Abstract
Determining pathogenicity of genomic variation identified by next-generation sequencing techniques can be supported by recurrent disruptive variants in the same gene in phenotypically similar individuals. However, interpretation of novel variants in a specific gene in individuals with mild–moderate intellectual disability (ID) without recognizable syndromic features can be challenging and reverse phenotyping is often required. We describe 24 individuals with a de novo disease-causing variant in, or partial deletion of, the F-box only protein 11 gene (FBXO11, also known as VIT1 and PRMT9). FBXO11 is part of the SCF (SKP1-cullin-F-box) complex, a multi-protein E3 ubiquitin-ligase complex catalyzing the ubiquitination of proteins destined for proteasomal degradation. Twenty-two variants were identified by next-generation sequencing, comprising 2 in-frame deletions, 11 missense variants, 1 canonical splice site variant, and 8 nonsense or frameshift variants leading to a truncated protein or degraded transcript. The remaining two variants were identified by array-comparative genomic hybridization and consisted of a partial deletion of FBXO11. All individuals had borderline to severe ID and behavioral problems (autism spectrum disorder, attention-deficit/hyperactivity disorder, anxiety, aggression) were observed in most of them. The most relevant common facial features included a thin upper lip and a broad prominent space between the paramedian peaks of the upper lip. Other features were hypotonia and hyperlaxity of the joints. We show that de novo variants in FBXO11 cause a syndromic form of ID. The current series show the power of reverse phenotyping in the interpretation of novel genetic variances in individuals who initially did not appear to have a clear recognizable phenotype.
Subject terms: Genetics of the nervous system, Disease genetics, Neurodevelopmental disorders
Introduction
Next-generation sequencing (NGS) in neurodevelopmental disorders, especially with regard to intellectual disability (ID), has increased the possibility to identify an underlying genetic defect as the cause of disease to provide a conclusive molecular diagnosis [1–5]. NGS-based techniques such as whole-exome sequencing (WES) and whole-genome sequencing have shown to be efficient for individuals with ID and lead to an overall diagnostic yield of 55–70% [6]. WES is rapidly becoming the first-tier test in individuals with ID due to increased diagnostic yield and reduction of costs [7].
Since the DNA of each individual contains thousands of genic variants, many of which are difficult to interpret [8], different clinical and molecular approaches have been used to prioritize variants [9]. It has, for example, been shown that the identification of recurrently mutated genes in individuals with an overlapping phenotype add to the interpretation of variants. This depends, however, on the rarity and specificity of the clinical feature(s) involved [10]. For instance, a combination of rare congenital abnormalities or dysmorphic features is more specific than non-syndromic ID and hypotonia on its own [10]. Also, the inheritance pattern can be dependent on the severity of the ID and this may help interpretation. Severe ID often occurs sporadic and is caused by de novo variants [6], whereas mild ID can be explained by either a de novo variant, an inherited variant from a more mildly affected parent, or may have a multifactorial origin [11]. Reverse phenotyping, which refers to clinical comparison of individuals after a variant in the same gene has been found [12], can be of great value for interpretation of genetic variation [13–15].
In this report we show the value of reverse phenotyping in multiple individuals with variants in FBXO11. FBXO11 (also known as VIT1 and PRMT9) encodes the substrate-recognition component of the SCF (SKP1-cullin-F-box) complex, which is responsible for ubiquitination and subsequent degradation of substrates and plays a role in the maintenance of genome stability [16]. In addition, FBXO11 has been reported to be involved in regulating alternative splicing [17]. Downregulation of FBXO11 has previously been associated with vitiligo [18] and somatic variants have been found in diffuse large B cell lymphoma [19]. Germline variants in FBXO11 were not reported until recently when a frameshift and missense variant were found in a cohort of individuals with autism spectrum disorder (ASD) [5] and one nonsense variant in a cohort of individuals with ID [20]. Whereas overlap between various neurodevelopmental disorders, such as ID and ASD, has been described before, detailed phenotypic information of additional individuals is needed to specify the phenotypic spectrum associated with FBXO11 variants. In this study, we collected clinical information on 24 individuals with a de novo disease-causing variant in or partial deletion of FBXO11 and used reverse phenotyping to establish the related phenotypic spectrum. We show that reverse phenotyping in our patient cohort delineates a novel ID syndrome caused by de novo disease-causing variants in FBXO11.
Methods
See Supplementary Information.
Results
Identification of individuals with FBXO11 variants
Individuals 1, 2, 3, 18, and 20 were identified by routine diagnostic WES at the Radboudumc, Nijmegen and Maastricht University Medical Center, Maastricht, and were performed as previously described [21]. Based on the identification of three individuals with a de novo disease-causing variant in FBXO11, in combination with FBXO11 function, we considered FBXO11 an interesting candidate gene for ID, warranting further studies. Hence, we performed trio-based targeted re-sequencing using molecular inversion probes (MIPs [22, 23]) in 1031 ID and/or ASD individuals and their healthy parents: 512 individuals from Antwerp and 519 from Nijmegen. Individuals 4 and 5 were identified by this MIP assay.
In addition, we collected clinical information on 17 individuals (Individuals 6–17, 19, 21–24) with a variant in, or deletion of, FBXO11 through direct contact with (inter)national colleagues and by using Genematcher [24, 25] and Decipher [26].
Using data from seven laboratories contributing patients to this study, which included 21 individuals with a de novo disease-causing variant in FBXO11 in a total cohort of 14,088 individuals with ID/ASD, we estimated the frequency of a de novo disease-causing variant in FBXO11 in an unbiased ID/ASD cohort to be in the range of 1:500 to 1:1000 (Supplemental Table 1). In addition, if we compare the frequency of these de novo disease-causing variants in FBXO11 in our cohorts to the gene-specific mutation rate [21, 27], we find significantly more such disease-causing variants than expected (p = 1.879082e−19, Bonferroni correction for multiple testing).
Of note, Individuals 23 and 24 were previously published as part of a large-scale study reporting on de novo disease-causing variants in ASD using the Simons Simplex Collection cohort [5].
All 24 individuals were re-evaluated by reverse phenotyping, and clinical data were collected and compared.
Spectrum of disease-causing variants
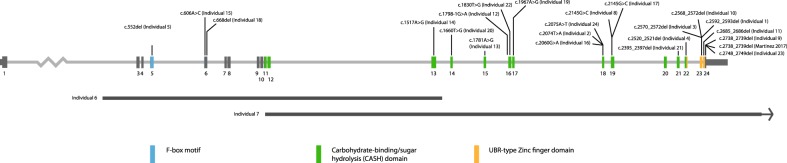
Twenty-four individuals carried 23 unique de novo disease-causing variants in the gene, comprising ten missense variants, one canonical splice site variant, two in-frame single amino acid deletions, two partial gene deletions, two nonsense variants, and six frameshift variants (Fig. 1; Supplemental Tables 2 and 3). Two individuals had an identical missense variant (Individuals 8 and 17). Interestingly, 16 of the 21 de novo variants are located in important protein domain structures (Fig. 1; Supplemental Table 2). Nine missense variants are in the carbohydrate-binding/sugar hydrolysis (CASH) domain, which is characterized by internal repeats of glycines and hydrophobic residues forming the β-helix repeat structures of the protein. Prediction of the pathogenicity of these missense variants using various classification tools, including SIFT (scale-invariant feature transform) and MutationTaster, indicate that these variants are likely pathogenic (Supplemental Table 2 and Supplemental Fig. 1). Three frameshift variants, one nonsense variant, and an in-frame deletion affect a UBR-type zinc-finger domain, which contains an N-degron sequence serving as a degradation signal that can be recognized by a unique class of E3 proteins for ubiquitin-dependent proteolysis [28] (Fig. 1).
Fig. 1.
FBXO11 with deletions and variants identified in our cohort. FBXO11 is expressed in two different isoforms (NM_001190274.1 and NM_0251433.4), which differ by an alternative exon 1, and of which NM_001190274.1 represents the longest transcript. Variants are annotated based on NM_0011190274.1. FBXO11 protein domains are depicted in color. Individuals 6 and 7 have a partial deletion of FBXO11 (gray bars)
In two individuals, a partial deletion of FBXO11 was identified by array-comparative genomic hybridization: a 21-kb intragenic deletion of exons 2–13 of FBXO11 (Individual 6), and an ~170-kb deletion disrupting the 3′ end of FBXO11 by deleting exons 12–24 (Individual 7) (Fig. 1).
Summary phenotypes
All 24 individuals had developmental delay ranging from borderline to severe ID. In addition, 16 individuals (67%) had behavioral problems, mostly consisting of ASD (n = 7), but attention-deficit/hyperactivity disorder (ADHD) (n = 5), aggression (n = 3), and anxiety (n = 3) were also observed. More than half of the individuals had hypotonia and a fraction had epilepsy (n = 3). Imaging of the brain was performed in 17 individuals, which showed enlarged ventricles in six of them. Three individuals had additional brain abnormalities (hypoplasia of anterior pituitary; bilateral hippocampal malformations, borderline large cerebellum; Chiari 2 malformation, syringomyelia).
Seven individuals were obese, which was not observed in the remaining 17 individuals. Although individuals did have dysmorphic features, there was no consistent facial gestalt (Fig. 2). The most consisted facial features were a high broad forehead (6/17, 35%), long palpebral fissures (7/20, 35%), and a thin upper lip with broad space between the paramedian peeks (15/19, 79%) (Figs. 2 and 3). Twelve individuals had minor hand abnormalities like brachydactyly or tapering fingers and eight individuals had hyperlaxity of the joints. Gastrointestinal problems such as feeding difficulties and vomiting was reported in 11 individuals (52%) and 9/21 (43%) had vision problems. Individuals encountered several other health problems, such as atrial septum defect (n = 1), cardiomyopathy (n = 1), diabetes mellitus (n = 1), and hypertrichosis (n = 2), but these only appeared in a minority. An overview of the most frequent features is shown in Table 1. Detailed clinical information is provided in the Supplemental Table 3 and the Supplemental Case Reports.
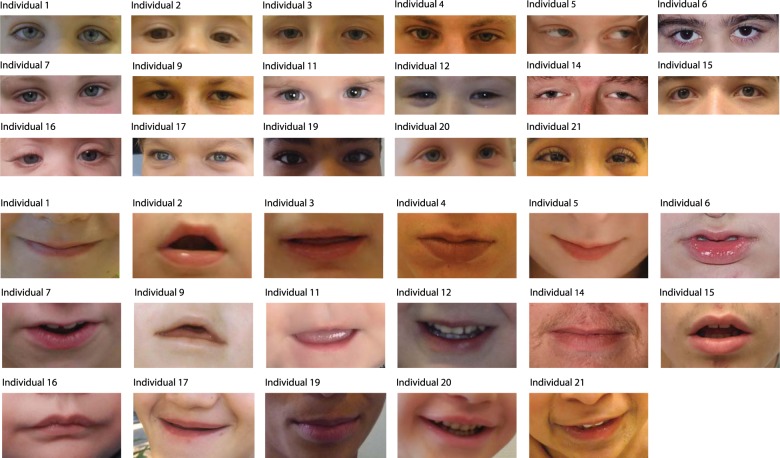
Fig. 2.
Photographs of individuals with a variant in FBXO11. The first row shows the individuals at a younger age, and the second row shows the respective individual at an older age. Facial features comprise a broad forehead, long palpebral fissures, and a thin upper lip with a broad space between the paramedian peaks of the upper lip. Some individuals show coarsening at an older age. The third row shows the hands of the respective individual with brachydactyly and tapering fingers in several cases
Fig. 3.
a Detailed photographs of the eyes of individuals with a variant in FBXO11. Eyes show long palpebral fissures. b Detailed photographs of the lips of individuals with a variant in FBXO11. There is a thin upper lip with a broad space between the paramedian peaks of the upper lip
Table 1.
Summary of clinical features of individuals with a variant in FBXO11
| Number | Percentage | |
|---|---|---|
| Gender | ||
| Male | 18/24 | 75% |
| Neurological | ||
| DD/ID | 24/24 | 100% |
| Behavioral problems | 16/24 | 67% |
| Hypotonia | 12/21 | 57% |
| Facial | ||
| High broad forehead | 6/17 | 35% |
| Long palpebral fissures | 7/20 | 35% |
| Thin upper lip with broad space between the paramedian peeks | 15/19 | 79% |
| Skeletal | ||
| Mild hand abnormalities | 12/20 | 60% |
| Joint hyperlaxity | 8/18 | 44% |
| Other | ||
| Gastrointestinal problems | 11/21 | 52% |
| Vision problems | 9/21 | 43% |
Discussion
Here we report on 24 individuals with a de novo disease-causing variant in or partial deletion of, FBXO11. We have provided a detailed clinical description showing that these individuals had borderline to severe ID, various behavioral problems (ASD, ADHD, aggression, anxiety), hypotonia, hyperlaxity of the joints, and some overlapping dysmorphisms (thin upper lip, broad space between the paramedian peaks of the upper lip, high broad forehead, long palpebral fissures). Six individuals had enlarged ventricles, of which three had additional brain abnormalities. A minority of individuals had other health problems, such as atrial septum defect, cardiomyopathy, diabetes mellitus, and hypertrichosis.
The 22 de novo disease-causing variants comprised 21 unique variants, including ten missense variants, two in-frame single amino acid deletions, one canonical splice site variant, two nonsense variants, and six frameshift variants.
FBXO11 is part of the F-box protein family, consisting of at least 69 different members in humans, divided over three protein subfamilies, which all contain at least one F-box motif, and, depending on the subfamily, another motif, that is, FBXL (leucine-rich repeats), FBXW (WD repeats), and FBXO (O = only or other motif) [29–32]. F-box proteins are part of SCF complexes, which are RING domain-containing E3 ubiquitin ligases [16, 30] degrading cellular proteins by ubiquitination during post-translational modification. This type of protein degradation is a quick and definitive way of (in)activating processes important for genome stability and maintenance [16]. In this complex, the F-box protein functions as a docking site for binding the protein requiring degradation [29, 30].
Of note, ubiquitination defects have previously been reported in ID [33, 34], and dysregulation of other SCF complex members, like Skp1 and Cul1, have also been associated with neurological disorders [35]. Moreover, pathogenic germline variants in other F-box protein family members have been linked to neurodevelopmental and neurodegenerative disorders [36–38].
FBXO11 is expressed in the (fetal) brain (https://gtexportal.org/home/, accessed date 19 January 2018; https://www.proteomicsdb.org/proteomicsdb/#human/proteinDetails/Q86XK2/expression, accessed date 19 January). Fbxo11 homozygous mutant mice were born with a cleft palate and died during the neonatal period [39, 40], whereas heterozygote mice (Jeff mouse) did not show a cleft palate, but did suffer from otitis media [40]. However, although one individual had a submucosal cleft palate, we did not see a relation between the phenotype in mice and the individuals with a de novo variant in FBXO11.
The occurrence of two deletions disrupting the coding sequence of FBXO11 suggests that haploinsufficiency is the molecular pathophysiological mechanism. This might also be the case for at least eight of 19 de novo disease-causing variants, including six frameshift variants and two nonsense variants. Whereas two frameshift variants are in the pen-ultimate exon, they occur beyond the 50–55 nucleotides from the last exon–exon junction, thereby still predicted to be degraded by nonsense-mediated decay (NMD). However, two nonsense variants and one frameshift variant are in the ultimate exon, escaping NMD. This might result in a stable, truncated protein with an altered function or an unstable protein with shortened protein half-life. Either way, these variants all lead to a shorter UBR-type zinc-finger domain by premature protein termination of FBXO11. Since the UBR-type zinc-finger domain has an important function in binding proteins with a N-terminal residue for ubiquitination [41], shortening of this domain results in functional loss of the SCF complex, similar to direct loss-of-function variants.
The nine missense variants located in the CASH domain affect amino acids strongly conserved in orthologs and it could therefore be speculated that these variants severely compromise normal protein function. Interestingly, we observed two recurrent identical de novo variants: c.2145G>C; p.(Lys715Asn) in Individuals 8 and 17, and c.2738_2739del; p.(Tyr913*) in Individual 9 and the Individual in the study of Martínez et al. [20]. At the moment, a clear genotype–phenotype correlation could not be established.
Population constraint metrics indicative for variation tolerance of FBXO11 as reported in, for instance, ExAC show that FBXO11 is extremely intolerant for loss-of-function variation (pLi = 1.00) and that there is an intolerance for missense variants (z-score = 4.03) [42]. Also, its residual variation intolerance score of −0.69 indicates that FBXO11 belongs to the 15% most intolerant human protein-coding genes [43] and we found more de novo disease-causing variants in FBXO11 than expected based on its gene-specific mutation rate [21, 27]. These findings, and the absence of de novo disease-causing variants in FBXO11 in over 2000 controls [5, 44], corroborate the pathogenicity of these variants.
Whereas these population metrics point towards the importance of de novo disease-causing variants in FBXO11 in ID, genotype–phenotype correlation at the individual level may be more complex. That is, 2 of 22 de novo disease-causing variants occurred as a postzygotic event and 5 of 22 individuals carried more than one de novo protein-coding disease-causing variants. Whereas postzygotic disease-causing variants are increasingly recognized as cause for early-onset genetic diseases [45–47], it is currently not clear to what extent the frameshift variant of Individual 10, which occurred in 35% of lymphocytes, and the missense variant in Individual 20, which occurred in 20–40% of lymphocytes, contribute to the phenotype. Unfortunately, other tissues of these individuals were not available for testing. Notably, Individual 10 also has a de novo missense variant in FGR, which is in a conserved amino acid (NM_001042747.1:c.1261C>T; p.(Pro421Ser)). The function of FGR and its relation to human disease has thus far not been reported, but a role for this variant in the individual’s phenotype cannot be formally excluded. Individual 20 also had a de novo in-frame deletion of five amino acids in LINGO2 (NM_001258282.1:c.1543_1557del; (p.(Pro515_Thr519del)). LINGO2 deletions have previously been reported in individuals with developmental delay, autistic behavior, and craniofacial abnormalities [48, 49], and a contribution of this variant to the individual’s phenotype cannot be excluded.
In Individual 7, the de novo 170-kb deletion not only included FBXO11, but also MSH6. Whereas heterozygous MSH6 whole gene deletions have not yet been reported, single-nucleotide variants in MSH6, including loss-of-function variants, are a known cause of Lynch syndrome (hereditary non-polyposis colon cancer) and result in an increased risk of colon cancer and other types of tumors [50–53]. The parents of the individual have been informed about Lynch syndrome as well as the related screening programs.
Given the per generation de novo mutation rate and the size of the exome [54], it is not uncommon that individuals have more than one protein-coding de novo variant. Most of these additional de novo variants are in genes that are unlikely to be related to ID, whereas others are in genes related to autosomal recessive disease, but for which no second mutated allele was present (Supplemental Case Reports). However, dominant disease-causing variants in ANK3, LINGO2, NF1, and CIC have previously been described in individuals with DD/ID and might have an additional effect on the phenotype [48, 49, 55–58]. Future (functional) research will resolve the role of each individual variant and further examine the role of FBXO11 in ID.
The individuals reported here with a de novo disease-causing variant in FBXO11 share their developmental and behavioral problems, although variable, but differ in other areas like gastrointestinal or vision problems. Although we observe recurrent facial dysmorphisms, a typical facial gestalt could not be established. FBXO11-related ID syndrome shows variability and is not highly recognizable. However, variability is seen frequently in many, also established, ID syndromes since we moved from phenotype-first to genotype-first approach. For instance, a broad phenotypic spectrum is also increasingly observed for syndromes such as ARID1B-related ID and ARID1B-related Coffin–Siris syndrome [59]. Future clinical research is needed and might reveal a genotype–phenotype relation, which could not be established now. We therefore founded the website http://www.humandiseasegenes.com/fbxo11 to collect additional clinical data.
In conclusion, we presented 24 cases that either had de novo disease-causing variants in FBXO11 or de novo disruptive deletions of FBXO11. All individuals showed ID with variable severity, behavioral problems, and dysmorphisms. While initial identification of these variants did not lead to immediate conclusive diagnoses, our reverse phenotyping strategy has been successful in determining pathogenicity of FBXO11 genomic variation and identifying a novel ID syndrome with a broad clinical spectrum.
Electronic supplementary material
Overview of clinical features in individuals with a variant in or partial deletion of FBXO11
Acknowledgements
We thank the individuals and their parents for participating in the study. We also thank Megan Cho, from GeneDx, Gaithersburg, MD, USA, for her help in contacting referring clinicians. We would like to thank Marisa Andrews and Dustin Baldridge, from the Department of Pediatrics, Washington University School of Medicine, and John N. Constantino, from the Department of Psychiatry and Pediatrics, Washington University School of Medicine, for their clinical support. We would also like to thank Christian Gilissen, from the Department of Human Genetics, Nijmegen, The Netherlands, for his support in comparing the frequency of de novo variants in FBXO11 to the gene-specific mutation rate. This work was financially supported by grants from the Netherlands Organization for Health Research and Development (912-12-109 to JAV, LELMV, and BBAdV). This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the NIH under award number U54HD087011, the Intellectual and Developmental Disabilities Research Center at Washington University. This study makes use of data generated by the DECIPHER community. A full list of centers that contributed to the generation of the data are available from http://decipher.sanger.ac.uk and via email from decipher@sanger.ac.uk. Funding for the project was provided by the Wellcome Trust. We would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about.
Compliance with ethical standards
Conflict of interest
M.J.G.S., K.G.M., and R.E.S. are employees of GeneDx Inc. H.R. is an employee of Impact Genetics Inc. The other authors declare that they have no conflict of interest.
Supplementary information
The online version of this article (10.1038/s41431-018-0292-2) contains supplementary material, which is available to authorized users.
References
- 1.Rauch A, Wieczorek D, Graf E, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet (Lond, Engl) 2012;380:1674–82. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 2.Gilissen C, Hehir-Kwa JY, Thung DT, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–7. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 3.Wright CF, Fitzgerald TW, Jones WD, et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet (Lond, Engl) 2015;385:1305–14. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grozeva D, Carss K, Spasic-Boskovic O, et al. Targeted next-generation sequencing analysis of 1,000 individuals with intellectual disability. Hum Mutat. 2015;36:1197–204. doi: 10.1002/humu.22901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iossifov I, O'Roak BJ, Sanders SJ, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–21. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vissers LE, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet. 2016;17:9–18. doi: 10.1038/nrg3999. [DOI] [PubMed] [Google Scholar]
- 7.Thevenon J., Duffourd Y., Masurel-Paulet A., Lefebvre M., Feillet F., El Chehadeh-Djebbar S., St-Onge J., Steinmetz A., Huet F., Chouchane M., Darmency-Stamboul V., Callier P., Thauvin-Robinet C., Faivre L., Rivière J.B. Diagnostic odyssey in severe neurodevelopmental disorders: toward clinical whole-exome sequencing as a first-line diagnostic test. Clinical Genetics. 2016;89(6):700–707. doi: 10.1111/cge.12732. [DOI] [PubMed] [Google Scholar]
- 8.de Ligt J, Willemsen MH, van Bon BW, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–9. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 9.Seaby Eleanor G., Pengelly Reuben J., Ennis Sarah. Exome sequencing explained: a practical guide to its clinical application. Briefings in Functional Genomics. 2015;15(5):374–384. doi: 10.1093/bfgp/elv054. [DOI] [PubMed] [Google Scholar]
- 10.Vulto-van Silfhout AT, Gilissen C, Goeman JJ, et al. Hum Mutat. 2017;38:594–9. doi: 10.1002/humu.23176. [DOI] [PubMed] [Google Scholar]
- 11.Ba W, Yan Y, Reijnders MR, et al. TRIO loss of function is associated with mild intellectual disability and affects dendritic branching and synapse function. Hum Mol Genet. 2016;25:892–902. doi: 10.1093/hmg/ddv618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schulze TG, McMahon FJ. Defining the phenotype in human genetic studies: forward genetics and reverse phenotyping. Hum Hered. 2004;58:131–8. doi: 10.1159/000083539. [DOI] [PubMed] [Google Scholar]
- 13.de Goede C, Yue WW, Yan G, et al. Role of reverse phenotyping in interpretation of next generation sequencing data and a review of INPP5E related disorders. Eur J Paediatr Neurol. 2016;20:286–95. [DOI] [PubMed]
- 14.Nambot Sophie, Masurel Alice, El Chehadeh Salima, Mosca-Boidron Anne-Laure, Thauvin-Robinet Christel, Lefebvre Mathilde, Marle Nathalie, Thevenon Julien, Perez-Martin Stéphanie, Dulieu Véronique, Huet Frédéric, Plessis Ghislaine, Andrieux Joris, Jouk Pierre-Simon, Billy-Lopez Gipsy, Coutton Charles, Morice-Picard Fanny, Delrue Marie-Ange, Heron Delphine, Rooryck Caroline, Goldenberg Alice, Saugier-Veber Pascale, Joly-Hélas Géraldine, Calenda Patricia, Kuentz Paul, Manouvrier-Hanu Sylvie, Dupuis-Girod Sophie, Callier Patrick, Faivre Laurence. 9q33.3q34.11 microdeletion: new contiguous gene syndrome encompassing STXBP1, LMX1B and ENG genes assessed using reverse phenotyping. European Journal of Human Genetics. 2015;24(6):830–837. doi: 10.1038/ejhg.2015.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Au PY, You J, Caluseriu O, et al. GeneMatcher aids in the identification of a new malformation syndrome with intellectual disability, unique facial dysmorphisms, and skeletal and connective tissue abnormalities caused by de novo variants in HNRNPK. Hum Mutat. 2015;36:1009–14. doi: 10.1002/humu.22837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silverman JS, Skaar JR, Pagano M. SCF ubiquitin ligases in the maintenance of genome stability. Trends Biochem Sci. 2012;37:66–73. doi: 10.1016/j.tibs.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y, Hadjikyriacou A, Xia Z, et al. PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145. Nat Commun. 2015;6:6428. doi: 10.1038/ncomms7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Le Poole IC, Sarangarajan R, Zhao Y, et al. “VIT1”, a novel gene associated with vitiligo. Pigment cell research/sponsored by the European Society for Pigment Cell Research and the International Pigment Cell. Society. 2001;14:475–84. doi: 10.1034/j.1600-0749.2001.140608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan S, Cermak L, Pagan JK, et al. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2012;481:90–3. doi: 10.1038/nature10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez F, Caro-Llopis A, Rosello M, et al. High diagnostic yield of syndromic intellectual disability by targeted next-generation sequencing. J Med Genet. 2017;54:87–92. doi: 10.1136/jmedgenet-2016-103964. [DOI] [PubMed] [Google Scholar]
- 21.Lelieveld SH, Reijnders MR, Pfundt R, et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci. 2016;19:1194–6. doi: 10.1038/nn.4352. [DOI] [PubMed] [Google Scholar]
- 22.O'Roak BJ, Vives L, Fu W, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science (New York, NY) 2012;338:1619–22. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coe BP, Witherspoon K, Rosenfeld JA, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet. 2014;46:1063–71. doi: 10.1038/ng.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sobreira N, Schiettecatte F, Boehm C, et al. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum Mutat. 2015;36:425–31. doi: 10.1002/humu.22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sobreira N, Schiettecatte F, Valle D, et al. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–30. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Firth HV, Richards SM, Bevan AP, et al. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet. 2009;84:524–33. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samocha KE, Robinson EB, Sanders SJ, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–50. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tasaki T, Mulder LC, Iwamatsu A, et al. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol. 2005;25:7120–36. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kipreos ET, Pagano M. The F-box protein family. Genome Biol. 2000;1:3002. Reviews. [DOI] [PMC free article] [PubMed]
- 30.Nelson DE, Randle SJ, Laman H. Beyond ubiquitination: the atypical functions of Fbxo7 and other F-box proteins. Open Biol. 2013;3:130131. doi: 10.1098/rsob.130131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borg NA, Dixit VM. Ubiquitin in cell-cycle regulation and dysregulation in cancer. Annu Rev Cancer Biol. 2017;1:59–77. doi: 10.1146/annurev-cancerbio-040716-075607. [DOI] [Google Scholar]
- 32.Zheng N, Zhou Q, Wang Z, et al. Recent advances in SCF ubiquitin ligase complex: clinical implications. Biochim Biophys Acta. 2016;1866:12–22. doi: 10.1016/j.bbcan.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tarpey PS, Raymond FL, O'Meara S, et al. Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit, cause an X-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus, and tremor. Am J Hum Genet. 2007;80:345–52. doi: 10.1086/511134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15:70–3. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 35.Bhutani S, Das A, Maheshwari M, et al. Dysregulation of core components of SCF complex in poly-glutamine disorders. Cell Death Dis. 2012;3:e428. doi: 10.1038/cddis.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Fonzo A, Dekker MC, Montagna P, et al. FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology. 2009;72:240–5. doi: 10.1212/01.wnl.0000338144.10967.2b. [DOI] [PubMed] [Google Scholar]
- 37.Mir A, Sritharan K, Mittal K, et al. Truncation of the E3 ubiquitin ligase component FBXO31 causes non-syndromic autosomal recessive intellectual disability in a Pakistani family. Hum Genet. 2014;133:975–84. doi: 10.1007/s00439-014-1438-0. [DOI] [PubMed] [Google Scholar]
- 38.Au PY, Argiropoulos B, Parboosingh JS, et al. Refinement of the critical region of 1q41q42 microdeletion syndrome identifies FBXO28 as a candidate causative gene for intellectual disability and seizures. Am J Med Genet A. 2014;164a:441–8. doi: 10.1002/ajmg.a.36320. [DOI] [PubMed] [Google Scholar]
- 39.Jin Y, Shenoy AK, Doernberg S, et al. FBXO11 promotes ubiquitination of the Snail family of transcription factors in cancer progression and epidermal development. Cancer Lett. 2015;362:70–82. doi: 10.1016/j.canlet.2015.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hardisty-Hughes RE, Tateossian H, Morse SA, et al. A mutation in the F-box gene, Fbxo11, causes otitis media in the Jeff mouse. Hum Mol Genet. 2006;15:3273–9. doi: 10.1093/hmg/ddl403. [DOI] [PubMed] [Google Scholar]
- 41.Xie Y, Varshavsky A. The E2–E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chain. EMBO J. 1999;18:6832–44. doi: 10.1093/emboj/18.23.6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrovski S, Wang Q, Heinzen EL, et al. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turner TN, Yi Q, Krumm N, et al. Denovo-db: a compendium of human de novo variants. Nucleic Acids Res. 2017;45(D1):D804–d11. doi: 10.1093/nar/gkw865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Halvorsen M, Petrovski S, Shellhaas R, et al. Mosaic mutations in early-onset genetic diseases. Genet Med. 2016;18:746–9. doi: 10.1038/gim.2015.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Acuna-Hidalgo R, Bo T, Kwint MP, et al. Post-zygotic point mutations are an underrecognized source of de novo genomic variation. Am J Hum Genet. 2015;97:67–74. doi: 10.1016/j.ajhg.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Lange IM, Koudijs MJ, van 't Slot R, et al. Mosaicism of de novo pathogenic SCN1A variants in epilepsy is a frequent phenomenon that correlates with variable phenotypes. Epilepsia. 2018;59:690–703. doi: 10.1111/epi.14021. [DOI] [PubMed] [Google Scholar]
- 48.Koufaris C, Papagregoriou G, Kousoulidou L, et al. Haploinsufficiency of the miR-873/miR-876 microRNA cluster is associated with craniofacial abnormalities. Gene. 2015;561:95–100. doi: 10.1016/j.gene.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 49.Gazzellone MJ, Zhou X, Lionel AC, et al. Copy number variation in Han Chinese individuals with autism spectrum disorder. J Neurodev Disord. 2014;6:34. doi: 10.1186/1866-1955-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miyaki M, Konishi M, Tanaka K, et al. Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet. 1997;17:271–2. doi: 10.1038/ng1197-271. [DOI] [PubMed] [Google Scholar]
- 51.Smith MJ, Urquhart JE, Harkness EF, et al. The contribution of whole gene deletions and large rearrangements to the mutation spectrum in inherited tumor predisposing syndromes. Hum Mutat. 2016;37:250–6. doi: 10.1002/humu.22938. [DOI] [PubMed] [Google Scholar]
- 52.Xiong HY, Alipanahi B, Lee LJ, et al. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science (New York, NY) 2015;347:1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wijnen J, de Leeuw W, Vasen H, et al. Familial endometrial cancer in female carriers of MSH6 germline mutations. Nat Genet. 1999;23:142–4. doi: 10.1038/13773. [DOI] [PubMed] [Google Scholar]
- 54.Acuna-Hidalgo R, Veltman JA, Hoischen A. New insights into the generation and role of de novo mutations in health and disease. Genome Biol. 2016;17:241. doi: 10.1186/s13059-016-1110-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iqbal Z, Vandeweyer G, van der Voet M, et al. Homozygous and heterozygous disruptions of ANK3: at the crossroads of neurodevelopmental and psychiatric disorders. Hum Mol Genet. 2013;22:1960–70. doi: 10.1093/hmg/ddt043. [DOI] [PubMed] [Google Scholar]
- 56.Ferner RE, Gutmann DH. Neurofibromatosis type 1 (NF1): diagnosis and management. Handb Clin Neurol. 2013;115:939–55. doi: 10.1016/B978-0-444-52902-2.00053-9. [DOI] [PubMed] [Google Scholar]
- 57.Lu HC, Tan Q, Rousseaux MW, et al. Disruption of the ATXN1-CIC complex causes a spectrum of neurobehavioral phenotypes in mice and humans. Nat Genet. 2017;49:527–36. doi: 10.1038/ng.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vissers LE, de Ligt J, Gilissen C, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–12. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 59.Santen GW, Clayton-Smith J. The ARID1B phenotype: what we have learned so far. Am J Med Genet C. 2014;166c:276–89. doi: 10.1002/ajmg.c.31414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of clinical features in individuals with a variant in or partial deletion of FBXO11