Significance
The role of cellular metabolism in conventional T cell fate and functions has been established, but little is understood about how metabolic pathways and nutrient requirements are controlled in natural killer T (NKT) cells. The present study shows that, unlike CD4 T cells, activation of NKT cells metabolize glucose to the pentose phosphate pathway and TCA cycle instead of converting into lactate. In line with this, a high lactate microenvironment is detrimental for NKT cell homeostasis and effector function. Glucose metabolism and IFN-γ expression is increased in NKT cells in response to bacterial infection. Taking these data together, our study demonstrates unique glucose metabolic regulation necessary for NKT cell survival, proliferation and cytokine expression.
Keywords: NKT, glucose, OXPHOS, PLZF
Abstract
Cellular metabolism and signaling pathways are key regulators to determine conventional T cell fate and function, but little is understood about the role of cell metabolism for natural killer T (NKT) cell survival, proliferation, and function. We found that NKT cells operate distinct metabolic programming from CD4 T cells. NKT cells are less efficient in glucose uptake than CD4 T cells with or without activation. Gene-expression data revealed that, in NKT cells, glucose is preferentially metabolized by the pentose phosphate pathway and mitochondria, as opposed to being converted into lactate. In fact, glucose is essential for the effector functions of NKT cells and a high lactate environment is detrimental for NKT cell survival and proliferation. Increased glucose uptake and IFN-γ expression in NKT cells is inversely correlated with bacterial loads in response to bacterial infection, further supporting the significance of glucose metabolism for NKT cell function. We also found that promyelocytic leukemia zinc finger seemed to play a role in regulating NKT cells’ glucose metabolism. Overall, our study reveals that NKT cells use distinct arms of glucose metabolism for their survival and function.
Invariant natural killer T (NKT) cells are lymphocytes sharing characteristics of innate and adaptive immune cells and acting as a bridge between the two immune responses. The hallmark of NKT cells is their ability to produce a copious amount of proinflammatory and immune modulatory cytokines very rapidly after antigenic stimulation (1, 2). NKT cells are CD1d-restricted T cells that respond to a variety of glycolipids (3). These cells express a highly restricted T cell receptor (TCR) repertoire and develop in the thymus from a common precursor of CD4 CD8 double-positive thymocytes (4). Compared with conventional T cells, NKT cells require distinct signaling for development in the thymus (5, 6). NKT cells develop to produce IFN-γ, IL-4, and IL-17, designated as NKT1, NKT2, and NKT17 subsets, respectively, which can be distinguished based on the expression of the transcription factors promyelocytic leukemia zinc finger (PLZF), T-bet, and RORγt (7–9). In the peripheral organs like liver, spleen, lymph node, and adipose tissue, NKT cells differentiate further and gain specific functional characteristics to become proinflammatory or immunomodulatory (10). Thus, NKT cells are a heterogeneous population that show a high degree of phenotypic and functional specialization and their role in autoimmune diseases (11, 12), infectious diseases (13–15), asthma (16), and antitumor immune responses in both experimental models and humans (10, 17–19) has been well established.
PLZF (encoded by Zbtb16) is a BTB-zinc finger transcription factor expressed by innate-like T cells, including NKT cells but not by conventional T cells or NK cells (20, 21). PLZF regulates early developmental and functional differentiation of NKT cells. Zbtb16 mutation or deletion leads to abrogation of expansion and effector-memory differentiation of NKT cells, resulting in reversal to a naive phenotype (22). Similarly, the ectopic expression of PLZF in CD4 T cells induces the phenotype and functions similar to those of NKT cells (21, 23). We have recently shown that PLZF also regulates the level of reactive oxygen species (ROS) in NKT cells, suggesting its role in cellular metabolism in NKT cells (24).
The signaling pathways that control cellular metabolism play a critical role in dictating the outcome of T cell activation and effector functions (25–30). Antigen-dependent T cell activation triggers the PI3K pathway and subsequently activates the mammalian target of rapamycin (mTOR) complex 1 (C1) and complex 2 (C2) (31). mTORC1, a serine-threonine kinase, integrates various environmental cues and regulates cell growth, cell proliferation, and metabolism (31). mTORC1 and mTORC2 deletions lead to defective NKT cell development at an early stage and impair their functions (5, 6, 32). Additionally, myelocytomatosis oncogene (c-myc), which is downstream of mTORC1, has been shown to play a role in early NKT cell development and in progression beyond the immature NKT stage (33, 34). Furthermore, unlike CD4 T cells, NKT cells are sensitive to changes in signaling pathways mediated by CD28 (35), ICOS (36), PI3K (37, 38), and Wnt/β-Catenin (39); most of them are known to regulate the metabolic pathways.
Upon activation, T cells proliferate and increase their biomass for which they not only require an increased amount of energy but also need to facilitate nutrient uptake for the generation of daughter cells (40). To fulfill these high demands of activated cells, T cell metabolism is reprogramming from preferentially utilizing OXPHOSto glycolysis (27, 41). Glycolysis produces biosynthetic materials to support proliferation and this shift regulates the acquisition of effector function in certain T cell subsets (25, 42). Despite the fact that activated T cells primarily use glycolysis, mitochondrial metabolism is also important for T cell activation (43). Decreasing the metabolic rate in T cells leads to immunological anergy that is associated with diminished cytokine production, growth, and proliferation (44, 45).
In contrast to classic T cells, little is known about the metabolic needs of NKT cells. A study has shown that during thymic development, NKT cells increase glucose uptake and expression of glucose transporter 1 (Glut1) at stages 0 and 1, the highly proliferating cells, but decreases subsequently at stages 2 and 3, the low proliferating cells (46). Recently, we have shown that NKT cells have lower mitochondrial ROS during steady state than CD4 T cells, suggesting distinct metabolic requirements in NKT cells (24). However, none of these studies have yet demonstrated a succinct overview of cellular metabolism in NKT cells.
In the present study, we report that resting NKT cells are less efficient in glucose uptake than resting CD4 T cells. Even after activation, NKT cells remain less glycolytic than CD4 T cells and rely more on oxidative phosphorylation (OXPHOS) for their survival, and the repurposed glucose carbon is used for optimal cytokine expression. Furthermore, we demonstrated that high lactate environment is detrimental for NKT cell growth and homeostasis. Finally, PLZF correlates negatively with glycolytic potential in NKT cells. Taken together, these data show tht distinct cellular metabolism governs NKT cells’ homeostasis and immune functions.
Results
NKT Cells Take Up Less Glucose than CD4 T Cells.
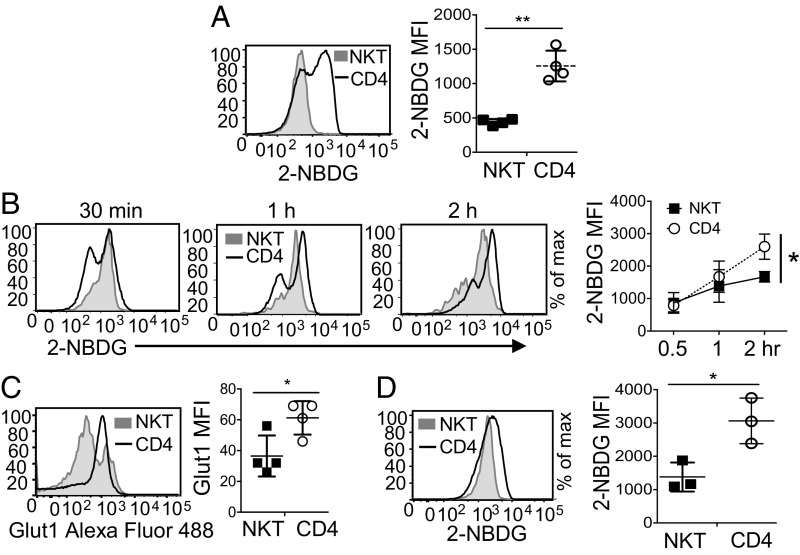
We have previously shown that peripheral NKT cells have higher levels of ROS at the steady state compared with CD4 T cells, suggesting that NKT cells are metabolically different from CD4 T cells (24). To investigate this, we first measured and compared the uptake of glucose in NKT and CD4 T cells in vivo after intravenous injection of a fluorescent analog of glucose, 2-(N-(7-nitrobenz-2-oxa-1,3-diaxol-4-yl) Amino)-2-deoxyglucose (2-NBDG), into C57BL/6 mice. At steady state, NKT cells showed less glucose uptake than CD4 T cells (Fig. 1A). Because it is possible that NKT and CD4 T cells may not have the same accessibility to 2-NBDG in vivo, we performed an in vitro assay where total splenocytes were incubated with 2-NBDG for different time periods. NKT cells again showed lower glucose uptake compared with CD4 T cells (Fig. 1B). To further elucidate whether this difference in glucose uptake at the steady state is due to a different expression level of a glucose transporter, we measured the surface expression of Glut1, a major glucose transporter in T cells (47). The expression of Glut1 was significantly lower in NKT cells compared with CD4 T cells (Fig. 1C), suggesting that low glucose uptake is likely due to low Glut1 expression. T cell activation and their subsequent phenotypic adaptation into different effector states are characterized by distinct metabolic pathways (25, 26). Therefore, we further investigated the glucose uptake in response to activation of NKT cells. In vivo activation of NKT cells by anti-CD3 antibody led to an increase in their glucose uptake but the levels were still lower compared with activated CD4 T cells (Fig. 1D). Altogether, these data indicate that NKT cells have lower potential for glucose uptake at steady-state and after activation compared with CD4 T cells.
Fig. 1.
NKT cells take up glucose less efficiently than CD4 T cells. (A) Representative histogram comparing in vivo glucose uptake in NKT and CD4 T cells from C57BL/6 mice injected with 100 µL of 2-NBDG (12.5-mg/g body weight) intravenously. The graph shows cumulative of four independent experiments. (B) The representative histograms show comparative glucose uptake between NKT and CD4 T cells. Freshly isolated splenocytes from C57BL/6 mice were incubated with 2-NBDG (20 µM) in glucose-free media for indicated time periods. The graph is cumulative of three independent experiments. (C) The histogram represents Glut1 expression between NKT and CD4 T cells from total splenocytes. The graph is cumulative of four independent experiments. (D) C57BL/6 mice were injected intraperitoneally with 100 µL of anti-CD3 antibody (2.5-mg/g body weight) 20–22 h before the intravenous injection of 2-NBDG, as mentioned in A. The representative histogram shows glucose uptake in NKT and CD4 T cells. The graph is cumulative of three independent experiments. Error bars represent mean ± SEM; *P < 0.05, **P < 0.01.
Limiting Glucose Reduces the Cytokine Expression of NKT Cells.
Our observation that NKT cells have increased glucose uptake after activation indicates that NKT cells might use glucose for cellular functions triggered as a result of activation. To determine the importance of glucose for the survival, proliferation, and function of NKT cells, we sorted NKT cells and stimulated them with α-Galactosylceramide (α-GalCer) for 3 d in media containing different concentrations of glucose. Of note, when NKT cells were stimulated with anti-CD3 and anti-CD28 antibodies that were used to stimulate CD4 T cells, we observed similar responses by the two stimulation methods (SI Appendix, Fig. S1). Therefore, we used both stimulation methods interchangeably. We found that NKT cell survival was comparable regardless of glucose concentration (Fig. 2A). Furthermore, NKT cell proliferation was not affected by low concentrations of glucose, although their cell division was slightly reduced in media without glucose (Fig. 2B, Left). In contrast to NKT cells, CD4 T cells were much more sensitive to glucose deficiency for their proliferation (Fig. 2B, Right). We also tested the responses of both cell types by treating cells with an inhibitor of glucose metabolism, 2-deoxyglucose (2-DG). NKT cell survival was not significantly different but proliferation was inhibited by 2-DG treatment (SI Appendix, Fig. S2A). It is likely that NKT cells rely on glucose for proliferation, which is inhibited by 2-DG. In the absence of glucose, however, NKT cells switch their metabolic preference and are able to proliferate.
Fig. 2.
Glucose is essential for optimal cytokine expression but not for survival and proliferation of NKT cells. (A) NKT cells were sorted from spleens of C57BL/6 mice and stimulated with α-GalCer (100 ng/mL) in the presence of indicated concentrations of glucose in the media. The graph shows the percentage of live NKT cells on day 3 of stimulation measured by PI exclusion (n = 3). (B) NKT and CD4 T cells were sorted, labeled with CellTrace Violet and then stimulated in the indicated concentrations of glucose-containing media. α-GalCer or anti-CD3 with anti-CD28 antibodies (details are described in Materials and Methods) were used to stimulate NKT or CD4 T cells, respectively. Representative histograms show cell proliferation measured after 3 d of stimulation. (C) Sorted NKT cells were stimulated as in A in the presence of 10 mM glucose or 2 mM glutamine alone, or with both in the media. The graph shows the fold-change in percent of live NKT cells after 3 d (n = 3). (D) Representative plots show cytokine expression in NKT cells stimulated for 3 d in the indicated glucose concentrations and restimulated with PMA and ionomycin in the same media conditions. Error bars represent mean ± SEM; *P < 0.05.
These observations prompted us to test if NKT cells use glutamine as a source of energy for their survival similar to CD4 T cells (48, 49). Sorted NKT cells were stimulated in the presence of either glucose or glutamine alone and compared with media containing both. Cell survival and proliferation were drastically reduced with glutamine deprivation compared with glucose deficiency, suggesting that glutamine is a critical carbon source for NKT cell survival and proliferation (Fig. 2C and SI Appendix, Fig. S2B). Although our data revealed an essential role of glutamine for NKT cell proliferation, the present study focuses on glucose metabolism because little is known about how NKT cells utilize glucose.
Glycolysis supports cytokine production in CD4 T cells (50). Therefore, we asked if glucose is necessary for cytokine production in NKT cells. We found that NKT cells displayed a dose-dependent decrease in IL-4+, IFN-γ+, and IL-17+ NKT cells under glucose-limiting conditions (Fig. 2D). To test the response of NKT cells to glucose deprivation and replenishment, we stimulated them without glucose for 3 d and then added glucose during the PMA/ionomycin restimulation step. While glucose depletion for 3 d reduced the frequency of NKT cells expressing IL-4, IFN-γ, and IL-17, replenishment with glucose for 4 h increased the levels of all cytokines (SI Appendix, Fig. S2C). Similarly, we investigated the role of glucose for the expression of cytokines in freshly isolated NKT cells by stimulating them with PMA/ionomycin for 4 h in the presence or absence of glucose. In the absence of glucose, the frequency of IFN-γ+ NKT cells was reduced (SI Appendix, Fig. S2D). In conclusion, glucose is indispensable for cytokine production by NKT cells but is less significant for either their survival or proliferation.
In Vivo Immune Response of NKT Cells to Bacterial Infection Correlates with Glucose Uptake.
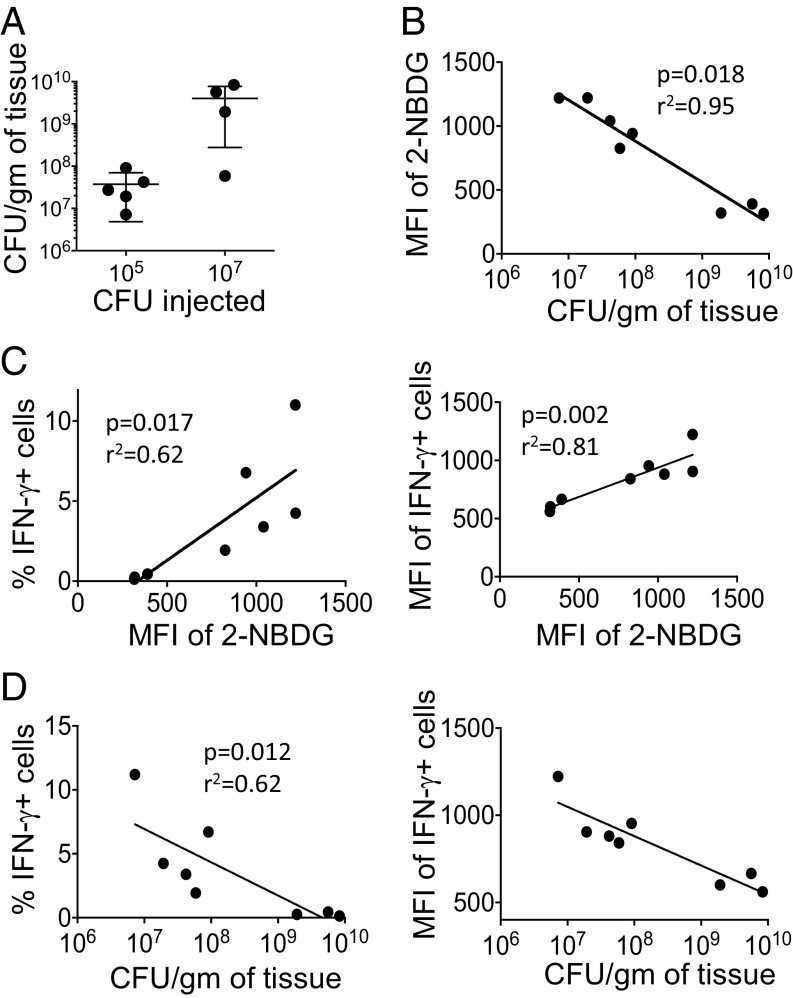
NKT cells constitute an important component of the innate immune response to Listeria monocytogenes infection (51). To understand the functional significance of glucose metabolism in NKT cells in response to bacterial infection, we infected C57BL/6 mice with two different doses (105 and 107 CFU per mouse) of L. monocytogenes. Two days after infection, we analyzed bacterial loads as well as glucose uptake and IFN-γ expression in splenic NKT cells. As expected, the mice infected with a higher dose of bacteria showed more bacterial titers in the spleen than those infected with the lower dose (Fig. 3A). In response to bacterial infection, glucose uptake by NKT cells was negatively correlated with bacterial growth (Fig. 3B). We also observed that the higher the glucose uptake, the more IFN-γ expressing NKT cells (Fig. 3C, Left). In addition, the amount of IFN-γ per cell, reflected by mean fluorescence intensity (MFI) values, was higher with increased glucose uptake (Fig. 3C, Right). Additionaly, data showed that increased expression of IFN-γ by NKT cells led to suppression of bacterial growth (Fig. 3D). Taken together, these observations suggest that, upon bacterial infection, the cytokine production of NKT cells is regulated by glucose consistent with the data shown in Fig. 2.
Fig. 3.
Glucose uptake of NKT cells correlates with their function in response to bacterial infection. A group of C57BL/6 mice (four to five mice per group) were infected with 105 or 107 CFU per mouse LM-Ova intraperitoneally. Two days after infection, spleens were used for bacterial enumeration, 2-NBDG uptake, and IFN-γ expression. (A) Bacterial loads in the spleens of mice that were infected with the two different doses of Listeria are shown. (B) Glucose uptake measured by 2-NBDG inversely correlates with the bacterial loads. The 2-NBDG uptake assay was performed using freshly isolated splenocytes from infected mice. (C and D) Total splenocytes were incubated in the presence of Golgi Plug for 2 h followed by intracellular cytokine staining for IFN-γ expression in NKT cells. The frequency of IFN-γ+ (L) as well as the MFI of IFN-γ+ (Right) NKT cells show a positive and negative correlation with glucose uptake (C) and bacterial loads (D), respectively. Two independent experiments showed similar results. Spearman correlation was used for correlation calculation.
Increased Glucose Uptake upon TCR Stimulation in NKT Cells Is Mediated by mTORC.
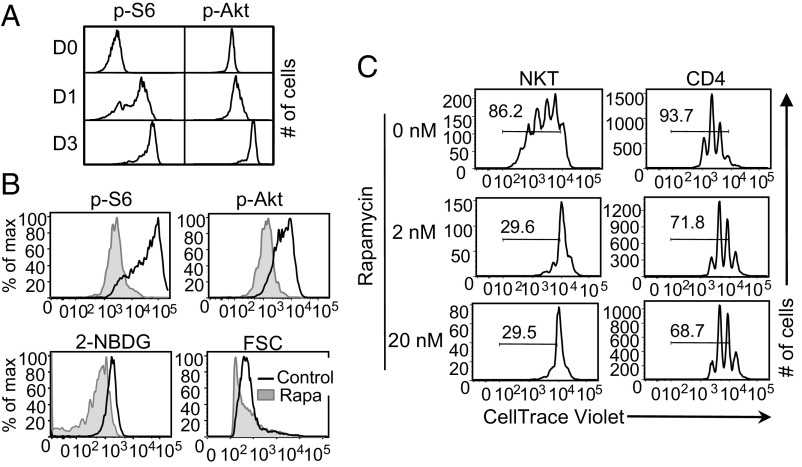
mTORC regulates various cellular aspects of metabolism and is involved in TCR activation-mediated induction of glucose uptake and glycolysis (31). Thus, we first assessed if the mTORC1 and mTORC2 activities are increased in NKT cells upon activation by measuring the expression of pS6Ser235/236 and pAktSer473, respectively. The level of phosphorylation of both molecules increased progressively at day 1 and day 3 of stimulation over day 0, demonstrating increased mTORC1 and mTORC2 signaling activity in NKT cells upon activation (Fig. 4A). Furthermore, compared with CD4 T cells, NKT cells have a higher mTORC1 expression on day 3 of stimulation (SI Appendix, Fig. S3A).
Fig. 4.
mTORC regulates glucose uptake in activated NKT cells. (A) NKT cells were sorted from spleens of C57BL/6 mice and stimulated with anti-CD3 and anti-CD28 antibodies. Representative histograms from three independent experiments show the level of pS6Ser235/236 and pAktSer473 in NKT cells at day 0 (D0), day 1 (D1), and day 3 (D3) of stimulation. (B) Sorted NKT cells were stimulated for 3 d with α-GalCer in the presence of either DMSO (Control) or rapamycin (20 nM). Representative histograms show the amounts of pS6Ser235/236, pAktSer473, 2-NBDG uptake (20 µM), and FSC of NKT cells on day 3. (C) NKT cells were stimulated as mentioned in B and CD4 T cells as described in Materials and Methods in the presence of rapamycin (2 nM and 20 nM) or DMSO for 3 d. Cell proliferation was measured using CellTrace Violet dilutions. Histograms are representative of at least three independent experiments.
Having observed that glucose uptake and mTORC activity are increased in NKT cells upon stimulation, we investigated if mTORC signaling regulates glucose uptake in these cells. For this, we treated NKT cells with rapamycin during stimulation. Rapamycin treatment decreased the level of pS6Ser235/236 and pAktSer473, confirming mTORC1 and mTORC2 inhibition, respectively (Fig. 4B, Upper). Data showed that rapamycin treatment decreased the glucose uptake in NKT cells, which was accompanied by decreased cell size as evidenced by lower FSC in the rapamycin-treated NKT cells than control cells (Fig. 4B, Lower). Therefore, mTORC activity is required for optimal glucose uptake and subsequently for proper cell growth of NKT cells. One of the main functions of mTORC activity is to regulate cell proliferation (45). We also found that mTORC inhibition affected cell proliferation of NKT cells more severely compared with CD4 T cells (Fig. 4C). Moreover, the frequencies of IL-4+, IFN-γ+, and IL-17+ NKT cells were decreased when mTORC was inhibited (SI Appendix, Fig. S3B), supporting the earlier studies showing that mTORC activity is important for effector functions of NKT cells (5). In summary, these data demonstrate that mTORC up-regulates glucose uptake, which in turn controls NKT cell growth upon activation.
NKT Cells Primarily Use OXPHOS for Their Survival, Proliferation, and Selective Cytokine Production.
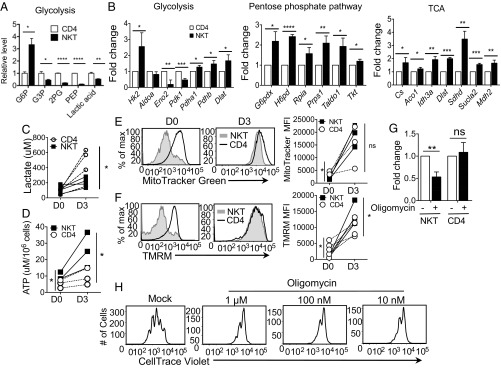
To gain a better understanding of metabolism in NKT cells, we next used liquid chromatography (LC) -coupled tandem mass spectrometry (LC-MS/MS)–based metabolomics approaches. We observed a higher level of glucose 6-phosphate (G6P) but lower levels of other metabolites from glycolysis in NKT cells compared with CD4 T cells (Fig. 5A). Levels of metabolites from the TCA cycle were comparable between NKT and CD4 T cells (SI Appendix, Fig. S4A). These data support the observation that glycolytic activity is lower in NKT than CD4 T cells. We also compared the expression of genes encoding key enzymes involved in these metabolic pathways between NKT and CD4 T cells. The first rate-limiting enzymatic gene of glycolysis, hexokinase 2 (Hk2), was up-regulated in NKT cells compared with CD4 T cells before stimulation supporting the increase of G6P (SI Appendix, Fig. S4B and Table S1). Expression of downstream enzyme genes was lower in NKT than CD4 T cells. Stimulated NKT cells showed a similar pattern by greatly inducing Hk2 expression but not other genes encoding enzymes leading to the glycolytic pathway (Fig. 5B, Left). In contrast, the genes involved in the pentose phosphate pathway (PPP) were up-regulated in NKT cells compared with CD4 T cells after activation (Fig. 5B, Center, and SI Appendix, Table S1). These results suggest that G6P is differentially funneled into the PPP in NKT cells relative to CD4 T cells.
Fig. 5.
Maintenance of optimum NKT cell survival, proliferation and function requires elevated OXPHOS. (A) Freshly sorted NKT and CD4 T cells from C57BL/6 mice were subjected to metabolomic analysis through LC-MS/MS. The graph shows relative levels of metabolites representing glycolysis between NKT and CD4 T cells. Values are from three replicas. (B) Sorted NKT and CD4 T cells were stimulated with anti-CD3 and anti-CD28 antibodies for 3 d, and RNA was prepared. Expression of genes involved in the indicated pathways of glucose metabolism was measured using RT2 PCR array. Fold-change was calculated as described in Materials and Methods and the values were compared between stimulated NKT and CD4 T cells. The graphs are cumulative of three independent experiments. (C and D) The graphs show intracellular levels of lactate (C) and ATP (D) in NKT and CD4 T cells with (D3) or without (D0) stimulation with anti-CD3 and anti-CD28 antibodies for 3 d. (E and F) Representative histograms show mitochondrial mass measured by staining with MitoTracker (E) and mitochondrial potential measured by TMRM staining (F) in NKT and CD4 T cells without (D0) and with stimulation (D3), as mentioned in B. The graphs are cumulative of three to four experiments. (G and H) Sorted NKT and CD4 T cells were stimulated with α-GalCer and anti-CD3 and anti-CD28 antibodies, respectively, in the presence of DMSO (0 mM) or oligomycin at indicated concentrations. (G) The graph shows fold-changes in percent live NKT and CD4 T cells measured by PI exclusion after 3 d of stimulation in the presence of 10 nM oligomycin (n = 3). (H) Representative histograms show NKT cell proliferation measured using CellTrace Violet dilutions on day 3 of stimulation of NKT cells. All of the results are representative of at least three independent experiments. Error bars represent mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Pyruvate dehydrogenase kinases (Pdk1 and Pdk2) phosphorylate and negatively inhibit the activity of the pyruvate dehydrogenase (Pdh) complex, which oxidizes pyruvate and channels it into the TCA cycle. Pdk1 and Pdk2 were down-regulated in NKT cells, whereas Pdh genes (Pdha1 and Pdhb) were highly increased, suggesting that NKT cells oxidize pyruvate in the mitochondria to a greater degree than do CD4 T cells (Fig. 5B, Left, and SI Appendix, Table S1). These observations were further supported by higher levels of TCA cycle enzymatic gene expression in NKT cells than in CD4 T cells (Fig. 5B, Right, and SI Appendix, Table S1). To further confirm the low glycolytic potential of NKT cells, we measured the intracellular lactate level in NKT and CD4 T cells with or without activation. We observed similar levels of intracellular lactate in freshly isolated NKT and CD4 T cells (Fig. 5C). After activation, lactate levels were increased in NKT and CD4 T cells, but the levels were lower in NKT cells than CD4 T cells (Fig. 5C). Finally, consistent with no known role for gluconeogenesis or glycogen metabolism in T cells, genes in these pathways were not altered in NKT cells upon activation (SI Appendix, Table S1). Based on this summation of data, we conclude that glucose carbon is differentially directed to the PPP and TCA cycle in NKT cells, relative to CD4 T cells (SI Appendix, Fig. S4C).
Low intracellular levels of lactate and high expression of TCA cycle genes led us to hypothesize that NKT cells rely on OXPHOS instead of glycolysis for their survival and homeostasis. Because mitochondria are best known for their role as energy powerhouses, we speculated that NKT cells generate high amounts of ATP. Indeed, NKT cells had higher amounts of intracellular ATP compared with CD4 T cells both before and after stimulation, suggesting higher OXPHOS in NKT cells (Fig. 5D). Because OXPHOS occurs in the mitochondria, we studied the mitochondrial activity in NKT cells. Activated NKT cells showed increased mitochondrial mass and mitochondrial function as measured by MitoTracker and tetramethylrhodamine methyl ester perchlorate (TMRM), respectively, relative to CD4 T cells (Fig. 5 E and F).
To further investigate the role of mitochondrial OXPHOS in NKT cell metabolism, we activated NKT cells in the presence or absence of oligomycin for 3 d. Oligomycin targets ATP synthase, leading to inhibition of electron flow through the electron transport chain and stopping respiration (50). NKT cell survival was decreased in the presence of oligomycin, indicating that, unlike CD4 T cells, they cannot readily switch to generating ATP from glycolysis (Fig. 5G). NKT cell proliferation was also reduced with oligomycin treatment (Fig. 5H). The frequencies of IL-4+ and IL-17+ NKT cells were also decreased in the presence of oligomycin (SI Appendix, Fig. S4D). Therefore, OXPHOS seems to be critical for NKT cell survival and proliferation, as well as for optimal cytokine production.
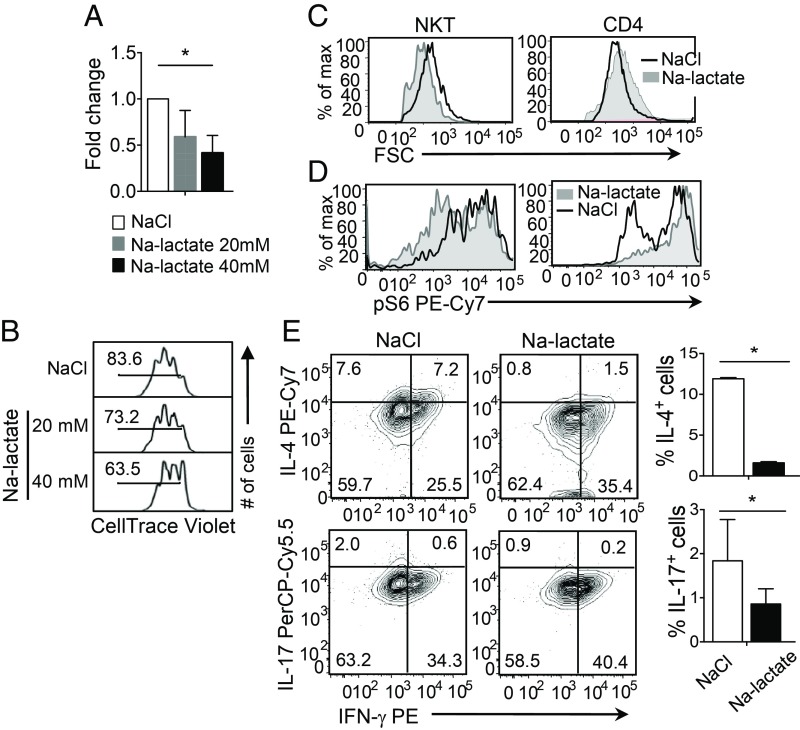
High Lactate Microenvironment Is Detrimental for NKT Cell Homeostasis and Their Effector Function.
Because conventional T cells are resistant to high lactate environment-mediated cell apoptosis (52), we asked if the less glycolytic NKT cells would be more sensitive to increased extracellular lactate. To test this, we cultured NKT cells in the presence of sodium lactate (20 mM and 40 mM) or sodium chloride, and then measured NKT cell survival, proliferation, and cell size. The data showed that high lactate exerted a negative effect on survival, proliferation, and cell size of NKT cells (Fig. 6 A–C), while high lactate increased the size of CD4 T cells (Fig. 6C, Right). In addition, high lactate reduced mTORC activity, as evidenced by the reduced level of pS6Ser235/236 in NKT but not in CD4 T cells (Fig. 6D). High-lactate environment also reduced the frequencies of IL-4+ and IL-17+ but not IFN-γ+ NKT cells, suggesting that cytokine functions of NKT cells are altered in a high-lactate environment (Fig. 6E). Taken together, the data suggest that a low level of lactate in NKT cells is important for their homeostasis and optimal cytokine expression.
Fig. 6.
A high-lactate environment is detrimental for NKT cell homeostasis and effector functions. Sorted NKT and CD4 T cells from of C57BL/6 spleens were stimulated for 3 d, as in Fig. 2, in the presence of NaCl or Na-lactate. (A) The graph shows fold-changes in surviving NKT cells under the indicated conditions at day 3 using PI exclusion assay (n = 3). (B) Representative histograms show proliferation of NaCl and Na-lactate (20 mM and 40 mM) treated NKT cells using CellTrace Violet dilutions (n = 3). (C and D) Representative histograms comparing the cell size measured using FSC (C) and levels of pS6Ser235/236 (D) in NKT and CD4 T cells treated with NaCl and Na-lactate (40 mM) (n = 3). (E) Representative plots show the cytokine expressions in NKT cells stimulated for 3 d in the presence of either NaCl or Na-lactate (40 mM) and restimulated with PMA and ionomycin on day 3. Summary graphs on the Right show percentages of IL-4+ and IL-17+ NKT cells. All of the data are representative of at least three independent experiments. Error bars represent mean ± SEM; *P < 0.05.
PLZF Correlates with Low Glycolysis in NKT Cells.
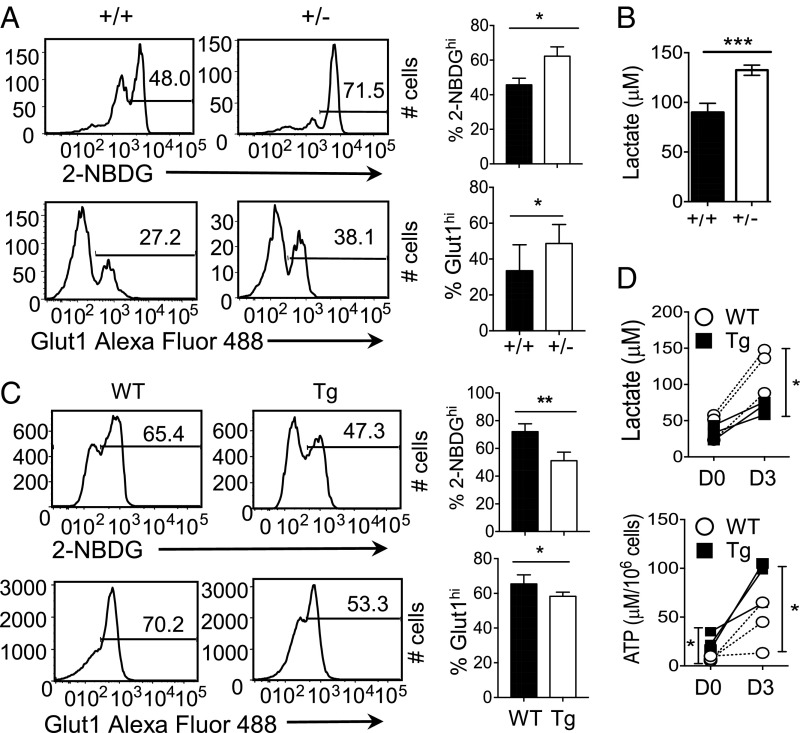
Previously, we have shown that PLZF controls the level of ROS in NKT cells (24), indicating that it might play a role in the metabolism of NKT cells. To test this, we used two approaches. First, we examined NKT cells from PLZF haplodeficient (PLZF+/−) mice. Because PLZF−/− mice are devoid of NKT cells, these mice are not suitable for the study (20). We observed that the percentages of 2-NBDGhi and Glut1-expressing NKT cells were significantly increased in PLZF+/− mice compared with the WT mice (Fig. 7A). In addition, the intracellular lactate levels, mitochondrial ROS, mitochondrial mass, and mitochondrial potential were increased in PLZF+/− NKT cells compared with WT NKT cells (Fig. 7B and SI Appendix, S5A). Next, we asked if PLZF expression is sufficient to reduce glucose uptake and subsequent processes. CD4 T cells from mice expressing the PLZF transgene (PLZFTg) showed reduction of both the frequency of 2-NBDGhi cells and the Glut1-expressing cells (Fig. 7C). Activated PLZFTg and WT CD4 T cells, however, had similar levels of glucose uptake and Glut1 expression (SI Appendix, Fig. S5B). To test if PLZF levels affect glycolysis, we measured intracellular lactate levels and found significantly lower lactate levels in PLZFTg than WT CD4 T cells (Fig. 7D, Upper). We also found that ATP levels were significantly higher in PLZFTg than WT CD4 T cells (Fig. 7D, Lower), indicating that CD4 T cells expressing PLZF have increased mitochondrial activity.
Fig. 7.
PLZF levels negatively correlate with glycolysis rate in NKT and PLZFTg CD4 T cells. (A) Representative histograms show 2-NBDG uptake (Upper) and Glut1 expression (Lower) in splenic NKT cells from PLZF haplodeficient (+/−) and WT littermate (+/+) mice. The graphs (Right) are cumulative of four independent experiments. (B) The graph shows intracellular lactate levels in freshly sorted NKT cells from PLZF haplodeficient (+/−) and WT littermate mice (n = 3). (C) Representative histograms compare 2-NBDG uptake (Upper) and Glut1 expression (Lower) in splenic CD4 T cells from PLZFTg (Tg) and WT littermate mice. The bar graphs are cumulative of four independent experiments. (D) The graphs show intracellular lactate (Upper) and ATP levels (Lower) in enriched CD4 T cells from PLZFTg and WT littermate mice before (D0) and after 3 d of stimulation (D3) with anti-CD3 and anti-CD28 antibodies. The graphs are cumulative of three independent experiments. Error bars represent mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001.
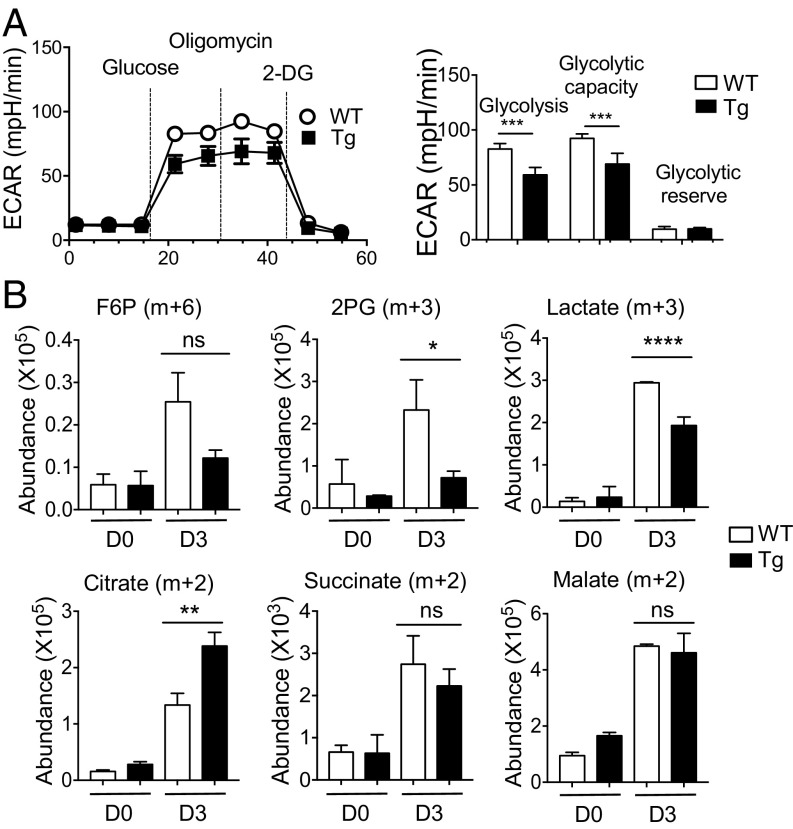
To further study PLZF-mediated metabolic changes, we used the Seahorse bioanalyzer to analyze the rate of glycolysis by measuring extracellular acidification rate (ECAR). In this experiment, we used WT and PLZFTg CD4 T cells because it is challenging to get a sufficient number of NKT cells for this assay. Moreover, PLZFTg CD4 T cells share similar metabolic characteristics to NKT cells (Fig. 7). In line with low intracellular lactate level, ECAR was lower in CD4 T cells from PLZFTg compared with WT mice (Fig. 8A). Thus, expression of PLZF reduced the rate of glycolysis. Glucose is a major carbon source for the TCA cycle. To test its utilization in a PLZF-dependent manner, we traced glucose carbon incorporation into TCA cycle intermediates using nonradioactive carbon-13 isotope labeled glucose ([U-13C]-glucose) and LC/MS-based metabolomics in PLZFTg and WT CD4 T cells for 4 h. PLZFTg CD4 T cells exhibited a lower abundance of glucose-derived 13C3-phosphoglycerate (2-PG) (m+3) and 13C3-lactate (m+3) (Fig. 8B), further illustrating that PLZF limits glycolytic flux. Furthermore, we found that PLZFTg CD4 T cells had a higher abundance of glucose-derived 13C2-citrate (m+2) compared with WT CD4 T cells, indicating a greater diversion of glucose-derived pyruvate into the TCA cycle (Fig. 8B). However, the abundance of glucose-derived 13C2-succinate (m+2) and 13C2-malate (m+2) were similar in WT and PLZFTg CD4 T cells (Fig. 8B). These results suggest that, in the PLZFTg CD4 T cells, glucose-derived carbon preferentially enters the TCA cycle to make citrate, and that other carbon sources also contribute to TCA cycle anaplerosis. Based on the results in Fig. 2, this is likely accounted for by glutamine-derived carbon.
Fig. 8.
PLZF expression correlates with levels of OXPHOS. (A) CD4 T cells from PLZFTg and WT littermate mice were enriched and stimulated as described in Materials and Methods for 3 d. The representative graph (Left) shows the comparison of ECAR using the Seahorse assay. (Right) Graph showing basal glycolysis, glycolytic capacity, and glycolytic reserve capacity in PLZFTg and WT CD4 T cells from three independent experiments. (B) WT and PLZFTg CD4 T cells were stimulated for 3 d, washed, and cultured in glucose-free RPMI containing glutamine (2 mM) and [13C6]-glucose (15 mM) for 4 h before subjecting to LC-MS analysis. Representative graphs show the abundance of 13C6-glucose-derived carbon in glycolysis (F6P, 2PG, and lactate) and TCA cycle (citrate, succinate, and malate) intermediate metabolites from cells before (D0) and after stimulation for 3 d (D3). Two independent experiments showed similar results. Error bars represent mean ± SEM; ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Discussion
The metabolic demands of naïve T cells are lower because they require energy only for survival, cell migration, and preventing atrophy. This demand increases in activated T cells, which requires energy for rapid growth, proliferation, and production of effector molecules (53). Thus, it is essential for T cells to switch their cellular metabolism to meet the biosynthetic needs of the cells. NKT cells are different from conventional T cells in that they are able to rapidly elicit effector functions in response to activation. As such, understanding the metabolic programming of NKT cells was the primary goal of this study. Here, we report that NKT cells in the resting state use less glucose than conventional CD4 T cells, suggesting that these cells are metabolically quiescent, like memory T cells (54). Due to the marked disparity in the energy requirement of resting versus activated T cells, regulation of glucose uptake is considerably different. In the resting state, glucose uptake is not regulated through TCR-mediated activation signals. On the other hand, activation of TCR-mediated signaling increases surface expression of key nutrient receptors and cellular metabolism (55). The present study showed that activated NKT cells up-regulate the surface expression of the glucose transporter Glut1 and increase their glucose uptake. However, activated NKT cells are still less efficient than activated CD4 T cells in their glucose uptake.
T cells switch metabolism from OXPHOS to glycolysis for their proliferation and effector functions as soon as they are activated (56). A previous study indicated that glycolysis affects the translation of cytokines at the posttranscriptional level (50). In agreement, our data show that glucose is required for proper expression of cytokines by NKT cells. This observation is further corroborated by the fact that the effect of 3 d of starvation on cytokine synthesis in NKT cells was reversed by replenishment of glucose for 4 h during PMA/ionomycin restimulation. These findings were also supported by our in vivo infection data, which showed that glucose uptake was directly correlated with cytokine production in NKT cells in response to the bacterial infection. Furthermore, NKT cells with high glucose uptake suppressed the bacterial growth more efficiently. However, glucose does not seem to be necessary for NKT cell proliferation. Instead, we found that glutamine is an essential carbon source for NKT cells’ survival and proliferation, and further investigation of glutamine metabolism in NKT cells is warranted.
Recently, metabolic regulators, such as Folliculin-interacting protein 1 (Fnip1), mTORC1, mTORC2, and c-myc have been shown to regulate NKT cell development, suggesting a possible role of signaling molecules in regulating metabolism in NKT cells (5, 6, 33, 57). mTORC is activated in response to signals from extracellular growth factors, nutrient availability, and the energy status of T cells. mTORC1 regulates various aspects of cellular metabolism, including glycolysis, protein synthesis, and lipid synthesis. mTORC1 regulates glycolysis through hypoxia-inducible factor-1α (HIF-1α) and c-myc (58, 59). These transcription factors control the expression of glucose transporters and multiple enzymes controlling glycolysis. Both the mTORC1 target pS6Ser235/236 and the mTORC2 target pAktSer473 were induced in activated NKT cells, and blocking these signals by rapamycin treatment decreased glucose uptake. The fact that the effects of mTORC inhibition are more severe than glucose depletion during stimulation suggests that NKT cells rely more on other metabolic pathways controlled by mTORC than glycolysis. NKT cell proliferation was more sensitive to rapamycin treatment than CD4 T cells, which could be explained by higher mTORC activity in NKT than CD4 T cells after 3 d of stimulation.
The results from metabolomic and gene-expression analyses for glucose metabolism demonstrated differences between NKT and CD4 T cells. NKT cells have lower levels of glycolytic metabolites in the steady state. Unlike CD4 T cells, activation of NKT cells substantially increased the expression of genes encoding enzymes involved in the PPP and the TCA cycle. Pdk increases the oxidation of pyruvate, the end product of glycolysis into lactate by inhibiting the activity of Pdha, the enzyme responsible for oxidizing pyruvate into acetyl-CoA in the mitochondria (60). NKT cells expressed reduced Pdk1 but increased Pdha1 genes, suggesting that pyruvate may be primarily converting into acetyl-CoA and then enters the mitochondria rather than converting into lactate. This was further corroborated by the finding of lower levels of intracellular lactate in NKT cells than CD4 T cells upon activation. High levels of lactate in the extracellular environment decreased mTORC signaling activity, which was detrimental for NKT cell survival but favorable for CD4 T cells. In addition, NKT cells reduced IL-4 and IL-17 production but not IFN-γ production, suggesting that IFN-γ expression is more resistant to cellular stress caused by high lactate. In contrast to this, CD4 T cells produce more IL-17 and less IFN-γ in high lactate environment (61).
In this study, we observed that the level of PLZF correlates with the glycolysis rate in NKT cells. PLZF suppresses the initiation of c-myc transcription in non-T cells (62). C-myc controls not only key cell cycle regulators, but also is responsible for the metabolic switch from OXPHOS to glycolysis in T cells upon activation (59). It is possible that the decreased glucose uptake that we observed in NKT cells is a consequence of low c-myc expression. PLZF is known to suppress mTORC1 activity by inducing expression of the mTORC1 inhibitor Redd1 in stem cells (63). In contrast to an earlier study showing that PLZF overexpression in brown adipose tissue increases mitochondrial respiration (64), we observed reduced mitochondrial mass and potential in PLZFTg CD4 T cells. The difference is likely due to the cell type-specific function of PLZF. Additionally, we provided supporting evidence that glucose is oxidized in the mitochondria as glucose-derived citrate levels are higher in PLZFTg than WT CD4 T cells. However, this citrate might be metabolizing in the fatty acid synthesis pathway instead of circulating in the TCA cycle, as revealed by similar levels of abundance of 13C2-succinate and malate. It should be noted that due to the limited number of NKT cells, metabolic assays requiring a high number of cells were performed with PLZFTg CD4 T cells as a model to study the role of PLZF. Nevertheless, the data from PLZF+/− NKT cells align with PLZFTg CD4 T cells, supporting the idea that PLZF might regulate low glycolysis in NKT cells. To investigate how PLZF regulates these metabolic differences, we reanalyzed the ChIP-sequencing data of NKT and PLZFTg thymic cells (GSE81772) archived in databases (22). The analysis showed that PLZF does not seem to bind to genes involved in glucose metabolism that expressed higher or lower levels in NKT cells than CD4 T cells before stimulation (SI Appendix, Fig. S6). Therefore, the data suggest an indirect role of PLZF in glucose metabolism. However, we cannot rule out the possibility that PLZF’s binding could be changed in stimulated NKT cells. Unfortunately, this type of experiment is highly challenging to do with stimulated NKT cells due to the limited cell number. Therefore, further mechanistic studies are warranted to understand how PLZF controls the metabolic activity in NKT cells.
In summary, the present study shows that NKT cells rely primarily on OXPHOS for their survival and proliferation. When NKT cells encounter antigen, they up-regulate mTORC signaling and glycolysis, both of which are important for proper cytokine expression.
Materials and Methods
Mice.
Male and female mice at 8–12 wk of age on the C57BL/6 background were used in this study. Mice were purchased from Jackson Laboratories and housed in specific pathogen-free conditions. PLZF transgenic (referred to as PLZFTg) mice driven by the lck promoter (23) and PLZF-haplodeficient mice (referred to as PLZF+/−) (65) have been kindly provided by Derek Sant’Angelo, Rutgers University, New Brunswick, NJ. In all of the experiments with these PLZFTg and PLZF+/− mice, WT littermate mice were used as a control. All animal experiments were performed in accordance with the Institutional Animal Care and Use Committee of the University of Michigan.
Cell Isolation and Purification.
The single-cell suspensions from spleens were prepared as per standard protocol (24). To sort NKT and CD4 T cells, splenocytes were incubated in anti-CD19 beads (Miltenyi Biotec) in MACS buffer for 30 min to exclude B cells, followed by passaging through MiniMACS columns (Miltenyi Biotec) and collection of the flow through. NKT and CD4 T cells were sorted using FACS Aria II/FACS Synergyhead1 (BD Biosciences). APC- or Pacific Blue-conjugated murine CD1d tetramers loaded with PBS-57 were kindly provided by the National Institute of Health Tetramer Facility, Atlanta, GA. For CD4 T cells enrichment, a positive selection enrichment kit was used according to the manufacturer’s instructions (Miltenyi Biotec).
Cell Culture.
To compare the responses of NKT and CD4 T cells after activation, sorted cells were activated with plate bound anti-CD3 (2.5 µg/mL) and soluble anti-CD28 (1 µg/mL) antibodies (eBioscience) for an indicated time in the presence of IL-2 (10 U/mL) in RPMI medium 1640 supplemented with 10% FBS, 2 mM glutamine, and penicillin/streptomycin at 37 °C. To study activated NKT cells without involving CD4 T cells, we used α-GalCer (100 ng/mL). For the treatment with 2-DG, Na-lactate, rapamycin, and oligomycin, NKT cells were stimulated with soluble α-GalCer (100 ng/mL). For glucose and glutamine limitation assay, sorted NKT and CD4 T cells were stimulated in glucose and glutamine-free RPMI 1640 media supplemented with 10% dialyzed FBS (Sigma Aldrich).
Flow Cytometry Assays.
The following fluorescently conjugated antibodies were used for surface and intracellular staining in the presence of anti-FcγR mAb (2.4G2): anti-mouse TCR-β (H57-597) Pacific Blue/APC, PBS-57-loaded CD1d tetramer APC/PE/Pacific Blue, anti-mouse CD4 (GK1.5) APC-Cy7, anti-mouse IFN-γ (XMG1.2) PE/FITC, anti-mouse IL-4 (BV D6-2462) PE-Cy7, and anti-mouse IL-17 (TC11-18H10) PerCP-Cy5.5 (all from eBioscience).
For intracellular cytokine expressions, unstimulated or stimulated cells were restimulated with 50 ng/mL of PMA (Sigma Aldrich) and 1.5 µM of ionomycin (Sigma Aldrich) in the presence of Golgi Plug (BD Biosciences) for 4 h, followed by intracellular cytokine staining according to manufacturer’s instruction (BD Biosciences). mTORC activity was measured using phospho-S6 Ribosomal protein (pS6Ser235/236) (Cell Signaling) and pAktSer473 (Cell Signaling) staining in permeabilization buffer. Cell death was measured by the exclusion of propidium iodide (PI; 1 μg/mL) or live/dead Fixable Aqua Dead Cell Stain (Invitrogen). Cells were acquired on FACS Canto II (BD Bioscience) and data were analyzed using FlowJo (TreeStar software v9.9).
Mitochondrial Parameters Detection.
To measure mitochondrial parameters, total splenocytes (3 × 106 cells) or stimulated NKT and CD4 T cells (1 × 105) cells were incubated with potentiometric dye TMRM (60 nM; Invitrogen), MitoTracker green (30 nM; Invitrogen), and MitoSox (2.5 µM; Invitrogen) for 30 min at 37 °C in RPMI media for flow cytometry.
Glucose Uptake Assay.
For in vitro glucose uptake assay, total splenocytes (3 × 106 cells) were incubated in 2-NBDG (20 µM; Invitrogen) for 1 h or as indicated at 37 °C in glucose-free RPMI 1640 media containing 10% dialyzed FBS. For in vivo glucose uptake, mice were injected with 2-NBDG (100 µg per mouse) intravenously. After 15 min of 2-NBDG injection, mice were killed, and spleens were removed. Total splenocytes were stained for the surface markers, as mentioned.
Glut1 Expression Detection.
To study Glut1 expression, total splenocytes (3 × 106 cells) were stained for surface markers and then fixed in 4% paraformaldehyde. The staining for the expression of Glut1 for flow cytometry was performed at room temperature for 30 min in the dark using anti-Glucose Transporter (Glut1) antibody (EPR3915; Abcam).
ATP and Lactate Assays.
ATP levels were measured using CellTiter-Glo Luminescent Cell Viability reagent (Promega). For intracellular lactate level, plate-based fluorometric measurement kit (excitation 530–540 nm, emission 585–595 nm) was used according to the manufacturer’s instructions (Cayman Chemicals).
Metabolic Seahorse Assay.
CD4 T cells from WT and PLZFTg mice were enriched from freshly prepared splenocytes and stimulated as indicated. In the XF96 well microplate coated with polylysine, 5 × 105 cells per well were plated in glucose-free Seahorse media (Sigma Aldrich) and the plate was briefly spun to affix the cells to the bottom of the wells. The plate was then incubated for 30 min in a non-CO2 incubator to equilibrate. ECAR was measured in stimulated CD4 T cells using glucose (10 mM; Sigma Aldrich), oligomycin (2 mM, ATP coupler; Sigma Aldrich), and 2-DG (100 mM; Sigma Aldrich) in Seahorse assay medium using Seahorse XFe96 bioanalyzer (Agilent Technologies).
MS Analysis of [U13C6]-Glucose.
For heavy isotope flux analysis, 2 × 106 unstimulated and stimulated enriched CD4 T cells from WT and PLZFTg mice were incubated with [U-13C6]-glucose (15 mM; Sigma Aldrich) and glutamine (2 mM) in glucose-free media containing 10% dialyzed serum for 4–5 h at 37 °C. Cells were washed with PBS and frozen in liquid nitrogen. Cells were further subjected to LC-MS/MS. Mass isotopomer distribution was determined using a custom algorithm.
Targeted Metabolites Measurements.
Lysate was prepared from sorted NKT and CD4 T cells (5 × 105 cells per replicate) by incubating the cells with 80% methanol and following a series of vigorous mixing steps. Cells were spun down at maximum speed to remove membranous debris, and the lysate was collected for drying using a SpeedVac. Dried metabolites were collected and analyzed using Agilent 6470 Triple Quadrupole LC-MS/MS metabolomics with dynamic multiple reaction monitoring detection method.
Cell Proliferation.
Sorted NKT and CD4 T cells were labeled with CellTrace Violet (5 µM; Invitrogen) in 1× PBS containing 0.1% BSA for 30 min at 37 °C. Cells were stimulated as indicated and analyzed by flow cytometry on day 3 of stimulation for CTV dilutions.
RT2 Gene PCR Array.
Total RNA was isolated from unstimulated and stimulated NKT and CD4 T cells using RNeasy Plus mini kit (Qiagen) according to the manufacturer’s instructions. RNA samples were transported to the University of Michigan DNA Sequencing Core to perform the Glucose Metabolism PCR Array, and gene array was performed using Mouse Glucose Metabolism RT2 Profiles PCR Array according to the manufacturer’s instructions (Qiagen) using Applied Biosystem’s 7900HT Sequence Detection System. Fold-changes were calculated from ΔCt values (gene of interest Ct value – an average of all housekeeping gene Ct values) using the ΔΔCt method. Gene expression of target genes was normalized to the average of the housekeeping genes β-actin, β-2 microglobulin, and heat-shock protein 90.
L. monocytogenes Infection.
L. monocytogenes expressing Ovalbumin (LM-Ova) strain 10403s (originally obtained from M. L. Gray, Montana State University, Bozeman, MT) was a kind gift from Mary O’Riordan, University of Michigan, Ann Arbor, MI. L. monocytogenes was grown in BHI broth media and the bacteria were collected in a midlog phase for infection. C57BL/6 mice were injected intraperitoneally with 105 or 107 CFU per mouse of LM-Ova in 200 μL of PBS or PBS alone. On day 2 of infection, bacterial burden was enumerated from homogenized spleen using colony forming unit determination by culturing serially diluted samples on LB agar plates. Intracellular cytokine expression and glucose uptake by NKT cells were measured as described above.
Statistical Analysis.
All of the graphs were prepared and data analyzed using Prism software (Prism v7; Graphpad Software). For comparison among multiple groups, data were analyzed using one-way ANOVA with the multicomparison post hoc test. For correlation determination, Spearman correlation was calculated. For comparison between two groups, unpaired and paired Student t tests were used. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. Derek B. Sant’Angelo (Rutgers University) for providing PLZFTg and PLZF+/− mice; the laboratory members for their critical reading of the manuscript and insightful comments; Drs. Charles Burant and Maureen Kachman, from the Metabolomics Core for carrying out LC-MS/MS analysis and for their critical interpretation of the data and helpful discussions; Dr. Mary O’Riordan for providing Listeria monocytogenes (10403s LM-Ova); and the National Institutes of Health Tetramer Facility for providing CD1d-tetramers. This work was supported in part by National Institutes of Health Grant AI121156 (to C.-H.C.). Carbon-tracing experiments were supported by a pilot and feasibility grant from the Michigan Regional Comprehensive Metabolomics Resource Core (U24-DK097153).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901376116/-/DCSupplemental.
References
- 1.Arase H, Arase N, Nakagawa K, Good RA, Onoé K. NK1.1+ CD4+ CD8- thymocytes with specific lymphokine secretion. Eur J Immunol. 1993;23:307–310. doi: 10.1002/eji.1830230151. [DOI] [PubMed] [Google Scholar]
- 2.Bendelac A, Rivera MN, Park SH, Roark JH. Mouse CD1-specific NK1 T cells: Development, specificity, and function. Annu Rev Immunol. 1997;15:535–562. doi: 10.1146/annurev.immunol.15.1.535. [DOI] [PubMed] [Google Scholar]
- 3.Zeng Z, et al. Crystal structure of mouse CD1: An MHC-like fold with a large hydrophobic binding groove. Science. 1997;277:339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 4.Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol. 2001;2:971–978. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- 5.Zhang L, et al. Mammalian target of rapamycin complex 1 orchestrates invariant NKT cell differentiation and effector function. J Immunol. 2014;193:1759–1765. doi: 10.4049/jimmunol.1400769. [DOI] [PubMed] [Google Scholar]
- 6.Prevot N, et al. Mammalian target of rapamycin complex 2 regulates invariant NKT cell development and function independent of promyelocytic leukemia zinc-finger. J Immunol. 2015;194:223–230. doi: 10.4049/jimmunol.1401985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watarai H, et al. Development and function of invariant natural killer T cells producing T(h)2- and T(h)17-cytokines. PLoS Biol. 2012;10:e1001255. doi: 10.1371/journal.pbio.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013;14:1146–1154. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buechel HM, Stradner MH, D’Cruz LM. Stages versus subsets: Invariant natural killer T cell lineage differentiation. Cytokine. 2015;72:204–209. doi: 10.1016/j.cyto.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Viale R, Ware R, Maricic I, Chaturvedi V, Kumar V. NKT cell subsets can exert opposing effects in autoimmunity, tumor surveillance and inflammation. Curr Immunol Rev. 2012;8:287–296. doi: 10.2174/157339512804806224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Illés Z, et al. Differential expression of NK T cell V alpha 24J alpha Q invariant TCR chain in the lesions of multiple sclerosis and chronic inflammatory demyelinating polyneuropathy. J Immunol. 2000;164:4375–4381. doi: 10.4049/jimmunol.164.8.4375. [DOI] [PubMed] [Google Scholar]
- 12.Beaudoin L, Laloux V, Novak J, Lucas B, Lehuen A. NKT cells inhibit the onset of diabetes by impairing the development of pathogenic T cells specific for pancreatic beta cells. Immunity. 2002;17:725–736. doi: 10.1016/s1074-7613(02)00473-9. [DOI] [PubMed] [Google Scholar]
- 13.Baron JL, et al. Activation of a nonclassical NKT cell subset in a transgenic mouse model of hepatitis B virus infection. Immunity. 2002;16:583–594. doi: 10.1016/s1074-7613(02)00305-9. [DOI] [PubMed] [Google Scholar]
- 14.Durante-Mangoni E, et al. Hepatic CD1d expression in hepatitis C virus infection and recognition by resident proinflammatory CD1d-reactive T cells. J Immunol. 2004;173:2159–2166. doi: 10.4049/jimmunol.173.3.2159. [DOI] [PubMed] [Google Scholar]
- 15.Crosby CM, Kronenberg M. Invariant natural killer T cells: Front line fighters in the war against pathogenic microbes. Immunogenetics. 2016;68:639–648. doi: 10.1007/s00251-016-0933-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lisbonne M, et al. Cutting edge: Invariant V alpha 14 NKT cells are required for allergen-induced airway inflammation and hyperreactivity in an experimental asthma model. J Immunol. 2003;171:1637–1641. doi: 10.4049/jimmunol.171.4.1637. [DOI] [PubMed] [Google Scholar]
- 17.Cui J, et al. Requirement for Valpha14 NKT cells in IL-12-mediated rejection of tumors. Science. 1997;278:1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- 18.Dhodapkar MV, et al. A reversible defect in natural killer T cell function characterizes the progression of premalignant to malignant multiple myeloma. J Exp Med. 2003;197:1667–1676. doi: 10.1084/jem.20021650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metelitsa LS, et al. Natural killer T cells infiltrate neuroblastomas expressing the chemokine CCL2. J Exp Med. 2004;199:1213–1221. doi: 10.1084/jem.20031462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savage AK, Constantinides MG, Bendelac A. Promyelocytic leukemia zinc finger turns on the effector T cell program without requirement for agonist TCR signaling. J Immunol. 2011;186:5801–5806. doi: 10.4049/jimmunol.1100119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao AP, et al. Multiple layers of transcriptional regulation by PLZF in NKT-cell development. Proc Natl Acad Sci USA. 2016;113:7602–7607. doi: 10.1073/pnas.1601504113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovalovsky D, et al. PLZF induces the spontaneous acquisition of memory/effector functions in T cells independently of NKT cell-related signals. J Immunol. 2010;184:6746–6755. doi: 10.4049/jimmunol.1000776. [DOI] [PubMed] [Google Scholar]
- 24.Kim YH, Kumar A, Chang CH, Pyaram K. Reactive oxygen species regulate the inflammatory function of NKT cells through promyelocytic leukemia zinc finger. J Immunol. 2017;199:3478–3487. doi: 10.4049/jimmunol.1700567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michalek RD, et al. Cutting edge: Distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerriets VA, et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J Clin Invest. 2015;125:194–207. doi: 10.1172/JCI76012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. 2013;31:259–283. doi: 10.1146/annurev-immunol-032712-095956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pearce EL, Pearce EJ. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollizzi KN, Powell JD. Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nat Rev Immunol. 2014;14:435–446. doi: 10.1038/nri3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei J, Yang K, Chi H. Cutting edge: Discrete functions of mTOR signaling in invariant NKT cell development and NKT17 fate decision. J Immunol. 2014;193:4297–4301. doi: 10.4049/jimmunol.1402042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dose M, et al. Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc. Proc Natl Acad Sci USA. 2009;106:8641–8646. doi: 10.1073/pnas.0812255106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mycko MP, et al. Selective requirement for c-Myc at an early stage of V(alpha)14i NKT cell development. J Immunol. 2009;182:4641–4648. doi: 10.4049/jimmunol.0803394. [DOI] [PubMed] [Google Scholar]
- 35.Williams JA, et al. Regulation of thymic NKT cell development by the B7-CD28 costimulatory pathway. J Immunol. 2008;181:907–917. doi: 10.4049/jimmunol.181.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akbari O, et al. ICOS/ICOSL interaction is required for CD4+ invariant NKT cell function and homeostatic survival. J Immunol. 2008;180:5448–5456. doi: 10.4049/jimmunol.180.8.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eberl G, Lowin-Kropf B, MacDonald HR. Cutting edge: NKT cell development is selectively impaired in Fyn- deficient mice. J Immunol. 1999;163:4091–4094. [PubMed] [Google Scholar]
- 38.Kishimoto H, et al. The Pten/PI3K pathway governs the homeostasis of Valpha14iNKT cells. Blood. 2007;109:3316–3324. doi: 10.1182/blood-2006-07-038059. [DOI] [PubMed] [Google Scholar]
- 39.Pyaram K, Sen JM, Chang CH. Temporal regulation of Wnt/β-catenin signaling is important for invariant NKT cell development and terminal maturation. Mol Immunol. 2017;85:47–56. doi: 10.1016/j.molimm.2017.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frauwirth KA, et al. The CD28 signaling pathway regulates glucose metabolism. Immunity. 2002;16:769–777. doi: 10.1016/s1074-7613(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 42.Cham CM, Driessens G, O’Keefe JP, Gajewski TF. Glucose deprivation inhibits multiple key gene expression events and effector functions in CD8+ T cells. Eur J Immunol. 2008;38:2438–2450. doi: 10.1002/eji.200838289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sena LA, et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Powell JD, Lerner CG, Schwartz RH. Inhibition of cell cycle progression by rapamycin induces T cell clonal anergy even in the presence of costimulation. J Immunol. 1999;162:2775–2784. [PubMed] [Google Scholar]
- 45.Powell JD, Delgoffe GM. The mammalian target of rapamycin: Linking T cell differentiation, function, and metabolism. Immunity. 2010;33:301–311. doi: 10.1016/j.immuni.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salio M, et al. Essential role for autophagy during invariant NKT cell development. Proc Natl Acad Sci USA. 2014;111:E5678–E5687. doi: 10.1073/pnas.1413935112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macintyre AN, et al. The glucose transporter Glut1 is selectively essential for CD4 T cell activation and effector function. Cell Metab. 2014;20:61–72. doi: 10.1016/j.cmet.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carr EL, et al. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J Immunol. 2010;185:1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson MO, et al. Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell. 2018;175:1780–1795.e19. doi: 10.1016/j.cell.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang CH, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ranson T, et al. Invariant V alpha 14+ NKT cells participate in the early response to enteric Listeria monocytogenes infection. J Immunol. 2005;175:1137–1144. doi: 10.4049/jimmunol.175.2.1137. [DOI] [PubMed] [Google Scholar]
- 52.Angelin A, et al. Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab. 2017;25:1282–1293 e7. doi: 10.1016/j.cmet.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finlay DK. Regulation of glucose metabolism in T cells: New insight into the role of phosphoinositide 3-kinases. Front Immunol. 2012;3:247. doi: 10.3389/fimmu.2012.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sukumar M, et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J Clin Invest. 2013;123:4479–4488. doi: 10.1172/JCI69589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jacobs SR, et al. Glucose uptake is limiting in T cell activation and requires CD28-mediated Akt-dependent and independent pathways. J Immunol. 2008;180:4476–4486. doi: 10.4049/jimmunol.180.7.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Menk AV, et al. Early TCR signaling induces rapid aerobic glycolysis enabling distinct acute T cell effector functions. Cell Rep. 2018;22:1509–1521. doi: 10.1016/j.celrep.2018.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park H, Tsang M, Iritani BM, Bevan MJ. Metabolic regulator Fnip1 is crucial for iNKT lymphocyte development. Proc Natl Acad Sci USA. 2014;111:7066–7071. doi: 10.1073/pnas.1406473111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Düvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39:171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patel MS, Nemeria NS, Furey W, Jordan F. The pyruvate dehydrogenase complexes: Structure-based function and regulation. J Biol Chem. 2014;289:16615–16623. doi: 10.1074/jbc.R114.563148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haas R, et al. Lactate regulates metabolic and pro-inflammatory circuits in control of T cell migration and effector functions. PLoS Biol. 2015;13:e1002202. doi: 10.1371/journal.pbio.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McConnell MJ, et al. Growth suppression by acute promyelocytic leukemia-associated protein PLZF is mediated by repression of c-myc expression. Mol Cell Biol. 2003;23:9375–9388. doi: 10.1128/MCB.23.24.9375-9388.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hobbs RM, Seandel M, Falciatori I, Rafii S, Pandolfi PP. Plzf regulates germline progenitor self-renewal by opposing mTORC1. Cell. 2010;142:468–479. doi: 10.1016/j.cell.2010.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Plaisier CL, et al. Zbtb16 has a role in brown adipocyte bioenergetics. Nutr Diabetes. 2012;2:e46. doi: 10.1038/nutd.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barna M, Hawe N, Niswander L, Pandolfi PP. Plzf regulates limb and axial skeletal patterning. Nat Genet. 2000;25:166–172. doi: 10.1038/76014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.