Significance
Plant bioengineering will be necessary to sustain plant biology and agriculture, where the delivery of biomolecules such as DNA, RNA, or proteins to plant cells is at the crux of plant biotechnology. Here, we show that DNA nanostructures can internalize into plant cells and deliver siRNA to mature plant tissues without external aid. Furthermore, we demonstrate that nanostructure size, shape, compactness, and stiffness affect both nanostructure internalization into plant cells and subsequent gene silencing efficiency. Interestingly, we also find that the siRNA attachment locus affects the endogenous plant gene silencing pathway. Our work demonstrates programmable delivery of biomolecules to plants and details the figures of merit for future implementation of DNA nanostructures in agriculture.
Keywords: plant biotechnology, DNA nanotechnology, siRNA gene silencing, RNA delivery, DNA origami
Abstract
Delivery of biomolecules to plants relies on Agrobacterium infection or biolistic particle delivery, the former of which is amenable only to DNA delivery. The difficulty in delivering functional biomolecules such as RNA to plant cells is due to the plant cell wall, which is absent in mammalian cells and poses the dominant physical barrier to biomolecule delivery in plants. DNA nanostructure-mediated biomolecule delivery is an effective strategy to deliver cargoes across the lipid bilayer of mammalian cells; however, nanoparticle-mediated delivery without external mechanical aid remains unexplored for biomolecule delivery across the cell wall in plants. Herein, we report a systematic assessment of different DNA nanostructures for their ability to internalize into cells of mature plants, deliver siRNAs, and effectively silence a constitutively expressed gene in Nicotiana benthamiana leaves. We show that nanostructure internalization into plant cells and corresponding gene silencing efficiency depends on the DNA nanostructure size, shape, compactness, stiffness, and location of the siRNA attachment locus on the nanostructure. We further confirm that the internalization efficiency of DNA nanostructures correlates with their respective gene silencing efficiencies but that the endogenous gene silencing pathway depends on the siRNA attachment locus. Our work establishes the feasibility of biomolecule delivery to plants with DNA nanostructures and both details the design parameters of importance for plant cell internalization and also assesses the impact of DNA nanostructure geometry for gene silencing mechanisms.
Plant bioengineering may generate high-yield and stress-resistant crops amid a changing climate and a growing population (1–3). However, unlike mammalian cells, plant cells have a cell wall which poses the dominant barrier to exogenous biomolecule delivery. Biological delivery (using bacteria or viruses) and particle bombardment are the two preferred methods of biomolecule delivery to plant cells. However, biological delivery methods are highly cargo and host specific (4), whereas particle bombardment can result in tissue damage (5). Nanomaterial-mediated biomolecule delivery has facilitated genetic engineering and biosynthetic pathway mapping in animal systems (6, 7) but has only recently been explored for plants. Specifically, two recent studies have shown that carbon nanotubes (8, 9) and clay nanosheets (10) enable intracellular delivery of DNA and RNA through surface-grafting or encapsulation strategies, circumventing the use of biolistics (external force). Biological cargo delivery to plants without external aid is an exciting development that warrants an understanding of how nanomaterials can internalize into plant cells, so nanotechnology can be logically designed for future applications in plant biotechnology.
DNA nanotechnology leverages the programmability of DNA Watson–Crick base pairing to assemble DNA nanostructures into custom predesigned shapes via sequence-specific hybridization of template and staple DNA strands (11). To date, a plethora of different DNA nanostructures of variable sizes and shapes have been synthesized (12–14) and have shown functionality in biotechnology for drug, DNA, RNA, and protein delivery applications in animal systems (15–19). However, to date, DNA nanostructures have not been explored for use in plant systems, despite their utility in other sectors of biotechnology.
Herein, we explore DNA nanotechnology as a biomolecule delivery platform in plants. We designed DNA nanostructures of controllable size, shape, stiffness, and compactness with attachment loci onto which DNA, RNA, or protein cargoes may be conjugated. By hybridizing fluorophore-conjugated DNA strands onto the loci of DNA nanostructures, we tracked nanostructure internalization into the plant cell cytoplasm of several plant species [Nicotiana benthamiana (Nb), Nicotiana tabacum, Eruca sativa, and Nasturtium officinale] and found that stiffness and size are important design elements for nanostructure internalization into plant cells. DNA nanostructures with sizes below ∼10 nm and higher stiffness or compactness showed higher cellular internalization, although size or stiffness alone is not a mutually exclusive contributor to internalization. DNA nanostructures were next loaded with siRNA targeting a GFP gene and infiltrated into plant leaves, revealing that DNA nanostructures enable gene silencing in plant leaves with efficiencies that match nanostructure internalization trends. Interestingly, the plant endogenous gene silencing mechanism can be affected by the DNA nanostructure shape and the siRNA attachment locus, affecting whether silencing occurs dominantly through transcriptional or posttranscriptional gene silencing. Our study confirms that DNA nanostructures can be designed to internalize into plant cells and that DNA nanostructures may be a promising tool set for the delivery of exogenous biomolecules to plants, as has proven valuable in animal systems.
Results
DNA Nanostructure Design, Synthesis, and Characterization.
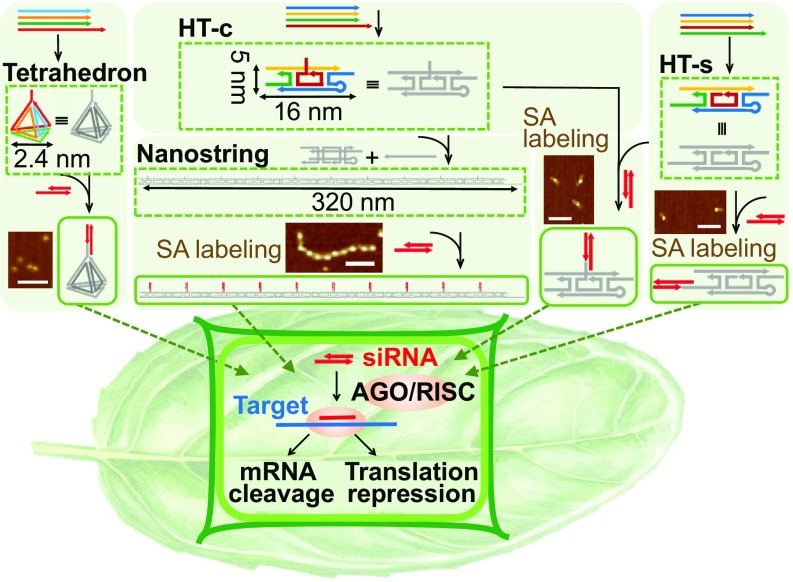
We report the synthesis and systematic assessment of different DNA nanostructures for their ability to internalize into plant cells and their subsequent utility for delivery of siRNAs to mature plants. Three DNA nanostructures with programmed sizes and shapes were synthesized: a 3D tetrahedron, a 1D hairpin-tile (HT) monomer, and a high-aspect-ratio 1D nanostring, as illustrated in Fig. 1 (SI Appendix, Table S1). Both the HT monomer and tetrahedron were assembled through four ssDNA oligonucleotides. Briefly, the HT monomer structure was designed to contain a sticky end and a stem–loop hairpin structure for copolymerization with another monomer to assemble into the length-controlled 1D nanostring by introduction of an initiator (SI Appendix, Fig. S1). The tetrahedron was also assembled through annealing of four predesigned ssDNA oligonucleotides. Based on B form double-helix DNA dimensions (2 nm diameter, 0.33 nm per base in the direction of the helical axis), the sizes of the nanostructures are 2 × 5 × 16 nm for the HT monomer, 2 × 5 × 320 nm for the 10-unit nanostring, and 2.4 nm for all edges of the tetrahedron. Atomic force microscopy (AFM) characterization in SI Appendix, Figs. S2 and S3 shows proper formation of the DNA nanostructures.
Fig. 1.
DNA nanostructure synthesis and plant infiltration workflow. The tetrahedron and HT monomer were synthesized from four ssDNA sequences, and the 1D nanostring structure was synthesized by polymerization of HT monomers with the introduction of an initiator strand. The cargo attachment locus was designed at the apex of the tetrahedron, along the nanostring, and at the side (HT-s) or center (HT-c) of each HT nanostructure. (Inset) AFM images of streptavidin-bound biotinylated HT monomers (HT-c and HT-s) and nanostring show attachment loci of the siRNA cargo. DNA nanostructures loaded with Cy3 or siRNA at each locus are infiltrated into the transgenic mGFP5 Nb plant leaves for downstream studies. (Scale bars, 100 nm.)
Each nanostructure was programmed to attach a biological cargo—DNA, RNA, or protein—to a predefined locus or loci through complementary base pair hybridization. As visualized in Fig. 1, the tetrahedron contained one attachment locus at its apex, the nanostring contained 10 attachment loci at the center of each of its constituent monomers, and the HT monomer contained one attachment locus either at its center (HT-c) or, for a separate construct, an attachment locus at its side (HT-s). To confirm the accessibility of the attachment loci, streptavidin protein was attached to the siRNA attachment locus to visualize the conjugation site in the HT monomer and nanostring. AFM imaging revealed the predicted attachment of one streptavidin protein in the center or side of the HT-c or HT-s monomer, respectively, and 10 streptavidin proteins per nanostring at the center of each constituent HT monomer (Fig. 1).
Internalization of DNA Nanostructures into Plant Cells.
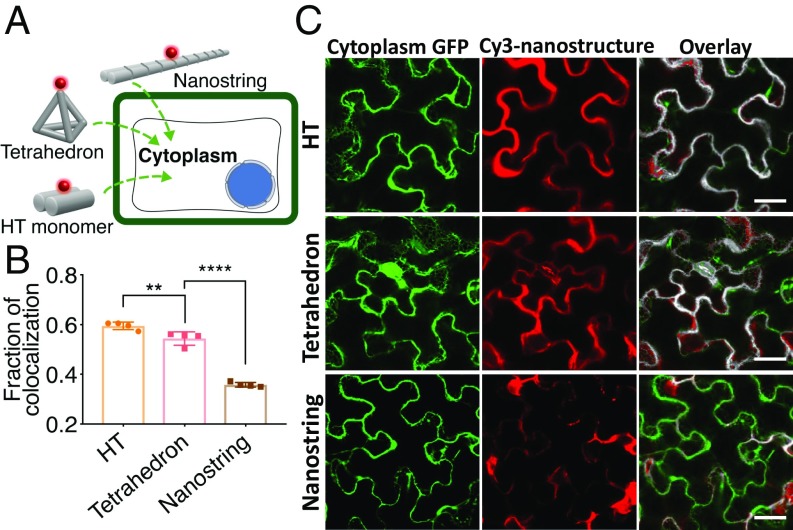
While the size exclusion limit set by the cell membrane is estimated to be around 500 nm, the plant cell wall has been reported to exclude particles larger than 5–20 nm (20). Motivated by this figure of merit, we tested whether DNA nanostructures could internalize into the cells of Nb leaves without external aid (such as a gene gun, electroporation, and ultrasound). DNA nanostructures were fluorescently labeled via attachment of Cy3-labeled DNA strands to nanostructure attachment loci and infiltrated into the leaf abaxial side to assess cellular uptake in mGFP5 Nb transgenic plants (Fig. 2Aand SI Appendix, Fig. S4). Confocal microscopy imaged the Cy3 fluorescence of the nanostructures concurrently with the intrinsic cytosolic GFP fluorescence generated by the plant cells and provided a metric by which to assess relative internalization efficiencies of different nanostructures into plant cells. Colocalization of the Cy3 fluorescence (nanostructure) with the GFP fluorescence (plant cell cytosol) 12 h postinfiltration was used to determine the extent of nanostructure internalization into the cell cytosol. Colocalization analysis in Fig. 2B shows that the HT monomer and tetrahedron nanostructures exhibit a high degree of colocalization with the plant cell cytosol (59.5 ± 1.5% and 54.4 ± 2.7%, mean ± SD, respectively), while the nanostring showed a lower degree of colocalization (35.8 ± 0.9%, mean ± SD). Representative colocalization images are shown in Fig. 2C and SI Appendix, Fig. S5, suggesting HT monomers and tetrahedrons internalize into plant cells significantly more than nanostrings. We observe that a large proportion of the Cy3 fluorescence from the Cy3-nanostring-infiltrated leaves originates from nanostrings that are putatively stuck in the guard cells, which are the dominant contribution to the colocalization fraction calculated for nanostring internalization. Conversely, we observe that most of the Cy3 fluorescence recovered from Cy3-HT-infiltrated leaves follows the cytosolic cell contour (the center of the cell is occupied by the vacuole, which occupies up to 80% of the cell volume in plants) (21), identified by cytosolic GFP expression (Fig. 2C and SI Appendix, Fig. S5). Free Cy3 oligonucleotides alone infiltrated into the leaves did not show significant colocalization with the cell cytoplasm (18.0 ± 4.6%, mean ± SD, SI Appendix, Fig. S6). Moreover, we found that Cy3-labeled HT monomers can internalize into tobacco, arugula, and watercress plant leaf cells, where the nanostring again does not show significant internalization in these plant species (SI Appendix, Fig. S7).
Fig. 2.
DNA nanostructure internalization into and colocalization with mGFP5 Nb cytoplasm. (A) Internalization of Cy3-tagged DNA nanostructures into mGFP5 Nb cells. (B) Colocalization of Cy3 fluorescence (nanostructure) with GFP fluorescence (plant cell cytosol) 12 h postinfiltration into mGFP5 Nb leaves. **P = 0.0041 and ****P < 0.0001 in one-way ANOVA. Error is SEM (n = 4). (C) Representative confocal images from data in B for HT, tetrahedron, and nanostring. (Scale bars, 40 µm.)
We next tested whether the cellular uptake mechanism is predominantly an energy-dependent or -independent process by infiltrating the Cy3-labeled HT monomer into mGFP5 Nb plant leaves at either 20 °C or 4 °C, where at 4 °C, energy-dependent cellular uptake is reduced (22). As shown in SI Appendix, Fig. S8, most of the Cy3-labeled monomer nanostructure is retained around the leaf stomata (guard cells) at 4 °C, whereas Cy3-labeled monomer nanostructures enter and diffuse uniformly into the cell cytoplasm if the infiltration and incubation are performed at 20 °C. Therefore, we propose that HT monomer nanostructures are taken up through the plant cell membrane by an energy-dependent mechanism. Furthermore, 3D z stack analysis (SI Appendix, Fig. S9) shows the monomer diffuses ∼50 µm in the z direction and 2–3 cm in the x–y direction with a 100-µL infiltration, where the lateral diffusion length can be increased by increasing the infiltration volume. Subcellular localization analysis (SI Appendix, Fig. S10) of Cy3-labeled HT monomers in single plant cells (protoplasts) indicates that DNA nanostructures are located in the cell cytosol and are excluded from the nucleus, a phenomenon that is also observed in plant leaf cells with high-resolution confocal imaging.
Prior work probing nanomaterial uptake in mammalian systems suggests uptake across the lipid bilayer is dependent on nanoparticle size, shape, aspect ratio, and mechanical stiffness (23, 24). We posit these parameters may also affect DNA nanostructure uptake across the plant cell wall. To better understand nanostructure parameters enabling plant cell internalization, we compiled and compared the size, compactness, aspect ratio (after conjugation with siRNA), and relative stiffness of the DNA nanostructures. Regarding size, as shown in SI Appendix, Fig. S11 and Table S2, we find that smaller nanostructures of the same shape internalize into plant cells significantly more than their larger-sized counterparts (tetrahedrons, 2.4, 8.8, or 12.6 nm), possibly because smaller nanostructures can remain below the plant cell wall size exclusion limit (25, 26). Specifically, we find an abrupt decline in the internalization efficiencies between the 8.8- and 12.6-nm tetrahedrons; thus, we estimate that the size cutoff for nanostructure internalization is ∼10 nm.
Regarding compactness (SI Appendix, Table S2), we calculated that the tetrahedron and HT monomer exhibit higher compactness than the nanostring (0.55 and 0.45 vs. 0.11, respectively). Our results indicate that nanostructures with higher compactness enable higher cellular uptake efficiency in mGFP5 Nb plants (59.5 ± 1.5% for HT monomer and 54.4 ± 2.7% for tetrahedron, compared with 35.8 ± 0.9% for nanostring), consistent with internalization results of HT and nanostring into tobacco, arugula, and watercress leaves (SI Appendix, Fig. S7) and some mammalian cell studies (27). We also simulated the bending stiffness of the aforementioned DNA nanostructure constructs and again found that higher calculated bending stiffness correlates with higher plant cell uptake (SI Appendix, Fig. S12 and Table S2). We therefore hypothesize that in addition to nanostructure size, the mechanical stiffness of the nanostructure plays an important role in nanostructure internalization into plant cells.
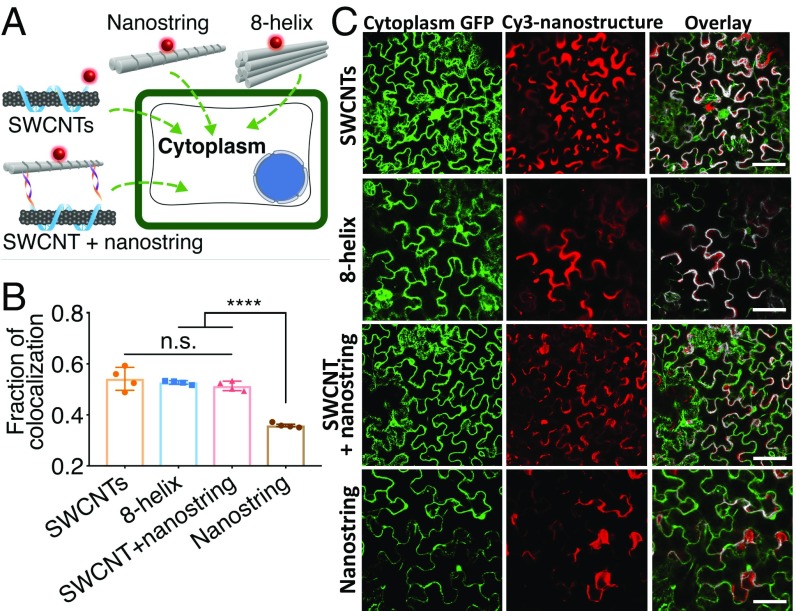
To further explore this hypothesis, we tested the effect of nanostructure stiffness—with similar nanostructure shape—on plant cell internalization by synthesizing nanostructures with different mechanical stiffnesses (Fig. 3A). We synthesized an eight-helix bundle DNA origami (28) with a similar length but higher stiffness than the nanostring (SI Appendix, Table S2) and tested their relative internalization efficiencies upon infiltration in Nb leaves. Additionally, we tethered the nanostring to single-walled carbon nanotubes (SWCNTs) to test the effect of nanostructure stiffness for internalization into Nb plant cells. AFM, agarose gel characterization, and near-infrared spectroscopy confirmed the successful assembly of eight-helix bundles (SI Appendix, Fig. S13) and nanostring–SWCNT conjugation (SI Appendix, Fig. S14). We chose to tether nanostrings to SWCNTs because SWCNTs have a small (∼1 nm) diameter but exhibit a stiffness on the order of hundreds of gigapascals to terapascals (29, 30), approximately 1,000 times stiffer than the nanostring nanostructure (several gigapascals) (31). Furthermore, SWCNTs have been shown to internalize into cells of a variety of plant species (32–34), with a leading hypothesis that the larger tensile strength of the SWCNT compared with that of the plant cell wall facilitates needlelike plant cell internalization (8, 35). As shown in Fig. 3 B and C, colocalization analysis indicates that Cy3-labeled SWCNTs (54.2 ± 4.5%, mean ± SD), eight-helix bundles with higher stiffness (52.7 ± 1.1%, mean ± SD), and the nanostring–SWCNT conjugate (51.4 ± 4.5%, mean ± SD) showed significantly higher plant cell internalization efficiency than the nanostring alone (35.8 ± 0.9%). These results suggest that higher DNA nanostructure stiffness or tethering a flexible nanostring to the stiffer SWCNT enables internalization into plant cells, despite the fact that the eight-helix bundles and nanostring–SWCNT conjugates are both slightly larger in diameter than either the nanostring or the SWCNT alone (SI Appendix, Table S2). Thus, we conclude that the nanostructure stiffness is an important design element for nanostructure internalization into plant cells, in addition to smaller size (at least one dimension below ∼10 nm; SI Appendix, Fig. S11), with all contributing figures of merit for nanostructure internalization being summarized in SI Appendix, Table S2.
Fig. 3.
Internalization of nanostructures with different mechanical stiffnesses into mGFP5 Nb cells. (A) Internalization of four different nanostructures postinfiltration into plant cells: Cy3-labeled GT15-SWCNTs, nanostrings labeled with Cy3 and hybridized onto SWCNT, Cy3 labeled eight-helix bundle origami, and Cy3-labeled nanostring alone. (B) Colocalization analysis of Cy3 fluorescence (nanostructure) with the GFP fluorescence (plant cell cytosol) after 12 h (nanostring and eight-helix bundles) or 6 h (SWCNTs and SWCNTs + nanostring) postinfiltration into mGFP5 Nb leaves. ****P < 0.0001 in one-way ANOVA; n.s., not significant. Error is SEM (n = 4). (C) Representative postinfiltration confocal images showing different internalization behaviors of different nanostructures shown in B. (Scale bars, 70 µm.)
Gene Silencing Efficiency of DNA Nanostructures.
We next examined whether DNA nanostructures could be loaded with a functional biological cargo, siRNA, to accomplish gene silencing in plants. RNAi is a phenomenon in which double-stranded RNA (dsRNA) induces gene silencing and has expedited discoveries in genomics and therapeutics (36). A key conserved feature of RNAi in plants is processing of dsRNA into siRNAs by the activity of Dicer-like enzymes (36, 37). siRNAs are subsequently incorporated into an RNA-induced silencing complex (RISC), resulting in sequence-specific blocking of mRNA translation (38).
To ascertain whether DNA nanostructures can deliver siRNA to achieve gene silencing in plants, we targeted the silencing of a GFP gene in transgenic mGFP5 Nb, which exhibits constitutive GFP expression from the nuclear genome. We designed a 21-bp siRNA sequence that inhibits GFP expression in a variety of monocot and dicot plants (39) and hybridized this duplex oligonucleotide to a complementary strand programmed into the site-specific loci on the DNA nanostructures. Native PAGE or agarose gel electrophoresis analysis (SI Appendix, Figs. S15 and S16) was performed to validate conjugation of siRNA to each DNA nanostructure. Furthermore, we confirmed that loading on DNA nanostructures protects the siRNA from degradation inside the cells compared with free siRNA (SI Appendix, Fig. S17) and that the DNA nanostructures remain stable in various biological media for at least 12 h (SI Appendix, Fig. S18), motivating their use in plant tissues.
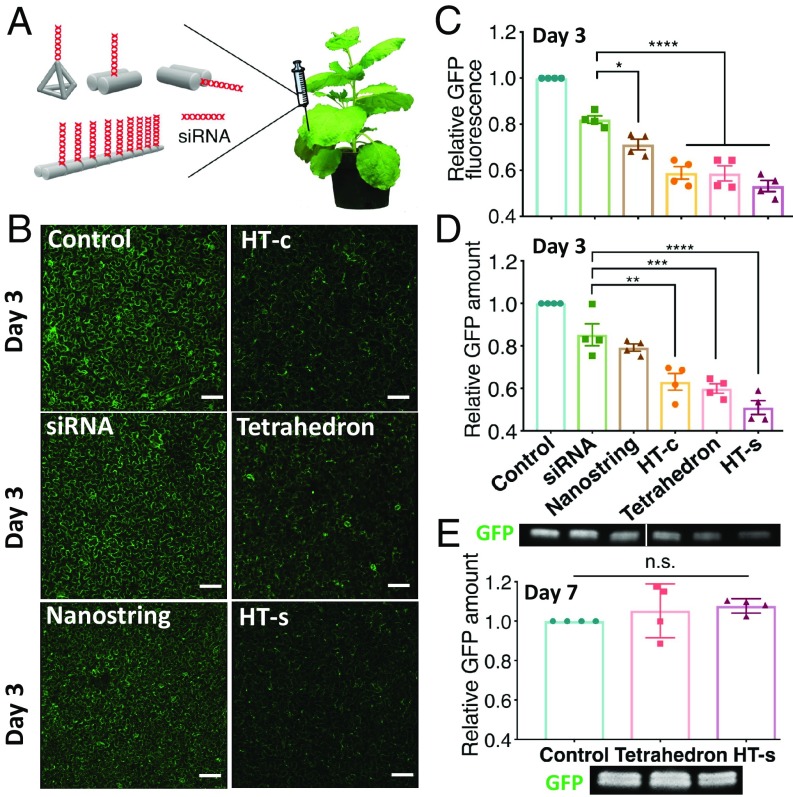
Following siRNA loading, each nanostructure with its linked active siRNA duplex(es) was introduced into the leaves of mGFP5 Nb via infiltration to the leaf abaxial side with an siRNA concentration of 100 nM (Fig. 4A). Confocal microscopy was performed to image GFP expression in infiltrated leaves, and Western blotting was utilized as a second method to confirm and quantify GFP expression changes. As shown in representative confocal images in Fig. 4B, untreated control leaves or leaves treated with free siRNA alone showed strong GFP fluorescence (low or no gene silencing), as expected, due to constitutive expression of GFP in the transgenic plant. Conversely, leaves infiltrated with siRNA-linked DNA nanostructures showed varying degrees of reduced GFP fluorescence. As shown in Fig. 4C, leaves infiltrated with siRNA-functionalized nanostrings showed an ∼29 ± 4.6% (mean ± SD) decrease of GFP fluorescence compared with the untreated leaf. Leaves infiltrated with the HT monomer showed a 41 ± 5.4% or 47 ± 4.7% (mean ± SD) reduction in GFP fluorescence for constructs in which the siRNA was linked at the center or side of the nanostructure, respectively. Last, leaves infiltrated with siRNA conjugated to the tetrahedron showed a 42 ± 6.5% (mean ± SD) decrease in GFP fluorescence intensity compared with untreated leaves. Notably, all leaves infiltrated with siRNA-functionalized DNA nanostructures exhibited a significantly larger fluorescence decrease compared with leaves infiltrated with free siRNA, suggesting that DNA nanostructures can serve as a nucleotide delivery tool in plant systems.
Fig. 4.
Transient gene silencing with siRNA tethered on DNA nanostructures. GFP silencing efficiency of siRNA-linked nanostructures quantified by confocal imaging and Western blotting. (A) Infiltration of siRNA-linked DNA nanostructures into mGFP5 Nb leaves. (B) Representative confocal images of leaves infiltrated with siRNA nanostructures 3 d postinfiltration, with nontreated control leaves. (Scale bars, 100 µm.) (C) Fluorescence intensity analysis of confocal images. *P = 0.0151 and ****P < 0.0001 in one-way ANOVA. (D) Representative Western blot gel of GFP extracted from nanostructure-treated leaves 2 d postinfiltration. **P = 0.0013, ***P = 0.0003, and ****P < 0.0001 in one-way ANOVA. (E) Representative Western blot of GFP extracted from leaves treated with siRNA linked to tetrahedron or HT-s 7 d postinfiltration. Control vs. tetrahedron = not significant (n.s.; P = 0.5806), control vs. HT-s = not significant (P = 0.3444). Error is SEM (n = 4).
We note that the degree of nanostructure internalization (Fig. 2) is proportional to the silencing efficiency achieved with each nanostructure (Fig. 4), suggesting that nanostructure internalization into the plant cell determines its ability to induce siRNA-based gene silencing. Interestingly, we observe higher (47 ± 4.7%, mean ± SD) gene silencing efficiency when siRNA is linked to the side of the HT monomer (HT-s, aspect ratio 5:1), compared with a lower (41 ± 5.4%, mean ± SD) silencing efficiency when siRNA is instead linked to the center of the HT monomer (HT-c, aspect ratio 1:1). These results are congruent with prior studies suggesting that higher-aspect ratio nanostructures facilitate nanoparticle entry into cells (20). However, the nanostring, which has the highest aspect ratio (20:1), surprisingly shows the lowest silencing efficiency (29 ± 4.6%, mean ± SD) and internalization efficiency (35.8 ± 0.9%, mean ± SD), corroborating our above findings that nanostructure shape is not the only parameter affecting internalization into plant cells. In particular, the above internalization assays show nanostring internalization into plant cells only if the nanostring is first conjugated to a high-stiffness nanostructure such as a SWCNT, confirming that nanostructure stiffness is an important parameter for both nanostructure internalization and gene silencing efficiency.
To further confirm siRNA-induced gene silencing, GFP expression in each plant leaf was quantified by Western blotting 3 d postinfiltration with the siRNA-linked nanostructure. As shown in Fig. 4D and Fig. S19, the HT monomer and tetrahedron nanostructures linked with siRNA show a significant decrease in GFP compared with untreated leaves: 37 ± 4.3% GFP decrease for HT-c, 49 ± 3.8% decrease for HT-s, and 40 ± 1.9% decrease for the tetrahedron (mean ± SD). Interestingly, the siRNA conjugated to the end of the HT monomer nanostructure showed the best silencing efficiency and the most GFP decrease, which was significantly higher than when the siRNA was instead conjugated to a locus on the center of the HT monomer. Moreover, we observed no statistically significant silencing by the siRNA-loaded nanostring compared with siRNA alone. We also tested the transience of the nanostructure-enabled siRNA-mediated gene silencing. Confocal imaging shows that GFP fluorescence for all siRNA-loaded DNA nanostructure-treated leaves recovers to preinfiltration or noninfiltration (control) levels by 7 d postinfiltration (SI Appendix, Fig. S20). Transience of siRNA-mediated gene silencing was also verified by quantifying GFP expression with quantitative Western blot analysis. As shown in Fig. 4E, the amount of GFP expressed in the leaves infiltrated with the HT-s monomer and tetrahedron nanostructures, which had induced the largest GFP silencing on day 3, returned to baseline protein expression levels by day 7. Notably, DNA nanostructures and their relevant chemistries for gene silencing are significantly cheaper (less than a dollar per infiltration) than biolistic RNA delivery and hence could be scaled up for large-scale experiments or periodically reapplied to sustain the silencing if needed (SI Appendix, Table S3). Additionally, we demonstrated that DNA nanostructures do not induce a stress response (SI Appendix, Fig. S21) in plants and are thus a biocompatible mode of siRNA delivery to plants.
siRNA Attachment Locus on Nanostructures Affects Endogenous Gene Silencing Pathways.
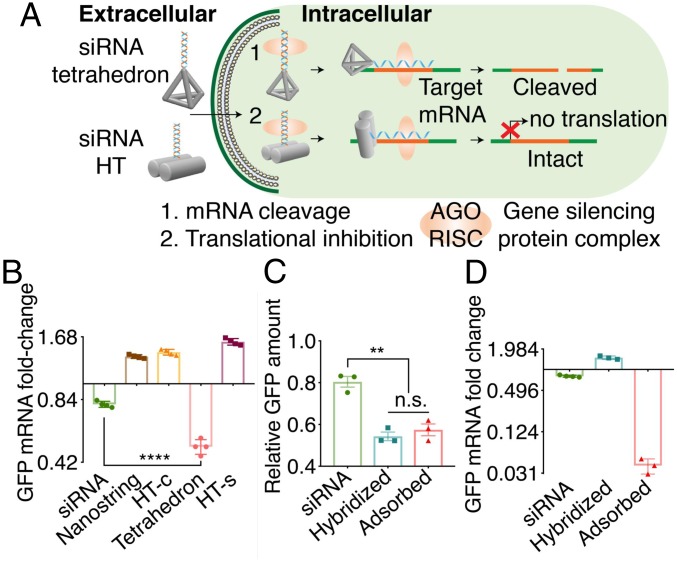
siRNA-mediated gene silencing in plants is a well-known sequence-specific gene regulation mechanism. However, RNA silencing can undergo different gene silencing pathways. Specifically, posttranscriptional gene silencing employs microRNA and siRNA pathways for mRNA cleavage or translation repression (40–42). We tested the siRNA silencing mechanism of DNA nanostructures as illustrated in Fig. 4A. Because degradation of transcriptional mRNA is the typical mechanism for gene silencing with exogenously introduced siRNA, we quantified changes in GFP mRNA with qPCR. GFP mRNA of mGFP5 Nb leaf tissues infiltrated with siRNA-linked nanostructures was quantified with qPCR 2 d postinfiltration. Interestingly, as shown in Fig. 5B, only siRNA alone and siRNA tethered to the tetrahedron showed a significant (22.3 ± 2.2% and 50.3 ± 4.9%, respectively, mean ± SD) reduction in GFP mRNA. In contrast, the siRNA tethered to HT monomer or nanostring nanostructures showed a significant increase in GFP mRNA of 59.1 ± 6.5% for the side-linked monomer HT-s, 45.2 ± 1.9% for the center-linked monomer HT-c, and 35.1 ± 3.2% for the nanostring (mean ± SD). We further confirmed that the DNA nanostructure alone (HT monomer without siRNA) does not induce a change in leaf GFP mRNA levels (SI Appendix, Fig. S22).
Fig. 5.
Gene silencing pathways for siRNA-linked nanostructures. (A) Proposed silencing pathways induced by siRNA–DNA nanostructures. (B) qPCR of leaves infiltrated with free siRNA, siRNA–nanostring, HT-c, tetrahedron, or HT-s 2 d postinfiltration. ****P < 0.0001 in one-way ANOVA. Error is SEM (n = 4). (C) Western blot of GFP extracted from siRNA–SWCNT–treated leaves 2 d postinfiltration. **P = 0.0041 in one-way ANOVA; n.s., not significant. Error is SEM (n = 3). (D) qPCR of leaves infiltrated with free siRNA, hybridized RNA–SWCNTs, or adsorbed RNA–SWCNTs 2 d postinfiltration. Error is SEM (n = 3).
The above results suggest that siRNAs conjugated to different DNA nanostructures employ different silencing pathways. Tetrahedron-mediated gene silencing appears to undergo an mRNA-targeted degradation pathway, as does free siRNA, while siRNAs linked to either locus on the HT monomer may undergo translation inhibition based on the observed increase and accumulation in mRNA (Fig. 5A). Of note, the observed trend of increasing GFP mRNA was consistent with the silencing efficiency trends of the three nanostructures: the side-linked monomer (HT-s) showed the largest mRNA increase and also the largest GPF decrease as measured by Western blotting and confocal microscopy. We thus hypothesize that steric and conformational hindrance of the siRNA, determined by the siRNA attachment locus, affects the resulting gene silencing pathway. Specifically, we find that siRNA tethered to the 1D nanostructures (HT monomer, nanostring) has greater steric hindrance than when tethered to the apex of the 3D tetrahedron nanostructure.
To further probe the effect of siRNA linking geometry on gene silencing and to test the above hypothesis, we probed GFP-targeted siRNA silencing efficiency under two different siRNA linking geometries. Because SWCNTs have been previously shown to internalize into plant cells without external aid (8) and it is possible to control the attachment geometry of siRNA to SWCNTs, we attached siRNA to the surface of a 1D SWCNT with two different attachment configurations: as shown in SI Appendix, Fig. S23, siRNA was either tethered to the surface of a 1D SWCNT (RNA–SWCNTs hybridized) with greater steric hindrance resembling the case of the HT monomer or reversibly loaded on the SWCNT in a releasable manner (RNA–SWCNTs adsorbed), exhibiting less or no conformational hindrance and thus resembling the case of the tetrahedron. Both constructs were introduced into mGFP5 Nb leaves and assessed for GFP silencing efficiency. As shown in Fig. 5C and SI Appendix, Fig. S23, both siRNA attachment configurations show similar levels of GFP decrease as quantified by Western blotting: siRNA hybridized to SWCNT decreased GFP expression by 54.3 ± 1.7% (mean ± SD), and siRNA absorbed onto but releasable from the 1D SWCNT decreased GFP expression by 48 ± 4.8% (mean ± SD). However, qPCR assessment reveals that GFP mRNA increases by 48.4 ± 8.5% if the siRNA is hybridized to the SWCNT, whereas the GFP mRNA decreases by 92 ± 1.0% if the siRNA is releasable from the SWCNT surface (Fig. 5D). These results suggest both the silencing efficiency and silencing pathway are affected by the siRNA loading geometry on the nanostructure carrier and the availability of siRNA to the requisite endogenous gene silencing proteins.
Discussion
DNA nanostructures have been extensively studied in animal systems for cell internalization, intracellular delivery, and downstream diagnostic and therapeutic applications owing to their unique sequence–structure programmability and inherent biocompatibility (16–18, 43, 44). Analogous work in plant systems is lacking, although a few studies have reported the biolistic or nonmechanical uptake, translocation, or localization of engineered nanoparticles (carbon nanotubes, SiO2, quantum dots, TiO2 nanoparticles) to plants (8, 10, 20, 25, 26, 45), while DNA nanostructure use in plants remains unexplored. Orthogonally, gene silencing through the introduction of siRNA has become a broadly adopted tool to inactivate gene expression, to probe biosynthetic pathways, and to serve as an exogenous regulator of developmental and physiological phenotypes in plants (46, 47).
Herein, we demonstrate that DNA nanostructures can be designed to internalize into plant cells through infiltration and that siRNA can be controllably tethered to specific loci on the DNA nanostructures for effective gene silencing in Nb leaves. We show that siRNA delivered by DNA nanostructures silences a transgene more effectively than siRNA delivered alone. We further find that structural and mechanical properties (size, shape, compactness, and stiffness) of DNA nanostructures and siRNA conjugation loci affect not only nanostructure internalization into plant cells but also subsequent gene silencing efficiencies and pathways.
In this work, we find that the likely gene silencing mechanisms undertaken by siRNA linked to DNA nanostructures depend on the siRNA attachment locus and steric availability of the attached siRNA. Interestingly, siRNA tethered to small 3D nanostructures shows gene silencing at both the transcript (mRNA) and protein levels, whereby siRNA attached to 1D nanostructures shows gene silencing at the protein level but shows an increase in mRNA transcript levels. This phenomenon of increased mRNA implies a possible silencing pathway and mechanism for siRNA delivered with HT or nanostring DNA nanostructures, in which translational inhibition of GFP expression is preferred over direct mRNA cleavage. We hypothesize that protein translation inhibition leads to continuous production and accumulation of repressed mRNAs, as we observe through qPCR of leaves treated with select nanostructure carriers. Specifically, we hypothesize that the steric accessibility of siRNA conjugated to different DNA nanostructures by endogenous silencing proteins plays a dominant role in determining the silencing mechanism, whereby formation of the RISC protein complex that leads to mRNA cleavage may be hindered by the proximity of a nanostructure scaffold for 1D nanostructures but absent for small 3D nanostructures.
In summary, DNA nanostructures can serve as effective scaffolds and nanoscale vehicles for siRNA delivery to plants for efficient gene silencing. This work establishes DNA nanostructures as a programmable toolset for the delivery of exogenous biomolecules such as siRNA to plants and establishes guidelines for the design of DNA nanostructures for effective uptake into plant cells for various applications in plant biotechnology.
Materials and Methods
Plant Growth.
Transgenic mGFP5 Nb (from the Staskawicz Lab, University of California, Berkeley), tobacco, arugula, and watercress seeds were germinated and kept in SunGro Sunshine LC1 Grower soil mixture and were grown to 3–4 wk of age within the chamber before experimental use.
Infiltration of Leaves with Nanomaterials.
Four-week-old mGFP5 Nb plants were punctured on the abaxial surface of the leaf lamina, and 100 μL of DNA nanostructure solutions were infiltrated with a 1-mL needleless syringe. Nanostructure internalization or GFP gene silencing efficiency was quantified after a set period of time, depending on experiment (6 or 12 h, 2–3 d, or 7 d postinfiltration). Nanostructure internalization efficiencies were determined as described in SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
We acknowledge the support of the Burroughs Wellcome Fund (Career Awards at the Scientific Interface), a US Department of Agriculture (USDA) Agriculture and Food Research Initiative grant with Award 2018-67021-27964, a Foundation for Food and Agriculture Research New Innovator Award, an NSF-USDA-Biotechnology and Biological Sciences Research Council Grant, and the Berkeley Molecular Imaging Center and QB3 Shared Stem Cell facilities. H.Z. acknowledges the National Natural Science Foundation of China (Grant 21605153). G.S.D. is supported by the Schlumberger Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818290116/-/DCSupplemental.
References
- 1.Liu Y, et al. A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice. Nat Biotechnol. 2015;33:301–305. doi: 10.1038/nbt.3069. [DOI] [PubMed] [Google Scholar]
- 2.Li T, Liu B, Spalding MH, Weeks DP, Yang B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol. 2012;30:390–392. doi: 10.1038/nbt.2199. [DOI] [PubMed] [Google Scholar]
- 3.Goswami R, Dasgupta P, Saha S, Venkatapuram P, Nandi S. Resource integration in smallholder farms for sustainable livelihoods in developing countries. Cogent Food Agric. 2016;2:1272151. [Google Scholar]
- 4.Binns AN. Agrobacterium‐mediated gene delivery and the biology of host range limitations. Physiol Plant. 1990;79:135–139. [Google Scholar]
- 5.Altpeter F, et al. Advancing crop transformation in the era of genome editing. Plant Cell. 2016;28:1510–1520. doi: 10.1105/tpc.16.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rudramurthy GR, Swamy MK. Potential applications of engineered nanoparticles in medicine and biology: An update. J Biol Inorg Chem. 2018;23:1185–1204. doi: 10.1007/s00775-018-1600-6. [DOI] [PubMed] [Google Scholar]
- 7.Rizvi SAA, Saleh AM. Applications of nanoparticle systems in drug delivery technology. Saudi Pharm J. 2018;26:64–70. doi: 10.1016/j.jsps.2017.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demirer GS, et al. High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat Nanotechnol. 2019 doi: 10.1038/s41565-019-0382-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demirer GS, Zhang H, Goh NS, Chang R, Landry MP. 2019. Nanotubes effectively deliver siRNA to intact plant cells and protect siRNA against nuclease degradation. BioRxiv:10.1101/564427. Preprint, posted March 1, 2019.
- 10.Mitter N, et al. Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat Plants. 2017;3:16207. doi: 10.1038/nplants.2016.207. [DOI] [PubMed] [Google Scholar]
- 11.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 12.Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 13.Lin C, Liu Y, Yan H. Designer DNA nanoarchitectures. Biochemistry. 2009;48:1663–1674. doi: 10.1021/bi802324w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Douglas SM, et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Fan C, Pei H, Shi J, Huang Q. Smart drug delivery nanocarriers with self-assembled DNA nanostructures. Adv Mater. 2013;25:4386–4396. doi: 10.1002/adma.201300875. [DOI] [PubMed] [Google Scholar]
- 16.Schüller VJ, et al. Cellular immunostimulation by CpG-sequence-coated DNA origami structures. ACS Nano. 2011;5:9696–9702. doi: 10.1021/nn203161y. [DOI] [PubMed] [Google Scholar]
- 17.Li J, et al. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides. ACS Nano. 2011;5:8783–8789. doi: 10.1021/nn202774x. [DOI] [PubMed] [Google Scholar]
- 18.Sun W, et al. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew Chem Int Ed Engl. 2015;54:12029–12033. doi: 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H, et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat Nanotechnol. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cunningham FJ, Goh NS, Demirer GS, Matos JL, Landry MP. Nanoparticle-mediated delivery towards advancing plant genetic engineering. Trends Biotechnol. 2018;36:882–897. doi: 10.1016/j.tibtech.2018.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stefano G, Renna L, Brandizzi F. Plant cell vacuoles: Staining and fluorescent probes. Methods Mol Biol. 2018;1789:55–63. doi: 10.1007/978-1-4939-7856-4_5. [DOI] [PubMed] [Google Scholar]
- 22.Horn MA, Heinstein PF, Low PS. Receptor-mediated endocytosis in plant cells. Plant Cell. 1989;1:1003–1009. doi: 10.1105/tpc.1.10.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gratton SEA, et al. The effect of particle design on cellular internalization pathways. Proc Natl Acad Sci USA. 2008;105:11613–11618. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartmann R, Weidenbach M, Neubauer M, Fery A, Parak WJ. Stiffness-dependent in vitro uptake and lysosomal acidification of colloidal particles. Angew Chem Int Ed Engl. 2015;54:1365–1368. doi: 10.1002/anie.201409693. [DOI] [PubMed] [Google Scholar]
- 25.Wang P, Lombi E, Zhao FJ, Kopittke PM. Nanotechnology: A new opportunity in plant sciences. Trends Plant Sci. 2016;21:699–712. doi: 10.1016/j.tplants.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Schwab F, et al. Barriers, pathways and processes for uptake, translocation and accumulation of nanomaterials in plants–Critical review. Nanotoxicology. 2016;10:257–278. doi: 10.3109/17435390.2015.1048326. [DOI] [PubMed] [Google Scholar]
- 27.Bastings MMC, et al. Modulation of the cellular uptake of DNA origami through control over mass and shape. Nano Lett. 2018;18:3557–3564. doi: 10.1021/acs.nanolett.8b00660. [DOI] [PubMed] [Google Scholar]
- 28.Pfitzner E, et al. Rigid DNA beams for high-resolution single-molecule mechanics. Angew Chem Int Ed Engl. 2013;52:7766–7771. doi: 10.1002/anie.201302727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu MF, Files BS, Arepalli S, Ruoff RS. Tensile loading of ropes of single wall carbon nanotubes and their mechanical properties. Phys Rev Lett. 2000;84:5552–5555. doi: 10.1103/PhysRevLett.84.5552. [DOI] [PubMed] [Google Scholar]
- 30.Merli R, Lazaro C, Monleon S, Domingo A. A molecular structural mechanics model applied to the static behavior of single-walled carbon nanotubes: New general formulation. Comput Struct. 2013;127:68–87. [Google Scholar]
- 31.Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 32.Liu Q, et al. Carbon nanotubes as molecular transporters for walled plant cells. Nano Lett. 2009;9:1007–1010. doi: 10.1021/nl803083u. [DOI] [PubMed] [Google Scholar]
- 33.Serag MF, et al. Trafficking and subcellular localization of multiwalled carbon nanotubes in plant cells. ACS Nano. 2011;5:493–499. doi: 10.1021/nn102344t. [DOI] [PubMed] [Google Scholar]
- 34.Serag MF, Kaji N, Habuchi S, Bianco A, Baba Y. Nanobiotechnology meets plant cell biology: Carbon nanotubes as organelle targeting nanocarriers. RSC Adv. 2013;3:4856–4862. [Google Scholar]
- 35.Wong MH, et al. Lipid exchange envelope penetration (LEEP) of nanoparticles for plant engineering: A universal localization mechanism. Nano Lett. 2016;16:1161–1172. doi: 10.1021/acs.nanolett.5b04467. [DOI] [PubMed] [Google Scholar]
- 36.Mahmood-ur-Rahman, Ali I, Husnain T, Riazuddin S. RNA interference: The story of gene silencing in plants and humans. Biotechnol Adv. 2008;26:202–209. doi: 10.1016/j.biotechadv.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 37.Taochy C, et al. A genetic screen for impaired systemic RNAi highlights the crucial role of DICER-LIKE 2. Plant Physiol. 2017;175:1424–1437. doi: 10.1104/pp.17.01181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herr AJ, Baulcombe DC. RNA silencing pathways in plants. Cold Spring Harb Symp Quant Biol. 2004;69:363–370. doi: 10.1101/sqb.2004.69.363. [DOI] [PubMed] [Google Scholar]
- 39.Tang W, et al. Post-transcriptional gene silencing induced by short interfering RNAs in cultured transgenic plant cells. Genomics Proteomics Bioinformatics. 2004;2:97–108. doi: 10.1016/S1672-0229(04)02015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molnar A, Melnyk C, Baulcombe DC. Silencing signals in plants: A long journey for small RNAs. Genome Biol. 2011;12:215. doi: 10.1186/gb-2010-11-12-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brodersen P, et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 42.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 43.Wang P, et al. Visualization of the cellular uptake and trafficking of DNA origami nanostructures in cancer cells. J Am Chem Soc. 2018;140:2478–2484. doi: 10.1021/jacs.7b09024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kocabey S, et al. Cellular uptake of tile-assembled DNA nanotubes. Nanomaterials (Basel) 2014;5:47–60. doi: 10.3390/nano5010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pérez-de-Luque A. Interaction of nanomaterials with plants: What do we need for real applications in agriculture? Front Environ Sci. 2017;5:12. [Google Scholar]
- 46.Dunoyer P, et al. Small RNA duplexes function as mobile silencing signals between plant cells. Science. 2010;328:912–916. doi: 10.1126/science.1185880. [DOI] [PubMed] [Google Scholar]
- 47.Sarkies P, Miska EA. Small RNAs break out: The molecular cell biology of mobile small RNAs. Nat Rev Mol Cell Biol. 2014;15:525–535. doi: 10.1038/nrm3840. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.