Significance
Proton-pumping V-ATPases maintain the pH of intracellular compartments in all eukaryotic cells and the extracellular environment of specialized cells. V-ATPase dysfunction is associated with diseases including osteopetrosis, renal tubular acidosis, and cancer. Many subunits of mammalian V-ATPases exist as multiple isoforms that are expressed in an organelle-, cell-, or tissue-dependent manner, but the consequences of this diversity are not understood. In the yeast Saccharomyces cerevisiae, there are only two V-ATPase isoforms, making it an ideal system to study isoform-specific characteristics. Identifying similarities and differences in structure and function between V-ATPase isoforms improves our understanding of the enzyme’s diverse cellular functions.
Keywords: V-ATPase, STV1, VPH1, cryo-EM, membrane protein
Abstract
Proton-translocating vacuolar-type ATPases (V-ATPases) are necessary for numerous processes in eukaryotic cells, including receptor-mediated endocytosis, protein maturation, and lysosomal acidification. In mammals, V-ATPase subunit isoforms are differentially targeted to various intracellular compartments or tissues, but how these subunit isoforms influence enzyme activity is not clear. In the yeast Saccharomyces cerevisiae, isoform diversity is limited to two different versions of the proton-translocating subunit a: Vph1p, which is targeted to the vacuole, and Stv1p, which is targeted to the Golgi apparatus and endosomes. We show that purified V-ATPase complexes containing Vph1p have higher ATPase activity than complexes containing Stv1p and that the relative difference in activity depends on the presence of lipids. We also show that VO complexes containing Stv1p could be readily purified without attached V1 regions. We used this effect to determine structures of the membrane-embedded VO region with Stv1p at 3.1-Å resolution, which we compare with a structure of the VO region with Vph1p that we determine to 3.2-Å resolution. These maps reveal differences in the surface charge near the cytoplasmic proton half-channel. Both maps also show the presence of bound lipids, as well as regularly spaced densities that may correspond to ergosterol or bound detergent, around the c-ring.
Vacuolar-type ATPases (V-ATPases) are proton pumps found in the endomembranes of all eukaryotic cells and the plasma membrane of specialized cells. V-ATPase activity is responsible for the acidification of intracellular compartments such as endosomes, lysosomes, the Golgi apparatus, and secretory vesicles (1, 2), as well as for acid secretion by osteoclasts (3), intercalated cells of the kidney (4, 5), and some tumor cells (6, 7). The enzyme consists of a soluble catalytic V1 region, containing subunits A3B3CDFH; three peripheral stalk structures, each comprising a heterodimer of subunits E and G; and a membrane-embedded VO region responsible for proton translocation (1). In the yeast Saccharomyces cerevisiae, the VO region consists of subunits ac8c′c″de with additional subunits f (8) and Voa1p (9) identified recently.
In mammals, there are four isoforms of subunit a, three isoforms of subunit G, and two isoforms each of subunits B, C, E, and d (1, 10). In contrast, only subunit a in S. cerevisiae is encoded by multiple genes: VPH1 (11) and STV1 (12). Vph1p, the major isoform, is found in the vacuole, while Stv1p is found in the Golgi apparatus and endosomes (13). The soluble N-terminal domain of subunit a is required for targeting V-ATPase to its appropriate cellular location (13–15). The membrane-embedded C-terminal domain contains two half-channels for proton translocation and, together with the c8c′c″-ring (c-ring), carries out transmembrane proton transport by a mechanism similar to that of the F-type ATP synthase (1, 8, 16–18). The N-terminal domain of subunit a connects the V1 and VO regions through interactions with two of the peripheral stalks, subunit H, and subunit C, the latter interacting with the third peripheral stalk (19, 20). These interactions are required for coupling ATP hydrolysis in the V1 region to proton translocation through the VO region (21). V-ATPase containing Vph1p (Vph1-V1VO) is reported to have a higher ratio of proton translocation to ATP hydrolysis than V-ATPase containing Stv1p (Stv1-V1VO), suggesting that the Golgi isoform of the enzyme has a lower coupling efficiency or is not fully coupled (22). In yeast, V-ATPase is regulated by reversible dissociation of the V1 and VO regions upon glucose starvation, with both regions becoming autoinhibited after separation (23). However, Stv1-V1VO was reported to resist glucose-dependent dissociation except when overexpressed in the absence of Vph1p, which leads to its mislocalization to the vacuole (22). Cryo-EM has revealed three different rotational states of the S. cerevisiae V-ATPase at resolutions between 6.9 and 8.3-Å (20). A crystal structure of the autoinhibited V1 region from yeast was determined at 6.2–6.5-Å resolution (24). This structure showed that subunit H assumes an inhibitory conformation in the isolated V1 complex, which abolishes ATPase activity (19, 24, 25). A 3.9-Å resolution structure of the autoinhibited VO region was determined by cryo-EM, defining the proton translocation pathway (8). Subsequent cryo-EM of the VO complex in lipid nanodiscs improved the resolution of the VO region structure to 3.5-Å resolution (9).
To understand functional differences between the vacuolar and Golgi isoforms of V-ATPase in yeast, we purified complexes containing either Vph1p or Stv1p and subjected them to ATPase activity assays. These experiments showed that purified Stv1-V1VO has lower ATPase activity than Vph1-V1VO and that the relative difference in activity depends strongly on the presence of lipids in the assay buffer. ATP hydrolysis could be inhibited completely by the addition of bafilomycin-A1 to both preparations, suggesting that ATP hydrolysis in the V1 region is fully coupled to proton transport through the VO region in both Vph1-V1VO and Stv1-V1VO. To investigate the structural basis for the difference in ATPase activity between Vph1-V1VO and Stv1-V1VO, we examined the Golgi V-ATPase by cryo-EM. Structures for the intact Stv1-V1VO complex were determined at 6.6–8.7-Å resolution and showed that the Golgi enzyme also exists in three rotational states, similar to what was observed previously for the vacuolar complex (20). By taking advantage of improved imaging and image analysis methods, structures of the isolated VO region containing Vph1p (Vph1-VO) and Stv1p (Stv1-VO) were determined at 3.2-Å and 3.1-Å resolution, respectively, to facilitate comparison of the two isoforms. Striking differences between electrostatic surface charges near the cytoplasmic half-channels of Vph1p and Stv1p may influence proton pumping and thus explain the lower ATPase activity observed for Stv1-V1VO. The Vph1-VO and Stv1-VO structures also revealed the presence of specifically bound phospholipids within the c-ring and around the transmembrane α-helices of subunit a. Surprisingly, both structures also show regularly spaced densities around the c-ring that may correspond to ergosterol, the major sterol species in yeast (26) which has been shown to interact with V-ATPase and enhance its activity (27), or the steroid-derived hydrophobic group of GDN, the detergent used to solubilize the complex.
Results and Discussion
ATPase Activity Assays.
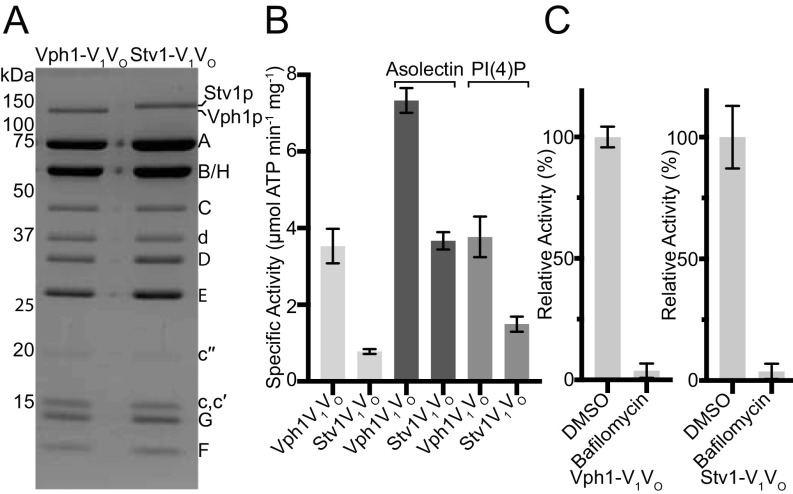
Intact V-ATPase containing either Vph1p (Vph1-V1VO) or Stv1p (Stv1-V1VO) was affinity purified from yeast membranes by a 3×FLAG tag fused to the C terminus of subunit A (Vma1p). The yeast strains used for intact V-ATPase purification had both endogenous subunit a genes knocked out and either VPH1 or STV1 was carried on a plasmid. SDS/PAGE of the purified complexes shows the slight difference in molecular weight between Vph1p (95.5 kDa) and Stv1p (101.6 kDa) (Fig. 1A).
Fig. 1.
Biochemical comparison of Vph1-V1VO and Stv1-V1VO. (A) Coomassie-stained SDS/PAGE gel of Vph1-V1VO and Stv1-V1VO. (B) Specific activity of V-ATPase with no lipids, 100 μg/mL asolectin, or 25 μg/mL PI(4)P. (C) Relative ATPase activity of Vph1-V1VO and Stv1-V1VO in the absence and presence of 1 μM bafilomycin-A1. Data shown are mean ± SD; n = 6 (two biological replicates, three measurements on each).
The specific activities of Vph1-V1VO and Stv1-V1VO were determined by enzyme-coupled spectrophotometric assay (28–30) (Fig. 1B). We found that the specific activity of V-ATPase is sensitive to pH, the presence of lipids, the time from purification to assay, and any freeze-thaw cycles of the enzyme. In the presence of asolectin, the specific activities of Vph1-V1VO and Stv1-V1VO were 7.3 ± 0.3 and 3.7 ± 0.2 μmol ATP min−1mg−1, respectively. Previously measured specific activities for Vph1-V1VO range from 0.22 (concanamycin A-sensitive activity for isolated vacuolar membranes) to 18 μmol ATP min−1mg−1 (13, 31, 32). The specific activity of Vph1-V1VO measured here is the same as the 7.3 ± 0.3 μmol ATP min−1mg−1 reported for lipid-nanodisc reconstituted Vph1-V1VO (32). In the presence of asolectin, Stv1-V1VO has ∼50% the activity of Vph1-V1VO. Previous measurements of concanamycin-sensitive ATPase activity for isolated vacuolar membranes with complexes containing Vph1p (0.221 ± 0.038 μmol ATP min−1mg−1) or Stv1p (0.082 ± 0.026 μmol ATP min−1mg−1) showed that Stv1-V1VO has ∼37% the activity of Vph1-V1VO (13). Asolectin has been shown to increase V-ATPase activity in assays (31). The addition of asolectin from soybean to the assay buffers produced a twofold increase for Vph1-V1VO–specific activity and a 4.7-fold increase for Stv1-V1VO-specific activity (Fig. 1B). In the absence of asolectin, Stv1-V1VO has a specific activity of 0.78 ± 0.06 μmol ATP min−1mg−1, ∼22% the specific activity of Vph1-V1VO, 3.53 ± 0.45 μmol ATP min−1mg−1.
The phosphatidylinositol lipid PI(4)P is enriched in the Golgi membranes of yeast but not in the vacuolar membranes, and has been shown to interact specifically with Stv1p but not Vph1p (15). PI(4)P is required for appropriate localization of Stv1-containing V-ATPases, and reduction in intracellular PI(4)P levels has also been shown to impair V-ATPase activity (15). In ATPase assays of the purified protein, the addition of PI(4)P did not significantly affect the activity of Vph1-V1VO (3.8 ± 0.5 μmol ATP min−1mg−1) but resulted in a 1.9-fold increase in the activity of Stv1-V1VO (1.5 ± 0.2 μmol ATP min−1mg−1) compared with when no lipids were added to the assay (Fig. 1B). The Vph1-V1VO and Stv1-V1VO complexes used in the activity assays were purified from strains lacking endogenous Vph1p and Stv1p. Both Vph1-V1VO and Stv1-V1VO may be targeted to the vacuole under these conditions (22). The mislocalization of Stv1-V1VO, which may remove interactions with Golgi-specific lipids, could explain the substantially lower ATPase activity observed when the assay was not supplemented with lipids. Together, these results suggest that V-ATPase activity is lipid-dependent and that interaction between Stv1p and PI(4)P enhances Stv1-V1VO activity. The vacuole is the most acidic compartment in yeast, with a pH ranging from <5–6.5 (33), while the Golgi apparatus in most eukaryotic cells has a pH gradient from the cis to the trans Golgi of 6.0–6.7 (34). With three ATP hydrolysis sites and 10 subunits in the c-ring, both Vph1-V1VO and Stv1-V1VO maintain the same ATP:H+ ratio and can establish the same equilibrium pH difference across a membrane under equivalent conditions (20). Factors such as proton leak and density of V-ATPase complexes in an organelle membrane likely contribute to the different pH of different organelles (35). However, the lower activity of Stv1-V1VO, which would alter the balance between proton pumping and proton leaking, may also be partially responsible for the higher pH in intracellular compartments acidified by Stv1-V1VO.
ATP hydrolysis in the V1 region drives rotation of the central rotor subcomplex (consisting of subunits D, F, and d) and results in proton translocation through the VO region. Coupling of these two activities may be assessed by measuring the decrease in ATPase activity when proton translocation is blocked. The V-ATPase–specific inhibitor bafilomycin-A1 binds to the VO region and inhibits proton translocation (36, 37). For both V-ATPase isoforms, bafilomycin-A1 was able to completely inhibit ATP hydrolysis activity (Fig. 1C), demonstrating that ATPase activity in the V1 region is fully coupled to proton transport through the VO region regardless of subunit a isoform.
Structure Determination of Stv1-V1VO.
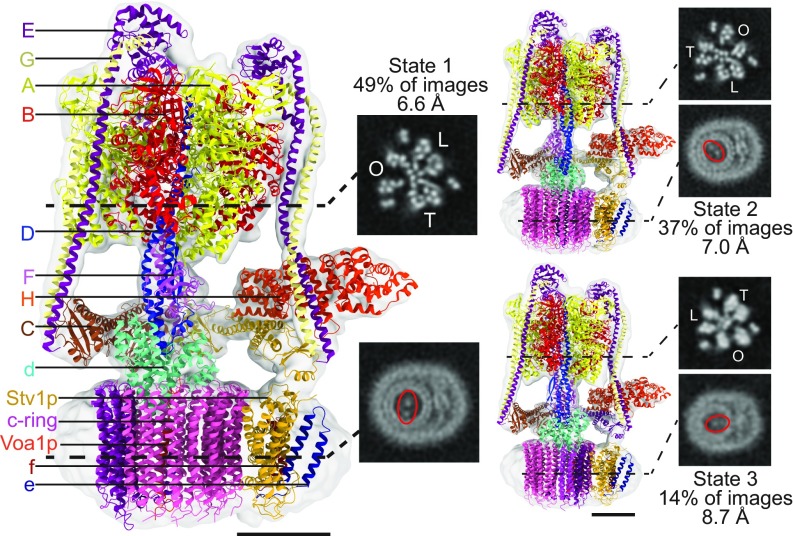
The difference in ATPase activity between the two yeast V-ATPase isoforms, with Vph1-V1VO having approximately twice the activity of Stv1-V1VO in the presence of asolectin, is remarkable considering that the two complexes differ only in the subunit a isoform that is present. The N-terminal domains of Vph1p and Stv1p are 33% identical and 61% similar, while the C-terminal domains, which form the proton transport channels through the membrane, are 66% identical and 84% similar (11, 12). In Vph1-V1VO, the N-terminal domain of Vph1p binds subunit H, which undergoes a conformational change to inhibit ATP hydrolysis in the dissociated V1 complex (19, 23, 24). We hypothesized that an altered binding site for subunit H on the N-terminal domain of Stv1p could lead to adoption of the inhibitory conformation by subunit H in Stv1-V1VO, thereby reducing the enzyme’s ATPase activity. To investigate this possibility, we determined structures of Stv1-V1VO by cryo-EM (Fig. 2 and SI Appendix, Fig. S2). Three rotational states were observed for Stv1-V1VO, corresponding to the previously described states 1 (49% of images), 2 (37% of images), and 3 (14% of images) at 6.6-, 7.0- and 8.7-Å resolution, respectively. These structures are remarkably similar to the previously determined structures of Vph1-V1VO (20) with map correlation values of 0.969, 0.972, and 0.960 between the two isoforms of states 1, 2, and 3, respectively. These cryo-EM maps did not provide any evidence to suggest that subunit H adopts an inhibitory conformation in Stv1-V1VO.
Fig. 2.
Cryo-EM of Stv1-V1VO. Cryo-EM structure of three rotational states of Stv1-V1VO with fitted atomic models and homology models. Cross-sections through the V1 region show the three catalytic AB heterodimers in open (O), loose (L), or tight (T) conformations. Cross sections through the VO region show the different position of the c-ring in each of the three rotational states. (Scale bars, 50 Å.)
Purification of the Isolated Stv1-VO Complex.
The lack of detectable structural differences between Vph1-V1VO and Stv1-V1VO at 6.6–8.7-Å resolution suggested that the difference in activity between the two complexes is due to subtle differences in the primary structure of Vph1p and Stv1p. The isolated VO complex is autoinhibited and thus less structurally heterogeneous than the intact complex, which facilitated high-resolutions structure determination of the VO complex containing Vph1p (Vph1-VO) (8, 9). To investigate the differences between Vph1p and Stv1p at the atomic level, we sought to purify the VO complex containing Stv1p (Stv1-VO). Long growth conditions induce glucose starvation and cause Vph1-V1VO dissociation, allowing purification of Vph1-VO. However, it has been reported that Stv1-V1VO can only undergo glucose-dependent dissociation when overexpressed and mislocalized to the yeast vacuole (22).
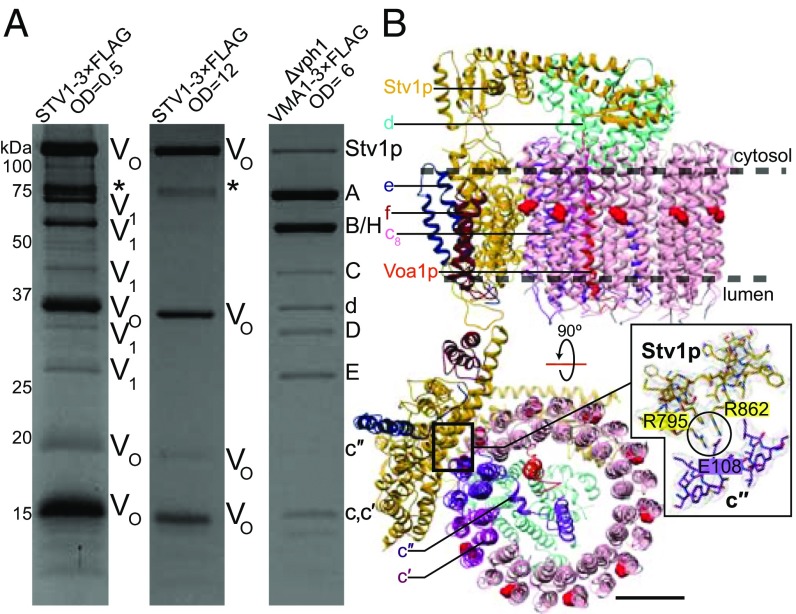
To isolate Stv1-VO, we prepared the yeast strain SABY64 with the sequence for a 3×FLAG tag introduced into the chromosomal DNA following the STV1 gene, producing a Stv1p-3×FLAG fusion protein expressed under the native promoter. The VPH1 gene was not deleted in this strain, maintaining the physiological ratio of Stv1p and Vph1p and proper localization of Stv1p-containing complexes. Yeast membranes were isolated and V-ATPase was purified from SABY64 via the 3×FLAG tag following growth in a fermenter for 12 h (∼OD660 = 0.5) or 48 h (∼OD660 = 12). We found that purification of V-ATPase containing Stv1p following growth to OD660 = 0.5 produced VO complexes with contaminating V1VO complexes, as indicated by the presence of both V1 and VO subunits by SDS/PAGE (Fig. 3A). In contrast, purification following growth to OD660 = 12 (48-h growth) resulted in isolation of VO complexes only, with no detectable contamination from intact V-ATPase by SDS/PAGE (Fig. 3A). This result suggests that almost exclusively dissociated Stv1-VO complexes exist in the membranes of yeast under long growth conditions, due to either disassembly of intact Stv1-V1VO or failure to assemble Stv1-V1VO. Additional bands observed in the gel (Fig. 3A, marked with *) that do not correspond to known V-ATPase subunits are contaminants that can be removed by size exclusion chromatography (SI Appendix, Fig. S1A). While this result does not show that V-ATPases containing Stv1p dissociate in response to glucose starvation, it does show that even under its native promoter most VO complexes are dissociated from V1 complexes when yeast is grown to high OD, a phenomenon that allows purification of isolated Stv1-VO complexes for structural analysis.
Fig. 3.
Purification and EM of Stv1-VO. (A) Coomassie-stained SDS/PAGE gel shows a mixture of intact V-ATPase and VO complexes when purified by a 3×FLAG tag on Stv1p at OD660 = 0.5, isolated VO complexes when purified by a 3×FLAG tag on Stv1p at OD660 = 12, and intact V-ATPase when purified by a 3×FLAG tag on Vma1p (subunit A) at OD660 = 6. (B) Atomic model of the Stv1-VO complex. Essential proton-carrying glutamate residues on the c-ring are shown in red. R795 and R862 from Stv1p are in contact with the essential glutamate residue E108 on subunit c″. (Scale bar, 25 Å.)
Structure Determination of Stv1-VO and Vph1-VO.
The ability to prepare isolated VO complexes of both isoforms enabled us to investigate the differences between Vph1-VO and Stv1-VO at the atomic level by determining high resolution cryo-EM structures. Following purification of Stv1-VO from yeast strain SABY64, the complex was exchanged from the detergent n-Dodecyl β-D-maltoside (DDM) into glycodiosgenin (GDN), which we previously found to be amenable to high-resolution structure determination (38). We subjected this sample to cryo-EM analysis, resulting in a map at 3.1-Å resolution that was used to build an atomic model of Stv1-VO (Fig. 3B and SI Appendix, Figs. S3 and S4 and Table S1). The improved resolution of this map compared with previous structures of Vph1-VO is likely due to improved specimen preparation, imaging, and image analysis methods. To facilitate comparison of Stv1-VO and Vph1-VO and to avoid interpretation due to differences in map resolutions, we purified Vph1-VO in DDM (8), exchanged it into GDN, and determined its structure to 3.2-Å resolution (SI Appendix, Fig. S3 and Table S1). As expected, the Vph1-VO atomic model is extremely similar to earlier Vph1-VO atomic models (8, 9) (SI Appendix, Fig. S5). The soluble N-terminal domain of the first Vph1p atomic model (8) was built as poly-alanine. Of the 570 residues that were built in the first model, the rmsd of 462 mutually modeled Cα atoms is 1.39 Å. The two models differ most at the linker region between the N- and C-terminal domains. Compared with the more recent lipid-nanodisc–embedded structure (9), the present Vph1p model is even more similar, with an rmsd between 730 mutually modeled Cα atoms of 1.22 Å.
Overall Structures of Stv1-VO and Vph1-VO.
The overall arrangement and stoichiometry of subunits in Stv1-VO is identical to that of Vph1-VO (Fig. 3B). The position of the c-ring in the autoinhibited Stv1-VO and Vph1-VO complexes matches the position of the c-ring in rotational state 3 of the intact enzyme. Upon dissociation, the N-terminal domain of Stv1p swings inward toward the center of the complex and interacts with subunit d, as was seen previously with the isolated Vph1-VO complex (8, 9, 39). The recently identified VO subunits f (8) and Voa1p (9) are also components of the Stv1-VO complex, and their presence was confirmed by mass spectrometry (Dataset S1). The presence of subunits f and e in the Stv1-VO complex is particularly notable, as the interaction between these proteins and V-ATPase are entirely mediated by subunit a, showing that binding sites are conserved between Vph1p and Stv1p.
Interestingly, a second 3D class of the Stv1-VO complex was obtained, also at 3.1-Å resolution (SI Appendix, Fig. S3H). In this class, the C-terminal domain of Stv1p is shifted, resulting in a slight difference in position for subunits a, e, and f relative to the c-ring. When the structures are aligned by the C-terminal domain of Stv1p, the rotational difference in the c-ring between these two conformations is ∼5°. The sidechain contacts between R795 and R862 of Stv1p and E108 of subunit c″ are maintained in both structures. This additional 3D class shows that there is flexibility in the linker between the N- and C-terminal domains of Stv1p and that the autoinhibited complex is not completely rigid. This flexibility may be important for reassembly of the V1 and VO regions following dissociation. A similar class was not observed for Vph1-VO, but this discrepancy may be due to a slightly lower quality in the dataset of images.
Similarities Between Vph1p and Stv1p.
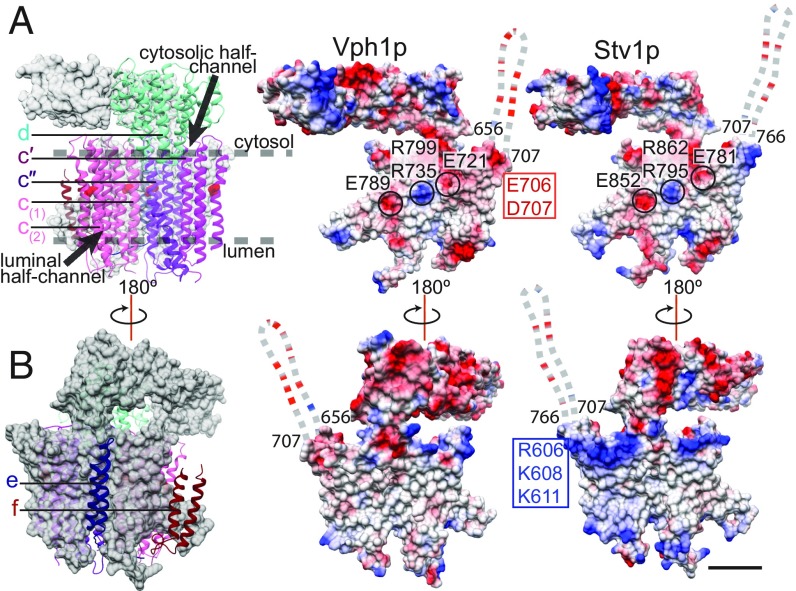
Like Vph1p, Stv1p contains a total of eight transmembrane α-helices, two of which are highly tilted relative to the plane of the membrane. The two subunit a isoforms have an identical fold (SI Appendix, Fig. S5), with an RMSD between 673 mutually modeled Cα atoms of 1.42 Å. In both the Vph1-VO and Stv1-VO structures, the essential arginine residue (R735 in Vph1p and R795 in Stv1p) is in contact with the conserved glutamate (E108) of subunit c″ (Fig. 3B, Inset). Both subunit a isoforms also display the pattern of electrostatic potentials identified for the mitochondrial ATP synthase (38) at the c-ring interaction interface: two negatively charged regions at the cytoplasmic and luminal half-channels with a strong positive charge in between from the essential arginine residue and the nearby conserved arginine (R799 in Vph1p and R862 in Stv1p) (Fig. 4A).
Fig. 4.
Comparison of Stv1p and Vph1p. (A) Surface representation of subunit a (gray), with atomic models for subunits d, c′, c″, c(1), and c(2), and electrostatic surface charge of Vph1p and Stv1p at the c-ring interaction interface. (B) Surface representation of subunit a (gray) with atomic model for subunits e, and f, and electrostatic surface charge of Vph1p and Stv1p at the surface facing the lipid bilayer. Negatively charged residues are highlighted in red; positively charged residues, in blue. The disordered loop near the opening of the cytosolic half-channel is shown as a dashed line. (Scale bars, 25 Å.)
The positions of many residues that are critical for V-ATPase activity (18) are conserved in both subunit a isoforms and explain how both isoforms can support a similar mechanism of proton translocation (8, 9, 40) (SI Appendix, Fig. S6A). Protons enter Stv1p through the cytoplasmic half-channel between Stv1p and the c-ring (8), which is lined with polar and charged residues (E781, N785, and H789) that are conserved in Vph1p (E721, N725, and H729) and decrease ATPase activity when mutated (18). A conserved glutamate residue from a c-ring subunit (Figs. 3B and 4A, red spheres) becomes protonated as rotation of the ring, driven by ATP hydrolysis in the V1 region, drags the residue from the aqueous environment of the cytoplasmic half-channel into the hydrophobic environment of the lipid bilayer (8, 41). In this way, ATP hydrolysis in the V1 region induces protonation of the c-ring in the VO region. Rotation of the ring delivers a protonated glutamate residue to the opening of the luminal half-channel. Formation of a salt bridge with the essential R795 (R735 in Vphp1) causes the conserved c-ring glutamate to lose its proton to the luminal half-channel (8), which is lined with charged and polar residues including D471, D527, and H803 (D425, D481, and H743 in Vph1p) (SI Appendix, Fig. S6A).
Differences Between Vph1p and Stv1p.
The N-terminal domain of Stv1p contains the W83KY sequence, which is necessary and sufficient for targeting Stv1-V1VO to the Golgi apparatus (14). K84 of this sequence has been shown to interact specifically with the phosphatidylinositol lipid PI(4)P, which is required for efficient localization and activity of the Golgi V-ATPase (15). Interestingly, no density was observed in the Stv1-VO map for the region corresponding to H78 to L111, which contains the Golgi targeting sequence, suggesting that this region is flexible (SI Appendix, Fig. S6B). In contrast, despite the slightly lower map quality, there is some density in the Vph1-VO map for the corresponding region (H79–V100), which has not been described as interacting with PI(4)P (15) (SI Appendix, Fig. S6B).
The main difference between the C-terminal domains of the two subunit a isoforms is seen in the electrostatic surface charge near the cytoplasmic half-channel (Fig. 4). Vph1p has a more negatively charged surface in this region due the presence of acidic residues E706 and D707, which are not conserved in Stv1p (Fig. 4A). The opening of the cytoplasmic half-channel in Stv1p has a more positively charged surface due to the presence of basic residues R606, K608, and K611, which are not conserved in Vph1p (Fig. 4B). A group of positively charged residues is conserved at the same position in the Golgi subunit a isoforms of the yeast Candida albicans (K644, R646) but not in the corresponding vacuolar isoform (SI Appendix, Fig. S8). Additionally, the disordered cytoplasmic loop near the opening of the cytoplasmic half-channel (Fig. 4, dashed lines; SI Appendix, Fig. S8, yellow box) has a higher proportion of negatively charged residues in Vph1p than in Stv1p (SI Appendix, Fig. S8, red box). A similar pattern is observed in the corresponding sequence of the vacuolar subunit a isoform in C. albicans (SI Appendix, Fig. S8). Additionally, this unmodeled cytosolic loop is the region with the lowest sequence similarity between the C-terminal domains of Vph1p and Stv1p, suggesting that it may have a role in the functional differences between the isoforms. The more negative charge near the opening of the cytoplasmic half-channel of Vph1p may enhance proton translocation activity relative to Stv1p by increasing the local concentration of protons.
Binding of Lipids.
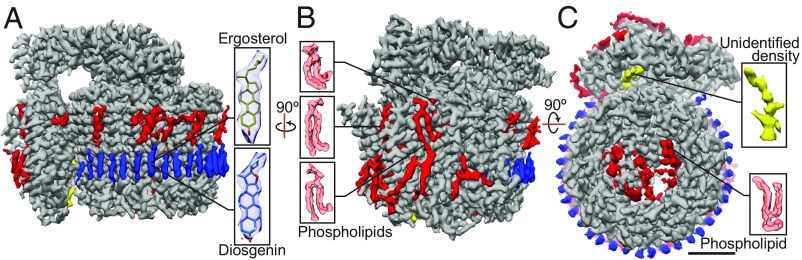
Improved resolution for the maps of both Vph1-VO and Stv1-VO revealed the presence of additional densities that could not be accounted for by protein subunits (shown in Fig. 5 for Stv1-VO and SI Appendix, Fig. S7 for Vph1-VO). These densities are located in the membrane-embedded regions of the complex and likely correspond to bound lipid or detergent molecules. Because of the higher quality map, these densities are clearer in the Stv1-VO map than in the Vph1-VO map. A ring of densities can be seen around the outside of the c-ring, in the region corresponding to the luminal leaflet of the membrane for both Stv1-VO and Vph1-VO (Fig. 5, blue densities; SI Appendix, Fig. S7, blue densities). These densities show a good fit for the size and shape of ergosterol, the major sterol species in yeast (26, 42), or diosgenin, the sterol-like moiety of GDN. If they correspond to GDN, the remainder of the GDN molecule may not be visible because it is flexible and does not average coherently in the cryo-EM map. Four of these sterol-like densities associate with each subunit of the c-ring. Liquid chromatography mass spectrometry (LC-MS) analysis of extracted lipids identified phospholipids and ergosterol in the sample (SI Appendix, Fig. S1). Functional studies of V-ATPase showed that extraction of sterols from membranes with methyl-β-cyclodextrin led to loss of vacuolar acidification in yeast (27) and reduced vacuolar acidification in mammalian cells (43), suggesting that the bound sterols are required for optimal V-ATPase activity. If these densities correspond to ergosterol molecules, they are reminiscent of cardiolipin molecules that are proposed to bind trimethylated lysine residues in the c-rings of metazoan F-type ATP synthases and are thought to have a role in stabilizing the ring and lubricating its rotation, or even enhancing proton translocation by the complex (44, 45). They would have to bind the c-ring with high affinity to copurify with the V-ATPase.
Fig. 5.
Lipids in the Stv1-VO map. (A) Stv1-VO map with densities corresponding to protein (gray) and a ring of densities around the luminal region of the c-ring, possibly corresponding to ergosterol or diosgenin, the sterol-like moiety of GDN (blue); unidentified regularly spaced densities are also seen around the cytosolic region of the c-ring (red). (B) Bound phospholipids around subunit a, modeled with phosphatidylcholine (red). (C) An unidentified density (yellow) is seen in the Stv1-VO map; also seen are phospholipids in the center of the c-ring (red). (Scale bar, 25 Å.)
LC-MS/MS analysis of extracted lipids from the Stv1-VO and Vph1-VO preparations also revealed the presence of phosphatidylethanolamine, phosphatidylserine, phosphatidylinositol, and phosphatidylcholine in the samples, with the latter two in greatest abundance (SI Appendix, Fig. S1). Several densities resembling phospholipids are seen around subunit a (Fig. 5B, red densities). Long, well-ordered densities are also observed lining the center of the c-ring, some of which clearly resemble phospholipids (Fig. 5C, red densities). These bound phospholipids cannot be identified specifically but have been tentatively modeled as phosphatidylcholine. The function of these phospholipids is not clear, but they may be important for stability of the VO complex, for maintaining intersubunit interactions, or for V-ATPase activity. An additional unidentified density that was seen previously in the 3.5-Å nanodisc-reconstituted Vph1-VO map (9) is also seen in both the Stv1-VO and Vph1-VO maps in GDN (Fig. 5C, yellow density; SI Appendix, Fig. S6, yellow density). The nature of this density remains unclear, although its structure appears more like a short stretch of polypeptide than a lipid or carbohydrate. Regularly spaced densities of unknown identity, possibly corresponding to bound detergent, are also observed around the c-ring in the region corresponding to the cytosolic leaflet of the membrane (Fig. 5A, red densities).
Together, these structures illustrate that lipids play a central part in the structure and dynamics of V-ATPase complexes. The ATPase activity of both Vph1-V1VO and Stv1-V1VO is affected by lipids, and both possess fully coupled ATP hydrolysis and proton translocation. However, the Golgi/endosomal V-ATPase has substantially lower activity than its vacuolar counterpart. The structures of Vph1-V1VO and Stv1-V1VO reveal that this difference in V-ATPase activity is likely due to subtle differences in the structure of the different subunit a isoforms.
Materials and Methods
Yeast Strains and Protein Purification.
The yeast strain SABY47 (VMA1-3×FLAG, Δvph1Δstv1) (29) was transformed with plasmids encoding either wild-type Vph1p (pSAB19) (29) or wild-type Stv1p (pTV6) to produce yeast strains TVY1 and TVY7, respectively. Plasmid pTV6 was made by conventional restriction-based cloning to place the STV1 gene (from pRS316-HA-STV1, a gift from Patricia Kane) in the p413ADH1 vector. Restriction-free cloning was used to remove the HA tag. TVY1 was used to purify Vph1-V1VO, and TVY7 was used to purify Stv1-V1VO, via the 3×FLAG tag fused to Vma1p (subunit A) of the V1 region. The yeast strain CACY1 (VPH1-3×FLAG) was used for isolating Vph1-VO (8). The yeast strain SABY64 was prepared by integrating the sequence for a 3×FLAG tag by homologous recombination at the 3′ end of the STV1 gene in the protease-deficient background strain BJ2168. SABY64 was used to purify Stv1-VO. Yeast were grown in 11 L YPD media in a Microferm fermenter (New Brunswick Scientific) at 30 °C, with aeration of 34 cubic feet per hour, and stirring at 300 rpm. For isolation of intact V-ATPase complexes, yeast were harvested after 20–24 h (OD660 = 6–8). For isolation of the dissociated VO complex, yeast were harvested after 48 h (OD660 = 12). Protein was purified with M2 Affinity agarose gel (Sigma-Aldrich) as described previously (19). For cryo-EM, the sample was concentrated to ∼300 μL with a 100-kDa MWCO Vivaspin 6 centrifugal concentrator (Sartorius). The protein was subsequently washed with 4 mL buffer containing GDN (50-mM Tris⋅HCl, pH 7.4, 150 mM NaCl, 0.004% [wt/vol] GDN) and concentrated back to ∼300 μL. The sample was washed two more times in this way to exchange it into the GDN buffer, transferred to a Vivaspin 500 centrifugal concentrator (Sartorius), and concentrated to ∼2 mg/mL. Protein concentrations were determined by bicinchoninic acid (BCA) assay (Pierce).
Mass Spectrometry.
Lipids copurifying with the protein samples (Vph1-VO or Stv1-VO, ∼50 μg) were extracted using the Folch method (46). The extracted lipids were dissolved with 2 μL ethanol, then diluted to a final volume of 20 μL with methanol. To avoid interference from detergents in the LC-MS, copurified ergosterol was extracted and analyzed directly by reverse-phase LC-MS without chemical labeling. Ergosterol does not ionize as readily as phospholipids and, as a result, has a low signal-to-noise ratio in the mass spectra. For LC-MS analysis of the extracted lipids, the buffers and gradient were adapted from a previously described protocol (47), which is detailed in the SI Appendix, Methods. Identification of subunits in the Stv1-VO complex by mass spectrometry was done as described previously (8) and as detailed in the SI Appendix, Methods.
ATPase Activity Assay.
Enzyme-coupled ATPase activity assays (28–30) were performed in a 96-well plate with a total reaction volume of 160 μL. Purified Vph1-V1VO or Stv1-V1VO was added to the ATPase assay reaction buffer (50 mM Tris pH 7.5, 3-mM magnesium chloride, 0.2-mM NADH disodium salt, 3.2 units pyruvate kinase, 8 units L-lactic dehydrogenase, and 0.02% [wt/vol] DDM). Where used, asolectin from soybean (Sigma) was added to 100 μg/mL; 16:0–18:1 PI(4)P [1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-myo-inositol-4′-phosphate)] (Avanti Polar Lipids) was added to 25 μg/mL. To initiate the reaction, ATP disodium salt (2 mM) and phosphoenol pyruvic acid monopotassium salt (1 mM) were added. Absorbance at 340 nm was monitored at 37 °C to measure the signal from NADH, which was converted to the concentration of NADH with a standard curve. The rate of ATP hydrolysis for Vph1-V1VO and Stv1-V1VO was linear for at least 100 s, before NADH in the assay buffer was depleted. When used, bafilomycin-A1 in DMSO was added to the reaction from a 1 mM stock solution to a final concentration of 1 μM and compared with a negative control where only DMSO was added.
Cryo-EM.
Cryo-EM, image analysis, and construction of atomic models was done as described previously (48) and as detailed in the SI Appendix, Methods.
Supplementary Material
Acknowledgments
This work is supported by Operating Grant MOP81294 from the Canadian Institutes of Health Research (to J.L.R.), the Canada Research Chairs Program (J.L.R.), Grant 69551-ENABLE from the European Research Council (to C.V.R.), Wellcome Trust Investigator Award 104633/Z/14/Z (to C.V.R.), and the Engineering and Physical Sciences Research Council (V.B.-E.). Titan Krios cryo-EM data were collected at the Toronto High-Resolution High-Throughput Cryo-EM facility supported by the Canadian Foundation for Innovation and Ontario Research Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: EM maps have been deposited in the Electron Microscopy Data Bank (EMDB), https://www.ebi.ac.uk/pdbe/emdb/ (accession nos. EMD-0644–EMD-0648); coordinates have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6O7T–6O7X).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814818116/-/DCSupplemental.
References
- 1.Forgac M. Vacuolar ATPases: Rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol. 2007;8:917–929. doi: 10.1038/nrm2272. [DOI] [PubMed] [Google Scholar]
- 2.Kane PM. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol Mol Biol Rev. 2006;70:177–191. doi: 10.1128/MMBR.70.1.177-191.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin A, et al. V-ATPases in osteoclasts: Structure, function and potential inhibitors of bone resorption. Int J Biochem Cell Biol. 2012;44:1422–1435. doi: 10.1016/j.biocel.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Brown D, Paunescu TG, Breton S, Marshansky V. Regulation of the V-ATPase in kidney epithelial cells: Dual role in acid-base homeostasis and vesicle trafficking. J Exp Biol. 2009;212:1762–1772. doi: 10.1242/jeb.028803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner CA, et al. Renal vacuolar H+-ATPase. Physiol Rev. 2004;84:1263–1314. doi: 10.1152/physrev.00045.2003. [DOI] [PubMed] [Google Scholar]
- 6.Stransky L, Cotter K, Forgac M. The function of V-ATPases in cancer. Physiol Rev. 2016;96:1071–1091. doi: 10.1152/physrev.00035.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGuire C, Cotter K, Stransky L, Forgac M. Regulation of V-ATPase assembly and function of V-ATPases in tumor cell invasiveness. Biochim Biophys Acta. 2016;1857:1213–1218. doi: 10.1016/j.bbabio.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mazhab-Jafari MT, et al. Atomic model for the membrane-embedded VO motor of a eukaryotic V-ATPase. Nature. 2016;539:118–122. doi: 10.1038/nature19828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roh S-H, et al. The 3.5-Å cryoEM structure of nanodisc-reconstituted yeast vacuolar ATPase Vo proton channel. Mol Cell. 2018;69:993–1004.e3. doi: 10.1016/j.molcel.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toei M, Saum R, Forgac M. Regulation and isoform function of the V-ATPases. Biochemistry. 2010;49:4715–4723. doi: 10.1021/bi100397s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manolson MF, et al. The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivo assembly and activity of the yeast vacuolar H(+)-ATPase. J Biol Chem. 1992;267:14294–14303. [PubMed] [Google Scholar]
- 12.Manolson MF, et al. STV1 gene encodes functional homologue of 95-kDa yeast vacuolar H(+)-ATPase subunit Vph1p. J Biol Chem. 1994;269:14064–14074. [PubMed] [Google Scholar]
- 13.Kawasaki-Nishi S, Bowers K, Nishi T, Forgac M, Stevens TH. The amino-terminal domain of the vacuolar proton-translocating ATPase a subunit controls targeting and in vivo dissociation, and the carboxyl-terminal domain affects coupling of proton transport and ATP hydrolysis. J Biol Chem. 2001;276:47411–47420. doi: 10.1074/jbc.M108310200. [DOI] [PubMed] [Google Scholar]
- 14.Finnigan GC, et al. Sorting of the yeast vacuolar-type, proton-translocating ATPase enzyme complex (V-ATPase): Identification of a necessary and sufficient Golgi/endosomal retention signal in Stv1p. J Biol Chem. 2012;287:19487–19500. doi: 10.1074/jbc.M112.343814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banerjee S, Kane PM. Direct interaction of the Golgi V-ATPase a-subunit isoform with PI(4)P drives localization of Golgi V-ATPases in yeast. Mol Biol Cell. 2017;28:2518–2530. doi: 10.1091/mbc.E17-05-0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vik SB, Antonio BJ. A mechanism of proton translocation by F1F0 ATP synthases suggested by double mutants of the a subunit. J Biol Chem. 1994;269:30364–30369. [PubMed] [Google Scholar]
- 17.Jungle W, Lill H, Engelbrecht S. ATP synthase: An electrochemical transducer with rotatory mechanics. Trends Biochem Sci. 1997;22:420–423. doi: 10.1016/s0968-0004(97)01129-8. [DOI] [PubMed] [Google Scholar]
- 18.Toei M, Toei S, Forgac M. Definition of membrane topology and identification of residues important for transport in subunit a of the vacuolar ATPase. J Biol Chem. 2011;286:35176–35186. doi: 10.1074/jbc.M111.273409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benlekbir S, Bueler SA, Rubinstein JL. Structure of the vacuolar-type ATPase from Saccharomyces cerevisiae at 11-Å resolution. Nat Struct Mol Biol. 2012;19:1356–1362. doi: 10.1038/nsmb.2422. [DOI] [PubMed] [Google Scholar]
- 20.Zhao J, Benlekbir S, Rubinstein JL. Electron cryomicroscopy observation of rotational states in a eukaryotic V-ATPase. Nature. 2015;521:241–245. doi: 10.1038/nature14365. [DOI] [PubMed] [Google Scholar]
- 21.Qi J, Forgac M. Function and subunit interactions of the N-terminal domain of subunit a (Vph1p) of the yeast V-ATPase. J Biol Chem. 2008;283:19274–19282. doi: 10.1074/jbc.M802442200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawasaki-Nishi S, Nishi T, Forgac M. Yeast V-ATPase complexes containing different isoforms of the 100-kDa a-subunit differ in coupling efficiency and in vivo dissociation. J Biol Chem. 2001;276:17941–17948. doi: 10.1074/jbc.M010790200. [DOI] [PubMed] [Google Scholar]
- 23.Kane PM. Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo. J Biol Chem. 1995;270:17025–17032. [PubMed] [Google Scholar]
- 24.Oot RA, Kane PM, Berry EA, Wilkens S. Crystal structure of yeast V1-ATPase in the autoinhibited state. EMBO J. 2016;35:1694–1706. doi: 10.15252/embj.201593447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parra KJ, Keenan KL, Kane PM. The H subunit (Vma13p) of the yeast V-ATPase inhibits the ATPase activity of cytosolic V1 complexes. J Biol Chem. 2000;275:21761–21767. doi: 10.1074/jbc.M002305200. [DOI] [PubMed] [Google Scholar]
- 26.Zinser E, Paltauf F, Daum G. Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism. J Bacteriol. 1993;175:2853–2858. doi: 10.1128/jb.175.10.2853-2858.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y-Q, et al. Requirement for ergosterol in V-ATPase function underlies antifungal activity of azole drugs. PLoS Pathog. 2010;6:e1000939. doi: 10.1371/journal.ppat.1000939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lötscher H-R, deJong C, Capaldi RA. Modification of the F0 portion of the H+-translocating adenosinetriphosphatase complex of Escherichia coli by the water-soluble carbodiimide 1-ethyl-3-[3-(dimethylamino)propyl]carbodiimide and effect on the proton channeling function. Biochemistry. 1984;23:4128–4134. doi: 10.1021/bi00313a018. [DOI] [PubMed] [Google Scholar]
- 29.Bueler SA, Rubinstein JL. Vma9p need not be associated with the yeast V-ATPase for fully-coupled proton pumping activity in vitro. Biochemistry. 2015;54:853–858. doi: 10.1021/bi5013172. [DOI] [PubMed] [Google Scholar]
- 30.Kiianitsa K, Solinger JA, Heyer W-D. NADH-coupled microplate photometric assay for kinetic studies of ATP-hydrolyzing enzymes with low and high specific activities. Anal Biochem. 2003;321:266–271. doi: 10.1016/s0003-2697(03)00461-5. [DOI] [PubMed] [Google Scholar]
- 31.Uchida E, Ohsumi Y, Anraku Y. Purification and properties of H+-translocating, Mg2+-adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J Biol Chem. 1985;260:1090–1095. [PubMed] [Google Scholar]
- 32.Sharma S, Wilkens S. Biolayer interferometry of lipid nanodisc-reconstituted yeast vacuolar H+ -ATPase. Protein Sci. 2017;26:1070–1079. doi: 10.1002/pro.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li SC, Kane PM. The yeast lysosome-like vacuole: Endpoint and crossroads. Biochim Biophys Acta. 2009;1793:650–663. doi: 10.1016/j.bbamcr.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casey JR, Grinstein S, Orlowski J. Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol. 2010;11:50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 35.Paroutis P, Touret N, Grinstein S. The pH of the secretory pathway: Measurement, determinants, and regulation. Physiology (Bethesda) 2004;19:207–215. doi: 10.1152/physiol.00005.2004. [DOI] [PubMed] [Google Scholar]
- 36.Bowman EJ, Graham LA, Stevens TH, Bowman BJ. The bafilomycin/concanamycin binding site in subunit c of the V-ATPases from Neurospora crassa and Saccharomyces cerevisiae. J Biol Chem. 2004;279:33131–33138. doi: 10.1074/jbc.M404638200. [DOI] [PubMed] [Google Scholar]
- 37.Bowman EJ, Siebers A, Altendorf K. Bafilomycins: A class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc Natl Acad Sci USA. 1988;85:7972–7976. doi: 10.1073/pnas.85.21.7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo H, Bueler SA, Rubinstein JL. Atomic model for the dimeric FO region of mitochondrial ATP synthase. Science. 2017;358:936–940. doi: 10.1126/science.aao4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Couoh-Cardel S, Milgrom E, Wilkens S. Affinity purification and structural features of the yeast vacuolar ATPase Vo membrane sector. J Biol Chem. 2015;290:27959–27971. doi: 10.1074/jbc.M115.662494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schep DG, Zhao J, Rubinstein JL. Models for the a subunits of the Thermus thermophilus V/A-ATPase and Saccharomyces cerevisiae V-ATPase enzymes by cryo-EM and evolutionary covariance. Proc Natl Acad Sci USA. 2016;113:3245–3250. doi: 10.1073/pnas.1521990113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacCallum JL, Bennett WFD, Tieleman DP. Distribution of amino acids in a lipid bilayer from computer simulations. Biophys J. 2008;94:3393–3404. doi: 10.1529/biophysj.107.112805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ejsing CS, et al. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci USA. 2009;106:2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoshinaka K, Kumanogoh H, Nakamura S, Maekawa S. Identification of V-ATPase as a major component in the raft fraction prepared from the synaptic plasma membrane and the synaptic vesicle of rat brain. Neurosci Lett. 2004;363:168–172. doi: 10.1016/j.neulet.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 44.Duncan AL, Robinson AJ, Walker JE. Cardiolipin binds selectively but transiently to conserved lysine residues in the rotor of metazoan ATP synthases. Proc Natl Acad Sci USA. 2016;113:8687–8692. doi: 10.1073/pnas.1608396113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watt IN, Montgomery MG, Runswick MJ, Leslie AGW, Walker JE. Bioenergetic cost of making an adenosine triphosphate molecule in animal mitochondria. Proc Natl Acad Sci USA. 2010;107:16823–16827. doi: 10.1073/pnas.1011099107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 47.Headley JV, Peru KM, Verma B, Robarts RD. Mass spectrometric determination of ergosterol in a prairie natural wetland. J Chromatogr A. 2002;958:149–156. doi: 10.1016/s0021-9673(02)00326-6. [DOI] [PubMed] [Google Scholar]
- 48.Vahidi S, et al. Reversible inhibition of the ClpP protease via an N-terminal conformational switch. Proc Natl Acad Sci USA. 2018;115:E6447–E6456. doi: 10.1073/pnas.1805125115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.