Significance
Functional interpretation of noncoding genetic variants identified by genome-wide association studies is a major challenge in human genetics and gene regulation. We generated epigenomics data using primary cells from type 1 diabetes patients. Using these data, we identified and validated multiple novel risk variants for this disease. In addition, our ranked list of candidate risk SNPs represents the most comprehensive annotation based on T1D-specific T-cell data. Because many autoimmune diseases share some genetic underpinnings, our dataset may be used to understand causal noncoding mutations in related autoimmune diseases.
Keywords: enhancer, noncoding variant, diabetes, GWAS, epigenomics
Abstract
Genome-wide association studies (GWASs) have revealed 59 genomic loci associated with type 1 diabetes (T1D). Functional interpretation of the SNPs located in the noncoding region of these loci remains challenging. We perform epigenomic profiling of two enhancer marks, H3K4me1 and H3K27ac, using primary TH1 and TREG cells isolated from healthy and T1D subjects. We uncover a large number of deregulated enhancers and altered transcriptional circuitries in both cell types of T1D patients. We identify four SNPs (rs10772119, rs10772120, rs3176792, rs883868) in linkage disequilibrium (LD) with T1D-associated GWAS lead SNPs that alter enhancer activity and expression of immune genes. Among them, rs10772119 and rs883868 disrupt the binding of retinoic acid receptor α (RARA) and Yin and Yang 1 (YY1), respectively. Loss of binding by YY1 also results in the loss of long-range enhancer–promoter interaction. These findings provide insights into how noncoding variants affect the transcriptomes of two T-cell subtypes that play critical roles in T1D pathogenesis.
Type 1 diabetes (T1D) is an autoimmune disease caused by immune-mediated destruction of the insulin-producing pancreatic beta cells. There is overwhelming evidence that imbalance between effector and regulatory T cells is a major cause of autoimmunity. Of all effector T cells, T-helper 1 (TH1) cells play a critical role in the pathogenesis of T1D by producing IFN-γ. The pathogenic role of TH1 cells is demonstrated by the transfer of disease through transplantation of pathogenic TH1 cells (1). TH1-initiated destruction of beta cells can be modulated by Foxp3+ regulatory T cells (TREG), which play an important role in the maintenance and regulation of immune tolerance and prevention of autoimmunity. They can modulate T-cell activation and promote immune tolerance by direct cell–cell interactions and production of immune modulatory cytokines such as transforming growth factor β (TGF-β) and interleukin 10 (IL-10). The protective role of TREG cells is impaired in susceptible population, and studies have demonstrated a deficiency in number and function of TREG cells in T1D (2).
Despite years of study, the molecular mechanisms responsible for the loss of immune tolerance remain to be fully elucidated. Genome-wide association studies (GWASs) have revealed 119 lead SNPs (59 genomic loci) that are associated with T1D (ImmunoBase; https://www.immunobase.org), but very few etiologic SNPs have been demonstrated. One challenge is that many associated SNPs are located in noncoding regions, which represent 98.5% of the human genome. Although several risk noncoding SNPs have been reported for the autoimmunity-associated genes DEX1 (3), GLUT1 (4), and IL6 (5), our knowledge of risk noncoding SNPs for autoimmune diseases remains limited.
Although multiple studies have demonstrated enrichment of T1D GWAS variants at T-cell–specific transcription enhancers in healthy donors (6, 7), to date, no study has examined the enhancer repertoire in primary TH1 and TREG cells from T1D patients, despite of the pivotal roles of TH1 and TREG cells in the pathogenesis of T1D. In this study, we conducted epigenomic and transcriptomic profiling of TH1 and TREG cells isolated from a cohort of five healthy donors and six newly diagnosed T1D patients. Our data (8) reveal significant alteration in the enhancer repertoire and transcriptional regulatory circuitry in TH1 and TREG cells of T1D patients. Intersecting our epigenomic data with a catalog of SNPs located in previously reported T1D-associated genomic loci, we identified several novel risk SNPs located in TH1 and TREG enhancers. We validated the functional roles of four candidate TREG SNPs using a combination of luciferase reporter assay, genome-editing, transcription factor chromatin immunoprecipitation (ChIP), and chromosome conformation capture (3C) assays.
Results
Transcriptome Changes in TH1 and TREG Cells of T1D Patients.
Using a panel of established cell surface markers, we purified effector memory TREG cells (CD3+ CD4+ CD25+ CD127dim/− CD45RO+) (9, 10) and effector memory TH1 cells (CD3+ CD4+ CXCR3+ CCR6− CCR7− CD45RO+) (9) from the peripheral blood of 11 subjects, including 6 T1D patients and 5 age-matched healthy controls (SI Appendix, Fig. S1 and Tables S1 and S2).
We profiled the transcriptome of patients and healthy donor subjects using RNA-seq (SI Appendix, Table S3) and found 370 and 250 differentially expressed transcripts between case and control groups for TH1 and TREG cells, respectively [false-discovery rate (FDR) < 0.1, Fig. 1A and SI Appendix, Table S4]. Consistent with the pathogenic role of TH1 in T1D, up-regulated genes are enriched for functions such as lymphocyte activation, cell growth, cell proliferation, and immune response, whereas down-regulated genes are enriched for functions such as cell death and apoptosis. For TREG cells, the up-regulated genes are involved in transcription, cell cycle, and chromosome organization, whereas down-regulated genes are involved in apoptosis, protein modification, targeting, and transport (Fig. 1C).
Fig. 1.
Differentially expressed genes between case and control subjects in TH1 and TREG cells. (A) Number of differentially expressed transcripts. FDR < 0.1, computed using EBSeq and corrected for multiple testing using the Benjamini–Hochberg (BH) method. Down-regulated, Expression is lower in the case group. Up-regulated, Expression is higher in the case group. (B) Volcano plot showing differentially expressed transcripts. Turquoise, Down-regulated in case group; magenta, up-regulated in case group. Genes associated with autoimmune disease are labeled. Genes associated with T1D are highlighted in orange. (C) Enriched GO terms among sets of differentially expressed genes in TH1 (Left) and TREG cells (adjusted hypergeometric test, P < 0.05, corrected for multiple testing using the BH method).
Several T1D-associated genes (obtained from ImmunoBase) are differentially expressed between case and control groups, including Rac family small GTPase 2 (RAC2) in both cell types, Ikaros 1 (IKZF1) and diacylglycerol kinase α (DGKA) in TH1 cells, and tyrosine kinase 2 (TYK2), IL10, and major histocompatibility complex, class II, DQ β1 (HLA-DQB1) in TREG cells (Fig. 1B). Besides those known T1D-associated genes, several genes associated with other autoimmune diseases are also differentially expressed, including cyclin-dependent kinase 6 (CDK6), Janus kinase 2 (JAK2), C-C motif chemokinase receptor 4 (CCR4), and signal transducer and activator of transcription 5A/5B (STAT5A/B) in TH1, and interleukin-5 (IL5), IFN regulatory factor 4 (IRF4), and signal transducer and activator of transcription 2 (STAT2) in TREG. STAT5A and STAT5B are reported to have a role in TH1 cell differentiation (11). SMAD3, which is down-regulated in the case group, has a critical role in TH1 inhibition and immune tolerance (12, 13). Together with FOXP3 and other transcription factors (TFs), IRF4 can form a transcriptional network that governs TREG cell differentiation (14). IL5 is reported to promote induction of antigen-specific TREG cells that suppress autoimmunity and reduced expression of IL5 will disrupt the immune balance (15) (Fig. 1B and SI Appendix, Table S4).
Deregulated Enhancers in TH1 and TREG Cells of T1D Patients.
Traditional ChIP followed by high-throughput sequencing (ChIP-seq) protocols require millions of cells. From a typical blood draw of 50 mL, it is only possible to purify approximately half a million TH1 cells and TREG cells, which is not sufficient for profiling multiple histone marks and the transcriptome. We thus developed a low-input ChIP-seq protocol using as low as 20,000 cells. We validated the protocol using the human lymphoblastoid cell line, GM12878. Data generated using our low-input protocol have an excellent agreement with data generated with conventional protocol using millions of cells (SI Appendix, Fig. S2).
Using our low-input ChIP-seq protocol, we profiled two histone modification marks, H3K4me1 and H3K27ac, the combination of which marks active transcriptional enhancers (SI Appendix, Table S5). By using normalized ChIP-seq signals in 1,000-bp windows genome-wide, we obtained a median genome-wide interindividual correlation ranging between 0.84 and 0.87 considering subject groups (control, case), cells (TH1, TREG), and histone marks (H3K4me1 and H3K27ac) (SI Appendix, Fig. S3A). These correlations are higher than a similar study performed using TREG cells from 11 healthy donors (16) (median, 0.70) (SI Appendix, Fig. S3C).
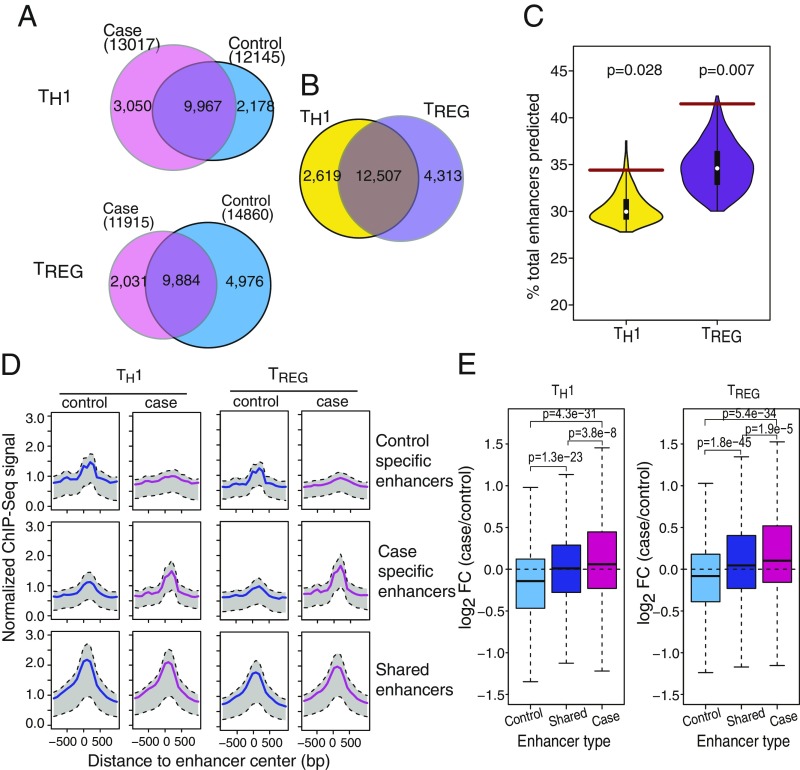
We predicted active enhancers for each cell type and cohort (case and control) separately using H3K4me1 and H3K27ac ChIP-seq data and the Chromatin Signature Identification by Artificial Neural Network (CSI-ANN) algorithm (17). For TH1 cell, we predicted 13,017 and 12,145 enhancers in case and control groups, respectively. For TREG cell, we predicted 11,915 and 14,860 enhancers in case and control groups, respectively (Fig. 2A and SI Appendix, Table S6). As expected, 83% of TH1 enhancers and 74% of TREG enhancers are shared between the two cell types, given their common origin from naive CD4+ T cells (Fig. 2B). However, within each cell type, a large fraction of the enhancers has altered activities between the case and control groups (21% for TH1 and 25% for TREG) (Fig. 2 A and D). The percentages of group-specific enhancers were significantly higher than expected (null distribution computed by permuting the sample labels in the dataset; Fig. 2C). TH1 cells in T1D patients appear to have an overall gain of active enhancers, whereas TREG cells in T1D patients appear to have an overall loss of active enhancers. Taken together, these data suggest a considerable change in the transcriptional regulatory networks (TRNs) of both cell types in T1D patients.
Fig. 2.
Enhancer repertoires of TH1 and TREG cells are significantly altered in T1D patients. (A) Overlap of enhancers between case and control subjects. Enhancers overlapping by >500 bp are considered to be overlapped. (B) Overlap of enhancers between TH1 and TREG cells. For each cell type, the total number of enhancers in case and control subjects is used. (C) P values for observed numbers of group-specific enhancers. Violin plots show the background distribution based on permutated ChIP-seq data. The brown horizontal lines show the observed percentage of group-specific enhancers. (D) Pile-up plots of histone mark H3K27ac signals associated with case-specific, control-specific, and shared enhancers. Signals are normalized by library size and input. (E) Enhancer activity profile is correlated with target gene expression profile (P values are calculated using two-sided Wilcoxon rank-sum test).
To understand the impact of case-specific enhancers on the transcriptomes, we need to know their target genes. We recently developed the Integrated Method for Predicting Enhancer Targets (IM-PET) algorithm (18). It predicts enhancer–promoter interactions by integrating four statistical features derived by integrating transcriptome, epigenome, and genome sequence data. Using IM-PET, on average, each gene is predicted to be targeted by 1.5 and 1.6 enhancers in TH1 and TREG cells, respectively. We compared our EP predictions with a recently published Capture-Hi-C data on CD4+ T cells (SI Appendix, Fig. S4A). Approximately 55% of our predictions are supported by Capture-Hi-C data, suggesting high quality of our predictions. Finally, for both cell types, there is a positive correlation between enhancer activity and gene expression level across the case and control groups (Fig. 2E), providing further support to the accuracy of our target gene prediction.
We next focused on target genes of case/control-specific enhancers (SI Appendix, Table S7). Many of the condition-specific enhancers are linked to genes already implicated in T1D, such as IKZF1, CCR5, CLEC16A, IL2RA, and UBASH3A for TH1 and IKZF1, IKZF4, RAC2, and RASGRP1 for TREG (7, 19). Gene ontology analysis of the enhancer targets suggests deregulation of specific pathways in TH1 and TREG cells of T1D patients, such as T-cell activation, lymphocyte activation, leukocyte activation, innate immune response, and cellular response to organic substances (SI Appendix, Table S8).
Key Transcription Factors Mediating Transcriptome Changes in TH1 and TREG Cells of T1D Patients.
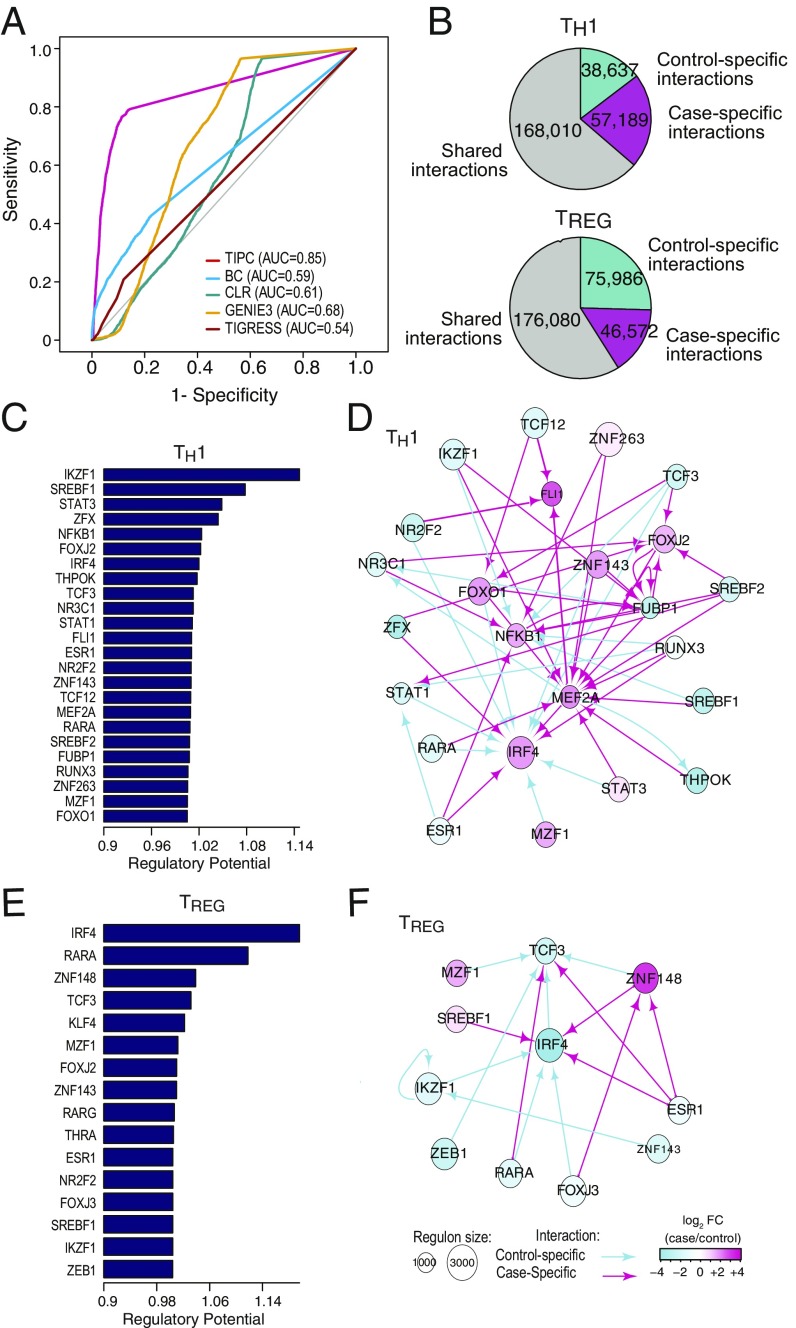
Our knowledge about the TRNs in TH1 and TREG cells of T1D patients is rather limited (20). Motivated by our knowledge of transcriptional regulation, we developed a method, target inference via physical connection (TIPC), to infer condition-specific TRNs. TIPC computes probability scores for three key components of transcriptional regulation, including probability of a DNA sequence being an enhancer, probability of a TF binding to an enhancer given the TF motif model and the enhancer sequence, and probability of enhancer–promoter interaction. The overall score for a TF regulating a target gene is the product of the three component probabilities and the expression level of the TF (see Methods for details). We evaluated the performance of TIPC using two approaches. First, using a set of gold-standard TF-target pairs in embryonic stem cells, we found that TIPC outperforms four state-of-the-art methods based on Pearson correlation (BC), mutual information [context likelihood ratio (CLR) (21)], decision trees [gene network inference with ensemble of trees (GENIE3) (22)], and regression [trustful inference of gene regulation with stability selection (TIGRESS) (23)] for predicting TF–target interactions (Fig. 3A).
Fig. 3.
Transcriptional regulatory networks (TRNs) and key transcription factors. (A) Benchmarking of our method for constructing TRN. BC, Pearson correlation between the gene pairs. Other methods are CLR, GENIE3, and TIGRESS (Methods). (B) Distribution of condition-specific and shared TF–target interactions for the TRNs constructed. (C and E) Predicted key TFs that play critical roles in T1D onset for TH1 (C) and TREG (E). Bars show the median proximity (1/distance) of TF to differentially expressed targets. (D and F) Condition-specific interactions between the key TFs for TH1 (D) and TREG (F). Control-specific interactions are denoted with turquoise, whereas case-specific interactions are shown with magenta. Shared interactions between case and control are not shown for better readability. The arrows show the direction of interaction (from TF to target). Node sizes are proportional to size of the TF regulon. Node colors show expression change of TFs between control and case.
We further evaluated the predicted TF–target interactions using publicly available TF ChIP-seq data (24) for T cells. We found that our predicted TF–target interactions have significant overlap with ChIP-seq data (SI Appendix, Fig. S4B). We predicted 263,836 and 298,638 TF–target pairs in TH1 and TREG cells, respectively. Thirty-six percent and 41% of the TF–target pairs are either control- or case-specific in TH1 and TREG cells, respectively (Fig. 3B), suggesting a significant degree of rewiring of the TRN in both cell types of T1D patients.
Next, we developed a method to systematically identify key TFs that play critical roles in the state transition of a TRN between two conditions. Our method is based on the assumption that a key TF can influence a larger set of differentially expressed targets compared with a nonkey TFs. To capture such signal, for two adjacent genes in the TRN, we convert their P values of differential expression into a distance measure such that the distance between two highly differentially expressed targets is very short. As a result, TFs that have shorter median distance to the set of differentially expressed targets are ranked higher (see Methods for details).
We identified 24 and 16 key TFs in TH1 and TREG cells, respectively (Fig. 3 C and E). Thirty-seven percent of TH1 TFs and 56% TREG TFs are shared between the two cell types. Consistent with their role as key regulators, these TFs target more other TFs than nonkey TFs (P < 0.001, t test; SI Appendix, Fig. S5). Most of the identified TFs have a reported role in either the biology of TH1 and TREG cells, or the pathogenesis of either T1D or other autoimmune diseases (SI Appendix, Table S9).
When ranked according to their regulatory potential (the inverse of median distance of TF to differentially expressed targets), the top key TFs are IKZF1 and SREBF1 for TH1 and IRF4 and RARA for TREG. IKZF1 plays a role in the TH1 versus TH2 polarization (25, 26). A SNP (rs10272724) in the 3′-UTR of IZKF1 has been shown to be protective from T1D (27). SREBF1 plays an important role in inducing genes that encode numerous genes in the lipid biosynthesis pathway, which controls full activation, proliferation, and differentiation of CD4+ T cells, including TREG cells (28, 29).
IRF4, the top key regulator in TREG, is reported to interact with FOXP3 and promote TREG function (14). RARA is critical for the normal differentiation and functions of both TREG and TH1 cells. All-trans retinoid acid (ATRA), an endogenous ligand of RARA, can prevent human natural TREG cells from converting to TH1/TH17 cells and sustains their suppressive function in inflammatory environments (30). Likewise, RARA can sustain TH1 cell lineage stability and prevents transition to a TH17 cell program in vivo (31). Consistent with the role of RARA in TH1 and TREG cells, RARA level decreases in both cell types (fold changes are 0.55 and 0.08 for TH1 and TREG, respectively; SI Appendix, Table S4) in T1D patients.
Fig. 3 D and F show the case/control-specific regulatory interactions among the key TFs. It highlights the complex and dynamic interactions among the key TFs during T1D development. For most of the key TFs, there are several case and control interactions. Moreover, although ∼47% of the TFs are shared between TH1 and TREG cells, the interaction partners of many of them are different in the two cell types, such as IKZF1, TCF3, and RARA.
Candidate Noncoding Risk Variants Affecting Enhancers in TH1 and TREG Cells.
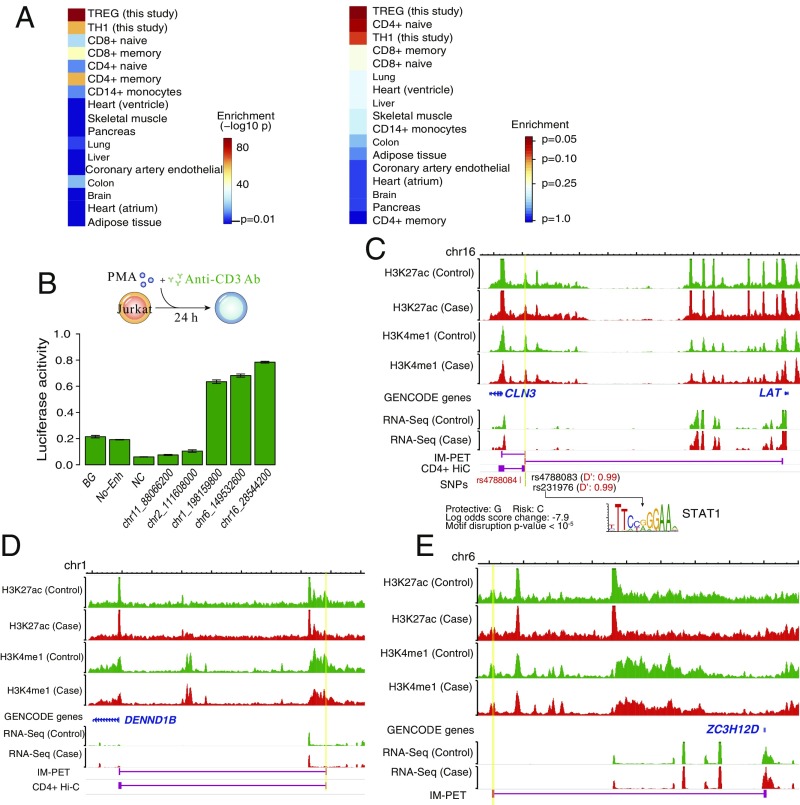
A recent study reported that T1D-associated SNPs are enriched in enhancers active in thymus, T and B cells, and CD34+ hematopoietic cells (7). However, that study is based on lymphocyte data from healthy individuals and thus may not fully capture the set of deregulated enhancers in T1D patients. We computed the overlap between sets of tissue- or cell-specific enhancers (including our TH1- and TREG-specific enhancers) and the set of T1D-associated SNPs, which consists of 119 lead GWAS SNPs (32–35) and 3,844 additional SNPs that are in the same linkage disequilibrium blocks with the lead SNPs. Consistent with the previous study, we found enrichment of T1D-associated SNPs in subsets of CD4+ and CD8+ T cells, especially memory T cells, using hypergeometric test. However, we found TREG and TH1 enhancers from this study have much higher enrichment for T1D-associated SNPs compared with enhancers identified in all subsets of CD4+ and CD8+ T cells used here from healthy individuals (Fig. 4A, Left). Since hypergeometric test does not take into account linkage disequilibrium, we used an alternative approach, linkage disequilibrium score regression (36, 37), to evaluate the statistical significance. We again observed that TREG and TH1 enhancers from this study tend to have higher statistical significance of overlapping GWAS SNPs than enhancers identified in all subsets of CD4+ and CD8+ T cells from healthy individuals (Fig. 4A, Right). We found no significant difference in overlapping GWAS SNPs between case and control-specific enhancers. This is not unexpected since enhancer SNPs may exert their effect on the phenotype via both loss or gain of function. This result provides additional support for the direct involvement of TREG and TH1 cells in T1D pathogenesis. It also highlights the importance of using primary cells from patients and control subjects for finding disease-associated genetic variants. The set of enhancers that overlap with T1D-associated SNPs is provided in SI Appendix, Table S10.
Fig. 4.
TH1 enhancers validated by luciferase reporter assay. (A) TH1 and TREG enhancers have higher degree of overlap with T1D-associated SNPs. (Left) Overlap P values were computed using hypergeometric test. (Right) Overlap P values were computed using linkage disequilibrium score regression. (B, Top) Schematic for the protocol for differentiating TH1 from Jurkat cells. (B, Bottom) Luciferase reporter activity of candidate enhancers. Enhancers are labeled by the genomic coordinate of the center of the enhancer. Enhancer activity is relative to cotransfected Renilla control. BG, No DNA vector control; NC, negative control, genomic region without H3K4me1 and H3K27ac signals. Data shown are means ± SEM of six replicates. (C) Genome browser tracks for the enhancer located at chr16: 28543200–28545200 (highlighted in yellow), including normalized histone modification ChIP-seq signals, RNA-seq signals of target genes of the enhancer, enhancer–promoter interactions predicted by IM-PET algorithm, supporting capture-Hi-C data [Javierre et al. (38)] for the enhancer–promoter interaction (if any), and SNP locations. In addition, TF motif and binding sites (if any) that are significantly affected by the SNP(s) are also shown. There are two SNPs, rs4788083 and rs231976, in this enhancer. rs231976 perturbs the binding site of STAT1 (BH-adjusted empirical value of P < 10−5; Methods). D′, Measurement of linkage disequilibrium between an enhancer SNP and the lead T1D GWAS SNP rs4708084 (shown in red). (D) Enhancer located at chr1: 198158800–198160800 targeting the DENND1B gene. (E) Enhancer located at chr6:149531600–149533600 targeting the ZC3H12D gene.
As the first step to identify noncoding risk SNPs that alter enhancer activity, we compared the normalized H3K4me1 and H3K27ac signals of each enhancer between case and control groups. We identified enhancers with significantly altered histone mark signals between case and control in TH1 and TREG cells, respectively (P < 0.1, Wilcoxon rank-sum test, two-sided, SI Appendix, Table S11). From this set of enhancers, using a combination of multiple orthogonal supporting evidence, including overlapping with super enhancers, expression quantitative trait loci (eQTL) and perturbation of TF binding sites (SI Appendix, Figs. S7 and S8 and Table S12), we selected five enhancers for each cell type to test their activity using luciferase reporter assay. We differentiated a Jurkat cell line as the model for TH1 cells (39) and a Foxp3-expressing Jurkat cell line as the model for TREG cells (40–42), hereby termed Foxp3+-Jurkat cells (Methods). Both cell models were further validated by checking the expression pattern of known signature genes for each cell type (SI Appendix, Fig. S9).
Of the five candidate TH1 enhancers, three of them showed significant activity compared with the negative controls (Fig. 4B, P < 0.01, Wilcoxon rank-sum test, one-sided). The first enhancer (chr16: 28543200–28545200) has a significant reduction in H3K4me1 signal (P < 0.01, Wilcoxon rank-sum test, two-sided; Fig. 4C) in T1D patients compared with healthy individuals. The predicted target genes of the enhancer are ceroid-lipofuscinosis, neuronal 3 (CLN3), and linker for activation of T cells (LAT). Both predictions are supported by published Capture-Hi-C data in CD4+ T cells (38). CLN3 is important for the normal function of lysosomes. Impaired lysosome function caused by mitochondrial respiration deficiency subverts T-cell differentiation toward proinflammatory subsets and exacerbates the in vivo inflammatory response (43). LAT is required for T-cell receptor-mediated signaling both in mature T cells and during T-cell development. Mice in which tyrosine 136 of LAT is constitutively mutated accumulate CD4+ T cells that trigger autoimmunity and inflammation (44). This enhancer harbors two T1D-associated SNPs, rs4788083 and rs231976. Using randomly selected SNPs from 1000 Genomes Project as the background null distribution, we found that rs231976 perturbs the binding site of STAT1 [empirical P < 10−5, corrected for multiple testing with Benjamini–Hochberg (BH) method], a key TF involved in the IL-27–mediated early commitment to the TH1 lineage (45).
The second validated enhancer (chr1:198158800–198160800) is predicted to target the gene DENN domain containing 1B (DENND1B) (Fig. 4D). The enhancer–promoter interaction is also supported by Capture-Hi-C data in CD4+ T cells (38). The H3K4me1 signal at the enhancer is significantly reduced in T1D patients compared with healthy individuals (P < 0.01, Wilcoxon rank-sum test, two-sided). Polymorphisms in DENND1B are associated with asthma and other immune disorders (46, 47).
The third validated enhancer (chr6: 149531600–149533600) targets the gene zinc finger CCCH-type containing 12D (ZC3H12D), and there is significant reduction in H3K4me1 level in T1D patients compared with healthy individuals (P < 0.01; Fig. 4E). ZC3H12D is an RNase that destabilizes a set of mRNAs. ZC3H12D−/− mice exhibit multiorgan inflammation (48) and autoimmune disease (49). A set of genes, including IL6, IL12, REL, TNFRSF4, and IL2, which are important for the activation of TH1 cells, are targets of ZC3H12D (49).
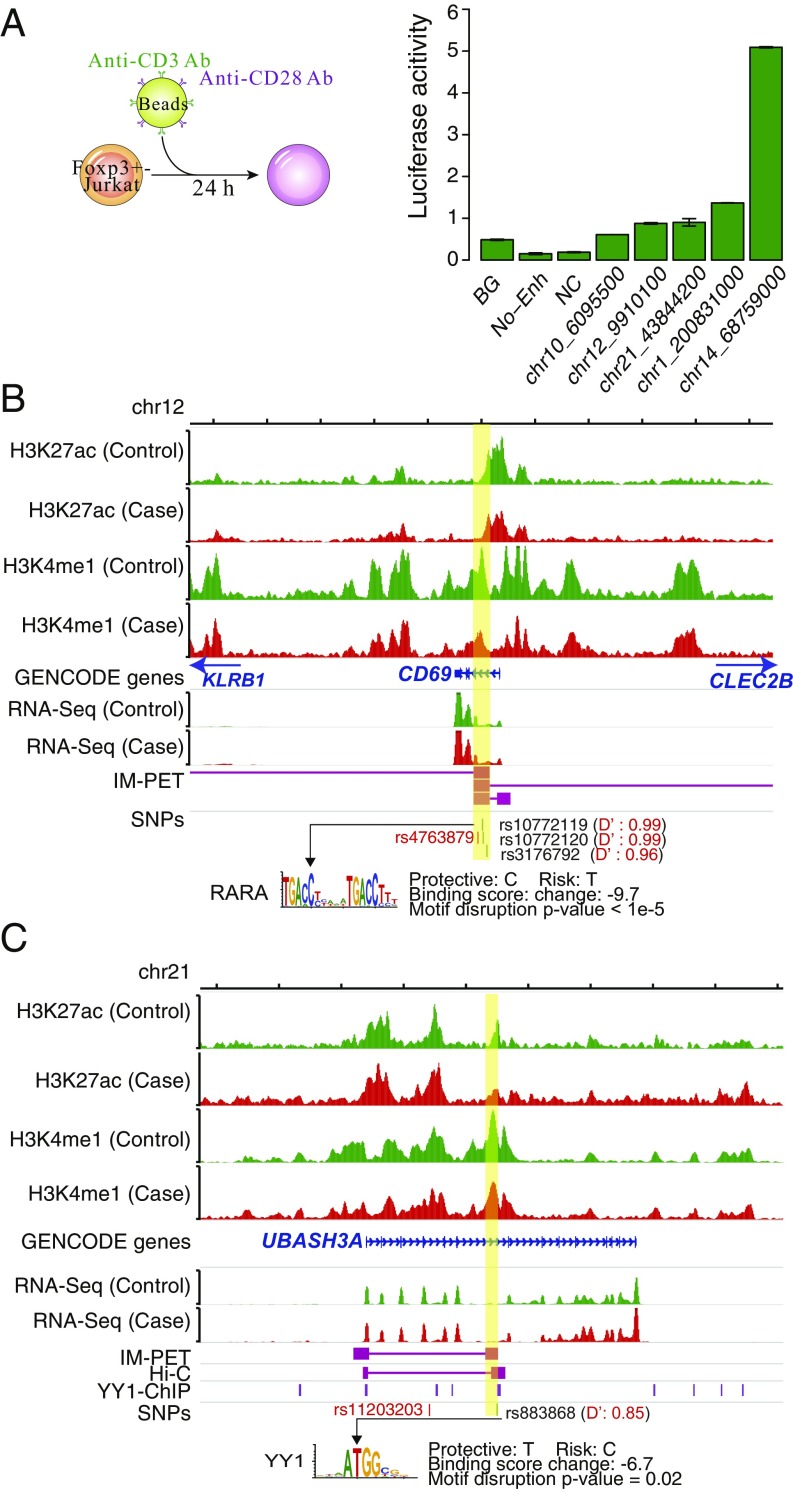
For TREG cells, all five enhancers show significant activity compared with the negative controls (Fig. 5A; P < 0.01, Wilcoxon rank-sum test). The first enhancer (chr12: 9908600–9911600) targets CD69, CLEC2B, KLRB1, and EIF2S3L, and harbors three SNPs (rs10772119, rs10772120, and rs3176792), which are in strong linkage disequilibrium with the T1D-associated GWAS lead SNP rs4763879 from ImmunoBase (D′ = 0.99, 0.99, and 0.96, respectively). The lead SNP is located in the CD69 gene body (50) (Fig. 5B). The second enhancer (chr21: 43843200–43845200) targets UBASH3A and harbors the SNP rs883868, which is in strong linkage disequilibrium with the T1D-associated GWAS lead SNP rs11203203. The lead SNP is located in the UBASHA gene body (50) (Fig. 5C).
Fig. 5.
TREG enhancers validated by reporter assay. (A, Left) Schematic for the protocol for differentiating TREG from Foxp3+-Jurkat cells. (A, Right) Luciferase reporter activity of candidate enhancers. Enhancers are labeled by the genomic coordinate of the center of the enhancer. Enhancer activity is relative to cotransfected Renilla control. BG, No DNA vector control; NC, negative control, genomic region without H3K4me1 and H3K27ac signals. Data shown are means ± SEM of six replicates. (B) Genome browser tracks for the enhancer located at chr12: 9908600–9911600 (highlighted with yellow). The SNP rs10772119 affects the binding site of RARA (BH-adjusted empirical value of P < 10−5). D′, Measurement of linkage disequilibrium between an enhancer SNP and the lead T1D GWAS SNP rs4763879 (shown in red). (C) Enhancer located at chr21: 43843200–43845200. The SNP rs883868 affects the binding site of YY1 (BH-adjusted empirical P value, 1.8 × 10−2). D′, Measurement of linkage disequilibrium between the enhancer SNP rs883868 and the lead T1D GWAS SNP rs11203203 (shown in red).
Of the five target genes by the first two enhancers, CD69 and UBASH3A are known T1D-associated genes (19), and CD69 is well studied for its role in T1D and TREG cells. CD69 is required for the repressive function of TREG cells (51). Diminished CD69 expression is associated with compromised function of TREG cells in systemic sclerosis (52). UBASH3A is primarily expressed in T cells and can negatively regulate T-cell signaling (53). Knockout of UBASH3A up-regulates inflammatory cytokine production and increase susceptibility to autoimmunity in a mouse model of multiple sclerosis (54). Roles of the other three genes in T1D and/or TREG have not yet been investigated. Nevertheless, KLRB1 could be of potential interest because it defines a subset of TREG cells capable of producing proinflammatory cytokines (55).
The third enhancer (chr10: 6094400–6096600) harbors the SNP rs10795763 and targets the gene IL2RA, which is associated with T1D, rheumatoid arthritis, and multiple sclerosis (19). Polymorphisms in IL2RA has been reported to negatively regulate the function of TREG cells (56). The SNP affects the binding of MBD2-interacting zinc finger protein (MIZF) (SI Appendix, Fig. S10). Methyl CpG binding protein 2 (MBD2) promotes demethylation of FOXP3 promoter and TREG function (57). The fourth enhancer (chr14: 68757800–68760200) harbors the SNP rs35763290 and targets the gene Actinin Alpha 1 (ACTN1). ACTN1 is an actin-binding protein and involved in the formation of immunological synapse, which is critical for T-cell activation, migration, and effector function at the interface between a T cell and its cognate antigen-presenting cell or target cell (58). The SNP affects the binding of nuclear respiratory factor 1 (NRF1). The fifth enhancer (chr1: 200830000–200832000) targets the PHLDA3 gene but does not harbor any SNP that disrupts any known TF binding motif. PHLDA3 is a p53-regulated repressor of Akt signaling, inhibition of which can suppress the activation and proliferation of TREG and results in a significant reduction of TREG cells in mouse (59).
Two Enhancer SNPs Affecting RARA and YY1 Binding at TREG Enhancers Regulating Immune Response Genes.
There are 25 SNPs in the five validated TREG enhancers described in the previous section. They are either a GWAS lead SNPs documented in the ImmunoBase or SNPs that are in linkage disequilibrium with a GWAS lead SNPs. We genotyped all 25 SNPs in the 11 study subjects using Sanger sequencing (SI Appendix, Table S13). Of these 25 SNPs, 9 have case-specific genotypes (the genotypes that are exclusively observed in T1D patients but not in healthy control), suggesting they are potential candidate risk SNPs. Among them, four SNPs (rs10772119, rs10772120, rs3176792, and rs883868), which are located in two enhancers, are associated with significant changes in histone modification and expression of the target genes of the host enhancer (Wilcoxon rank-sum test, P < 0.1; SI Appendix, Fig. S11). We therefore focused on these two enhancers and the four SNPs to further investigate their role in transcriptional regulation in TREG cells.
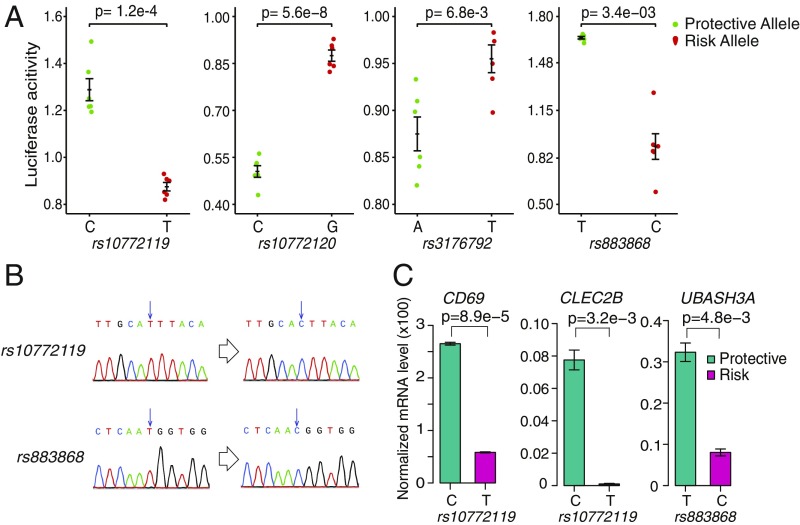
To test whether the SNPs can alter enhancer activity, we first mutated all four SNPs by site-directed mutagenesis and performed luciferase reporter assay in Foxp3+-Jurkat cells. We found that all four mutations cause significant change in the enhancer activity (P < 0.01, two-sided Student’s t test; Fig. 6A).
Fig. 6.
Four risk SNPs affect enhancer activity and target gene expression. (A) Luciferase reporter activity for enhancers containing risk SNPs with protective or risk allele. The green dots are replicates of protective genotype. The red dots represent risk genotype. Data shown are means ± SEM of six replicates. P values are computed using Wilcoxon rank-sum test, two-sided. (B) Sanger sequencing confirmation of genome-edited cells. (C) Effect of genotypes on the expression of target genes in TREG cells measured by RT-qPCR. CRISPR-mediated mutations of three SNPs in TREG cells affect the expression of target genes. The turquoise bars represent protective genotypes, and the magenta bars represent risk genotypes. mRNA level of each gene was normalized to ACTB gene, and relative levels were calculated using the ∆∆Ct method. Data shown are means ± SEM of two biological replicates. P values are computed using Wilcoxon rank-sum test, two-sided.
To investigate the role of the risk SNPs in target gene expression, we used CRISPR-Cas9–based genome editing to engineer Foxp3+-Jurkat cell lines with homozygous alleles at the SNPs (Fig. 6B). Using these cell lines, we found that cells with a homozygous T allele (risk allele) at rs10772119 have significantly reduced expression levels of CD69 and CLEC2B. Likewise, cells with a homozygous allele C (risk allele) at rs883868 have significantly reduced UBASH3A expression (Fig. 6C). Taken together, these results are consistent with the association between the genotype and gene expressions we found in our cohort.
To further establish the enhancer–target gene relationship, we tested the predicted enhancer–promoter interactions in Foxp3+-Jurkat cells using 3C coupled with quantitative PCR (3C-qPCR). As a negative control for the enhancer–promoter interaction, we used the human embryonic kidney 293FT (HEK 293FT) cell line. Compared with TREG cells, 293FT has very low expression of CD69 and UBASH3A genes, but the mRNA levels of CLEC2B in both cell types are comparable. Compared with negative control regions, the enhancer at CD69 locus significantly interacts with the promoters of CD69 and CLEC2B (P < 0.01, one-sided t test). The enhancer at UBASH3A locus interacts with the promoter of UBASH3A (Fig. 7A). Consistent with mRNA expression, we only found significant but weaker interaction between the enhancer with the CLEC2B promoter in HEK 293FT cells.
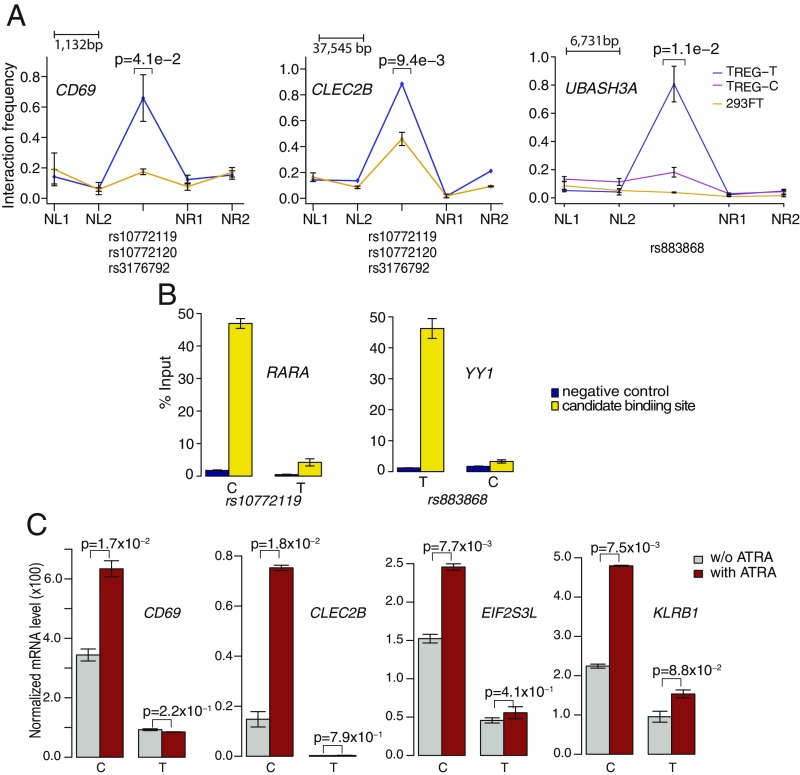
Fig. 7.
Two risk SNPs disrupt the binding of RARA and YY1 to TREG enhancers. (A) Chromatin interaction between SNP-bearing enhancers and promoters of predicted target genes. 3C-qPCR experiment was performed for each enhancer–promoter pair in Foxp3+-Jurkat cells. 293FT cells were used as negative control. For each pair of enhancer and promoter, six primers were designed, including two primers for the promoter and enhancer and four primers surrounding the enhancer (NL1, NL2, NR1, and NR2). Each consecutive primer pairs have roughly equal distance between them. Data shown are means ± SEM of two biological replicates. For enhancer bearing the YY1 binding site and the SNP rs883868, 3C-qPCR was performed using genome-edited cells having homozygous “T” (TREG-T) or “C” (TREG-C) allele at rs883868 to examine the effect of the SNP on enhancer–promoter interaction. Data shown are means ± SEM of two replicates. All P values are calculated with one-sided t test. (B) Change in TF binding at SNPs with homozygous alleles created by CRISPR-based genome editing of Foxp3+-Jurkat cells. Shown are normalized ChIP-qPCR value expressed as percent of input. Negative control site, Genomic region without any histone modification signal. Data shown are means ± SEM of four replicates. (C) Target genes of RARA enhancer are responsive to treatment of ATRA in Foxp3+-Jurkat cells. Genome-edited cells bearing homozygous alleles at rs10772119 were treated with ATRA and target gene expression was measured using RT-qPCR. Data shown are means ± SEM of two replicates. All P values are calculated with one-sided t test.
To identify TFs whose binding is affected by the two risk SNPs, we performed TF motif analysis using a collection of 1,772 motifs from the CIS-BP database (60). We found that rs10772119 exhibits allelic-specific binding by RARA (Fig. 5B) and rs883868 exhibits allelic-specific binding by YY1 (Fig. 5C). For both TFs, we performed ChIP-qPCR to confirm the binding of the TF to the predicted binding sites (Fig. 7B). Furthermore, ChIP-qPCR using cells with homozygous alleles at the SNP showed that the sites bearing the risk alleles has significantly lower binding affinity, confirming the result of our computational analysis.
To further test the role of the RARA binding site at rs10772119 in target gene expression, we treated Foxp3+-Jurkat cells with ATRA and measured the expression of the target genes using RT-qPCR. We found that only in cells with homozygous C alleles, the target genes are significantly induced by ATRA treatment (Fig. 7C), providing additional support to the role of rs10772119 in RARA binding and target gene expression.
YY1 has been reported to mediate enhancer–promoter interactions in a number of mammalian cell types (61). We hypothesized that reduced YY1 binding to the enhancer harboring rs883868 can result in reduced enhancer–promoter interaction. We therefore performed 3C-qPCR using Fox3+-Jurkat cells engineered to have homozygous T and C alleles at the SNP. Consistent with our hypothesis, we found significantly reduced enhancer–promoter interaction in cells with homozygous C allele at the SNP rs883868 (Fig. 7A).
In summary, the series of experiments above uncovered four candidate risk SNPs (rs10772119, rs10772120, rs3176792, and rs883868) that are in linkage disequilibrium with known T1D GWAS SNPs and alter enhancer activity and expression of immune response genes (Figs. 4–7). Among them, rs10772119 and rs883868 disrupt the binding of RARA and YY1, respectively. Moreover, allelic-specific binding by YY1 results in the allelic-specific long-range enhancer–promoter interaction involving UBASH3A (Fig. 7).
Discussion
GWASs have revealed 59 high-confidence genomic loci associated with T1D, which harbor 119 lead SNPs and 3,844 additional SNPs that are in the same linkage disequilibrium blocks with the lead SNPs. Among those SNPs, functional interpretation of noncoding SNPs remains particularly challenging. Toward this goal, we conducted an epigenomics-based fine map to identify candidate risk noncoding SNPs. We mapped the genome-wide distribution of two enhancer-associated histone modifications (H3K4me1 and H3K27ac) in TH1 and TREG cells from five healthy and six T1D patient subjects using a low-input ChIP-seq protocol.
We identified four new risk SNPs for T1D and two of them exhibit allele-specific binding by two TFs, RARA and YY1. ATRA induces the normal development of pancreas and affects the function of beta cells. RA signaling has a strong association with the onset of T1D (62). T1D patients (63, 64) are known to have vitamin A deficiency. Dietary RA reduces diabetes in diabetic-prone BB/Wor rats (65, 66). ATRA treatment significantly reduces diabetes incidence and delays the onset of diabetes transferred from NOD mice to NOD/SCID recipient mice. In summary, these findings suggest a protective role of ATRA against T1D. This protective role is due to induced TREG cell-dependent immune tolerance by suppressing both CD4+ and CD8+ T effector cells, while promoting TREG cell expansion (67). Supporting this mechanism, ATRA cannot exert the protective role in mice with abrogated TREG cells (68).
Here, we found that rs10772119 can disrupt the binding of RARA to the enhancer located at chr12: 9908600–9911600. Subjects with this risk SNP have lower expression of several target genes, including CD69, CELE2B, KLRB1, and EIF2S3L. CD69 is an early marker of T-cell activation. ATRA can increase the expression of CD69 in a dose-dependent fashion (69), and this effect is aborted if RARA was knockout (70). Consistent with this finding, we found that RA can induce the expression of CD69 and CLEC2B in TREG cell lines engineered to have a C allele at rs10772119 (Fig. 7C). Taken together, our finding provides a mechanistic explanation for which rs10772119 promotes the onset of T1D via ATRA signaling in TREG.
YY1 is a zinc finger protein that functions either as a transcriptional activator or repressor. Research in rats shows that YY1 may be associated with decreased incidence of T1D (71, 72). We found that the risk SNP rs883868 can disrupt the binding of YY1 in TREG cells, which leads to lower expression of the target gene UBASH3A. In support of the regulation of UBASH3A by YY1, YY1 knockout in pro-B cells can decrease the expression of UBASH3A (73). YY1 is one of a few proteins that has a reported role in mediating long-range chromatin interactions. Consistent with its role in chromatin looping, here we showed that a single SNP can disrupt YY1 binding and consequently leads to the loss of enhancer–promoter looping mediated by YY1. Our finding adds to increasing reports of genetic variants that can disrupt three-dimensional genome organization and gene expression (74–76).
None of the four risk SNPs identified in this study are lead GWAS SNPs. Instead, they are in linkage disequilibrium with lead SNPs. This result highlights the challenge of finding risk variants in the presence of linkage disequilibrium. Two of the four enhancer SNPs, rs10772120 and rs3176792, do not appear to overlap with any known TF binding motif, although mutating the SNPs showed an effect on enhancer activity and target gene expression. Additional studies are needed to uncover the mechanism by which the two SNPs affect gene expression in TREG cells.
Methods
Study Subjects.
Eleven subjects were recruited at The University of Iowa Children’s Hospital. Recent-onset T1D subjects (within 1 y from the first day of diagnosis; n = 6) were recruited through the Division of Pediatric Endocrinology and Diabetes at The University of Iowa Children’s Hospital. Diabetes was defined according to World Health Organization criteria and included blood glucose levels of 200 mg/dL with symptoms confirmed by a physician. Healthy subjects (n = 5) were recruited by posting flyers at The University of Iowa Children’s Hospital. The control criteria comprised fasting blood glucose of 100 mg/dL, no familial history of any autoimmune disorder, and lack of islet autoantibodies. All study subjects were free of known infection at the time of sample collection. For T1D subjects, no use of corticosteroids or glucocorticoids within the prior 6 mo was required. For healthy control subjects, no medications (especially steroids) and nonrelatives of T1D patients were required. At the time of each visit, the following clinical measurements were taken: HbA1c, autoantibodies, weight, and body mass index (subject information is shown in SI Appendix, Tables S1 and S2). The research protocol was approved by the IRB of the University of Iowa, and participants and/or their parents (guardians) provided written informed consent.
Purification of TREG and TH1 Cells from Peripheral Blood.
Fifty milliliters of peripheral blood were collected from each subject and processed within 0.5 h of collection. Blood was diluted with 1× DPBS (Gibco) and subjected to Ficoll-Hypaque centrifugation at 700 × g at 20 °C for 20 min in a swinging-bucket rotor without brake. The PBMC layer was transferred to a new 50-mL conical tube and cells were washed twice with 1× staining buffer 1 (1× DPBS plus 1% FBS). Cells were resuspended with 1× staining buffer 1 to 1 × 107 cells per 50 µL. After adding 5 µL of CD4+ T cell biotin–antibody mixture per 107 cells, cells were mixed and incubated for 5 min at 4 °C. Thirty-five microliters of 1× staining buffer per 107 cells and 10 µL of CD4+ T Cell MicroBead mixture per 107 cells were added and mixed. After incubation for 10 min at 4 °C, cells were diluted to 4 mL and applied to autoMACS Separator for negative selection of CD4+ cells using the “Deplete” program. In total, 0.2 × 106 cells were taken out as unstained cells, and the remaining cells were pelleted by centrifugation at 300 × g, 20 °C for 7 min. Supernatant was discarded and cells were resuspended in 1× staining buffer 1 to 2 × 107 cells per mL. Cells were incubated with an antibody mixture (anti-CD3, anti-CD4, anti-CD25, anti-CD45RO, anti-CD127, anti-CCR6, anti-CCR7, anti-CXCR3) [1:400 (vol/vol)] (see list of antibodies in SI Appendix, Table S14) at 20 °C in the dark for 30 min and washed with 1× staining buffer twice. Cells were resuspended in 1× staining buffer 1 to a final concentration of 1 × 107 cells per mL and sorted on an Aria flow cytometer (BD Biosciences) to obtain two cell populations: effector memory TREG cells (CD3+CD4+CD25+CD127dim/−CD45RO+) and effector memory TH1 cells (CD3+CD4+CXCR3+CCR6−CCR7−CD45RO+). Cells were collected in 0.5 mL of 1× staining buffer 2 [1× DPBS plus 1% human AB serum (Sigma)] within 1.5-mL siliconized low-retention microcentrifuge tubes (Fisher) coated with human AB serum before use. The purities of sorted cells were over 99% (SI Appendix, Fig. S1). Sorted cells were used for ChIP-seq and RNA-seq.
Low-Input ChIP-Seq.
Purified cells were crosslinked with 1% formaldehyde (Thermo Scientific) in 1× fixing buffer for 5 min at room temperature according to the vendor’s manual (Covaris). Cross-linked cells were then resuspended in 1× shearing buffer and sonicated using Covaris E220 for 780 s using the following settings: duty factor, 5%; peak incident power, 105; cycles per burst, 200. Five percent of sheared chromatin was used as the input, and the remaining chromatin was used for immunoprecipitation (IP). IP was performed using the ChIP-IT high-sensitivity kit (Active Motif). IP and input samples were treated with RNase A followed by proteinase K treatment. Cross-linking was reversed by incubation overnight at 65 °C. Reverse cross-linked DNA was purified using a MinElute PCR purification kit (Qiagen). All ChIPed DNA and 1 ng of input DNA were used for sequencing library preparation using the ThruPLEX-FD Prep Kit (Rubicon Genomics). Ten to 12 PCR cycles were used for IP DNA and 9 PCR cycles were used for input DNA at step 5 of the library preparation protocol. Libraries were sequenced on an Illumina HiSeq 2500 sequencer in single-end mode with a read length of 50 nt.
CRISPR/Cas9-Mediated Genome Editing.
Single-guide RNAs targeting the SNPs (rs10772119 and rs883868) were designed using CRISPOR (crispor.tefor.net) and cloned into the CRISPR vector pX459 (Addgene plasmid no. 448139). The 91-nt single-stranded DNA oligonucleotide (ssODN) repair templates were designed with homology arms centered around the SNPs (SI Appendix, Table S15). CRISPR/Cas9-mediated genome editing of Foxp3+-Jurkat cells was performed as described (77) with some modifications. Briefly, plasmids and ssODN templates were cotransfected to Foxp3+-Jurkat cells using TransIT-LT1 (Mirus Bio). Twenty-four hours posttransfection, cells were selected by puromycin for 72 h. Dead cells were removed by centrifugation at 300 × g, at room temperature for 5 min and discarding supernatant. Cell concentration and viability were measured using Countess II (Life Technologies). Live cells were resuspended at a concentration of five cells per mL. One hundred microliters of cell suspension was transferred to a 96-well plate and cultured for 2–3 wk for clonal cell line isolation. Genomic DNA was isolated using Quick-DNA 96 kit (ZYMO) and used for screening for desired mutations using allele-specific PCR and fluorescence melting curves as described (78). Genomic DNA of positive clones were further confirmed by Sanger sequencing.
Single-Nucleotide Variants Associated with T1D.
The list of T1D-associated SNPs was obtained from the ImmunoBase. The database reports 59 genomic loci containing 119 lead SNPs that are associated with T1D from GWAS studies. We identified linkage disequilibrium blocks using the Haploview software (79) and the genotyping data from 1000 Genomes project (80). We used the default parameter setting except for blockout, which was set to “ALL.” All SNPs that fall within the same linkage disequilibrium block as the T1D-associated GWAS lead SNP were also considered as candidate risk SNPs.
Code Availability.
All published software used for data processing are open source. Additional custom Perl, Python, R, and Unix shell scripts are developed for data processing and analysis. All custom scripts are available upon request.
Data Availability.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession no. GSE112342).
Supplementary Material
Acknowledgments
We thank the Research Information Services at the Children’s Hospital of Philadelphia for providing computing support. This work was supported by National Institutes of Health Grants GM104369, GM108716, HG006130, and AA024486 (to K.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE112342).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815336116/-/DCSupplemental.
References
- 1.Katz JD, Benoist C, Mathis D. T helper cell subsets in insulin-dependent diabetes. Science. 1995;268:1185–1188. doi: 10.1126/science.7761837. [DOI] [PubMed] [Google Scholar]
- 2.Brusko TM, Wasserfall CH, Clare-Salzler MJ, Schatz DA, Atkinson MA. Functional defects and the influence of age on the frequency of CD4+ CD25+ T-cells in type 1 diabetes. Diabetes. 2005;54:1407–1414. doi: 10.2337/diabetes.54.5.1407. [DOI] [PubMed] [Google Scholar]
- 3.Leikfoss IS, et al. Multiple sclerosis-associated single-nucleotide polymorphisms in CLEC16A correlate with reduced SOCS1 and DEXI expression in the thymus. Genes Immun. 2013;14:62–66. doi: 10.1038/gene.2012.52. [DOI] [PubMed] [Google Scholar]
- 4.Ng DP, et al. Minor effect of GLUT1 polymorphisms on susceptibility to diabetic nephropathy in type 1 diabetes. Diabetes. 2002;51:2264–2269. doi: 10.2337/diabetes.51.7.2264. [DOI] [PubMed] [Google Scholar]
- 5.Fishman D, et al. The effect of novel polymorphisms in the interleukin-6 (IL-6) gene on IL-6 transcription and plasma IL-6 levels, and an association with systemic-onset juvenile chronic arthritis. J Clin Invest. 1998;102:1369–1376. doi: 10.1172/JCI2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farh KK, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–343. doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Onengut-Gumuscu S, et al. Type 1 Diabetes Genetics Consortium Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015;47:381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng G, et al. 2018 Novel risk variants affecting enhancers of TH1 and TREG cells in type 1 diabetes. Gene Expression Omnibus. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE112342. Deposited March 26, 2018.
- 9.Mahnke YD, Brodie TM, Sallusto F, Roederer M, Lugli E. The who’s who of T-cell differentiation: Human memory T-cell subsets. Eur J Immunol. 2013;43:2797–2809. doi: 10.1002/eji.201343751. [DOI] [PubMed] [Google Scholar]
- 10.Rosenblum MD, Way SS, Abbas AK. Regulatory T cell memory. Nat Rev Immunol. 2016;16:90–101. doi: 10.1038/nri.2015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy KM. Permission to proceed: Jak3 and STAT5 signaling molecules give the green light for T helper 1 cell differentiation. Immunity. 2008;28:725–727. doi: 10.1016/j.immuni.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giroux M, et al. SMAD3 prevents graft-versus-host disease by restraining Th1 differentiation and granulocyte-mediated tissue damage. Blood. 2011;117:1734–1744. doi: 10.1182/blood-2010-05-287649. [DOI] [PubMed] [Google Scholar]
- 13.Takimoto T, et al. Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development. J Immunol. 2010;185:842–855, and erratum (2011) 186:636. doi: 10.4049/jimmunol.0904100. [DOI] [PubMed] [Google Scholar]
- 14.Fu W, et al. A multiply redundant genetic switch “locks in” the transcriptional signature of regulatory T cells. Nat Immunol. 2012;13:972–980. doi: 10.1038/ni.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran GT, et al. IL-5 promotes induction of antigen-specific CD4+CD25+ T regulatory cells that suppress autoimmunity. Blood. 2012;119:4441–4450. doi: 10.1182/blood-2011-12-396101. [DOI] [PubMed] [Google Scholar]
- 16.Arvey A, et al. Genetic and epigenetic variation in the lineage specification of regulatory T cells. eLife. 2015;4:e07571. doi: 10.7554/eLife.07571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Firpi HA, Ucar D, Tan K. Discover regulatory DNA elements using chromatin signatures and artificial neural network. Bioinformatics. 2010;26:1579–1586. doi: 10.1093/bioinformatics/btq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He B, Chen C, Teng L, Tan K. Global view of enhancer-promoter interactome in human cells. Proc Natl Acad Sci USA. 2014;111:E2191–E2199. doi: 10.1073/pnas.1320308111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bakay M, Pandey R, Hakonarson H. Genes involved in type 1 diabetes: An update. Genes (Basel) 2013;4:499–521. doi: 10.3390/genes4030499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fang D, Zhu J. Dynamic balance between master transcription factors determines the fates and functions of CD4 T cell and innate lymphoid cell subsets. J Exp Med. 2017;214:1861–1876. doi: 10.1084/jem.20170494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haned H, Benschop CCG, Gill PD, Sijen T. Complex DNA mixture analysis in a forensic context: Evaluating the probative value using a likelihood ratio model. Forensic Sci Int Genet. 2015;16:17–25. doi: 10.1016/j.fsigen.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 22.Huynh-Thu VA, Irrthum A, Wehenkel L, Geurts P. Inferring regulatory networks from expression data using tree-based methods. PLoS One. 2010;5:e12776. doi: 10.1371/journal.pone.0012776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haury AC, Mordelet F, Vera-Licona P, Vert JP. TIGRESS: Trustful inference of gene REgulation using stability selection. BMC Syst Biol. 2012;6:145. doi: 10.1186/1752-0509-6-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sánchez-Castillo M, et al. CODEX: A next-generation sequencing experiment database for the haematopoietic and embryonic stem cell communities. Nucleic Acids Res. 2015;43:D1117–D1123. doi: 10.1093/nar/gku895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gregory GD, Raju SS, Winandy S, Brown MA. Mast cell IL-4 expression is regulated by Ikaros and influences encephalitogenic Th1 responses in EAE. J Clin Invest. 2006;116:1327–1336. doi: 10.1172/JCI27227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomas RM, et al. Ikaros silences T-bet expression and interferon-gamma production during T helper 2 differentiation. J Biol Chem. 2010;285:2545–2553. doi: 10.1074/jbc.M109.038794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swafford AD, et al. An allele of IKZF1 (Ikaros) conferring susceptibility to childhood acute lymphoblastic leukemia protects against type 1 diabetes. Diabetes. 2011;60:1041–1044. doi: 10.2337/db10-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Angela M, et al. Fatty acid metabolic reprogramming via mTOR-mediated inductions of PPARγ directs early activation of T cells. Nat Commun. 2016;7:13683. doi: 10.1038/ncomms13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lochner M, Berod L, Sparwasser T. Fatty acid metabolism in the regulation of T cell function. Trends Immunol. 2015;36:81–91. doi: 10.1016/j.it.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Lu L, et al. Critical role of all-trans retinoic acid in stabilizing human natural regulatory T cells under inflammatory conditions. Proc Natl Acad Sci USA. 2014;111:E3432–E3440. doi: 10.1073/pnas.1408780111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown CC, et al. Retinoic acid is essential for Th1 cell lineage stability and prevents transition to a Th17 cell program. Immunity. 2015;42:499–511. doi: 10.1016/j.immuni.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burren OS, et al. T1DBase: Update 2011, organization and presentation of large-scale data sets for type 1 diabetes research. Nucleic Acids Res. 2011;39:D997–D1001. doi: 10.1093/nar/gkq912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hulbert EM, et al. T1DBase: Integration and presentation of complex data for type 1 diabetes research. Nucleic Acids Res. 2007;35:D742–D746, and erratum (2007) 35:6338. doi: 10.1093/nar/gkl933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smink LJ, et al. T1DBase, a community web-based resource for type 1 diabetes research. Nucleic Acids Res. 2005;33:D544–D549. doi: 10.1093/nar/gki095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrett JC, et al. Type 1 Diabetes Genetics Consortium Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finucane HK, et al. ReproGen Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; RACI Consortium Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bulik-Sullivan BK, et al. Schizophrenia Working Group of the Psychiatric Genomics Consortium LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Javierre BM, et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene ppromoters. Cell. 2016;167:1369–1384. doi: 10.1016/j.cell.2016.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smeets RL, et al. Molecular pathway profiling of T lymphocyte signal transduction pathways; Th1 and Th2 genomic fingerprints are defined by TCR and CD28-mediated signaling. BMC Immunol. 2012;13:12. doi: 10.1186/1471-2172-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allan SE, et al. Generation of potent and stable human CD4+ T regulatory cells by activation-independent expression of FOXP3. Mol Ther. 2008;16:194–202. doi: 10.1038/sj.mt.6300341. [DOI] [PubMed] [Google Scholar]
- 41.Kim JY, et al. Functional and genomic analyses of FOXP3-transduced Jurkat-T cells as regulatory T (Treg)-like cells. Biochem Biophys Res Commun. 2007;362:44–50. doi: 10.1016/j.bbrc.2007.07.187. [DOI] [PubMed] [Google Scholar]
- 42.Nie H, et al. Phosphorylation of FOXP3 controls regulatory T cell function and is inhibited by TNF-α in rheumatoid arthritis. Nat Med. 2013;19:322–328. doi: 10.1038/nm.3085. [DOI] [PubMed] [Google Scholar]
- 43.Baixauli F, et al. Mitochondrial respiration controls lysosomal function during inflammatory T cell responses. Cell Metab. 2015;22:485–498. doi: 10.1016/j.cmet.2015.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mingueneau M, et al. Loss of the LAT adaptor converts antigen-responsive T cells into pathogenic effectors that function independently of the T cell receptor. Immunity. 2009;31:197–208. doi: 10.1016/j.immuni.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 45.Takeda A, et al. Cutting edge: Role of IL-27/WSX-1 signaling for induction of T-bet through activation of STAT1 during initial Th1 commitment. J Immunol. 2003;170:4886–4890. doi: 10.4049/jimmunol.170.10.4886. [DOI] [PubMed] [Google Scholar]
- 46.Fiuza BSD, et al. Polymorphisms in DENND1B gene are associated with asthma and atopy phenotypes in Brazilian children. Mol Immunol. 2017;90:33–41. doi: 10.1016/j.molimm.2017.06.030. [DOI] [PubMed] [Google Scholar]
- 47.Nakamura M. Analysis of disease-pathway by identifying susceptible genes to primary biliary cirrhosis. Nihon Rinsho Meneki Gakkai Kaishi. 2012;35:503–510, and erratum (2013) 36:1. doi: 10.2177/jsci.35.503. [DOI] [PubMed] [Google Scholar]
- 48.Miao R, et al. Targeted disruption of MCPIP1/Zc3h12a results in fatal inflammatory disease. Immunol Cell Biol. 2013;91:368–376. doi: 10.1038/icb.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uehata T, et al. Malt1-induced cleavage of regnase-1 in CD4+ helper T cells regulates immune activation. Cell. 2013;153:1036–1049. doi: 10.1016/j.cell.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 50.MacArthur J, et al. The new NHGRI-EBI catalog of published genome-wide association studies (GWAS catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cortés JR, et al. Maintenance of immune tolerance by Foxp3+ regulatory T cells requires CD69 expression. J Autoimmun. 2014;55:51–62. doi: 10.1016/j.jaut.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radstake TR, et al. Increased frequency and compromised function of T regulatory cells in systemic sclerosis (SSc) is related to a diminished CD69 and TGFbeta expression. PLoS One. 2009;4:e5981. doi: 10.1371/journal.pone.0005981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ge Y, Paisie TK, Newman JRB, McIntyre LM, Concannon P. UBASH3A mediates risk for type 1 diabetes through inhibition of T-cell receptor-induced NF-κB signaling. Diabetes. 2017;66:2033–2043. doi: 10.2337/db16-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carpino N, et al. Regulation of ZAP-70 activation and TCR signaling by two related proteins, Sts-1 and Sts-2. Immunity. 2004;20:37–46. doi: 10.1016/s1074-7613(03)00351-0. [DOI] [PubMed] [Google Scholar]
- 55.Pesenacker AM, et al. CD161 defines the subset of FoxP3+ T cells capable of producing proinflammatory cytokines. Blood. 2013;121:2647–2658. doi: 10.1182/blood-2012-08-443473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garg G, et al. Type 1 diabetes-associated IL2RA variation lowers IL-2 signaling and contributes to diminished CD4+CD25+ regulatory T cell function. J Immunol. 2012;188:4644–4653. doi: 10.4049/jimmunol.1100272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L, et al. Mbd2 promotes foxp3 demethylation and T-regulatory-cell function. Mol Cell Biol. 2013;33:4106–4115. doi: 10.1128/MCB.00144-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ritter AT, Angus KL, Griffiths GM. The role of the cytoskeleton at the immunological synapse. Immunol Rev. 2013;256:107–117. doi: 10.1111/imr.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abu-Eid R, et al. Selective inhibition of regulatory T cells by targeting the PI3K-Akt pathway. Cancer Immunol Res. 2014;2:1080–1089. doi: 10.1158/2326-6066.CIR-14-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weirauch MT, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weintraub AS, et al. YY1 is a structural regulator of enhancer-promoter loops. Cell. 2017;171:1573–1588. doi: 10.1016/j.cell.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rhee EJ, Plutzky J. Retinoid metabolism and diabetes mellitus. Diabetes Metab J. 2012;36:167–180. doi: 10.4093/dmj.2012.36.3.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baena RM, et al. Vitamin A, retinol binding protein and lipids in type 1 diabetes mellitus. Eur J Clin Nutr. 2002;56:44–50. doi: 10.1038/sj.ejcn.1601279. [DOI] [PubMed] [Google Scholar]
- 64.Forga L, et al. Low serum levels of prealbumin, retinol binding protein, and retinol are frequent in adult type 1 diabetic patients. J Diabetes Res. 2016;2016:2532108. doi: 10.1155/2016/2532108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Driscoll HK, et al. Vitamin A status affects the development of diabetes and insulitis in BB rats. Metabolism. 1996;45:248–253. doi: 10.1016/s0026-0495(96)90062-1. [DOI] [PubMed] [Google Scholar]
- 66.Kadison A, et al. Retinoid signaling directs secondary lineage selection in pancreatic organogenesis. J Pediatr Surg. 2001;36:1150–1156. doi: 10.1053/jpsu.2001.25734. [DOI] [PubMed] [Google Scholar]
- 67.Van YH, et al. All-trans retinoic acid inhibits type 1 diabetes by T regulatory (Treg)-dependent suppression of interferon-gamma-producing T-cells without affecting Th17 cells. Diabetes. 2009;58:146–155. doi: 10.2337/db08-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stosić-Grujicić S, Cvjetićanin T, Stojanović I. Retinoids differentially regulate the progression of autoimmune diabetes in three preclinical models in mice. Mol Immunol. 2009;47:79–86. doi: 10.1016/j.molimm.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 69.Dawson HD, Collins G, Pyle R, Key M, Taub DD. The retinoic acid receptor-alpha mediates human T-cell activation and Th2 cytokine and chemokine production. BMC Immunol. 2008;9:16. doi: 10.1186/1471-2172-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hall JA, et al. Essential role for retinoic acid in the promotion of CD4+ T cell effector responses via retinoic acid receptor alpha. Immunity. 2011;34:435–447. doi: 10.1016/j.immuni.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klöting N, Klöting I. Congenic mapping of type 1 diabetes—protective gene(s) in an interval of 4 Mb on rat chromosome 6q32. Biochem Biophys Res Commun. 2004;323:388–394. doi: 10.1016/j.bbrc.2004.08.104. [DOI] [PubMed] [Google Scholar]
- 72.Klöting N, Klöting I. Genetic variation in the multifunctional transcription factor Yy1 and type 1 diabetes mellitus in the BB rat. Mol Genet Metab. 2004;82:255–259. doi: 10.1016/j.ymgme.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 73.Kleiman E, Jia H, Loguercio S, Su AI, Feeney AJ. YY1 plays an essential role at all stages of B-cell differentiation. Proc Natl Acad Sci USA. 2016;113:E3911–E3920. doi: 10.1073/pnas.1606297113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hnisz D, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351:1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smemo S, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Northcott PA, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Papp AC, Pinsonneault JK, Cooke G, Sadée W. Single nucleotide polymorphism genotyping using allele-specific PCR and fluorescence melting curves. Biotechniques. 2003;34:1068–1072. doi: 10.2144/03345dd03. [DOI] [PubMed] [Google Scholar]
- 79.Barrett JC. Haploview: Visualization and analysis of SNP genotype data. Cold Spring Harb Protoc. 2009;2009:pdb.ip71. doi: 10.1101/pdb.ip71. [DOI] [PubMed] [Google Scholar]
- 80.Auton A, et al. 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession no. GSE112342).