Dear Editor,
Histone marks deposited by post-translational modifications (PTMs) frequently occur in interrelated combinational patterns to create a complex and precise control on the chromatin structure and function.1 One of the landmark findings of histone PTM crosstalk is the trans-histone regulation of histone H3 lysine 79 (H3K79) methylation by the monoubiquitination of histone H2B on lysine 120 (H2BK120).2 Mono-, di-, and tri-methylation of histone H3K79 serves as a prominent histone mark that participates in transcription regulation and DNA damage response.3 H2BK120 monoubiquitination (H2BK120ub1) is a prerequisite for the efficient methylation of H3K79 by the unique non-SET domain-containing histone methyltransferase DOT1L (Disrupter of telomere silencing protein 1-like) in vivo.2 Incorporation of chemically monoubiquitinated H2B into in vitro reconstituted nucleosome directly stimulates the catalytic activity of DOT1L4 (Supplementary information, Figs. S1 and S2). It still remains poorly understood how the H3K79 methyl marks are deposited and how the associated PTM crosstalk occurs on nucleosome.
Here we present the cryo-electron microscopy (cryo-EM) structure of the catalytic domain of human DOT1L (residues 1–416) in complex with an H2BK120ub1 nucleosome core particle (abbreviated as ubNCP) at an overall resolution of 4.1 Å (Supplementary information, Figs. S3-S5 and Table S1). The structure of DOT1L-ubNCP reveals that DOT1L extensively interacts with core histones on the disk-face of nucleosome (buried surface area, ~2020 Å2), with its C-terminal region (residues 269–331) sandwiched between ubiquitin and the histone H2A-H2B dimer (Fig. 1a). The direct association of DOT1L with the H2BK120-conjugated ubiquitin extends the recognition interface between DOT1L and histone surface, and probably increases the binding affinity of DOT1L toward the H2BK120ub1 nucleosome. We used a DOT1L construct (residues 1–351), which lacks a positively charged region that binds the nucleosomal DNA, to compare its binding affinity to the histone surface of NCP and ubNCP. GST-pull-down assay clearly showed that DOT1L1–351 only interacts with the ubiquitinated nucleosome, indicating that H2BK120ub1 dramatically increases the weak association of DOT1L with the histone surface of nucleosome (Supplementary information, Fig. S6b, lanes 7–9). The increased binding affinity toward the H2BK120ub1 nucleosome results in the enhanced catalytic efficiency of DOT1L and the accumulation of higher levels of H3K79 di- and tri-methylation (Supplementary information, Fig. S2).
Fig. 1.
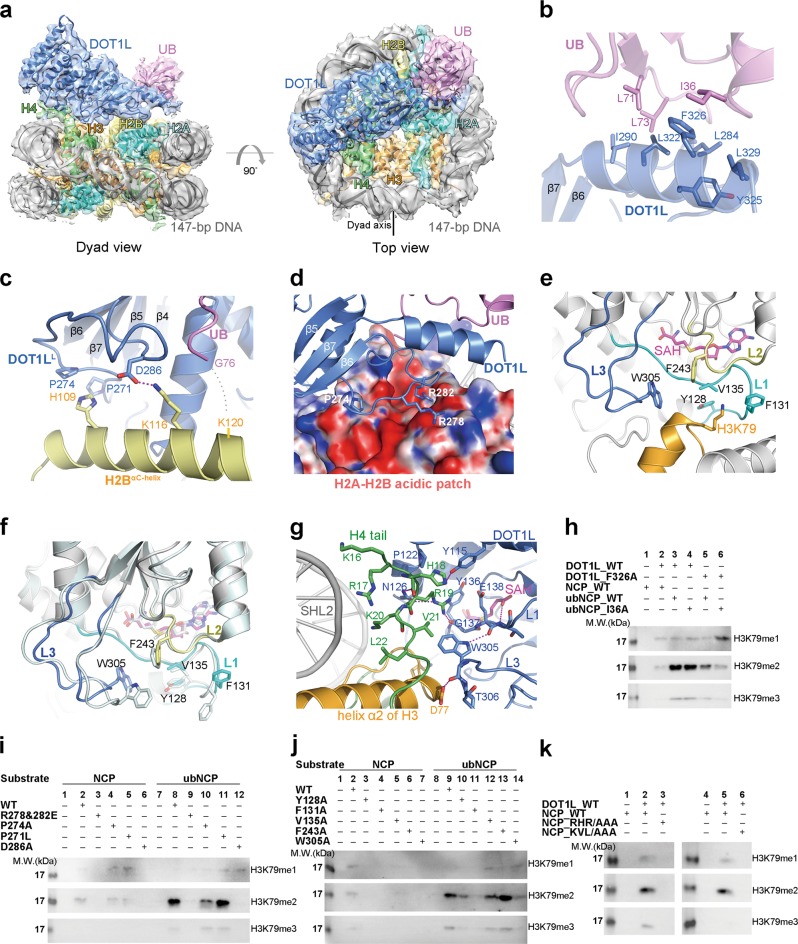
Structural characterization of the recognition of the H2BK120ub1 nucleosome by DOT1L. a Overall structure of the DOT1L-ubNCP complex shown from two orthogonal views. The cryo-EM map was segmented and colored according to the respective components of the DOT1L-ubNCP complex. b Detailed view of the DOT1L-ubiquitin interface, the related EM density map is shown in Supplementary information, Fig. S8b. c The recognition interface between DOT1LL and the ubiquitin-conjugated C-terminal helix of histone H2B, the related EM density map is shown in Supplementary information, Fig. S8c. d DOT1L Arg278 and Arg282 interact with the acidic patch of histone H2A-H2B (shown with electrostatic surface potential), the related EM density map is shown in Supplementary information, Fig. S8e. e Detailed view of the interface between the active site of DOT1L and the nucleosomal H3K79, the related EM density map is shown in Supplementary information, Fig. S9a-e. f Comparison of the crystal structure of DOT1L alone (PDB code: 1NW3, colored in pale cyan) and the cryo-EM structure of DOT1L in complex with ubNCP (colored as in e) reveals conformational changes of the active site of DOT1L upon binding to nucleosome. g Detailed interactions between the histone H4 tail and the active site of DOT1L. The related EM density map is shown in Supplementary information, Fig. S9f. The hydrogen-bonds are shown as magenta dash lines. h Mutational studies on the function of the interacting Phe326DOT1L-Ile36Ub residues in the H3K79 methylation of ubNCP through the HMT assay. Input of the HMT reactions is shown in Supplementary information, Fig. S8f. i HMT assays carried out with DOT1L mutants that disrupt the interactions between DOT1L and the histone H2A-H2B dimer. Input of the HMT reactions is shown in Supplementary information, Fig. S8h. j Mutations of the DOT1L residues that make direct contact with the adjacent region of H3K79 abrogate the HMT activity of DOT1L to varying degrees. Input of the HMT reactions is shown in Supplementary information, Fig. S9g. k DOT1L fails to methylate the mutant nucleosomes that have alanine substitutions of the DOT1L-interacting H4 residues. Input of the HMT reactions is shown in Supplementary information, Fig. S9h
We also determined the cryo-EM structure of DOT1L in complex with an unmodified NCP at an overall resolution of 5.0 Å (Supplementary information, Fig. S7 and Table S1). The density of DOT1L in this 3D reconstruction is not clear enough to build an atomic model. However, docking of the DOT1L-ubNCP complex structure into this map shows a good fit of the DOT1L-ubNCP structure with the DOT1L-NCP density map, indicating that monoubiquitination of H2BK120 does not change the overall location of DOT1L on the histone surface of nucleosome (Supplementary information, Fig. S7g and h). EM density maps of NCP from the two structures reach to a similar range of resolution (3.5–4.5 Å), whereas the density of DOT1L from the unmodified complex is severely dampened and blurred (Supplementary information, Figs. S3 and S7). This observation suggested that the interaction of DOT1L with the histone surface might be very dynamic, whereas ubiquitin attachment to H2BK120 could rigidly constrain the mobility of DOT1L and lead to an unambiguous 3D reconstruction of the DOT1L-ubNCP complex.
Ubiquitin is a highly conserved 76-amino-acid protein with several well-characterized surface patches, such as the I44 patch (residues Leu8, Ile44, His68, and Val70) and the I36 patch (residues Ile36, Leu71, and Leu73), which are frequently recognized by ubiquitin-binding proteins (Supplementary information, Fig. S8a).5 The DOT1L-ubNCP complex structure indicates that the I36 patch is in close proximity to a hydrophobic surface composed of residues Leu284, Ile290, Leu322, Tyr325, Phe326, and Leu329 within the C-terminal region (residues 269–331) of DOT1L (Fig. 1b). Residue Ile36 of ubiquitin stacks on DOT1L Phe326, which serves as an important recognition interface between ubiquitin and DOT1L (Fig. 1b). Substitution of Phe326DOT1L with alanine impaired the stimulatory effect of H2BK120ub1 on DOT1L activity (Fig. 1h, lane 5). By contrast, it had no impact on the catalysis of DOT1L toward the unmodified nucleosome (Supplementary information, Fig. S8g, lane 5). Double mutations of F326ADOT1L and I36AUB resulted in a further decrease in the activity of DOT1L toward the mutant ubNCP (Fig. 1h, lane 6). Previously, an alanine scan of the ubiquitin surface revealed that H2BK120ub1 with alanine substitutions of the ubiquitin residues Leu71 and Leu73 failed to stimulate the DOT1L activity.6 This is consistent with our structure that the I36 patch of ubiquitin participates in the recognition of DOT1L.
In addition to binding to ubiquitin, the C-terminal region of DOT1L also interacts with the histone H2A-H2B dimer (Fig. 1a). A long loop (DOT1LL, residues 269–290) that connects two parallel strands (β5 and β6) of the central β sheet of DOT1L confers specific recognition of the H2A-H2B surface (Fig. 1c, d). DOT1LL sits on the C-terminal helix (αC) of histone H2B and is juxtaposed with the C-terminus of ubiquitin on the αC helix (Fig. 1c). At the DOT1LL-H2BαC interface, DOT1L Pro274 constrains the conformation of DOT1LL around residue His109 of H2BαC, and DOT1L Asp286 forms a salt bridge with residue Lys116 of H2BαC (Fig. 1c). Mutation of Pro274 or Asp286 severely impaired the HMT (histone methyltransferase) activity of DOT1L toward NCP and ubNCP (Fig. 1i). P271L, a melanoma-associated mutation of DOT1L,7 is located near to the Pro274DOT1L-His109H2B interface (Fig. 1c). It decreased the activity of DOT1L on NCP, but had no distinguishable defect in the methylation of ubNCP (Fig. 1i). Therefore, the P271L mutation might disturb the Pro274DOT1L-His109H2B interaction and lead to the moderate defect in DOT1L activity.
The H2A-H2B acidic patch is a histone surface that is frequently involved in the recognition of nucleosome-binding partners.8 The DOT1L-ubNCP structure revealed that Arg278 and Arg282 of DOT1LL contact the acidic patch, and Arg282DOT1L forms multiple salt bridges and hydrogen bonds with Glu56H2A and Gln44H2B of the acidic patch (Fig. 1d; Supplementary information, Fig. S8d). Glutamate substitutions of Arg278 and Arg282 completely abolished the methyltransferase activity of DOT1L on NCP and ubNCP (Fig. 1i). Collectively, DOT1LL binds to both the αC helix of H2B and the acidic patch of H2A-H2B, forming an essential recognition interface between DOT1L and nucleosome.
The active site of DOT1L, consisting of an S-adenosyl-L-methionine (SAM) binding pocket and a lysine binding channel,9 is precisely positioned above residue lysine 79 of histone H3 in the cryo-EM structure of the DOT1L-ubNCP complex (Fig. 1e). Three loops of DOT1L (L1: residues 127–140, L2: residues 241–246, and L3: residues 296–312) form the lysine binding channel that connects the side chain of H3K79 to the methyl group of SAM (Fig. 1e). Within the loops, several aromatic and hydrophobic residues (Tyr128, Phe131, Val135, Phe243, and Trp305) surround the entrance of the channel and make direct contact with the adjacent region of H3K79 (Fig. 1e). Structural comparison of DOT1L alone and DOT1L in complex with ubNCP revealed that substrate-binding induces the conformational change of loop L3 at the active site of DOT1L and leads to the rearrangement of Trp305 at the catalytic interface (Fig. 1f). Phe131DOT1L also adopts altered conformation when associated with nucleosome (Fig. 1f). Mutagenesis analysis revealed that mutations of either Tyr128, Phe131, or Trp305 nearly completely abrogated the catalytic activity of DOT1L, suggesting that these residues play a pivotal role in the catalysis of histone H3K79 methylation (Fig. 1j). Alanine substitutions of DOT1L Val135 resulted in a moderate decrease of the H3K79 methylation, and an F243A mutation only showed a defect in the methylation of NCP but not ubNCP (Fig. 1j). In the lysine-binding channel of DOT1L, a weak density that possibly corresponds to the side chain of H3K79 could be observed in the EM density map of the DOT1L-ubNCP complex (Supplementary information, Fig. S9e).
Previous studies indicate that a short basic patch (residues 16–20) of the histone H4 tail interacts with yeast Dot1 and is required for the methylation of nucleosomal histone H3K79.10 The cryo-EM structure of the DOT1L-ubNCP complex reveals how the histone H4 tail regulates H3K79 methylation at the structural level. Within the complex, the H4 tail sits on helix α2 of histone H3 and extends to the N-terminal region of DOT1L as well as the superhelix location 2 (SHL2) of the nucleosomal DNA (Fig. 1a, g). H4 tail becomes folded through residue K16 due to the associations with DOT1L (Fig. 1g). Residues His18, Arg19, and Val21 of the H4 tail point to the active site of DOT1L and mediate extensive electrostatic and hydrophobic interactions with multiple residues at the active site of DOT1L (Fig. 1g). This interaction network stabilizes the conformations of loops L1 and L3 at the catalytic site. NCP mutants with alanine substitutions of H4 residues 17RHR19 or 20KVL22 failed to be methylated by DOT1L, underscoring the importance of these residues in the methylation of H3K79 (Fig. 1k).
In summary, our structural and biochemical studies provide mechanistic insights into the catalytic specificity of DOT1L toward the nucleosomal H3K79 and the activity regulation of DOT1L by the H2BK120 monoubiquitination (Supplementary information, Fig. S10). Our findings shed light on the functional mechanism of histone monoubiquitination in histone PTM crosstalk and are insightful for understanding other histone modifications regulated by monoubiquitination. DOT1L is associated with mixed lineage leukemia and its inhibitors that target the SAM-binding pocket have been in the clinical trials of treating acute leukemia. Structural elucidation of the DOT1L-nucleosome interface provides possible strategies for developing novel targeted therapies against DOT1L.
Supplementary information
Acknowledgements
We thank staffs from the Electron Microscopy System and the Database and Computation System of the National Facility for Protein Science in Shanghai (NFPS), Zhangjiang Lab for their assistance with the EM instruments and data pre-processing. We are grateful to the NFPS Large-scale Protein Preparation System and Mass Spectrometry System for instrument support and technical assistance. This work was supported by the National Key R&D Program of China (2016YFA0501803 and 2017YFA0504504), the National Natural Science Foundation of China (31570766 and U1632130), the Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (2017YZ004), the SHIPM-sigma fund (BJ1–7009–18–1303 and JY201805) from Shanghai Institute of Precision Medicine, Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, and Chinese Academy of Sciences Facility-based Open Research Program. J.H. is a recipient of the Thousand Young Talents Program of China and a recipient of the Hundred Talents Program of Shanghai Jiao Tong University School of Medicine.
Author contributions
T.Y. performed sample preparation, EM data collection and biochemical analyses; W.J. and Z.H. helped with protein purifications; G.Y. helped with nucleosome preparation; M.T., M.C., Q.W., and Y.L. helped with EM data analyses; M.L. and J.H. initiated the project; J.H. designed and supervised all the research, and wrote the manuscript with help from all other authors.
Competing interests
The authors declare no competing interests.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41422-019-0146-7.
References
- 1.Kouzarides T. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Briggs SD, et al. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen AT, Zhang Y. Genes Dev. 2011;3:1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajalingam K, Dikic I. Cell. 2016;164:1074. doi: 10.1016/j.cell.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 6.Holt MT, et al. Proc. Natl Acad. Sci. USA. 2015;112:10365–10370. doi: 10.1073/pnas.1504483112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu B, et al. Nat. Commun. 2018;9:1–13. doi: 10.1038/s41467-017-02088-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson MD, et al. Nature. 2016;536:100–103. doi: 10.1038/nature18951. [DOI] [PubMed] [Google Scholar]
- 9.Min J, et al. Cell. 2003;112:711–723. doi: 10.1016/S0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 10.Altaf M, et al. Mol. Cell. 2007;28:1002–1014. doi: 10.1016/j.molcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.