reads, and these were aligned to the Mouse Reference Assembly 38 from the NCBI database. Variant discovery was calibrated against the whole-genome sequencing from a small set of CFW mice, SNPs and indels from the Wellcome Trust Sanger Mouse Genome project and SNPs in dbSNPs release 137. Finally, IMPUTE2 was used to estimate missing genotypes. A complete description of the genotyping process can be found in Pallares et al. (2015a).
The raw genomic data in this study consisted of the gene dosages of a set of genomic markers (Pallares et al. 2015b). We performed the analysis on these data in order to study the genomic additive effects. To run a dominance model we designed a second dataset, where each marker represents the probability of heterozygosity. Following Pallares et al. (2015a), we discarded SNPs with low imputation quality based on genotype probabilities. We kept SNPs with a maximum genotype probability greater than 0.5 and a minor allele frequency greater than 2%. Thus we were left with 79,787 SNPs in the additive model and 79,352 in the dominance model.
Mapping methods
Although our mice sample was conceived to have low levels of population structure (Nicod et al. 2016), we controlled for population structure by using linear mixed models (Speed and Balding 2015). We applied a leave-one-chromosome-out approach to estimate the genomic relationship matrix, which reflects relatedness among individuals. This approach is designed to diminish the problems caused by the inclusion of tested SNPs in the relationship matrix (Cheng et al. 2013; Listgarden et al. 2012; Pallares et al. 2015a; Parker et al. 2014). Then, we assessed the association between the SNPs and FA using a two-step linear mixed model. First, the variance components and a phenotypic covariance matrix were estimated for each chromosome (based on its genomic relationship matrix). Second, the Mahalanobis distances (phenotype) and gene dosages (genotypes) were corrected by the inverse of the squared root of the phenotypic covariance matrix, removing the correlation among errors. We could then fit efficiently the association between FA and SNPs in a linear model. We ran 1000 permutations to obtain two significance thresholds (5 and 10%) from a distribution of the maximum negative log10 p (Churchill and Doerge 1994). Analyses were run in R v3.3.1 (R Core Team 2013) using custom scripts (available from the authors).
Epistasis: genome scan
We performed an epistasis scan to assess non-heritable genetic associations with FA (previously corrected for overall DA). Using the method recently developed by Crawford et al. (2017) (https://github.com/lorinanthony/MAPIT/; Downloaded 15/05/18). Because there is a huge number of pairwise comparisons among all the genomic markers, the marginal epistasis test (MAPIT) selects first a small number of genomic markers to study in detail later. It runs a linear model to associate each genomic marker with a variance parameter composed of the epistatic contribution of the marker with all other markers. Here we followed this approach using the Davies method, which is more precise at the expense of more computational demands (Crawford et al. 2017). We selected all the markers with p < 2.5 × 10−4 for further comparisons in pairwise analyses. Although arbitrary, such a relatively high threshold allows the inclusion of more markers potentially involved in significant pairwise interactions. Pairwise interactions were considered significant when p < 0.001. Because population structure is not an issue in our population (see results) and MAPIT deals reasonably well with it (Crawford et al. 2017), the epistasis tests were run without relatedness correction.
Candidate genes
Significant results were explored using the annotated Mus musculus reference genome (version m38.92) from Ensembl (Yates et al. 2016) and Mouse Genome Informatics (Blake et al. 2017). A window of 200 kb around each SNP was considered to obtain an overlapping set of candidate genes. This window size was chosen based on the genomic linkage disequilibrium of this population, which is much lower than for other stocks of mice (Nicod et al. 2016; Parker et al. 2016). This size would correspond to an r2 of ~0.6. In the cases where no protein-coding genes were found we reported the processed transcripts, RNA genes and pseudogenes of interest. To further explore the results we also merged the markers where all the candidate genes were the same, and explored the interactions involving each of these different genomic regions.
Over-representation of known craniofacial or growth genes
Two reference gene lists were collected from the MGI HMDC database based on ‘Craniofacial’ and ‘Growth/Size/Body’ queries (http://www.informatics.jax.org/humanDisease.shtml; Accessed 04/04/2018). We removed genes absent from the mouse genome (i.e., human-specific genes) and redundancies between the two lists. This correction yielded 1044 and 2829 genes, respectively, from both groups. The over-representation of candidate genes within these two lists was assessed with Fisher exact tests.
To simplify the interaction representation, we merged the SNPs whose likely best candidate gene was the same. Each of these genes was then classified as either ‘Craniofacial’ or ‘Growth/Size/Body’ according to the reference list where it belonged. Protein-coding candidate genes absent from both lists were classified as ‘Other’ and candidate genes associated with regulatory elements as ‘Regulatory’.
Craniofacial enhancers
We assessed the potential importance of craniofacial enhancers among the interacting markers. Low distances between the SNPs involved in significant pairwise interactions and their nearest transcription start site might suggest an important role of regulatory SNPs. To test whether these distances were significantly lower than expected, we estimated these distances in 10,000 random samples with the same number of genomic regions. For each set of distances, we inferred how many of them were shorter than the 95th percentile of the distribution of genomic distances between the craniofacial enhancers in Attanasio et al. (2013) and their nearest transcription start site (236 kb). Finally, we estimated how many random samples showed a larger number of short distances (<236 kb) than our set of significant SNPs. We repeated this procedure after merging SNPs with the same set of candidate genes.
Two strategies were followed to perform the random sampling. First, random samples were collected uniformly across the genome for which regions were assumed to be independent. Second, we tested a sampling with stratification by taking into account the number of observed SNPs on each chromosome. This second sampling considers the presence of a single causal locus and therefore a strong dependence between significant loci. For this second case we considered the minimal distance from the significant SNPs to their most proximal craniofacial enhancer as the observed value for the gene region once we merged all the markers with the same candidate genes.
Gene networks
The complete list of candidate genes obtained from the significant epistasis pairwise tests was curated against the STRING databases (https://string-db.org) of known and predicted protein-protein interactions (Szklarczyk et al. 2017). Results were represented as a network. We built a second network after merging the genomic regions with the same list of candidate genes, and a third one representing the epistasis interactions among the best candidate genes for each region.
Results
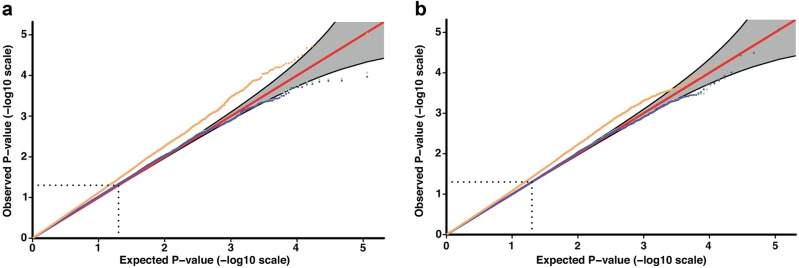
The results obtained are virtually the same with and without population structure correction (r > 0.99), as expected from previous studies (Nicod et al. 2016; Pallares et al. 2015a) and the low-heritable nature of FA (Leamy and Klingenberg 2005; Leamy et al. 2015; Van Dongen 2006). These low estimates are not surprising because the sample was designed to standardise the relatedness among individuals as much as possible (Parker et al. 2014). ASDA correction had a larger influence on the results (r > 0.87), those results are presented here. Given the 95% significance threshold of 6.61 × 10−7, no genomic marker showed a significant additive association with shape FA. With a dominance model (significance threshold of 6.31 × 10−7), we did not obtain any significant marker either (Fig. 2).
Fig. 2.
Quantile-quantile (QQ) plots for the datasets in the study of additive and dominant associations. The correction for population structure (green) does not influence the raw Procrustes data (violet) (r > 0.99). Procrustes distances corrected for DA (orange) are not influenced by the population structure either (not shown; r > 0.99). Raw and DA-corrected Procrustes distances are also highly related (r > 0.87). No significant marker is identified with either of the datasets. a QQ plot for additive effects. b QQ plot for dominant effects
Epistatic variance and pairwise interactions
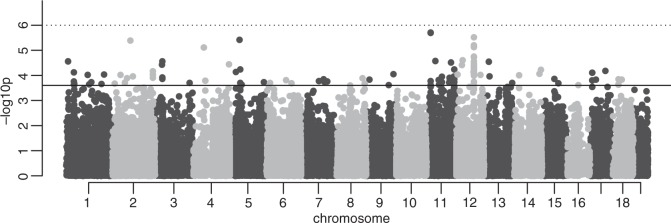
MAPIT provided individual SNPs, as well as pairwise SNP interactions significantly associated with FA (Fig. 3). Considering a selection threshold of 2.5 × 10−4 (the −log10(p) = 3.6, Fig. 3) on marker probabilities from the Davies method, a total of 118 markers were selected for further analyses. Of these, the 9 best-associated SNPs (p < 10−5) are clustered in six regions of 1–3 SNPs (Table 1). One region includes Hspa2 (Hsp70), one of the most studied genes for DS and canalisation (Geiler-Samerotte et al. 2016; Takahashi et al. 2010). Only one of these SNPs does not overlap with protein-coding genes previously associated with either craniofacial or size development (‘Growth/Size/Body’). The others overlap genes encoding for the transcription factor Max or the epidermal growth factor receptor (Egfr). Detailed information can be found in Supplementary Information S1.
Fig. 3.
Manhattan plot from the marginal epistasis test. The negative log10 of the probabilities from the Davies method (−log10(p)) is shown for each marker. This test estimates the epistatic variance between a given marker and all other markers. The plain red line represents the chosen threshold of −log10(2.5 × 10−4), all the markers with a LOD scores above the threshold are included in the pairwise comparisons
Table 1.
SNPs best associated to FA (p < 10−5)
| Marker | Chr | Pos (Mb) | p | Candidate genesa |
|---|---|---|---|---|
| cfw-2-78028827 | 2 | 78.03 | 4.15 × 10−6 | Cwc22 |
| cfw-4-46702257 | 4 | 46.70 | 7.60 × 10−6 | Tbc1d2, Gabbr2 |
| rs33490187 | 5 | 40.18 | 3.88 × 10−6 | |
| rs29463914 | 11 | 16.94 | 2.01 × 10−6 | Egfr |
| rs26873798 | 11 | 16.94 | 1.98 × 10−6 | Egfr |
| rs32442879 | 12 | 76.62 | 3.07 × 10−6 | Sptb, Fntb, Hspa2b |
| rs47669499 | 12 | 77.00 | 6.42 × 10−6 | Fntb, Max |
| rs29130163 | 12 | 77.05 | 7.60 × 10−6 | Fntb, Max, Fut8 |
| cfw-12-77225182 | 12 | 77.22 | 6.93 × 10−6 | Fut8 |
Candidate genes in bold are annotated for ‘Craniofacial’, whereas underlined ones are annotated for ‘Growth/Size/Body’
Chr chromosome, Pos position in Mb, p probability from Davies method
aCandidate genes are based on gene content in a 200 kb window
bHspa2 is 214 kb from the associated SNP
MAPIT found 162 significant pairwise comparisons (Table 2 and Table S2). These 162 significant interactions were composed of 68 SNPs overlapping with 377 candidate genes, of which 201 are protein-coding genes. Based on gene contents, these SNPs correspond to 50 genomic regions. After merging those with equivalent candidate genes, we were left with 109 interactions composed of 38 differing genomic regions. Six of the 38 equivalent regions were involved in 10 or more interactions (Table 3). The genomic region showing the majority of interactions overlaps Mecom, which is a well-known craniofacial gene causing an abnormal skull morphology (Hardisty-Hughes et al. 2010). We may note also that Hspa2 is in a region of 1 Mb, presenting the most of interactions at the SNP level.
Table 2.
Most significant pairwise interactions associated to FA in marginal epistasis test
| Marker | Chr | Candidate | Marker | Chr | Candidate Genes | p |
|---|---|---|---|---|---|---|
| rs31197155 | 3 | Gng5 | cfw-12-104797355 | 12 | Dicer1 | 5.64 × 10−8 |
| rs31926426 | 1 | Prex2 | rs27620389 | 2 | Bmp7 | 4.20 × 10−7 |
| rs33490187 | 5 | Reg-Chr5 | rs31272574 | 15 | Khdrbs3 | 6.10 × 10−7 |
| rs46801958 | 11 | Sdk2 | rs29827444 | 13 | Hfe | 9.35 × 10−7 |
Chr chromosome, Candidate Candidate gene within a 200 Kb window, p probability associated to the epistasis test
Only interactions with p < 10–6 are shown. Candidate genes in bold are annotated for ‘Craniofacial’ or for ‘Growth/Siz
Table 3.
The 10 most interacting genomic regions within the 109 significant pairwise interactions
| Candidate gene | Chr | Classification | Number of appearances |
|---|---|---|---|
| Mecom | 3 | Craniofacial | 20 |
| Pabpc6 | 17 | Other | 19 |
| Sdk2 | 11 | Other | 19 |
| Prex2 | 1 | Growth/size/body | 14 |
| Kcnmb2 | 3 | Other | 13 |
| Reg-Chr5 | 5 | Regulatory | 11 |
| Bmp7 | 2 | Craniofacial | 9 |
| Trim71 | 9 | Craniofacial | 9 |
| Cwc22 | 2 | Other | 8 |
| Gng5 | 9 | Craniofacial | 8 |
Each row shows the candidate genes within a 200 Kb window, the position within the genome (Chr chromosome), the most likely classification of the best candidate gene assessed in Mouse Genome Informatics (Blake et al. 2017) and the number of times the genomic region (identified with the candidate gene) appears within a significant pairwise interaction with one of the other 38 regions
Over-representation of craniofacial genes
Most of the SNPs involved in these pairwise interactions have one or more candidate genes involved in craniofacial development. We found that these SNPs are related to shape or skeleton growth via one or more protein-coding genes (see Table S3). Craniofacial genes were significantly over-represented (11 genes, p = 1.78 × 10−6), although for ‘growth’ genes the test is just marginally significant (9 genes, p = 0.06).
Contrary to the 68 epistatic SNPs to the 7283 enhancers reported by Attanasio et al (2013), we found that 54 SNPs were located within the 95th percentile of distances between enhancers and transcription start sites. This proportion is significantly higher than in a random sampling of 68 SNPs along the genome (p = 0.005) or stratified by chromosome (p = 0.01). However, when only the 38 best candidate genes were assessed, the association with craniofacial enhancers vanished (p = 0.37). This shows that the initial association was probably due to a set of 21 SNPs within a 741 kb region on chromosome 12, which overlaps the same candidate genes: the Fntb Max Fut8 cluster.
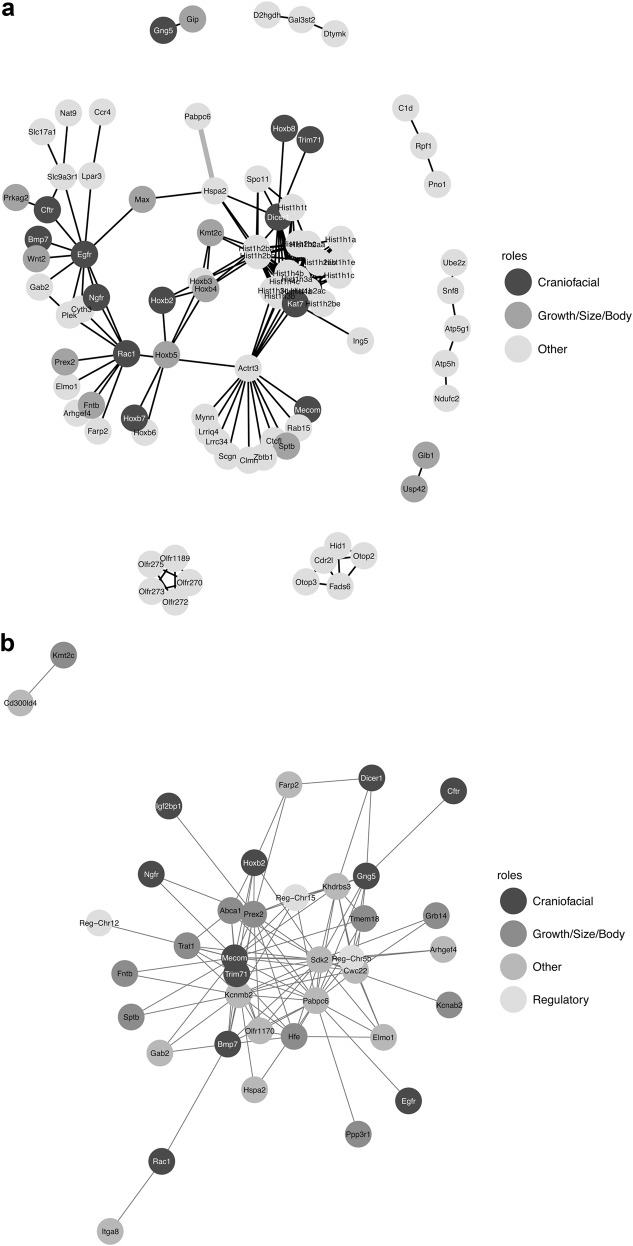
Gene networks
The literature-curated network obtained from SPRINT (Fig. 4a) connected about 44.3% of the 201 protein-coding genes. This gene network had more interactions than expected from a random set of proteins (p < 0.0001) even when the large histone cluster was removed. The network appears to be enriched for embryonic skeletal system morphogenesis and development (GO:0048704 and GO:0048706, fdr = 0.008) and embryonic organ development (GO:0048568, fdr = 0.03). Some of these edges represent significant interactions associated with FA (thicker edges in Fig. 4a). Interestingly, one of them represents the interaction between Pabpc6 and Hspa2: the former is the second most frequent candidate as it appears in 33 of the 162 epistatic interactions, and Hspa2 appears as the central gene in the network. Histone Cluster 1 (corresponds to the Hfe candidate gene in the epistatis network, Fig. 4b) also appears as an important hub in this network. When only the 38 best candidate genes were considered, five edges were common with the epistatic network without the regulatory regions (Figure S4). They link a set of six regions: Bmp7, Pabpc6, Hspa2, Max, Mecom/Actrt3 and Hfe/Histone Cluster1.
Fig. 4.
Gene network proposed to be involved in the development of the mouse skull. a Protein-interaction network based on the list of candidate genes and estimated with the SPRINT database. Edges in grey are between genes located within the same genomic region. b Network based on the 38 best candidate genes identified in the pairwise interactions. Each node represents the most likely candidate gene for each SNP (Table 2) and is connected to other nodes according to the set of significant interactions (Table 1). The node colours relate to the functional classification of the gene (‘Craniofacial’ or ‘Growth/Size/Body’) obtained from the HDMC/MGO databases
Discussion
Our results precluded a major role of single SNPs in FA and confirmed epistasis as the main driver of FA variation. The epistasis test detected significant epistatic associations among markers, as well as specific genomic regions that account for a relatively large amount of the epistatic variance. These results confirm empirically the previous suggestions about the genetic architecture of FA (Burgio et al. 2009; Leamy et al. 2005; Leamy et al. 2015; Leamy et al. 2002). Our population also exhibits some of the previously described features, such as a very low structure of the mapping population (Nicod et al. 2016; Pallares et al. 2015a) and weak patterns of genetic DA (Mikula et al. 2010; Mikula and Macholán 2001).
An epistatic basis for FA variation
A major role for additive effects in FA was unexpected (Leamy et al. 2015). However, the fact that no genomic marker at all has been additively associated to FA may be surprising to some extent. Other genomic processes, such as epistasis, could show linear associations between individual markers and the phenotypic trait under certain circumstances (e.g., when one of the associated genotypes has a big effect overall). On the other hand, the molecular roots of asymmetry are relatively complex (Davies et al. 1996) and a wide range of different developmental processes can originate FA (Scheiner et al. 1991): from subtle ecological fluctuations to serious genetic disorders. Therefore, the heterogeneity of the processes originating FA (i.e., non-linear interactions between genome, environment and development) might confound one or several linear associations when a consistent measure of FA over the whole genome is considered. Indeed, nonlinear developmental processes have been associated with phenotypic robustness, although these have not been measured with FA (Green et al. 2017). The lack of susceptibility of the head development to environmental fluctuations (Beardmore 1960) or a constrained expression towards common axes of variation (Klingenberg 2013) might also be among the possible explanations to the absolute lack of additive effects.
Some studies have suggested that specific proteins may control phenotypic robustness, such as Hsp90 (Takahashi et al. 2010). Heat shock proteins commonly figure in the literature associated with DS and canalisation (Takahashi et al. 2010), but mixed results have tempered the initial enthusiasm for this kind of deterministic association. Recent results led to the idea that heat shock proteins are contributing to, rather than controlling, DS (Debat et al. 2006; Geiler-Samerotte et al. 2016): the current view is that heat shock genes are regular epistatic genes involved in DS, with differing roles and weight depending on the structure. Here we mapped Hspa2 (Hsp70), as one of the best-associated SNPs but also as a very proximal candidate (~200 kb) on chromosome 12, to a 1 Mb region showing most of the significant interaction and also containing an important transcription factor. Hsp70 also appears to be a central node in the protein interaction network built from the literature, which is coherent with the role attributed to Hsp90 in yeast experiments (Geiler-Samerotte et al. 2016).
Epistasis is generally accepted as an important pattern widespread in nature (Moore 2003) and in the evolution of multivariate traits (Jones et al. 2014; Weinreich and Knies 2013). Previous studies have shown that epistatic relationships may hide the genomic variation associated with complex traits (Shorter et al. 2015). Our findings confirmed this idea. The epistatic associations found here constitute a starting point for the study of the genetic elements involved in differing development processes (Carlborg et al. 2006; Cheverud et al. 1983). The identification of specific genes is beyond the scope of our study, even though our population has a low rate of linkage disequilibrium that allows a better resolution (Nicod et al. 2016) than classical QTL mapping (Maga et al. 2015; Navarro and Maga 2016; Pallares et al. 2015a). The preferential choice of protein-coding genes as candidate genes is classic in GWAS, but the generation of facial shape variation is known to integrate heterogeneous genetic pathways (Hallgrímsson et al. 2014), as well as distant-acting enhancers (Attanasio et al. 2013). Therefore regulatory elements may well be behind some of the significant regions, but disentangling them will require much-higher-resolution data.
Skull DS is associated gene networks in craniofacial and skeletal development
Our results did not involve any common SNP or candidate gene suggested by the GWAS of the symmetric component of the skull (Pallares et al. 2015a). This might suggest that mean and variance differences in shape are not associated with the same genomic regions. However, a note of caution is needed here. The original analyses on the symmetric component of shape implemented a suboptimal strategy for a multivariate trait, i.e., mapping one principal component at a time. Such strategy lowers the statistical power (Galesloot et al. 2014) and may lead to a reduced list of associated markers. Indeed, the enrichment for craniofacial regions and embryonic skeletal development in both candidate-gene and literature networks suggests a likely common ground for mean shape and FA.
Our results indicate with reasonable confidence that genomic regions involved in the development of the craniofacial skeleton and in cellular processes, as well as regulatory elements are behind DS. It is also not surprising that many of the significant markers are expressed within the brain, since it is in turn associated with skull shape (Perrine et al. 2017; Richtsmeier et al. 2006). For instance, Dicer1 is required to develop a normal forebrain and its absence causes significant facial abnormalities (Hall 2014). In addition, some of the candidate genes are sometimes associated with brain disorders, e.g., Sdk2 (Otowa et al. 2009), which leaves open the possibility of an association between the expression of that protein, the development of a brain disorder and skull developmental instability (Egan et al. 2017).
This complex network of a relatively small group of genes is congruent with previous suggestions about genomic coadaptation (Clarke 1993) and the description of DS as an emergent property of networks (Barkoulas et al. 2013). Such a developmental network would be composed of a set of protein-coding genes directly involved in the formation of the skull and a set of regulatory genes that can modulate their effects through direct (Nadeau 2001) or indirect (Gjuvsland et al. 2007) pathways. This pattern might be common in nature (Flint and Mackay 2009; Gilchrist and Partridge 2001; Shook and Johnson 1999) although evidence against this hypothesis is also available (Polak and Starmer 2001). Our results are therefore coherent with the general notion about the genotype–phenotype map (Wagner and Zhang 2011), although specific experimental set-ups would be required to confirm our suggestions (Lehner 2011; Mackay 2013).
Large overall effects of some interactive hubs
Some individual markers are highly associated with the general epistatic variance, but they are not common within the most significant pairwise interactions, e.g., the Egfr region. This apparent contradiction is due to the fact that MAPIT does not analyse all the possible pairwise interactions within the genome. However, our results might also suggest the presence of some complexity within the network: interactions might involve more than two genes at a time and different degrees of intensity, especially those where regulatory elements are involved.
Regulatory elements are candidates to be high-interacting nodes. Indeed, the SNP that is best associated with the general epistatic variance, rs33490187, is in an area where no protein-coding gene has been reported. The fact that DS is a consequence of the buffering of environmental fluctuations could suggest an optimal configuration of the network. An optimal configuration would imply a base of DNA and RNA regulatory elements, which are less sensitive to subtle microenvironmental changes (i.e., temperature and pH) than proteins. Indeed, recent studies have acknowledged the importance of these regulatory elements (Fatica and Bozzoni 2014). Our results support the importance of these elements, although evidence against this hypothesis is also pervasive (Hartmann 2009). Here we found that some of the candidate genes we have identified are strongly regulated by miRNAs. For instance, it is well characterised how Mir669k interacts with Dicer1 to define the role of the latter in development (Calabrese et al. 2007). However, this miRNA also has as predicted targets Bbs1, Cdk2 (Blake et al. 2017) and Pabpc6 (targetscan.org) among others. This adds an extra layer of complexity to the interacting network of genes that may be behind DS. Further studies would be needed to study how the interaction of these miRNAs happens with many genes simultaneously, in order to accurately predict the structure of the network.
Conclusions
The non-additive nature of FA proposed here suggests that skull development is a complex interactive network of protein-coding genes and genomic regulators (Hallgrímsson et al. 2014). These epistatic relationships also support the idea of craniofacial DS as an emergent property of this network complexity (Barkoulas et al. 2013). Evolution has shaped morphological variation via pleiotropy and other processes involving trade-offs (e.g., genomic coadaptation) (Klingenberg 2013). The resulting complexity of the genotype–phenotype map can explain the differing role of some important protein-coding genes in DS (Debat et al. 2000; Debat et al. 2009). As in other domains within genetic regulation, a full understanding of these epistastic networks in DS is still far from complete.
Data archiving
This article does not report new empirical data or software.
Electronic supplementary material
Acknowledgements
We thank Luisa Pallares and Abraham Palmer for kindly sharing the data with us. The data collection in this project was supported by a National Institute of Health grant (R01GM097737) received by Abraham A. Palmer from the University of California San Diego. We would like to thank the editor, two anonymous reviewers, and Mihaela Pavlicev for their suggestions and comments on earlier drafts of the manuscript. We thank Catalina Onofrei (Université de Bourgogne) for the language editing. CVG would also like to thank JPM Camacho for his insightful teaching on population genetics and evolution, which surely has contributed to some of the positive achievements of this paper. CVG was supported by a postdoctoral grant of the Conseil Régional Bourgogne (PARI AGREE 2016-9201AAO049S01670). Calculations were performed using HPC resources from PSIUN CCUB (Centre de Calcul de l’Université de Bourgogne).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41437-018-0140-8) contains supplementary material, which is available to authorised users.
References
- Atchley WR, Rutledge JJ. Genetic components of size and shape. I. Dynamics of components of phenotypic variability and covariability during ontogeny in the laboratory rat. Evolution. 1980;34:1161–1173. doi: 10.1111/j.1558-5646.1980.tb04061.x. [DOI] [PubMed] [Google Scholar]
- Attanasio C, Nord AS, Zhu Y, Blow MJ, Li Z, Liberton DK, et al. Fine tuning of craniofacial morphology by distant-acting developmental enhancers. Science. 2013;342:1241006. doi: 10.1126/science.1241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkoulas M, van Zon JS, Milloz J, van Oudenaarden A, Félix MA. Robustness and epistasis in the C. elegans vulval signaling network revealed by pathway dosage modulation. Dev Cell. 2013;24:64–75. doi: 10.1016/j.devcel.2012.12.001. [DOI] [PubMed] [Google Scholar]
- Beardmore JA. Developmental stability in constant and fluctuating temperatures. Heredity. 1960;14:411–422. [Google Scholar]
- Blake JA, Eppig JT, Kadin JA, Richardson JE, Smith CL, Bult CJ, et al. Mouse Genome Database (MGI)-2017: community knowledge resource for the laboratory mouse. Nucleic Acids Res. 2017;4:D723–D729. doi: 10.1093/nar/gkw1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock MT, Bowman AW. On the measurement and analysis of asymmetry with applications to facial modelling. J R Stat Soc Ser C. 2006;55:77–91. [Google Scholar]
- Breuker CJ, Patterson JS, Klingenberg CP. A single basis for developmental buffering of Drosophila wing shape. PLoS ONE. 2006;1:e7. doi: 10.1371/journal.pone.0000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgio G, Baylac M, Heyer E, Montagutelli X. Genetic analysis of skull shape variation and morphological integration in the mouse using interspecific recombinant congenic strains between C57BL/6 and mice of the Mus spretus species. Evolution. 2009;63:2668–2686. doi: 10.1111/j.1558-5646.2009.00737.x. [DOI] [PubMed] [Google Scholar]
- Calabrese JM, Seila AC, Yeo GW, Sharp PA. RNA sequence analysis defines Dicer’s role in mouse embryonic stem cells. Proc Natl Acad Sci. 2007;104:18097–18102. doi: 10.1073/pnas.0709193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlborg Ouml, Jacobsson L, Åhgren P, Siegel P, Andersson L. Epistasis and the release of genetic variation during long-term selection. Nat Genet. 2006;38:418–420. doi: 10.1038/ng1761. [DOI] [PubMed] [Google Scholar]
- Cheng R, Parker CC, Abney M, Palmer AA. Practical considerations regarding the use of genotype and pedigree data to model relatedness in the context of genome-wide association studies. G3: Genes, Genomes, Genet. 2013;3:1861–1867. doi: 10.1534/g3.113.007948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud JM, Rutledge JJ, Atchley WR. Quantitative genetics of development: genetic correlations among age-specific trait values and the evolution of ontogeny. Evolution. 1983;37:895–905. doi: 10.1111/j.1558-5646.1983.tb05619.x. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke GM. The genetic basis of developmental stability. I. Relationships between stability, heterozygosity and genomic coadaptation. Genetica. 1993;89:15–23. [Google Scholar]
- Coutelis JB, González-Morales N, Géminard C, Noselli S. Diversity and convergence in the mechanisms establishing L/R asymmetry in metazoa. EMBO Rep. 2014;15:926–937. doi: 10.15252/embr.201438972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford L, Zeng P, Mukherjee S, Zhou X. Detecting epistasis with the marginal epistasis test in genetic mapping studies of quantitative traits. PLoS Genet. 2017;13:e1006869. doi: 10.1371/journal.pgen.1006869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AG, Game AY, Chen Z, Williams TJ, Goodall S, Yen JL, et al. Scalloped wings is the Lucilia cuprina Notch homologue and a candidate for the Modifier of fitness and asymmetry of diazinon resistance. Genetics. 1996;143:1321–1337. doi: 10.1093/genetics/143.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coster G, Van Dongen S, Malaki P, Muchane M, Alcántara-Exposito A, Matheve H, et al. Fluctuating asymmetry and environmental stress: understanding the role of trait history. PLoS ONE. 2013;8:e57966. doi: 10.1371/journal.pone.0057966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Visser JAGM, Hermisson J, Wagner GP, Meyers LA, Bagheri-Chaichian H, Blanchard JL, et al. Evolution and detection of genetic robustness. Evolution. 2003;57:1959–1972. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- Debat V, Alibert P, David P, Paradis E, Auffray JC. Independence between developmental stability and canalization in the skull of the house mouse. Proc R Soc London B. 2000;267:423–430. doi: 10.1098/rspb.2000.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debat V, Debelle A, Dworkin I. Plasticity, canalization, and developmental stability of the Drosophila wing: Joint effects of mutations and developmental temperature. Evolution. 2009;63:2864–2876. doi: 10.1111/j.1558-5646.2009.00774.x. [DOI] [PubMed] [Google Scholar]
- Debat V, Milton CC, Rutherford S, Klingenberg CP, Hoffmann AA. Hsp90 and the quantitative variation of wing shape in Drosophila melanogaster. Evolution. 2006;60:2529–2538. [PubMed] [Google Scholar]
- Dryden IL, Mardia KV. Statistical shape analysis. New York: John Wiley & Sons; 1998. [Google Scholar]
- Egan PJ, Mitrovics TC, Tomandl BF. Alternate ventricular asymmetry could suggest a psychiatric diagnosis. Clin Anat. 2017;30:50–52. doi: 10.1002/ca.22789. [DOI] [PubMed] [Google Scholar]
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- Fernández Iriarte P, Céspedes W, Santos M. Quantitative-genetic analysis of wing form and bilateral asymmetry in isochromosomal lines of Drosophila suboscura using Procrustes methods. J Genet. 2003;82:95–113. doi: 10.1007/BF02715812. [DOI] [PubMed] [Google Scholar]
- Flint J, Eskin E. Genome-wide association studies in mice. Nat Rev Genet. 2012;13:807–817. doi: 10.1038/nrg3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Mackay TFC. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 2009;19:723–733. doi: 10.1101/gr.086660.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galesloot TE, van Steen K, Kiemeney LALM, Janss LL, Vermeulen SH. A Comparison of Multivariate Genome-Wide Association Methods. PLOS ONE. 2014;9:e95923. doi: 10.1371/journal.pone.0095923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiler-Samerotte KA, Zhu YO, Goulet BE, Hall DW, Siegal ML. Selection transforms the landscape of genetic variation interacting with Hsp90. PLoS Biol. 2016;14:e2000465. doi: 10.1371/journal.pbio.2000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist AS, Partridge L. The contrasting genetic architecture of wing size and shape in Drosophila melanogaster. Heredity. 2001;86:144–152. doi: 10.1046/j.1365-2540.2001.00779.x. [DOI] [PubMed] [Google Scholar]
- Gjuvsland AB, Hayes BJ, Omholt SW. Statistical epistasis is a generic feature of gene regulatory networks. Genetics. 2007;175:411–420. doi: 10.1534/genetics.106.058859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez PN, Pavlicev M, Mitteroecker P, Pardo-Manuel de Villena F, Spritz RA, Marcucio RS, et al. Genetic structure of phenotypic robustness in the collaborative cross mouse diallel panel. J Evol Biol. 2016;29:1737–1751. doi: 10.1111/jeb.12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodall C. Procrustes methods in the statistical analysis of shape. J R Stat Soc Ser B. 1991;53:285–339. [Google Scholar]
- Green RM, Fish JL, Young NM, Smith FJ, Roberts B, Dolan K, et al. Developmental nonlinearity drives phenotypic robustness. Nat Commun. 2017;8:1970. doi: 10.1038/s41467-017-02037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadalupe T, Zwiers MP, Teumer A, Wittfeld K, Arias Vasquez A, Hoogman M, et al. Measurement and genetics of human subcortical and hippocampal asymmetries in large datasets. Hum Brain Mapp. 2014;35:3277–3289. doi: 10.1002/hbm.22401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BK. Summarizing craniofacial genetics and developmental biology (SCGDB) Am J Med Genet A. 2014;164A:884–891. doi: 10.1002/ajmg.a.35288. [DOI] [PubMed] [Google Scholar]
- Hallgrímsson B, Mio W, Marcucio RS, Spritz R. Let’s face it - Complex traits are just not that simple. PLoS Genet. 2014;10:e1004724. doi: 10.1371/journal.pgen.1004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardisty-Hughes RE, Parker A, Brown SDM. A hearing and vestibular phenotyping pipeline to identify mouse mutants with hearing impairment. Nat Protoc. 2010;5:177–190. doi: 10.1038/nprot.2009.204. [DOI] [PubMed] [Google Scholar]
- Hartmann C. Transcriptional networks controlling skeletal development. Curr Opin Genet Dev. 2009;19:437–443. doi: 10.1016/j.gde.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Jones AG, Bürger R, Arnold SJ. Epistasis and natural selection shape the mutational architecture of complex traits. Nat Commun. 2014;5:3709. doi: 10.1038/ncomms4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingenberg CP. MorphoJ: an integrated software package for geometric morphometrics. Mol Ecol Resour. 2011;11:353–357. doi: 10.1111/j.1755-0998.2010.02924.x. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP. Cranial integration and modularity: insights into evolution and development from morphometric data. Hystrix. 2013;24:43–58. [Google Scholar]
- Klingenberg CP. Analyzing fluctuating asymmetry with geometric morphometrics: concepts, methods, and applications. Symmetry. 2015;7:843–934. [Google Scholar]
- Klingenberg CP, Barluenga M, Meyer A. Shape analysis of symmetric structures: quantifying variation among individuals ans asymmetry. Evolution. 2002;56:1909–1920. doi: 10.1111/j.0014-3820.2002.tb00117.x. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP, McIntyre GS. Geometric morphometrics of developmental instability: analyzing patterns of fluctuating asymmetry with procrustes methods. Evolution. 1998;52:1363–1375. doi: 10.1111/j.1558-5646.1998.tb02018.x. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP, Monteiro LR. Distances and directions in multidimensional shape spaces: implications for morphometric applications. Syst Biol. 2005;54:678–688. doi: 10.1080/10635150590947258. [DOI] [PubMed] [Google Scholar]
- Leamy L, Pomp D, Eisen EJ, Cheverud JM. Quantitative trait loci for directional but not fluctuating asymmetry of mandible characters in mice. Genet Res. 2000;76:27–40. doi: 10.1017/s0016672300004559. [DOI] [PubMed] [Google Scholar]
- Leamy L, Workman MS, Routman EJ, Cheverud JM. An epistatic genetic basis for fluctuating asymmetry of tooth size and shape in mice. Heredity. 2005;94:316–325. doi: 10.1038/sj.hdy.6800637. [DOI] [PubMed] [Google Scholar]
- Leamy LJ. Morphological integration of fluctuating asymmetry in the mouse mandible. Genetica. 1993;89:139–153. [Google Scholar]
- Leamy LJ, Klingenberg CP. The genetics and evolution of fluctuating asymmetry. Annu Rev Ecol, Evol Syst. 2005;36:1–21. [Google Scholar]
- Leamy LJ, Klingenberg CP, Sherratt E, Wolf JB, Cheverud JM. The genetic architecture of fluctuating asymmetry of mandible size and shape in a population of mice: another look. Symmetry. 2015;7:146–163. [Google Scholar]
- Leamy LJ, Meagher S, Taylor S, Carroll L, Potts WK. Size and fluctuating asymmetry of morphometric characters in mice: their associations with inbreeding and t-haplotype. Evolution. 2001;55:2333–2341. doi: 10.1111/j.0014-3820.2001.tb00746.x. [DOI] [PubMed] [Google Scholar]
- Leamy LJ, Routman EJ, Cheverud JM. An epistatic genetic basis for fluctuating asymmetry of mandible size in mice. Evolution. 2002;56:642–653. doi: 10.1111/j.0014-3820.2002.tb01373.x. [DOI] [PubMed] [Google Scholar]
- Lehner B. Molecular mechanisms of epistasis within and between genes. Trends Genet. 2011;27:323–331. doi: 10.1016/j.tig.2011.05.007. [DOI] [PubMed] [Google Scholar]
- Levin M. Left-right asymmetry in embryonic development: a comprehensive review. Mech Dev. 2005;122:3–25. doi: 10.1016/j.mod.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Listgarden J, Lippert C, Kadie CM, Davidson RI, Eskin E, Heckerman D. Improved linear mixed models for genome-wide association studies. Nat Methods. 2012;9:525–526. doi: 10.1038/nmeth.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC. Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat Rev Genet. 2013;15:22–33. doi: 10.1038/nrg3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga AM, Navarro N, Cunningham ML, Cox TC. Quantitative trait loci affecting the 3D skull shape and size in mouse and prioritization of candidate genes in-silico. Front Physiol. 2015;6:92. doi: 10.3389/fphys.2015.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather K. Genetical control of stability in development. Heredity. 1953;7:297–336. [Google Scholar]
- Mikula O, Auffray JC, Macholán M. Asymmetric size and shape variation in the Central European transect across the house mouse hybrid zone. Biol J Linn Soc. 2010;101:13–27. [Google Scholar]
- Mikula O, Macholán M. There is no heterotic effect upon developmental stability in the ventral side of the skull within the house mouse hybrid zone. J Evol Biol. 2001;21:1055–1067. doi: 10.1111/j.1420-9101.2008.01539.x. [DOI] [PubMed] [Google Scholar]
- Miller SF, Weinberg SM, Nidey NL, Defay DK, Marazita ML, Wehby GL, et al. Exploratory genotype-phenotype correlations of facial form and asymmetry in unaffected relatives of children with non-syndromic cleft lip and/or palate. J Anat. 2014;224:688–709. doi: 10.1111/joa.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JH. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum Hered. 2003;56:73–82. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- Nadeau JH. Modifier genes in mice and humans. Nat Rev Genet. 2001;2:165–174. doi: 10.1038/35056009. [DOI] [PubMed] [Google Scholar]
- Navarro N, Maga AM. Does 3D phenotyping yield substantial insights in the genetics of the mouse mandible shape? G3: Genes Genomes Genet. 2016;6:1153–1163. doi: 10.1534/g3.115.024372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicod J, Davies RW, Cai N, Hassett C, Goodstadt L, Cosgrove C, et al. Genome-wide association of multiple complex traits in outbred mice by ultra-low-coverage sequencing. Nat Genet. 2016;48:912–918. doi: 10.1038/ng.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otowa T, Yoshida E, Sugaya N, Yasuda S, Nishimura Y, Inoue K, et al. Genome-wide association study of panic disorder in the Japanese population. J Hum Genet. 2009;54:122–126. doi: 10.1038/jhg.2008.17. [DOI] [PubMed] [Google Scholar]
- Pallares LF, Carbonetto P, Gopalakrishnan S, Parker CC, Ackert-Bicknell CL, Palmer AA, et al. Mapping of craniofacial traits in outbred mice identifies major developmental genes involved in shape determination. PLoS Genet. 2015;11:e1005607. doi: 10.1371/journal.pgen.1005607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallares LF, Carbonetto P, Gopalakrishnan S, Parker CC, Ackert-Bicknell CL, Palmer AA et al. (2015b). Dryad Digital Repository. [DOI] [PMC free article] [PubMed]
- Palmer AR, Strobeck C. Fluctuating asymmetry: measurement, analysis, patterns. Annu Rev Ecol Syst. 1986;17:391–421. [Google Scholar]
- Parker CC, Carbonetto P, Sokoloff DD, Park YJ, Abney M, Palmer AA. High-resolution genetic mapping of complex traits from a combined analysis of F2 and advanced intercross mice. Genetics. 2014;198:103–116. doi: 10.1534/genetics.114.167056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CC, Gopalakrishnan S, Carbonetto P, Gonzales NM, Leung E, Park YJ, et al. Genome-wide association study of behavioral, physiological and gene expression traits in outbred CFW mice. Nat Genet. 2016;48:919–926. doi: 10.1038/ng.3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrine SMM, Stecko T, Neuberger T, Jabs EW, Ryan TM, Richtsmeier JT. Integration of brain and skull in prenatal mouse models of apert and Crouzon syndromes. Front Human Neurosci. 2017;11:369. doi: 10.3389/fnhum.2017.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polak M, Starmer WT. The quantitative genetics of fluctuating asymmetry. Evolution. 2001;55:498–511. doi: 10.1554/0014-3820(2001)055[0498:tqgofa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Queitsch C, Carlson KD. Lessons from model organisms: phenotypic robustness and missing heritability in complex disease. PLoS Genet. 2012;8:e1003041. doi: 10.1371/journal.pgen.1003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2016). R: A language and environment for statistical computing.R Foundation for Statistical Computing, Vienna, Austria.
- Reeve ECR. Some genetic tests on asymmetry of sternopleural chaeta number in Drosophila. Genet Res. 1960;1:151–172. [Google Scholar]
- Rice SH. The evolution of canalization and the breaking of Von Baer’s laws: modeling the evolution of development with epistasis. Evolution. 1998;52:647–656. doi: 10.1111/j.1558-5646.1998.tb03690.x. [DOI] [PubMed] [Google Scholar]
- Richtsmeier JT, Aldridge K, DeLeon VB, Panchal J, Kane AA, Marsh JL, et al. Phenotypic integration of neurocranium and brain. J Exp Zool Part B. 2006;306:360–378. doi: 10.1002/jez.b.21092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richtsmeier JT, Cole IIITM, Lele SR. An invariant approach to the study of fluctuating asymmetry: Developmental instability in a mouse model for Down syndrome. In: Slice DE, editor. Modern morphometrics in physical anthropology. New York: Kluwer Academic / Plenum Publishers; 2005. pp. 187–212. [Google Scholar]
- Scheiner SM, Caplan RL, Lyman RF. The genetics of phenotypic plasticity. III. Genetic correlations and fluctuating asymmetries. J Evol Biol. 1991;4:51–68. [Google Scholar]
- Shook DR, Johnson TE. Quantitative trait loci affecting survival and fertility-related traits in Caenorhabditis elegans show genotype-environment interactions, pleiotropy and epistasis. Genetics. 1999;153:1233–1243. doi: 10.1093/genetics/153.3.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J, Couch C, Huang W, Carbone MA, Peiffer J, Anholt RRH, et al. Genetic architecture of natural variation in Drosophila melanogaster aggressive behavior. Proc Natl Acad Sci USA. 2015;112:E3555–E356o3. doi: 10.1073/pnas.1510104112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegal ML, Leu JY. On the nature and evolutionary impact of phenotypic robustness mechanisms. Annu Rev Ecol, Evol Syst. 2014;45:495–517. doi: 10.1146/annurev-ecolsys-120213-091705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed D, Balding DJ. Relatedness in the post-genomic era: is it still useful? Nat Rev Genet. 2015;16:33–44. doi: 10.1038/nrg3821. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, von Mering C. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Research. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadayon SH, Vaziri-Pashkam M, Kahali P, Ansari Dezfouli M, Abbassian A. Common genetic variant in VIT is associated with human brain asymmetry. Front Hum Neurosci. 2016;10:236. doi: 10.3389/fnhum.2016.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi KH, Rako L, Takano-Shimizu T, Hoffmann AA, Lee SF. Effect of small Hsp genes on developmental stability and microenvironmental canalization. BMC Evol Biol. 2010;10:284. doi: 10.1186/1471-2148-10-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoday JM. Homeostasis in a selection experiment. Heredity. 1958;12:401–415. [Google Scholar]
- Van Dongen S. Fluctuating asymmetry and developmental instability in evolutionary biology: past, present and future. J Evol Biol. 2006;19:1727–1743. doi: 10.1111/j.1420-9101.2006.01175.x. [DOI] [PubMed] [Google Scholar]
- Van Valen L. A study of fluctuating asymmetry. Evolution. 1962;16:125–142. [Google Scholar]
- Vanderberg L, Levin M. A unified model for left-right asymmetry? Comparison and synthesis of molecular models of embryonic laterality. Dev Biol. 2013;379:1–15. doi: 10.1016/j.ydbio.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, et al. 10 years of GWAS discovery: Biology, function and translation. Am J Hum Genet. 2017;101:5–22. doi: 10.1016/j.ajhg.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. The strategy of the genes. London, Great Britain: George Allen & Unwin LTD; 1957. [Google Scholar]
- Wagner GP, Zhang J. The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat Rev Genet. 2011;12:204. doi: 10.1038/nrg2949. [DOI] [PubMed] [Google Scholar]
- Weinreich DM, Knies JL. Fisher’s geometric model of adaptation meets the functional synthesis: Data on pairwise epistasis for fitness yields insights into the shape and size of phenotype space. Evolution. 2013;67:2957–2972. doi: 10.1111/evo.12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, et al. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–D716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article does not report new empirical data or software.