Significance
Ubiquitination is an important posttranslational modification that regulates almost every aspect of cellular functions. Ubiquitin can form chains of different topology; each has a distinctive role in dictating the function and fate of the modified proteins. Deubiquitinases (DUBs) reverse ubiquitination. How DUBs recognize ubiquitin chains is a topic of immense interest due to the therapeutic potentials of human DUBs. We obtained the atomic details of the USP9X catalytic core, a DUB involved in cancers and developmental disorders, and revealed its unusual mechanisms of action using a set of activity-based ubiquitin probes. These probes will propel future investigation of how DUBs recognize and process ubiquitin chains and identify potential new sites on DUBs for drug discovery.
Keywords: deubiquitinase, activity-based probes, zinc finger, linkage specificity, USP9X
Abstract
USP9X is a conserved deubiquitinase (DUB) that regulates multiple cellular processes. Dysregulation of USP9X has been linked to cancers and X-linked intellectual disability. Here, we report the crystal structure of the USP9X catalytic domain at 2.5-Å resolution. The structure reveals a canonical USP-fold comprised of fingers, palm, and thumb subdomains, as well as an unusual β-hairpin insertion. The catalytic triad of USP9X is aligned in an active configuration. USP9X is exclusively active against ubiquitin (Ub) but not Ub-like modifiers. Cleavage assays with di-, tri-, and tetraUb chains show that the USP9X catalytic domain has a clear preference for K11-, followed by K63-, K48-, and K6-linked polyUb chains. Using a set of activity-based diUb and triUb probes (ABPs), we demonstrate that the USP9X catalytic domain has an exo-cleavage preference for K48- and endo-cleavage preference for K11-linked polyUb chains. The structure model and biochemical data suggest that the USP9X catalytic domain harbors three Ub binding sites, and a zinc finger in the fingers subdomain and the β-hairpin insertion both play important roles in polyUb chain processing and linkage specificity. Furthermore, unexpected labeling of a secondary, noncatalytic cysteine located on a blocking loop adjacent to the catalytic site by K11-diUb ABP implicates a previously unreported mechanism of polyUb chain recognition. The structural features of USP9X revealed in our study are critical for understanding its DUB activity. The new Ub-based ABPs form a set of valuable tools to understand polyUb chain processing by the cysteine protease class of DUBs.
Deubiquitinases (DUBs) remove a key posttranslational modification by ubiquitin (Ub) from target proteins. Of the seven classes of DUBs, Ub-specific proteases (USPs) comprise the largest subfamily and account for over half of the DUBs encoded by the human genome (1). The USPs are cysteine proteases characterized by a conserved USP catalytic domain (CD) fold, which resembles an overall hand-like structure with fingers, thumb, and palm subdomains. The core fold can be divided into six conserved boxes, where the N-terminal box 1 contains a Cys of the catalytic triad, and the C-terminal boxes 5 and 6 contain a His and an Asp/Asn residue, respectively (2). The USP CD architecture is structurally conserved, although insertions at the loop regions between the boxes can drastically alter the size of the USP CDs (300–800 residues) (1–3).
Previous structural and biochemical studies of USP CDs, both in apo forms and in complex with Ub substrates, have provided insights into the molecular mechanism governing the diversity of USP activity regulation and substrate specificity (3, 4). The most-studied USP7 is regulated by both its Ub substrate and its C-terminal Ub-like domains (5, 6). Apo-USP7 adopts an inactive conformation, with loops near the active site blocking Ub binding to a misaligned catalytic triad. The binding of Ub induces conformational changes that promote substrate engagement and realignment of the catalytic residues into a productive state (5). A C-terminal peptide of USP7 further activates the CD activity by binding to an adjacent activation cleft to stabilize the active conformation of USP7 (7). Unlike other subfamilies of DUBs, USPs generally show a limited preference for a polyUb chain of different linkages. Only a few USP structures in complex with a diUb were reported (8–11). The crystal structures of the zebrafish CYLD CD in complex with M1- and K63-linked diUbs revealed the important contributions of the insertions to influence the proximal Ub binding (8). A zinc finger (ZnF) motif in the fingers subdomain of USP21 CD accommodates the distal Ub to allow specificity for a linear polyUb chain, as well as a diUb-like modifier ISG15 (9). A recent structure of USP18 CD in complex with ISG15 provides a molecular basis for its unique specificity toward ISG15 instead of Ub (10). More recently, the cocrystal structure of USP30 CD with K6-linked diUb revealed the engagement of a proximal Ub binding site responsible for K6-linkage specificity (11).
USP9X was first identified as the fat facets (faf) gene in Drosophila with key roles in eye and embryo development (12). Subsequent homolog analyses revealed a highly conserved protein from Drosophila to mammals (>90%) with important functions in both development and diseases (13, 14). As the X-linked ortholog of faf, human USP9X escapes from X-chromosome inactivation. A highly related paralog, USP9Y (91% sequence identity), is found on the Y chromosome, the function of which remains uncharacterized. USP9X protein regulates numerous cellular processes, including cell polarity, apoptosis, migration, and stem cell self-renewal (15). The importance of USP9X is further highlighted by a growing list of identified substrates and binding partners, including E3 ligases ITCH, SMURF1, and MARCH7 (15–18). USP9X is up-regulated in cervical, colorectal cancers, as well as myeloma, and proposed as a therapeutic target for cancer treatment (19, 20). Dysregulation of USP9X is also linked to neurodevelopmental disorders and neurodegenerative diseases, including epilepsy, autism, Parkinson’s, and Alzheimer’s disease (20–22). Multiple genetic studies have established the causal link between USP9X mutations and X-linked intellectual disability (15, 23, 24). Despite the importance of USP9X, our understanding of USP9X structure and activity remains limited.
We determined the X-ray crystal structure of the USP9X CD and characterized its activity using a set of novel activity-based di- and triUb probes (ABPs) and revealed the role of key structural elements responsible for polyUb chain recognition and cleavage. Comparison of the structure of USP9X with those of other USPs in complex with diUbs suggests that USP9X CD harbors three potential sites (S2, S1, and S1′) that could bind to distal, central, and proximal Ubs of a polyUb chain, respectively. A unique β-hairpin insertion in sequence box 6 contributes to the recognition of the proximal Ub in a linkage-specific manner. Furthermore, labeling with triUb ABPs revealed linkage-dependent endo- or exo-cleavage by USP9X. Unexpectedly, besides the catalytic cysteine, we also identified that a noncatalytic cysteine close to the catalytic cleft reacts with a K11-linked diUb probe, which implicates that USP9X samples the conformational space of diUb before a productive substrate engagement. Taking these data together, we propose a model in which the USP9X CD serves as an extensive platform to accommodate multiple Ub moieties of a polyUb chain to achieve linkage-specific cleavage.
Results
Structure of the USP9X CD.
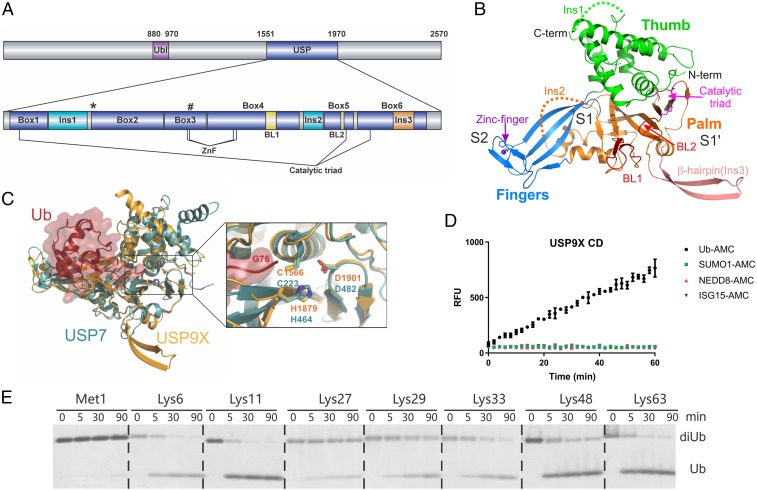
USP9X is a large protein of over 2,500 aa, with little domain annotation outside the CD (Fig. 1A). The crystal structure of the USP9X CD (amino acids 1,551–1,970) was solved at 2.5-Å resolution in space group P21 using the USP7 CD as a molecular replacement search model. The asymmetric unit contains four copies of USP9X that show a noncrystallographic pseudo C2 symmetry. Electron density for residues 1,593–1,631, 1,839–1,858, 1,891–1,895, and 1,959–1,960 of chain A and similar regions in chains B, C, and D is not observed, and these regions are presumably disordered (Fig. 1 A and B). Chain A in the asymmetric unit was used in structural analysis unless otherwise specified. A Dali search (25) revealed the core structure of USP9X is closest to the USP7 CD, with an rmsd of about 2.4 Å over 350 aligned amino acids and a z-score of 34. The overall structure represents a canonical USP fold, comprising thumb, palm, and fingers subdomains (Fig. 1B). Additional features common to other USP CDs are also found in USP9X. For example, the tip of the fingers subdomain contains a CHC2-type ZnF motif with a zinc ion coordinated by residues Cys1727, His1729, Cys1771, and Cys1774 arranged in a tetrahedron (SI Appendix, Fig. S1A). Unexpectedly, a nearby Cys1724 is not involved, as seen in other known USP structures, where the zinc is coordinated by four cysteines.
Fig. 1.
Structure and activity of USP9X. (A) Domain architecture of the full-length USP9X and the CD. Sites for surface entropy reduction mutations (K1637E1638) and ZnF mutations (Cys1727 or H1729) are marked with an asterisk (*) and pound sign (#), respectively. Blocking loops are colored yellow. (B) Crystal structure of the USP9X CD. The fingers, palm, and thumb subdomains are colored blue, orange, and green, respectively. The β-hairpin insertion (Ins3) is salmon-colored, and the two blocking loops are colored in red. Residues in the catalytic triad and the ZnF are shown in sticks. The zinc ion is shown as a sphere. S2, S1, and S1′ indicate three potential Ub binding sites. (C) Superposition of the USP9X and USP7/Ub-aldehyde (PDB ID code 1NBF) structures. Ub-aldehyde is shown in surface and ribbon representations (red), whereas USP7 (teal) and USP9X (orange) are shown in ribbon. (Right) A zoomed-in view of the overlay of the active sites of USP7 and USP9X. The catalytic triad residues in USP9X and USP7 and Ub Gly76 are labeled. (D) Activity assay using fluorogenic substrates showing that USP9X CD hydrolyzes Ub-AMC substrate efficiently but not SUMO1-, NEDD8-, or ISG15-AMC. (E) Gel-based cleavage assay of the USP9X CD using native M1-, K6-, K11-, K27-, K29-, K33-, K48-, and K63-diUb substrates.
Structural alignment of USP9X with a USP7/Ub-aldehyde covalent complex (PDB ID code 1NBF) reveals that the catalytic triad (Cys1566, His1879, Asp1901) superimposes well with the catalytic triad of USP7 in the active conformation (Fig. 1C). Notably, a Gln1683 residue is positioned close (3.6 Å) to the catalytic Cys1566 in the apo structure (SI Appendix, Fig. S1B). It must adopt a different rotamer conformation to accommodate the C terminus of a Ub substrate as observed for an equivalent Gln293 in the USP7–Ub complex structure (SI Appendix, Fig. S1C). Interestingly, Gln1683 is conserved among USP DUBs and often with a juxtaposed second Gln residue. Close to the active site, USP9X contains two blocking loops, BL1 (1,801–1,810) connecting β13 and β14 and BL2 (1,872–1,879) connecting β16 and β17 (Fig. 1B and SI Appendix, Fig. S2), similar to those in USP7 (5–7, 26) that form the binding cleft for the C-terminal tail of Ub. A unique USP9X β-hairpin insertion is located next to the two blocking loops (Fig. 1B), suggesting the insertion may have some effect on the recognition of the Ub tail. Interestingly, a loop connecting helices α4 and α5, which corresponds to the switching loop of USP7 (6), is 2 aa longer than that of USP7, and protrudes into space otherwise occupied by the C-terminal activating tail in USP7 (SI Appendix, Fig. S1D). The extension may exclude a regulatory mechanism similar to that of USP7 (6).
The USP9X CD contains three large insertions in the sequence. Two of them are located at the common insertion points of the core domain (2). A 39-aa insertion (1,593–1,631) between boxes 1 and 2 (helices α2 and α3), and a 22-aa insertion (1,838–1,859) between boxes 4 and 5 (helix α8 and strand β16) (Fig. 1A and SI Appendix, Fig. S2) cannot be traced in the electron density map and are presumably disordered. The importance of these loops remains unclear, but a high percentage of negatively charged Asp and Glu residues in the α2–α3 loop may be of significance. A third 20-aa insertion (1,924–1,943) between α9 and β22 within box 6 forms an unexpected β-hairpin structure in the palm subdomain of the USP fold. Sequence alignment revealed the β-hairpin insertion is completely conserved among USP9X from different species, as well as in the related USP9Y, whereas the two other insertions are not (SI Appendix, Fig. S2). This suggests the β-hairpin insertion may be functionally important. Analysis of the evolutionary conservation profile of USP9X CD using ConSurf algorithm (27) indicates the active site, the Ub C-terminal tail binding region, and a patch of the fingers subdomain, are highly conserved (SI Appendix, Fig. S3).
A survey of USP domain structural features reveals the ZnF and β-hairpin insertion in USP9X may be functionally significant (SI Appendix, Fig. S4). Both USP9X and the K63 and M1 dual-specific USP, CYLD, have a large insertion within sequence box 6. The insertion in CYLD forms helices/bend structural elements (28) located next to the two blocking loops and also extends a β-hairpin in the palm subdomain that contributes to proximal Ub recognition (8). Similarly, the insertion in USP9X may also be involved in polyUb chain specificity. Furthermore, the fingers subdomain of some USPs is known to bind to a distal Ub of a polyUb chain, as shown in the USP21-linear diUb complex (9). Based on structural modeling and comparison with other USPs, we postulate that the USP9X CD may harbor three Ub binding sites (S2, S1, and S1′) (Fig. 1B) and cleave a polyUb chain linkage-dependently.
DUB Activity of USP9X.
We first measured the DUB activity of USP9X CD against fluorogenic 7-amido-4-methylcoumarin (AMC)-derived Ub and Ub-like modifiers (UBLs). Unlike some other USPs that show cross-activities toward UBLs, such as SUMO and ISG15 (29, 30), USP9X is only active toward Ub, with no measurable activity for SUMO1-, NEDD8-, or ISG15-AMC (Fig. 1D). Using the Ub-AMC substrate, we determined the kinetic parameters for USP9X CD with kcat = 0.25 ± 0.01 min−1 and Km = 0.10 ± 0.02 µM (SI Appendix, Fig. S5A and Table S1). The kcat and Km of USP9X CD are vastly different from those of the USP7 CD (kcat = 4.6 min−1 and Km = 44.2 µM) (31). The specificity constant (kcat/Km) of USP9X (2.5 min−1µM−1) is 25-fold higher than that of USP7 CD (0.10 min−1µM−1). We next assessed the activity of USP9X CD toward eight diUb substrates of different linkages. USP9X CD displayed various cleavage efficiencies, with a preference for K11-, K63-, K48-, and K6-linkages (Fig. 1E and SI Appendix, Fig. S5B). Low level of cleavage was observed for K27-, K29-, and K33-diUbs and no cleavage observed for M1-linked diUb (Fig. 1E). We further assessed the cleavage of K6-, K11-, K63-, K48-linked tetraUb substrates and observed that K11-linked tetraUb was processed more efficiently than K63- or K48-linked chains, with poor cleavage of K6-linked tetraUb (SI Appendix, Fig. S5C).
PolyUb Probes for Interrogating USPs.
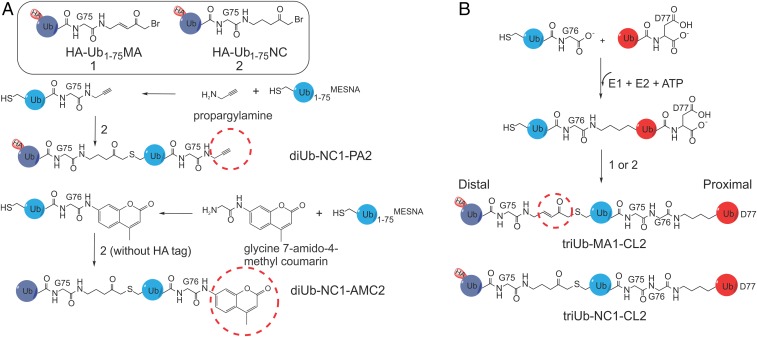
ABPs capture DUBs in action. To study the three potential binding sites of USP9X, we generated a set of ABPs. We systematically name the ABPs based on the chemical bond subjected to DUB cleavage, which can be a cleavable native peptide bond (CL), a noncleavable linker (NC), a Michael acceptor (MA) warhead, or the abbreviation of a specific reactive group, followed by a number to indicate the location of the bond of interest. We previously reported K48- and K63-diUb probes with an internal MA (32), which are now denoted K48- or K63-diUb-MA1. In this study, we additionally generated K6- and K11-diUb-MA1 ABPs (SI Appendix, Fig. S6). These diUb-MA1 ABPs interrogate the Ub binding to the S1 site and a potential S1′ (proximal) site in the DUB catalytic core. To probe the Ub binding at a potential S2 site in DUBs, we prepared K6-, K11-, K48-, and K63-linked diUb probes that contain an NC linker (33) and a terminal propargylamine (PA) warhead (diUb-NC1-PA2) (Fig. 2A and SI Appendix, Fig. S6). For enzymatic kinetics quantification, we generated fluorogenic AMC derivatives of diUbs (diUb-NC1-AMC2) (Fig. 2A and SI Appendix, Fig. S7).
Fig. 2.
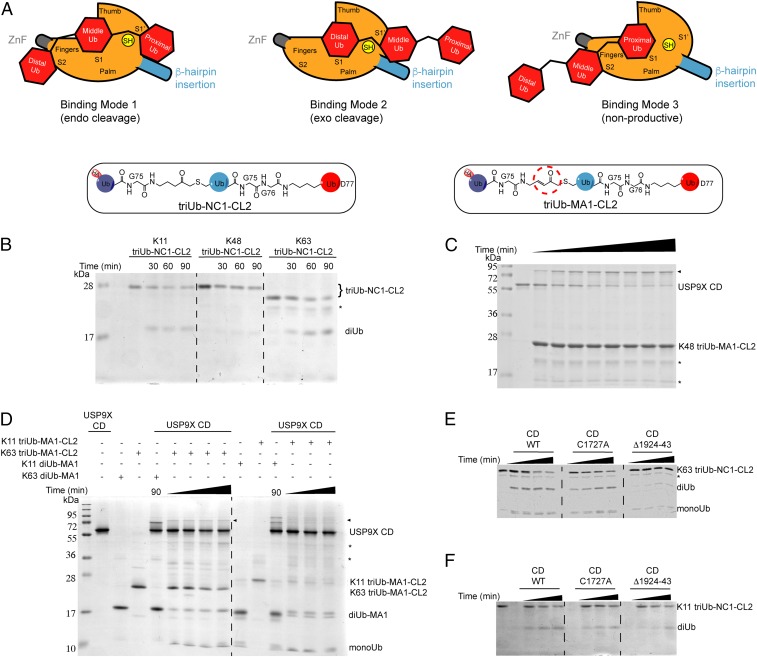
Activity-based di- and triUb probes. (A) Structures and synthesis of the diUb-NC1-PA2 probes and diUb-NC1-AMC2 fluorogenic substrates. (B) Generation of two types of hybrid triUb probes using a chemoenzymatic approach. TriUb-NC1-CL2 contains a noncleavable bond between the distal (light purple) and middle (blue) Ubs and a native isopeptide linkage between the middle and proximal (red) Ubs. TriUb-MA1-CL2 contains an MA warhead between the distal and middle Ubs and a native isopeptide linkage between the middle and proximal Ubs.
We also designed and synthesized two classes of hybrid triUb probes (Fig. 2B and SI Appendix, Fig. S8) (see Methods for details). TriUb-NC1-CL2 (Fig. 2B) ensures that DUB-catalyzed cleavage only occurs when the middle Ub binds to the DUB S1 site, while the distal and proximal Ubs occupy the potential S2 and S1′ sites, respectively. TriUb-MA1-CL2 (Fig. 2B) differentiates two binding modes in one assay. If the DUB S1 site engages the middle Ub of a triUb-MA1-CL2, we expect a cleavage at the native isopeptide bond to produce a diUb-MA1 and a Ub. If the DUB S1 site engages the distal Ub, we expect labeling of the DUB active site cysteine by the probe’s MA that gives a triUb conjugated product. These ABPs were synthesized and the identity characterized by mass spectrometry (SI Appendix, Fig. S8).
The USP9X Zinc-Finger Is a Crucial Structural Motif.
A ZnF motif in the fingers subdomain is found in many USPs, although its function is not entirely clear. The ZnF of USP21 is important for binding the distal Ub in a linear diUb (SI Appendix, Fig. S9A) as well as the distal UBL domain in ISG15 (9). To evaluate the role of the ZnF of USP9X, we mutated residues chelating the zinc ion, C1727A, or H1729A, and measured their activities.
For Ub-AMC substrate, the C1727A and H1729A mutants showed a five- and sevenfold increase in Km compared with WT, respectively, without substantial change in kcat (SI Appendix, Fig. S9B and Table S1). We next used internally quenched fluorescence (IQF)-diUb substrates of K11- and K63-linkage to monitor the continuous isopeptide cleavage and observed reduced cleavage activity of the C1727A mutant (SI Appendix, Fig. S9C), again supporting the importance of the ZnF in the S1 site Ub binding. This is also consistent with the reduced labeling of the ZnF mutants by monoUb-based ABP (SI Appendix, Fig. S10A).
To measure the effect of the ZnF on S2-S1 diUb binding, we interrogated the USP9X CD using diUb-NC1-PA2 (Fig. 2A) and observed reduced labeling of the ZnF mutants compared with WT at a single time point (2 h) by K6-, K11-, and K48-probes, but with little change for the K63-linked probe (SI Appendix, Fig. S10B). To confirm the result, a time-dependent labeling was carried out. The results show a clear reduction in the labeling efficiency for the K11-diUb-NC1-PA2 probe but little change for the K63-diUb probe (SI Appendix, Fig. S10C). These results reveal that the ZnF in USP9X plays an important role in chain linkage-specific recognition of diUb.
Role of the USP9X β-Hairpin Insertion on Catalysis.
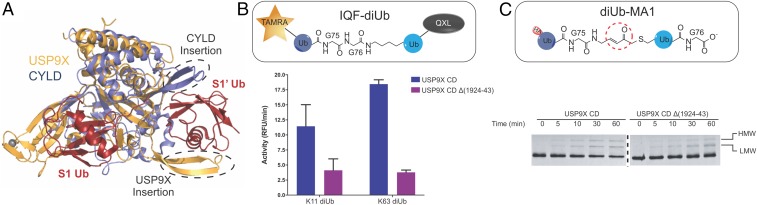
An insertion in CYLD sequence box 6 is important for the recognition of the proximal Ub in M1- and K63-diUbs (8). The insertion in box 6 of the USP9X CD could play a similar role. Unlike the CYLD insertion that is partially disordered in the apo structure and induced into an extended β-hairpin upon substrate binding (8, 28), the USP9X β-hairpin insertion is ordered in the apo structure. Crystal packing analysis revealed the β-hairpin pairs with the β-strands of the fingers subdomain of a symmetry-related molecule (SI Appendix, Fig. S11A). Secondary structure (34) and disorder (35) predictions suggest this insertion has no strong propensity to adopt an α-helix, β-strand, or disordered conformation (SI Appendix, Fig. S11 B and C). This implicates that the observed conformation of the β-hairpin could have been influenced by crystal packing, and it may adopt a different conformation upon substrate binding. Structure overlay of USP9X CD with CYLD CD in complex with diUb (PDB ID code 3WXE) shows that the USP9X β-hairpin is close to, but on the opposite side of a modeled S1′ Ub compared with the CYLD β-hairpin (Fig. 3A).
Fig. 3.
Interrogation of the role of β-hairpin in proximal Ub recognition (A) Superposition of the USP9X CD apo structure with CYLD in complex with M1-diUb (PDB ID code 3WXE). USP9X, CYLD, and diUb are colored orange, purple, and red, respectively. The CYLD and USP9X insertions are circled and labeled. (B) Cleavage activity of WT USP9X CD and deletion mutant Δ(1924–1943) assayed using IQF-diUb substrates of K11 and K63 linkages. (C) Labeling of WT and β-hairpin deletion mutant USP9X CD by K11 diUb-MA1 probe at different time points.
To understand the role of the USP9X β-hairpin insertion, we generated a deletion mutant Δ(1924–1943) and assessed its activity. We only observed a slight change of the mutant in kcat and Km toward Ub-AMC (SI Appendix, Table S1), and monoUb-based Ub-PA probe labeling compared with WT (SI Appendix, Fig. S12 A and B). Furthermore, labeling with diUb-NC1-PA2 probes, which measures S2-S1 binding, also only showed slightly reduced labeling efficiency for the deletion mutant (SI Appendix, Fig. S12C). Together, these observations suggest that removal of the β-hairpin insertion has a minimal impact on S2-S1 Ub binding.
We next accessed S1-S1′ site engagement using diUb-MA1 probes. The WT USP9X CD is most strongly labeled by K48- followed by K63- and K11-diUb-MA1 probes, but not by K6-diUb-MA1 in the time frame treated (1 h) (SI Appendix, Fig. S12D). Labeling of USP9X Δ(1924–1943) showed similar linkage preference (SI Appendix, Fig. S12D). Using a gel-based DUB assay, the deletion mutant showed reduced cleavage of K11-, K48-, and K63-linked diUb (SI Appendix, Fig. S5B) and triUb substrates (SI Appendix, Fig. S12E) compared with WT. To quantify the activity, we used the IQF-diUb substrates for continuous monitoring of diUb cleavage. With K11- and K63-linked IQF-diUb, the β-hairpin deletion reduced the cleavage efficiency significantly (Fig. 3B). Overall, the β-hairpin insertion is important for the S1-S1′ cleavage activity.
Labeling by K11 DiUb-MA1 Probe Reveals the Reactivity of a Noncatalytic Cysteine.
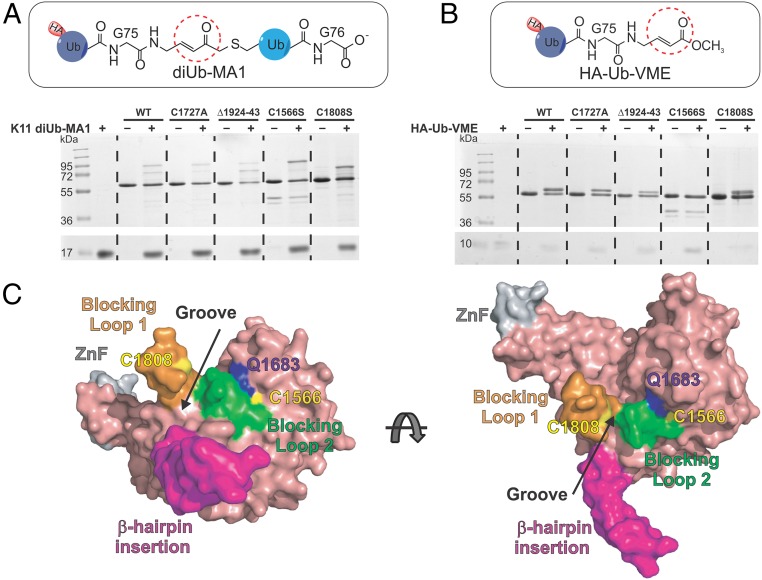
In the WT USP9X CD labeling experiment with diUb-MA1 probes of different linkages, we observed an interesting difference in the labeling pattern. As expected, the K48- and K63-diUb-MA1 probes yield one upshifted product band. Surprisingly, with the K11-diUb-MA1 probe, we observed two labeling bands with comparable intensities (Fig. 3C), which we denoted HMW and LMW bands (for high- and low-molecular weights).
To understand the nature of the labeling products, we subjected the HMW and LMW bands to tryptic digestion and MS/MS analysis. The results suggest that conjugation of Cys1808 on the blocking loop BL1 by the K11 diUb-MA1 probe results in the HMW band, while the labeling of the catalytic Cys1566 leads to the LMW band. Subsequent mutational analysis confirmed the finding (Fig. 4A). Specifically, labeling of the active site mutant C1566S by the K11-diUb-MA1 probe showed only the HMW band, whereas the C1808S mutant showed only the LMW band. Notably, when the C1566S mutant was incubated with HA-Ub-vinyl methyl ester (VME), which bears the same MA as that in the diUb-MA1 probes, no labeling band was observed (Fig. 4B). This indicates the proximal Ub in the K11-diUb-MA1 is required for the alternate labeling of Cys1808 in USP9X CD.
Fig. 4.
A noncatalytic Cys1808 in USP9X CD is labeled by the K11-diUb-MA1 probe. (A) Labeling band pattern of the WT and mutant USP9X CD by K11 diUb-MA1 probe. C1566S is a catalytically dead mutant but is labeled at C1808 position. Mutation of the noncatalytic cysteine Cys1808 to Ser abolished the HMW labeling band but retained the LMW labeling band. (B) HA-Ub-VME only labels the catalytic Cys1566 but not Cys1808 and is not affected by ZnF mutation. (C) Surface representation of USP9X CD revealing a groove between the blocking loops BL1 (orange) and BL2 (green). Noncatalytic Cys1808 (yellow) labeled by K11-diUb-MA1 is on BL1 facing the groove.
We also subjected the ZnF mutant C1727A and the β-hairpin deletion mutant Δ(1924–1943) to the same labeling assay with a K11 diUb-MA1 probe. Interestingly, of the two labeling bands, the LMW band was diminished for the ZnF mutant (Fig. 4A). The opposite was observed for the deletion mutant Δ(1924–1943) with a reduced HMW band.
We generated a Q1683A mutation to unblock the access to the catalytic triad (SI Appendix, Fig. S1C) and subjected the mutant to labeling with the K11-diUb-MA1 probe. Notably, the LMW band (labeling of catalytic Cys1566) showed a more increased intensity than the HMW band (SI Appendix, Fig. S13A), suggesting improved access to the active site. The Q1683A mutant also showed a large increase in Km and little decrease in kcat for Ub-AMC substrate compared with the WT (SI Appendix, Fig. S13B and Table S1). However, no significant change in kcat and Km values was observed for the IQF-K11-diUb substrate (SI Appendix, Fig. S13C).
USP9X Displays Linkage-Specific Endo- and Exo-DUB Activities.
The multiple Ub binding sites and linkage specificity of USP9X CD prompted us to investigate its endo- and exo-Ub chain cleavage activity. Using the diUb-NC1-AMC2 substrates (Fig. 2A), we performed a kinetic analysis of S2-S1 diUb cleavage and found that USP9X CD possesses robust endo-DUB activity for K11- and K63-diUb substrates. USP9X CD showed a twofold higher catalytic activity (kcat/Km) for K11- compared with K63-linked diUb-NC1-AMC2 (SI Appendix, Table S2). Notably, USP9X CD is much less active against K48-diUb-NC1-AMC2 with an eightfold lower kcat/Km, suggesting that the positioning of the distal Ub influences the endo-cleavage activity.
We then used the hybrid triUb probes that mimic polyUb chain substrates (Fig. 2B). Three binding modes are possible with two of them leading to chain cleavage (endo and exo) (Fig. 5A). To probe the effect of simultaneous binding of a polyUb substrate to all three potential DUB Ub binding sites (S2, S1, and S1′, mode 1), we used the triUb-NC1-CL2 probes (Fig. 2B), cleavage of which indicates an endo-mode. We found that USP9X CD endo-cleaved K11- and K63-triUb-NC1-CL2 into diUb and monoUb efficiently (Fig. 5B and SI Appendix, Fig. S14 A and B). However, little cleavage of K48 triUb-NC1-CL2 was detected (Fig. 5B and SI Appendix, Fig. S14C). These assays were run in parallel with native diUb substrates, and for all linkages (K11, K48, and K63) efficient cleavage by the USP9X CD was observed (SI Appendix, Fig. S14). We also used USP2 CD, which is permissive for different Ub chain linkages, as a comparison and observed efficient cleavage of triUb-NC1-CL2 of all three linkages (K11, K48, and K63) (SI Appendix, Fig. S14).
Fig. 5.
USP9X CD exhibits linkage-specific endo/exo recognition of polyUb chains. (A) Illustration of the three potential binding modes of a triUb to the S2, S1, and S1′ binding sites of USP9X CD. (B) Cleavage of the K11-, K48-, and K63-linked triUb-NC1-CL2 by WT USP9X CD at three different time points. (C) Incubation of WT USP9X CD with K48 triUb-MA1-CL2 probe at 0-, 5-, 10-, 15-, 30-, 45-, 60-, and 90-min time points. (D) Labeling of WT USP9X CD by K63- and K11-linked triUb-MA1-CL2 probes at 15-, 30-, 60-, and 90-min time points and 30-, 60-, and 90-min time points, respectively. Labeling of WT USP9X CD by K63- and K11-linked diUb-MA1 probes is included for comparison. (E) Comparison of cleavage of K63- and K11- (F) triUb-NC1-CL2 by USP9X WT, ZnF mutant (C1727A) and deletion mutant (Δ1924–43) up to 120 min and 90 min, respectively. The reaction products were analyzed by SDS/PAGE and Coomassie brilliant blue staining. The asterisk denotes contaminating band in the probe preparations and triangle denotes labeling band.
Next, the triUb-MA1-CL2 probes (Fig. 2B) were used to detect both endo- and exo-cleavage modes by DUBs in one assay. We envisioned three possible outcomes (SI Appendix, Fig. S15A) In scenario 1, the DUB engages the triUb substrate in a binding mode that leads to endo-cleavage of the triUb-MA1-CL2 to yield di- and monoUb species. The resulting diUb-MA1 probe may in turn label the DUB. In scenario 2, the DUB engages the triUb substrate exclusively in an exo-binding mode that leads to the labeling of DUB by the triUb probe. In scenario 3, both endo- or exo-binding modes are possible, and it will lead to simultaneous cleavage of the triUb at the native isopeptide bond and labeling by the warhead in the triUb probe. For K48 triUb-MA1-CL2, we observed no cleavage by USP9X CD but a robust labeling product in a time-course experiment (Fig. 5C). The identity of the upshifted labeling product band was confirmed by comparison with labeling of USP9X CD using K48-diUb-MA1 probe (SI Appendix, Fig. S15B), consistent with scenario 2. For K63-triUb-MA1-CL2, both cleavages of and labeling by the triUb were observed (Fig. 5D), agreeing with scenario 3. For K11-triUb-MA1-CL2, a robust cleavage of the triUb probe was observed with a low level of labeling product (Fig. 5D), suggesting a dominant scenario 2.
We further compared the cleavage of K11- and K63-triUb-NC1-CL2 by WT, the ZnF mutant (C1721A), and the deletion mutant Δ(1924–1943) of USP9X CD. Although little effect was observed for the ZnF mutation, we observed a clear reduction in cleavage efficiency for the β-hairpin deletion mutant for both linkages of triUb (Fig. 5 E and F). In contrast, the effect of the β-hairpin deletion was less pronounced toward K11- and K63-diUb substrates (SI Appendix, Fig. S5B).
Discussion
The USP family of deubiquitinating enzymes show an extraordinary diversity of sequences and structural elements. The consensus sequence of USP CD can be divided into six boxes, with possible insertions in several regions (2). Our crystal structure of the USP9X CD confirms a conserved, canonical core USP fold with additional structural features that include a CHC2-type ZnF in the fingers subdomain and a unique β-hairpin insertion in sequence box 6. The structure of USP9X CD and the functional studies using novel Ub-based probes implicate three potential Ub binding sites (S1, S2, and S1′) in USP9X CD. Some USPs contain a C4-type ZnF at the tip of the fingers subdomain. Only in USP21 was it shown structurally that the ZnF is involved in binding the S2 Ub in a polyUb chain. Our ZnF mutagenesis and ABP labeling data suggest that an S2 Ub binding site likely exists in USP9X. CYLD contains a distinctive truncated fingers subdomain without a ZnF motif and a large insertion in box 6. The reduced binding surface of the fingers subdomain and the insertion render specificities for the more flexible linear and K63-linked polyUbs through S1 and S1′ sites. Similarly, a large, conserved insertion of USP9X within box 6 is also involved in the specific recognition of the proximal Ub of a polyUb chain. There exist two other less conserved, large insertions in the USP9X CD that are disordered in the crystal structure, located farther away from the catalytic binding cleft and may have other unknown functions.
In contrast to the broadly conserved ZnF motif in USPs, a structured β-hairpin insertion within box 6 of the USP catalytic core was somewhat unexpected. Previous bioinformatics analyses revealed such insertion of about 20 residues exist in USP9X, USP9Y, USP24, USP34, USP47, USP48, and CYLD (2). However, only CYLD crystal structures were known, and the insertion plays an important role in binding to a proximal Ub. It remains to be determined whether the box 6 insertion in other USPs also plays a role in proximal Ub recognition or is unique to CYLD and USP9X (and likely USP9Y).
In assessing USP9X activity, we observed that USP9X is specific for Ub but not for other UBL modifiers. Conversely, several other USPs showed cross-reactivity toward ISG15 and SUMO (29, 30). The recent crystal structure of USP18 in complex with ISG15 provided a structure-based explanation (10). A hydrophobic patch in ISG15 centered on Trp121, Pro128, and His149 interacts with complementary hydrophobic residues in USP18. Unlike ISG15, Ub contains polar residues (Arg42, Gln49, and Val70) at these ISG15 equivalent sites, which would complement polar residues found in Ub-reactive USPs such as USP7 (10). Like USP7, USP9X also contains polar residues at this binding interface that would be unfavorable for ISG15 (SI Appendix, Fig. S16). Neither is USP9X active against NEDD8, the most homologous UBL to Ub. The complex structure of USP21 with linear diUb provided insight into how USP21 distinguishes Ub from NEDD8 (9). A conserved Glu304 residue in USP21 interacts specifically with the guanidinium group of Arg72 of Ub C-terminal tail. The equivalent position in NEDD8 is an Ala. A secondary polar patch of USP21 (Glu427 and Lys448) in the fingers subdomain interact with the β1/β2 loop of Ub but would clash sterically with and repel the larger, charged residues in NEDD8. Not surprisingly, USP9X is conserved at both sites (Glu1688 and Asp1761/Lys1782), which explains why NEDD8 is not a substrate for USP9X.
Although the catalytic triad in the apo USP9X structure is properly aligned, both the turnover number kcat and the Michaelis constant Km of USP9X CD toward Ub-AMC are much lower than those of USP7 CD (4). This suggests additional structural restraints may play a role. We noticed the side chain of Gln1683 in the switching loop forms hydrogen bonds with the side chain of the catalytic Cys1566 and the main-chain carbonyl group of Gly1878 in the neighboring blocking loop BL2 and blocks the entrance of Ub C terminus to the active site cleft (SI Appendix, Fig. S1B). For comparison, in the USP7/Ub-aldehyde complex structure, the switching loop in USP7 adopts an “out-conformation” (6), and the equivalent Gln293 is positioned away from the catalytic cysteine (SI Appendix, Fig. S1C). Gln1683Ala mutation did not increase the kcat of USP9X CD, but rather significantly increased Km toward monoUb substrate. Kinetics of cleavage assay of IQF-K11 diUb substrate did not show any significant differences between the Q1683A mutant and the WT (SI Appendix, Fig. S13C). These observations suggest that Gln1683 is more important for the binding of the Ub tail of a monoUb to the catalytic cleft than for the native isopeptide in diUb or polyUb substrates. Instead, the interaction between the proximal Ub and the potential USP9X CD S1′ site may help to orient the isopeptide of a diUb in the catalytic cleft for cleavage.
Another notable finding in this study is that a noncatalytic cysteine 1808 in USP9X was specifically labeled by an MA warhead introduced in the K11-diUb-MA1 probe. Cys1808 is in blocking loop BL1 and 19 Å away from the catalytic cysteine. A groove between BL1 and BL2 loops likely traps the diUb isopeptide in a nonproductive conformation (Fig. 4C) and places Cys1808 in proximity to the warhead for labeling. This specific labeling event by a noncatalytic cysteine residue revealed a possible search motion of the flexible S1 Ub C-terminal tail and the attached proximal Ub during active site engagement (Fig. 4C). The elongated β-hairpin insertion likely restricts the motion of the proximal Ub and entraps the isopeptide into the groove between BL1 and BL2, leading to the labeling of Cys1808 by the MA warhead in the K11-diUb-MA1 probe (Fig. 4C). By this notion, the removal of β-hairpin insertion in USP9X CD resulted in reduced labeling efficiency of Cys1808 by the K11-diUb-MA1 probe (Fig. 4A), likely due to reduced contact time of the MA warhead with Cys1808. Furthermore, a Gln1683Ala mutation in USP9X, intended to unblock the active site cleft, improved the active site Cys1566 labeling markedly (SI Appendix, Fig. S13A), mimicking the effect of β-hairpin deletion albeit with greater efficiency. Differently, we observed preference of Cys1808 labeling over the catalytic Cys1566 by K11-diUb-MA1 probe in the USP9X ZnF mutant (Fig. 4A). This observation suggests that the interaction between the USP9X ZnF and the Ub N-terminal region may influence the binding mode of the S1 Ub C-terminal tail and the attached proximal Ub.
Although DUBs are thought to possess endo- or exo-Ub chain cleavage specificity, few tools were available to interrogate the two cleavage modes. To address this question, we designed and generated two types of triUb probes (Fig. 2B). We observed efficient endo-cleavage of the K11- and K63-, but not the K48-linked triUb-NC1-CL2 substrates (Fig. 5B). When comparing the endo- and exo-modes of Ub chain processing using the triUb-MA1-CL2 probes, we found that an exo-cleavage is exclusively preferred for K48-polyUb chain. For K11-triUb chain, USP9X CD prefers endo- over exo-cleavage. These observations revealed linkage-dependent polyUb cleavage modes of the USP9X CD. Our results also suggest that the unique β-hairpin in box 6 facilitates the efficient endo-cleavage of K63-polyUb chain based on the reduced cleavage of K63-linked triUb-NC1-CL2 by the USP9X β-hairpin deletion mutant (Fig. 5E).
Interestingly, multiple E3 ligases are known to interact with USP9X, including HECT-type E3 ligases ITCH and SMURF1, and RING-type E3 ligases MARCH7, Mind bomb 1, and TRIM25 (16–18, 36, 37). The formation of an E3 ligase and DUB complex is a recurring theme observed for many DUBs and probably plays a role in more precise control of substrate ubiquitination and the chain linkages through trimming and editing. OTU-type DUB A20 contains both a ligase domain and a DUB domain and shows an exo-cleavage preference for K48-chain and endo-cleavage activity for the K63-chain, presumably allowing the assembly of an intact degradation signal (K48) while maintaining rapid control of signaling event through the K63-polyUb chain (38). USP9X plays multiple roles in cell cycle, endocytosis, and degradation. The endo-cleavage preference for K11- and exo-cleavage preference for K48-polyUb chains of USP9X may fit in its role in the different cellular processes, which needs to be further tested in a cellular system.
In summary, while monoUb and diUbs provide a snapshot of substrate recognition by DUBs, our activity-based hybrid triUb probes allow deeper interrogation of the polyUb chain binding modes of USP9X and reveal interesting details. Linkage-dependent endo- and exo-Ub chain cleavage modes were clearly defined using the two types of hybrid triUb probes. We propose a model where the USP9X CD accommodates a chain of three Ubs, with the distal and proximal Ubs occupying the S2 and S1′ sites close to the ZnF motif and β-hairpin insertion, respectively (Fig. 5A). A structure-based triUb recognition model has previously been proposed for the human OTU subfamily DUBs and SARS PLpro (39, 40). In our USP9X model, whereas the ZnF motif is rigid, the extended β-hairpin insertion is presumably flexible and may impart specificity for different chain topologies. PolyUb chains can bind USP9X in three distinct modes, which could dictate the endo- versus exo-cleavage specificity (Fig. 5A). A potential binding groove between BL1 and BL2, revealed by the specific labeling of the noncatalytic Cys1808, suggests an intriguing possibility that USP9X CD may recognize a branched polyUb chain, such as K11/K63 (41). This remains to be tested in future studies. Overall, the USP9X CD provides a fascinating example of small, yet important structural features that may dictate the polyUb chain linkage specificity and influence the dynamics of polyUb binding and recognition by the USP catalytic core.
Methods
Cloning and Protein Purification.
A human USP9X (amino acids 1,551–1,970) construct was cloned into the pET28-MHL vector (GenBank EF456735), which encodes a tobacco etch virus (TEV) protease cleavable N-terminal His6-tag. Mutants were generated by site-directed mutagenesis. All USP9X proteins were overexpressed in Escherichia coli BL21-CodonPlus (DE3)-RIL strain grown in Terrific Broth using a LEX bioreactor (Epiphyte3) or in LB media using a shaker incubator. Cells were grown at 37 °C until the OD600 reached 1.5 and protein expression was induced with 0.25 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 18 °C overnight using the LEX bioreactor. Alternatively, shaker flask culture was grown at 37 °C until the OD600 reached 0.6 and protein expression was induced with 0.4 mM IPTG at 16 °C overnight. For USP9X purification, all buffers were adjusted to pH 8.0 at room temperature. Harvested cells were resuspended in a lysis buffer (25 mM Tris, 500 mM NaCl, 5% glycerol) supplemented with 1 mM PMSF and benzamidine and lysed using sonication for 10 min with 5-/7-s on/off cycles (Sonicator 3000; Misonix). The crude extract was then centrifuged at 36,000 × g for 1 h at 4 °C. The protein was batch-absorbed using Ni-NTA resin (Qiagen), washed with 25 mM Tris (pH 8.0), 500 mM NaCl, 25 mM imidazole, and eluted with an elution buffer (25 mM Tris, 250 mM NaCl, 250 mM imidazole, 1 mM TCEP). The His6-tag was removed by overnight incubation with His6-tagged TEV protease during dialysis in 25 mM Tris, 200 mM NaCl, 5 mM β-mercaptoethanol. Uncleaved protein and the TEV protease were removed by passing through Ni-NTA resin, and the target protein was purified by anion exchange chromatography (HiTrap Q; GE Healthcare). The protein was further purified using a Superdex 200 column (GE Healthcare) preequilibrated in 25 mM Tris, 150 mM NaCl, 1 mM TCEP. Pooled fractions were concentrated using centrifugal concentrators (Amicon Ultra, molecular mass cut-off of 30 kDa) to 20 mg/mL, flash frozen, and stored at −80 °C. USP2 CD was used as a control, and its expression and purification were described previously (42).
Ub-β-mercaptoethanesulfonic acid sodium derivative (Ub1–75-MESNA) was generated as previously described (32). Briefly, Ub1–75 was expressed as an intein fusion using a Ub1–75-pTYB1 plasmid transformed into BL21(DE3) strain. Cells were cultured in LB media supplemented with 100 μg/mL ampicillin at 37 °C and the overexpression was induced with 0.4 mM IPTG at an OD600 of 0.6 for 16–18 h at 16 °C. Cells were harvested via centrifugation and resuspended in 20 mM Tris (pH 7.5), 200 mM NaCl, 1 mM EDTA, 5% glycerol. Cells were lysed on the ice, and the lysate was cleared by centrifugation and incubated with chitin resin (New England Biolabs) at 4 °C for 2 h. Subsequently, the chitin resin was washed with 20 mM Tris (pH 7.5), 1 M NaCl, 1 mM EDTA, 5% glycerol, and equilibrated with 20 mM MES (pH 6.5), 100 mM NaCl. The resin was then incubated with 50-mL cleavage buffer containing 20 mM MES (pH 6.5), 100 mM NaCl, 75 mM β-MESNA salt for 12 h at room temperature to yield activated Ub1–75-MESNA. Ub1–76 and its Lys-to-Cys mutants were expressed in BL21(DE3) cells using a Ub1–76-pET3a plasmid. Cells were lysed in 50 mM ammonium acetate (pH 4.5), 50 mM NaCl and heat treated to precipitate E. coli proteins. Subsequently, the supernatant was saturated with ammonium sulfate (80%) and centrifuged to isolate precipitate, which was redissolved in a buffer containing 20 mM MES (pH 6.5), 50 mM NaCl, and further purified on a HiLoad Superdex 200-pg column (GE Healthcare). Protein purity was assessed by SDS/PAGE, and the protein identity was confirmed using a Xevo Q-TOF mass spectrometer.
Crystallization.
A surface entropy reduction (43) mutation (K1637E1638 to two alanines) was introduced to crystalize USP9X CD. Crystal used for X-ray data collection was grown at room temperature using the sitting-drop vapor diffusion method by mixing 0.5-µL protein (20 mg/mL) with 0.5-µL reservoir solution containing 20% PEG3350 and 0.2 M diammonium tartrate. Crystal was cryoprotected by first immersion in the well solution supplemented with 10% ethylene glycol, then in Paratone-N (Hampton Research) before being flash frozen.
Structure Determination and Refinement.
X-ray diffraction data for USP9X were collected at temperature 100 K at beamline 19ID of Advanced Photon Source (APS), Argonne National Laboratory. The datasets were processed using the HKL-3000 suite (44) and indexed in the P21 space group. Imposition of C2 symmetry incurred a significantly higher penalty than P2 and was not pursued. The USP9X structure was solved by molecular replacement using PDB ID code 1NBF as a search template with BALBES (45). The graphics program COOT (46) was used for model building and visualization. Restrained refinement was performed using REFMAC v5.8 (47) incorporating NCS restraints. The final structure was validated by MOLPROBITY (48) and deposited in the PDB under ID code 5WCH (49). Raw diffraction images were deposited in the Integrated Resource for Reproducibility in Macromolecular Crystallography (50) under doi:10.18430/m35wch. X-ray data collection and refinement statistics are listed in Table 1. The two mutations K1637E1638AA are located on helix α3 and did not impact structure analysis.
Table 1.
X-ray data collection and refinement statistics
| Data collection and refinement | USP9X (5WCH) |
| Data collection | |
| Space group | P 21 |
| Cell dimensions | |
| a, b, c, Å | 79.4, 79.0, 132.0 |
| α, β, γ, ° | 90.0, 131.8, 90.0 |
| Resolution, Å | 50.0–2.50 |
| Rsym or Rmerge | 0.125 (0.863) |
| I/σ, I | 13.0 (1.6) |
| Completeness, % | 96.4 (95.6) |
| Redundancy | 4.5 (4.2) |
| Refinement | |
| Resolution, Å | 50.0–2.50 |
| No. reflections | 51,658 |
| Rwork/Rfree | 0.207/0.261 |
| No. atoms | |
| Protein | 10,757 |
| Ligand/ion | 4 |
| Water | 40 |
| B-factors | |
| Protein | 46.8 |
| Ligand/ion | 32.5 |
| Water | 35.7 |
| Rmsd | |
| Bond lengths, Å | 0.008 |
| Bond angles, ° | 1.226 |
| Ramachandran plot | |
| Favored regions, % | 97.2 |
| Allowed regions, % | 2.8 |
| Disallowed regions, % | 0.0 |
Values in parentheses are for the highest-resolution shell.
Di- and PolyUb DUB Cleavage Assays.
DiUb cleavage assay was performed using 5 μM diUb and 0.5 μM USP9X CD in a 25 mM Tris buffer (pH 8.0) containing 50 mM NaCl and 5 mM DTT at 37 °C. Reactions were quenched at specified time points using NuPAGE LDS sample buffer (ThermoFisher) and the products resolved using SDS/PAGE. TriUb and tetraUb assays were performed using 2.5 μM polyUb and 0.5 μM USP9X CD in 25 mM Tris buffer (pH 8.0) containing 50 mM NaCl and 2 mM DTT. All di-, tri-, and tetraUb substrates were purchased from UBPBio. For Western blotting, Ub was probed using P4D1 monoclonal antibody (BioLegend). IRDye secondary antibodies (LI-COR) were used for indirect detection, and the results were visualized using an Odyssey CLx infrared imaging system.
Generation of ABPs and Assays Using the ABPs.
The monoUb-based probe was synthesized by reacting the Ub1–75-MESNA with PA or Gly-VME for 40 min followed by HiTrap SP HP cation exchange (GE Healthcare) column purification. To make diUb-NC1-PA2 probes, HA-tagged Ub1–75-MESNA was first reacted with an NC molecule followed by deprotection, as previously described to obtain HA- Ub1–75-NC (33). In parallel, a mutant Ub1–75-MESNA with a Lys-to-Cys mutation at the selected lysine site was reacted with PA as described above to generate Cys-Ub-PA. After cation-exchange purification, equal molar amounts of HA-Ub-NC and Cys-Ub-PA were incubated at 0.5 mg/mL each in a 20 mM MES (pH 6.5) buffer containing 0.03 mM EDTA for 1 h at room temperature. Subsequently, the reaction mixture was purified using an SP column followed by buffer exchange to a 20 mM MES (pH 6.5) buffer to yield the diUb-NC1-PA2 probes.
To make diUb-MA1 probes, HA-Ub1–75-MESNA was first reacted with a MA linker molecule followed by deprotection as previously reported for generating HA-Ub1–75-MA (32). Concurrently, full-length Ub1–76 with a Lys-to-Cys mutation for the desired linkage was purified as described above. HA-Ub1–75-MA was then incubated with Cys-Ub1–76 on ice at 0.5 mg/mL concentration each in a 20 mM MES (pH 6.5) buffer containing 0.03 mM EDTA. The reaction mixture was then purified using an SP column followed by buffer exchange to a 20 mM MES (pH 6.5) solution to yield diUb-MA1 probes.
Assays of WT USP9X CD and mutants using monoUb and diUb-NC1-PA2 ABPs were performed in 20 mM Tris (pH 8.0), 50 mM NaCl and 1 mM DTT at room temperature for 2 h with 2-μM probes. DiUb-MA1 labeling assays with USP9X CD and mutants were conducted in 20 mM Hepes (pH 7.0) buffer with no additional DTT present for 1 h at room temperature with 4-μM probes. For comparison, labeling assays using K6-, K11-, K48-, and K63-linked diUb-MA1 and diUb-NC1-PA2 probes (∼4 μM) were performed alongside HA-Ub-VME and HA-Ub-PA against USP9X CD and USP9X Δ(1924–1943) in 20 mM Hepes (pH 7) buffer with no additional DTT present for 1 h at room temperature. For all labeling assays, the WT and mutant USP9X CD concentrations were 1.4 μM.
Generation of DiUb-AMC Fluorescent Substrates and Fluorescence-Based DUB Assays.
AMC-derived Ub, and Ub-like proteins (SUMO1, NEDD8, ISG15) (Boston Biochem) were used in fluorescence-based DUB assays to measure the activity of USP9X CD. Assays were carried out in 10-μL reaction volumes in 384-well black polypropylene PCR plates (PCR-384-BK; Axygen) and the fluorescence was measured on a Synergy plate reader (Biotek). For AMC assays, 10 nM USP9X was mixed with 0.5 μM AMC substrate in 20 mM Tris (pH 8.0), 50 mM NaCl, 0.005% Triton X-100, and 2 mM DTT, and the excitation and emission wavelengths were set at 360 and 460 nm, respectively, with a 40-nm bandwidth filter. All kinetic parameters were determined using Michaelis–Menten regression analysis in Prism 6 software (GraphPad).
Continuous diUb cleavage assays were performed using 50 nM USP9X CD and 500 nM K11- (Pos4, #UF-440; Boston Biochem) or 200 nM K63- (K63-2, #DU6302; LifeSensors) IQF-diUb substrates in an optimized USP9X buffer (20 mM Bicine, pH 8.0, 0.004% Triton X-100, 0.6 mM DTT), with excitation and emission wavelengths at 530 and 575 nm, respectively. Alternatively, for full kinetic analysis of K11 IQF-diUb and Ub-AMC substrate, 10–25 nM WT and mutant USP9X CD was used. Assays were performed in 50 mM Hepes (pH 7.8), 50 mM NaCl, 0.5 mM EDTA, 0.1 mg/mL BSA, and 1 mM DTT at 25 °C. Fluorescence was measured using a Fluoromax-4 fluorescence spectrometer (Horiba). For Ub-AMC, excitation and emission wavelengths at 355 and 440 nm were used, respectively. For IQF-diUb, excitation and emission wavelengths at 544 nm and 572 nm were used, respectively.
K11-, K48-, and K63-linked diUb-NC1-AMC2 fluorescent substrates were generated by first reacting Ub1–75-MESNA containing a respective cysteine mutation at K11, K48, or K63 with 32 mM glycine-AMC in the presence of 0.5 M MESNA, 0.17 M collidine and 5 mg/mL N-hydroxysuccinimide for 40 h. The resulting reaction mixture was treated with 10 mM DTT overnight, purified using an SP column with a salt gradient (50–500 mM NaCl) and buffer exchanged into a 20 mM MES (pH 6.5) buffer. Following purification, the cysteine-containing Ub-AMC was reacted with Ub-NC to generate diUb-NC1-AMC2 substrate (Fig. 2A) and further purified using an SP column with a salt gradient from 100 mM to 1 M NaCl. Fluorogenic assay was performed in a reaction buffer containing 50 mM Hepes (pH 7.8), 50 mM NaCl, 0.5 mM EDTA, 0.1 mg/mL BSA, and 1 mM DTT at 25 °C. Fluorescence was measured using a Fluoromax-4 fluorescence spectrometer (Horiba) with excitation and emission wavelength at 355 and 440 nm, respectively.
Generation of Hybrid TriUb Probes.
K11-, K48-, and K63-linked triUb probes were generated chemoenzymatically in two steps as shown in Fig. 2B. First, diUbs containing a native isopeptide and appropriate Lys-to-Cys mutation were generated enzymatically. Specifically, mouse E1 and E2-25K at 0.5 μM and 5 μM, respectively, were incubated with ∼7 mg/mL of Ub K48C and Ub D77 in the presence of 2 mM ATP, and 0.5 mM DTT to generate K48-diUb. Similarly, mouse E1, E2 complex Ubc13/Mms2, Ub K63C and Ub D77 were used to generate K63C diUb. Mouse E1, Ube2s-UBD, and AMSH at 0.5 μM, 5 μM, and 0.5 μM were incubated with Ub K11C and Ub D77 to generate K11-diUb adapted from the previously reported method (51). All reactions were performed overnight in a reaction volume of 500 μL with buffer containing 50 mM Tris (pH 7.6), 5 mM MgCl2, 10 mM creatine, 0.6 unit inorganic phosphatase, and 0.6 unit creatine phosphokinase at 37 °C. The reaction mixture was buffer exchanged to a buffer containing 50 mM ammonium acetate (pH 4.5) and 50 mM NaCl, and purified using an SP column.
To generate hybrid triUb probes, the enzymatically synthesized diUb with a position-specific cysteine introduced in the distal Ub was incubated with an equal molar amount of HA-Ub1–75-NC or HA-Ub1–75-MA for 3 h at room temperature to generate triUb-NC1-CL2 and triUb-MA1-CL2 probes, respectively. The reaction mixture was then buffer exchanged into ammonium acetate (pH 4.5) and purified using an SP column. Alternatively, the reaction mixture was centrifuged at 16,000 × g for 10 min and directly injected into a Superdex 75 pg size-exclusion column in 20 mM MES (pH 6.5), 100 mM NaCl, and 5 mM EDTA. The probes were characterized using SDS/PAGE and Xevo Q-TOF-MS. Protein concentration was determined using a combination of Bradford assay and SDS/PAGE based quantification. Hybrid triUb probe assays were conducted in a 20 mM Hepes (pH 7.0) buffer containing 1 mM DTT for triUb-NC1-CL2 cleavage assay while no additional DTT was present for triUb-MA1-CL2 assay. All diUb-MA1 labeling assays were performed at room temperature for 1 h unless otherwise noted, and all triUb-MA1-CL2 labeling assays were performed at room temperature for up to 2 h. Assays with triUb-MA1-CL2 were performed with ∼1.4 µM enzyme and 4-µM probe, while cleavage assays with triUb-NC1-CL2 were performed with 2.8-μM enzyme and 2-μM probe.
In-Gel Digestion and High-Resolution MS/MS Analysis.
To identify the cysteine residues responsible for labeling with the K11-diUb-MA1 probe, large-scale labeling of USP9X CD mutants was performed. Labeling bands were excised and cut into 1-mm3 pieces and transferred to microcentrifuge tubes. Gel pieces were destained at 37 °C in a 50% acetonitrile solution containing 100 mM ammonium bicarbonate for 30 min and repeated until all stain was removed. The proteins in the gel pieces were reduced by incubation with 10 mM DTT at 60 °C for 10 min and alkylated with 100 mM iodoacetamide (IAA) in a 100 mM ammonium bicarbonate buffer. After removal of the IAA solution, gel pieces were shrunk with acetonitrile, dried, and incubated in 50 μL of 0.01 mg/mL trypsin at 37 °C overnight. Following digestion and extraction, samples were lyophilized. Lyophilized samples were redissolved in 0.1% TFA solution and introduced into Q-Exactive Orbitrap via UPLC column. Subsequent proteomics analysis was performed with Proteome Discoverer v1.4 (ThermoFisher).
Supplementary Material
Acknowledgments
The Structural Genomics Consortium is a United Kingdom registered charity (no. 1097737) that receives funds from AbbVie, Bayer, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada through the Ontario Genomics Institute (OGI-055), Innovative Medicines Initiative (EU/EFPIA) (ULTRA-DD Grant 115766), Janssen, Merck KGaA, MSD, Novartis, Ontario Ministry of Research, Innovation and Science, Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and the Wellcome Trust. X-ray data were collected at the Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source, which is operated by the University of Chicago Argonne, LLC, for the US Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357. This work was supported in part by National Institutes of Health Grants R01GM097468 and R21NS085509 (to Z.Z.); National Sciences and Engineering Research Council of Canada Grant RGPIN-2017-06520 (to Y.T.); and a joint Janssen–Ontario Centre of Excellences Neuroscience Catalyst grant (to Y.T. and M.V.). It was also supported by the Delaware COBRE program for instrumentation facilities, with a grant from the National Institute of General Medical Sciences (1 P30 GM110758−01). Y.G. is a visiting scholar sponsored by the Excellent Young Teachers program and Innovation Fund of Guangdong Ocean University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. H.O. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 5WCH).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815027116/-/DCSupplemental.
References
- 1.Nijman SM, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Ye Y, Scheel H, Hofmann K, Komander D. Dissection of USP catalytic domains reveals five common insertion points. Mol Biosyst. 2009;5:1797–1808. doi: 10.1039/b907669g. [DOI] [PubMed] [Google Scholar]
- 3.Komander D, Clague MJ, Urbé S. Breaking the chains: Structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 4.Faesen AC, et al. The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem Biol. 2011;18:1550–1561. doi: 10.1016/j.chembiol.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 5.Hu M, et al. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- 6.Faesen AC, et al. Mechanism of USP7/HAUSP activation by its C-terminal ubiquitin-like domain and allosteric regulation by GMP-synthetase. Mol Cell. 2011;44:147–159. doi: 10.1016/j.molcel.2011.06.034. [DOI] [PubMed] [Google Scholar]
- 7.Rougé L, et al. Molecular understanding of USP7 substrate recognition and C-terminal activation. Structure. 2016;24:1335–1345. doi: 10.1016/j.str.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 8.Sato Y, et al. Structures of CYLD USP with Met1- or Lys63-linked diubiquitin reveal mechanisms for dual specificity. Nat Struct Mol Biol. 2015;22:222–229. doi: 10.1038/nsmb.2970. [DOI] [PubMed] [Google Scholar]
- 9.Ye Y, et al. Polyubiquitin binding and cross-reactivity in the USP domain deubiquitinase USP21. EMBO Rep. 2011;12:350–357. doi: 10.1038/embor.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basters A, et al. Structural basis of the specificity of USP18 toward ISG15. Nat Struct Mol Biol. 2017;24:270–278. doi: 10.1038/nsmb.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gersch M, et al. Mechanism and regulation of the Lys6-selective deubiquitinase USP30. Nat Struct Mol Biol. 2017;24:920–930. doi: 10.1038/nsmb.3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer-Vize JA, Rubin GM, Lehmann R. The fat facets gene is required for Drosophila eye and embryo development. Development. 1992;116:985–1000. doi: 10.1242/dev.116.4.985. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Overstreet E, Wood SA, Fischer JA. On the conservation of function of the Drosophila fat facets deubiquitinating enzyme and Fam, its mouse homolog. Dev Genes Evol. 2000;210:603–610. doi: 10.1007/s004270000109. [DOI] [PubMed] [Google Scholar]
- 14.Wood SA, et al. Cloning and expression analysis of a novel mouse gene with sequence similarity to the Drosophila fat facets gene. Mech Dev. 1997;63:29–38. doi: 10.1016/s0925-4773(97)00672-2. [DOI] [PubMed] [Google Scholar]
- 15.Homan CC, et al. Mutations in USP9X are associated with X-linked intellectual disability and disrupt neuronal cell migration and growth. Am J Hum Genet. 2014;94:470–478. doi: 10.1016/j.ajhg.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nathan JA, et al. The ubiquitin E3 ligase MARCH7 is differentially regulated by the deubiquitylating enzymes USP7 and USP9X. Traffic. 2008;9:1130–1145. doi: 10.1111/j.1600-0854.2008.00747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mouchantaf R, et al. The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J Biol Chem. 2006;281:38738–38747. doi: 10.1074/jbc.M605959200. [DOI] [PubMed] [Google Scholar]
- 18.Xie Y, et al. Deubiquitinase FAM/USP9X interacts with the E3 ubiquitin ligase SMURF1 protein and protects it from ligase activity-dependent self-degradation. J Biol Chem. 2013;288:2976–2985. doi: 10.1074/jbc.M112.430066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson LF, et al. Targeting deubiquitinase activity with a novel small-molecule inhibitor as therapy for B-cell malignancies. Blood. 2015;125:3588–3597. doi: 10.1182/blood-2014-10-605584. [DOI] [PubMed] [Google Scholar]
- 20.Murtaza M, Jolly LA, Gecz J, Wood SA. La FAM fatale: USP9X in development and disease. Cell Mol Life Sci. 2015;72:2075–2089. doi: 10.1007/s00018-015-1851-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rott R, et al. α-Synuclein fate is determined by USP9X-regulated monoubiquitination. Proc Natl Acad Sci USA. 2011;108:18666–18671. doi: 10.1073/pnas.1105725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaltenbach LS, et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3:e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reijnders MR, et al. DDD Study De novo loss-of-function mutations in USP9X cause a female-specific recognizable syndrome with developmental delay and congenital malformations. Am J Hum Genet. 2016;98:373–381. doi: 10.1016/j.ajhg.2015.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Au PYB, et al. Two females with mutations in USP9X highlight the variable expressivity of the intellectual disability syndrome. Eur J Med Genet. 2017;60:359–364. doi: 10.1016/j.ejmg.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 25.Holm L, Rosenstrom P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Molland K, Zhou Q, Mesecar AD. A 2.2 Å resolution structure of the USP7 catalytic domain in a new space group elaborates upon structural rearrangements resulting from ubiquitin binding. Acta Crystallogr F Struct Biol Commun. 2014;70:283–287. doi: 10.1107/S2053230X14002519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ashkenazy H, et al. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:W344–W350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Komander D, et al. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol Cell. 2008;29:451–464. doi: 10.1016/j.molcel.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 29.Lecona E, et al. USP7 is a SUMO deubiquitinase essential for DNA replication. Nat Struct Mol Biol. 2016;23:270–277. doi: 10.1038/nsmb.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Catic A, et al. Screen for ISG15-crossreactive deubiquitinases. PLoS One. 2007;2:e679. doi: 10.1371/journal.pone.0000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernández-Montalván A, et al. Biochemical characterization of USP7 reveals post-translational modification sites and structural requirements for substrate processing and subcellular localization. FEBS J. 2007;274:4256–4270. doi: 10.1111/j.1742-4658.2007.05952.x. [DOI] [PubMed] [Google Scholar]
- 32.Li G, Liang Q, Gong P, Tencer AH, Zhuang Z. Activity-based diubiquitin probes for elucidating the linkage specificity of deubiquitinating enzymes. Chem Commun (Camb) 2014;50:216–218. doi: 10.1039/c3cc47382a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang K, et al. Chemical protein ubiquitylation with preservation of the native cysteine residues. ChemBioChem. 2016;17:995–998. doi: 10.1002/cbic.201600042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Källberg M, et al. Template-based protein structure modeling using the RaptorX web server. Nat Protoc. 2012;7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oates ME, et al. D2P2: Database of disordered protein predictions. Nucleic Acids Res. 2013;41:D508–D516. doi: 10.1093/nar/gks1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mertz J, et al. Sequential elution interactome analysis of the Mind bomb 1 ubiquitin ligase reveals a novel role in dendritic spine outgrowth. Mol Cell Proteomics. 2015;14:1898–1910. doi: 10.1074/mcp.M114.045898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, et al. The ubiquitin ligase TRIM25 targets ERG for degradation in prostate cancer. Oncotarget. 2016;7:64921–64931. doi: 10.18632/oncotarget.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mevissen TET, Komander D. Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem. 2017;86:159–192. doi: 10.1146/annurev-biochem-061516-044916. [DOI] [PubMed] [Google Scholar]
- 40.Bekes M, et al. Recognition of Lys48-linked di-ubiquitin and deubiquitinating activities of the SARS coronavirus papain-like protease. Mol Cell. 2016;62:572–585. doi: 10.1016/j.molcel.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stolz A, Dikic I. Heterotypic ubiquitin chains: Seeing is believing. Trends Cell Biol. 2018;28:1–3. doi: 10.1016/j.tcb.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 42.Bozza WP, Liang Q, Gong P, Zhuang Z. Transient kinetic analysis of USP2-catalyzed deubiquitination reveals a conformational rearrangement in the K48-linked diubiquitin substrate. Biochemistry. 2012;51:10075–10086. doi: 10.1021/bi3009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldschmidt L, Cooper DR, Derewenda ZS, Eisenberg D. Toward rational protein crystallization: A Web server for the design of crystallizable protein variants. Protein Sci. 2007;16:1569–1576. doi: 10.1110/ps.072914007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 45.Long F, Vagin AA, Young P, Murshudov GN. BALBES: A molecular-replacement pipeline. Acta Crystallogr D Biol Crystallogr. 2008;64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 48.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong A, et al. 2018 Crystal structure of the catalytic domain of human USP9X. Protein Data Bank. Available at https://www.rcsb.org/structure/5WCH. Deposited June 30, 2017.
- 50.Grabowski M, et al. A public database of macromolecular diffraction experiments. Acta Crystallogr D Struct Biol. 2016;72:1181–1193. doi: 10.1107/S2059798316014716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bremm A, Freund SM, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol. 2010;17:939–947. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.