Abstract
Magnesium lithospermate B (MLB) is a new drug marketed in China to treat angina, but its low oral bioavailability limits its clinical application to the intravenous route. Paradoxically, orally administered low-dose MLB was found to alleviate kidney injury in diabetic nephropathy (DN) rats, but its mechanism of action remains unknown. In recent years, the kidney-gut axis has been suspected to be involved in kidney damage pathogenesis, potentially representing a non-classical pathway for pharmacologic intervention. To ascertain whether MLB targets the kidney-gut axis, streptozotocin (STZ)-treated mice were prepared as a mouse model of DN. The STZ mice were treated with MLB (50 mg kg−1 d−1, p.o.) for 8 weeks. Twenty-four-hour urinary albumin was detected to mirror kidney function. At week 4, 6, 8, feces were collected; bile acids (BAs) were quantified to examine the alterations in the BA metabolic profiles, and bacterial 16S rRNA gene fragments were sequenced to identify alterations in gut microbial composition. In STZ mice, 24-h urinary albumin levels and total fecal BAs, especially cholic acids (CAs) and deoxycholic acids (DCAs) were greatly increased, and the gut microbiome was dramatically shifted compared with control mice. Oral administration of MLB significantly decreased 24-h urinary albumin levels and total BAs, CAs and DCAs, and reversed CA:TCA (taurocholic acid) and DCA:CA ratios. It also changed the microbiome composition in STZ mice based on operational units. Thus the therapeutic effect of MLB on kidney injury might be attributed (at least partially) to its ability to modulate the disordered gut microbiome and BA metabolism.
Keywords: magnesium lithospermate B, diabetic nephropathy, intestinal microbiota, bile acid.
Introduction
Diabetes mellitus (DM) is one of the most prevalent causes of chronic kidney disease (CKD) [1] and end-stage-renal disease [2]. In the United States, ~20–40% of DM patients develop diabetic nephropathy (DN) [3]. Decreased kidney function in animal DN models is characterized by albuminuria [4]; however, despite conventional therapies, such as ameliorating hyperglycemia and hypertension, an effective DN treatment/drug is lacking [5].
In recent years, a correlation between CKD and gut microbiota, termed the “kidney–gut axis” [6], has attracted considerable attention in CKD pathogenesis. The gut microbiome can shift markedly in CKD patients and uremic rats [7]. Gut microbiota-derived metabolites, such as ammonia [8, 9], indoxyl sulfate, p-cresyl sulfate [10], short-chain fatty acids [11, 12], and lipopolysaccharides [13] may be associated with kidney injury and DM progression.
Bile acids (BAs) are also metabolites derived from gut microbiota. Primary and secondary BAs are transformed by gut microbiota in the intestines, and most are reabsorbed to the liver, with a few translocating into the circulation [14]. In CKD patients, the serum BA concentration is increased [15]. BA accumulation in the plasma and its accompanying oxidative stress have been proposed as pathogenic factors of kidney injury [16]; however, the relationship between intestinal BA biotransformation and hyperglycemia-induced kidney damage is unknown.
“Danshen”, the root of Radix Salviae Miltiorrhizae, is a traditional Chinese medicine that is also used in Western countries for its excellent activity against cardiovascular disease. Magnesium lithospermate B (MLB) is the major component (Fig. 1) of danshen water extracts and is marketed to treat angina in China. Unfortunately, MLB’s extremely low oral bioavailability (0.02%) [17] limits its clinical application to the intravenous route. Conversely, orally administering MLB has been found to alleviate kidney damage in some animal models. In streptozotocin (STZ)-treated rats, orally administering MLB (10 mg/kg per day for 8 weeks) significantly suppressed kidney injury [18]. In an adenine-induced kidney failure model, orally administering MLB (10 mg/kg per day for 24 days) reduced serum urea nitrogen [19]. The paradox between MLB’s poor bioavailability and significant oral activity cannot be explained by canonical pharmacokinetics/pharmacodynamics.
Fig. 1.
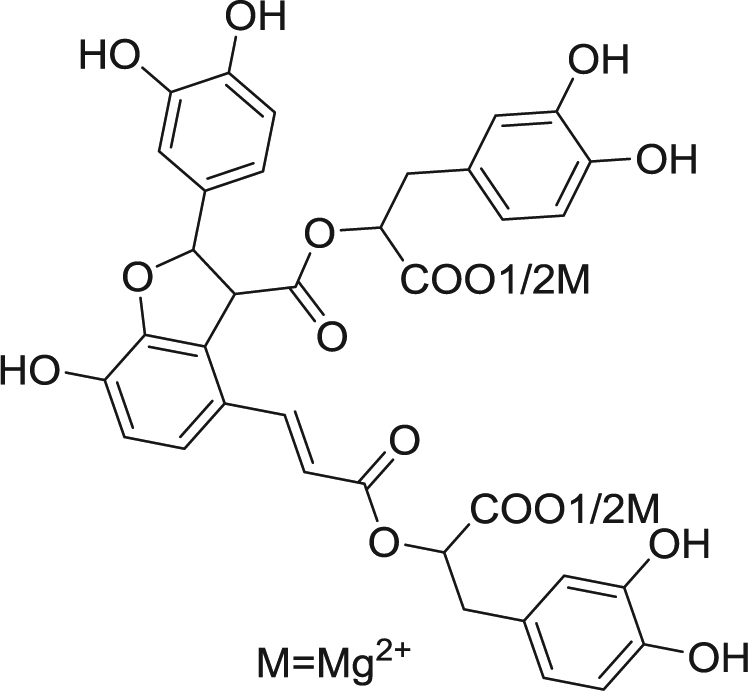
MLB chemical structure
Gut microbiota were shown to be closely related to DM, obesity and insulin resistance in several studies [20, 21]. Metformin [22], berberine [23], and resveratrol [24] were demonstrated to alleviate DM through the gut microbiota. Thus, we hypothesized that the kidney–gut axis might participate in MLB’s action. Raw extract of danshen (a complex mixture) was reported to restore the gut microbiota balance in the liver injury model [25]; however, whether MLB is the main component of danshen that causes this effect remains unverified.
Here, we used STZ-induced diabetic DBA/2J mice (a validated DN model [4]) to test MLB’s effects on kidney function, gut microbiome, and fecal BAs. We aimed to ascertain how MLB changes the gut microbiome and BA metabolism and whether these changes would illustrate MLB’s efficacy in protecting the kidneys.
Materials and methods
Ethical approval of the study protocol
The study protocol was approved by the Animal Care and Use Committee of the Shanghai Institute of Materia Medica (2017-02-YY-05; Shanghai, China). Animals were raised per guidelines set by Ministry of Science and Technology of the People’s Republic of China (Beijing, China).
Magnesium lithospermate B preparation
Lithospermate B (LAB) crude powder was purchased from Yuanye Biological Technology (Nanjing, China). LAB was acidified and extracted using ethyl acetate and subjected to chromatography using an MCI GEL CHP20P column (Mitsubishi Chemicals, Tokyo, Japan). After washing the column with water and 25% methanol in water, the purified LAB solution was yielded with 40% methanol elution in water. And then this solution was treated with newly prepared Mg(HCO3)2 before being applied to the MCI GEL CHP20P column. Elution with 25% methanol in water and solvent removal under reduced pressure yielded a light-yellow powder. The purity of MLB was greater than 95%.
Animals
Male DBA/2J mice (specific pathogen-free; 10 weeks old; 24.3 ± 2.8 g) were obtained from Shanghai SLAC Laboratory Animal Co. Ltd. (Shanghai, China) and were housed in a room with a 12-h dark-light cycle at 22 ± 2 °C and relative humidity at 50–60%. Food and water were provided ad libitum.
Experimental design
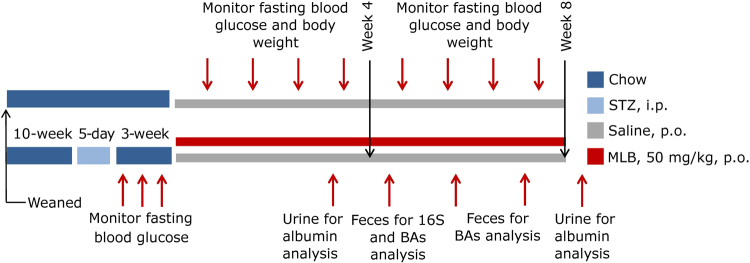
The DN model was established, as described previously [4]. Briefly, DBA/2J mice were injected with STZ (40 mg/kg, i.p.; Sigma-Aldrich, Saint Louis, MO, USA) in citrate buffer solution (pH 4.5) for 5 consecutive days. Fasting blood glucose (FBG) levels were monitored weekly using a blood glucose analyzer (AccuChek; Roche, Basel, Switzerland) to ascertain whether the DN was set. Three weeks after modeling, these mice were randomly assigned to two groups: STZ (orally administered saline once daily for 8 weeks, n = 7), or STZ-MLB (orally administered 50 mg/kg MLB solution once daily for 8 weeks, n = 8). Besides, mice those had not suffer from STZ injection (n = 7) were assigned into control group and administered saline daily for 8 weeks (Fig. 2).
Fig. 2.
Animal experiment design
FBG levels and body weight were monitored once per week. Feces were collected at weeks 4, 6, and the final week, and stored at −80 °C. Urine was collected after 24 h at week 4 and the final week and stored at −80 °C for further analyses.
Twenty-four-hour urinary albumin
Before the assay, urine samples were centrifuged at 1000 × g for 20 min at room temperature. The supernatants were used in an enzyme-linked immunosorbent assay (DL-ALB-Mu, DLdevelop, Shanghai, China).
Bile acid quantification
Fecal BAs were extracted using acetonitrile (1:5 mg/µL), thoroughly homogenized, and ultrasonically processed on ice. All samples were centrifuged at 14,000 × g for 5 min at 4 °C before transferring the supernatants to a 384-well microplate. BA concentration was quantified using ultra-high-performance liquid chromatography-tandem mass spectrometry [26].
DNA extraction
DNA suitable for the polymerase chain reaction (PCR) of bacterial 16S rRNA gene fragments was obtained from the feces using the QIAamp Fast DNA Stool Mini kit (Qiagen, Stanford, VA, USA).
PCR amplification and sequencing
The V3 and V4 regions of the bacterial 16S rRNA genes were amplified by the primers, 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). Each PCR reaction contained 4 µL of 5×Fast-Pfu buffer, 2 µL of deoxyribonucleotide triphosphate (2.5 mM), 0.8 µL of each primer (5 µM), 0.4 µL of Fast-Pfu polymerase, 0.2 µL of bovine serum albumin, and 10 ng of DNA template. We used 27 cycles of denaturation (95 °C for 30 s), annealing (50 °C for 30 s), and elongation (72 °C for 45 s). Agarose gel electrophoresis was used to semi-quantify the PCR products to ascertain whether the PCR products were available for subsequent two-pair end sequencing on an MiSeq Illumina platform (Majorbio BioTech, Shanghai, China).
Bioinformatics
Raw fastq data were qualified using bioinformatic software FLASH [27] (version 0.36) and Trimmomatic [28] (version 1.2.11). Resultant data were clustered by bioinformatic software Usearch [29] (version 7.0) to harvest operational taxonomic units (OTUs) with identities of >97% and mapped to the Silva database on the Qiime platform [30]. All processes were performed on the Majorbio BioTech platform (www.i-sanger.com/).
Statistical analyses
The alpha diversity (Chao and Shannon indexes) of the microbiome was calculated based on the OTU level by mother [31] (version 1.30.1) on the Majorbio BioTech platform (www.i-sanger.com/). Principal component analysis (PCA) was performed using R and visualized by the R package, ggplot2. To investigate potential biomarkers in STZ and STZ-MLB mice, linear discriminant analysis effect size (LEfSe) was evaluated from phylum to genus level using the one-against-all method, and the linear discriminant analysis (LDA) score was set at >2.0 (http://huttenhower.sph.harvard.edu/galaxy/root). The 100 most abundant OTUs that differed significantly among the control, STZ, and STZ-MLB groups (analyzed by the Kruskal–Wallis test and adjusted by false discovery rate (FDR < 0.05)) were correlated with the BA ratios by Spearman correlation using R. OTUs that correlated with the BA ratios (P < 0.05) and shared the same genus were added and correlated with the BA ratios and deoxycholic acids (DCAs), cholic acids (CAs), and taurocholic acid (TCA) abundances using R. Heatmaps were constructed using R (the pheatmap package), rows were clustered using Euclidean distance, and columns were clustered using the Pearson correlation distance. Other significant differences were assessed using the Mann–Whitney U-test or unpaired Student’s t-test in R.
Results
Mouse kidney function
In STZ mice, 24-h urinary albumin levels increased significantly with time. The 24-h urinary albumin level in the STZ-MLB mice was significantly lower than that in the STZ mice at week 8 (Table 1). MLB did not affect fasting blood gucose levels in the STZ mice (Table 1).
Table 1.
Kidney function and fasting blood glucose levels in the control, STZ and STZ-MLB groups at weeks 4 and 8
| Time | Group | 24-h urine ALB (μg) | Fasting blood glucose (mM) |
|---|---|---|---|
| 4th week | Control | 0.22 ± 0.15 | 7.06 ± 1.63 |
| STZ | 2.83 ± 2.19** | 23.48 ± 3.65** | |
| STZ-MLB | 2.76 ± 1.05** | 23.4 ± 6.38** | |
| 8th week | Control | 0.73 ± 0.42 | 6.19 ± 1.07 |
| STZ | 24.09 ± 16.02*** | 26.86 ± 2.47** | |
| STZ-MLB | 8.56 ± 3.96***,# | 27.01 ± 2.32** |
Significant differences were determined by unpaired t-tests corrected by FDR (**FDR < 0.01, ***FDR < 0.001 vs. control, #FDR < 0.05 vs. STZ group)
ALB albumin
Control, STZ, and STZ-MLB mice microbiomes
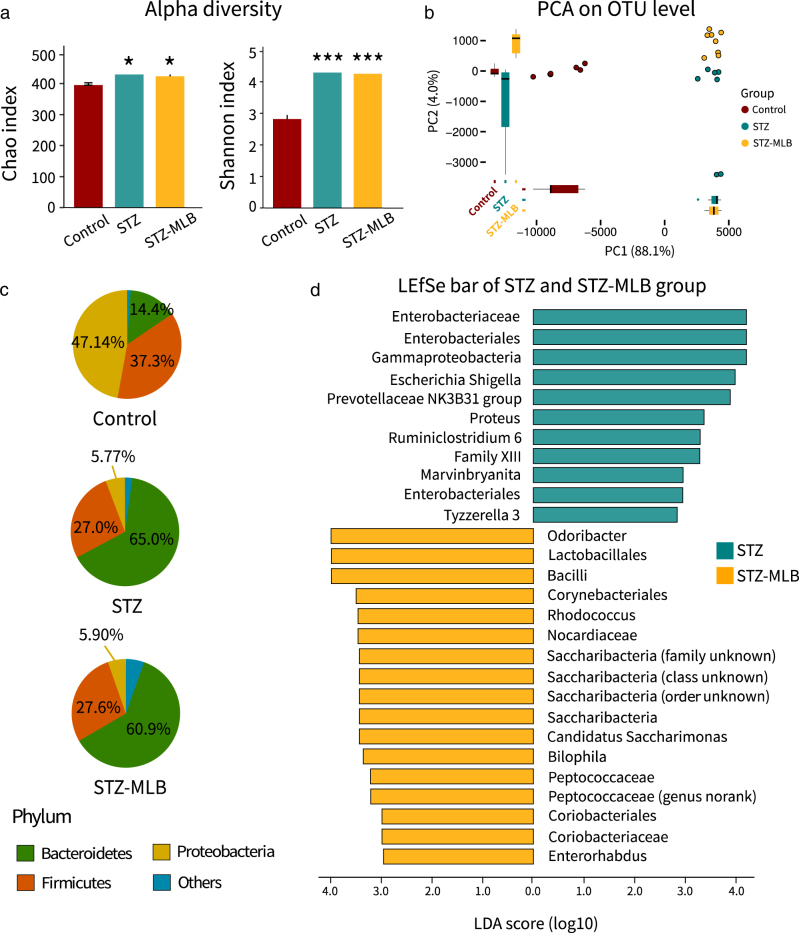
In the alpha diversity analyses, the STZ and STZ-MLB mice microbiomes were richer than those in the control group based on OTU levels (Fig. 3a). The STZ-MLB group’s gut microbiome differed from that in the STZ mice by OTU levels (Fig. 3b). Moreover, LEfSe identified potential biomarkers in the STZ and STZ-MLB mice relative to their gut microbiomes. Genus-level potential biomarkers in the STZ mice were Escherichia Shigella, Tyzzerella 3, Proteus, NK3B31, and unclassified genera in Bacteroidales, Marvinbryantia, and Ruminiclostridium 6 (Fig. 3d, Supplementary Table 1). Potential biomarkers in the STZ-MLB mice included Odoribacter, Peptococcaceae, Bilophila, Enterorhabdus, Rhodococcus, and Candidatus Saccharimonas (Fig. 3d, Supplementary Table 1).
Fig. 3.
Differences in the fecal bacterial microbiomes in the control, STZ and STZ-MLB groups. a The Chao and Shannon indexes of the three groups were calculated based on operational taxonomic unit (OTU) levels. Data are the means ± SEM. Significant differences were determined by the Mann–Whitney U-test and corrected by FDR (*FDR < 0.05, ***FDR < 0.001 vs. control group). b Principle component analysis (PCA) was executed based on OTU levels in R. c Pie plots of the three groups show the phylum level microbial composition. Data are the means, and microbiota <5% were assigned to other phyla. d LEfSe is represented as bar plots (LDA > 2). LDA linear discriminant analysis score
In the STZ and STZ-MLB groups, the Bacteroidetes phylum abundance increased, while that of the Proteobacteria phylum decreased greatly, including a relatively slight decrease in the Firmicutes phylum abundance (Fig. 3c, Supplementary Table 2). The Firmicutes:Bacteroidetes ratio decreased significantly in STZ and STZ-MLB mice compared with the control mice (Supplementary Table 2). The Firmicutes:Bacteroidetes ratio was negatively correlated with FBG levels (Supplementary Figure 1).
Control, STZ and STZ-MLB group bile acid metabolic profiles at weeks 4, 6, and 8
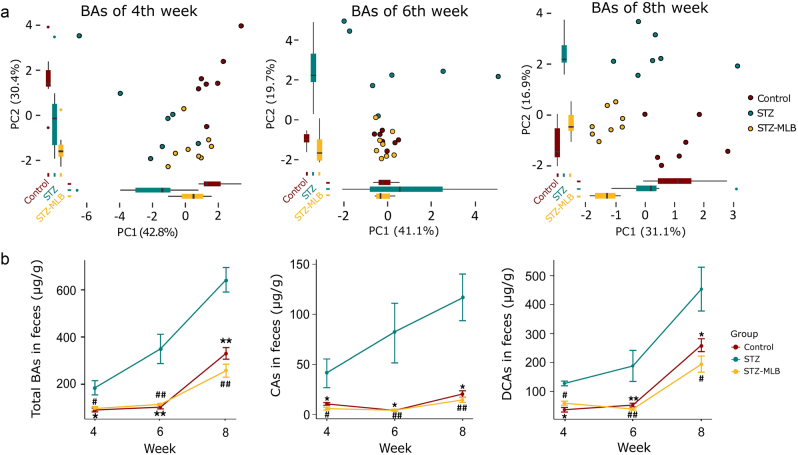
The BA metabolic profiles in fecal samples from the three groups were distinguished using PCA plots (Fig. 4a). The total BA levels increased considerably in STZ mice. In the STZ-MLB group, the total BA level was similar to that at baseline (Fig. 4b). Levels of CAs and DCAs, the most two prevalent BAs in fecal samples from the three groups (Fig. 4b), were also higher in the STZ group than in the control and STZ-MLB groups.
Fig. 4.
BA metabolic profiles in mouse feces from weeks 4 to 8. a PCA plot shows a shift of the BA metabolome in the control, STZ, and STZ-MLB groups from weeks 4 to 8. b Total BA, CA and DCA levels from weeks 4 to 8 in the three groups are represented as the means ± SEM. Significant differences were determined by the Mann–Whitney U-test and corrected by FDR (*FDR < 0.05, **FDR < 0.01 control vs. STZ group; #FDR < 0.05, ##FDR < 0.01 STZ-MLB vs. STZ group)
BA and microbiota correlational analyses
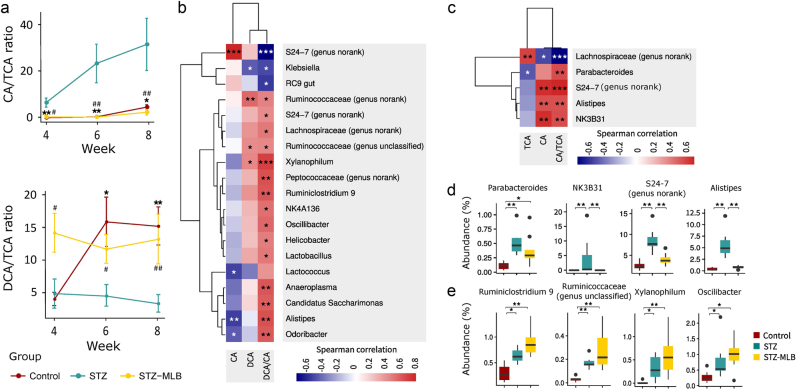
At week 4, the CA:TCA ratio increased in STZ mice and decreased in STZ-MLB mice (Fig. 5a). The DCA:CA ratio in the STZ group was lower than that in the STZ-MLB group (Fig. 5a). Twelve OTUs from five genera were potentially correlated with the CA:TCA ratios (Fig. 5c, d, Supplementary Table 3, Supplementary Table 4) In addition, 24 diverse OTUs from 18 genera were putatively related to the DCA:CA ratio (Fig. 5b, e, Supplementary Table 5, Supplementary Table 6).
Fig. 5.
Potential relationships between BA metabolism and gut microbiota. a The CA:TCA and DCA:CA ratios from weeks 4 to 8 are presented as the means±SEM. Significant differences were determined by the Mann–Whitney U-test and corrected by FDR (*FDR < 0.05, **FDR < 0.01 control vs. STZ group; #FDR < 0.05, ##FDR < 0.01 STZ-MLB vs. STZ group). b Heatmap of Spearman correlation analyses between the microbiota abundance and DCA:CA ratio. c Heatmap of Spearman correlation analyses between the microbiota abundance and CA:TCA ratio. d The microbiota abundance that potentially participated in biotransforming TCA to CA. Boxplots show the microbiota that correlate with the CA:TCA ratio. Significant differences were determined by the Mann–Whitney U-test corrected by FDR (*FDR < 0.05, **FDR < 0.01). e The microbiota abundance that potentially participated in biotransforming CA to DCA. Boxplots show the microbiota that correlated with the DCA:CA ratio. Significant differences were determined by the Mann–Whitney U-test corrected by FDR (*FDR < 0.05, **FDR < 0.01)
Discussion
An abnormal change in the gut microbiota composition suggests dysbiosis of the gut microenvironment. Gut microbiota diversity decreases significantly in people with type-2 diabetes mellitus (T2DM) [12], but gut-flora diversity only declines slightly in T2DM patients compared with healthy individuals [32]. The gut microbiota composition decreases in T1DM patients [33]. In an STZ-induced T1DM rat model, the diversity (as judged by the Shannon index) was reduced significantly compared with that in control rats [34]. Here, Proteobacteria was the prevalent phylum in the control mice, suggesting a special gut microbiome in DBA/2J mice because Bacteroidetes and Firmicutes are usually the more prevalent phyla [35]. Thus, in our DN mouse model, the gut microbiota diversity was increased as per the Shannon index, which may correlate with species differences. In addition to gut flora abundance, metabolites may reflect individual physiological statuses. In our study, the Firmicutes:Bacteroidetes ratio, potential microbiota biomarkers, and BA metabolic profiles were investigated. The V3-V4 region of the 16S rRNA gene was sequenced; the identity of which was assigned to the same OTU if it was >97%. All bacterial taxonomy was assigned based on OTU analyses.
Regarding DM, the Firmicutes:Bacteroidetes ratio is considered to potentially indicate glycemic levels [36–38]. We found that the FBG level was negatively correlated with the Firmicutes:Bacteroidetes ratio, a finding that is consistent with observations in children with T1DM [36]. In our study, the abundance of lower microbiota taxonomy, such as the S24-7, Prevotellaceae, Rikenellaceae, and Ruminococcaceae families, increased. In a CKD model using rats who underwent a 5/6 nephrectomy, the Rikenellaceae family abundance also increased [7]. The Lachnospiraceae family and Klebsiella genus abundances decreased in our study. These data were contrary to results in STZ-injected rats, wherein the Klebsiella abundance increased significantly, and Klebsiella were recommended as T1DM biomarkers [39]. Such data suggest that species differ when employing chemical stimulus-induced gut microbiome alterations.
LEfSe was used to determine potential biomarkers in STZ-treated and MLB-treated mice. Little information is available on using these potential microbial biomarkers to predict DN. Interestingly, the Odoribacter genus, whose abundance is reported to be negatively correlated with systolic blood pressure [37], was identified as a potential biomarker in STZ-MLB mice in our study. Hypertension is generally accepted as one of the main pathologic factors of CKD. We hypothesized that the improved kidney function in MLB-treated mice was related (at least in part) to the enriched Odoribacter genus via a cardiovascular pathway [37]. We did not monitor blood pressure during our study, but MLB’s antihypertensive effect has been reported [40, 41]. Our study observed, for the first time, a potential connection between MLB’s antihypertensive effect and the gut microbiota. Furthermore, Proteus, Escherichia and Shigella genera were putative biomarkers in DN mice in our study. Genetically related species such as Proteus mirabilis are uropathogenic bacteria that tend to infect DM mice [42]. Escherichia and Shigella species are related genetically to Escherichia coli, which would damage glomerular endothelial cells and tubular epithelial cells in the kidneys by producing one or more Shiga-like toxins [43]. In MLB-treated mice, Escherichia, Shigella, and Proteus species abundance was reduced, which partially accounted for MLB’s efficacy in preventing further kidney injuries.
In addition to the MLB-modulated change in the gut microbiome, we also investigated the metabolic functions of BA-related microbiota in fecal samples. The fluctuating BA concentration has been hypothesized to profoundly impact kidney function. BAs increase mucosal permeability and gut inflammation due to their detergent effect [44, 45], thus breaking down gut-barrier integrity. A “leaky” gut barrier allows endotoxins and bacterial metabolites to translocate into the systemic circulation, which contributes to progressing renal impairment [46]. Moreover, serum BA levels are increased in CKD patients [15]. BA accumulation in plasma damages tubular cell membranes by generating oxygen free radicals and promoting inflammatory cell release [16]. We found that total BA levels in STZ mice feces were higher than those in the control and STZ-MLB mice from week 4 to week 8, suggesting that hyperglycemia-induced kidney damage is accompanied by increased fecal BA levels. MLB could reduce fecal BA levels to near those observed at baseline without reducing blood glucose levels. Hence, MLB could affect BA metabolism without glycemic control. CAs and DCAs were the most prevalent BAs in the fecal samples. Their levels were reduced upon MLB treatment compared with the DM condition, implying that these two BAs may play more important roles in progressing kidney injuries.
BA contact time with bacteria, adequate bile flow, and diet also influence BA abundance [47]; thus, knowledge of the BA ratios is useful [48]. Bile salts are conjugated forms of BAs that flow into the intestinal tract to aid digestion and absorption after meals. Bile salt metabolism by gut microbiota consists mainly of deconjugation and dehydroxylation [14].
Bile salt hydrolase (BSH) deconjugates bile salts to primary BAs in the gut. BSH is present in many species across most phyla [49], one of which belongs to the Bacteroides genus [50]. In our study, the CA:TCA ratio, which may indicate the biotransformation level from TCA to CA, was increased significantly in STZ mice and reversed by MLB, suggesting that MLB may impact BA deconjugation. Per the Spearman correlation analyses in our study, OTUs from Parabacteroidetes, S24-7 (genus no rank), Alistipes and NK3B31 genera were positively correlated with the CA:TCA ratio, and all belonged to the same order (Bacteroidales) as the Bacteroides genus. MLB treatment decreased OTU abundance in these five genera, suggesting that a decrease in the CA:TCA ratio might be mediated (at least in part) by reducing the BSH activity in these bacteria.
7α-dehydroxylation produces secondary BAs, such as DCA and CA, from primary BAs and is an important form of BA transformation in the intestine. Few microbiota are reported to perform 7α-dehydroxylation (e.g., Clostridiales order) [14, 48]. At week 4, several OTUs that were positively correlated with the DCA:CA ratio belonged to the Clostridiales order (OTUs 403, 21, 482, 531, 399, 234, 461, 492, 240, 581, and 269), indicating that these bacteria can catalyze 7α-dehydroxylation. In STZ mice, the abundance of OTUs 240 and 581 (Ruminiclostridium 9), 269 (unclassified Ruminococcaceae), 403 (Xylanophilum), and 461 and 492 (Oscillibacter) increased significantly compared with those in the control group. These findings could reflect a stress response from the intestinal milieu to mobilize more microbiota capable of 7α-dehydroxylation to mediate the excessive supply of CAs. In addition, the DCA:CA ratio in the STZ group was lower than that in the STZ-MLB group from weeks 4 to 8, which was consistent with the reduced fecal CA concentration in the STZ-MLB group.
Due to its extremely low bioavailability, the oral efficacy of low-dose MLB for treating DN is unlikely to result from its direct action on the kidneys. We found that the fecal gut microbiome and BA profiles were significantly altered in MLB-administered DM mice. MLB’s local regulation of the gut microbiota and BA metabolism might be an important part of its mechanism in protecting kidney function.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the Youth Innovation Promotion Association CAS, the State Key Laboratory of Drug Research (SIMM1403ZZ-03), the Strategic Priority Research Program of the Chinese Academy of Sciences (No. XDA12050306), and the National Natural Science Foundation of China (81673302).
Author contributions
J-hS, KW, JL, and JZ design the study. JZ and Q-lZ performed the experiments. JZ wrote the paper. J-hS, KW, and JL reviewed and edited the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Kai Wang, Email: kwang@simm.ac.cn.
Jia Liu, Email: jia.liu@simm.ac.cn.
Electronic supplementary material
The online version of this article (10.1038/s41401-018-0029-3) contains supplementary material, which is available to authorized users.
References
- 1.Webster AC, Nagler EV, Morton RL, Masson P. Chronic kidney disease. Lancet. 2017;389:1238–52. doi: 10.1016/S0140-6736(16)32064-5. [DOI] [PubMed] [Google Scholar]
- 2.Recio C, Lazaro I, Oguiza A, Lopez-Sanz L, Bernal S, Blanco J, et al. Suppressor of cytokine signaling-1 peptidomimetic limits progression of diabetic nephropathy. J Am Soc Nephrol. 2017;28:575–85. doi: 10.1681/ASN.2016020237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dronavalli S, Duka I, Bakris GL. The pathogenesis of diabetic nephropathy. Nat Clin Pract Endoc. 2008;4:444–52. doi: 10.1038/ncpendmet0894. [DOI] [PubMed] [Google Scholar]
- 4.Qi ZH, Fujita H, Jin JP, Davis LS, Wang YH, Fogo AB, et al. Characterization of susceptibility of inbred mouse strains to diabetic nephropathy. Diabetes. 2005;54:2628–37. doi: 10.2337/diabetes.54.9.2628. [DOI] [PubMed] [Google Scholar]
- 5.Ahmad J. Management of diabetic nephropathy: Recent progress and future perspective. Diabetes Metab Syndr. 2015;9:343–58. doi: 10.1016/j.dsx.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Hu JJ, Luo HH, Wang JY, Tang WL, Lu JQ, Wu S, et al. Enteric dysbiosis-linked gut barrier disruption triggers early renal injury induced by chronic high salt feeding in mice. Exp Mol Med. 2017;49:e370. doi: 10.1038/emm.2017.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaziri ND, Wong J, Pahl M, Piceno YM, Yuan J, DeSantis TZ, et al. Chronic kidney disease alters intestinal microbial flora. Kidney Int. 2013;83:308–15. doi: 10.1038/ki.2012.345. [DOI] [PubMed] [Google Scholar]
- 8.Tang WHW, Wang ZN, Kennedy DJ, Wu YP, Buffa JA, Agatisa-Boyle B, et al. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ Res. 2015;116:448–55. doi: 10.1161/CIRCRESAHA.116.305360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shan ZL, Sun TP, Huang H, Chen SJ, Chen LK, Luo C, et al. Association between microbiota-dependent metabolite trimethylamine-N-oxide and type 2 diabetes. Am J Clin Nutr. 2017;106:888–94. doi: 10.3945/ajcn.117.157107. [DOI] [PubMed] [Google Scholar]
- 10.Yacoub R, Wyatt CM. Manipulating the gut microbiome to decrease uremic toxins. Kidney Int. 2017;91:521–3. doi: 10.1016/j.kint.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12:661–72. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 12.Maniar K, Moideen A, Mittal A, Patil A, Chakrabarti A, Banerjee D. A story of metformin-butyrate synergism to control various pathological conditions as a consequence of gut microbiome modification: genesis of a wonder drug? Pharmacol Res. 2017;117:103–28. doi: 10.1016/j.phrs.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 13.Chen HH, Zhu JF, Liu Y, Dong Z, Liu H, Liu YH, et al. Lipopolysaccharide induces chronic kidney injury and fibrosis through activation of mTOR signaling in macrophages. Am J Nephrol. 2015;42:305–17. doi: 10.1159/000441506. [DOI] [PubMed] [Google Scholar]
- 14.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–59. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Chu L, Zhang KK, Zhang YY, Jin XB, Jiang HX. Mechanism underlying an elevated serum bile acid level in chronic renal failure patients. Int Urol Nephrol. 2015;47:345–51. doi: 10.1007/s11255-014-0901-0. [DOI] [PubMed] [Google Scholar]
- 16.Bomzon A, Holt S, Moore K. Bile acids, oxidative stress, and renal function in biliary obstruction. Semin Nephrol. 1997;17:549–62. [PubMed] [Google Scholar]
- 17.Zhang Y, Akao T, Nakamura N, Duan CL, Hattori M, Yang XW, et al. Extremely low bioavailability of magnesium lithospermate B, an active component from Salvia miltiorrhiza, in rat. Planta Med. 2004;70:138–42. doi: 10.1055/s-2004-835854. [DOI] [PubMed] [Google Scholar]
- 18.Lee GT, Ha H, Jung MK, Li H, Hong SW, Cha BS, et al. Delayed treatment with lithospermate B attenuates experimental diabetic renal injury. J Am Soc Nephrol. 2003;14:709–20. doi: 10.1097/01.ASN.0000051660.82593.19. [DOI] [PubMed] [Google Scholar]
- 19.Yokozawa T, Lee TW, Oura H, Hattori M, Nonaka G, Nishioka I. Renal responses to magnesium lithospermate B in rats with adenine-induced renal failure. Phytother Res. 1993;7:235–9. doi: 10.1002/ptr.2650070305. [DOI] [PubMed] [Google Scholar]
- 20.Knip M, Siljander H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat Rev Endocrinol. 2016;12:154–67. doi: 10.1038/nrendo.2015.218. [DOI] [PubMed] [Google Scholar]
- 21.Yang JY, Lee YS, Kim Y, Lee SH, Ryu S, Fukuda S, et al. Gut commensal Bacteroides acidifaciens prevents obesity and improves insulin sensitivity in mice. Mucosal Immunol. 2017;10:104–16. doi: 10.1038/mi.2016.42. [DOI] [PubMed] [Google Scholar]
- 22.Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Manneras-Holm L, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23:850–8. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, Zhao Y, Xu J, Xue Z, Zhang M, Pang X, et al. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci Rep. 2015;5:14405. doi: 10.1038/srep14405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sung MM, Kim TT, Denou E, Soltys CM, Hamza SM, Byrne NJ, et al. Improved glucose homeostasis in obese mice treated with resveratrol is associated with alterations in the gut microbiome. Diabetes. 2017;66:418–25. doi: 10.2337/db16-0680. [DOI] [PubMed] [Google Scholar]
- 25.Xing HC, Li LJ, Xu KJ, Shen T, Chen YB, Chen Y, et al. Effects of Salvia miltiorrhiza on intestinal microflora in rats with ischemia/reperfusion liver injury. Hepatobiliary Pancreat Dis Int. 2005;4:274–80. [PubMed] [Google Scholar]
- 26.Sarafian MH, Lewis MR, Pechlivanis A, Ralphs S, McPhail MJW, Patel VC, et al. Bile acid profiling and quantification in biofluids using ultra-performance liquid chromatography tandem mass spectrometry. Anal Chem. 2015;87:9662–70. doi: 10.1021/acs.analchem.5b01556. [DOI] [PubMed] [Google Scholar]
- 27.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–63. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 30.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang XY, Shen DQ, Fang ZW, Jie ZY, Qiu XM, Zhang CF, et al. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS ONE. 2013;8:e71108. doi: 10.1371/journal.pone.0071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5:82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patterson E, Marques TM, O’Sullivan O, Fitzgerald P, Fitzgerald GF, Cotter PD, et al. Streptozotocin-induced type-1-diabetes disease onset in Sprague-Dawley rats is associated with an altered intestinal microbiota composition and decreased diversity. Microbiology. 2015;161:182–93. doi: 10.1099/mic.0.082610-0. [DOI] [PubMed] [Google Scholar]
- 35.Weldon L, Abolins S, Lenzi L, Bourne C, Riley EM, Viney M. The gut microbiota of wild mice. PLoS ONE. 2015;10:e0134643. doi: 10.1371/journal.pone.0134643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones FJ, Cardona F, Soriguer F, et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 2013;11:46. doi: 10.1186/1741-7015-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larsen N, Vogensen FK, van den Berg FWJ, Nielsen DS, Andreasen AS, Pedersen BK, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barlow GM, Yu A, Mathur R. Role of the gut microbiome in obesity and diabetes mellitus. Nutr Clin Pract. 2015;30:787–97. doi: 10.1177/0884533615609896. [DOI] [PubMed] [Google Scholar]
- 39.Wirth R, Bodi N, Maroti G, Bagyanszki M, Talapka P, Fekete E, et al. Regionally distinct alterations in the composition of the gut microbiota in rats with streptozotocin-induced diabetes. PLoS ONE. 2014;9:e110440. doi: 10.1371/journal.pone.0110440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yokozawa T, Chung HY, Oura H, Nonaka G-i, Nishioka I. Antihypertensive effect of magnesium lithospermate B, a component of Salviae Miltiorrhizae Radix, in spontaneously hypertensive rats. Nat Med. 1995;49:164–7. [Google Scholar]
- 41.Yokozawa T, Zhou JJ, Oura H, Tanaka T, Nonaka GI, Nishioka I. Effects on blood pressure of caffeic acid analogs isolated from Salviae Miltiorrhizae Radix in rats with adenine-induced renal hypertension. Phytother Res. 1995;9:105–9. doi: 10.1002/ptr.2650090205. [DOI] [Google Scholar]
- 42.Tsai YL, Chien HF, Huang KT, Lin WY, Liaw SJ. cAMP receptor protein regulates mouse colonization, motility, fimbria-mediated adhesion, and stress tolerance in uropathogenic Proteus mirabilis. Sci Rep. 2017;7:7282. doi: 10.1038/s41598-017-07304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tesh VL, Obrien AD. The pathogenic mechanisms of Shiga toxin and the Shiga-like toxins. Mol Microbiol. 1991;5:1817–22. doi: 10.1111/j.1365-2958.1991.tb00805.x. [DOI] [PubMed] [Google Scholar]
- 44.Munch A, Strom M, Soderholm JD. Dihydroxy bile acids increase mucosal permeability and bacterial uptake in human colon biopsies. Scand J Gastroenterol. 2007;42:1167–74. doi: 10.1080/00365520701320463. [DOI] [PubMed] [Google Scholar]
- 45.Islam KB, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141:1773–81. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 46.Ramezani A, Raj DS. The gut microbiome, kidney disease, and targeted interventions. J Am Soc Nephrol. 2014;25:657–70. doi: 10.1681/ASN.2013080905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Devkota S, Wang YW, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10(-/-) mice. Nature. 2012;487:104–8. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58:949–55. doi: 10.1016/j.jhep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Staley C, Weingarden AR, Khoruts A, Sadowsky MJ. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl Microbiol Biotechnol. 2017;101:47–64. doi: 10.1007/s00253-016-8006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawamoto K, Horibe I, Uchida K. Purification and characterization of new hydrolase for conjugated bile-acids, chenodeoxycholyltaurine hydrolase, from bacteroides-vulgatus. J Biochem. 1989;106:1049–53. doi: 10.1093/oxfordjournals.jbchem.a122962. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.