Significance
Parkinson’s disease (PD) is a chronic, progressive neurodegenerative disease with both familial and sporadic forms and a clear genetic component. In addition, underlying immunoregulatory dysfunction and inflammatory processes have been implicated in PD pathogenesis. In this study, deep sequencing of HLA genes, which encode highly variable cell surface immune receptors, reveals specific variants conferring either risk or protection in PD. Because a history of cigarette smoking is known to be protective in PD, we analyzed the interaction of these genetic variants with smoking history in PD patients and healthy controls and found that the genetic effects are modified by history of cigarette smoking. These results provide a molecular model that explains the unique epidemiology of smoking in PD.
Keywords: Parkinson’s disease, HLA, smoking, shared epitope
Abstract
Parkinson’s disease (PD) is a neurodegenerative disease in which genetic risk has been mapped to HLA, but precise allelic associations have been difficult to infer due to limitations in genotyping methodology. Mapping PD risk at highest possible resolution, we performed sequencing of 11 HLA genes in 1,597 PD cases and 1,606 controls. We found that susceptibility to PD can be explained by a specific combination of amino acids at positions 70–74 on the HLA-DRB1 molecule. Previously identified as the primary risk factor in rheumatoid arthritis and referred to as the “shared epitope” (SE), the residues Q/R-K/R-R-A-A at positions 70–74 in combination with valine at position 11 (11-V) is highly protective in PD, while risk is attributable to the identical epitope in the absence of 11-V. Notably, these effects are modified by history of cigarette smoking, with a strong protective effect mediated by a positive history of smoking in combination with the SE and 11-V (P = 10−4; odds ratio, 0.51; 95% confidence interval, 0.36–0.72) and risk attributable to never smoking in combination with the SE without 11-V (P = 0.01; odds ratio, 1.51; 95% confidence interval, 1.08–2.12). The association of specific combinations of amino acids that participate in critical peptide-binding pockets of the HLA class II molecule implicates antigen presentation in PD pathogenesis and provides further support for genetic control of neuroinflammation in disease. The interaction of HLA-DRB1 with smoking history in disease predisposition, along with predicted patterns of peptide binding to HLA, provide a molecular model that explains the unique epidemiology of smoking in PD.
Parkinson’s disease (PD; MIM: 168600) is a chronic, progressive neurodegenerative disease characterized by bradykinesia, tremor, rigidity, and postural instability plus a constellation of associated features including autonomic, sensory, cognitive, and psychological changes. With both familial and sporadic forms of the disease and a clear genetic component, numerous genome-wide association studies (GWAS), fine-mapping, and candidate gene studies over the past two decades have revealed dozens of risk-associated genetic variants. Consistent with underlying immunoregulatory dysfunction and inflammatory processes operating in PD, immune system genetic factors are consistently represented among replicated PD loci. Most notably, a clear association peak has been shown to map to the major histocompatibility complex (MHC) on chromosome 6 (6p21.3), which houses the HLA genes.
HLA class II gene products are expressed on the surface of professional antigen-presenting cells, which generally present peptide fragments of extracellular origin to helper (CD4+) T cells (1). HLA class I molecules are expressed on all nucleated cells and present antigens originated from intracellular sources to cytotoxic (CD8+) T cells for killing tumorous or infected cells (2). While an association of HLA class I with PD was first observed more than four decades ago (3), more contemporaneous single nucleotide polymorphism (SNP) associations point to the class II region of the locus (4, 5). In an early GWAS, the association peak in the MHC was found with rs3129882 within HLA-DRA (MIM: 142860) (6), although attempts to replicate this observation have generated varied and conflicting results (7–13).
Recent large studies have used SNP-based GWAS to fine-map the MHC association signal and to impute functional alleles of the HLA system (4, 5, 12, 14–17), but the complexity of the region presents major challenges for studies aimed at defining the causative variants. Extremely high levels of polymorphism, structural variation, an extensive history of gene duplication, conversion and recombination events, and unusually high levels of linkage disequilibrium (LD) confound attempts to infer precise allelic associations from SNP data (18). The few studies that have directly queried HLA variation through traditional genotyping methods have produced mixed results and have been hampered by a relatively small sample size, low-resolution HLA genotyping methodology, or both (19–22).
To unravel the immunogenetic underpinnings of the disease, we undertook a large-scale deep-sequencing study of the HLA genes in PD. We analyzed HLA variation at high resolution using a well-validated next-generation sequencing (NGS) method in 1,941 PD cases and 1,612 controls from the United States with European ancestry. By sequencing comprehensively across 11 full-length HLA genes— HLA-DRB1 (MIM: 142857), HLA-DRB3 (MIM: 612735), HLA-DRB4, HLA-DRB5 (MIM: 604776), HLA-DQA1 (MIM: 146880), HLA-DQB1 (MIM: 604305), HLA-DPA1 (MIM: 142880), HLA-DPB1 (MIM: 142858), HLA-A (MIM:142800), HLA-B (MIM: 142830), and HLA-C (MIM: 142840)—we were able to examine associations at the highest possible resolution, yielding the most complete representation to date of the complex immunogenetics underlying PD. We found that protection in PD can be explained by a specific epitope at positions 70–74 on the HLA-DRB1 molecule in combination with valine at position 11 (11-V), while risk is attributable to the identical epitope in the absence of 11-V, and that these effects are modified by a history of cigarette smoking. In identifying the source of association signals in the disease and interaction with a key environmental factor, we provide a framework for understanding the nature of the immune component in PD pathogenesis.
Results
High-Resolution Genotyping Demonstrates That Risk and Protection in PD Are Mediated by HLA-DRB1.
All 11 of the highly-polymorphic HLA class I and II genes were sequenced from 1,597 PD cases and 1,606 controls (SI Appendix, Table S1). We first analyzed data for the entire molecule at polypeptide sequence resolution, described in the HLA nomenclature by the first two fields of the allele name (23), to examine the entire functional unit of interest. In this analysis, we considered results sufficient to warrant follow-up when significant at a false discovery rate (FDR) (24) of 10%, corresponding to a cutoff of P = 0.02. We found strong protective effects for HLA-DRB1*04:01 [P = 0.007; odds ratio (OR), 0.78; 95% confidence interval (CI), 0.65–0.93] and HLA-DQB1*03:02 (P = 0.006; OR, 0.78; 95% CI, 0.66–0.93) (Fig. 1). These alleles are in strong LD, forming part of the HLA-DQA1*03:01∼HLA-DQB1*03:02∼HLA-DRB1*04:01 class II haplotype, which is also significantly associated with disease (P = 0.008; OR, 0.66; 95% CI, 0.54–0.84). Confirming HLA-DRB1 as the source of this signal, another HLA-DRB1*04:01–containing haplotype, HLA-DQA1*03:03∼HLA-DQB1*03:01∼HLA-DRB1*04:01, was observed at nearly equal frequency (Table 1) and is also significantly protective against developing PD (P = 0.005; OR, 0.66; 95% CI, 0.54–0.85). While both haplotypes are in LD with HLA-DRB4*01:03, this allele may be excluded as causative because it is also observed in some HLA-DRB1*07:01 haplotypes not associated with disease. Furthermore, no association remained for HLA-DQB1*03:02 on conditioning on HLA-DRB1*04:01.
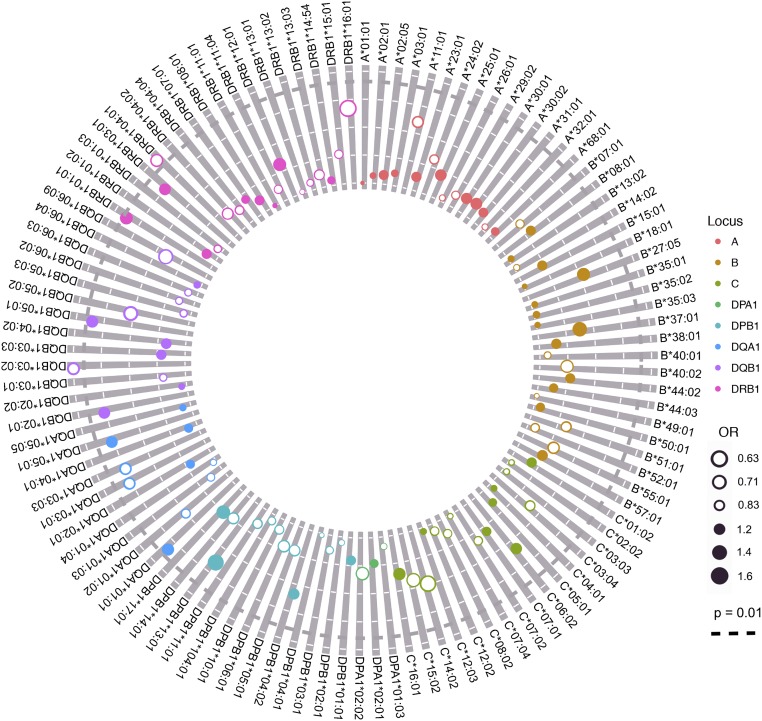
Fig. 1.
Association analysis results for all HLA class I and class II alleles. HLA alleles that were detected in the study population are shown around the perimeter. Decreasing P values are indicated by the distance of points from the center, with P = 0.01 indicated by the black dotted concentric line. The magnitude of ORs is indicated by the diameter of points, with filled points indicating OR >1 and open points indicating OR <1. The magnitude of global linkage disequilibrium between loci is indicated by the width of interior lines.
Table 1.
HLA-DQA1∼DQB1∼DRB1 haplotype frequencies in PD cases and controls
| DQA1∼DQB1∼DRB1 | Control frequency | Case frequency |
| 01:02∼06:02∼15:01 | 0.124 | 0.122 |
| 05:01∼02:01∼03:01 | 0.114 | 0.137 |
| 02:01∼02:02∼07:01 | 0.091 | 0.092 |
| 01:01∼05:01∼01:01 | 0.079 | 0.099 |
| 01:03∼06:03∼13:01 | 0.055 | 0.053 |
| 05:05∼03:01∼11:01 | 0.051 | 0.051 |
| 03:03∼03:01∼04:01 | 0.047 | 0.039 |
| 03:01∼03:02∼04:01 | 0.037 | 0.031 |
| 05:05∼03:01∼11:04 | 0.037 | 0.031 |
| 01:02∼06:04∼13:02 | 0.034 | 0.034 |
| 02:01∼03:03∼07:01 | 0.033 | 0.038 |
| 03:01∼03:02∼04:04 | 0.032 | 0.027 |
| 01:04∼05:03∼14:54 | 0.02 | 0.02 |
| 01:02∼05:02∼16:01 | 0.019 | 0.011 |
| 04:01∼04:02∼08:01 | 0.018 | 0.02 |
| 05:05∼03:01∼12:01 | 0.017 | 0.02 |
| 03:01∼03:02∼04:02 | 0.015 | 0.012 |
| 01:01∼05:01∼01:02 | 0.014 | 0.016 |
| 01:02∼06:09∼13:02 | 0.013 | 0.01 |
| 01:03∼06:01∼15:02 | 0.011 | 0.008 |
| 01:01∼05:01∼01:03 | 0.011 | 0.011 |
| 05:05∼03:01∼13:03 | 0.01 | 0.01 |
| 03:03∼03:01∼04:07 | 0.009 | 0.01 |
Complementing this finding, we observed borderline significance for PD risk mediated by HLA-DRB1*01:01 (P = 0.012; OR, 1.26; 95% CI, 1.05–1.50). This result remained significant after conditioning for HLA-DRB1*04:01 (P = 0.02; OR, 1.23; 95% CI, 1.03–1.49), confirming it as an independent association. HLA-DRB1*01:01 is observed in near-complete LD with HLA-DQA1*01:01 and HLA-DQB1*05:01 as a haplotype in populations with European ancestry (25, 26). Although we also found similar associations and significance levels for the HLA-DQA1 and HLA-DQB1 alleles of this haplotype (Fig. 1), their effect sizes were smaller, and on conditioning for HLA-DRB1*01:01, no effect remained for these alleles. This finding suggests that any observed HLA-DQA1/-DQB1 association is attributable to LD with the primary predisposing allele, HLA-DRB1*01:01. Taken together, our results strongly implicate HLA-DRB1 as the operative locus in disease.
Our genotyping methodology provides information for virtually all coding and noncoding variations at the HLA loci; however, we did not observe any appreciable differences between cases and controls with respect to noncoding variation at any locus. Likewise, there were no significant differences between cases and controls for alleles of any class I locus (HLA-A, -B, and -C) or class II loci HLA-DPA1, HLA-DPB1, HLA-DRB3, HLA-DRB4, and HLA-DRB5 after conditioning on the HLA-DRB1 associations. Similarly, tests for interaction between HLA alleles and between HLA and known PD risk SNPs, when available in our dataset, did not yield any significant results.
Specific Amino Acid Residues of HLA-DRB1 Mediate Risk and Protection in PD.
To decipher the precise structural elements mediating the suggested HLA-DRB1 associations with PD, we examined the contribution of individual amino acids of the HLA-DRB1 molecule to disease risk. Omnibus testing revealed four positions to be significantly associated with PD: 11 (P = 0.002; pcorr = 0.05), 13 (P = 0.001; pcorr = 0.04), 26 (P = 0.001; pcorr = 0.03), and 33 (P = 0.001; pcorr = 0.04). Of these, positions 11, 13, and 26 are highly polymorphic and occupy key positions in the peptide-binding groove of the class II molecule. Results of analysis of amino acid variation for each position are provided in SI Appendix, Table S2. The effect at position 26 is driven by phenylalanine, mediating protection (P = 3 × 10−5; OR, 0.79; 95% CI, 0.71–0.89). Position 11 is extremely polymorphic, with six different residues observed across this dataset; of these, significant associations are observed for valine, which mediates protection (P = 0.006; OR, 0.83; 95% CI, 0.73–0.95), and lysine, which mediates risk (P = 0.006; OR, 1.24; 95% CI, 1.06–1.44). Position 13 is also extremely polymorphic and is in very strong yet incomplete LD with position 11; however, the association at this position appears to be driven mainly by a protective effect of histidine (P = 0.005; OR, 0.82; 95% CI, 0.72–0.95). Examination of these amino acid associations in a replication cohort of an additional 462 controls and 536 PD patients confirms the findings for 11-V (P = 0.02; OR, 0.7; 95% CI, 0.51–0.97) and 13-H (P = 0.03; OR, 0.7; 95% CI, 0.5–0.97).
The protective effects observed for 11-V, 13-H, and 26-F can be attributed to association of these variants with alleles of the HLA-DRB1*04 lineage, aligning with our initial observation of protection mediated by HLA-DRB1*04:01. Likewise, 11-L is specific to alleles of the HLA-DRB1*01 lineage, in line with our observation of risk mediated by HLA-DRB1*01:01. Thus, while the extreme levels of polymorphism at HLA-DRB1 hampers our power to confirm high-resolution allelic associations beyond 10% FDR or to fully account for the burden of multiple comparisons, the totality and context of variation at individual amino acids support our initial observations in high-resolution alleles. Indeed, combining alleles of the HLA-DRB1*04 lineage, we find significant protection at this resolution (P = 0.001, pcorr = 0.01; OR, 0.79; 95% CI, 0.69–0.91). Confirming this observation, HLA-DRB1*04 is also significantly protective in our replication cohort, with a nearly identical effect size (P = 0.04; OR, 0.77; 95% CI, 0.60–0.99).
The “Shared Epitope” and Position 11 Explain HLA Associations in PD.
Notably, along with HLA-DRB1*01:01, which we find to be predisposing in PD, HLA-DRB1*04:01 and HLA-DRB1*10:01 share a common set of amino acids at positions 70–74 (70-Q/R, 71-R/K, 72-R, 73-A, and 74-A), referred to as the “shared epitope” (SE) and originally described with respect to predisposition to rheumatoid arthritis (RA; MIM: 180300) (27). In our dataset, seven HLA-DRB1 alleles have the SE, including HLA-DRB1*10:01 and additional alleles of the HLA-DRB1*04 and HLA-DRB1*01 lineages (Table 2). Looking more closely at the lower-frequency HLA-DRB1*04 alleles in our dataset, we observed that only HLA-DRB1*04 alleles with the SE are increased in controls compared with PD cases (Table 2), while those without the SE are not.
Table 2.
Amino acid residues for HLA-DRB1 allotype SE and position 11 and frequencies in PD cases and controls
| HLA-DRB1 | 11 | 70 | 71 | 72 | 73 | 74 | Controls | Cases |
| 01:01 | L | Q | R | R | A | A | 0.083 | 0.099 |
| 01:02 | L | Q | R | R | A | A | 0.014 | 0.016 |
| 04:01 | V | Q | K | R | A | A | 0.085 | 0.076 |
| 04:04 | V | Q | R | R | A | A | 0.034 | 0.027 |
| 04:05 | V | Q | R | R | A | A | 0.006 | 0.004 |
| 04:08 | V | Q | R | R | A | A | 0.006 | 0.004 |
| 10:01 | V | R | R | R | A | A | 0.009 | 0.008 |
The HLA-DRB1 SE is defined as residues Q/R-K/R-R-A-A at position 70–74.
Given these findings, we examined the association of HLA-DRB1 with PD with respect to the SE in combination with position 11-V. We found that the risk for PD is mediated by the SE in the absence of 11-V (P = 0.01; OR, 1.25; 95% CI, 1.05–1.50). While not statistically significant, a trend toward a risk for the SE without 11-V was seen in our replication cohort. Most striking, we found that protection in PD can be explained by the SE in combination with 11-V (P = 0.001; OR, 0.76; 95% CI, 0.64–0.89), and that this result is clearly reproducible in our replication cohort (P = 0.01; OR, 0.61; 95% CI, 0.42–0.90), confirming a role for this specific set of amino acids in disease.
Smoking History and the HLA SE Interact in Risk and Protection in PD.
A protective effect of cigarette smoking in PD is well established (28). Moreover, recent work suggests an interaction between smoking history and variation in HLA-DRB1 in disease (17), but that analysis examined only a single SNP in the locus. Having identified the SE as the structural variant of HLA-DRB1 associated with disease, we sought to examine in greater detail the role of smoking history in combination with HLA variation. We analyzed the interaction of smoking history with the HLA-DRB1 SE in 837 PD cases and 918 controls for whom data on cigarette smoking history were available. These individuals had self-reported as either a former or current smoker or having never smoked; for our analysis, we dichotomized these groups into those with any history of smoking and those with no history of smoking. HLA-DRB1 alleles were coded according to the presence of the SE with and without 11-V (SE+V11+ and SE+V11−), and the impact on disease occurrence was determined by multiple logistic regression with gender, principal component (PC) 1, PC2, PC3, and PC4 as covariates. No significant differences with respect to SE frequencies were observed between individuals for whom smoking data were available and those for whom it was not available (SI Appendix, Table S3).
To study whether the effect of the SE genotype on disease was modified by smoking history, an interaction term (SE × smoking history) was included in the model. The importance of each possible risk factor under consideration was assessed by the Wald test and confirmed by the likelihood ratio test on the deviance of the model. We found that the interaction between positive smoking history and homozygosity for SE+V11+ alleles was significant (P = 0.02), suggesting that protection mediated by SE+V11+ HLA-DRB1 alleles was further enhanced by a history of smoking. The data show that the combined effect of a history of smoking with the presence of one or two SE+V11+ alleles was highly protective in PD (P = 10−4; OR, 0.51; 95% CI, 0.36–0.72) (SI Appendix, Table S4), with an effect size greater than that when considering HLA-DRB1 alone in the same individuals (OR, 0.65; 95% CI, 0.52–0.82). Likewise, the interaction term for a negative history of smoking with homozygosity for SE+V11− alleles also showed a strong trend (P = 0.06), and we found that a history of having never smoked in combination with SE+V11− alleles are predisposing to PD (P = 0.01; OR, 1.51; 95% CI, 1.08–2.12), with a greater effect size than for HLA alone (OR, 1.29; 95% CI, 1.01–1.66). Thus, it appears that the protective effect in PD of a positive history of smoking can be explained in part by interaction with HLA variation, suggesting that molecular changes mediated by cigarette smoke may be involved in alteration of HLA antigen presentation.
Citrullination of α-Synuclein Peptides Is Predicted to Alter Binding Affinity to HLA SE Alleles.
Because α-synuclein aggregates in the PD brain, and T-cell recognition of α-synuclein peptides by PD patients has been recently reported and proposed as an autoantigen in disease (29), we explored the binding affinity of peptides derived from α-synuclein to the HLA SE allotypes using a bioinformatic prediction method. We used the NetMHCII3.2 server (30) to predict binding of peptides from α-synuclein to SE alleles associated with PD risk (V11−) or protection (V11+). We input the entire protein sequence of α-synuclein and predicted the probable peptides that bind to HLA-DRB1*01:01 and *01:02 (SE+V11−) and to HLA-DRB1*04:01, *04:04, and *04:05 (SE+V11+). Overall, we found that peptides derived from α-synuclein were predicted to bind with higher affinity to SE+V11− (risk) alleles compared with SE+V11+ (protective) alleles (P = 0.004); in fact, among the 126 peptides tested, none reached the threshold for predicted binding (<500 nM) to the SE+V11+ alleles (SI Appendix, Table S5).
Given our finding that a history of smoking in our cohort interacts with HLA-DRB1 in PD, and that smoking significantly increases posttranslational modifications such as citrullination and homocitrullination (31), which in turn may alter peptide-binding affinity for HLA class II (32), we performed replicate peptide-binding predictions with all lysine modified to homocitrulline residues (SI Appendix, Table S6). For SE+V11− alleles, peptide modification is predicted to reduce the binding affinity of α-synuclein (P = 2.2 × 10−16). Conversely, peptide modification is predicted to increase the overall binding affinity to the SE+V11+ alleles (P = 0.0001). Importantly, the predicted increase in affinity for the SE+V11+ alleles resulted in a number of α-synuclein–derived peptides meeting the affinity threshold for binding. These results suggest that posttranslational modification of peptides mediated by cigarette smoking may simultaneously increase peptide affinity for protective HLA-DRB1 alleles such as HLA-DRB1*04:01 and decrease peptide affinity for risk HLA-DRB1 alleles such as HLA-DRB1*01:01.
Discussion
Despite clear evidence for the role of HLA in genetic predisposition to PD, the complex and highly polymorphic character of the region has been a barrier to defining the precise nature of its contribution to disease. Here we have performed deep sequencing across entire genes of all polymorphic classical HLA loci. In doing so, we were able to identify the specific amino acids and elucidate structural features mediating both risk and protection in PD. Our data establish that the entirety of HLA risk and protection in PD can be explained by the SE of HLA-DRB1 at positions 70–74 in the β-chain α-helix defining the sides of the peptide-binding groove, in combination with position 11 in the β-pleated sheet on the floor of the groove.
While our findings are in agreement with numerous previous studies that have identified HLA-DRB1*04 alleles as protective in PD (12, 14, 17, 20), by examining the HLA genes at sequence level resolution we refine these observations to clarify that only alleles of this lineage bearing the SE are protective. Extending the understanding of the role of HLA in PD, we find that another SE allele, HLA-DRB1*01:01, is predisposing in disease. While HLA-DRB1*01:01 has not been previously associated with PD, a recent meta-analysis of genome-wide studies in PD identified HLA-DRB6, a pseudogene on the same haplotype, as a risk allele in PD (33).
Originally defined in rheumatoid arthritis, the “SE hypothesis” sought to explain the HLA-DRB1 predisposition to disease across several allelic lineages (27). Among Europeans, the most common SE alleles belong to the HLA-DRB1*01 and HLA-DRB1*04 lineages and are predisposing to RA. The crystal structures for the SE alleles HLA-DRB1*01:01 and HLA-DRB1*04:01 reveal many mutual features with respect to the antigen-binding groove and have been shown to present several peptides in common; for example, both alleles have been crystallized with influenza virus protein hemagglutinin residues 306–318 (34). In contrast to RA, our observation in PD is of opposing effects for these alleles, with the direction of effect mediated by the presence or absence of 11-V; however, it is within reason that both risk and protective alleles present the same peptide.
In support of that model, a landmark study examining HLA function in Goodpasture disease demonstrated that risk vs. protective HLA alleles presenting the same immunodominant autoreactive peptide can produce markedly different T-cell responses (35), with the protective allele producing a more regulatory phenotype. We propose that HLA peptide specificity in PD is determined by the SE, residues of which participate in the P4, P7, and P9 pockets of the peptide- binding groove (36), while the shift to a regulatory phenotype is mediated by 11-V, which defines the specificity for the P6 pocket (Fig. 2).
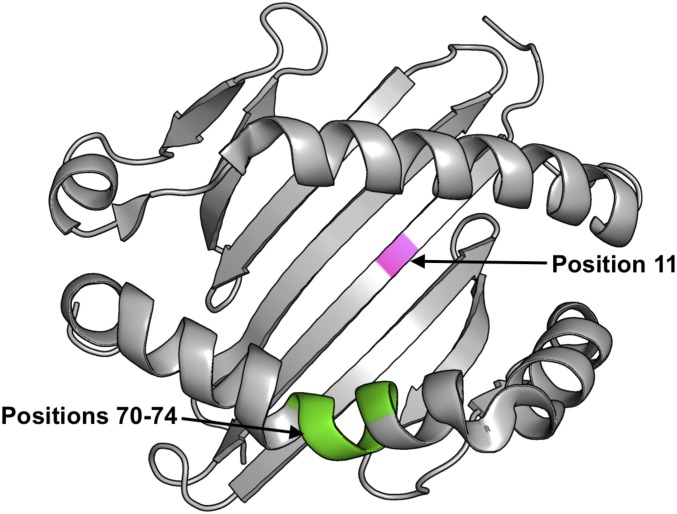
Fig. 2.
HLA-DRB1 amino acid positions mediating risk and protection in PD. Position 11 (pink) is located in the β-pleated sheet on the floor of the peptide-binding groove of the HLA-DR molecule. The SE at positions 70–74 (green) is located on the second α-helix, bordering the peptide-binding groove of HLA-DR.
Our observation that the protective effect of cigarette smoking interacts with the HLA-DRB1 SE draws another intriguing parallel to RA. In contrast to the protection afforded in PD, smoking is a well-established risk factor in RA (37). A common feature in RA is the presence of antibodies to citrullinated protein antigens (38), the generation of which has been associated with smoking (39). Further evidence suggests that citrullination enhances the binding affinity of some arithrogenic peptides to SE alleles (40), suggesting a pathway for interaction between smoking and HLA in RA. Our finding that citrullination increases the predicted affinity for HLA-DRB1 alleles protective in PD while decreasing the predicted affinity for HLA-DRB1 alleles predisposing to PD supports the notion that the epidemiologic association of smoking in both RA and PD is mediated by its posttranslational modification of antigens presented by HLA-DRB1. Finally, and in support of these observations, epidemiologic studies have shown that individuals with RA are at reduced risk of developing PD (41).
These results also lend credence to the notion that PD is, at least in part, autoimmune in nature. Further underscoring this notion, a recent study demonstrating T-cell autoreactivity to HLA class II presented peptides derived from α-synuclein, which accumulates in the PD brain (29), suggested that neuronal loss may be mediated by autoimmune mechanisms.
In summary, we find a clear and replicable role for HLA class II in predisposition and protection in PD and provide an explanation for the association of smoking with protection from disease. The association of specific combinations of amino acids that participate in critical peptide-binding pockets of the HLA class II molecule implies antigen presentation as a key factor in disease and strongly supports a role for immunogenetic variation in mediating the immunopathology of PD.
Methods
The study was approved by the University of California San Francisco (UCSF) Institutional Review Board.
Datasets.
Discovery cohort.
The discovery cohort consisted of the National Institutes of Health (NIH)-GWAS and NIH-NeuroX datasets derived from the National Institute of Neurological Disorders and Stroke-funded Neurogenetics Repository at the Coriell Institute for Medical Research, as well as the UCSF- GeneMSA dataset. After initial quality controls performed on HLA sequencing data, 3,647 samples with calling at all loci were received for analysis. Patients were removed from the discovery dataset if (i) they did not have available SNP data, (ii) they self-identified as nonwhite or non-Caucasian, (iii) they had an SNP genotyping rate <90% (see below), or (iv) the genetic population stratification analysis identified them as outliers (see below). The number of patients removed at each step of sample quality control is presented in SI Appendix, Table S7. This process resulted in a discovery dataset of 1,606 controls and 1,597 PD patients.
Replication cohort.
The replication cohort consisted of two Michael J. Fox Foundation for Parkinson’s Research datasets (BioFIND and DATATOP) and UCSF controls (UCSF1). Patients were removed from the replication dataset if (i) they did not have an allele call at all loci considered for replication (HLA-DQA1, HLA-DQB1, and HLA-DRB1) or (ii) they self-identified as nonwhite or non-Caucasian or of Hispanic origin (SI Appendix, Table S8). Because no SNP data were available for these datasets, genetic population stratification was not performed. This process resulted in a replication dataset of 462 controls and 536 PD patients.
Demographic data for the discovery and replication cohorts are presented in SI Appendix, Table S9. All patients were non-Hispanic whites from the United States with idiopathic PD. All controls are reported to be unrelated non-Hispanic white and free from neurologic disorders. Informed consent was obtained for each participant under locally approved protocols.
SNP Data.
Samples from the NIH dataset were genotyped on one of two Illumina arrays, the Immunochip or the NeuroX chip, which we refer to as the NIH-GWAS and NIH-NeuroX datasets, respectively. Some of the UCSF controls were genotyped on the HumanHap 550 BeadChip and are referred to as UCSF-GeneMSA dataset. For the UCSF-GeneMSA dataset, SNP positions were updated to the HG19 assembly using the LiftOver tool (https://genome.ucsc.edu/cgi-bin/hgLiftOver). Quality controls on samples included the genotyping rate per individuals and genetic outlier assessment, defined as individuals >6 SDs outside the mean for the first 10 PCs. Quality controls on SNPs included duplicated variant removal, Hardy–Weinberg equilibrium departure (P < 0.001), and low SNP call rate (<95%) (SI Appendix, Table S10). Quality control checks were performed at the dataset level, using R (42) and Plink v1.07 (43) software.
Control for Population Stratification.
Population stratification analysis was performed using a core set of 8,006 SNPs. These 8,006 SNPs remained after excluding those that (i) did not pass quality control checks as described above; (ii) were not genotyped in the NIH-GWAS, NIH-NeuroX, and UCSF-GeneMSA datasets; (iii) were located on nonautosomal chromosomes; (iv) were from the extended MHC region (all markers on chromosome 6 from 26 to 36 Mb); (v) were from other regions of extended LD (all markers on chromosome 8 from 6 to 16 Mb and on chromosome 17 from 40 to 45 Mb); (vi) had a minor allele frequency <1%; or (vii) were from LD pruning so that no marker had a pairwise r2 > 0.1 with any marker within a 100-SNP window. SNP data management was done using R (42) and Plink v1.07 (43). PCs were computed using the SNPRelate R package (44) on the final set of SNPs. Samples at >6 SDs from the mean for any of the first 10 PCs were considered population-specific outliers and removed (SI Appendix, Fig. S1). Using a final set of 7,918 SNPs, population stratification was also performed in regard to the 1000 Genomes white samples (SI Appendix, Fig. S2). A total of 12 samples were removed using these criteria.
HLA Next-Generation Sequencing and Genotyping.
DNA samples were typed for 11 HLA loci—HLA-A, -B, -C, -DPA1, -DPB1, -DQA1, -DQB1, -DRB1, -DRB3, -DRB4, and -DRB5—using the MIA FORA NGS high-throughput typing kit and analysis software (Immucor) and performed according to the manufacturer’s semiautomated protocol. Details are provided in SI Appendix, Methods.
Statistical Analysis.
All analyses were performed in R (42). Associations of HLA alleles with disease were tested at all levels of resolution using logistic regression. Conditioned analyses were performed by adding specific alleles/variants of interest as covariates. Only HLA alleles with a frequency >1% were analyzed. Frequencies of HLA alleles and an omnibus test for association with specific amino acids were computed using the BIGDAWG R package (45). Logistic regression was also used to test for interactions between HLA alleles and between HLA alleles and known PD risk SNPs. Haplotype estimations and association analyses with disease were performed using the “haplo.glm” function in the haplo.stats R package (46), with gender and the first four PCs as covariates. All analyses were adjusted for gender and the first four PCs to correct for population stratification (SI Appendix, Fig. S3).
To test for interaction between smoking history and HLA, the model parameters of logistic regression were estimated using the maximum likelihood method. The Wald test was used to test the mean difference between the estimated coefficient and the null parameter; typically the null hypothesis assumed it equal to 0. The significance of the interaction term was further confirmed by the likelihood ratio test on the deviance between the full model (with interaction term) and the reduced model (without interaction term). Power was assessed through simulation to obtain empirical distributions for P values. Data were simulated 100 times based on the distribution of the predictors. The effect (β) of each predictor was used to determine the outcome (0 or 1). A binary logistic regression was fit on the simulated dataset, and the P value of the interaction term was extracted. The power to detect significant interactions with a single copy of SE+V11+ was 0.96, and the power to detect interactions with two copies of SE+V11+ was 0.36. Likewise, the power to detect significant interactions with a single copy of SE+V11− was 0.82, and the power to detect interactions with two copies of SE+V11− was 0.70.
The FDR was calculated as described previously (47) and was set to 10%, giving a significance threshold of P < 0.02 for the primary association analysis. Correction for multiple testing was performed using the Bonferroni method.
The data supporting the findings of this study are available from the corresponding author on request.
Supplementary Material
Acknowledgments
We thank all members of the Parkinson Study Group who collected samples used in this study, as well as the patients and their families, whose help and participation made this work possible. Special thanks to Danillo Augusto and Yvonne Zhou for help with the figures. The samples used in this work were obtained from the Coriell Biorepository, the Deprenyl and Tocopherol Antioxidative Therapy of Parkinsonism (DATATOP) trial, and the Fox Investigation for New Discovery of Biomarkers (BioFIND; biofind.loni.usc.edu/). BioFIND is sponsored by the Michael J. Fox Foundation for Parkinson’s Research (MJFF) with support from the National Institute for Neurological Disorders and Stroke (NINDS). DATATOP samples were provided for this research by the Indiana University Genetics Biobank with support from the MJFF. This study was supported by National Institutes of Health Grant U19AI119350.
Footnotes
Conflict of interest statement: S.L.H. currently serves on the Science Advisory Boards of Symbiotix, Annexon, Bionure, and Molecular Stethoscope and on the Board of Trustees of Neurona, and has received travel reimbursement and writing assistance from F. Hoffman-La Roche for CD20-related meetings and presentations.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821778116/-/DCSupplemental.
References
- 1.Holling TM, Schooten E, van Den Elsen PJ. Function and regulation of MHC class II molecules in T-lymphocytes: Of mice and men. Hum Immunol. 2004;65:282–290. doi: 10.1016/j.humimm.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Bailey A, et al. Selector function of MHC I molecules is determined by protein plasticity. Sci Rep. 2015;5:14928. doi: 10.1038/srep14928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emile J, Truelle JL, Pouplard A, Hurez D. Association of Parkinson’s disease with HLA-B17 and B18 antigens. Nouv Presse Med. 1977;6:4144. [PubMed] [Google Scholar]
- 4.Hamza TH, Payami H. The heritability of risk and age at onset of Parkinson’s disease after accounting for known genetic risk factors. J Hum Genet. 2010;55:241–243. doi: 10.1038/jhg.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nalls MA, et al. International Parkinson Disease Genomics Consortium Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet. 2011;377:641–649. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamza TH, et al. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson’s disease. Nat Genet. 2010;42:781–785. doi: 10.1038/ng.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo Y, et al. HLA rs3129882 variant in Chinese Han patients with late-onset sporadic Parkinson disease. Neurosci Lett. 2011;501:185–187. doi: 10.1016/j.neulet.2011.05.245. [DOI] [PubMed] [Google Scholar]
- 8.Jamshidi J, et al. HLA-DRA is associated with Parkinson’s disease in Iranian population. Int J Immunogenet. 2014;41:508–511. doi: 10.1111/iji.12151. [DOI] [PubMed] [Google Scholar]
- 9.Puschmann A, et al. Human leukocyte antigen variation and Parkinson’s disease. Parkinsonism Relat Disord. 2011;17:376–378. doi: 10.1016/j.parkreldis.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, et al. Genome-wide association study identifies candidate genes for Parkinson’s disease in an Ashkenazi Jewish population. BMC Med Genet. 2011;12:104. doi: 10.1186/1471-2350-12-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Do CB, et al. Web-based genome-wide association study identifies two novel loci and a substantial genetic component for Parkinson’s disease. PLoS Genet. 2011;7:e1002141. doi: 10.1371/journal.pgen.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed I, et al. Association between Parkinson’s disease and the HLA-DRB1 locus. Mov Disord. 2012;27:1104–1110. doi: 10.1002/mds.25035. [DOI] [PubMed] [Google Scholar]
- 13.Ma ZG, Liu TW, Bo YL. HLA-DRA rs3129882 A/G polymorphism was not a risk factor for Parkinson’s disease in Chinese-based populations: A meta-analysis. Int J Neurosci. 2015;125:241–246. doi: 10.3109/00207454.2014.926349. [DOI] [PubMed] [Google Scholar]
- 14.Wissemann WT, et al. Association of Parkinson disease with structural and regulatory variants in the HLA region. Am J Hum Genet. 2013;93:984–993. doi: 10.1016/j.ajhg.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nalls MA, et al. International Parkinson’s Disease Genomics Consortium, Parkinson’s Study Group Parkinson’s Research: The Organized GENetics Initiative, 23andMe, GenePD, NeuroGenetics Research Consortium, Hussman Institute of Human Genomics, Ashkenazi Jewish Dataset Investigator, Cohorts for Health and Aging Research in Genetic Epidemiology, North American Brain Expression Consortium, United Kingdom Brain Expression Consortium, Greek Parkinson’s Disease Consortium, Alzheimer Genetic Analysis Group Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill-Burns EM, Factor SA, Zabetian CP, Thomson G, Payami H. Evidence for more than one Parkinson’s disease-associated variant within the HLA region. PLoS One. 2011;6:e27109. doi: 10.1371/journal.pone.0027109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chuang YH, et al. Pooled analysis of the HLA-DRB1 by smoking interaction in Parkinson disease. Ann Neurol. 2017;82:655–664. doi: 10.1002/ana.25065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pappas DJ, et al. Significant variation between SNP-based HLA imputations in diverse populations: The last mile is the hardest. Pharmacogenomics J. 2018;18:367–376. doi: 10.1038/tpj.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saiki M, et al. Association of the human leucocyte antigen region with susceptibility to Parkinson’s disease. J Neurol Neurosurg Psychiatry. 2010;81:890–891. doi: 10.1136/jnnp.2008.162883. [DOI] [PubMed] [Google Scholar]
- 20.Sun C, et al. HLA-DRB1 alleles are associated with the susceptibility to sporadic Parkinson’s disease in Chinese Han population. PLoS One. 2012;7:e48594. doi: 10.1371/journal.pone.0048594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lampe JB, et al. HLA typing and Parkinson’s disease. Eur Neurol. 2003;50:64–68. doi: 10.1159/000072500. [DOI] [PubMed] [Google Scholar]
- 22.Leheny WA, Davidson DL, deVane P, House AO, Lenman JA. HLA antigens in Parkinson’s disease. Tissue Antigens. 1983;21:260–261. doi: 10.1111/j.1399-0039.1983.tb00167.x. [DOI] [PubMed] [Google Scholar]
- 23.Robinson J, et al. The IMGT/HLA database. Nucleic Acids Res. 2013;41:D1222–D1227. doi: 10.1093/nar/gks949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 25.Maiers M, Gragert L, Klitz W. High-resolution HLA alleles and haplotypes in the United States population. Hum Immunol. 2007;68:779–788. doi: 10.1016/j.humimm.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Cao K, et al. Analysis of the frequencies of HLA-A, -B, and -C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum Immunol. 2001;62:1009–1030. doi: 10.1016/s0198-8859(01)00298-1. [DOI] [PubMed] [Google Scholar]
- 27.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis: An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 28.Quik M. Smoking, nicotine and Parkinson’s disease. Trends Neurosci. 2004;27:561–568. doi: 10.1016/j.tins.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 29.Sulzer D, et al. T cells from patients with Parkinson’s disease recognize α-synuclein peptides. Nature. 2017;546:656–661. doi: 10.1038/nature22815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen KK, et al. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology. 2018;154:394–406. doi: 10.1111/imm.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makrygiannakis D, et al. Smoking increases peptidylarginine deiminase 2 enzyme expression in human lungs and increases citrullination in BAL cells. Ann Rheum Dis. 2008;67:1488–1492. doi: 10.1136/ard.2007.075192. [DOI] [PubMed] [Google Scholar]
- 32.Sidney J, et al. Citrullination only infrequently impacts peptide binding to HLA class II MHC. PLoS One. 2017;12:e0177140. doi: 10.1371/journal.pone.0177140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang D, et al. International Parkinson’s Disease Genomics Consortium, 23andMe Research Team A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet. 2017;49:1511–1516. doi: 10.1038/ng.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones EY, Fugger L, Strominger JL, Siebold C. MHC class II proteins and disease: A structural perspective. Nat Rev Immunol. 2006;6:271–282. doi: 10.1038/nri1805. [DOI] [PubMed] [Google Scholar]
- 35.Ooi JD, et al. Dominant protection from HLA-linked autoimmunity by antigen-specific regulatory T cells. Nature. 2017;545:243–247. doi: 10.1038/nature22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Agudelo WA, Patarroyo ME. Quantum chemical analysis of MHC–peptide interactions for vaccine design. Mini Rev Med Chem. 2010;10:746–758. doi: 10.2174/138955710791572488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoovestol RA, Mikuls TR. Environmental exposures and rheumatoid arthritis risk. Curr Rheumatol Rep. 2011;13:431–439. doi: 10.1007/s11926-011-0203-9. [DOI] [PubMed] [Google Scholar]
- 38.Huizinga TW, et al. Refining the complex rheumatoid arthritis phenotype based on specificity of the HLA-DRB1 shared epitope for antibodies to citrullinated proteins. Arthritis Rheum. 2005;52:3433–3438. doi: 10.1002/art.21385. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z, et al. Protein carbamylation links inflammation, smoking, uremia and atherogenesis. Nat Med. 2007;13:1176–1184. doi: 10.1038/nm1637. [DOI] [PubMed] [Google Scholar]
- 40.Anderson KM, et al. A molecular analysis of the shared epitope hypothesis: Binding of arthritogenic peptides to DRB1*04 alleles. Arthritis Rheumatol. 2016;68:1627–1636. doi: 10.1002/art.39636. [DOI] [PubMed] [Google Scholar]
- 41.Sung YF, et al. Reduced risk of Parkinson disease in patients with rheumatoid arthritis: A nationwide population-based study. Mayo Clin Proc. 2016;91:1346–1353. doi: 10.1016/j.mayocp.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 42.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: 2015. [Google Scholar]
- 43.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng X, et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28:3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pappas DJ, Marin W, Hollenbach JA, Mack SJ. Bridging immunogenomic data analysis workflow gaps (BIGDAWG): An integrated case-control analysis pipeline. Hum Immunol. 2016;77:283–287. doi: 10.1016/j.humimm.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sinwell J, Schaid D. 2016 Haplo Stats: Statistical analysis of haplotypes with traits and covariates when linkage phase is ambiguous. Available at https://cran.r-project.org/web/packages/haplo.stats/vignettes/manualHaploStats.pdf. Accessed March 4, 2019.
- 47.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.