Significance
Chromatin dynamics play a central role in the DNA damage response (DDR). A longstanding experimental obstacle was the lack of technology capable of visualizing chromatin dynamics at double-strand break (DSB) sites. Here, we describe biophysical methods that quantify spatiotemporal chromatin compaction dynamics in living cells. Using these tools, we investigate dynamic remodeling of chromatin architecture at DSB loci and find that chromatin “opening” and “compacting” events facilitate repair factor access while demarcating the lesion from the surrounding nuclear environment. Furthermore, we identify regulatory roles for key DDR enzymes in this process. Finally, we demonstrate method utility through physical, pharmacological, and genetic manipulation of the chromatin environment, which collectively demonstrates the potential for its use in future studies of chromatin biology.
Keywords: chromatin organization, fluorescence lifetime imaging microscopy, DNA repair, spatiotemporal correlation spectroscopy, Förster resonance energy transfer
Abstract
To investigate how chromatin architecture is spatiotemporally organized at a double-strand break (DSB) repair locus, we established a biophysical method to quantify chromatin compaction at the nucleosome level during the DNA damage response (DDR). The method is based on phasor image-correlation spectroscopy of histone fluorescence lifetime imaging microscopy (FLIM)-Förster resonance energy transfer (FRET) microscopy data acquired in live cells coexpressing H2B-eGFP and H2B-mCherry. This multiplexed approach generates spatiotemporal maps of nuclear-wide chromatin compaction that, when coupled with laser microirradiation-induced DSBs, quantify the size, stability, and spacing between compact chromatin foci throughout the DDR. Using this technology, we identify that ataxia–telangiectasia mutated (ATM) and RNF8 regulate rapid chromatin decompaction at DSBs and formation of compact chromatin foci surrounding the repair locus. This chromatin architecture serves to demarcate the repair locus from the surrounding nuclear environment and modulate 53BP1 mobility.
The DNA damage response (DDR) is constantly scanning chromatin for genomic lesions (1). In the event that DNA damage is detected, the DDR signals the lesion’s presence, mediates downstream repair, and prevents cell division until chromatin restoration is complete (2). Following induction of a double-strand break (DSB), the ataxia–telangiectasia mutated (ATM) kinase localizes to the lesion to regulate cell-cycle arrest and signal recruitment of repair factors to the damaged chromatin (3–5). This includes the RNF8 E3 ubiquitin–protein ligase, which initiates ubiquitin signaling at the lesion that is essential for recruitment of additional repair factors, such as 53BP1 (6–9). In addition, the DDR also recruits proteins to the repair locus that contribute to chromatin decompaction or nucleosome eviction (10, 11). Paradoxically, factors mediating chromatin compaction, such as heterochromatin protein 1, also localize to DSBs to facilitate repair (12–14). Currently, it remains unclear how these contrasting chromatin dynamics coincide at a DSB repair locus and how this impacts chromatin macrostructure elsewhere in the nucleus. Interrogating DDR-dependent chromatin dynamics requires a technology capable of simultaneously probing chromatin structure at DNA lesions and throughout the entire nucleus, during the entirety of the DNA-repair process.
A basic feature of chromatin structure is the nanometer spacing (2–50 nm) between nucleosomes (15, 16). Given that the scale of this structural subunit is below the diffraction limit of optical and most superresolution imaging modalities, the rearrangements in chromatin structure during the DDR are rendered “invisible.” To get around this technical hurdle, fluorescence recovery after photobleaching (17), single-particle tracking (18, 19), and fluorescence correlation spectroscopy (20, 21) have been employed to measure molecular diffusion within the chromatin landscape and indirectly probe chromatin nanostructure (22, 23). These methods, however, do not directly observe chromatin compaction during DNA repair. What is needed to visualize changes in chromatin architecture is an experimental means to directly observe chromatin organization in live cells, at the level of nucleosome packaging, throughout the entirety of the nuclear volume and for the duration of the DDR. Furthermore, these data should be amenable to spatiotemporal image analysis to reveal underlying stability or dynamic alteration in the chromatin network.
To bridge this experimental gap, here, we establish the phasor approach to fluorescence lifetime imaging microscopy (FLIM) of Förster resonance energy transfer (FRET) interaction between fluorescently labeled histones. FRET is a phenomenon exquisitely sensitive to fluorophore proximity that can report on changes in nucleosome spacing when the core histone H2B is individually tagged to donor and acceptor fluorophores (24, 25). By coupling this readout of chromatin structure with DSB induction using near-infrared (NIR) laser microirradiation (26, 27), we are able to directly measure nanometer changes in chromatin compaction at a DNA damage site vs. globally throughout the nucleus. Furthermore, because phasor analysis enables fit-free quantification of spectroscopic properties recorded in each pixel of an image (28–30), FRET maps of chromatin foci are compatible with image-correlation spectroscopy (ICS) (31). ICS is a general approach to quantitate fluorophore distribution within an image (32, 33), and phasor transformation of local image correlation functions (PLICS) uncovers heterogeneity within fluorophore number, size, and spatial distribution (34). Therefore, in the context of histone FRET, phasor-based ICS enables nuclear-wide chromatin organization to be spatiotemporally characterized in live cells at the level of nucleosome proximity.
In this study, we use phasor-ICS analysis of histone FLIM-FRET in HeLa cells stably coexpressing low levels of H2B-eGFP and H2B-mCherry (HeLaH2B-2FP) to investigate how chromatin architecture is organized at DSBs by the DDR. We find that, following DSB induction with an NIR laser, ATM and RNF8 regulate chromatin decompaction at the damage site, coupled with formation of compact chromatin foci surrounding the repair locus. Correlation of chromatin architecture with measurements of 53BP1 accumulation and mobility reveal that DNA repair activity is restrained to the decompacted chromatin environment at the lesion and that the surrounding compact chromatin foci demarcate the DNA repair locus from the nuclear environment while controlling 53BP1 mobility. Collectively, these data reveal how chromatin architecture is organized by the DDR and demonstrate that phasor ICS analysis of histone FLIM-FRET is amenable to the study of local and global chromatin dynamics under conditions of physical, pharmacological, or genetic alteration.
Results
Phasor Analysis of Chromatin Compaction in Live Cells Using FLIM-FRET Microscopy.
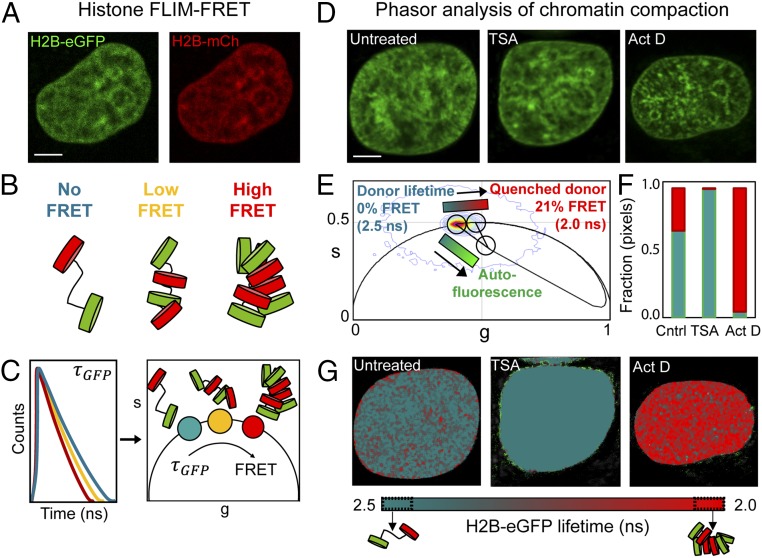
To assay chromatin compaction at the level of nucleosome proximity, we generated HeLaH2B-2FP cells stably coexpressing histone H2B-eGFP and histone H2B-mCherry (Fig. 1A and SI Appendix, Fig. S1). Tagged histone cDNAs were transduced via retrovectors and selected for transgene expression, and cells were sorted for low eGFP and mCherry expression to minimize the impact of exogenous transgene expression on cellular function. Stable coexpression of both histones into chromatin was confirmed by live cell imaging (SI Appendix, Fig. S1A), which also revealed no increase in underlying genome instability or mitotic abnormalities in HeLaH2B-2FP (SI Appendix, Fig. S1 B–F). Western blot analysis revealed that the cellular pool of H2B in the HeLaH2B-2FP cells was distributed as follows: 71.5 ± 2.7% endogenous, 9.7 ± 0.6% H2B-eGFP, and 18.8 ± 2.4% H2B-mCherry (mean ± SEM; SI Appendix, Fig. S2 A and B). Calculation of all nucleosome outcomes indicated that 51.1% of nucleosomes contained only endogenous H2B and 45.1% of the nucleosomes contained only one species of fluorescent tag (14.8% H2B-eGFP and 30.3% H2B-mCherry), while 3.7% of nucleosomes contained both an eGFP- and mCherry-tagged H2B as a potential source of intranucleosomal FRET (SI Appendix, Fig. S2C). Thus, on average, 18.5% of the nucleosomes in HeLaH2B-2FP contained a donor H2B-eGFP molecule.
Fig. 1.
Phasor approach to FLIM-FRET analysis of chromatin compaction. (A) HeLaH2B-2FP nucleus coexpressing H2B-eGFP and H2B-mCherry (H2B-mCh). (B) Graphical depiction of how increasing nucleosome proximity leads to increased FRET between fluorescent histones. (C) Graphical depiction of phasor transformation of HeLaH2B-2FP FLIM-FRET data. (C, Left) Fluorescence lifetime of H2B-eGFP reports on the degree of FRET interaction in each pixel. Each line represents the fluorescent lifetime from a different pixel. (C, Right) These data when phasor-transformed give rise to phasor coordinates (s, g). The donor phasor is right-shifted to shorter fluorescent lifetimes depending on the efficiency of FRET interaction. In HeLaH2B-2FP, decreasing lifetime and increasing FRET corresponds to more compact chromatin. (D) Untreated, TSA-treated, or Actinomycin D (Act D)-treated HeLaH2B-2FP nuclei, shown in the H2B-eGFP channel. (E) Combined phasor distribution of H2B-eGFP fluorescence lifetime from all conditions shown in D with the theoretical FRET trajectory superimposed to determine the range of FRET efficiencies in HeLaH2B-2FP. The linear combination of unquenched donor and background cellular autofluorescence (teal–bright green) (defined in SI Appendix, Fig. S3) follows a distinct trajectory from FRET (teal–red). (F) Fraction of pixels in a compact (red) vs. open (teal) chromatin state in control (Cntrl), TSA-, and Act D-treated cells. (G) Pseudocolored chromatin compaction maps of the cells in D according to the palette defined in the phasor plot data in E. (Scale bars, 5 μm.)
We then established a phasor lifetime-analysis workflow to quantify the local chromatin compaction status within each pixel of a FLIM-FRET image acquired in a HeLaH2B-2FP nucleus. FRET detects molecular interaction within 1–10 nm and the efficiency of energy transfer between a donor and acceptor molecule is exquisitely sensitive to distance (35). Thus, an increasing FRET efficiency in HeLaH2B-2FP corresponds to a reduced spacing between nucleosomes containing H2B-eGFP (donor) and nucleosomes containing H2B-mCherry (acceptor) (graphically depicted in Fig. 1B). FRET efficiency can be quantified by measurement of the fluorescence lifetime of H2B-eGFP, as this property is increasingly quenched upon closer interaction with H2B-mCherry. Therefore, in a FLIM-FRET image of HeLaH2B-2FP, which is diffraction limited, the fluorescence lifetime recorded in each pixel reports the ensemble average of FRET events occurring on a spatial scale of 1–10 nm, enabling us to sample chromatin architecture in terms of nucleosome proximity.
To measure the fluorescence lifetime of H2B-eGFP in HeLaH2B-2FP nuclei, we used two-photon excitation on a laser-scanning confocal microscope equipped with time-resolved detection. For translation of fluorescence lifetime into FRET efficiency, we employed phasor analysis (graphically depicted in Fig. 1C). Phasor analysis transformed the fluorescence lifetime recorded in each pixel of a FLIM image into a 2D coordinate system (g, s) (SI Appendix; refs. 28 and 29). When these coordinates are placed in a single phasor plot, the phasor coordinates derived from each pixel give rise to a series of clusters, which fingerprint all of the measured FRET efficiencies within a FLIM image. Specifically, pixels in a HeLaH2B-2FP nucleus that contain compacted chromatin, and therefore exhibit a reduced H2B-eGFP fluorescence lifetime, have an increasingly right-shifted phasor value with respect to the unquenched lifetime of H2B-eGFP. Thus, the phasor method enables quantitation of FRET in each pixel of a FLIM image and spatially maps the different chromatin states throughout a HeLaH2B-2FP nucleus.
To establish the dynamic range of chromatin compaction states in our experimental system, we treated HeLaH2B-2FP with the histone deacetylase inhibitor Trichostatin A (TSA), which has been shown to loosen chromatin structure (22), or the transcriptional inhibitor Actinomycin D to compact the chromatin network (36) (Fig. 1D). Phasor coordinates recorded from the FLIM data in untreated and treated cells were collated into a single phasor plot (Fig. 1E) and found to induce varying degrees of chromatin compaction between a fluorescence lifetime of 2.5 ns (TSA treatment, low compaction) and 2.0 ns (Actinomycin D treatment, high compaction). To translate this lifetime shift into FRET efficiency, we superimposed the theoretical FRET trajectory that H2B-eGFP would adopt upon interaction with H2B-mCherry over the measured phasor distribution (defined by SI Appendix, Eq. S7). From independent measurement of background autofluorescence in parental HeLa cells and the unquenched donor lifetime in HeLa transduced with H2B-eGFP (HeLaH2B-GFP) (SI Appendix, Fig. S3 A–E), the FRET trajectory presented in Fig. 1E enables us to: (i) quantify the phasor location of “open” vs. “compacted” chromatin to correspond to a 0–21% change in FRET efficiency; and, most importantly, (ii) resolve this readout of chromatin compaction from cellular autofluorescence.
Throughout this study, we employed a phasor-based cursor analysis centered on the FRET trajectory defined in Fig. 1E to quantify and pseudocolor the heterogeneous chromatin compaction states recorded in each pixel of a FLIM acquisition (teal–red) independently from background (teal–bright green) (Fig. 1 E–G). FLIM measurement of HeLaH2B-GFP treated with TSA and Actinomycin D confirmed that the local chromatin environment had negligible impact on the fluorescence lifetime of H2B-eGFP compared with HeLaH2B-2FP where H2B-mCherry is present (SI Appendix, Fig. S3F). FLIM analysis of the acceptor channel in HeLaH2B-2FP verified that the shortened H2B-eGFP lifetimes detected in our FRET maps (Fig. 1G) spatially correlated with a delay in the lifetime of H2B-mCherry (SI Appendix, Fig. S4 A–D), and thus our phasor cursor analysis does accurately report bona fide FRET in HeLaH2B-2FP.
Image Correlation Analysis of FRET Quantifies Global Chromatin Network Organization.
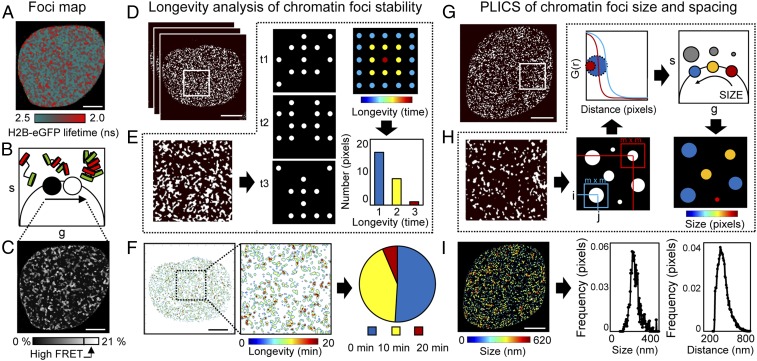
Global chromatin organization is dynamic and nonrandom (37, 38). Thus, next, we developed methodology to quantify the temporal stability and spatial reorganization of local chromatin compaction, as detected by live-cell phasor FLIM-FRET imaging, in a single cell over time. To quantify the stability, size, and spacing of compact chromatin foci, we first extracted the coordinates of pixels in a high-FRET state within a FLIM acquisition (16–21% FRET efficiency) and derived a map of compact chromatin localization (Fig. 2 A–C). To identify which compact chromatin foci persisted in a cell over time, we then developed an image-correlation analysis method termed longevity analysis (Fig. 2 D–F). In longevity analysis, a moving average (MAV) is applied across a time series of chromatin compaction maps derived from FLIM-FRET data (Fig. 2D) to produce a longevity map (graphically presented in Fig. 2E). The amplitude recorded in each pixel of a longevity map directly reports how long a structure persists in time. Longevity analysis therefore enabled us to extract and spatially map stable vs. dynamic compact chromatin foci (Fig. 2 F, Left) and provided a quantitative readout of chromatin network dynamics that can be tracked throughout the nucleus for the entirety of the DDR (Fig. 2 F, Right).
Fig. 2.
ICS applied to a phasor FLIM-FRET map measures nuclear-wide compact chromatin stability, size, and spacing. (A–C) Foci map. (A) FLIM-FRET map from an unirradiated HeLaH2B-2FP nucleus. (B and C) Pixel coordinates of the high FRET state (16–21% FRET) can be extracted from the phasor plot (B) to produce a localization map of compact chromatin (C). (D–F) Longevity analysis. (D) Localization map of compacted chromatin from the HeLaH2B-2FP cell in A at 0, 10, and 20 min (as detected by FRET). (E) Schematic of longevity analysis: Averaging three binary images gives rise to a heat map of pixel longevity, which contains structural information that is not evident in the source images and can be used to quantify the stability of detected structures. (F) Longevity map of the compacted chromatin foci detected and tracked in D, with digital enlargement shown for a ROI that contains foci present for 10 min (green pixels) and 20 min (red pixels). The fraction of foci persistence across the time course is calculated as a measure of overall chromatin network stability. (G–I) PLICS analysis. (G) A localization map of compacted chromatin from an unirradiated HeLaH2B-2FP nucleus as detected by FLIM-FRET. (H) Schematic of PLICS analysis: In a binary image showing different-sized structures, we can calculate localized 2D spatial correlation functions using an m × m matrix and by collapsing them into 1D correlation profiles. The resulting decay is characteristic of the size of a structure within each m × m matrix. By transforming each decay into phasor coordinates (g, s), we can graphically pseudocolor each pixel according to size. (I) Size map of compacted chromatin foci detected in G and PLICS/iPLICS analysis of the average size (305 nm) and spacing (485 nm). (Scale bars, 5 μm.)
Longevity analysis identifies when and where compact chromatin foci undergo a rearrangement. However, it does not reveal how compact chromatin spatially redistributes within a given time window. To visualize this, we established an additional method based on PLICS (34) to uncover the size and spacing between compact chromatin foci (Fig. 2 G–I). PLICS analysis started with a localized spatial autocorrelation analysis within square (m × m) matrices that surround each pixel (i, j) in a derived chromatin-localization map (graphically presented in Fig. 2H). This gave rise to a series of 2D autocorrelation functions (ACFs) that, when collapsed into a one-dimensional (1D) decay, described the size of the objects centered at that pixel. The local ACF in each pixel was then transformed into a set of phasor coordinates (g, s), which, when placed in a single phasor plot, identified the different-sized compact chromatin structures (Fig. 2 H, Upper). The phasor coordinates corresponding to different-sized compact chromatin foci were then pseudocolored and spatially mapped back to the source image (Fig. 2 H, Lower Right).
Next, we extended PLICS analysis to an “inverse PLICS” (iPLICS) approach that enabled us to identify the spacing between compact chromatin clusters (SI Appendix, Fig. S5A). Here, the negative of a chromatin compaction map was first derived. Then, when these data were analyzed through the PLICS workflow, we calculated localized 2D spatial correlation functions that, when collapsed into a 1D ACF, described the characteristic spacing between structures within a square (m × m) matrix. Transforming each decay into phasor coordinates (g, s) enabled us to identify spacing between compact chromatin foci and pseudocolor the source image. To demonstrate the feasibility of PLICS and iPLICS, we simulated an image containing circular binary structures with a diameter of three pixels that were evenly spaced by eight pixels and correctly recovered the size and spatial distribution of these structures (SI Appendix, Fig. S5 B and C). Thus, when PLICS and iPLICS were applied to the HeLaH2B-2FP nucleus shown in Fig. 2G, we were able to detect compact chromatin foci between 0 and 620 nm (Fig. 2 I, Left), with an average compact chromatin foci size of 305 nm and spacing of 485 nm between structures (Fig. 2 I, Right). Collectively, these parameters characterized in a single cell the nuclear-wide spatial organization of local chromatin compaction at the level of nucleosome proximity, with submicrometer resolution.
Chromatin Compaction Is Spatiotemporally Controlled During the DDR.
We next coupled our phasor FLIM-FRET workflow with NIR laser microirradiation to investigate chromatin compaction dynamics during the DDR in live HeLaH2B-2FP cells. This multiplexed approach enabled induction of DSBs within a defined region of interest (ROI) and measurement of local vs. global rearrangements in chromatin structure as a function of time. NIR laser microirradiation can be tuned to induce different types of DNA damage by modulating the laser frequency, wavelength, or power (10, 27). To determine conditions that generate DSBs in HeLaH2B-2FP, we identified laser settings that induced lesions which recruited eGFP–53BP1 to the irradiated ROI in transiently transfected parental HeLa cells (SI Appendix, Fig. S6) (26). These settings were used for the remainder of the experimentation described below.
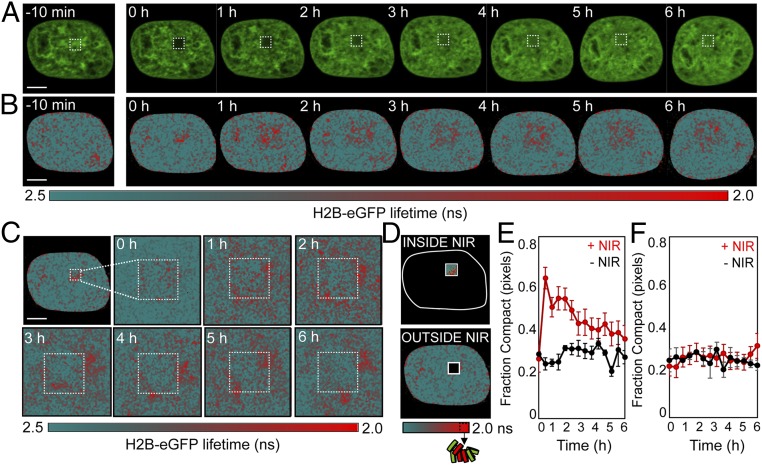
For all experiments, we measured the fluorescence intensity (Fig. 3A) and lifetime (Fig. 3B) of H2B-eGFP in each pixel of a HeLaH2B-2FP nucleus starting 10 min before NIR irradiation and then at regular time intervals from 0 min to 6 h after DSB induction. From the FLIM maps derived during the DDR (Fig. 3B), we observed that 0 min after NIR laser microirradiation, chromatin compaction was induced at the periphery of the DNA damage site (red pixels). Then, within the following 60 min, this compacted chromatin architecture rearranged outside the DNA damage site to form a border of compacted chromatin foci surrounding a central region of open chromatin at the DNA lesion (Fig. 3C). We noted that laser microirradiation did partially photobleach the NIR-treated ROI (Fig. 3A and SI Appendix, Fig. S7A), and, while FLIM is in theory concentration-independent, a reduction in H2B-eGFP fluorescence would result in an increase in the contribution of background signal in the NIR-treated ROI. However, because the linear combination of cellular autofluorescence and unquenched donor followed a distinct phasor trajectory from that of energy transfer between H2B-eGFP and H2B-mCherry, this artifact was resolvable from FRET (SI Appendix, Fig. S3).
Fig. 3.
FLIM-FRET analysis of chromatin compaction reveals chromatin architectural changes during the DDR. (A and B) Time series of H2B-eGFP fluorescence intensity images (A) and lifetime maps (B) acquired in a HeLaH2B-2FP 10 min before and at hourly intervals after NIR irradiation. The white square indicates the NIR laser-treated locus. (C) Digital enlargement of the DNA damage site selected in B and the corresponding time series of lifetime maps within this ROI. (D) Masks selected for analysis of the number of pixels in a compacted (high-FRET) vs. noncompacted (low-FRET) state at the DNA damage site vs. outside this ROI. (E and F) Fraction of pixels within (E) and outside (F) of the NIR-irradiated ROI that are in a compacted state during the DDR (red curve) vs. an unperturbed cell (black curve) (n = 10 cells, mean ± SEM). (Scale bars, 5 μm.)
To quantify the temporal progression of chromatin compaction at the DNA damage site, and any potential changes in global chromatin network organization, we next quantified the fraction of pixels in a compacted state inside and outside of the NIR-treated ROI in HeLaH2B-2FP (Fig. 3D). This analysis revealed: (i) the fraction of pixels with compact chromatin inside the NIR-treated locus sharply increased in the first 30 min after DSB induction and persisted for up to 3 h (Fig. 3E); and (ii) there was no significant change in the percentage of pixels with compact chromatin in the nuclear-wide undamaged chromatin network outside of the NIR-treated ROI (Fig. 3F). Comparison of this result with NIR treatment in HeLaH2B-GFP (SI Appendix, Fig. S7 B and C) confirmed that, in the absence of H2B-mCherry, the increase in autofluorescence at the DNA damage site had negligible impact on our capacity to detect FRET at this location in the hour following DSB induction in HeLaH2B-2FP (SI Appendix, Fig. S7 D and E).
Uniform Induction and Heterogenous Resolution of Chromatin Organization in the DDR.
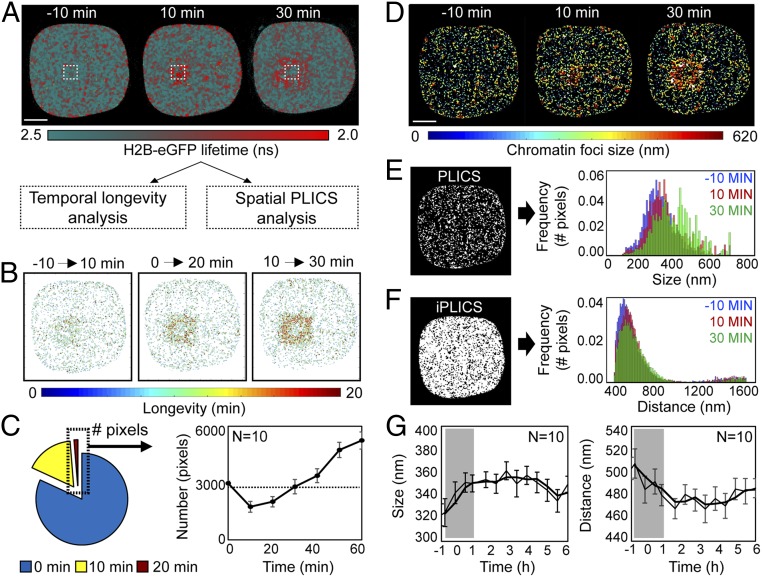
To reveal the underlying spatiotemporal dynamics of compact chromatin foci formation and propagation at induced DSBs, we applied the longevity, PLICS, and iPLICS workflow to phasor FLIM-FRET datasets from NIR-irradiated HeLaH2B-2FP cells (Fig. 4A). Longevity analysis was performed by using an MAV of 10 min during the first hour after NIR irradiation (Fig. 4B) and every hour for 6 h postirradiation (SI Appendix, Fig. S8 A and B). This revealed induction of stable compact chromatin foci around the damage site 10–30 min postirradiation (Fig. 4B). Plotting the number of stable compact chromatin foci formed as a function of time revealed the chromatin network to be dynamic during the first 30 min after DSB induction, before stabilizing by 60 min (Fig. 4C). While stable compact chromatin foci were uniformly generated in all cells analyzed within the first 30 min following irradiation, we observed intercell heterogeneity in resolution of this structure over the course of 6 h (SI Appendix, Fig. S8C).
Fig. 4.
A compact chromatin border forms around the lesion site within the first 30 min of the DDR. (A) FLIM-FRET maps acquired in a HeLaH2B-2FP cell during the first 30 min after NIR irradiation. (B) Longevity maps of compact chromatin foci taken at 10-min intervals over a 20-min duration. (C) Quantification of the number of stable compact chromatin pixels across multiple cells (n = 10, mean ± SEM) in the first hour following NIR-induced DSBs (dashed line represents average number of stable compact chromatin pixels in an unperturbed cell). (D) Chromatin compaction size maps derived by PLICS analysis 0–30 min after DSB induction. (E) PLICS analysis of the histogram of sizes induced by DSBs in the data presented in D. (F) iPLICS analysis of the histogram of distance changes induced by DSBs in the data presented in D. (G) Change in mean size and distance between compacted chromatin foci as a function of time during the DDR (n = 10 cells, mean ± SEM). (Scale bars, 5 μm.)
Applying PLICS and iPLICS to the same dataset revealed that, in the first 30 min following NIR laser irradiation, there was an increase in the size of compact chromatin foci surrounding the NIR-treated ROI (Fig. 4 D–G). This local chromatin reorganization gave rise to an overall increase in the detected compact chromatin foci size (Fig. 4E) and a subtle reduction in the global spatial distribution of compacted chromatin foci during the first 30 min following DSB induction (Fig. 4F). Similar to longevity analysis, PLICS and iPLICS demonstrated a uniform increase across multiple cells in compact chromatin foci size (320 nm up to 370 nm) and decrease in spacing between foci (505 nm down to 470 nm) within the first 30 min of the DDR (Fig. 4G). Thus, the stable border of compact chromatin detected by longevity analysis at the perimeter of the DNA damage site arose from a local increase in chromatin foci size.
ATM and RNF8 Regulate DDR-Dependent Chromatin Dynamics.
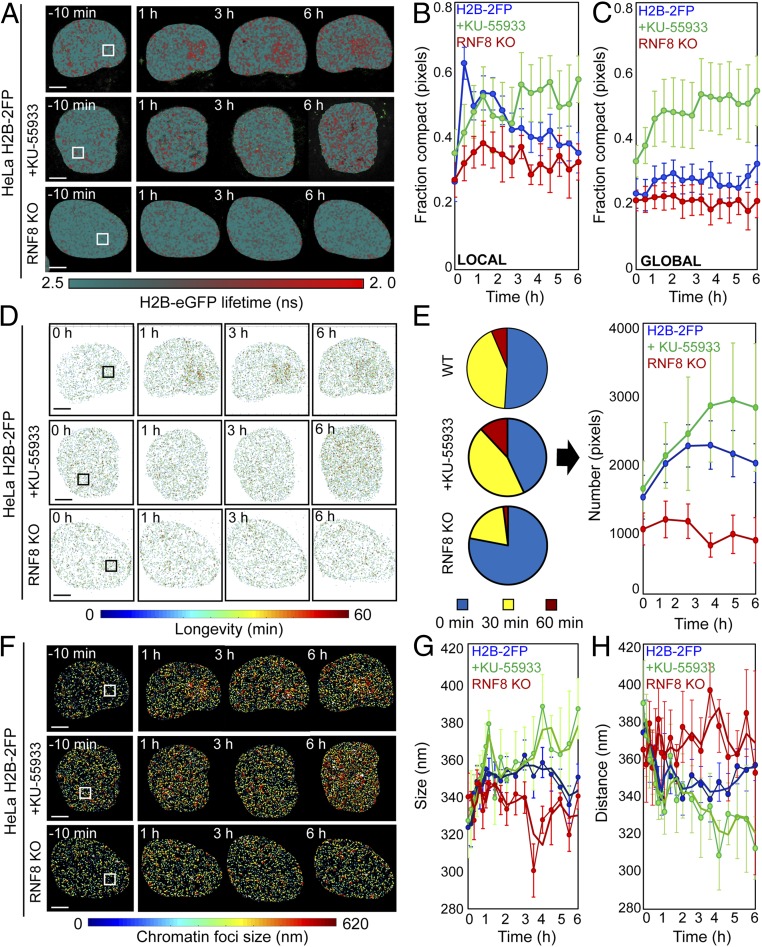
To probe DDR-dependent regulation of chromatin architecture, we inhibited key DDR enzymes and measured the impact on chromatin dynamics following DSB induction. For these experiments, we targeted ATM kinase with the potent and specific inhibitor KU-55933 (39). Additionally, we used CRISPR/Cas9 to genetically delete RNF8 in HeLaH2B-2FP. ATM inhibition in HeLaH2B-2FP cells was confirmed by Western blot (SI Appendix, Fig. S9A), and RNF8 deletion in HeLaH2B-2FP RNF8 knockout (KO) cells was confirmed by sequencing the targeted locus and visualizing suppressed 53BP1 recruitment to DSBs induced by ionizing or NIR laser irradiation (SI Appendix, Fig. S9 B–E). We acquired phasor FLIM-FRET data in KU-55933–treated HeLaH2B-2FP cultures and HeLaH2B-2FP RNF8 KO cells 10 min before and from 0 min to 6 h after NIR irradiation (Fig. 5A). ATM inhibition corresponded with an inhibition of acute chromatin compaction surrounding the lesion site within the first hour after break induction (green curve, Fig. 5B) and showed a gradual increase in global chromatin compaction throughout the experiment (green curve, Fig. 5C). RNF8 deletion suppressed both local rearrangement of chromatin architecture at the lesion (red curve, Fig. 5B) and inhibited alteration of global chromatin compaction following break induction (red curve, Fig. 5C).
Fig. 5.
ATM and RNF8 regulate chromatin architecture in the DDR. (A) FLIM-FRET maps of HeLaH2B-2FP, HeLaH2B-2FP treated with KU-55933, or HeLaH2B-2FP RNF8 KO cells 10 min before and 1, 3, and 6 h after microirradiation. (B and C) Fraction of compacted chromatin pixels within (B) and outside (C) of the NIR irradiation site in HeLaH2B-2FP (blue curve; n = 10), KU-55933-treated (green curve; n = 6), or RNF8 KO cells (red curve; n = 3; mean ± SEM). (D) Longevity maps of compact chromatin foci from the cells in A. (E) Quantification of compact chromatin foci stability (Left) throughout the DDR (Right) in NIR laser-treated HeLaH2B-2FP, KU-55933–treated HeLaH2B-2FP, and HeLaH2B-2FP RNF8 KO cells (n as indicated above, mean ± SEM). (F) Chromatin compaction size map derived by PLICS from the cells shown in A. (G and H) Change in the mean size of (G) and distance between (H) compacted chromatin foci during the DDR in NIR laser-treated HeLaH2B-2FP, KU-55933–treated HeLaH2B-2FP, and HeLaH2B-2FP RNF8 KO cells (n as indicated above, mean ± SEM). (Scale bars, 5 μm.)
Longevity analysis revealed that both ATM inhibition and RNF8 deletion disrupted formation of the stable chromatin border surrounding the damage site (Fig. 5D). ATM inhibition and RNF8 deletion also altered global compact chromatin stability over the 6-h imaging duration following DSB induction (Fig. 5E). Specifically, KU-55933 conferred the gradual accumulation of stable compact chromatin foci throughout the nucleus with no geographical connection to the NIR-treated ROI, while RNF8 deletion resulted in more dynamic chromatin throughout the experiment. Likewise, PLICS and iPLICS analysis revealed that both KU-55933 and RNF8 KO suppressed the increase in compacted chromatin foci size in the first 30 min after break induction (Fig. 5F). Over the course of the experiment, ATM inhibition corresponded with a persistent and gradual increase in compact chromatin foci size and reduced spacing throughout the nucleus, irrespective of the damage site (green curves, Fig. 5 G and H). Conversely, RNF8 KO cells fluctuated between significant increases and decreases in compact chromatin foci size and spacing, without exhibiting any trend (red curves, Fig. 5 G and H). Together, these results demonstrate that ATM and RNF8 spatiotemporally regulate chromatin decompaction and compaction activities at the DSB locus and the surrounding global chromatin network organization. We note that the nuclear-wide gradual increase in chromatin compaction with KU-55933 treatment is a phenotype that continued after DSB induction and occurred geographically, independent of damage location. We anticipate that this reflects an off-target effect of KU-55933 or a cellular response to acute ATM inhibition that is independent of the cellular response to DSBs.
Chromatin Architecture Demarcates DSB Repair Locus Boundaries.
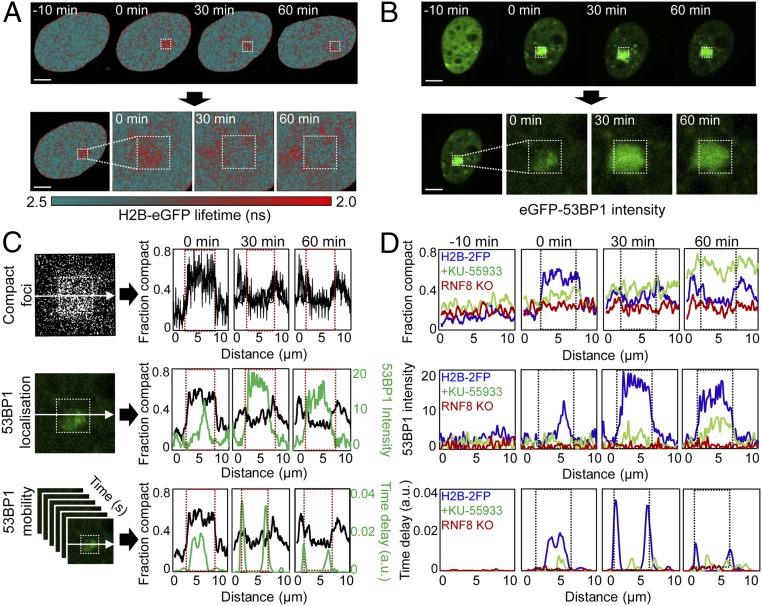
To determine how chromatin architecture at DSBs relates to the local DNA repair environment, we correlated the spatial localization of compact chromatin in HeLaH2B-2FP with eGFP-53BP1 localization and mobility in parental HeLa cells. To measure 53BP1 mobility, we employed pair-correlation analysis: a branch of spatiotemporal correlation spectroscopy demonstrated in a study of Ku-GFP dynamics during DSB repair (26). Pair-correlation analysis is based on the acquisition of fluctuations in fluorescent intensity in each pixel of a line or frame scan (40, 41) and then spatial cross-correlation of pairs of fluorescent fluctuations located outside and inside a DSB site, to determine the mobility of a fluorescently tagged DNA-repair factor within or adjacent to a lesion (26). For these experiments, HeLa parental cells were transiently transfected with eGFP-53BP1, and then eGFP-53BP1 mobility was monitored before and during the first hour after DSB induction via NIR laser microirradiation. By employing the exact same NIR laser microirradiation settings used for the FLIM-FRET experiments presented in Figs. 3–5, eGFP-53BP1 mobility data acquired in parental HeLa cells could be spatially linked with the observed chromatin dynamics in HeLaH2B-2FP (Fig. 6 A and B).
Fig. 6.
Chromatin architecture demarcates the repair locus. (A) FLIM-FRET maps acquired in HeLaH2B-2FP cells 10 min before and 0, 30, and 60 min after microirradiation (Upper) and expanded images of the DSB ROI (Lower). (B) eGFP-53BP1 intensity images acquired in HeLa cells 10 min before and 0, 30, and 60 min after microirradiation (Upper) and expanded images of the DSB ROI (Lower). (C) Correlation of compact chromatin foci localization along the horizontal axis as a function of time (Top; image from 0 min is shown), with 53BP1 localization (Middle; image from 0 min is shown), and mobility in terms of time delay (Bottom). eGFP-53BP1 localization and mobility are averaged along the horizontal axes (green plots). Red dashed box indicates laser microirradiation ROI. (D) Comparison of compact chromatin foci localization (Top), eGFP-53BP1 localization (Middle), and eGFP-53BP1 mobility (Bottom) in untreated HeLa cells (blue curve), KU-55933–treated HeLa cells (green curve), or RNF8 KO cells (red curve) 10 min before and 0, 30, and 60 min after microirradiation. Representative example of n = 3 shown. (Scale bars, 5 μm.)
Quantitation of chromatin compaction along a line intersecting the NIR laser-treated focus in HeLaH2B-2FP confirmed that by 30 min after DSB induction, compact chromatin foci adopted a localization at the periphery of the damage site (Fig. 6 C, Top). Starting within the first minute following break induction, eGFP-53BP1 accumulated at the NIR-laser–treated locus, reaching maximum intensity by 30 min (Fig. 6 C, Middle). Notably, the radial spread of eGFP-53BP1 was constrained to the decompacted interior of the DNA damage site. We then plotted the time delay of 53BP1 movement by performing pair correlation across the same line intersecting the NIR-laser–treated focus (SI Appendix, Fig. S10). These data revealed delayed eGFP-53BP1 mobility at the break 0 min after DNA damage (Fig. 6 C, Bottom). Then, by 30–60 min, delayed eGFP-53BP1 mobility was constrained to the interface of the compact chromatin border. Notably, the detected eGFP-53BP1 dynamics superimposed spatiotemporally with outward propagation of the compact chromatin foci and spread of eGFP-53BP1 accumulation. Also, ATM inhibition and RNF8 deletion in parental HeLa suppressed eGFP-53BP1 localization and delayed mobility at the damage site (Fig. 6D). Within the context of chromatin architecture, eGFP-53BP1 accumulated in the decompacted chromatin internal to the break while being excluded from the compact chromatin surrounding the laser-treated ROI. We interpret these data to indicate that establishment of a transient compact chromatin border demarcates the repair locus from the surrounding chromatin environment.
Discussion
In this study, we established a biophysical method to measure chromatin organization in live cells using the phasor approach to FLIM-FRET analysis of fluorescently labeled histones. Coupling this technology with ICS and laser microirradiation enabled us to identify DDR-dependent chromatin architectural changes that occur with DSBs. In doing so, we found that ATM and RNF8 regulate formation of compact chromatin foci at the periphery of a DSB-repair locus, and these chromatin dynamics modulate 53BP1 mobility.
A major advance of this study is that the imaging and analysis tools we have developed enable local chromatin structure to be measured on a nuclear-wide scale in a living cell, while maintaining sensitivity toward heterogeneity of individual chromatin foci dynamics during the DDR. We achieved this through phasor analysis of FLIM and image-correlation spectrums. Phasor provides a fit-free solution that does not require a priori knowledge of system complexity (28, 34). Instead, phasor analysis of histone FLIM-FRET directly maps the chromatin compaction status at each pixel, independent of the multitude of potential compaction states. This, in turn, enabled us to extract the localization of specific chromatin foci, assess their temporal stability by longevity analysis, and characterize spatial organization in terms of individual chromatin foci size or spacing by PLICS and iPLICS.
Together, our longevity and PLICS analysis revealed the uniform alteration of chromatin architecture within the first 30 min following damage induction, which is consistent with a highly regulated process. Resolution of chromatin architecture occurs thereafter with cell-to-cell heterogeneity over 3–6 h. This is consistent with the kinetics of DDR focus resolution, and in agreement with resolution of chromatin alteration at DSBs occurring with completion of repair. We anticipate that the observed resolution heterogeneity reflects differing cell-to-cell repair kinetics, dependent upon the genetic location where breaks occurred, the cell cycle, or other unknown factors (42, 43). Critically, it suggests that DDR-dependent changes in chromatin architecture are tightly regulated through the repair process from break induction to repair completion. Additionally, these observations identify that our analysis is sufficiently sensitive to monitor single-cell heterogeneity in chromatin macrostructure.
Correlation between our phasor FLIM-FRET and eGFP-53BP1 pair-correlation analysis provided insight into the relationship between chromatin architecture and establishment of the repair focus. Immediately following break induction, delayed 53BP1 mobility was observed at the damage site. By 30 min, 53BP1 accumulation had maximized, and mobility was not detected within the decompacted chromatin interior of the repair focus. At the same time, delayed 53BP1 mobility was observed only at the interface of the expanding compact chromatin border. These data are consistent with 53BP1 binding to the decompacted chromatin, as this substrate becomes available with outward expansion of the compact chromatin border. Mobility of 53BP1 is detected at the chromatin border as this protein continues to localize to the break, but its binding is restricted within the compact chromatin environment. We interpret these findings to signify that the compacted chromatin boundary serves to distinguish the decompacted repair locus from the remainder of the nucleus.
The significance of this physical separation of the repair locus from the surrounding environment may provide an important function to the DNA-repair process. Chromatin decompaction and nucleosome eviction is proposed to ease access of DNA-repair factors to their DNA substrate and increase repair kinetics (11, 44). Conversely, the compacted chromatin border excludes 53BP1 retention and may also inhibit the spurious binding of other DNA-repair factors. We also predict that the compacted chromatin border likely contributes to DDR-dependent transcriptional inhibition, which is predicted to halt passage of RNA polymerases through a DDR locus to enable efficient repair (45). DDR-dependent transcriptional silencing occurs in cis and is dependent upon ATM and RNF8 (5). This is consistent with our observation that formation of the compact chromatin border surrounding the damage focus is ATM- and RNF8-dependent.
Consistent with their key function in the DDR, we identified that ATM and RNF8 regulate chromatin architecture at NIR laser-induced DSBs. RNF8-dependent chromatin decompaction is also consistent with a role for this protein in regulating the local chromatin environment to promote assembly of checkpoint and repair machineries at DNA lesions (46). As RNF8 activity and chromatin ubiquitylation are essential for 53BP1 binding (7–9), our data suggest that the interior decompacted chromatin environment coincides with the domain of ubiquitylated chromatin at the break. Cumulatively, our data provide a macrostructural understanding of how chromatin architecture is arranged at the DDR focus. Furthermore, they imply that ATM- and RNF8-dependent regulation of a compact chromatin border serves to demarcate and arrange the repair locus to simultaneously balance DNA-repair factor accumulation and potentially transcriptional shutdown of neighboring genes.
In conclusion, phasor histone FLIM-FRET analysis coupled with laser microirradiation enabled us to study DDR-dependent chromatin dynamics with unprecedented spatiotemporal resolution at the single-cell level. While we focused on the DDR in this study, the methodology presented here is agnostic to research topic and, as we have shown, is easily combined with physical, pharmacological, or genetic manipulation of the underlying system. As chromatin dynamics underlie a wide range of biological functions, we anticipate that the analytical tools presented here are widely applicable and will contribute to advancing our understanding of how genomes work in the context of living cells.
Methods
Plasmids for Cell Line and Vector Generation.
pLXSN H2B-mCherry was a gift from Laure Crabbe, The Centre for Integrative Biology, Toulouse, France (47). pWZL H2B-eGFP was created by PCR-amplifying H2B-eGFP from eGFP-N1 H2B-eGFP (a gift from Geoff Wahl, The Salk Institute for Biological Sciences, La Jolla, CA; Addgene plasmid no. 11680) (48) using the BamHI H2B-eGFP forward and EcoRI H2B-eGFP reverse oligonucleotides (SI Appendix, Table S1), followed by in-fusion (Clontech) cloning into BamHI- and EcoRI-digested (New England Biolabs) pWZL vector (a gift from Scott Lowe, Memorial Sloan Kettering Cancer Center, New York; Addgene no. 18750). pLentiCRISPRv2 was a gift from Feng Zhang, Massachusetts Institute of Technology, Cambridge, MA (Addgene plasmid no. 52961) (49). pLentiCRISPRv2-RNF8-2 was generated by cloning an sgRNA sequence targeting RNF8 exon 1 by annealing the sgRNF8-2 sense and antisense oligonucleotides (SI Appendix, Table S1) and in-fusion cloning into BsmBI-digested pLentiCRISPRv2. pcDNA5-FRT/TO-eGFP-53BP1 was a gift from Dan Durocher, The Lunenfeld-Tanenbaum Research Institute, Toronto (Addgene plasmid no. 60813) (6). All plasmids were verified by Sanger sequencing (Australian Genome Research Facility).
Cell Culture and Treatment.
HeLa cells were originally provided by Jan Karlseder, Salk Institute, La Jolla, CA. Cells were grown in DMEM (Life Technologies) supplemented with 1% nonessential amino acids (Life Technologies), 1% glutamax (Life Technologies), and 10% bovine growth serum at 37 °C, 10% CO, and 3% O2. During FLIM-FRET imaging, cells were cultured in DMEM (Lonza) and supplemented with 10% FBS (Gibco), Hepes (Gibco), and 1x Pen-Strep (Lonza). Cell-line identity was confirmed by using tandem repeat profiling by Cell Bank Australia (Children's Medical Research Institute), and cell lines were confirmed as Mycoplasma negative (MycoAlert; catalog no. LT07-118; Lonza). To establish unquenched fluorescence lifetime of H2B-eGFP, identify laser microirradiation conditions, or measure DNA-repair factor mobility, HeLa cells were transiently transfected with pWZL H2B-eGFP or eGFP-53BP1 by using Lipofectamine 3000, according to manufacturer’s instructions. Where applicable, cells were treated with 10 µM KU-55933 (Merck Millipore), 5 μg⋅ml−1 Actinomycin D, 400 nM TSA (Abcam), or 1 gray ionizing radiation using a Gammacell 3000 Elan (Best Theratronics).
Cell-Line Generation.
pLXSN H2B-mCherry and/or pWZL H2B-wGFP retrovectors were created by transfecting Phoenix-AMPHO cells (purchased from ATCC) with Lipofectamine 2000, according to manufacturer’s instructions (ThermoFisher). Vector containing supernatant was collected at 12 and 24 h after transfection, passed through a 0.22-µm filter, and used to infect HeLa cells in the presence of 4 µg/mL polybrene (Sigma). Cultures were selected with 600 µg/mL G418 and/or 200 µg/mL Hygromycin before sorting at the Westmead Institute for Medical Research (WIMR) flow-cytometry center (Sydney). Cells sorted for low expression of H2B-mCherry and/or H2B-eGFP were then propagated in 600 µg/mL G418, 200 µg/mL Hygromycin, and 1x Pen/Strep for 10 d before experimentation.
RNF8 KO.
pLentiCRISPRv2-RNF8-2 lentivectors were created in the Children’s Medical Research Institute Vector and Genome Engineering Facility as described (50). RNF8 was deleted by transducing cells with pLentiCRISPRv2-RNF8-2 vectors and selecting with 1 µg/mL puromycin for 2 d. Single cells were sorted into individual wells of a 96-well plate at the WIMR Flow Cytometry center, and cultures were expanded for 14 d. Clones were screened for inhibited 53BP1 recruitment to γ-H2AX foci following 1-gray ionizing radiation and immunofluorescence staining as described below. Clones negative for 53BP1 recruitment were then screened by PCR-amplifying the targeted locus using the RNF8 exon 1 infusion forward and reverse primers and cloning the PCR products into PmeI-digested (New England Biolabs) pcDNA3.1 (ThermoFisher) using Infusion cloning. Plasmids were sequenced with RNF8 sequence forward oligonucleotide, and clones verified to contain gene-disruptive mutations in all three RNF8 alleles in HeLa were used for experimentation.
FLIM-FRET Microscopy.
All FLIM-FRET data were acquired with a Zeiss LSM 880 laser scanning microscope coupled to a 2-photon Ti:Sapphire laser (Spectra-Physics Mai Tai) producing 80-fs pulses at a repetition of 80 MHz and a Picoquant Picoharp (Picoquant) for time-resolved detection. A 60× water immersion objective of 1.2 NA (Zeiss) was used for all experiments. The two-photon excitation laser was tuned to 900 nm for selective excitation of the donor fluorophore and minimization of DNA damage during live cell imaging. A short-pass 760-nm dichroic mirror was used to separate the fluorescence signal from the laser light. The fluorescence signal was directed through a 550-nm-long pass eGFP/mCherry filter, and the donor and acceptor signal was split between two photomultiplier detectors (H7422P-40 of Hamamatsu), with the following bandwidth filters placed in front of each: eGFP 520/25 and mCherry 590/25, respectively. We simultaneously acquired intensity and lifetime data by raster-scanning the two-photon Ti:Sapphire laser beam (900 nm) across a selected HeLaH2B-2FP interphase nucleus at a digital zoom that resulted in a frame size of 30 μm. The pixel frame size was set to 512, which gave a pixel size of 0.06 μm. The pixel dwell time was set at 32.77 µs/pixel, which, for a 512-pixel frame size, resulted in a 13.2-s frame time. For each FLIM experiment 10–20 frames were integrated. Together, these conditions resulted in an acquisition time of ∼2–3 min. Calibration of the system and phasor plot space was performed by measuring fluorescein (pH 9.0), which has a known single-exponential lifetime of 4.04 ns. The FLIM data were processed by the SimFCS software developed at the Laboratory for Fluorescence Dynamics (LFD) (https://www.lfd.uci.edu/).
FLIM-FRET Analysis.
The fluorescence decay recorded in each pixel of a FLIM image was quantified by the phasor approach to lifetime analysis (SI Appendix, Text S1) (28, 29). Each pixel of the FLIM image gives rise to a single point (phasor) in the phasor plot, and, when used in reciprocal mode, enables each point of the phasor plot to be mapped to each pixel of the FLIM image. Since phasors follow simple vector algebra, it is possible to determine the fractional contribution of two or more independent molecular species coexisting in the same pixel (e.g., autofluorescence and a fluorescent protein). In the case of two independent species, all possible weightings give a phasor distribution along a linear trajectory that joins the phasors of the individual species in pure form (SI Appendix, Text S2). In the case of a FRET experiment where the lifetime of the donor molecule is changed upon interaction with an acceptor molecule, the realization of all possible phasors quenched with different efficiencies describe a curved trajectory in the phasor plot. The FRET trajectory follows the classical definition of FRET efficiency (SI Appendix, Text S3). As described in previous papers, the contribution of the background (cellular autofluorescence) and of the donor without acceptor were evaluated by using the rule of the linear combination, with the background phasor and donor unquenched determined independently (28, 29).
In the case of HeLaH2B-2FP, the FRET efficiency varied as a function of local nucleosome proximity; thus, the trajectory of phasor clusters between the donor phasor (2.5 ns, 0% FRET) and the highest FRET state (2.0 ns, 21% FRET) represent the different states of chromatin compaction in any one pixel. By linking two phasor cursors between these two terminal phasor locations, we can pseudocolor each pixel of a FLIM map according to the exact FRET efficiency detected at that location. Furthermore, from linking two additional cursors between the donor phasor and background phasor, we can also quantify the contribution of cellular autofluorescence in each pixel. To increase phasor accuracy, a 3 × 3 spatial median filter was applied to the FLIM maps presented in Results before FRET analysis. All FLIM-FRET quantitation was performed in the SimFCS software developed at the LFD.
Temporal Longevity Analysis.
A phasor FRET map (0–21% FRET efficiency) reports the localization of a range of chromatin compaction states detected within a FLIM image. Thus, by extracting the x–y coordinates of pixels in high FRET state (16–21% FRET efficiency), we can derive an image of compacted chromatin localization. If we are interested in the temporal persistence of each located chromatin foci, we can generate a time series of chromatin-localization maps and, from application of an MAV across them, derive a heat map of pixel longevity for the selected MAV temporal window. The number of frames employed for the MAV determines the range of amplitudes recorded in a longevity map and, thus, the threshold parameters for extracting the localization of stable vs. transient chromatin foci. For example, in Fig. 3, we average three frames acquired with a 10-min interval, and the resulting longevity map contains pixels with an amplitude of 0, 0.333, 0.666, or 1. These values correspond to chromatin foci that are either not present in a pixel or present for one, two, or three frames, which in turn corresponds to chromatin foci that persist from 0 up to 20 min. By counting the number of pixels present for 0–3 frames, we can obtain a histogram which reports overall chromatin network stability. The chromatin-localization maps were generated in SimFCS, and the longevity analysis routine, from which can extract when, where, and how many chromatin foci are stable within a time series of chromatin-localization maps, was implemented in MATLAB (The MathWorks).
PLICS Analysis.
To investigate the size and spacing between chromatin foci localized in a phasor FRET map, similarly to longevity analysis, we first generated an image of compact chromatin localization (FRET efficiency of 16–21%). Then, from the resulting images, we applied the PLICS approach (34). PLICS consists of computing the spatial ACF of a small region of the image, computing its angular average, and transforming the resulting curve into the phasor space. Successively, the phase coordinate of the transformed local ACF is stored and is used to characterize the ACF shape, which is linked to the size of the local structures. This approach is iteratively applied to analyze the entire image, providing a map of the size of the structures. If the source image is made binary, the distance between two structures is merely the amount of empty space between them. Therefore, applying PLICS to the negative of the binary image provides a direct measurement of the distance between structures. We will refer to this analysis as iPLICS. For the chromatin-localization maps analyzed in this work, a 12 × 12 PLICS moving mask was used to obtain the local size of the analyzed regions, while the negative of the binary image was analyzed with a 36 × 36 moving mask to characterize the distance between the structures. A dedicated calibration has been employed to extract the size of the binary regions by simulating binary circles with varying diameter; the details of the calibration procedure for an arbitrary function and the complete characterization of the technique are described in ref. 34. The chromatin-localization maps were generated from SimFCS, and the PLICS/iPLICS analysis routine, which can extract the size and spacing of chromatin foci within a time series of chromatin-localization maps, was implemented in MATLAB (The MathWorks).
Laser Microirradiation and FLIM-FRET Microscopy During the DDR.
For all microirradiation, experiments the two-photon Ti:Sapphire laser (80 fs, repetition rate 80 MHz; Spectra-Physics Mai Tai) was tuned to 780 nm and used in conjunction with the Zeiss LSM880 laser-scanning microscope (51). The laser beam was then focused on a small section of the nucleus (2 µm × 2 µm) which avoided the nucleolus or nuclear envelope, and a frame scan was acquired (256 × 256 pixels, 32.77 µs/pixel) at a power found to recruit DNA-repair factor eGFP-53BP1 (SI Appendix, Fig. S6). FLIM-FRET microscopy was performed in parallel by using the microscope acquisition settings described above. FLIM-FRET data were recorded before laser microirradiation and at the following time points after this treatment: (i) 10-min interval during the first hour of DDR (0, 10, 20, 30, 40, 50, and 60 min); (ii) 20-min interval during the second hour of DDR (80, 100, and 120 min); and (iii) 30-min interval for the remaining 4 h of DDR (150, 180, 210, 240, 270, 300, 330, and 360 min).
Laser Microirradiation and Live Cell Imaging of eGFP-53BP1 Mobility During the DDR.
The Zeiss LSM880 laser scanning microscope was also employed for live cell imaging of eGFP-53BP1 localization and mobility. Specifically, a 60× water-immersion objective of 1.2 NA (Zeiss) was used for all experiments, and then a time series of eGFP-53BP1 intensity frames was acquired by using the 488-nm emission line of an Argon laser and a 510- to 560-nm detection range. A total of 2,048 intensity frames were acquired with a pixel frame size of 64 and at a zoom that resulted in a 10-μm ROI (0.166 μm per pixel). The pixel dwell time was set at 8.19 µs per pixel, which for a 64-pixel frame size resulted in a 79-ms frame time (2–3 min acquisition time). NIR laser microirradiation was then performed in the center of this ROI by employing the exact same settings described above (2 µm × 2 µm square with a 256-pixel frame size). This acquisition enabled correlation of 53BP1-eGFP mobility at the DNA damage site with independent FLIM-FRET data recording chromatin compaction during the DDR. To quantify 53BP1-eGFP mobility, we employed pair-correlation analysis (SI Appendix, Fig. S10) (26). In this study, we used the 2D extension of the pair-correlation function described in published papers (41, 52). The 2D pair-correlation analysis was performed at a distance that enabled mobility in and out of the DNA damage site to be tested (δr = 4 pixels). In Fig. 6, mobility is expressed in terms of time delay, a parameter proportional to the time it takes for a particle to reach the pair-correlated distance. All pair-correlation analysis was performed in the SimFCS software developed at the LFD.
Wide-Field Live Cell Imaging of HeLaH2B-2FP.
Live cell imaging shown in SI Appendix, Fig. S1 was performed on a ZEISS Cell Observer inverted wide-field microscope with a ZEISS HXP 120C mercury short-arc lamp and compatible filter cubes, using a 20×, 0.8-NA air objective. Cells were plated on glass-bottom MatTek six-well plates, and images were captured every 6 min for 65 h at 37 °C, 10% CO2, and atmospheric oxygen. Mitotic duration and outcome were scored by eye using Zen software (Zeiss).
Immunofluorescence.
Cells cultured on Alcain blue-stained glass coverslips were fixed with 2% paraformaldehyde in 1× PBS at room temperature for 10 min. The cells were rinsed in 1× PBS, then permeabilized in KCM buffer for 10 min (120 mM KCl, 20 mM NaCl, 10 mM Tris, pH 7.5, and 0.1% Triton X-100). Samples were blocked, and RNase was treated in ABDIL (20 mM Tris, pH 7.5, 2% BSA, 0.2% fish gelatin, 150 mM NaCl, 0.1% Triton, and 0.1% sodium azide) containing 100 µg/mL RNAseA (Sigma) plus 2% normal goat serum (vol/vol) for 1 h at room temperature. Samples were overlaid with 1:1,000 primary antibody in ABDIL overnight at 4 °C in a humidity chamber. The following day, the cells were washed three times for 5 min with shaking in 1× PBS + 0.1% (vol/vol) Tween 20 (1× PBST), before overlaying with 1:1,000 secondary antibody in ABDIL plus 2% normal goat serum (vol/vol) for 1 h at room temperature. The slides were washed again as described above with DAPI added to the second wash at a concentration of 50 ng/mL Slides were dehydrated through a 70%, 90%, and 100% ethanol series by washing for 3 min in each condition, then air dried and mounded with prolong gold (ThermoFisher). Images were captured on a Zeiss AxioImager Z.2 with a 63× 1.4-NA oil objective, appropriate filter cubes, and an Axiocam 506 monochromatic camera using Zen software (Zeiss).
Western Blots.
Cells were collected with trypsin, and the reaction was quenched with growth medium containing serum. Cells were washed in 1× PBS and homogenized in NuPage 4× LDS sample buffer without EDTA (Invitrogen) supplemented with 2% (vol/vol) β-mercaptoethanol (Sigma) and 5% (vol/vol) Benzonase (Merck Millipore) at 1 × 104 cells per µL for 1 h at room temperature. Whole-cell lysates were denatured at 68 °C for 10 min before resolution at 10 µL per well on precast gels, according to manufacturer’s protocols (Life Technologies). Protein was transferred to nitrocellulose (Amersham) at 100 V for 1 h, and the membranes were blocked for 1 h with 5% (wt/vol) powdered skim milk in 1× PBS + 0.1% (vol/vol) Tween 20 (1× PBST). Membranes were probed overnight with primary antibody diluted in 1× PBST + 5% (wt/vol) powdered skim milk at 4 °C with gentle nutation. The following morning, blots were washed three times for 10 min with 1× PBST, then probed with horseradish peroxidase-conjugated secondary antibody diluted in 1× PBST + 5% (wt/vol) powdered skim milk for 1 h at room temperature, followed by three washes in 1× PBST. Membranes were rinsed in deionized water blotted dry and overlaid with 0.22-µm filtered Enhanced Chemiluminescent Western blotting substrate for 5 min (Pierce) before visualization with a Fujifilm LAS 4000 luminescent image analyzer and associated software (GE Health). Where applicable, blots were quantified by using ImageJ.
Oligonucleotides.
Please see SI Appendix, Table S1 for a list of the oligonucleotides used in this study. All oligonucleotides were produced by Integrated DNA Technologies.
Antibodies.
The following antibodies were used in this study: ATM (catalog no. 2873; Cell Signaling Technology), ATM-S1981 (catalog no. ab81292; Abcam), CHK2 (catalog no. 05-649; Millipore), CHK2-T68 (catalog no. 2661; Cell Signaling Technology), eGFP (catalog no. ab6556; Abcam), γ-H2AX (catalog no. 05-636; Millipore), H2B (catalog no. 8135; Cell Signaling Technology), mCherry (catalog no. STJ34373; St. John’s Laboratory), Vinculin (catalog no. V9131; Sigma), 53BP1 (catalog no. Sc-22760; Santa Cruz), goat anti-mouse Alexa 488 conjugate (catalog no. A11031; ThermoFisher), goat anti-rabbit Alexa 568 conjugate (catalog no. A-11011; ThermoFisher), polyclonal goat anti-rabbit immunoglobulins HRP-conjugate (catalog no. P0448; Dako), and polyclonal goat anti-mouse immunoglobulins HRP-conjugated (catalog no. P0447; Dako).
Statistics and Figure Preparation.
Statistical analysis was performed by using GraphPad Prism software. Sanger sequencing traces were exported from SnapGene. Figures were prepared by using Adobe Photoshop, Illustrator, MATLAB, and SimFCS.
Supplementary Material
Acknowledgments
We thank Elvis Pandzic, Alex Macmillan, Renee Whan, and the Biomedical Imaging Facility and the Mark Wainwright Analytical Centre at the University of New South Wales for enabling access to the Zeiss LSM 880; Scott Page and the Children’s Medical Research Institute (CMRI) Australian Cancer Research Foundation (ACRF) Telomere Analysis Centre (supported by the ACRF) for assistance with imaging infrastructure; Leszek Lisowski and the CMRI Vector and Genome Engineering Facility for making Lentivectors; the Westmead Institute for Medical Research Flow Cytometry Centre [supported by the Cancer Council NSW and the Australian National Health and Medical Research Council (NHMRC)] for cell sorting; and David Priest and Georgia Kafer for their helpful discussion and feedback on the manuscript. K.G. is supported by Australian NHMRC Grant APP1059278 and Australian Research Council (ARC) Grants CE140100011 and LP140100967. E.G. is supported by National Institutes of Health Grants P41-GM103540 and P50-GM076516. A.J.C. is supported by Australian NHMRC Grants 1053195, 1106241, and 1104461; Cancer Council NSW Grant RG 15-12; and Cancer Institute NSW Grant 11/FRL/5-02. E.H. is supported by Australian NHMRC Grants APP1104461 and APP1124762 and ARC Grant DP180101387.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Matlab code used for data analysis has been deposited on the GitHub repository (https://github.com/ehinde/histone-FLIM-FRET).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814965116/-/DCSupplemental.
References
- 1.Hauer MH, Gasser SM. Chromatin and nucleosome dynamics in DNA damage and repair. Genes Dev. 2017;31:2204–2221. doi: 10.1101/gad.307702.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clouaire T, Marnef A, Legube G. Taming tricky DSBs: ATM on duty. DNA Repair (Amst) 2017;56:84–91. doi: 10.1016/j.dnarep.2017.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Lee JH, Paull TT. ATM activation by DNA double-strand breaks through the Mre11-Rad50-Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- 5.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fradet-Turcotte A, et al. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 2013;499:50–54. doi: 10.1038/nature12318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huen MS, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolas NK, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mailand N, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 10.Luijsterburg MS, et al. PARP1 links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol Cell. 2016;61:547–562. doi: 10.1016/j.molcel.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hauer MH, et al. Histone degradation in response to DNA damage enhances chromatin dynamics and recombination rates. Nat Struct Mol Biol. 2017;24:99–107. doi: 10.1038/nsmb.3347. [DOI] [PubMed] [Google Scholar]
- 12.Klement K, et al. Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair. J Cell Biol. 2014;207:717–733. doi: 10.1083/jcb.201405077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalousi A, et al. The nuclear oncogene SET controls DNA repair by KAP1 and HP1 retention to chromatin. Cell Rep. 2015;11:149–163. doi: 10.1016/j.celrep.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 14.Thorslund T, et al. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature. 2015;527:389–393. doi: 10.1038/nature15401. [DOI] [PubMed] [Google Scholar]
- 15.Luger K, Hansen JC. Nucleosome and chromatin fiber dynamics. Curr Opin Struct Biol. 2005;15:188–196. doi: 10.1016/j.sbi.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 16.Ou HD, et al. ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science. 2017;357:eaag0025. doi: 10.1126/science.aag0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cremer T, et al. The 4D nucleome: Evidence for a dynamic nuclear landscape based on co-aligned active and inactive nuclear compartments. FEBS Lett. 2015;589:2931–2943. doi: 10.1016/j.febslet.2015.05.037. [DOI] [PubMed] [Google Scholar]
- 18.Woringer M, Darzacq X, Izeddin I. Geometry of the nucleus: A perspective on gene expression regulation. Curr Opin Chem Biol. 2014;20:112–119. doi: 10.1016/j.cbpa.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Levi V, Gratton E. Chromatin dynamics during interphase explored by single-particle tracking. Chromosome Res. 2008;16:439–449. doi: 10.1007/s10577-008-1240-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wachsmuth M, Knoch TA, Rippe K. Dynamic properties of independent chromatin domains measured by correlation spectroscopy in living cells. Epigenetics Chromatin. 2016;9:57. doi: 10.1186/s13072-016-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dross N, et al. Mapping eGFP oligomer mobility in living cell nuclei. PLoS One. 2009;4:e5041. doi: 10.1371/journal.pone.0005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baum M, Erdel F, Wachsmuth M, Rippe K. Retrieving the intracellular topology from multi-scale protein mobility mapping in living cells. Nat Commun. 2014;5:4494. doi: 10.1038/ncomms5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinde E, Cardarelli F, Digman MA, Gratton E. In vivo pair correlation analysis of EGFP intranuclear diffusion reveals DNA-dependent molecular flow. Proc Natl Acad Sci USA. 2010;107:16560–16565. doi: 10.1073/pnas.1006731107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Llères D, et al. Quantitative FLIM-FRET microscopy to monitor nanoscale chromatin compaction in vivo reveals structural roles of condensin complexes. Cell Rep. 2017;18:1791–1803. doi: 10.1016/j.celrep.2017.01.043. [DOI] [PubMed] [Google Scholar]
- 25.Llères D, James J, Swift S, Norman DG, Lamond AI. Quantitative analysis of chromatin compaction in living cells using FLIM-FRET. J Cell Biol. 2009;187:481–496. doi: 10.1083/jcb.200907029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinde E, Kong X, Yokomori K, Gratton E. Chromatin dynamics during DNA repair revealed by pair correlation analysis of molecular flow in the nucleus. Biophys J. 2014;107:55–65. doi: 10.1016/j.bpj.2014.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong X, et al. Laser microirradiation to study in vivo cellular responses to simple and complex DNA damage. J Vis Exp. January 31, 2018 doi: 10.3791/56213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Digman MA, Caiolfa VR, Zamai M, Gratton E. The phasor approach to fluorescence lifetime imaging analysis. Biophys J. 2008;94:L14–L16. doi: 10.1529/biophysj.107.120154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinde E, Digman MA, Welch C, Hahn KM, Gratton E. Biosensor Förster resonance energy transfer detection by the phasor approach to fluorescence lifetime imaging microscopy. Microsc Res Tech. 2012;75:271–281. doi: 10.1002/jemt.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ranjit S, Lanzano L, Gratton E. Mapping diffusion in a living cell via the phasor approach. Biophys J. 2014;107:2775–2785. doi: 10.1016/j.bpj.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hinde E, Digman MA, Hahn KM, Gratton E. Millisecond spatiotemporal dynamics of FRET biosensors by the pair correlation function and the phasor approach to FLIM. Proc Natl Acad Sci USA. 2013;110:135–140. doi: 10.1073/pnas.1211882110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kolin DL, Wiseman PW. Advances in image correlation spectroscopy: Measuring number densities, aggregation states, and dynamics of fluorescently labeled macromolecules in cells. Cell Biochem Biophys. 2007;49:141–164. doi: 10.1007/s12013-007-9000-5. [DOI] [PubMed] [Google Scholar]
- 33.Bidaux G, et al. FRET image correlation spectroscopy reveals RNAPII-independent P-TEFb recruitment on chromatin. Biophys J. 2018;114:522–533. doi: 10.1016/j.bpj.2017.11.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scipioni L, Gratton E, Diaspro A, Lanzanò L. Phasor analysis of local ICS detects heterogeneity in size and number of intracellular vesicles. Biophys J. 2016;111:619–629. doi: 10.1016/j.bpj.2016.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Day RN. Measuring protein interactions using Förster resonance energy transfer and fluorescence lifetime imaging microscopy. Methods. 2014;66:200–207. doi: 10.1016/j.ymeth.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bensaude O. Inhibiting eukaryotic transcription: Which compound to choose? How to evaluate its activity? Transcription. 2011;2:103–108. doi: 10.4161/trns.2.3.16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belmont AS. Large-scale chromatin organization: The good, the surprising, and the still perplexing. Curr Opin Cell Biol. 2014;26:69–78. doi: 10.1016/j.ceb.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erdel F, Rippe K. Formation of chromatin subcompartments by phase separation. Biophys J. 2018;114:2262–2270. doi: 10.1016/j.bpj.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hickson I, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 40.Digman MA, Gratton E. Imaging barriers to diffusion by pair correlation functions. Biophys J. 2009;97:665–673. doi: 10.1016/j.bpj.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Rienzo C, Cardarelli F, Di Luca M, Beltram F, Gratton E. Diffusion tensor analysis by two-dimensional pair correlation of fluorescence fluctuations in cells. Biophys J. 2016;111:841–851. doi: 10.1016/j.bpj.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hustedt N, Durocher D. The control of DNA repair by the cell cycle. Nat Cell Biol. 2016;19:1–9. doi: 10.1038/ncb3452. [DOI] [PubMed] [Google Scholar]
- 43.Kalousi A, Soutoglou E. Nuclear compartmentalization of DNA repair. Curr Opin Genet Dev. 2016;37:148–157. doi: 10.1016/j.gde.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 44.Adam S, et al. Real-time tracking of parental histones reveals their contribution to chromatin integrity following DNA damage. Mol Cell. 2016;64:65–78. doi: 10.1016/j.molcel.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polo SE. Switching genes to silent mode near DNA double-strand breaks. EMBO Rep. 2017;18:659–660. doi: 10.15252/embr.201744052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luijsterburg MS, et al. A new non-catalytic role for ubiquitin ligase RNF8 in unfolding higher-order chromatin structure. EMBO J. 2012;31:2511–2527. doi: 10.1038/emboj.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crabbe L, Cesare AJ, Kasuboski JM, Fitzpatrick JA, Karlseder J. Human telomeres are tethered to the nuclear envelope during postmitotic nuclear assembly. Cell Rep. 2012;2:1521–1529. doi: 10.1016/j.celrep.2012.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 49.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lisowski L, Sadelain M. Locus control region elements HS1 and HS4 enhance the therapeutic efficacy of globin gene transfer in beta-thalassemic mice. Blood. 2007;110:4175–4178. doi: 10.1182/blood-2007-08-108647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kong X, et al. Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nucleic Acids Res. 2009;37:e68. doi: 10.1093/nar/gkp221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malacrida L, Hedde PN, Ranjit S, Cardarelli F, Gratton E. Visualization of barriers and obstacles to molecular diffusion in live cells by spatial pair-cross-correlation in two dimensions. Biomed Opt Express. 2017;9:303–321. doi: 10.1364/BOE.9.000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.